Abstract
Two cases of conjunctivitis caused by adenovirus type 34 (Ad34) are reported. The isolates were identified as Ad34 by the neutralization test and the PCR-sequence method of the hexon gene but as Ad14 by PCR-restriction fragment length polymorphism analysis. The genome types of these two isolates were identical to that of Ad34a.
Adenovirus type 34 (Ad34) has been isolated from the urine of a renal transplant recipient and patients with AIDS (4–6). However, two cases of acute follicular conjunctivitis caused by Ad34 in patients without serious systemic disorder have been reported (11). These clinical isolates were identified as Ad34 by the standard neutralization test (NT) and PCR-sequence method (10) in 1998. The PCR-sequence method combines nested PCR, which amplifies hypervariable regions (HVRs) upstream of the hexon gene (40 to 1,847 bp) participating in type-specific neutralization (2, 3), with direct sequencing. It was suggested that HVR4, HVR5, and HVR7 were candidates for neutralization epitopes among seven HVRs (10).
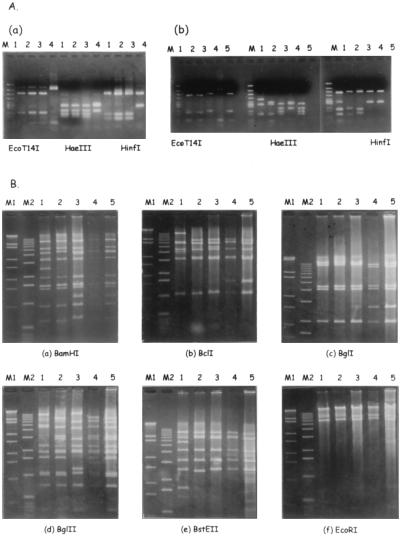
For comparison with NT and the PCR-sequence method, PCR-restriction fragment length polymorphism (RFLP) analysis was also performed to determine the serotypes of these two conjunctival scrapings, as previously described (8). Briefly, nested PCR was performed to amplify the 956-bp DNA fragment (20,743 to 21,698 bp) downstream of the hexon gene. Different restriction patterns were obtained with three restriction enzymes, EcoT14I, HaeIII, and HinfI (Takara Shuzo Co., Kyoto, Japan). Restriction enzyme digests were electrophoresed on 2% agarose gels in Tris-acetate buffer (pH 8.0) with 1 mM EDTA. After staining with ethidium bromide (1 μg/ml), the fragments were visualized under a UV transilluminator and photographed with a Polaroid camera. φX174 HincII digests (Takara Shuzo Co.) were run as a molecular weight standard. The clinical isolates were identified as Ad14 prototype (Ad14p) (Fig. 1A, panel a), which was shown to be Ad34/Ad14 by Takeuchi et al. (10). Then, it was evaluated whether prototypes of Ad3, Ad7, Ad14, Ad34, and Ad35 (Ad3p, Ad7p, Ad14p, Ad34p, and Ad35p) could be classified by PCR-RFLP analysis, because only prototypes of Ad1 through Ad8, Ad11, Ad14, Ad19, Ad37, Ad40, and Ad41 had been used to establish the PCR-RFLP analysis. Ad3, Ad4, Ad7, Ad8, Ad11, Ad19, and Ad37 are considered to be causative serotypes of acute conjunctivitis. It was shown that all of them could be classified with this method (Fig. 1A, panel b).
FIG. 1.
(A) (Panel a) Two clinical isolates were identified as Ad14p by PCR-RFLP analysis with EcoT14I, HaeIII, and HinfI. Lanes: M, φ X174/HincII; 1, isolate A; 2, isolate B; 3, Ad14p; 4, Ad34p. (Panel b) PCR-RFLP analysis was able to classify Ad3p, Ad7p, Ad14p, Ad34p, and Ad35p. Lanes: M, φ X174/HincII; 1, Ad3p; 2, Ad7p; 3, Ad14p; 4, Ad34p; 5, Ad35p. (B) Restriction patterns obtained after cleavage of Ad14p, Ad34p, Ad35p, and two clinical isolates with BamHI (a), BclI (b), BglI (c), BglII (d), BstEII (e), and EcoRI (f). Lanes: M1, λ/EcoT14I; M2, 1-kb ladder; 1, Ad14p; 2, Ad34p; 3, Ad35p; 4, isolate A; 5, isolate B.
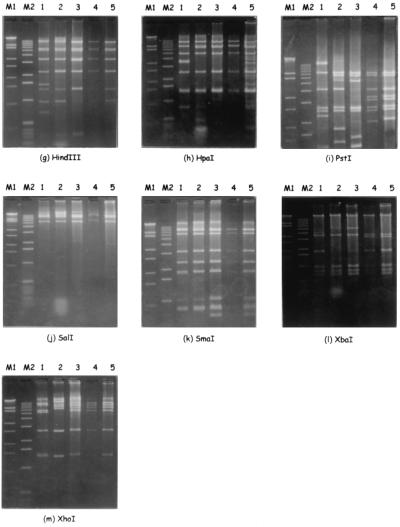
To investigate the reason for the discrepancy between the results of NT, the PCR-sequence method, and PCR-RFLP analysis, restriction enzyme digestion of adenoviral genomic DNA (genome typing) was carried out with these clinical isolates. Virus propagation was performed in 25-cm2 plastic flasks containing a monolayer of HEp-2 or A549 cells with incubation at 35°C. When a typical adenovirus cytopathic effect was evident, usually 48 to 72 h after infection, the viral DNA was extracted following the protocol described by Wadell and de Jong (1980) (12), with some modifications. The cells were pelleted and rinsed twice with phosphate-buffered saline and then suspended in 1 ml of Hirt lysis solution (10 mM Tris, 1 mM EDTA, 0.6% sodium dodecyl sulfate [pH 8.0]). Proteinase K was added at a final concentration of 50 μg/ml, and the samples were incubated at 37°C for 1 h. Cellular DNA was precipitated with NaCl (1 M) overnight at 4°C and discarded. The supernatant was cleaned with a mixture of ribonucleases A and T1, proteinase K (200 μg/ml), and phenol-chloroform extraction. Viral DNA was precipitated with ethanol and suspended in 50 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). For DNA restriction analysis, aliquots of 2 μl of viral DNA (approximately 1 μg) were digested with 10 units of restriction endonucleases BamHI, BclI, BglI, BglII, BstEII, EcoRI, HindIII, HpaI, PstI, SalI, SmaI, XbaI, and XhoI, under the conditions specified by the manufacturer (Takara Shuzo Co.). Restriction enzyme digests were loaded onto 1.5% agarose gels and run for 2 h at 100V in Tris-acetate buffer (pH 8.0) with 1 mM EDTA. After staining with ethidium bromide (1 μg/ml), the fragments were visualized under a UV transilluminator and photographed with a Polaroid camera. EcoT14I digests of lambda DNA (Takara Shuzo Co.) and a 1-kb DNA ladder (Gibco BRL, Rockville, Md.) were run as molecular weight standards. Genome type and serotype identification were performed by comparison of the resulting patterns with the published restriction patterns. The restriction patterns of both isolates were identical to that of Ad34a (7) and not to those of Ad14p and Ad34p. The patterns are shown in Fig. 1B, panels a through f, and in Fig. 2, panels g through m.
FIG. 2.
Restriction patterns obtained after cleavage of Ad14p, Ad34p, Ad35p, and two clinical isolates with HindIII (g), HpaI (h), PstI (i), SalI (j), SmaI (k), XbaI (l), and XhoI (m). Lanes: M1, λ/EcoT14I; M2, 1-kb ladder; 1, Ad14p; 2, Ad34p; 3, Ad35p; 4, isolate A; 5, isolate B.
Li et al. first reported Ad34a (isolated from a hemorrhagic cystitis patient) (7), for which the restriction pattern with BamHI and SmaI was different from that of Ad34p (1, 13). The patterns of our clinical isolates with BamHI, BstEII, EcoRI, HpaI, SalI, SmaI, and XhoI were similar to those of Ad35p, whereas those with BclI, BglI, BglII, HindIII, PstI, and XbaI were not. The BglII, EcoRI, and HindIII patterns were similar to those of Ad34p. From these results, the isolates were deemed to be identical to Ad34a.
Sawada et al. reported that Ad34/35 (isolated from a conjunctival swab), which was neutralized by both Ad34 and Ad35 antisera, showed a restriction pattern identical to that of Ad35p with BamHI, HpaI, and SmaI; however, with HindIII, Ad34/35 showed a pattern identical to that of Ad34p (9). It is possible that Ad34/35 could be similar to Ad34a. It is suggested that Ad34a seems to be associated with tropism for the conjunctiva, in addition to the urinary tract.
It was suggested that the result of PCR-RFLP analysis was different from those of the NT and PCR-sequence method because of adenoviral DNA mutation. Several cases in which there were difficulties in identifying Ad3, Ad4, and Ad7, or Ad8 and Ad9, are reported (data not shown). The targets of the PCR-sequence method are HVRs associated with NT, whereas PCR-RFLP analysis uses conserved regions that may not be consistent with these epitopes, since it is not clear whether neutralization epitopes exist within HVRs alone. At present, NT is the standard method for classifying Ad serotypes; therefore, the PCR-sequence method might be regarded as more accurate than PCR-RFLP analysis. However, there remains an advantage in genome typing using restriction enzymes, because there have been many comparative reports for reference, and it is a commonly used differentiating method, as described above.
Acknowledgments
We thank T. Inada from the National Institute of Infectious Diseases for kindly providing Ad34 antiserum. We also thank A. Ikeda for help with correspondence.
REFERENCES
- 1.Adrian T, Wadell G, Hierholzer J C, Wigand R. DNA restriction analysis of adenovirus prototypes 1 to 41. Arch Virol. 1986;91:277–290. doi: 10.1007/BF01314287. [DOI] [PubMed] [Google Scholar]
- 2.Crawford-Miksza L, Schnurr D P. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crawford-Miksza L K, Schnurr D P. Adenovirus serotype evolution is driven by illegitimate recombination in the hypervariable regions of the hexon protein. Virology. 1996;224:357–367. doi: 10.1006/viro.1996.0543. [DOI] [PubMed] [Google Scholar]
- 4.de Jong P J, Valderrama G, Spigland I, Horwitz M S. Adenovirus isolates from urine of patients with acquired immunodeficiency syndrome. Lancet. 1983;i:1293–1296. doi: 10.1016/s0140-6736(83)92411-x. [DOI] [PubMed] [Google Scholar]
- 5.Hierholzer J C, Atuk N O, Gwaltney J M., Jr New human adenovirus isolated from a renal transplant recipient: description and characterization of candidate adenovirus type 34. J Clin Microbiol. 1975;1:366–376. doi: 10.1128/jcm.1.4.366-376.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keller E W, Rubin R H, Black P H, Hirsch M S, Hierholzer J C. Isolation of adenovirus type 34 from a renal transplant recipient with interstitial pneumonia. Transplantation. 1977;23:188–191. doi: 10.1097/00007890-197702000-00020. [DOI] [PubMed] [Google Scholar]
- 7.Li Q G, Hambraeus J, Wadell G. Genetic relationship between thirteen genome types of adenovirus 11, 34, and 35 with different tropisms. Intervirology. 1991;32:338–350. doi: 10.1159/000150218. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh-Inagawa W, Oshima A, Aoki K, Itoh N, Isobe K, Uchio E, Ohno S, Nakajima H, Hata K, Ishiko H. Rapid diagnosis of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J Clin Microbiol. 1996;34:2113–2116. doi: 10.1128/jcm.34.9.2113-2116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sawada H, Aoki K, Kawana R, Matsumoto I, Shinagawa M, Guo D F, Fajardo R V. Molecular epidemiology of adenoviral conjunctivitis in Sapporo, Japan, and Manila, the Philippines. Jpn J Ophthalmol. 1987;31:538–546. [PubMed] [Google Scholar]
- 10.Takeuchi S, Itoh N, Uchio E, Aoki K, Ohno S. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J Clin Microbiol. 1999;37:1839–1845. doi: 10.1128/jcm.37.6.1839-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchio E, Matsuura N, Takeuchi S, Itoh N, Ishiko H, Aoki K, Ohno S. Acute follicular conjunctivitis caused by adenovirus type 34. Am J Ophthalmol. 1999;128:680–686. doi: 10.1016/s0002-9394(99)00238-x. [DOI] [PubMed] [Google Scholar]
- 12.Wadell G, de Jong J C. Restriction endonucleases in identification of a genome type of adenovirus 19 associated with keratoconjunctivitis. Infect Immun. 1980;27:292–296. doi: 10.1128/iai.27.2.292-296.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wadell G, Hammarskjold M L, Winberg G, Varsanyi T M, Sundell G. Genetic variability of adenoviruses. Ann N Y Acad Sci. 1980;354:16–42. doi: 10.1111/j.1749-6632.1980.tb27955.x. [DOI] [PubMed] [Google Scholar]