Abstract
A mutation (C-to-T transition) at position 491 of the rrs gene was identified in a Mycobacterium tuberculosis strain family (n = 208 isolates) that was predominant in a suburb of Cape Town, South Africa. This nucleotide change is not involved in streptomycin resistance, and we suggest caution in assuming that all mutations in genes targeted by antituberculosis drugs confer drug resistance.
Multiple-drug-resistant Mycobacterium tuberculosis is a major concern to health authorities worldwide (1, 7, 8, 12). Unlike the antibiotic resistance in many bacterial species, which is acquired by gene transduction, conjugation, or transformation, the drug resistance in M. tuberculosis is genomically based. Resistance to first-line antituberculosis drugs has been linked to mutations in nine genes, viz., katG, inhA, aphC, and kasA for isoniazid resistance, rpoB for rifampin resistance, rpsL and rrs for streptomycin (SM) resistance, embB for ethambutol resistance, and pncA for pyrazinamide resistance (6). Mutations identified in these genes have been associated with drug resistance based on their absence in drug-susceptible isolates (6). Multiple-drug resistance results from the accumulation of mutations in different genes (2, 5). Analysis of these mutations has been proposed as a powerful tool for rapid prediction of drug resistance (6), thereby enhancing the efficiency of diagnosis and limiting the spread of drug-resistant strains.
In this study we used mutational analysis (14) and DNA fingerprinting (11) to analyze M. tuberculosis isolates collected from two high-incidence communities in Cape Town, South Africa. Mutation analysis was done using a PCR-based dot blot hybridization technique to identify mutations associated with resistance to isoniazid, rifampin, SM, and ethambutol (6). To ensure accurate genotypeclassification of drug resistance, amplified products of the reference strain H37Rv and those from fully susceptible isolates and resistant isolates (characterized by culture testing and gene sequencing) were included on each blot as wild-type and mutant controls (13, 15). Using the dot blot hybridization method in combination with an oligonucleotide complementary to the mutant sequence, we detected a mutation in the rrs gene of an SM-susceptible clinical isolate (isolate 208). Subsequent automated sequence analysis with an ABI PRISM (model 3100; Applied Biosystems) analyzer confirmed a C-to-T transition at position 491 of the rrs gene, a mutation which has previously been associated with resistance to SM (4, 12).
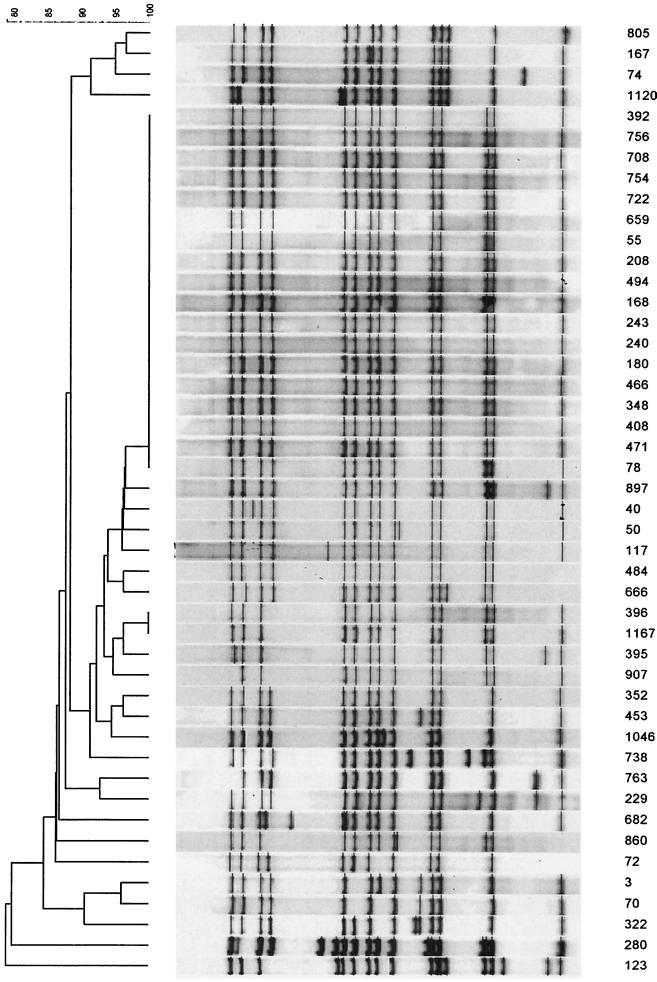
Characterization of isolate 208 by IS6110 DNA fingerprinting showed the presence of 14 hybridizing bands. Analysis of the local M. tuberculosis restriction fragment length polymorphism (RFLP) database (16) of the two communities in the study classified the isolate as belonging to a predominant strain family designated family 11, a family representing 21.4% of all infecting strains (n = 208 individual patients) (17). The RFLP pattern of representative isolates of family 11 is given in Fig. 1. Dot blot hybridization analysis of 71 selected representative isolates of this strain family showed that they all had the C-to-T transition at position 491 in the rrs gene. In contrast, dot blot hybridization analysis of isolates (n = 184) representative of other strain families (16, 17) failed to identify this mutation. This suggests that the C-to-T transition at position 491 in the rrs gene is specific to family 11. Previously this strain family was classified as belonging to pathogenic group 2, according to the sequences of the katG and gyrA genes (17). Interestingly, family 11 isolates are shown to form part of an independently evolving group, which has branched recently from a large clade of strain families all classified as pathogenic group 2. This would suggest that the nucleotide change at position 491 of the rrs gene is more recent than the polymorphism at position 463 of the katG gene used for the group classification (9). The nucleotide change at position 491 of the rrs gene may therefore be useful to further subclassify pathogenic group 2 isolates.
FIG. 1.
RFLP pattern of representative isolates of family 11.
To study the relationship between the C-to-T transition at position 491 of the rrs gene and drug susceptibility, tests at critical concentrations of 0.1, 1.5, and 2 μg of SM per ml were performed by the 1% proportion method (3) on 10 randomly selected isolates of family 11. All of the isolates tested were susceptible at these concentrations. Review of records from previous drug susceptibility testing of the 208 patients infected with a family 11 strain indicated that the infecting strain was resistant to SM in three patients. Retesting of the phenotypic susceptibilities of these three isolates demonstrated that they were susceptible to concentrations of 0.1, 1.5, and 2 μg of SM per ml. Thus, all of the isolates tested were fully susceptible to SM at a concentration 20 times lower than the standard critical concentration for discriminating between drug-resistant and -susceptible isolates.
Previous reports described nucleotide changes at position 491 of the rrs gene in two clinical isolates resistant to SM (4, 10). However, no information was given about the MICs for these two isolates. Based on the results presented in this study and in contrast to the previous reports, we conclude that the nucleotide change at position 491 is a polymorphism that is not associated with drug resistance in this strain family. Although numerous studies have shown associations between different mutations and drug resistance, there is limited data to indicate that such mutations are the causative mechanisms for resistance. This study not only further questions the assumption that all mutations confer resistance in genes in which mutations have been associated with antituberculosis drug resistance but also highlights the importance of establishing the causal relationship between any given mutation and drug resistance. This is particularly relevant in view of numerous reports proposing the use of molecular techniques to identify drug resistance.
Acknowledgments
We thank Tygerberg Hospital, the Harry Crossley Foundation, and the IAEA (projects SAF6/003 and CRP 9925) for financial assistance.
REFERENCES
- 1.Edlin B R, Tokars J I, Grieco M H, Crawford J T, Williams J, Sordillo E M, Ong K R, Kilburn J O, Dooley S W, Castro K G, Jarvis W R, Holmberg S D. An outbreak of multidrug-resistant tuberculosis among hospitalized patients with the acquired immunodeficiency syndrome. N Engl J Med. 1992;326:1514–1521. doi: 10.1056/NEJM199206043262302. [DOI] [PubMed] [Google Scholar]
- 2.Heym B, Honore N, Truffat P C, Banerjee A, Schurra C, Jacobs W R, van Embden J D, Grosset J H, Cole S T. Implications of multidrug resistance for the future of short-course chemotherapy of tuberculosis: a molecular study. Lancet. 1994;344:293–298. doi: 10.1016/s0140-6736(94)91338-2. [DOI] [PubMed] [Google Scholar]
- 3.Kleeberg H H, Blacklock Z, Boulahbal F, David H L, Fink H, Gatner E M S, Grosset J, Juhlin J, Kallich R, Kawamura I, Kilburn J O, Kleeberg C G, Mandler F, Pattyn S R, Petersen K F, Reutgen H, Runon E H, Saito H, Schröder K H, Stander M F, Szabo I, Takahashi S, Tripathy S P, Tinka L, Vergmann B. A simple method of testing drug susceptibility of Mycobacterium tuberculosis: a report of an international collaborative study. Bull Int Union Tuberc. 1985;60:147–150. [Google Scholar]
- 4.Meier A, Kirschner P, Bange F C, Vogel U, Böttger E C. Genetic alterations in streptomycin-resistant Mycobacterium tuberculosis: mapping of mutations conferring resistance. Antimicrob Agents Chemother. 1994;38:228–233. doi: 10.1128/aac.38.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morris S, Han Bai G, Suffys P, Portillo-Gomez L, Fairchok M, Rouse D. Molecular mechanisms of multiple drug resistance in clinical isolates of Mycobacterium tuberculosis. J Infect Dis. 1995;171:954–960. doi: 10.1093/infdis/171.4.954. [DOI] [PubMed] [Google Scholar]
- 6.Ramaswamy S, Musser J M. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis. Tuber Lung Dis. 1998;79:3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 7.Rittaco V, Di Lonardo M, Reniero A, Ambroggi M, Barrera L, Dambrosi A, Lopez B, Isola N, de Kantor I N. Nosocomial spread of human immunodeficiency virus-related multidrug-resistant tuberculosis in Buenos Aires. J Infect Dis. 1997;176:637–642. doi: 10.1086/514084. [DOI] [PubMed] [Google Scholar]
- 8.Rullan J V, Herrera D, Cano R, Moreno V, Godoy P, Peiro E F, Castell J, Ibanez C, Ortega A, Agudo L S, Pozo F. Nosocomial transmission of multidrug-resistant Mycobacterium tuberculosis in Spain. Emerg Infect Dis. 1996;2:125–129. doi: 10.3201/eid0202.960208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sreevatsan S, Pan X, Stockbauer K E, Cornell N D, Kreiswirth B N, Whittam T C, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sreevatsan S, Pan X, Stockbauer K E, Williams D L, Kreiswirth B N, Musser J M. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob Agents Chemother. 1996;40:1024–1026. doi: 10.1128/aac.40.4.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T M, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Rie A, Warren R M, Beyers N, Gie R P, Classen C N, Richardson M, Sampson S L, Victor T C, van Helden P D. Transmission of a multidrug-resistant Mycobacterium tuberculosis strain among non-institutionalized, human immunodeficiency virus-seronegative patients. J Infect Dis. 1999;180:1174–1179. doi: 10.1086/315054. [DOI] [PubMed] [Google Scholar]
- 13.Van Rie A, Warren R, Mshanga I, Jordaan A M, van der Spuy G D, Richardson M, Simpson J, Gie R P, Enarson D A, Beyers N, van Helden P D, Victor T C. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J Clin Microbiol. 2001;39:636–641. doi: 10.1128/JCM.39.2.636-641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Victor T C, Jordaan A M, Van Rie A, van Der Spuy G D, Richardson M, van Helden P D, Warren R. Detection of mutations in drug resistance genes of Mycobacterium tuberculosis by a dot-blot hybridization strategy. Tuber Lung Dis. 1999;79:343–348. doi: 10.1054/tuld.1999.0222. [DOI] [PubMed] [Google Scholar]
- 15.Victor T C, Warren R, Butt J L, Jordaan A M, Felix J V, Venter A, Sirgel F A, Schaaf H S, Donald P R, Richardson M, Cynamon M H, van Helden P D. Genome and MIC stability in Mycobacterium tuberculosis and indications for continuation of use of isoniazid in multidrug-resistant tuberculosis. J Med Microbiol. 1997;46:847–857. doi: 10.1099/00222615-46-10-847. [DOI] [PubMed] [Google Scholar]
- 16.Warren R, Richardson M, van der Spuy G, Victor T, Sampson S, Beyers N, van Helden P. DNA fingerprinting and molecular epidemiology of tuberculosis: use and interpretation in an epidemic setting. Electrophoresis. 1999;20:1807–1812. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1807::AID-ELPS1807>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 17.Warren R M, Sampson S L, Richardson M, Van der Spuy G D, Lombard C J, Victor T C, Van Helden P D. Mapping of IS6110 flanking regions in clinical isolates of Mycobacterium tuberculosis demonstrates genome plasticity. Mol Microbiol. 2000;37:1405–1416. doi: 10.1046/j.1365-2958.2000.02090.x. [DOI] [PubMed] [Google Scholar]