Abstract
A novel coronavirus (COVID-19) recently emerged as an acute respiratory syndrome, and has caused a pneumonia outbreak world-widely. As the COVID-19 continues to spread rapidly across the world, computed tomography (CT) has become essentially important for fast diagnoses. Thus, it is urgent to develop an accurate computer-aided method to assist clinicians to identify COVID-19-infected patients by CT images. Here, we have collected chest CT scans of 88 patients diagnosed with COVID-19 from hospitals of two provinces in China, 100 patients infected with bacteria pneumonia, and 86 healthy persons for comparison and modeling. Based on the data, a deep learning-based CT diagnosis system was developed to identify patients with COVID-19. The experimental results showed that our model could accurately discriminate the COVID-19 patients from the bacteria pneumonia patients with an AUC of 0.95, recall (sensitivity) of 0.96, and precision of 0.79. When integrating three types of CT images, our model achieved a recall of 0.93 with precision of 0.86 for discriminating COVID-19 patients from others. Moreover, our model could extract main lesion features, especially the ground-glass opacity (GGO), which are visually helpful for assisted diagnoses by doctors. An online server is available for online diagnoses with CT images by our server (http://biomed.nscc-gz.cn/model.php). Source codes and datasets are available at our GitHub (https://github.com/SY575/COVID19-CT).
Keywords: COVID-19, deep learning, pneumonia diagnosis, weakly supervised learning
1. Introduction
In late December, 2019, a cluster of pneumonia cases were reported with unknown etiology [1]. From their lower respiratory tract samples, one kind of novel coronavirus was revealed through deep sequencing, which resembled severe acute respiratory syndrome coronavirus (SARS-CoV) [3], and is currently named as the 2019 novel coronavirus (COVID-19) by the World Health Organization (WHO) [2]. The COVID-19 is a beta-CoV of group 2B with at least 70 percent similarity in genetic sequence to SARS-CoV [5] and is the seventh member of the family of enveloped RNA coronavirus to infect humans [6]. Outbreaks in healthcare workers and families indicated human-to-human transmissions [1], [4]. Since January 17, the confirmed cases dramatically increased, and COVID-19 has been designated as a public health emergency of international concerns by WHO. As of Oct 10, 2020, China has documented over 90,000 confirmed cases with the death toll rose to 4,700, and the world totally identified more than 36 million confirmed cases with over one million deaths. The COVID-19 has posed significant threats to international health [7].
The spectrum of this disease in humans is yet to be fully determined. Signs of infection are nonspecific, including respiratory symptoms, fever, cough, dyspnea, and viral pneumonia [1]. With the daily increase in the number of newly diagnosed and suspected cases, the diagnosis has become a growing problem in major hospitals due to the insufficient supply of nucleic acid detection boxes and limited detection rates in the epidemic area [8]. Computed tomography (CT) and radiography techniques have thus emerged as integral players in the preliminary identification and diagnosis of COVID-19 [9], [10], [11]. However, the overwhelming patients and relatively insufficient radiologists led to high false positive rates [11]. Advanced computer-aided lung CT diagnosis systems are urgently needed for accurately confirming suspected cases, screening patients, and conducting virus surveillance.
With the rapid development of artificial intelligence, computer vision techniques, originally used for classifications of general images, have been applied to medical images including CT images [21]. Among existing techniques, convolutional neural network (CNN) [27] is a widely used feed-forward artificial neural network, base on which many models have shown great promise in capturing feature representations [13]. However, these general models are not satisfactory for CT images classification because CT images are fine-grained that have low inter-class variances, and thus are difficult to distinguish [28]. Recently, there are encouraging developments in fine-grained image representation learning methods that have been proven effective in CT image deep learning [23]. More importantly, these methods required only image-level annotations and have been shown able to achieve close accuracy to models trained on refined expert annotations [24].
Generally, these fine-grained image learning methods follow a two-stage strategy that first locates key objects in the images and then discriminates their belonging subclasses based on the identified objects [22]. One key to the success of these methods is to accurately identify the objects in the images. In consideration that the objects might be of different sizes, Feature Pyramid Network (FPN) method was proposed to recognize objects (sub-images) at different scales with corresponding confidence scores [14]. Though the method has shown great power in several fine-grained image classification datasets [21], existing studies usually extracted the features of the detected sub-images separately without considering connections between sub-images. Therefore, it should be beneficial to extract relations between these sub-images.
Another bottleneck of current deep learning techniques is its lack of interpretability. High interpretability is important in both theory and practice, especially for medical diagnoses that has low endurance to wrong predictions. The interpretability of model has recently been significantly advanced by several studies through visualizing the trained CNN models [25], [26]. The combination of the visualization methods will be essential for constructing an interpretable model for COVID diagnosis.
In this study, we have collected chest CT scans of 88 patients diagnosed as COVID-19 from hospitals of two provinces in China, along with 100 patients infected by bacteria pneumonia and 86 healthy persons for comparison and modeling. To effectively capture the subtle differences in medical images, we have constructed a new deep learning architecture DRENet. For a given CT image slice, we first detected potential lesion regions at different scales by integrating the pre-trained ResNet50 with the FPN network. According to the detected regions, ResNet50 was employed again to extract local features at each region and relational features between regions. These features were then concatenated with global features extracted from the original image, and input to a multiple layer perception (MLP) for image-level prediction. Finally, the prediction over each CT image slice in one patient were aggregated for the person-level diagnoses. By training and validation on the collected dataset, our model was shown to accurately discriminate the COVID-19 patients from the bacteria pneumonia patients with an AUC of 0.95, recall (sensitivity) of 0.96, and precision of 0.79. When integrating three types of CT images, the model achieved a recall of 0.93 with precision of 0.86 for discriminating COVID-19 patients from others. More importantly, our model could extract main lesion features, especially the ground-glass opacity (GGO), which visually assist diagnoses by doctors.
2. Materials and Methods
Our institutional reviewing board waived written informed consent for this study that evaluated de-identified data and involved no potential risk to patients.
2.1. Data Acquisition
This study was based on reliable resources provided by the Renmin Hospital of Wuhan University and two affiliated hospitals (the Third Affiliated Hospital and Sun Yat-Sen Memorial Hospital) of the Sun Yat-sen University in Guangzhou. We obtained CT images of totally 88 COVID-19 infected patients, which comprised of 76 and 12 patients from the Renmin Hospital of Wuhan University and the Third Affiliated Hospital, respectively. All COVID-19 patients’ nasopharyngeal swabs were subjected to nucleic acid kit lysis extraction and calculation by the respective laboratories. In the tests, the fluorescence RT-PCR was performed to detect the viral nucleic acid sequences, which were then compared with the novel coronavirus nucleocapsid protein gene (nCoV-NP) and the novel coronavirus open reading coding frame lab (nCoV ORFlab) sequence. The patients were selected only when the nucleic acid results were positive and the HRCT images of chest were of acceptable qualities with no significant artifacts or missing images. For comparison, we also retrieved CT underwent chest images of 86 healthy persons and 100 patients with bacterial pneumonia from the Renmin Hospital of Wuhan University and Sun Yat-Sen Memorial Hospital.
For persons in Wuhan, chest CT examinations were performed with a 64-section scanner (Optima 680, GE Medical Systems, Milwaukee, WI, USA) without the use of contrast materials. The CT protocol was as follows: 120 kV; automatic tube current; detector, 35 mm; rotation time, 0.35 second; section thickness, 5 mm; collimation, 0.75 mm; pitch, 1–1.2; matrix, 512 × 512; and inspiration breath hold. The images were photographed at lung (window width, 1000–1500 HU; window level, –700 HU) and mediastinal (window width, 350 HU; window level, 35–40 HU) settings. The reconstructions were made at 0.625 mm slice thickness on lung settings. These data consisted of only transverse plane images of lung. For persons in Guangzhou, the chest CT examinations were performed with a 64-slice spiral scanner (Somatom Sensation 64; Siemens, Germany). All CT parameters were the same as the above settings except the followings: section thickness, 8 mm, and collimation, 0.6 mm.
2.2. Model Architecture and Model Training
We have developed a deep learning-based CT diagnosis system to detect the COVID-19 causing pneumonia and to localize the main lesions. As shown in Fig. 1, the fully automated lung CT diagnosis system was developed by three main steps. First, we extracted the main region of the lung and filled the blank of lung segmentation with the lung itself to avoid noises caused by different lung contours. Then, we designed a Details Relation Extraction neural network (DRENet) to obtain the image-level predictions. Finally, the image-level predictions were aggregated to achieve person-level diagnoses.
Fig. 1.
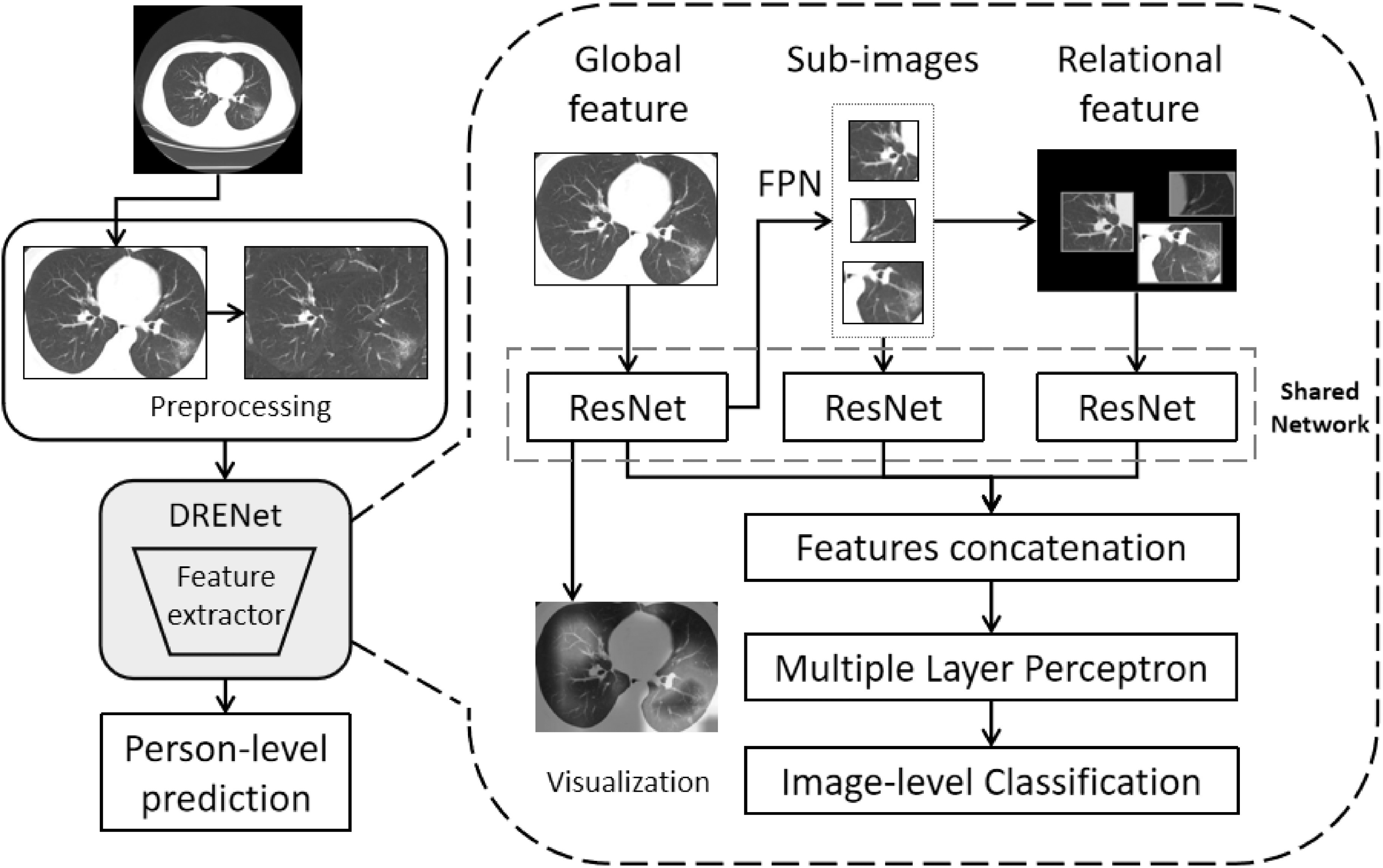
The proposed training architecture including (1) Preprocessing: the CT images are first preprocessed to remove boundary regions around the lungs, and to further fill the blank regions around two lungs with its rotational lung areas (for model training only). (2) Image-level classifications by DRENet: each CT image is input to the pre-trained ResNet50 to extract global features, from which FPN module is employed to identify top-K lesion regions. According to the top-K regions, the shared ResNet is utilized again to extract local features within the sub-images and relational features between the sub-images. These features are concatenated with the learned global features to input into MLP for the image-level prediction. (3) Person-level prediction: the image-level predictions will be aggregated for person-level prediction.
2.2.1. Data Preprocessing
As 3D CT images of one patient may contain more than 200 images and neighbored images are highly similar, we only selected 15 representative images equidistantly, i.e., images index [i*ntot/15, i = 0..14] with ntot as the total number images. The removal of redundant images could speed up the calculation and reduce the impact of different numbers of scanned images between hospitals. As many CT images contain incomplete lungs, we further detected lungs through the OpenCV package[12], and removed the images with lung regions occupying less than 50 percent of the total image. However, there is a problem that the lung contours are substantially different between humans, which may cause deep learning model to over fit the features of lung contour. To solve this problem, we simply filled the blank area of image with its rotational lung areas. Finally, we kept 88 COVID-19 patients with 777 CT images, 100 bacterial pneumonia patients with 505 slices, and 86 healthy people with 708 slices in this study.
2.2.2. DRENet
The DRENet was constructed based on the pre-trained ResNet50 that has been proven to be robust to detect objects in images [13]. We further added the Feature Pyramid Network (FPN) [14] to extract the top-K details from each image. Based on the extracted details, an attention module is coupled to learn the importance [15].
Concretely, as shown in Fig. 1, an image is input to ResNet50 to extract the feature map. The feature map is then input to a pooling layer and a dense layer to extract global features, and to the FPN module to extract local details. Inspired by the recent modification of FPN [21] to utilize a top-down architecture for detecting multi-scale regions, we modified the FPN module to identify multi-scale lesion area, where convolutional layers are used at all layers to compute hierarchical features. Moreover, we used feature maps of sizes {14x14, 7x7} corresponding to regions of scales {48x48, 96x96}, emphasizing on small regions because lesion areas are generally small and the use of the large size of feature maps will bring noises to the model. By this way, the FPN will identify important regions with confidence scores.
According to the identified regions, top-K sub-images are then cropped from the original image. At the same time, the original image was scaled to generate a new image by multiplying the corresponding regions of the top-K sub-images with their confidence scores. The regions not including top-K images were set to zero. These K sub-images and the newly generated image were input to ResNet50 to extract respective features.
Finally, we concatenated the features extracted from the original image, top-K sub-images, and the generated image to a 1-D vector and sent them into a multi layer perceptron to predict the image-level state. Based on FPN, our model can not only provide person-level prediction score for the doctor but also interpret the prediction by weighing each pixel of the original image. More details of the implementation and parameters are attached in the Supplementary File, which can be found on the Computer Society Digital Library at http://doi.ieeecomputersociety.org/10.1109/TCBB.2021.3065361.
2.2.3. Aggregation
For each patient, the predictions were made on each image slice, and the image-level scored results were aggregated for person-level prediction. Here, the mean pooling was used to integrate the image-level scores into the morbidity of each person as the person-level prediction.
2.3. Implementation and Evaluation
In our experiments, we fixed K = 3 meaning to extract 3 sub-images from each input image. For practical purposes, we designed the models with two tasks: discriminating COVID-19-infected patients from the healthy people, and separating COVID-19 patients from bacterial patients and healthy controls. For each task, we employed the person-level split strategy following the LUNA16 competition[16] by using random splits of 60/10/30 percent for training, validation, and test sets, respectively. The training set was used to train models, and the validation set was used to optimize the hyper-parameters for the best performance. The final optimal model was independently assessed on the test set. We computed the measured metrics as
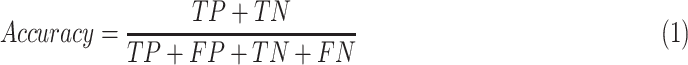
|

|

|

|
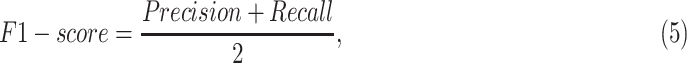
|
where TP, TN, FP, and FN are the numbers of true positives, true negatives, false positives, and false negatives, respectively. We also computed Area under the receiver-operating characteristics Curve (AUC) by the scikit-learn 0.19 (scikit-learn.org). For the tri-classification task, we computed the precision, recall, F1-score for each type, and reported the average in default.
3. Results
3.1. Deep Learning Model for COVID-19 Diagnosis
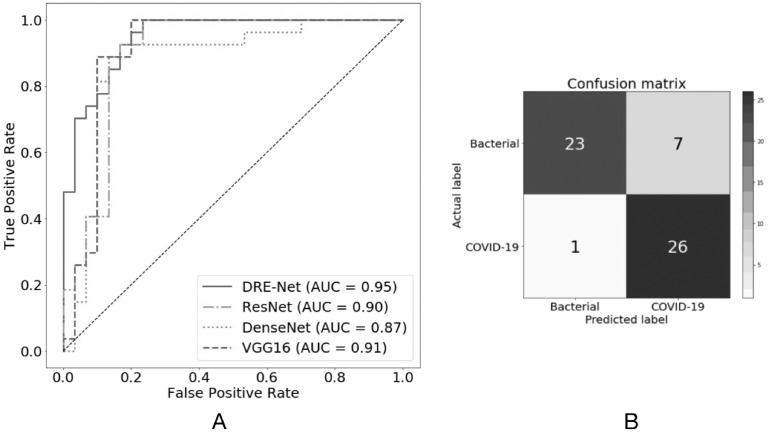
We first performed experiments by discriminating 88 COVID-19 patients and 100 bacteria pneumonia patients, from which 777 and 505 images were collected. By tuning hyperparameters on the 10 percent validation set, we achieved AUC of 0.91 in the image level, essentially the same as the AUC of 0.92 in the test set. Aggregation of the image-level predictions led to slight increases of AUC values in person level to 0.93 and 0.95 for the validation and independent test, respectively. The consistent results between the validation and test indicated the robustness of our model. To indicate the effectiveness of our proposed DRENet architecture, we also employed the classical deep residual network (Resnet) [13], DenseNet [17], and VGG16 [18] for comparisons. As shown in Table 1, DRENet achieved the highest AUC score among all methods, VGG16 and ResNet were close, and DenseNet had the lowest score. When measured by F1-score, another balanced measurement, DRENet ranked the first and DenseNet had the lowest score, while ResNet was slightly higher than VGG16. As DRENet was mostly based on ResNet, the improvement should result from the further learning of extracted important regions.
TABLE 1. Comparisons of Different Models by Person-Level Performances on the Independent Test.
| Model | AUC | Recall | Precision | Specificity | F1-score | Accuracy |
|---|---|---|---|---|---|---|
| VGG16 | 0.91 | 0.89 | 0.80 | 0.80 | 0.84 | 0.84 |
| DenseNet | 0.87 | 0.93 | 0.76 | 0.73 | 0.83 | 0.82 |
| ResNet | 0.90 | 0.93 | 0.81 | 0.80 | 0.86 | 0.86 |
| DRE-Net | 0.95 | 0.96 | 0.79 | 0.77 | 0.87 | 0.86 |
TABLE 2. Performance Comparisons on Pneumonia Three-Class Classification Task.
| Model | Recall | Precision | Specificity | F1-score | Accuracy |
|---|---|---|---|---|---|
| VGG16 | 0.91 | 0.91 | 0.91 | 0.91 | 0.91 |
| DenseNet | 0.88 | 0.89 | 0.88 | 0.88 | 0.88 |
| ResNet | 0.91 | 0.92 | 0.91 | 0.91 | 0.91 |
| DRE-Net | 0.93 | 0.93 | 0.93 | 0.93 | 0.93 |
Fig. 2A showed the receiver-operating characteristic curves for comparisons with other baseline models. Obviously, DRENet has higher true positive rates at regions of low false positive rate (FPR < 0.02), which are also the most important regions because of the much higher number of bacterial pneumonia than COVID-19 patients. At a higher FPR (>0.2), all methods except DenseNet can correctly recognize COVID-19 patients. When using a cutoff of 0.5, the DRENet predictions have one false positive and two false negatives as shown in the confusion matrix (Fig. 2B), corresponding to a precision of 0.79 and a recall of 0.96.
Fig. 2.
Performances of different networks on the pneumonia diagnosis with A) Receiver operating characteristic curves for the diagnosis of COVID-19, and B) Confusion matrix of the DRENet on the test set.
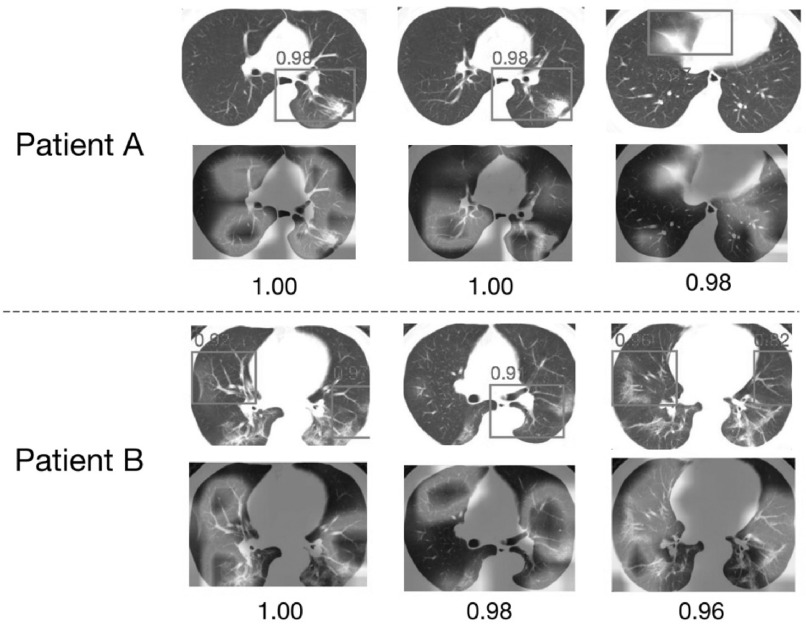
Another advantage of DRENet is its ability to detect potential lesion regions for interpretation. For visualization, we selected two successfully predicted pneumonia patients from the test set (Fig. 3), and showed three slice images with the highest scores for each patient. The detected regions by DRENet contained ground-glass opacity (GGO) abnormality, which has been reported as the most important character for COVID-19 patients by recent studies [9], [10], [11]. To quantify the potential lesion regions, we also drew the heat maps through the Gradient-weighted Class Activation Mapping (Grad-CAM) as proposed by [25]. The visualizations by Grad-CAM were essentially consistent to detect potential lesion regions while providing more information. These findings indicate that DRENet could detect key features. The detected features provide reasonable clues for the judgements, which are especially helpful for diagnoses by doctors.
Fig. 3.
Visualization of two correctly diagnosed nCOV-19 pneumonia patients. For each patient, we showed the top 3 predicted slices and the extracted details (bounding boxes with red color) with normalized predicted scores above 0.8 (ranging from 0 to 1). Moreover, we also used the method proposed by [25] to draw the heat map.
3.2. Deep Learning Model for Pneumonia Three-Class Classification
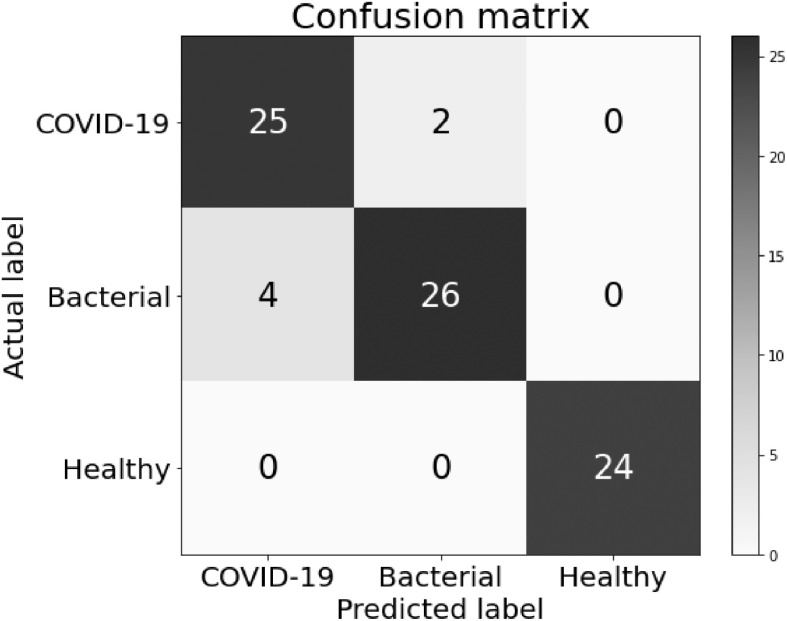
In order for real-world assisted diagnoses in hospitals, we further performed three-class classification experiments by adding pneumonia patients caused by bacteria. To this end, we built a model by the proposed DRENet learning architecture based on 88 COVID-19, 100 bacterial pneumonia patients, and 86 healthy persons, consisted of 777, 505 and 708 CT images, respectively. As shown in Table 1, DRENet achieved a precision of 0.93 with a recall of 0.93 in average, corresponding to F1-score of 0.93 as well. By comparison, the F1-score of DRENet is 2 percent higher than the next best ResNet and VGG16. DenseNet performed the worst with F1-score of 0.88. Fig. 4 shows the confusion matrix of the DRENet on the pneumonia three-class classification. We observed that the model can discriminate all healthy persons from pneumonia patients while make only six wrong predictions in classifying the bacterial pneumonia and viral pneumonia (COVID-19). When considering the separation of COVID-19 patients from others, the model achieved a recall of 0.93 and precision of 0.86.
Fig. 4.
Confusion matrix of the DRENet on the pneumonia three-class classification.
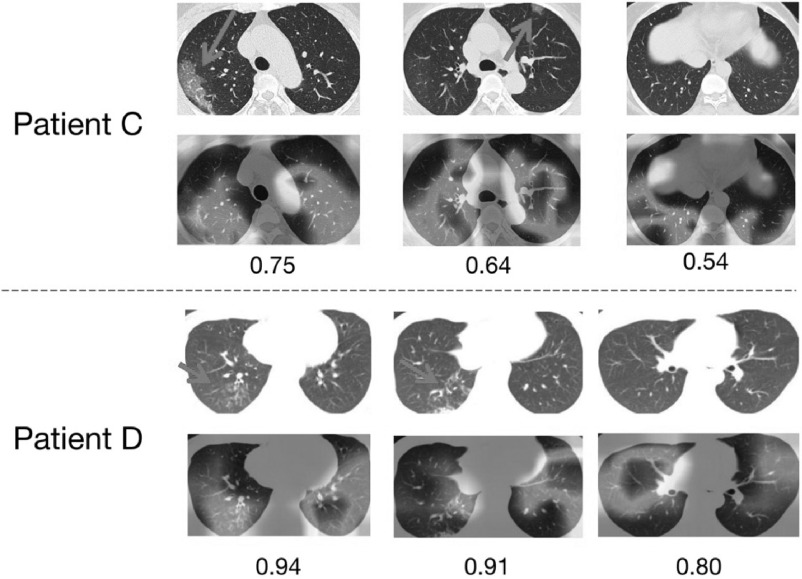
By examining two COVID-19 patients who were wrongly predicted to be caused by bacteria, we found their respective predicted scores were 0.46 and 0.39 in the patient level. These scores were both marginally lower than the threshold of 0.5. As shown in Fig. 5, although the patient C has a score of 0.46 in the patient level, three most suspected slice images (the highest scores in the image level) all have scores greater than 0.5 in the image level. Therefore, DRENet could still assist doctors to make correct decisions by visualizing key images in occasionally wrong cases. On the other hand, the patient D of bacterial pneumonia was wrongly predicted as the COVID-19 patient. The misjudgements might be caused by the GGO abnormalities appearing in the patient of bacterial pneumonia for unknown reasons, as shown in the rectangles.
Fig. 5.
Visualization of two wrongly predicted cases on the pneumonia classification for A) one patient of bacterial pneumonia wrongly predicted as the COVID-19 patient, and B) one COVID-19 patient wrongly predicted as bacterial pneumonia. Arrows were added by hand to point to regions containing GGO abnormality.
4. Discussion
The outbreak of COVID-19 has resulted in a world-wide panic recently. There are no specific signs for the infected patients. Those who have fallen into ills were reported with the ground-glass opacity. Thus, CT becomes the most convenient and fastest diagnosis approach. However, difficulties were met due to lacks of quantified diagnosis criteria and of experienced radiologists. It is essential to develop a publicly available model to assist the diagnosis by artificial intelligence.
In this study, we have developed a new deep learning architecture to construct the prediction model based on 88 patients diagnosed with the COVID-19, 100 patients infected with bacteria pneumonia, and 86 healthy persons from three hospitals in China. Our models achieved a recall of 96 percent and a precision of 93 percent for COVID-19 patients against bacteria pneumonia patients. When training a model to separate three classes simultaneously, the tri-classification model achieved a recall of 93 percent and a precision of 86 percent for the COVID-19 patients. Due to the high infectivity of COVID-19 virus, a high recall is essential for correctly recognizing COVID-19 patients to prevent the spreading of virus. The model developed here is appropriate for assisting clinical diagnoses considering the 60–70 percent recall rate by nucleic acid examinations. More importantly, our method also provides interpretation of the prediction through recognized lesion regions and detailed heat map. Since clinical diagnosis is of low endurance to wrong predictions, such interpretation is essential for assisting doctors to make decisions.
Since the training data is still a small data, the model can't effectively solve the batch effect to make accurate predictions for other sources of data, a well known problem in AI diagnosis through medical images [CITE]. One possible way for this problem is to accumulate data from multiple hospitals. Alternatively, the model can be fine-tuned or retrained by using data from hospitals that need to use the model.
Additionally, we performed an experiment to distinguish the COVID-19 infected patients from bacterial pneumonia. The independent test indicated a recall of 0.93 and a precision of 0.79 (Table S1), available online. The same recall but lower precision than the tri-class model (recall = 0.86) suggests that adding other types might help the training in tasks with small number of samples. Therefore, the performance might be further improved by adding more CT images, or even other types of pneumonia [20].
5. Conclusion
In conclusion, our study demonstrated the feasibility of a deep learning approach to assist doctors in detecting patients with COVID-19 and automatically to identify the potential lesions from CT images. With the accurate discrimination of the COVID-19 patients, the proposed system may enable a rapid and accurate identification of patients.
There are also several weakness points in our work. One major problem is the dataset. We accomplished the work at the very early stage of the COVID pandemic, where there are only very few samples to build such a deep learning prototype. Second, due to the issue of batch effect, our method achieved good results on the datasets from original source data collection hospital but was not able to predict the external data directly. Fortunately, it can be alleviated by finetuning the model with the images from target sources. In the future, we may include a pre-trained model with more image sources for more accurate diagnosis of COVID-19.
Acknowledgments
The work was supported in part by the National Key R&D Program of China under Grant 2018YFC1315405, the National Natural Science Foundation of China under Grants U1611261, 61772566, 81801132 and 81871332, Guangdong Frontier and Key Tech Innovation Program under Grants 2018B010109006 and 2019B020228001, the Natural Science Foundation of Guangdong, China under Grant 2019A1515012207, and Introducing Innovative and Entrepreneurial Teams under Grant 2016ZT06D211. Ying Song, Shuangjia Zheng, Liang Li, and Xiang Zhang are contributed equally.
Funding Statement
The work was supported in part by the National Key R&D Program of China under Grant 2018YFC1315405, the National Natural Science Foundation of China under Grants U1611261, 61772566, 81801132 and 81871332, Guangdong Frontier and Key Tech Innovation Program under Grants 2018B010109006 and 2019B020228001, the Natural Science Foundation of Guangdong, China under Grant 2019A1515012207, and Introducing Innovative and Entrepreneurial Teams under Grant 2016ZT06D211.
Contributor Information
Ying Song, Email: songy75@mail2.sysu.edu.cn.
Shuangjia Zheng, Email: zhengshj9@mail2.sysu.edu.cn.
Liang Li, Email: liliang_082@163.com.
Xiang Zhang, Email: zhangx345@mail.sysu.edu.cn.
Xiaodong Zhang, Email: zhangxd2@mail.sysu.edu.cn.
Ziwang Huang, Email: huangzw26@mail.sysu.edu.cn.
Jianwen Chen, Email: chenjw48@mail2.sysu.edu.cn.
Ruixuan Wang, Email: wangruix5@mail.sysu.edu.cn.
Huiying Zhao, Email: zhaohy8@mail.sysu.edu.cn.
Yutian Chong, Email: chongyt@mail.sysu.edu.cn.
Jun Shen, Email: shenjun@mail.sysu.edu.cn.
Yunfei Zha, Email: zhayunfei999@126.com.
Yuedong Yang, Email: yangyd25@mail.sysu.edu.cn.
References
- [1].Huang C., et al. , “Clinical features of patients infected with 2019 novel Coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].WHO website WM, Accessed: Feb. 5, 2020. [Online]. Available: https://www.who.int
- [3].Lu R., et al. , “Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding,” Lancet, vol. 395, 565–574, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li Q., et al. , “Early transmission dynamics in Wuhan, China, of novel Coronavirus–infected pneumonia,” New Engl. J. Med., vol. 382, pp. 1199–1207, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hui D. S., et al. , “The continuing 2019-nCoV epidemic threat of novel Coronaviruses to global health–The latest 2019 novel Coronavirus outbreak in Wuhan, China,” Int. J. Infect. Dis., vol. 91, pp. 264–266, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhu N., et al. , “A novel Coronavirus from patients with pneumonia in China,” New Engl. J. Med., vol. 382, pp. 727–733, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].WHO reports, WHONC-ns, Assessed: Feb. 6th, 2020. [Online]. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/
- [8].Chu D. K., et al. , “Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of Pneumonia,” Clin. Chem., vol. 66, pp. 549–555, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lei J., Li J., Li X., and Qi X., “CT imaging of the 2019 novel coronavirus (2019-nCoV) Pneumonia,” Radiology, vol. 295, 2020, Art. no. 200236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shi H., Han X., and Zheng C., “Evolution of CT manifestations in a patient recovered from 2019 novel Coronavirus (2019-nCoV) Pneumonia in Wuhan, China,” Radiology, vol. 295, 2020, Art. no. 200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Song F., et al. , “Emerging Coronavirus 2019-nCoV Pneumonia,” Radiology, vol. 295, 2020, Art. no. 200274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Bradski G. and Kaehler A., Learning OpenCV: Computer Vision With the OpenCV Library, Newton, MA, USA: O’Reilly Media, Inc, 2008. [Google Scholar]
- [13].He K., Zhang X., Ren S., and Sun J., “Deep residual learning for image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2016, pp. 770–778. [Google Scholar]
- [14].Lin T.-Y., Dollár P., Girshick R., He K., Hariharan B., and Belongie S., “Feature pyramid networks for object detection,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2017, pp. 936–944. [Google Scholar]
- [15].Fu J., Zheng H., and Mei T., “Look closer to see better: Recurrent attention convolutional neural network for fine-grained image recognition,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2017, pp. 4476–4484. [Google Scholar]
- [16].LUNA16, Accessed: Feb. 6th, 2020. [Online]. Available: https://luna16.grand-challenge.org/
- [17].Iandola F., Moskewicz M., Karayev S., Girshick R., Darrell T., and Keutzer K., “DenseNet: Implementing efficient ConvNet descriptor pyramids,” 2014, arXiv:14041869.
- [18].Simonyan K. and Zisserman A., “Very deep convolutional networks for large-scale image recognition,” 2014, arXiv:14091556.
- [19].Liu X., Faes L., Kale A. U., et al. , “A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: A systematic review and meta-analysis,” Lancet Digit Health, vol. 1, pp. e271–e297, 2019. [DOI] [PubMed] [Google Scholar]
- [20].E. L., et al. , “Using deep-learning techniques for pulmonary-thoracic segmentations and improvement of pneumonia diagnosis in pediatric chest radiographs,” Pediatric Pulmonol., vol. 54, no. 10, pp. 1617–1626, 2019. [DOI] [PubMed] [Google Scholar]
- [21].Yang Z., et al. , “Learning to navigate for fine-grained classification,” in Proc. Eur. Conf. Comput. Vis., 2018, pp. 438–454. [Google Scholar]
- [22].Peng Y., He X., and Zhao J., “Object-part attention model for fine-grained image classification,” IEEE Trans. Image Process., vol. 27, no. 3, pp. 1487–1500, Mar. 2018. [DOI] [PubMed] [Google Scholar]
- [23].Yan K., Peng Y., Sandfort V., Bagheri M., Lu Z., and Summers R. M., “Holistic and comprehensive annotation of clinically significant findings on diverse CT images: Learning from radiology reports and label ontology,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2019, pp. 8523–8532. [Google Scholar]
- [24].Zhou Y., Zhu Y., Ye Q., Qiu Q., and Jiao J., “Weakly supervised instance segmentation using class peak response,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., 2018, pp. 3791–3800. [Google Scholar]
- [25].Selvaraju R. R., Cogswell M., Das A., Vedantam R., Parikh D., and Batra D., “Grad-CAM: Visual explanations from deep networks via gradient-based localization,” in Proc. IEEE Int. Conf. Comput. Vis., 2017, pp. 618–626. [Google Scholar]
- [26].Zhang Q., Wang W., and Zhu S. C., “Examining CNN representations with respect to dataset bias,” 2017, arXiv: 1710.10577.
- [27].Krizhevsky A., Sutskever I., and Hinton G. E., “ImageNet classification with deep convolutional neural networks,” in Proc. 25th Int. Conf. Neural Inf. Process. Syst., 2012, pp. 1097–1105. [Google Scholar]
- [28].Zhao B., et al. , “A survey on deep learning-based fine-grained object classification and semantic segmentation,” Int. J. Autom. Comput., vol. 14. no. 2, pp. 119–135, 2017. [Google Scholar]