Abstract
Methylation of adenosine in RNA to N6-methyladenosine (m6A) is widespread in eukaryotic cells with his integral RNA regulation. This dynamic process is regulated by methylases (editors/writers), demethylases (remover/erasers), and proteins that recognize methylation (effectors/readers). It is now evident that m6A is involved in the proliferation and metastasis of cancer cells, for instance, altering cancer cell metabolism. Thus, determining how m6A dysregulates metabolic pathways could provide potential targets for cancer therapy or early diagnosis. This review focuses on the link between the m6A modification and the reprogramming of metabolism in cancer. We hypothesize that m6A modification could dysregulate the expression of glucose, lipid, amino acid metabolism, and other metabolites or building blocks of cells by adaptation to the hypoxic tumor microenvironment, an increase in glycolysis, mitochondrial dysfunction, and abnormal expression of metabolic enzymes, metabolic receptors, transcription factors as well as oncogenic signaling pathways in both hematological malignancies and solid tumors. These metabolism abnormalities caused by m6A’s modification may affect the metabolic reprogramming of cancer cells and then increase cell proliferation, tumor initiation, and metastasis. We conclude that focusing on m6A could provide new directions in searching for novel therapeutic and diagnostic targets for the early detection and treatment of many cancers.
Keywords: M6A, methylation, reprogramming, metabolism, metabolite, oncogenic, cancer
Introduction
Adenosine methylation is the most common modification of RNA in eukaryotes. The methyl group is attached to the nitrogen-6 position of adenosine, creating N6-methyladenosine or m6A (Wang et al., 2017). This modification is highly dynamic and reversible, as it involves enzymes that methylate adenosine (writers), remove methylation (erasers), or recognize it (readers) (Chen et al., 2019b). Moreover, the m6A modification is integral to the regulation of RNA, as it affects mRNA processing, mRNA translation, mRNA decay, mRNA export to the cytoplasm, and miRNA maturation (Roundtree et al., 2017a). In the past several years, compelling evidence has witnessed the implication of m6A in RNA modification. Recent work has uncovered that m6A plays an important role in gene expression regulation emerged as critical post-transcriptional modifications. Currently, Shi et al., (2019) review advances progress in understanding the mechanisms which specific cellular contexts and molecular function of N6-methyladenosine and highlight the importance of RNA modification regulation, including mRNA, tRNA, rRNA, and other non-coding RNA. They conclude that the recent biological outcome of m6A methylation could be promising for translational medicine. Previously, the roles of m6A modifications in modulating gene expression throughout cell differentiation and animal development were reviewed by Frye et al., (2018). Their study illustrates that m6A methylation plays a critical role by regulating various aspects of RNA metabolism, physiological processes, and stress response (Frye et al., 2018). More interestingly, others recent evidence indicates that the modification of m6A also regulates physiology and metabolism in tumors (Faubert et al., 2017; Choe et al., 2018).
Metabolic reprogramming in cancer cells was discovered to promote tumorigenesis (Frezza, 2020). Biochemical and molecular studies have suggested several possible mechanisms for its evolution during cancer development (Hanahan and Weinberg, 2011). Recently, m6A’s function in oncology and its involvement in the regulation of cancer metabolism has received growing attention. As a result, our understanding of the metabolic mechanisms regulated by the m6A’s modification in carcinogenesis and their potential therapeutic implications have progressed significantly.
Interestingly, m6A can act as a suppressor or promoter in the proliferation (Liu et al., 2018; Shen, 2020), differentiation (Chen et al., 2019a), and metastasis of tumor cells (Ma et al., 2017) in various cancers. It also appears to reprogram cancer cell metabolism (Shen et al., 2020), as it can regulate metabolic enzymes, transporters, pathways, and transcription factors that promote cancer progression (Li et al., 2020c; Chen et al., 2021a). Here, we discuss the current understanding of how the m6A modification affects cancer metabolism and the potential for regulating it to provide new targets for cancer therapy.
M6A Regulation
Modification of m6A is regulated by: methyltransferases that catalyze methylation (writers), demethylases that remove (erasers) the methyl group from m6A, then m6A recognition proteins (readers) recognize the modification (Lewis et al., 2017). Interestingly, m6A methyltransferase, m6A demethylases, and m6A recognition proteins play essential roles in gene regulation.
m6A Methyltransferase
Methyltransferase-like3 (METTL3) and Methyltransferase-like14 (METTL14) are the critical components of the m6A methyltransferase complex (MTC). These two methyltransferases colocalize in the nucleus (Liu et al., 2014), forming a heterodimer. METTL3 transfers the methyl of the S-adenosyl methionine (SAM) to produce S-adenosyl homocysteine (SAH) and leads to global miRNA downregulation. By binding with eIF3h in the cytoplasm, METTL3 can also promote oncogenic mRNAs translation (Choe et al., 2018). METTL3 could be modulated through post-transcriptional modifications, affecting protein stability, localization, writer complex formation, and writer catalytic activity (Shi et al., 2019). In comparison, METTL14 identifies specific RNA sequences as a target and stabilizes the structure of MTC (Liu et al., 2014; Lin et al., 2016; Sledz and Jinek, 2016). For example, METTL14 can methylate target miRNA by cooperating with HNRNPA2B1 and DGCR8, promoting miRNA maturation (Alarcon et al., 2015).
Wilms’ tumor associating protein (WTAP), another writer protein, plays a role in localizing the methylase complex in the nucleus by interaction with heterodimer (Ping et al., 2014; Knuckles et al., 2018). Recently, other components, such as HAKAI, ZC3H13, and VIRMA/KIAA1429, have been identified to interact with other parts of the MTC (Yue et al., 2018), while ZCCHC4 is a ribosomal RNA-28S methyltransferase (Ma et al., 2019). Other methyltransferase components like METTL5 have been found to be independent m6A writers. It catalyzes the attachment of m6A onto specific structure RNAs, including U6-small nuclear RNA (snRNA), 18S rRNA, and 28S rRNA (Wang et al., 2017; Ignatova et al., 2020). METTL16 catalyzes m6A of the U6- spliceosomal small nuclear RNA and MAT2A 3′-UTR mRNA (Pendleton et al., 2017).
m6A Demethylases
The m6A remover proteins erase the m6A modification by increasing the level of iron ferrous (Fe2+) (co-factor) and α-ketoglutarate (co-substrate) dependent oxygenase family (Fedeles et al., 2015). Two erasers that catalyze m6A demethylation ALKB homolog 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) can recognize adenine and cytosine methylation in RNA (Fu et al., 2013). ALKBH5 and FTO are members of the Fe2+/α-ketoglutarate-dependent dioxygenases family. The first RNA demethylase identified FTO was reported to remove the methyl group of N6 - methyladenosine (m6A) in RNA. m6A erasers may exhibit different expression levels, post-translational modifications, and cellular localization, depending on cell types. For instance, m6A demethylase FTO is predominantly nucleus localized and regulates 5–10% of total mRNA m6A demethylation (Wei et al., 2018). In contrast, FTO is also highly abundant in the cell cytoplasm and can mediate up to 40% m6A demethylation of total mRNA in certain leukemia cells (Shi et al., 2019). Additionally, FTO regulates alternative splicing via m6A by interacting with Serine-rich splicing factor 2(SRSF2) (Bartosovic et al., 2017). Interestingly, FTO may control metabolic disorders. ALKBH5 another m6A demethylase, affects mRNA export and processing factors (Zheng et al., 2013). ALKB homolog 3 (ALKBH3) was found to demethylate only tRNAs (Ueda et al., 2017; Yang et al., 2020).
m6A Recognition
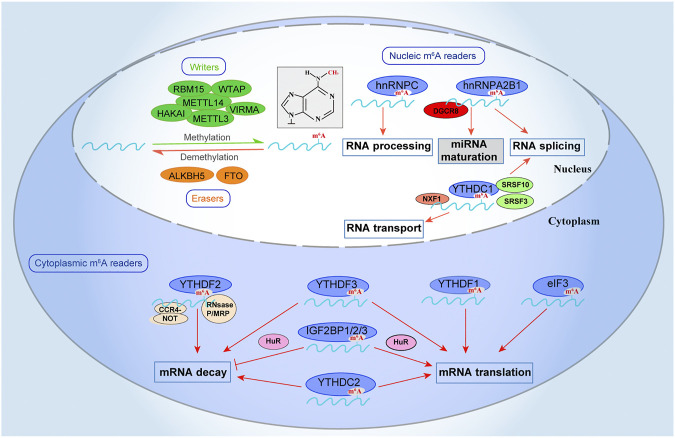
The m6A recognition proteins (readers) control the destiny of RNAs that have been modified. Readers/effectors are distributed in the nucleus and cytoplasm, indicating their functional diversity. While writers and erasers carry out methylation and demethylation, the readers determine the functional consequences of modification. m6A recognition proteins characterization has provided valuable insights into the molecular mechanisms of the m6A-mediated post-transcriptional gene regulation (Shi et al., 2019). Furthermore, RNA binding proteins (RBPs) could regulate the interactions between m6A effectors and RNA substrates.
YTHDF1/2/3 and YTHDC1 recognize the m6A change and alter mRNA’s splicing, translation, and decay (Xu et al., 2014; Wu et al., 2017a). Intriguingly, these proteins also play crucial roles in mRNA metabolism (Wang et al., 2015). For instance, YTHDF1 binds to mRNA, including eukaryotic translation initiation factor 3 (eIF3) and poly-A- binding protein (PABP) complex to promote RNA translation (Wang et al., 2015). YTHDF2 recognizes mRNAs not destined for translation, accelerating their destruction. Interestingly, it identifies specific m6A -modified binds to CCR4-NOT transcription complex subunit 1 (CNOT1). However, it recruits the CCR4-NOT complex of the m6A -tagged RNA P-body to promote its destruction (Du et al., 2016). YTHDF3 by interaction with YTHDF1 accelerates mRNA translation, affecting YTHDF2-mediated degradation of mRNAs labeled with m6A (Li et al., 2017a).
YTHDC1 mediates mRNA export marked with m6A by interaction with the nuclear export adaptor protein SRSF3 (Roundtree et al., 2017b). Importantly, YTHDC1 regulates splicing events by inhibiting SRSF10 or activating splicing factor SRSF3. In conjunction with nuclear RNA export factor 1 (NXF1), YTHDC1 can also mediate mRNA export to the cytoplasm. Unlike the rest of the family, YTHDC2, an RNA helicase. Its helicase domain contributes to RNA binding (Hsu et al., 2017). Significantly, YTHDC2 and YTHDF3 can facilitate RNA degradation or enhance RNA translation depending on the context (Shi et al., 2017).
hnRNPs, another m6A recognition family, is localized in the nucleus where heterogeneous nuclear ribonucleoprotein C (hnRNPC) can bind with nascent RNA transcripts and control their processing (Alarcon et al., 2015). For instance, the lncRNA MALAT1 facilitates a change in the m6A site for recognition and binding by hnRNPC (Liu et al., 2015). Interestingly, m6A regulates RNA binding motifs (RBMs) accessibility by altering mRNA and long noncoding RNA (lncRNA) structure to promote hnRNPC interaction. These changes influence RNA-protein interactions in human cells. This mechanism is called the “m6A -switch” (Liu et al., 2015). hnRNPC-binding regulated by the m6A -switch regulates RNA alternative splicing, indicating that the switch helps regulate gene expression and RNA maturation (Liu et al., 2015).
Another component of the m6A recognition family, Heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1), regulates RNA alternative splicing and microRNA processing (Alarcon et al., 2015; Liu et al., 2015). Further, it interacts with DiGeorge syndrome critical region gene 8 (DGCR8) for miRNA maturation and recognizes the m6A signals of microRNA (Zhao et al., 2017). Eukaryotic initiation factor 3 (eIF3), another effector/reader, could initiate protein translation in a cap on its 5′-UTR (Meyer et al., 2015). In conjunction with Hu antigen R (HuR), these proteins recognize m6A’s modification and stabilize their RNA transcripts (Meyer et al., 2015).
Insulin growth factor-2 binding proteins 1, 2, and 3 (IGF2BP 1/2/3) were identified as another m6A recognition. After co-localizing with HuR, these proteins protect mRNA decay and enhance mRNA translation (Huang et al., 2018). These findings demonstrated that m6A methyltransferases (editors/writers) and m6A demethylases (remover/erasers) cooperate to modulate the distribution of m6A on RNA by adding (writer) or removing (erasers) the methyl. While the m6A recognition (effectors/readers) proteins recognize the m6A modified transcripts and determine their fate regulate functions (Figure 1).
FIGURE 1.
m6A-mediated RNA regulation. The m6A modification is integral to the regulation of RNA. m6A can be installed by “writers” (METTL3/14, WTAP, RBM15, VIRMA, and HAKAI), removed by “erasers” (FTO and ALKBH5), and recognized by “readers” (YTHDF1/2/3, YTHDC1/2, IGF2BP1/2/3, eIF3, and HNRNPC/A2B1). m6A methyltransferases (writers) catalyze methylation while the m6A demethylases (erasers) remove the methyl in m6A. The m6A recognition (readers) proteins bind the m6A modified transcripts and determine their fate. The modification of “writers,” “erasers,” and “readers” proteins affect RNA processing, including RNA splicing, mRNA translation, mRNA decay, mRNA export to the cytoplasm, and miRNA maturation.
M6A Regulates Cancer Metabolism
Cancer cells need abundant energy and raw materials to grow and divide; therefore, they substantially alter their metabolic pathways (Hanahan and Weinberg, 2011; Li and Zhang, 2016). Importantly, biochemical and molecular studies suggest several possible mechanisms for the evolution of aberrant metabolism during cancer development (Hanahan and Weinberg, 2011). For example, proliferating cancer cells can enhance the synthesis of glucose of carbohydrates, lipids, and proteins to obtain an ample and uninterrupted supply of molecules needed for biosynthesis (Khan et al., 2020). Moreover, most cancer cells depend on aerobic glycolysis rather than the TCA cycle (Vander Heiden et al., 2009). The preference for glycolysis over mitochondrial oxidative phosphorylation seems to be a hallmark of cancer cells (Garber, 2006).
However, aerobic glycolysis transports chemical generates ATP. This ATP and its breakdown product adenosine are widespread throughout the body, and both have been shown to regulate cell proliferation and differentiation. Therefore, metabolic reprogramming is widely utilized during oncogenesis, and the m6A modification can regulate metabolism in cancer progression (Figure 2).
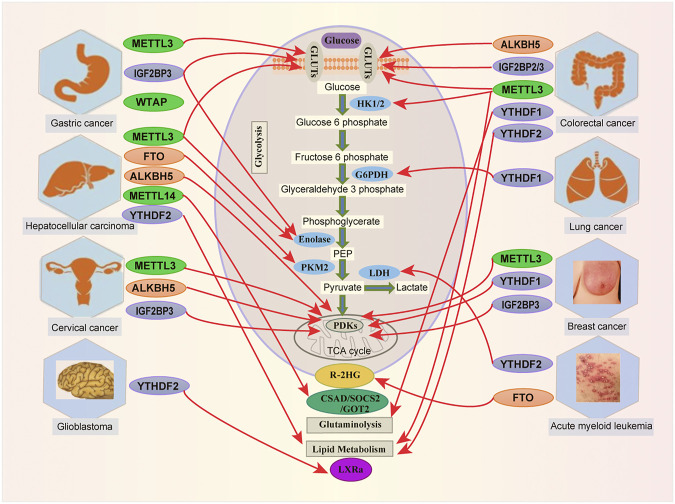
FIGURE 2.
Links between m6A modification and metabolites in human cancer. m6A RNA modification by targeting metabolic pathways is involved in various tumorigenesis, including Acute Myeloid Leukemia (AML), Breast Cancer (BC), Cervical Cancer (CC), Colorectal Cancer (CRC), Glioblastoma (GBM), Hepatocellular Carcinoma (HCC), Gastric Cancer (GC) and Lung Cancer (LC).
m6A Regulates Glucose Metabolism
Glucose, an essential nutrient in blood, is the main energy source for cells (Shaw, 2006). However, several studies have found that hyperglycemia increases the overall risk of cancer (Stattin et al., 2007). Cancer cells enhanced glucose uptake has also been implicated in metastasis and poor prognosis (Macheda et al., 2005). Aerobic glycolysis in cancer can increase the m6A modification genes associated with glycolysis (Fry et al., 2017).
Recent evidence demonstrated that cancer reprograms glucose metabolism (Li et al., 2020c); thus, aerobic glycolysis exemplifies an evolutionary change in cancer cells. Not surprisingly, glycolytic transporters like glucose transporter (GLUT), glycolytic enzymes such as pyruvate kinase isozyme M1/2 (PKM1/2), pyruvate dehydrogenase kinase (PDK), lactate dehydrogenase (LDH), and hexokinase (HK) is important targets to understand cancer metabolism (Doherty and Cleveland, 2013; Viale et al., 2014). The relationship between m6A and glucose metabolism is crucial for understanding cancer progression because glucose is the most important metabolite associated with many enzymes and transporters. Additionally, glycolysis is an essential pathway involved in cancer progression, metastasis, and chemotherapy resistance (Ganapathy-Kanniappan and Geschwind, 2013).
In Colorectal Cancer (CRC), the METTL3-HK2/GLUT1-MYC-IGF2BP is involved in cells proliferation and metastasis (Shen et al., 2020; Xiang et al., 2020; Chen et al., 2021a). Hexokinase (HK) catalyzed the first step of anaerobic glycolysis and oxidative phosphorylation, which converts glucose to glucose 6-phosphate (G6P) (Wilson, 2003). Many investigations reveal the implication of HK in cancers. For instance, HK2 bound to mitochondria enable cancer cells to become more glycolytic (Chen et al., 2014; Wu et al., 2017b). GLUT1, a glucose transporter, mediates the first step of glucose inside cells (Cheeseman, 2008). Overexpression of GLUTs facilitates glucose consumption in cancer progression (Ancey et al., 2018). METTL3 stabilizes GLUT1 and HK2 mRNA in colorectal cancer by directly interacting with the 3′ UTR mRNA of GLUT1 and the 5'/3′-UTRs mRNA of HK2. This enhanced HK2 and GLUT1 expression, promoting CRC progression (Shen et al., 2020). One recent study established that METTL3 enhanced CRC growth and identified GLUT1-mTORC1 as the primary target of METTL3 in that disease (Chen et al., 2021a). More interestingly, METTL3 induced GLUT1 translation in m6A to promote glucose uptake and lactate production, leading to mTORC1 activation. These findings indicate that METTL3 promotes CRC via the m6A-mediated GLUT1-mTORC1 signaling activation.
In Cervical Cancer, (Li et al., 2020c) demonstrated that m6A regulates glycolysis in cancer cells through pyruvate dehydrogenase kinase 4 (PDK4). PDKs are the gatekeeper enzymes involved in altered glucose metabolism in tumors (Patel and Korotchkina, 2006; Devedjiev et al., 2007). They are remarkably overexpressed in multiple human tumor samples. Among them, PDK4 was noticed as one critical metabolic factor of metabolism control because it can divert carbon flux from oxidative phosphorylation into glycolysis (OXPHOS) (Stacpoole, 2017). According to Li and collaborators, the extracellular acidification rate (ECAR) was decreased in Mettl3Mut/- HeLa cells, While the oxygen consumption rate (OCR) was increased (Li et al., 2020c), demonstrating that METTL3 promotes glycolysis. Additionally, PDK4 can reverse lactate production level, glucose consumption, and ATP rate in Mettl3-depleted cells. More importantly, overexpression of PDK4 to an endogenous level attenuated the metabolic phenotypes of SiHa cells that had lost METTL3. Also, overexpression of ALKBH5 suppressed PDK4 expression in HeLa cells (Li et al., 2020c). Moreover, compared with negative control samples, IGF2BP3 and YTHDF1 were significantly higher in cervical cancer samples (Li et al., 2020c). Li et al. (2020b) further determined whether the m6A modification can regulate PDK4 expression in addition to affecting the stability of the kinase’s mRNA. Modifying the PDK4 mRNA at its 5′-UTR by m6A positively regulated its elongation during translation and the stability of its mRNA because m6A is bound to the YTHDF1 and IGF2BP3 (Li et al., 2020c). In HeLa cells, IGF2BP3 inhibition can suppress PDK4 expression and alter the suppressive effect of METTL3 on PDK4 expression (Li et al., 2020c). More interestingly, YTHDF1 and IGF2BP3-targeting PDK4 with d m6A CRISPR significantly downregulated PDK4 mRNA and protein levels (Li et al., 2020c). Thus, targeting m6A on PDK4 mRNA with dm6ACRISPR appears to regulate glycolysis and ATP generation in cancer (Li et al., 2020c). These studies suggest that PDK4 is a critical metabolic agent of glycolysis and ATP generation regulated by m6A in cervical cancer progression.
In Hepatocellular Carcinoma (HCC), hepatic FTO helps regulate the expression of the gluconeogenic gene. Recent evidence indicates that demethylation of m6A by FTO affects glucose metabolism via hepatic gluconeogenesis (Shen et al., 2015). On the other hand, the FTO level may be affected by insulin in HCC (Mizuno et al., 2017). Pyruvate kinase isozymes M1 (PKM1) and M2 (PKM2) are glycolytic enzymes (Doherty and Cleveland, 2013). They mediate the final steps of glycolysis by dephosphorylation of phosphoenolpyruvate (PEP), producing pyruvate and ATP. According to (Li et al., 2019a), FTO promotes HCC tumorigenesis by demethylating m6A on PKM2 mRNA. This demethylation accelerates translation, leading to tumorigenesis in HCC (Li et al., 2019a). The demethylation of PKM2 mRNA by FTO suggests that FTO also regulates the expression of PKM2. Knocking down FTO repressed HCC progression (Li et al., 2019a). This finding revealed that FTO could demethylate PKM2 mRNA, thereby upregulating the kinase’s expression. Upregulated PDK4 was found to reduce hepatic chemotherapy-induced colorectal liver metastasis (Strowitzki et al., 2019). PDK4 collaborates with METTL3 to induce proliferation and hepatic chemosensitivity cancer cells (Li et al., 2020c). Regarding the link between PDK4 and m6A, Li and collaborators found that m6A -PDK4 plays an essential role in liver cancer progression. Consistent with this finding, knocking down METTL3 inhibited PDK4 antibodies in Huh7 cells. Moreover, overexpression of the demethylase ALKBH5 (another m6A eraser) decreased glucose, lactate, and ATP abundance in Huh7 HCC cells (Li et al., 2020c). Li and collaborators also provided evidence that METTL3 regulates glycolytic activity in HCC. Downregulation of METTL3 cooperates with the 2-deoxyglucose (2-DG) to inhibit HCC proliferation, suggesting that suppressing glycolysis by inhibiting METTL3 might be a potential strategy for treating HCC (Lin et al., 2020).
In Acute Myeloid Leukemia (AML), α-ketoglutarate, produced by isocitrate dehydrogenase in the TCA cycle, interacts with m6A demethylase proteins (Losman et al., 2020). R-2HG (R-2-hydroxyglutarate) inhibited FTO activity by stimulating the modification of m6A -RNA in cells. Moreover, through targeting the FTO/MYC/CEBPA axis, R-2HG inhibited the proliferation of leukemia cells (Su et al., 2018). It was reported that knocking down FTO or LDHB (lactate dehydrogenase B) inhibits R-2HG in leukemia cells (Qing et al., 2021). Additionally, R-2HG abrogated FTO/m6A/YTHDF2-mediated upregulation of LDHB, suppressing aerobic glycolysis (Qing et al., 2021). These findings show that R-2HG attenuates aerobic glycolysis by inhibiting FTO in leukemia cells. Lactate dehydrogenase (LDH) converts pyruvate to lactate, and this enzyme is frequently upregulated in multiple cancers (Wang et al., 2012). Lactate, ketone, and pyruvate are monocarboxylates that play essential roles in cancer metabolism (Halestrap, 2013).
In Gastric Cancer (GC), overexpression of METTL3 (a writer) promoted metastasis to the liver in vitro and in vivo, and it also stimulated the modification of adenosine to m6A, enhancing mRNA stability (Wang et al., 2020c). Tumor angiogenesis was promoted by Hepatoma-derived growth factor (HDGF) upregulation, while nuclear HDGF activated GLUT4 and ENO2 expression and increased metastasis in GC cells (Wang et al., 2020c). WTAP (a writer) promoted GC cell proliferation and glycolytic capacity and enhanced HK2 expression through interacting with the m6A modified 3′-UTR of HK2 mRNA (Yu et al., 2021).
In Glioblastoma (GBM), Li et al. recently showed that long noncoding RNA just proximal to X-inactive specific transcript (JPX) interacted with N6-methyladenosine (m6A) demethylase FTO and enhanced FTO-mediated PDK1 mRNA demethylation. Additionally, JPX exerted its GBM-promotion effects through the FTO/PDK1 axis (Li et al., 2021). These outcomes reveal the critical role of JPX in promoting GBM aerobic glycolysis-m6A demethylase FTO.
In Lung Cancer (LC), YTHDF2 expression is increased in tumor tissues, promoted proliferation, and bound to 3′-UTR of 6-phosphogluconate dehydrogenase (G6PD) mRNA (Sheng et al., 2020). This binding facilitates G6PD mRNA translation in LC and promotes tumorigenesis. Recently, Yang and collaborators showed that FTO is declined in lung adenocarcinoma, which correlates with poor patient overall survival. Moreover, downregulated FTO expression enhanced m6A levels in mRNAs of genes involved in metabolic pathways such as MYC (Yang et al., 2021). Interestingly, the enhanced levels recruited the binding of YTHDF1, which promoted the translation of MYC mRNA and increased glycolysis and cancer progression (Yang et al., 2021).
In Breast Cancer (BC), METTL3 overexpression enhanced the PDK4 protein expression in breast cancer cells (Li et al., 2020c). Interestingly, the m6A -modified 5′-UTR of PDK4 regulated the kinase’s elongation during translation and the stability of its mRNA through interaction with YTHDF1 and IGF2BP3. Further, clinical data confirm that m6A/PDK4 is implicated in breast cancer progression (Li et al., 2020c). These findings suggest that proteins associated with m6A regulate glycolysis in breast cancer cells.
m6A Regulates Lipid Metabolism
Recently, elevated lipid levels were recognized as an important aberration of cancer metabolism (Swierczynski et al., 2014). Moreover, previous studies have noticed that lipid metabolism is reprogrammed in tumors (Schulze and Harris, 2012; Nath and Chan, 2016). Dysregulation of lipid metabolism is an essential feature of cancer cells (Murai, 2015; Gaida et al., 2016).
There is also a link between m6A proteins and lipid metabolism in cancer. After observing that knocking down METTL3 and YTHDF2 decreased lipid accumulation in hepatocellular carcinoma cells, Zhong et al. (2018) proposed that the presence of m6A in mRNA mediates crosstalk between the circadian clock and lipid metabolism (Zhong et al., 2018). Kang et al. (2018) showed that FTO increased triglyceride (TG) deposition and decreased mitochondrial content. FTO regulates lipid metabolism in hepatocytes by modulating RNA m6A levels (Kang et al., 2018). These studies revealed that FTO’s demethylating is an important actor in the lipid metabolism of hepatocytes. By linking the epigenetic modification of RNA with fat deposition, they suggested a new m6A target for regulating hepatic fat metabolism (Kang et al., 2018). FTO overexpression in HepG2 cells also reduced m6A levels, enhancing stearoyl CoA desaturase (SCD), monoacylglycerol O acyltransferase 1 (MOGAT1), and fatty acid synthase (FAS), which contribute to cell growth (Kang et al., 2018). Numerous studies demonstrated that METTL3-mediated m6A modification and inhibition of mRNA decay promoted the miR-3619-5p/HDGF axis, enhancing lipogenesis in Hepatocellular Carcinoma (Zhong et al., 2019; Zuo et al., 2020).
In GBM, Fang et al. (2021) recently showed that YTHDF2 facilitates m6A -dependent mRNA decay, impacting glioma patients’ survival. Moreover, YTHDF2 inhibited cholesterol homeostasis in GBM cells. These outcomes highlight the critical function of YTHDF2 regulated cholesterol homeostasis in GBM (Fang et al., 2021). Other reported studies showed that YTHDF2 could also regulate lipogenic genes, including acetyl CoA carboxylase 1 (ACC1), fatty acid synthase (FAS), and stearoyl-CoA desaturase 1(SCD1), to decrease their mRNA stability (Zhou et al., 2021).
As a lipid, sphingolipids also regulate cancer proliferation, migration, invasion, and metastasis. Among this class of lipid, delta 4 desaturase sphingolipid 2 (DEGS2) catalyzes the conversion of dhCers to phytoceramides (Casasampere et al., 2016). Recently, Guo and collaborators found the role of m6A modification on DEGS2 in colorectal cancer and suggested that inhibited m6A promotes DEGS2 expression and dysregulated lipid metabolites, contributing to colorectal cancer (Guo et al., 2021). Furthermore, overexpression of DEGS2 promoted cell growth, while depletion of DEGS2 inhibited cell growth (Casasampere et al., 2016). Regarding the molecular mechanism, Guo and collaborators found that METTL3 depletion promoted the DEGS2 mRNA, increased DEGS2 expression in HCT116 cells, suggesting that METTL3 is essential for the stability and translation of DEGS2. YTHDF2 knockdown induced the level of DEGS2 mRNA expression, meaning that YTHDF2 contributes to the DEGS2 mRNA decay (Guo et al., 2021). Collectively, this recent evidence suggests that m6A regulates lipid metabolism in cancer.
m6A Regulates Amino Acid Metabolism
To proliferate, cancer cells need large amounts of amino acids (Sivanand and Vander Heiden, 2020), which are essential building blocks of proteins (Murugan, 2019; Vettore et al., 2020). Moreover, there is much evidence for specific degradation in amino acid metabolism in cancers (Li and Zhang, 2016). Glutamine, which regulates the expression of many genes related to metabolism (Curi et al., 2005), is carried into cancer cells by multiple transporters, such as Na + -coupled neutral amino acid transporters (SNATs) and Na + -dependent transporters (Kandasamy et al., 2018).
To renew the TCA cycle, many tumor cells highly need glutamine (Matés et al., 2013). Glutamate dehydrogenase (GLUD1) and transaminases can transform glutamine to α-KG to reconstruct the TCA cycle (Vander Heiden et al., 2009). FTO and ALKBH5 were identified as α-KG-dependent dioxygenases (Zhu et al., 2020). METTL14 may promote HCC progression by modulating m6A -regulated genes, including glutamic oxaloacetic transaminase 2 (GOT2), cysteine sulfonic acid decarboxylase (CSAD), and suppressor of cytokine signaling 2 (SOCS2) (Li et al., 2020b). In colon cancer, Chen and collaborators demonstrated YTHDF1-mediated as a positive association between glutamine metabolism and cisplatin resistance (Chen et al., 2021b).
Recently, reports have indicated that AMP-activated protein kinase (AMPK) could act as a beneficial target for treating cancer patients (Wang et al., 2016b). AMPK can act to inhibit tumorigenesis through the regulation of cell proliferation. AMP-activated protein kinase-alpha2 (AMPKα2) was inversely correlated with FTO (Wang et al., 2016a). FTO is upregulated in colorectal cancer and interacts with MYC to accelerate cell proliferation and migration (Zou et al., 2019). In colorectal cancer, Yue and collaborators reveal that AMPKα2 inhibits CRC cell growth and promotes apoptosis through altering FTO (Yue et al., 2020). More interestingly, miR-96 could retard cancerogenesis by inactivating the FTO-mediated MYC AMPKα2-dependent manner in CRC cells (Yue et al., 2020). Together, these findings elucidate links between m6A and metabolic changes in cancers (Table 1 and Figure 2).
TABLE 1.
Regulation of metabolites by m6A associated proteins in cancer.
| Metabolic pathways | Metabolites/Enzymes/Signaling pathways | m6A associated proteins | Cancer type | Role in cancer | References |
|---|---|---|---|---|---|
| Glycolysis | GLUT1-mTORC1 | METTL3 | CRC | Oncogene | Chen et al. (2021a) |
| GLUT1 | METTL3/IGF2BP2/3 | CRC | Oncogene | Shen et al. (2020) | |
| PDK4 | METTL3 | Breast cancer | Oncogene | Li et al. (2020c) | |
| HK2 | METTL3 | CRC | Oncogene | Shen et al. (2020) | |
| GLUT4/Enolase | METTL3 | Liver cancer | Oncogene | Wang et al. (2020c) | |
| GLUT4/HDGF/ENO2 | METTL3/IGF2BP3 | Gastric cancer | Oncogene | Wang et al. (2020c) | |
| PDK4 | METTL3/IGF2BP3/ALKBH5 | Cervical cancer | Oncogene | Li et al. (2020c) | |
| MYC | FTO/YTHDF1 | Lung cancer | Oncogene | Yang et al. (2021) | |
| PDK4 | METTL3 | Liver cancer | Oncogene | Li et al. (2020c) | |
| HK2 | WTAP | Gastric cancer | Oncogene | Yu et al. (2021) | |
| PDK4 | YTHDF1/IGF2BP3 | Breast cancer | Oncogene | Li et al. (2020c) | |
| PKM2 | FTO | HCC | Oncogene | Li et al. (2019a) | |
| PDK4 | ALKBH5 | Cervical cancer | Oncogene | Li et al. (2020c) | |
| PDK4 | ALKBH5 | HCC | Oncogene | Li et al. (2020c) | |
| GLUT1 | ALKBH5 | CRC | Oncogene | Shen et al. (2020) | |
| G6PD | YTHDF2 | Lung cancer | Oncogene | Sheng et al. (2020) | |
| 2-deoxyglucose | METTL3 | HCC | Oncogene | Lin et al. (2020) | |
| MYC | METTL3 | CRC | Oncogene | Lin et al. (2020) | |
| LDHB | YTHDF2 | AML | Oncogene | Qing et al. (2021) | |
| Lipid metabolism | Lipid | METTL3/YTHDF2 | Liver cancer | Oncogene | Zhong et al. (2018) |
| Cholesterol | YTHDF2 | Glioblastoma cancer | Oncogene | Fang et al. (2021) | |
| Triglyceride | METTL3 | Liver cancer | Oncogene | Kang et al. (2018) | |
| Sphingolipid (DEGS2) | METTL3/YTHDF2 | CRC | Oncogene | Guo et al. (2021) | |
| Glutaminolysis | R-2HG-MYC | FTO | Leukemia | Oncogene | Su et al. (2018) |
| CSAD/GOT2/SOCS2 | METTL14 | HCC | Oncogene | Li et al. (2020b) | |
| R-2HG | FTO | AML | Oncogene | Qing et al. (2021) | |
| Glutamine | YTHDF1 | Colon cancer | Oncogene | Chen et al. (2021b) | |
| Other metabolic | Iron and ferritin metabolism | YTHDF1 | HPSCC | Oncogene | Ye et al. (2020) |
Other Metabolic Processes Regulated by m6A in Cancer
Emerging evidence demonstrates that m6A can also regulate metabolic processes in carcinogenesis that do not involve glucose, lipids, or amino acids. For example, iron metabolism plays a key role in tumorigenesis (Jung et al., 2019). Therefore, pathways that acquire, export, or store iron are often perturbed in cancer (Jung et al., 2019). The tumor microenvironment exerts selective pressure that renders the cancer cells adopt altered metabolism, supporting these cells’ energy and metabolic demands, thereby facilitating tumor growth. Recent evidence showed that tumor-associated macrophages (TAMs) could provide iron to impact metabolism within the tumor microenvironment. When Ye and collaborators evaluated the correlation between the m6A modification and iron metabolism, they found that YTHDF1 regulates growth and iron metabolism in hypopharyngeal squamous cell carcinoma (HPSCC) (Ye et al., 2020). YTHDF1 was also associated with intratumoral iron and ferritin levels in hypopharyngeal squamous cell carcinoma (HPSCC) patients. They further demonstrated that HPSCC tumorigenesis induced by YTHDF1 is dependent on iron metabolism and regulates transferrin receptor protein (TFRC) expression in this cancer (Ye et al., 2020). Regarding the molecular mechanism, YTHDF1 binds to the UTR of TFRC mRNA to regulate mRNA translation of TFRC (Ye et al., 2020). Targeting TFRC-mediated iron metabolism and YTHDF1 could become potential candidates for early diagnosis or treatment for HPSCC patients (Ye et al., 2020).
Control of M6A by Metabolites in Cancer
In cancer, metabolism is often regulated by the m6A modification. But could certain metabolites regulate m6A? This controversial idea is supported by the finding that proteins that regulate m6A associate highly with many types of cancer. Also, Wang et al. (2020b) showed that nicotinamide adenine dinucleotide phosphate (NADP) binds to FTO, decreases m6A methylation in RNA, and promotes adipogenesis. Furthermore, NADP regulated mRNA m6A via FTO in vivo, and deletion of FTO blocked adipogenesis caused by enhanced NADP in 3T3-L1 pre-adipocytes.
Succinate prevents α-ketoglutarate-dependent dioxygenase from regulating critical factors of tumorigenesis, including hypoxia responses and histone demethylation. Additionally, hypoxia in tumors broadly increases levels of m6A in GLUT1 and MYC mRNAs (Priolo et al., 2014). ALKBH5 and FTO m6A demethylases require α-KG, Fe(II), and O2 for total enzymatic activity (Zhang et al., 2019; Xu and Bochtler, 2020; Losman et al., 2020). The TCA cycle produces other metabolites that regulate m6A demethylation. Interestingly, citrate, another critical metabolite in the TCA, was noticed with an α-KG-binding site in ALKBH5 (Feng et al., 2014). Citrate by binding to α-KG/FTO complex can inhibit the enzyme’s activity (Aik et al., 2013).
In AML cells, the FTO’s enzymatic activity is inhibited, carrying the IDH (isocitrate dehydrogenase) mutation, which correlates with significantly increased m6A levels (Li et al., 2017b). IDHs are critical enzymes that catalyze isocitrate to α-ketoglutarate (α-KG) and CO2 in the TCA cycle. They also epigenetically control gene expression through effects on α-KG-dependent dioxygenases. R-2HG was recently reported to exhibit antitumor activity. It attenuates aerobic glycolysis and downregulates the expression of FTO/LDHB/PFKP in leukemia cells (Qing et al., 2021). Moreover, it increases m6A modification of RNA by inhibiting FTO activity, destabilizing CEBPA/MYC transcripts in leukemia cells (Su et al., 2018). These findings, therefore, indicate that certain metabolites can drive the m6A modification of RNA in cancer (Table 2).
TABLE 2.
Control of m6A by metabolites in cancer.
| m6A implicated proteins | Metabolites | Effects | References |
|---|---|---|---|
| FTO | NADP | NADP decreases m6A methylation in RNA and promotes adipogenesis | Wang et al. (2020b) |
| FTO | R-2HG | R-2HG attenuates aerobic glycolysis and downregulates the expression of FTO in leukemia cells | Qing et al. (2021) |
| FTO | R-2HG | R-2HG increases m6A modification of RNA by inhibiting FTO activity, destabilizing MYC transcripts in leukemia cells | Su et al. (2018) |
| FTO | Isocitrate | Isocitrate increases m6A levels of RNA by inhibiting FTO’s activity in leukemia cells | Li et al. (2017b) |
Potential Clinical Applications of M6A and Targeting the Modification in Cancer
As proteins that create, erase and recognize m6A play a role in cancer metabolism, targeting altered metabolic pathways by focusing on m6A modification has become a promising anticancer strategy. Survival analysis of patients showed that METTL3 (a writer) is a prognostic factor for poor outcomes in HCC (Lin et al., 2020), thyroid carcinoma (Wang et al., 2020a), pancreatic cancer (Xia et al., 2019), CRC (Li et al., 2019b), gastric cancer (Wang et al., 2020c), and colorectal cancer (Chen et al., 2021a). WTAP (another writer) predicts the survival of patients with high-grade serous ovarian carcinoma (Yu et al., 2019), HCC (Chen et al., 2019c), RCC, and GC (Li et al., 2020a). As METTL3 depletion can decline oncogenes’ expression and reduce CRC proliferation (Shen et al., 2020), breast cancer (Li et al., 2020c), cervical cancer (Li et al., 2020c), and liver cancer (Wang et al., 2020c), METTL3 offers an alternative therapeutic target in colorectal cancer patients with high glucose levels (Shen et al., 2020). It also could promote colorectal tumorigenesis via the m6A-GLUT1-mTORC1 axis. Combined targeting of METTL3 and mTORC1 showed promise for suppressing CRC proliferation, suggesting that METTL3 could also be an alternative therapeutic target in that disease (Chen et al., 2021a). Deleting METTL3 from HeLa cells also decreased PDK4 expression and increased the cells’ sensitivity to doxorubicin (DOX) treatment (Li et al., 2020c). However, ectopic overexpression of PDK4 attenuated this effect and reduced DOX sensitivity in cervical cancer cells. This suggests that PDK4 is involved in the proliferation and chemosensitivity of METTL3-cells (Li et al., 2020c). Moreover, METTL3-silenced pancreatic cancer cells and glioma stem cells (GSCs) showed enhanced irradiation sensitivity (Visvanathan et al., 2018) (Taketo et al., 2018). High level of R-2HG expressed by mutant isocitrate dehydrogenase, was demonstrated to play important antitumor effect in glioma and leukemia cells by inhibiting FTO activity (Su et al., 2018).
Recently, Yankova et al. (2021) showed that STM2457, the small-molecule inhibitor targeting METTL3, might be a strategy for treating myeloid leukemia. Pharmacological METTL3 inhibition prolonged survival in AML mouse models (Yankova et al., 2021). Intriguingly, treating tumors with STM2457 increased apoptosis and reduced AML growth (Yankova et al., 2021). These results identified METTL3 inhibition as a promising therapeutic strategy for AML treatment and demonstrated that targeting enzymes that modify RNA is a new approach promising anticancer therapy (Yankova et al., 2021). Depleting METTL3 from cells induced resistance to cisplatin, gemcitabine, and 5-fluorouracil in pancreatic cancer and non-small cell lung cancer (Jin et al., 2019). Also, FTO inhibitors (FB23 and FB23-2) provide a therapeutic strategy for treating leukemia. Targeting regulators of RNA methylation have also shown promise in preclinical models, which are effective against AML, as exemplified by FB23 and FB23-2 (small-molecule inhibitors) of the m6A eraser FTO (Huang et al., 2019).
By pharmacological approaches, FTO is broadly viewed as an attractive biological target. Peng et al. (2019) found a small molecular inhibitor of FTO and selected m6A demethylase FTO as a potential target by developing a new strategy. By studying the molecular function of FTO in metabolism, they identified entacapone (FDA-approved drug) as a selective inhibitor of FTO activity involved in the regulation of metabolic homeostasis (Peng et al., 2019). Entacapone bound to FTO and inhibited FTO activity. They conclude that the FTO-entacapone complex may be promising for designing new drug-like FTO inhibitors as translational medicine (Peng et al., 2019). Furthermore, they discovered that the transcription factor forkhead box protein O1 (FOXO1) mRNA as a substrate of FTO, which Knockdown of FOXO1 through the inhibition of FTO could be used to treat metabolic dysregulation (Peng et al., 2019).
Targeting YTHDF1 (a reader) might be another promising therapeutic approach, as (Liu et al., 2020) identified the YTHDF1-EIF3C axis as a critical translational factor involved in ovarian cancer progression (Liu et al., 2020). Chen and collaborators (2021) recently reported that YTHDF1 is associated positively with cisplatin resistance in colon cancer (Chen et al., 2021a; Chen et al., 2021b). Furthermore, inhibition of GLS1 synergized with cisplatin to induce cell death of colon cancer cells (Chen et al., 2021b). Recently, Kumar et al., 2021 reviewed how components of EEE (Editor/Eraser/Effector) could become potential candidates for treating leukemia (Kumar et al., 2021).
Regarding immunotherapy against cancer cells, FTO was identified as an essential regulator of glycolytic metabolism that tumors could use to escape immune surveillance (Liu et al., 2021). Consistent with this idea, depleting FTO impaired the glycolytic activity of tumor cells to restore the CD8+ T cell function needed to inhibit tumor growth (Liu et al., 2021). Moreover, Dac51 (a small molecule) can block FTO-mediated immune evasion and control immunity, suggesting that RNA epitranscriptome could promise a new strategy for immunotherapy against cancer cells (Liu et al., 2021).
On the other hand, Yang et al. (2019) demonstrate that the effect of FTO knockdown on melanoma response to anti-PD-1 (a novel immunotherapies for the patient with melanomas) immunotherapy is dependent on the immune system. The combination of m6A demethylase FTO inhibition with anti-PD-1 blockade may reduce the resistance to immunotherapy in melanoma (Yang et al., 2019). Additionally, FTO depletion sensitizes melanoma cells to interferon-gamma (IFNγ) and sensitizes melanoma to anti-PD-1 treatment (Yang et al., 2019). Their findings suggest a crucial role of FTO, which increases FTO’s level, decreases response to anti-PD-1 blockade immunotherapy, and enhances tumor growth in melanoma (Yang et al., 2019). One other recent study demonstrates that the YTHDF1 reader regulated antitumor immunity, a synergetic effect on immunotherapy by improving the therapeutic effect of PD-L1 inhibitors (Han et al., 2019). Yan and collaborators demonstrate that FTO-m6A axis deregulation induces response to tyrosine kinase inhibitor (TKI) treatment in leukemia cells (Yan et al., 2018). Cells with FTO upregulation have more TKI tolerance and higher growth rates in mice (Yan et al., 2018). Currently, Li and collaborators demonstrated that the JPX/FTO/PDK1 axis could facilitate aerobic glycolysis in GBM cells, which was correlated with GBM cells’ sensitivity to temozolomide (TMZ). These findings provide valuable information for understanding that blocking the JPX/FTO/PDK1 axis may serve as a promising strategy for mitigating the efficacy of TMZ in GBM(Li et al., 2021).
By elucidating the biological roles of m6A’s modification in natural killer (NK) cells, Ma and collaborators uncovered a new direction for harnessing NK Cell antitumor immunity. YTHDF2 deficiency in NK Cells impaired NK Cells’ antitumor and antiviral activity in vivo. Upon activation by cytokines, YTHDF2 is upregulated in NK Cells. More interestingly, YTHDF2 promoted NK Cell effector function by inhibiting a STAT5-YTHDF2-positive feedback loop involved in tumor progression (Ma et al., 2021). These findings suggested that m6A and its regulatory or associated proteins are involved in cancer progression. The development of new applicable inhibitors or the translation of existing inhibitors into clinical practice may provide innovative and effective therapeutic strategies for treatment (Table 3).
TABLE 3.
Non-exhaustive list of Potential alternative therapeutic agents offers by m6A targeting modifications in cancer.
| m6A proteins involved | Drugs/Therapeutic agents | Metabolites Pathways/Immune system | Underlying mechanism and Key results | References |
|---|---|---|---|---|
| METTL3 | Doxorubicin (DOX) | Glycolytic metabolism/Antitumor | METTL3 depletion decreased PDK4 expression and increased sensitivity to doxorubicin treatment in cervical cancer cells | Li et al. (2020c) |
| METTL3 | STM2457 | Antitumor | STM2457 by targeting METTL3 increased apoptosis and reduced AML growth treating myeloid leukemia | Yankova et al. (2021) |
| METTL3 | Cisplatin, Gemcitabine, 5-fluorouracil | Antitumor | Depleting METTL3 from cells induced resistance to cisplatin, gemcitabine, and 5-fluorouracil in pancreatic cancer and non-small cell lung cancer | Jin et al. (2019) |
| METTL3 | Gamma-irradiation | Antitumor | METTL3-silenced pancreatic cancer cells and glioma stem cells showed enhanced irradiation sensitivity | Visvanathan et al. (2018) |
| FTO | R-2HG | Metabolic regulation/ Antitumor | R-2HG, highly expressed by isocitrate dehydrogenase, inhibit FTO and act an antitumor in glioma and leukemisa cells | Su et al. (2018) |
| FTO | Entacapone | Metabolic regulation/Antitumor | Entacapone bound to FTO and inhibited FTO activity involved in the regulation of metabolic homeostasis and amino acid metabolism | Peng et al. (2019) |
| FTO | FB23 and FB23-2 | Antitumor | Targeting FTO, FB23 and FB23-2 are effective promise in preclinical models against acute myeloid leukemia | Huang et al. (2019) |
| FTO | Tyrosine kinase inhibitor (TKI) | Immunity control | Disregulated FTO help tumor cells to escape TKI-mediated killing and broad defense mechanism by which leukemia cells develop resistance mechanism to TKI | Yan et al. (2018) |
| FTO | Dac51 | Antitumor/Immunity control | Small molecule Dac51 can block FTO-mediated immune evasion and control immunity against cancer cells | Liu et al. (2021) |
| FTO | Glycolytic agents | Immunity control | Disregulated complex FTO - glycolytic agents help tumor cells to escape immune surveillance | Liu et al. (2021) |
| FTO | Anti-PD-1 blockade | Antitumor immunity | Knockdown of FTO sensitizes melanoma cells to interferon-gamma (IFNγ) and sensitizes melanoma to anti-PD-1 treatment in mice | Yang et al. (2019) |
| FTO | Temozolomide (TMZ) | Glycolytic metabolism/Antitumor | JPX/FTO/PDK1 axis facilitate aerobic glycolysis in GBM cells, and correlated with GBM cells' sensitivity to temozolomide | Li et al. (2021) |
| YTHDF1 | Cisplatin | Amino acid metabolism/Antitumor | YTHDF1 is associated with cisplatin resistance in colon cancer.Inhibition of GLS1 synergized with cisplatin to induce cell death of colon cancer cells | Chen et al. (2021b) |
| YTHDF1 | PD-L1 inhibitor | Antitumor immunity | YTHDF1 regulate antitumor immunity and have synergetic effect on immunotherapy by improving the therapeutic effect of PD-L1 inhibitor | Han et al. (2019) |
| YTHDF2 | STAT5 | Immune response | Upon activation by cytokines, YTHDF2 is upregulated in NK Cells. YTHDF2 promoted NK Cell effector function by inhibiting a STAT5-YTHDF2-positive feedback loop involved in tumor progression | Ma et al. (2021) |
Conclusion and Perspectives
The connection between metabolism and tumorigenesis is attracting attention, and many gratifying results have revealed the link between the m6A modification and oncometabolite in cancer progression. The data demonstrates that the m6A modification regulators could act as promising candidates for diagnosis, prognosis, or treatment against cancer. Thus, designing a diagnostic profile for cancer is possible based on oncometabolite regulated by m6A. In this review, the potential crosstalk between m6A RNA methylation and metabolic control in tumorigenesis was described. These findings build a link between metabolic reprogramming and the m6A modification. As investigators have focused mostly on glucose metabolism and performed in vitro studies with cell lines, their investigations need to be validated in animal models and clinical studies.
As integrated regulation of metabolism in cancers, the network of several major anabolic and catabolic pathways are important co-factors or substrates of the critical enzymes for RNA modifications. Since many of the metabolic alterations and consequently aberrated RNA regulation are common to a wide range of cancer types, they can serve as promising targets for anti-cancer therapies. Considering current efforts to target both cancer metabolism and regulation of the epigenome, it is still elusive to fully clarify the critical downstream factors functions mediated by some oncometabolite in cancer cells. Understanding the integrated metabolism in cancer cells may open new avenues for anti-cancer strategies. Therefore, determining metabolic differences between normal proliferating and cancer cells will be of great interest. Nevertheless, heterogeneity of tumors is yet another challenge, which is multiples phenotypes metabolic in multi-cellular systems. In addition, more researches should be conducted to better understand the molecular mechanisms among metabolic enzymes, transporters, transcription factors, and their pathways regulated by the m6A modification in cancer metabolism.
By pharmacological approach, evidence has shown that characterization of m6A writers and erasers proteins have provided valuable insights promising anti-cancer drugs targeting modification in cancer. While several small-molecule inhibitors targeting writers or erasers are either approved drugs or are currently being evaluated in clinical trials, the targeting m6A recognition proteins have lagged behind. After writers and erasers carry out methylation and demethylation, the readers determine the functional consequences of modification. Thus, more investigations and pharmacological research needs to target m6A readers in cancer progression to yield meaningful results.
Most importantly, attempts to target m6A pathways and their associated metabolic pathways need to consider immune cells, as m6A was recently reported to play roles in antitumor immunity, immune responses, and immunotherapy in cancers (Liu et al., 2021; Ma et al., 2021). Such an approach will help us better understand and fully clarify how the dysregulation of metabolism by m6A in tumorigenesis jeopardizes immune surveillance. As well as regulating glucose, amino acids, and lipids, m6A can regulate other metabolites, such as SAM, SAH, IDH, R-2HG, vitamin C, and iron. It will be interesting to understand how the m6A modification affects those compounds and how that knowledge could enhance cancer treatment. As m6A often alters metabolism, some metabolites might also regulate the production, editing, and recognition of m6A to affect cancer progression. Due to this controversial idea, it will also be interesting to discover how metabolite signaling networks regulate m6A in cancer and how they, in turn, could be regulated.
Author Contributions
PY, JY and TL conceived the review. YM and XL wrote the manuscript. PY, JY and TL. revised the manuscript with feedback from YM and XL All authors approved the manuscript for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Aik W., Demetriades M., Hamdan M. K. K., Bagg E. A. L., Yeoh K. K., Lejeune C., et al. (2013). Structural Basis for Inhibition of the Fat Mass and Obesity Associated Protein (FTO). J. Med. Chem. 56, 3680–3688. 10.1021/jm400193d [DOI] [PubMed] [Google Scholar]
- Alarcón C. R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. F. (2015). HNRNPA2B1 Is a Mediator of m6A-dependent Nuclear RNA Processing Events. Cell 162, 1299–1308. 10.1016/j.cell.2015.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancey P. B., Contat C., Meylan E. (2018). Glucose Transporters in Cancer - from Tumor Cells to the Tumor Microenvironment. Febs J. 285, 2926–2943. 10.1111/febs.14577 [DOI] [PubMed] [Google Scholar]
- Bartosovic M., Molares H. C., Gregorova P., Hrossova D., Kudla G., Vanacova S. (2017). N6-methyladenosine Demethylase FTO Targets Pre-mRNAs and Regulates Alternative Splicing and 3′-end Processing. Nucleic Acids Res. 45, 11356–11370. 10.1093/nar/gkx778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casasampere M., Ordoñez Y. F., Pou A., Casas J. (2016). Inhibitors of Dihydroceramide Desaturase 1: Therapeutic Agents and Pharmacological Tools to Decipher the Role of Dihydroceramides in Cell Biology. Chem. Phys. Lipids 197, 33–44. 10.1016/j.chemphyslip.2015.07.025 [DOI] [PubMed] [Google Scholar]
- Cheeseman C. (2008). GLUT7: a New Intestinal Facilitated Hexose Transporter. Am. J. Physiology-Endocrinology Metab. 295, E238–E241. 10.1152/ajpendo.90394.2008 [DOI] [PubMed] [Google Scholar]
- Chen H., Gao S., Liu W., Wong C.-C., Wu J., Wu J., et al. (2021a). RNA N6-Methyladenosine Methyltransferase METTL3 Facilitates Colorectal Cancer by Activating the m6A-GLUT1-mTORC1 Axis and Is a Therapeutic Target. Gastroenterology 160, 1284–1300. e1216. 10.1053/j.gastro.2020.11.013 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang S., Li Y., Tang Z., Kong W. (2014). Hexokinase 2 Overexpression Promotes the Proliferation and Survival of Laryngeal Squamous Cell Carcinoma. Tumor Biol. 35, 3743–3753. 10.1007/s13277-013-1496-2 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang Y.-C., Huang C., Shen H., Sun B., Cheng X., et al. (2019a). m6A Regulates Neurogenesis and Neuronal Development by Modulating Histone Methyltransferase Ezh2. Genomics, Proteomics & Bioinformatics 17, 154–168. 10.1016/j.gpb.2018.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Liu X.-q., Lin X., Gao L.-y., Zhang S., Huang X. (2021b). Targeting YTHDF1 Effectively Re-sensitizes Cisplatin-Resistant colon Cancer Cells by Modulating GLS-Mediated Glutamine Metabolism. Mol. Ther. - Oncolytics 20, 228–239. 10.1016/j.omto.2021.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-Y., Zhang J., Zhu J.-S. (2019b). The Role of m6A RNA Methylation in Human Cancer. Mol. Cancer 18, 103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Peng C., Chen J., Chen D., Yang B., He B., et al. (2019c). WTAP Facilitates Progression of Hepatocellular Carcinoma via m6A-HuR-dependent Epigenetic Silencing of ETS1. Mol. Cancer 18, 127. 10.1186/s12943-019-1053-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., et al. (2018). mRNA Circularization by METTL3-eIF3h Enhances Translation and Promotes Oncogenesis. Nature 561, 556–560. 10.1038/s41586-018-0538-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curi R., Lagranha C. J., Doi S. Q., Sellitti D. F., Procopio J., Pithon-Curi T. C., et al. (2005). Molecular Mechanisms of Glutamine Action. J. Cel. Physiol. 204, 392–401. 10.1002/jcp.20339 [DOI] [PubMed] [Google Scholar]
- Devedjiev Y., Steussy C. N., Vassylyev D. G. (2007). Crystal Structure of an Asymmetric Complex of Pyruvate Dehydrogenase Kinase 3 with Lipoyl Domain 2 and its Biological Implications. J. Mol. Biol. 370, 407–416. 10.1016/j.jmb.2007.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty J. R., Cleveland J. L. (2013). Targeting Lactate Metabolism for Cancer Therapeutics. J. Clin. Invest. 123, 3685–3692. 10.1172/jci69741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., et al. (2016). YTHDF2 Destabilizes m(6)A-Containing RNA Through Direct Recruitment of the CCR4-NOT Deadenylase Complex. Nat. Commun. 7, 12626. 10.1038/ncomms12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang R., Chen X., Zhang S., Shi H., Ye Y., Shi H., et al. (2021). EGFR/SRC/ERK-stabilized YTHDF2 Promotes Cholesterol Dysregulation and Invasive Growth of Glioblastoma. Nat. Commun. 12, 177. 10.1038/s41467-020-20379-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faubert B., Li K. Y., Cai L., Hensley C. T., Kim J., Zacharias L. G., et al. (2017). Lactate Metabolism in Human Lung Tumors. Cell 171, 358–371. 10.1016/j.cell.2017.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedeles B. I., Singh V., Delaney J. C., Li D., Essigmann J. M. (2015). The AlkB Family of Fe(II)/α-Ketoglutarate-dependent Dioxygenases: Repairing Nucleic Acid Alkylation Damage and beyond. J. Biol. Chem. 290, 20734–20742. 10.1074/jbc.r115.656462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Liu Y., Wang G., Deng Z., Zhang Q., Wu W., et al. (2014). Crystal Structures of the Human RNA Demethylase Alkbh5 Reveal Basis for Substrate Recognition. J. Biol. Chem. 289, 11571–11583. 10.1074/jbc.m113.546168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C. (2020). Metabolism and Cancer: the Future Is Now. Br. J. Cancer 122, 133–135. 10.1038/s41416-019-0667-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry N. J., Law B. A., Ilkayeva O. R., Holley C. L., Mansfield K. D. (2017). N6-methyladenosine Is Required for the Hypoxic Stabilization of Specific mRNAs. Rna 23, 1444–1455. 10.1261/rna.061044.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., Harada B. T., Behm M., He C. (2018). RNA Modifications Modulate Gene Expression during Development. Science 361, 1346–1349. 10.1126/science.aau1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Jia G., Pang X., Wang R. N., Wang X., Li C. J., et al. (2013). FTO-mediated Formation of N6-Hydroxymethyladenosine and N6-Formyladenosine in Mammalian RNA. Nat. Commun. 4, 1798. 10.1038/ncomms2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaida M. M., Mayer C., Dapunt U., Stegmaier S., Schirmacher P., Wabnitz G. H., et al. (2016). Expression of the Bitter Receptor T2R38 in Pancreatic Cancer: Localization in Lipid Droplets and Activation by a Bacteria-Derived Quorum-sensing Molecule. Oncotarget 7, 12623–12632. 10.18632/oncotarget.7206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganapathy-Kanniappan S., Geschwind J.-F. H. (2013). Tumor Glycolysis as a Target for Cancer Therapy: Progress and Prospects. Mol. Cancer 12, 152. 10.1186/1476-4598-12-152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. (2006). Energy Deregulation: Licensing Tumors to Grow. Science 312, 1158–1159. 10.1126/science.312.5777.1158 [DOI] [PubMed] [Google Scholar]
- Guo W., Zhang C., Feng P., Li M., Wang X., Xia Y., et al. (2021). M6A Methylation of DEGS2, a Key Ceramide-Synthesizing Enzyme, Is Involved in Colorectal Cancer Progression through Ceramide Synthesis. Oncogene. 10.1038/s41388-021-01987-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halestrap A. P. (2013). Monocarboxylic Acid Transport. Compr. Physiol. 3, 1611–1643. 10.1002/cphy.c130008 [DOI] [PubMed] [Google Scholar]
- Han D., Liu J., Chen C., Dong L., Liu Y., Chang R., et al. (2019). Anti-tumour Immunity Controlled through mRNA m6A Methylation and YTHDF1 in Dendritic Cells. Nature 566, 270–274. 10.1038/s41586-019-0916-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of Cancer: the Next Generation. Cell 144, 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hsu P. J., Zhu Y., Ma H., Guo Y., Shi X., Liu Y., et al. (2017). Ythdc2 Is an N6-Methyladenosine Binding Protein that Regulates Mammalian Spermatogenesis. Cell Res 27, 1115–1127. 10.1038/cr.2017.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., et al. (2018). Recognition of RNA N6-Methyladenosine by IGF2BP Proteins Enhances mRNA Stability and Translation. Nat. Cel Biol 20, 285–295. 10.1038/s41556-018-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Su R., Sheng Y., Dong L., Dong Z., Xu H., et al. (2019). Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 35, 677–691. e610. 10.1016/j.ccell.2019.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova V. V., Stolz P., Kaiser S., Gustafsson T. H., Lastres P. R., Sanz-Moreno A., et al. (2020). The rRNA m6A Methyltransferase METTL5 Is Involved in Pluripotency and Developmental Programs. Genes Dev. 34, 715–729. 10.1101/gad.333369.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D., Guo J., Wu Y., Du J., Yang L., Wang X., et al. (2019). m6A mRNA Methylation Initiated by METTL3 Directly Promotes YAP Translation and Increases YAP Activity by Regulating the MALAT1-miR-1914-3p-YAP axis to Induce NSCLC Drug Resistance and Metastasis. J. Hematol. Oncol. 12, 135. 10.1186/s13045-019-0830-6 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Jung M., Mertens C., Tomat E., Brüne B. (2019). Iron as a Central Player and Promising Target in Cancer Progression. Int. J. Mol. Sci. 20. 10.3390/ijms20020273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy P., Gyimesi G., Kanai Y., Hediger M. A. (2018). Amino Acid Transporters Revisited: New Views in Health and Disease. Trends Biochemical Sciences 43, 752–789. 10.1016/j.tibs.2018.05.003 [DOI] [PubMed] [Google Scholar]
- Kang H., Zhang Z., Yu L., Li Y., Liang M., Zhou L. (2018). FTO Reduces Mitochondria and Promotes Hepatic Fat Accumulation through RNA Demethylation. J. Cel. Biochem. 119, 5676–5685. 10.1002/jcb.26746 [DOI] [PubMed] [Google Scholar]
- Khan M. A., Zubair H., Anand S., Srivastava S. K., Singh S., Singh A. P. (2020). Dysregulation of Metabolic Enzymes in Tumor and Stromal Cells: Role in Oncogenesis and Therapeutic Opportunities. Cancer Lett. 473, 176–185. 10.1016/j.canlet.2020.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P., Lence T., Haussmann I. U., Jacob D., Kreim N., Carl S. H., et al. (2018). Zc3h13/Flacc Is Required for Adenosine Methylation by Bridging the mRNA-Binding Factor Rbm15/Spenito to the m6A Machinery Component Wtap/Fl(2)d. Genes Dev. 32, 415–429. 10.1101/gad.309146.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nagpal R., Kumar A., Ashraf M. U., Bae Y. S. (2021). Immunotherapeutic Potential of m6A-Modifiers and MicroRNAs in Controlling Acute Myeloid Leukaemia. Biomedicines 9. 10.3390/biomedicines9060690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis C. J. T., Pan T., Kalsotra A. (2017). RNA Modifications and Structures Cooperate to Guide RNA-Protein Interactions. Nat. Rev. Mol. Cel Biol 18, 202–210. 10.1038/nrm.2016.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Chen Y.-S., Ping X.-L., Yang X., Xiao W., Yang Y., et al. (2017a). Cytoplasmic m6A Reader YTHDF3 Promotes mRNA Translation. Cel Res 27, 444–447. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Su Q., Li B., Lan L., Wang C., Li W., et al. (2020a). High Expression of WTAP Leads to Poor Prognosis of Gastric Cancer by Influencing Tumour‐associated T Lymphocyte Infiltration. J. Cel Mol Med 24, 4452–4465. 10.1111/jcmm.15104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhu L., Shi Y., Liu J., Lin L., Chen X. (2019a). m6A Demethylase FTO Promotes Hepatocellular Carcinoma Tumorigenesis via Mediating PKM2 Demethylation. Am. J. Transl Res. 11, 6084–6092. [PMC free article] [PubMed] [Google Scholar]
- Li T., Hu P.-S., Zuo Z., Lin J.-F., Li X., Wu Q.-N., et al. (2019b). METTL3 Facilitates Tumor Progression via an m6A-igf2bp2-dependent Mechanism in Colorectal Carcinoma. Mol. Cancer 18, 112. 10.1186/s12943-019-1038-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. D., Wang M. J., Zheng J. L., Wu Y. H., Wang X., Jiang X. B. (2021). Long Noncoding RNA Just Proximal to X‐inactive Specific Transcript Facilitates Aerobic Glycolysis and Temozolomide Chemoresistance by Promoting Stability of PDK1 mRNA in an m6A‐dependent Manner in Glioblastoma Multiforme Cells. Cancer Sci. 112, 4543–4552. 10.1111/cas.15072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Li F., Peng Y., Fang J., Zhou J. (2020b). Identification of Three m6A‐related mRNAs Signature and Risk Score for the Prognostication of Hepatocellular Carcinoma. Cancer Med. 9, 1877–1889. 10.1002/cam4.2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng Y., Li J., Chen Z., Chen F., Tu J., et al. (2020c). N6-methyladenosine Regulates Glycolysis of Cancer Cells through PDK4. Nat. Commun. 11, 2578. 10.1038/s41467-020-16306-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Weng H., Su R., Weng X., Zuo Z., Li C., et al. (2017b). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N 6 -Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. 10.1016/j.ccell.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Zhang H. (2016). Reprogramming of Glucose, Fatty Acid and Amino Acid Metabolism for Cancer Progression. Cell. Mol. Life Sci. 73, 377–392. 10.1007/s00018-015-2070-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Choe J., Du P., Triboulet R., Gregory R. I. (2016). The M 6 A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cel 62, 335–345. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Wei X., Jian Z., Zhang X. (2020). METTL3 Expression Is Associated with Glycolysis Metabolism and Sensitivity to Glycolytic Stress in Hepatocellular Carcinoma. Cancer Med. 9, 2859–2867. 10.1002/cam4.2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Eckert M. A., Harada B. T., Liu S.-M., Lu Z., Yu K., et al. (2018). m6A mRNA Methylation Regulates AKT Activity to Promote the Proliferation and Tumorigenicity of Endometrial Cancer. Nat. Cel Biol 20, 1074–1083. 10.1038/s41556-018-0174-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., et al. (2014). A METTL3-METTL14 Complex Mediates Mammalian Nuclear RNA N6-Adenosine Methylation. Nat. Chem. Biol. 10, 93–95. 10.1038/nchembio.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. (2015). N6-methyladenosine-dependent RNA Structural Switches Regulate RNA-Protein Interactions. Nature 518, 560–564. 10.1038/nature14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wei Q., Jin J., Luo Q., Liu Y., Yang Y., et al. (2020). The m6A Reader YTHDF1 Promotes Ovarian Cancer Progression via Augmenting EIF3C Translation. Nucleic Acids Res. 48, 3816–3831. 10.1093/nar/gkaa048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liang G., Xu H., Dong W., Dong Z., Qiu Z., et al. (2021). Tumors Exploit FTO-Mediated Regulation of Glycolytic Metabolism to Evade Immune Surveillance. Cell Metab. 10.1016/j.cmet.2021.04.001 [DOI] [PubMed] [Google Scholar]
- Losman J.-A., Koivunen P., Kaelin W. G., Jr. (2020). 2-Oxoglutarate-dependent Dioxygenases in Cancer. Nat. Rev. Cancer 20, 710–726. 10.1038/s41568-020-00303-3 [DOI] [PubMed] [Google Scholar]
- Ma H., Wang X., Cai J., Dai Q., Natchiar S. K., Lv R., et al. (2019). N6-Methyladenosine Methyltransferase ZCCHC4 Mediates Ribosomal RNA Methylation. Nat. Chem. Biol. 15, 88–94. 10.1038/s41589-018-0184-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. z., Yang F., Zhou C. c., Liu F., Yuan J. h., Wang F., et al. (2017). METTL14 Suppresses the Metastatic Potential of Hepatocellular Carcinoma by Modulating N 6 ‐methyladenosine‐dependent Primary MicroRNA Processing. Hepatology 65, 529–543. 10.1002/hep.28885 [DOI] [PubMed] [Google Scholar]
- Ma S., Yan J., Barr T., Zhang J., Chen Z., Wang L. S., et al. (2021). The RNA m6A Reader YTHDF2 Controls NK Cell Antitumor and Antiviral Immunity. J. Exp. Med. 218. 10.1084/jem.20210279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheda M. L., Rogers S., Best J. D. (2005). Molecular and Cellular Regulation of Glucose Transporter (GLUT) Proteins in Cancer. J. Cel. Physiol. 202, 654–662. 10.1002/jcp.20166 [DOI] [PubMed] [Google Scholar]
- Matés J. M., Segura J. A., Martín-Rufián M., Campos-Sandoval J. A., Alonso F. J., Márquez J. (2013). Glutaminase Isoenzymes as Key Regulators in Metabolic and Oxidative Stress against Cancer. Curr. Mol. Med. 13, 514–534. [DOI] [PubMed] [Google Scholar]
- Meyer K. D., Patil D. P., Zhou J., Zinoviev A., Skabkin M. A., Elemento O., et al. (2015). 5′ UTR m6A Promotes Cap-independent Translation. Cell 163, 999–1010. 10.1016/j.cell.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T. M., Lew P. S., Luo Y., Leckstrom A. (2017). Negative Regulation of Hepatic Fat Mass and Obesity Associated (Fto) Gene Expression by Insulin. Life Sci. 170, 50–55. 10.1016/j.lfs.2016.11.027 [DOI] [PubMed] [Google Scholar]
- Murai T. (2015). Lipid Raft-Mediated Regulation of Hyaluronan-CD44 Interactions in Inflammation and Cancer. Front. Immunol. 6, 420. 10.3389/fimmu.2015.00420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan A. K. (2019). mTOR: Role in Cancer, Metastasis and Drug Resistance. Semin. Cancer Biol. 59, 92–111. 10.1016/j.semcancer.2019.07.003 [DOI] [PubMed] [Google Scholar]
- Nath A., Chan C. (2016). Genetic Alterations in Fatty Acid Transport and Metabolism Genes Are Associated with Metastatic Progression and Poor Prognosis of Human Cancers. Sci. Rep. 6, 18669. 10.1038/srep18669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M. S., Korotchkina L. G. (2006). Regulation of the Pyruvate Dehydrogenase Complex. Biochem. Soc. Trans. 34, 217–222. 10.1042/bst0340217 [DOI] [PubMed] [Google Scholar]
- Pendleton K. E., Chen B., Liu K., Hunter O. V., Xie Y., Tu B. P., et al. (2017). The U6 snRNA M 6 A Methyltransferase METTL16 Regulates SAM Synthetase Intron Retention. Cell 169, 824–835. e814. 10.1016/j.cell.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S., Xiao W., Ju D., Sun B., Hou N., Liu Q., et al. (2019). Identification of Entacapone as a Chemical Inhibitor of FTO Mediating Metabolic Regulation through FOXO1. Sci. Transl Med. 11. 10.1126/scitranslmed.aau7116 [DOI] [PubMed] [Google Scholar]
- Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., et al. (2014). Mammalian WTAP Is a Regulatory Subunit of the RNA N6-Methyladenosine Methyltransferase. Cel Res 24, 177–189. 10.1038/cr.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priolo C., Pyne S., Rose J., Regan E. R., Zadra G., Photopoulos C., et al. (2014). AKT1 and MYC Induce Distinctive Metabolic Fingerprints in Human Prostate Cancer. Cancer Res. 74, 7198–7204. 10.1158/0008-5472.can-14-1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing Y., Dong L., Gao L., Li C., Li Y., Han L., et al. (2021). R-2-hydroxyglutarate Attenuates Aerobic Glycolysis in Leukemia by Targeting the FTO/m6A/PFKP/LDHB axis. Mol. Cel 81, 922–939. e929. 10.1016/j.molcel.2020.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I. A., Luo G. Z., Zhang Z., Wang X., Zhou T., Cui Y., et al. (2017b). YTHDC1 Mediates Nuclear export of N6-Methyladenosine Methylated mRNAs. Elife 6. 10.7554/eLife.31311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree I. A., Evans M. E., Pan T., He C. (2017a). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze A., Harris A. L. (2012). How Cancer Metabolism Is Tuned for Proliferation and Vulnerable to Disruption. Nature 491, 364–373. 10.1038/nature11706 [DOI] [PubMed] [Google Scholar]
- Shaw R. J. (2006). Glucose Metabolism and Cancer. Curr. Opin. Cel. Biol. 18, 598–608. 10.1016/j.ceb.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Shen C., Xuan B., Yan T., Ma Y., Xu P., Tian X., et al. (2020). m6A-dependent Glycolysis Enhances Colorectal Cancer Progression. Mol. Cancer 19, 72. 10.1186/s12943-020-01190-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F., Huang W., Huang J.-T., Xiong J., Yang Y., Wu K., et al. (2015). DecreasedN6-Methyladenosine in Peripheral Blood RNA from Diabetic Patients Is Associated WithFTOExpression rather ThanALKBH5. J. Clin. Endocrinol. Metab. 100, E148–E154. 10.1210/jc.2014-1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R. (2020). Commentary on 'Metabolic Reprogramming‐associated Genes Predict Overall Survival for Rectal Cancer'. J. Cel. Mol. Med. 24, 12862–12863. 10.1111/jcmm.15938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng H., Li Z., Su S., Sun W., Zhang X., Li L., et al. (2020). YTH Domain Family 2 Promotes Lung Cancer Cell Growth by Facilitating 6-phosphogluconate Dehydrogenase mRNA Translation. Carcinogenesis 41, 541–550. 10.1093/carcin/bgz152 [DOI] [PubMed] [Google Scholar]
- Shi H., Wang X., Lu Z., Zhao B. S., Ma H., Hsu P. J., et al. (2017). YTHDF3 Facilitates Translation and Decay of N6-Methyladenosine-Modified RNA. Cel Res 27, 315–328. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wei J., He C. (2019). Where, when, and How: Context-dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol. Cel 74, 640–650. 10.1016/j.molcel.2019.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanand S., Vander Heiden M. G. (2020). Emerging Roles for Branched-Chain Amino Acid Metabolism in Cancer. Cancer Cell 37, 147–156. 10.1016/j.ccell.2019.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledz P., Jinek M. (2016). Structural Insights into the Molecular Mechanism of the M(6)A Writer Complex. Elife 5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacpoole P. W. (2017). Therapeutic Targeting of the Pyruvate Dehydrogenase Complex/Pyruvate Dehydrogenase Kinase (PDC/PDK) Axis in Cancer. J. Natl. Cancer Inst. 109. 10.1093/jnci/djx071 [DOI] [PubMed] [Google Scholar]
- Stattin P., Björ O., Ferrari P., Lukanova A., Lenner P., Lindahl B., et al. (2007). Prospective Study of Hyperglycemia and Cancer Risk. Diabetes care 30, 561–567. 10.2337/dc06-0922 [DOI] [PubMed] [Google Scholar]
- Strowitzki M. J., Radhakrishnan P., Pavicevic S., Scheer J., Kimmer G., Ritter A. S., et al. (2019). High Hepatic Expression of PDK4 Improves Survival upon Multimodal Treatment of Colorectal Liver Metastases. Br. J. Cancer 120, 675–688. 10.1038/s41416-019-0406-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 172, 90–105. e123. 10.1016/j.cell.2017.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swierczynski J., Hebanowska A., Sledzinski T. (2014). Role of Abnormal Lipid Metabolism in Development, Progression, Diagnosis and Therapy of Pancreatic Cancer. Wjg 20, 2279–2303. 10.3748/wjg.v20.i9.2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taketo K., Konno M., Asai A., Koseki J., Toratani M., Satoh T., et al. (2018). The Epitranscriptome m6A Writer METTL3 Promotes Chemo- and Radioresistance in Pancreatic Cancer Cells. Int. J. Oncol. 52, 621–629. 10.3892/ijo.2017.4219 [DOI] [PubMed] [Google Scholar]
- Ueda Y., Ooshio I., Fusamae Y., Kitae K., Kawaguchi M., Jingushi K., et al. (2017). AlkB Homolog 3-mediated tRNA Demethylation Promotes Protein Synthesis in Cancer Cells. Sci. Rep. 7, 42271. 10.1038/srep42271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009). Understanding the Warburg Effect: the Metabolic Requirements of Cell Proliferation. Science 324, 1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vettore L., Westbrook R. L., Tennant D. A. (2020). New Aspects of Amino Acid Metabolism in Cancer. Br. J. Cancer 122, 150–156. 10.1038/s41416-019-0620-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A., Pettazzoni P., Lyssiotis C. A., Ying H., Sánchez N., Marchesini M., et al. (2014). Oncogene Ablation-Resistant Pancreatic Cancer Cells Depend on Mitochondrial Function. Nature 514, 628–632. 10.1038/nature13611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visvanathan A., Patil V., Arora A., Hegde A. S., Arivazhagan A., Santosh V., et al. (2018). Essential Role of METTL3-Mediated m6A Modification in Glioma Stem-like Cells Maintenance and Radioresistance. Oncogene 37, 522–533. 10.1038/onc.2017.351 [DOI] [PubMed] [Google Scholar]
- Wang K., Jiang L., Zhang Y., Chen C. (2020a). Progression of Thyroid Carcinoma Is Promoted by the m6A Methyltransferase METTL3 through Regulating m6A Methylation on TCF1. Ott 13, 1605–1612. 10.2147/ott.s234751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Song C., Wang N., Li S., Liu Q., Sun Z., et al. (2020b). NADP Modulates RNA m6A Methylation and Adipogenesis via Enhancing FTO Activity. Nat. Chem. Biol. 16, 1394–1402. 10.1038/s41589-020-0601-2 [DOI] [PubMed] [Google Scholar]
- Wang Q., Chen C., Ding Q., Zhao Y., Wang Z., Chen J., et al. (2020c). METTL3-mediated m6A Modification of HDGF mRNA Promotes Gastric Cancer Progression and Has Prognostic Significance. Gut 69, 1193–1205. 10.1136/gutjnl-2019-319639 [DOI] [PubMed] [Google Scholar]
- Wang S., Sun C., Li J., Zhang E., Ma Z., Xu W., et al. (2017). Roles of RNA Methylation by Means of N6-Methyladenosine (m6A) in Human Cancers. Cancer Lett. 408, 112–120. 10.1016/j.canlet.2017.08.030 [DOI] [PubMed] [Google Scholar]
- Wang X., Zhao B. S., Roundtree I. A., Lu Z., Han D., Ma H., et al. (2015). N6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Buyse J., Song Z., Decuypere E., Everaert N. (2016a). AMPK Is Involved in the Differential Neonatal Performance of Chicks Hatching at Different Time. Gen. Comp. Endocrinol. 228, 53–59. 10.1016/j.ygcen.2016.02.008 [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Loo T. Y., Shen J.-G., Wang N., Wang D.-M., Yang D.-P., et al. (2012). LDH-A Silencing Suppresses Breast Cancer Tumorigenicity through Induction of Oxidative Stress Mediated Mitochondrial Pathway Apoptosis. Breast Cancer Res. Treat. 131, 791–800. 10.1007/s10549-011-1466-6 [DOI] [PubMed] [Google Scholar]
- Wang Z., Wang N., Liu P., Xie X. (2016b2012). AMPK and Cancer. Experientia supplementum 107, 203–226. 10.1007/978-3-319-43589-3_9 [DOI] [PubMed] [Google Scholar]
- Wei J., Liu F., Lu Z., Fei Q., Ai Y., He P. C., et al. (2018). Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cel 71, 973–985. 10.1016/j.molcel.2018.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson J. E. (2003). Isozymes of Mammalian Hexokinase: Structure, Subcellular Localization and Metabolic Function. J. Exp. Biol. 206, 2049–2057. 10.1242/jeb.00241 [DOI] [PubMed] [Google Scholar]
- Wu B., Li L., Huang Y., Ma J., Min J. (2017a). Readers, Writers and Erasers of N6-Methylated Adenosine Modification. Curr. Opin. Struct. Biol. 47, 67–76. 10.1016/j.sbi.2017.05.011 [DOI] [PubMed] [Google Scholar]
- Wu J., Hu L., Wu F., Zou L., He T. (2017b). Poor Prognosis of Hexokinase 2 Overexpression in Solid Tumors of Digestive System: a Meta-Analysis. Oncotarget 8, 32332–32344. 10.18632/oncotarget.15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T., Wu X., Cao M., Zhang P., Shi G., Zhang J., et al. (2019). The RNA m6A Methyltransferase METTL3 Promotes Pancreatic Cancer Cell Proliferation and Invasion. Pathol. - Res. Pract. 215, 152666. 10.1016/j.prp.2019.152666 [DOI] [PubMed] [Google Scholar]
- Xiang S., Liang X., Yin S., Liu J., Xiang Z. (2020). N6-methyladenosine Methyltransferase METTL3 Promotes Colorectal Cancer Cell Proliferation through Enhancing MYC Expression. Am. J. Transl Res. 12, 1789–1806. [PMC free article] [PubMed] [Google Scholar]
- Xu C., Wang X., Liu K., Roundtree I. A., Tempel W., Li Y., et al. (2014). Structural Basis for Selective Binding of m6A RNA by the YTHDC1 YTH Domain. Nat. Chem. Biol. 10, 927–929. 10.1038/nchembio.1654 [DOI] [PubMed] [Google Scholar]
- Xu G.-L., Bochtler M. (2020). Reversal of Nucleobase Methylation by Dioxygenases. Nat. Chem. Biol. 16, 1160–1169. 10.1038/s41589-020-00675-5 [DOI] [PubMed] [Google Scholar]
- Yan F., Al-Kali A., Zhang Z., Liu J., Pang J., Zhao N., et al. (2018). A Dynamic N6-Methyladenosine Methylome Regulates Intrinsic and Acquired Resistance to Tyrosine Kinase Inhibitors. Cel Res 28, 1062–1076. 10.1038/s41422-018-0097-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Sun Z., Zhang N. (2020). Reshaping the Role of m6A Modification in Cancer Transcriptome: a Review. Cancer Cel Int 20, 353. 10.1186/s12935-020-01445-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Wei J., Cui Y.-H., Park G., Shah P., Deng Y., et al. (2019). m6A mRNA Demethylase FTO Regulates Melanoma Tumorigenicity and Response to Anti-PD-1 Blockade. Nat. Commun. 10, 2782. 10.1038/s41467-019-10669-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Shao F., Guo D., Wang W., Wang J., Zhu R., et al. (2021). WNT/β-catenin-suppressed FTO Expression Increases m6A of C-Myc mRNA to Promote Tumor Cell Glycolysis and Tumorigenesis. Cell Death Dis 12, 462. 10.1038/s41419-021-03739-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankova E., Blackaby W., Albertella M., Rak J., De Braekeleer E., Tsagkogeorga G., et al. (2021). Small Molecule Inhibition of METTL3 as a Strategy against Myeloid Leukaemia. Nature. 10.1038/s41586-021-03536-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Wang Z., Chen X., Jiang X., Dong Z., Hu S., et al. (2020). YTHDF1-enhanced Iron Metabolism Depends on TFRC m6A Methylation. Theranostics 10, 12072–12089. 10.7150/thno.51231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.-L., Ma X.-D., Tong J.-F., Li J.-Q., Guan X.-J., Yang J. (2019). WTAP Is a Prognostic Marker of High-Grade Serous Ovarian Cancer and Regulates the Progression of Ovarian Cancer Cells. Ott 12, 6191–6201. 10.2147/ott.s205730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., Zhao K., Zeng H., Li Z., Chen K., Zhang Z., et al. (2021). N6-methyladenosine (m6A) Methyltransferase WTAP Accelerates the Warburg Effect of Gastric Cancer through Regulating HK2 Stability. Biomed. Pharmacother. 133, 111075. 10.1016/j.biopha.2020.111075 [DOI] [PubMed] [Google Scholar]
- Yue C., Chen J., Li Z., Li L., Chen J., Guo Y. (2020). microRNA-96 Promotes Occurrence and Progression of Colorectal Cancer via Regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 39, 240. 10.1186/s13046-020-01731-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., et al. (2018). VIRMA Mediates Preferential m6A mRNA Methylation in 3′UTR and Near Stop Codon and Associates with Alternative Polyadenylation. Cell Discov 4, 10. 10.1038/s41421-018-0019-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wei L.-H., Wang Y., Xiao Y., Liu J., Zhang W., et al. (2019). Structural Insights into FTO's Catalytic Mechanism for the Demethylation of Multiple RNA Substrates. Proc. Natl. Acad. Sci. USA 116, 2919–2924. 10.1073/pnas.1820574116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. S., Roundtree I. A., He C. (2017). Post-transcriptional Gene Regulation by mRNA Modifications. Nat. Rev. Mol. Cel Biol 18, 31–42. 10.1038/nrm.2016.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G., Dahl J. A., Niu Y., Fedorcsak P., Huang C.-M., Li C. J., et al. (2013). ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cel 49, 18–29. 10.1016/j.molcel.2012.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L., Liao D., Zhang M., Zeng C., Li X., Zhang R., et al. (2019). YTHDF2 Suppresses Cell Proliferation and Growth via Destabilizing the EGFR mRNA in Hepatocellular Carcinoma. Cancer Lett. 442, 252–261. 10.1016/j.canlet.2018.11.006 [DOI] [PubMed] [Google Scholar]
- Zhong X., Yu J., Frazier K., Weng X., Li Y., Cham C. M., et al. (2018). Circadian Clock Regulation of Hepatic Lipid Metabolism by Modulation of m6A mRNA Methylation. Cel Rep. 25, 1816–1828. e1814. 10.1016/j.celrep.2018.10.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Liu C., Xu L., Yuan Y., Zhao J., Zhao W., et al. (2021). N 6 ‐Methyladenosine Reader Protein YT521‐B Homology Domain‐Containing 2 Suppresses Liver Steatosis by Regulation of mRNA Stability of Lipogenic Genes. Hepatology 73, 91–103. 10.1002/hep.31220 [DOI] [PubMed] [Google Scholar]
- Zhu S., Wang J.-Z., Chen D., He Y.-T., Meng N., Chen M., et al. (2020). An Oncopeptide Regulates m6A Recognition by the m6A Reader IGF2BP1 and Tumorigenesis. Nat. Commun. 11, 1685. 10.1038/s41467-020-15403-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou D., Dong L., Li C., Yin Z., Rao S., Zhou Q. (2019). The m6A Eraser FTO Facilitates Proliferation and Migration of Human Cervical Cancer Cells. Cancer Cel Int 19, 321. 10.1186/s12935-019-1045-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X., Chen Z., Gao W., Zhang Y., Wang J., Wang J., et al. (2020). M6A-mediated Upregulation of LINC00958 Increases Lipogenesis and Acts as a Nanotherapeutic Target in Hepatocellular Carcinoma. J. Hematol. Oncol. 13, 5. 10.1186/s13045-019-0839-x [DOI] [PMC free article] [PubMed] [Google Scholar]