Abstract
Since the end of 2019, novel coronavirus disease (COVID-19) has brought about a plethora of unforeseen changes to the world as we know it. Despite our ceaseless fight against it, COVID-19 has claimed millions of lives, and the death toll exacerbated due to its extremely contagious and fast-spreading nature. To control the spread of this highly contagious disease, a rapid and accurate diagnosis can play a very crucial part. Motivated by this context, a parallelly concatenated convolutional block-based capsule network is proposed in this article as an efficient tool to diagnose the COVID-19 patients from multimodal medical images. Concatenation of deep convolutional blocks of different filter sizes allows us to integrate discriminative spatial features by simultaneously changing the receptive field and enhances the scalability of the model. Moreover, concatenation of capsule layers strengthens the model to learn more complex representation by presenting the information in a fine to coarser manner. The proposed model is evaluated on three benchmark datasets, in which two of them are chest radiograph datasets and the rest is an ultrasound imaging dataset. The architecture that we have proposed through extensive analysis and reasoning achieved outstanding performance in COVID-19 detection task, which signifies the potentiality of the proposed model.
Keywords: COVID-19, capsule network, dynamic routing, normalized contrast limited adaptive histogram equalization (N-CLAHE), point of care ultrasound (POCUS), ultrasound (US) imaging
I. Introduction
Severe acute respiratory syndrome coronavirus 2 mainly known as coronavirus disease or COVID-19, for its extremely contagious nature, has been declared as a global pandemic by the World Health Organization (WHO), which has threatened the healthcare system all around the world on the verge of complete collapse. Since its emergence in Wuhan, China [1]–[3], virtually no country has been spared by this pandemic, which perplexedly deteriorated the world health system in a short window of time. Lack of proper medication and vaccine has made the situation worse, and every day, the number of COVID-19-affected patients is rising rapidly, especially in low-income countries. The issues related to manufacturing infrastructure, distribution burden, limited supply, and public trust are further impeding peoples’ access to the COVID-19 vaccine, consequently affecting the health sectors [53]. In these circumstances, the WHO has emphasized mass testing and isolation of the COVID-19 patients to flatten the epidemic curve. Hence, detection of COVID-19-affected patients is considered as the most vital step of COVID-19 prevention process, and reverse transcription-polymerase chain reaction (RT-PCR) is the worldwide accepted method for COVID-19 detection [4], [5]. By monitoring the amplification reaction of specific DNA targets using polymerase chain reaction, RT-PCR is used to analyze the gene expression and quantification of viral RNA in clinical settings [54]. However, this method is time consuming, and specific material and equipment that are not easily accessible are required [4]. On the other hand, medical imaging has shown great potential in disease detection and diagnosis [6], which is less time consuming as well as it requires less materials and equipment. Computed tomography (CT) scans, ultrasound (US) imaging, and X-ray imaging are being used for disease detection for a long time, and now, introduction of an automatic detection procedure has made disease detection task easier for the physicians [7]–[9]. US is considered one of the most used imaging modalities because of its relatively safe, low cost, noninvasive nature, and real-time display [27], [28]. It is widely used to detect breast cancer, classify breast lesions, and classify carotid artery intima media thickness [29]–[31]. Using deep learning in medical imaging such as X-ray and CT, detection of COVID-19 is more accurate and quicker [10]–[12]. Recent work has shown that CT scans can detect COVID-19 at higher sensitivity rate (98%, respectively 88%) compared to RT-PCR (71% and 59%) in cohorts of 51 [18] and 1014 patients [19]. Furthermore, a cheap diagnosis method, i.e., chest X-rays, is also being used for detecting COVID-19 patients, and a twofold accuracy of 96% and 98% has been reported in detecting generic pulmonary disease and COVID-19, respectively, by evaluating 6523 chest X-rays [20]. But this diagnosis method involves high level of expertise and manual observation, which underscores the need of automatic detection. Although this signifies the success of deep learning models in automating COVID-19 detection, a lot of factors are left unanswered here about the generalization of this study and application in clinical use.
A convolutional neural network (CNN) has gained much popularity due to its superior performance in automatic disease detection tasks [13]–[15]. With remarkable improvement in computational ability and availability, a large amount of training data have made deep learning suitable for many disease detection tasks. Another reason for the current popularity of deep learning is that deep leaning architectures do not need manual feature extraction procedure, rather it automatically extracts features from the training data [16], [17]. Deep learning techniques, especially CNN, have great potential to complement the conventional diagnostic techniques of COVID-19. Various methods based on deep learning have also been applied in X-ray and CT images, and substantial results have been achieved [21]–[24]. Moreover, transfer-learning-based approaches have gained popularity in this field, where pretrained networks are utilized and integrated with different learning factors and selection algorithms to determine the best models [51], [52]. Almost all of these COVID-19-related studies have used CNNs of different form, which despite being very powerful have some major drawbacks associated with them. On this basis, capsule networks have been introduced as an alternative to CNNs, which have the ability to overcome the shortcomings of CNNs [25], [26].
Recently, several capsule-network-based architectures have been proposed for COVID-19 detection from chest X-ray [32], [33]. Although a capsule network represents a breakthrough in the field of artificial intelligence, especially in the field of image classification task, study shows that the performance of capsule network degrades with increasing dimensionality of the input data [60]. To solve this issue, all aforementioned literature used a sequential convolutional encoder block that gradually reduces the dimension of the input data and focuses only on the discriminative features. Then, the output of the convolutional block is fed into the primary capsule block for further analysis. However, deep sequential convolutional encoder block does not always provide satisfying results as there is always a chance of losing contextual information while using them [61]. To address this issue, we have proposed a parallel convolutional encoder block prior to the primary capsule block to transform the high-dimensional data into lower dimensions using different kernel sizes. This parallel convolutional block is implemented with gradually increased kernel size, which not only helps to reduce the input data size but also helps to incorporate more fine-grained and discriminative features by using larger receptive fields. Moreover, we have used an interconnection between capsule layers so that meaningful and discriminative features can be learned at the class capsule layer.
The main contributions of this article are the following.
-
1)
A parallelly concatenated convolutional block-based capsule network is introduced to detect patients affected by COVID-19, which has the advantage of integrating more discriminative fine to coarser spatial features than conventional deep learning architectures.
-
2)
To handle images of large spatial dimension, we have extended the number of capsule layers and routing number. At the same time, a concatenation method is applied in the capsule layers to extract more competent set of features from a particular area of images. The motivation behind this approach is because, with the escalation of the dimensionality of images, the performance of the baseline capsule network decreases significantly due to the inefficacy in capturing underlying complex features present in the high spatial dimension. Although our proposed parallel convolution operation prior to the capsule network efficiently reduces the dimension of the features fed into the primary capsule block to handle the high dimensionality problem stated before, increment of capsule layers within a certain range has been found very useful in extracting useful features from high-dimensional feature space resulting in higher accuracy [60]. Additionally, concatenated connections between capsule layers strengthen the coupling coefficient and increase the learning ability of the capsule layers and prevent the gradient flow from being damped that makes the model more capable of extracting complex features from images of large spatial dimensions, eventually results in a higher score in the validation metrics. Moreover, another considerable issue is the selection of optimal routing numbers, which plays an imperative role to tackle the overfitting and underfitting issues in the training phase. In this article, optimal performance of the network is achieved by extending the number of routing and then choose the optimal routing number accordingly.
-
3)
A pretrained method is also introduced, which is more capable of handling a small dataset1.
II. Limitations of a CNN
CNNs are devised such that layer 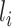 is fed from layer
is fed from layer 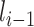 and then layer
and then layer 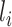 feeds its outputs to layer
feeds its outputs to layer 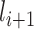 . Hence, whatever layer
. Hence, whatever layer 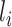 learns is a composition of features from the initial layers to layer
learns is a composition of features from the initial layers to layer 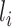 . While learning from its previous layers, layer
. While learning from its previous layers, layer 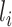 completely ignores the spatial relations between input data instances. Hence, CNNs have to be trained with same data having different orientations and transformations such as rotation, zoom in or zoom out, flipping, spatial shifting, color perturbation, quantization, etc. Another limitation impedes its performance when a max-pooling layer is used. Location information of features, which gives translation invariance quality, is lost when the max-pooling layer is used [25]. To mitigate these limitations, a large amount of data are used to train neural networks, and different augmentation techniques are also used to limit the loss of important spatial relationship information. Capsule networks are alternative models of CNN models, which can capture spatial information by storing information at the neuron level as vectors rather than scalars like CNNs. And max-pooling is replaced by the routing-by-agreement mechanism, which prevents information loss [25].
completely ignores the spatial relations between input data instances. Hence, CNNs have to be trained with same data having different orientations and transformations such as rotation, zoom in or zoom out, flipping, spatial shifting, color perturbation, quantization, etc. Another limitation impedes its performance when a max-pooling layer is used. Location information of features, which gives translation invariance quality, is lost when the max-pooling layer is used [25]. To mitigate these limitations, a large amount of data are used to train neural networks, and different augmentation techniques are also used to limit the loss of important spatial relationship information. Capsule networks are alternative models of CNN models, which can capture spatial information by storing information at the neuron level as vectors rather than scalars like CNNs. And max-pooling is replaced by the routing-by-agreement mechanism, which prevents information loss [25].
III. General Structure of a Capsule Network
To overcome the aforementioned shortcomings of CNNs, in a capsule network, a group of capsules is considered, and each capsule is a collection of neurons that store different information about any object it is trying to identify. In a high-dimensional vector space, it stores information mostly about its position, rotation, scale, and so on, with each dimension representing special features about the object that can be understood intuitively. The main concept that works as an important building block of the capsule network depends on deconstruction of different hierarchical subparts of an object and developing a relationship between these internal parts of the whole object. The architecture of a simple capsule network can be described into three main blocks:
-
1)
primary capsules;
-
2)
higher layer capsules;
-
3)
loss function.
Each main block has suboperations in it, which is briefly discussed in the following subsections.
A. Primary Capsules
In this step, the input image is fed into a series of convolutional layers to extract array of feature maps. Then, a reshaping function is applied to reshape these feature maps into vectors. Now, in the last step of this block, a nonlinear Squash function is applied to keep the length of each vector within value 1. This nonlinear Squash function can be described as follows:
 |
Here, 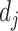 is considered as the vector output of capsule
is considered as the vector output of capsule 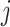 and
and 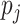 is its total output. The input capsule
is its total output. The input capsule 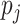 is a weighted sum of prediction vectors
is a weighted sum of prediction vectors 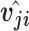 , which is calculated from the output of previous capsules by multiplying the output
, which is calculated from the output of previous capsules by multiplying the output 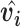 of previous capsule’s output by a weighted matrix
of previous capsule’s output by a weighted matrix 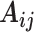
 |
B. Higher Layer Capsule
In this block, coupling coefficient 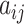 is calculated through routing by an agreement technique, which indicates the coupling or bonding between the higher level capsule and the lower capsule by using the following softmax function:
is calculated through routing by an agreement technique, which indicates the coupling or bonding between the higher level capsule and the lower capsule by using the following softmax function:
 |
where 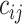 is the log probability, and it indicates whether lower level capsule
is the log probability, and it indicates whether lower level capsule 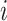 would be coupled with higher level capsule
would be coupled with higher level capsule 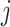 . At the beginning of the routing by the agreement process, its initial value is set to 0. The log probabilities are updated in the routing process based on the agreement between
. At the beginning of the routing by the agreement process, its initial value is set to 0. The log probabilities are updated in the routing process based on the agreement between 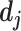 and
and 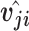 , and they produce a large inner product, which is calculated as follows:
, and they produce a large inner product, which is calculated as follows:
 |
There is an additional decoder network connected to the higher layer capsule, which learns to reconstruct the input image by minimizing the squared difference between the reconstructed image and the input image. This decoder network consists of three fully connected layers: two of them are rectified linear unit activated units, and the last one is the sigmoid activated layer.
C. Loss Function
Capsules use a separate loss function called margin loss 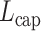 for each digit capsule
for each digit capsule 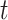 , which provides intraclass compactness and interclass separability. It is defined as
, which provides intraclass compactness and interclass separability. It is defined as
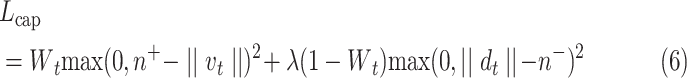 |
where 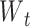 is 1 whenever class
is 1 whenever class 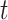 is actually present and is 0 otherwise. Terms
is actually present and is 0 otherwise. Terms 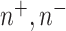 , and
, and 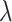 are hyperparameters that are tuned before the learning process. While trying to reconstruct the input image, the decoder network provides a reconstruction loss
are hyperparameters that are tuned before the learning process. While trying to reconstruct the input image, the decoder network provides a reconstruction loss 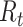 .
. 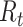 is computed as the mean square error between the input image and the reconstructed image, as follows:
is computed as the mean square error between the input image and the reconstructed image, as follows:
 |
Here, 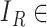 { reconstructed image} and
{ reconstructed image} and 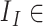 { input image data}. Hence, the total loss is
{ input image data}. Hence, the total loss is
 |
Here, 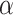 is used to minimize the effect of reconstruction loss in the total loss so as to give more importance to the margin loss so that the margin loss can dominate the training process. Basically, in the highest layer of capsule, images are reconstructed using the information that is preserved using the reconstruction unit and the reconstruction loss. During the training process, this loss acting as a regularizer helps to avoid overfitting.
is used to minimize the effect of reconstruction loss in the total loss so as to give more importance to the margin loss so that the margin loss can dominate the training process. Basically, in the highest layer of capsule, images are reconstructed using the information that is preserved using the reconstruction unit and the reconstruction loss. During the training process, this loss acting as a regularizer helps to avoid overfitting.
IV. Proposed Convolutional Capsule Network for COVID-19 Detection From Chest X-Ray and US Imaging
The proposed CapsCovNet consists of three blocks—a deep parallel convolutional network, a concatenated capsule network, and a decoder network. These blocks are described as follows.
-
1)
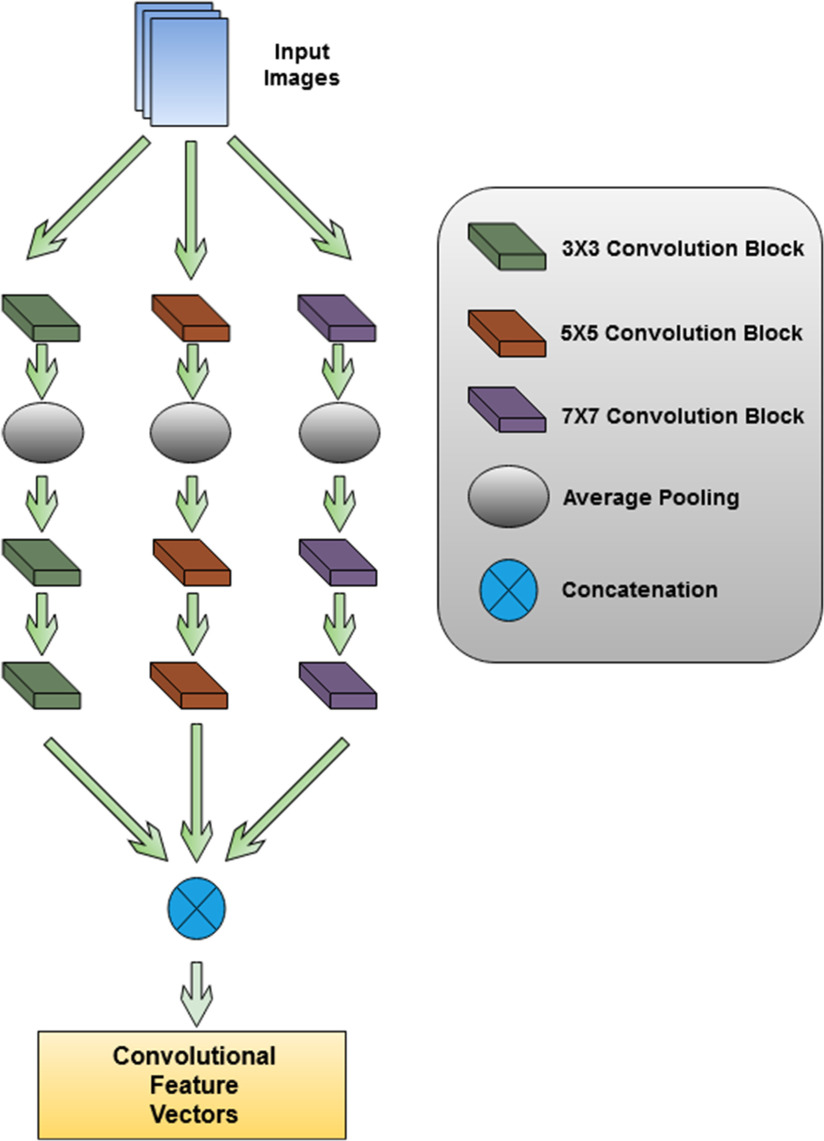
Parallel CNN Block: A graphical representation of parallel convolutional block is presented in Fig. 1. In this block, a deep parallel convolutional network is used to extract features from input images. This convolutional block consists of three parallelly connected convolutional blocks. Each convolutional block has three convolutional layers and one average pooling layer. The filter size of the convolutional layers is 32, 64, and 128. The kernel size of each block is varied to extract unique features from each convolutional block. The first convolutional block has a kernel size of
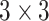 . The second convolutional block has a kernel size of
. The second convolutional block has a kernel size of 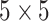 , and the last convolutional block has a kernel size of
, and the last convolutional block has a kernel size of 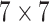 . Strides 1 is used in each convolutional layer. At first, a batch normalization layer is added after convolutional layer of each block. Then, a global average pooling layer is added in the architecture. To prevent overfitting, a dropout layer with 50% frequency rate is used. At the end of the convolutional layer block, all the features from the three convolutional arms are concatenated.
. Strides 1 is used in each convolutional layer. At first, a batch normalization layer is added after convolutional layer of each block. Then, a global average pooling layer is added in the architecture. To prevent overfitting, a dropout layer with 50% frequency rate is used. At the end of the convolutional layer block, all the features from the three convolutional arms are concatenated. -
2)
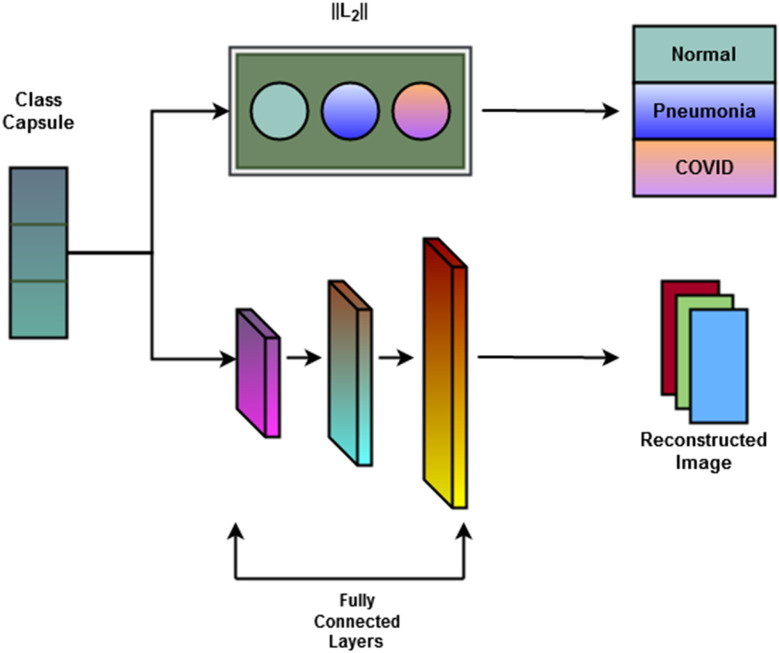
Concatenated Capsule Network: In Fig. 2, a schematic diagram of the concatenated capsule network is presented. This capsule network block consists of a primary capsule layer and a higher capsule layer. First, a reshaped layer is used to form the first primary capsule layer, which is a convolutional capsule layer that consists of 32 channels of 8-D convolutional capsules. The output of the primary capsule is a [
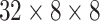 ] tensor of capsules, and each output is an 8-D vector. The output of this primary capsule layer is then passed into the higher capsule layer. Finally, the output vectors from primary capsules and higher layer capsules are concatenated and passed to the classification capsule layer. This layer has one 16-D capsule per class, and the lower layer capsules pass input features to each of these higher layer capsules. The routing-by-agreement algorithm is used between the capsules to pass lower layer capsule features into the higher layer capsules. The last capsule layer is a class capsule layer, which contains the instantiation parameters of the three classes, which are normal, pneumonia, and COVID-19 having a dimension of 16. The length of these three capsules represents the probability of each class being present.
] tensor of capsules, and each output is an 8-D vector. The output of this primary capsule layer is then passed into the higher capsule layer. Finally, the output vectors from primary capsules and higher layer capsules are concatenated and passed to the classification capsule layer. This layer has one 16-D capsule per class, and the lower layer capsules pass input features to each of these higher layer capsules. The routing-by-agreement algorithm is used between the capsules to pass lower layer capsule features into the higher layer capsules. The last capsule layer is a class capsule layer, which contains the instantiation parameters of the three classes, which are normal, pneumonia, and COVID-19 having a dimension of 16. The length of these three capsules represents the probability of each class being present. -
3)
Decoder Network: Class capsule layer is connected to three dense layers, which are composed of 512, 1024, and 49 152 neurons, respectively, which are then reshaped to
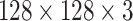 image size. A pictorial representation of the decoder block and the final output block is presented in Fig. 3.
image size. A pictorial representation of the decoder block and the final output block is presented in Fig. 3.
Fig. 1.
Parallel convolutional feature extractor, which extracts diverse feature vectors using variable kernel size.
Fig. 2.
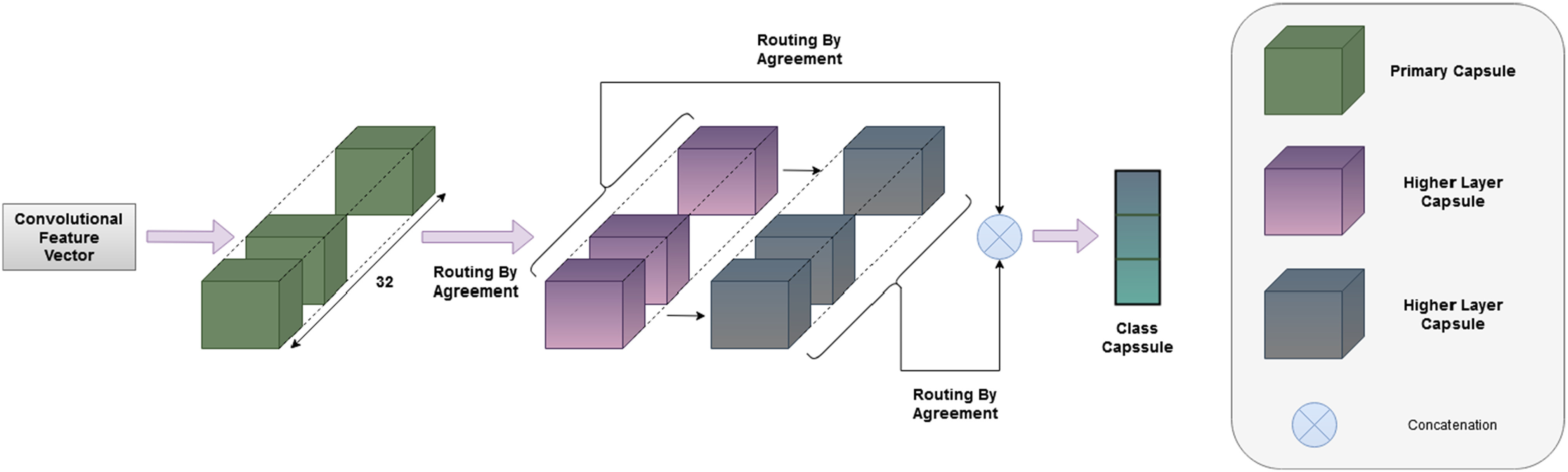
Graphical representation of the proposed convolutional capsule network. Features extracted from parallel convolutional blocks are used as input for the capsule block.
Fig. 3.
Schematic of the decoder block. Decoder block is composed of three fully connected dense layers, which have 512, 1024, and 49 152 neurons, respectively, which are then reshaped to 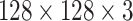 image size.
image size.
A. Loss Function
Loss is calculated using the loss function in (8). Since the dataset is very small, there is a chance of overfitting the model. Hence, reconstruction loss with a high weight value 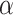 is associated with the margin loss, which prevents the model to become overfitted.
is associated with the margin loss, which prevents the model to become overfitted.
B. Hyperparameter
For the task of COVID-19 detection, we have trained the model using 50 epochs. The number of routings and the batch size is set to 3 and 64, respectively. The Adam optimizer has been used with a learning rate of 0.001. For total loss calculation, we have used weight 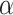 = 0.6 for reconstruction loss. For margin loss calculation, both
= 0.6 for reconstruction loss. For margin loss calculation, both 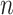 + and
+ and 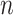
 are set to 0.1, and
are set to 0.1, and 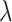 is set to 0.5.
is set to 0.5.
V. Experimental Setup
A. Dataset
In this article, three publicly available datasets are used for finding the efficiency of the proposed network. The first one is an open-source dataset of US imaging called point of care ultrasound (POCUS) dataset [35]. It contains 64 videos taken from various sources. Hence, the format and illumination of US images differ significantly.
The remaining datasets are two open-source chest X-ray datasets, which are used for training and testing the proposed model. The first dataset contains chest X-ray images of COVID-affected patients collecting from six different publicly available databases. These data were collected from the Italian Society of Medical and Interventional Radiology COVID-19 DATABASE, Novel Corona Virus 2019 Dataset, COVID-19 positive chest X-ray images from different articles, COVID-19 Chest imaging at thread reader, RSNA-Pneumonia-Detection-Challenge, and Chest X-ray Images (pneumonia) [39], [43]–[46], [48]. The second dataset is an aggregation of two publicly available chest X-ray datasets [22], [40]–[42]. In the rest of this article, these two datasets will be addressed as dataset-1 and dataset-2 for simplicity.
B. Pretraining Process and Pretraining Dataset
Usually, to train any deep CNN, a large dataset is needed. A large dataset helps a neural network to avoid overfitting and underfitting problems. Our proposed deep convolutional capsule network has a deep parallel convolutional neural block. Hence, a large dataset can significantly improve the performance of our proposed network. However, no such large medical dataset is available for COVID-19. Especially, US data of COVID-19 patients are very limited. Hence, we adopt a pretraining and fine-tuning technique. Normally, in transfer learning, a deep neural network is first trained using a large source dataset, and then, early layers of the trained model are kept frozen. Next, some deeper layers of the network are again retrained using a target dataset. This pretraining technique has been proven to be an efficacious process in different fields of computer vision tasks especially in those cases where aggregating a large dataset is not possible [49]. In this article, a dataset of X-ray image containing 94 323 images distributed into five classes has been used as the source dataset in the pretraining process [47]. The capsule block and the decoder block are fine-tuned by freezing the convolutional block after pretraining. This pretraining process improved the detection accuracy in our work to a great extent.
C. Data Processing and Classification Pipeline
US images are extracted from the US videos at the frame rate of 30 frame/s, as it is done in [35]. After the image extraction process, a total of 1103 images are found, where 654 are of COVID-19, 277 are of pneumonia, and the rest 172 are of healthy types. Since capsule networks are equivariant to pose changes, the data augmentation technique does not add any improvement in the performance of the capsule network. Hence, data augmentation techniques have not been used in the training phase. To minimize the effect of sampling bias, histogram equalization was applied to images using the normalized contrast limited adaptive histogram equalization (N-CLAHE) algorithm [36]. This method first globally normalizes the images and then applies contrast limited adaptive histogram equalization (CLAHE), which normalizes images and enhances small details, textures, and local contrast [37], [38]. The classification pipeline of our proposed method is shown in Fig. 4. At the preprocessing step, N-CLAHE has been applied to enhance smaller details, and then, images are resized to 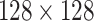 pixels. Finally, training images are fed to the network.
pixels. Finally, training images are fed to the network.
Fig. 4.
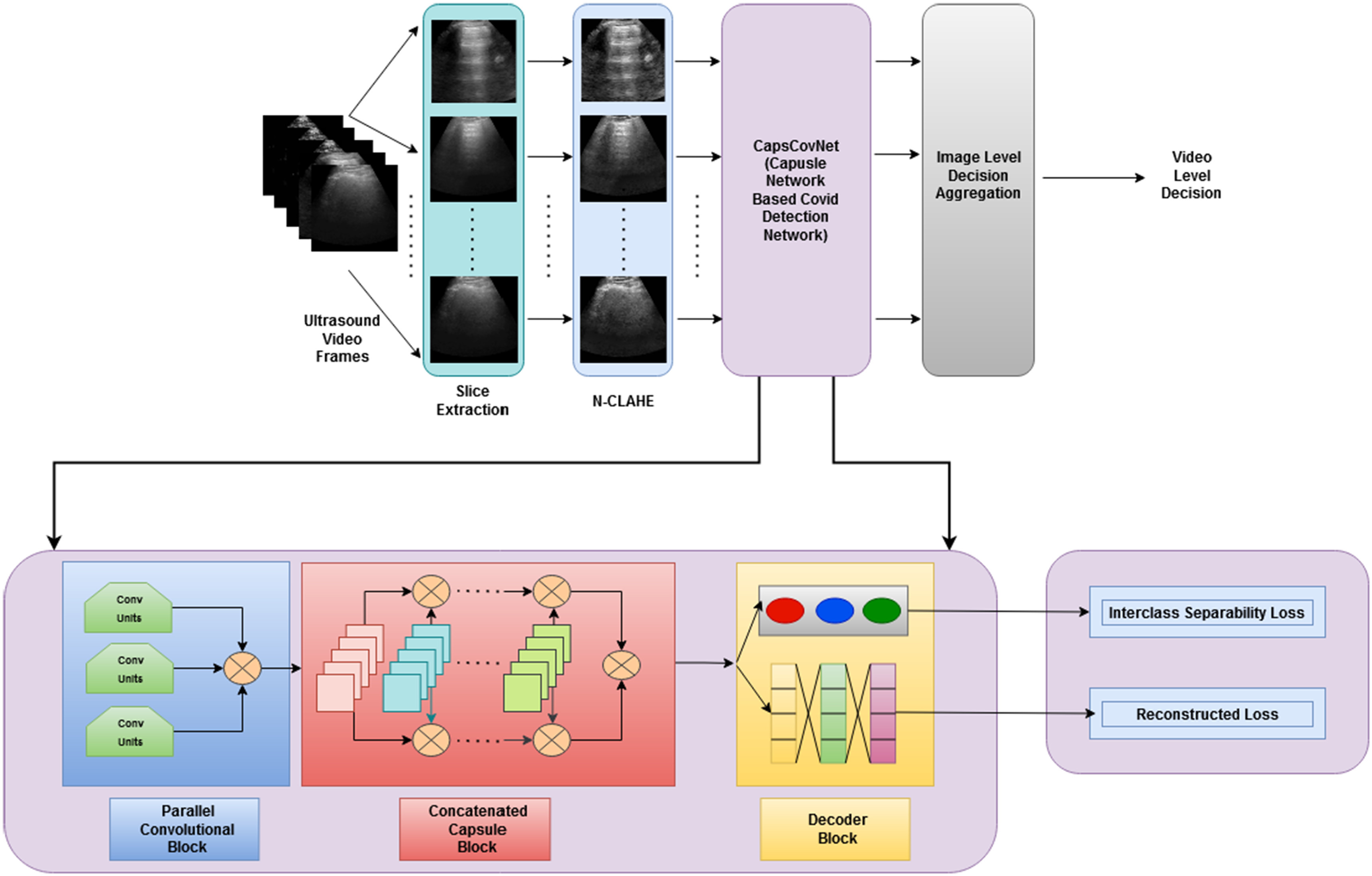
Classification pipeline of the proposed method showing the main data preprocessing steps and deep convolutional and capsule blocks.
D. Performance Evaluation Matrix
Five performances metrics such as accuracy, sensitivity or recall, specificity, precision (positive predictive value), and 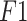 score have been used to compare the classification performance of the proposed method with the existing deep learning algorithms. They are defined as follows:
score have been used to compare the classification performance of the proposed method with the existing deep learning algorithms. They are defined as follows:
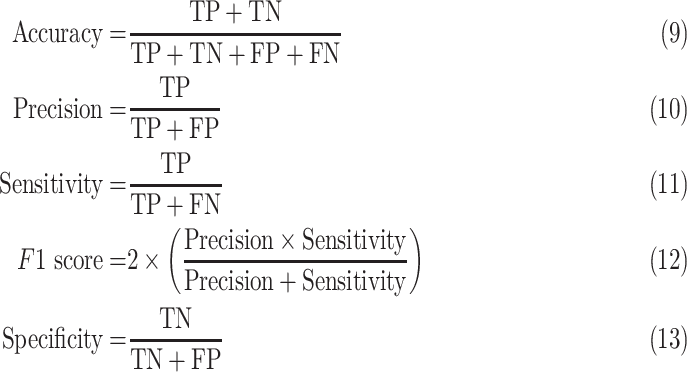 |
where
-
1)
TP = true positive;
-
2)
TN = true negative;
-
3)
FP = false positive;
-
4)
FN = false negative.
VI. Results and Discussion
In this section, the results obtained from rigorous experimentations on three publicly available datasets are discussed in three diverse perspectives. For evaluation purposes, a fivefold cross-validation technique is used for the US dataset and dataset-1, and a tenfold cross-validation technique is used for dataset-2. To ensure that training and testing data are completely disjoint, extracted frames from a single video are included within a single fold only.
A. Ablation Study
The ablation study is conducted to validate the contribution of each of the steps included in the proposed methodology. The performance improvement achieved from including the preprocessing and pretraining step is summarized in Table IV, where it is found that applying these steps significantly improves the performance of the proposed method. The N-CLAHE technique, which is applied as the main preprocessing step, enhances the image quality significantly, and 1.6–2.13% improvement in evaluation metrics has been achieved by applying it. Due to technical fault, image quality can be degraded significantly, and as a result, the performance of the machine learning technique can deteriorate. Using the N-CLAHE approach, the quality of raw medical images is enhanced and eventually contributes to performance improvement. Moreover, for performance improvement, as mentioned earlier, the pretraining technique is used for cases where the aggregation of large-scale labeled data is not possible. As the proposed method has a deep feature extractor block that requires an extensive amount of training data, pretraining this block is found to be an effective step contributing notably toward the improved performance of the proposed method.
TABLE IV. Comparison Between Capsule Network With Sequential Convolutional Blocks and Capsule Network With Parallely Concatenated Convolutional Blocks.
| Dataset | Evaluated metrics | Without n-CLAHE | With n-CLAHE | |
|---|---|---|---|---|
| Without Pretraining | With Pretraining | |||
| POCUS dataset | Accuracy | 0.945 | 0.960 | 0.983 |
| Precision | 0.943 | 0.959 | 0.981 | |
| Sensitivity | 0.951 | 0.956 | 0.985 | |
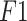 score score |
0.942 | 0.959 | 0.987 | |
| Specificity | 0.954 | 0.962 | 0.991 | |
| Dataset-1 | Accuracy | 0.951 | 0.968 | 0.990 |
| Precision | 0.953 | 0.963 | 0.992 | |
| Sensitivity | 0.952 | 0.965 | 0.997 | |
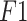 score score |
0.952 | 0.968 | 0.996 | |
| Specificity | 0.948 | 0.967 | 0.996 | |
| Dataset-2 | Accuracy | 0.940 | 0.959 | 0.989 |
| Precision | 0.945 | 0.961 | 0.986 | |
| Sensitivity | 0.943 | 0.962 | 0.99 | |
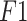 score score |
0.945 | 0.96 | 0.993 | |
| Specificity | 0.944 | 0.964 | 0.99 | |
For comparison purpose, kernel size of 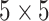 is used for sequential convolutional blocks. Kernel size of
is used for sequential convolutional blocks. Kernel size of 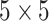 gives the maximum values in the evaluation matrices in capsule network with sequential convolutional blocks.
gives the maximum values in the evaluation matrices in capsule network with sequential convolutional blocks.
B. Quantitative Analysis
In Table II, comparison between existing state-of-the-art methods and proposed method in COVID-19 detection from the chest X-ray dataset is summarized. In dataset-1, our proposed method has scored 1.3–3% higher in different comparison metrics than the existing state-of-the-art method [39]. In dataset-2, our proposed method performs better than the current state-of-the-art method [42]. In this case, the improvement is between 0.71% and 23% in different performance evaluation metrics.
TABLE II. Comparison Between the Proposed Method and the Existing State-of-the-Art Methods in the Chest X-Ray Dataset.
| Dataset | Methods | Accuracy | Precision | Sensitivity |
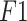 Score Score
|
Specificity |
| Dataset-1 | Chowdhury et al. | 0.9774 | 0.9661 | 0.9661 | 0.9661 | 0.9831 |
| Asnaoui et al. | 0.922 | 0.924 | 0.921 | 0.921 | 0.960 | |
| Ucar et al. | 0.983 | – | 0.983 | – | 0.991 | |
| Apostolopoulos et al. | 0.947 | – | 0.986 | – | 0.964 | |
| Proposed | 0.990 | 0.982 | 0.997 | 0.996 | 0.996 | |
| Dataset-2 | Afshar et al. | 0.983 | – | 0.8 | – | 0.986 |
| Goel et al. | 0.978 | 0.929 | 0.977 | 0.953 | 0.962 | |
| Panahi et al. | 0.96 | – | 0.96 | – | – | |
| Proposed | 0.989 | 0.986 | 0.99 | 0.993 | 0.99 |
In Table III, comparison between the existing state-of-the-art method and our proposed is summarized in the US image dataset. Our proposed method has outperformed the state-of-the-art method in US images also. We have a performance increase of 3.12–20.2% in performance metrics. Then, we have classified the US videos using a process, where the class occupying most of the frames in one video indicates the class of the entire US video. Using this process, 100% accuracy is obtained in US COVID-19 video classification.
TABLE III. Comparison Between the Proposed Method and the Existing State-of-the-Art Method in the US Dataset.
| Methods | Class | Sensitivity | Specificity | Precision |
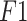 Score Score
|
|---|---|---|---|---|---|
| Pocovid Net | COVID-19 | 0.96 | 0.79 | 0.88 | 0.92 |
| Pneumonia | 0.93 | 0.98 | 0.95 | 0.94 | |
| Normal | 0.55 | 0.98 | 0.78 | 0.62 | |
| Muhammad et al. | COVID-19 | 0.902 | – | 0.952 | – |
| Pneumonia | 0.958 | – | 0.969 | – | |
| Normal | 0.936 | – | 0.832 | – | |
| Proposed | COVID-19 | 0.99 | 0.95 | 0.99 | 0.99 |
| Pneumonia | 0.98 | 0.99 | 0.96 | 0.96 | |
| Normal | 0.97 | 0.99 | 1.00 | 0.99 |
C. Qualitative Analysis
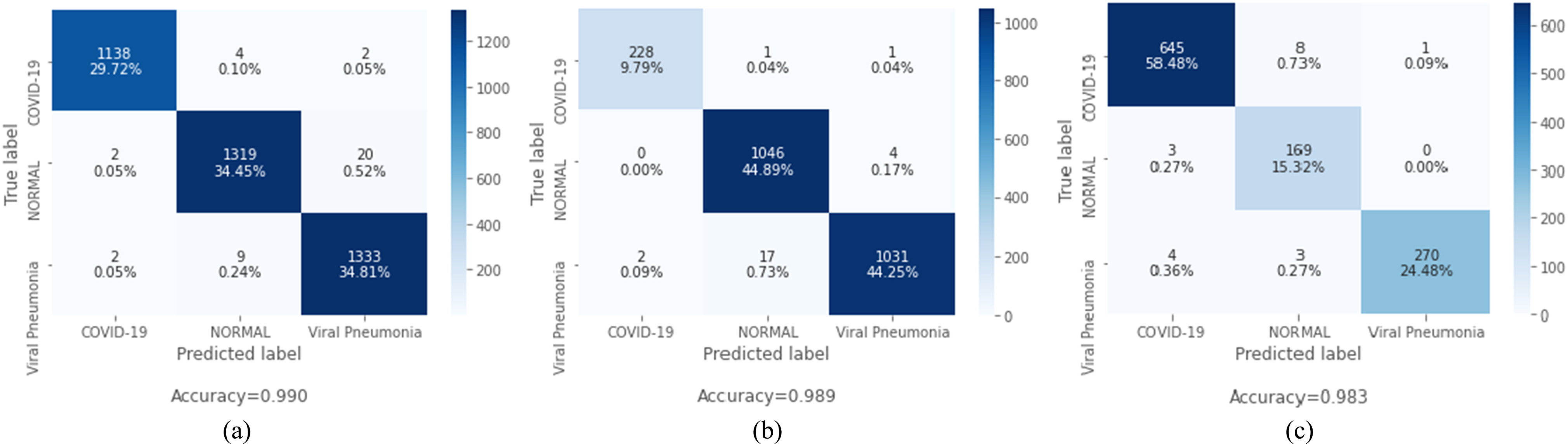
Our proposed method significantly reduces false positive and false negative cases over the existing state-of-the-art methods. As COVID-19 and pneumonia patients have similar symptoms, there is a high chance to misclassify these classes. But our proposed method can handle these cases very efficiently. Most importantly, from Fig. 5(a), the superior performance of this method in detecting COVID-19 cases from the chest X-ray can also be observed. Among 1144 COVID-19 chest X-ray cases, only six cases are misclassified, and among these six cases, only two of them are classified as pneumonia. Again from Fig. 5(b), we can see that our proposed method performs exceptionally, and among 230 COVID-19 cases, our proposed method can correctly identify 228 cases and misclassify only two cases, where only one COVID-19 case is misclassified as pneumonia. Fig. 5(c) has depicted the confusion matrix of our proposed method using US images. Out of 654 COVID-19 US frames, our proposed method has accurately classified 645 frames and misclassified only nine frames, and only eight frames are misclassified as pneumonia. Furthermore, to express the capability of the proposed model in distinguishing between the classes, the areas under the curve (AUCs) of receiver operating characteristics (ROC) are illustrated in Fig. 6, where the AUCs are calculated using the one versus all approach, in which the ROC curve for a specific class will be generated as classifying that class against the other classes. Analyzing each of the ROC curves for three separate datasets depicted in Fig. 6(a)–(c), it can be concluded that the proposed model can differentiate each of the classes with a negligible amount of false positive cases. In Table I, comparison between the capsule network with sequential convolutional blocks and the capsule network with parallely concatenated convolutional blocks is summarized. It should be noticed that the proposed capsule network with parallely concatenated convolutional blocks outperforms the capsule network with sequential convolutional blocks compared by a considerable margin in all the metrics. The capsule network with sequential convolutional blocks directly operates on the whole image to extract features for COVID-19 detection, whereas the proposed capsule network with parallely concatenated convolutional blocks effectively integrates features from its parallel convolutional hands. These convolutional blocks are comprised of dissimilar kernel size, which provides a group of features that are more capable of differentiating multimodal medical images than the capsule network with sequential convolutional blocks and that results in higher accuracy.
Fig. 5.
Confusion matrix from the proposed CapsCovNet. (a) Confusion matrix for dataset-1. (b) Confusion matrix for dataset-2. (c) Confusion matrix for POCUS dataset.
Fig. 6.
ROC curve from the proposed CapsCovNet. (a) ROC curve for dataset-1. (b) ROC curve for dataset-2. (c) ROC curve for POCUS dataset.
TABLE I. Performance Evaluation Matrices for the Proposed Deep Parallel Convolutional Capsule Network With and Without N-CLAHE and Pretraining.
| Dataset | Evaluated metrics | Capsule Network with Sequential Convolutional Blocks | Capsule Network with Parallely Concatenated Convolutional Blocks |
|---|---|---|---|
| POCUSDdataset | Accuracy | 0.943 | 0.983 |
| Precision | 0.939 | 0.981 | |
| Sensitivity | 0.931 | 0.985 | |
| F1 score | 0.932 | 0.987 | |
| Specificity | 0.932 | 0.991 | |
| Dataset-1 | Accuracy | 0.945 | 0.990 |
| Precision | 0.949 | 0.992 | |
| Sensitivity | 0.951 | 0.997 | |
| F1 score | 0.950 | 0.996 | |
| Specificity | 0.951 | 0.996 | |
| Dataset-2 | Accuracy | 0.939 | 0.989 |
| Precision | 0.944 | 0.986 | |
| Sensitivity | 0.946 | 0.99 | |
| F1 score | 0.941 | 0.993 | |
| Specificity | 0.944 | 0.99 |
By applying data preprocessing techniques, significant improvement in results has been achieved. More improvement in results is achieved by applying the pretraining technique.
D. Clinically Relevant Information Obtained by CapsCovNet
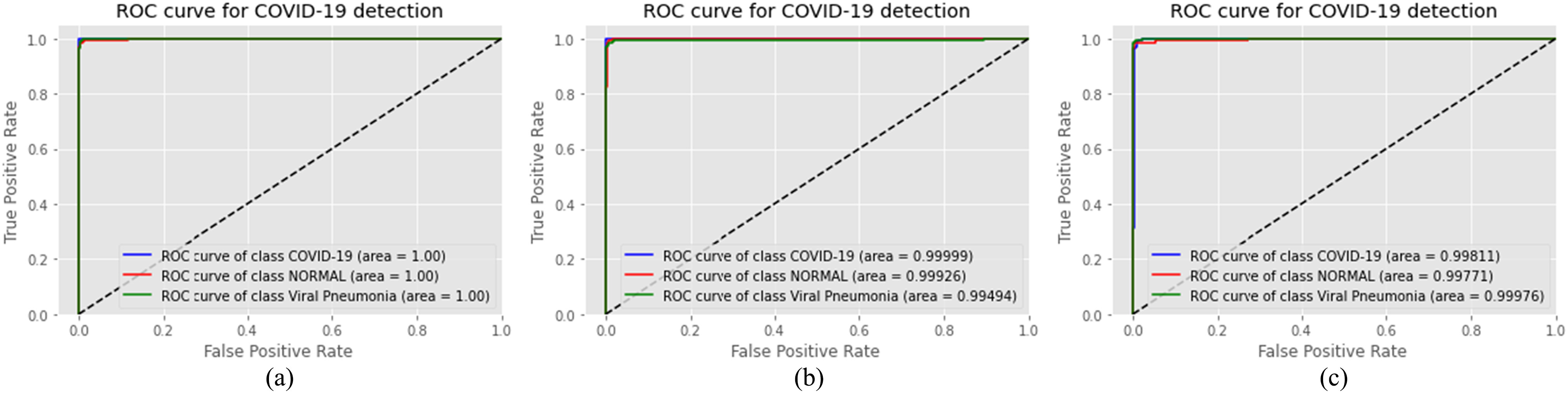
A very arduous challenge while working on COVID-19 detection from chest X-ray is the unavailability of a large publicly available dataset to train and test deep learning models, which can solidify the claim of generalization and low biased nature of the model. For this reason, while moving onward with this limitation, we have put utmost emphasis on checking whether our model is learning clinically relevant features or not. The surmisable issues that impede the generalization of models proposed for COVID-19 detection are bias introduced by distinction between different datasets and source of acquisition, presence of medical equipment, and various text data that are embedded in the scan [62]. To visualize the class activation mapping for localizing particular areas of multimodal medical imaging (US and X-ray in this case) that assists the decision process, the gradient-based class activation mapping (Grad-CAM) algorithm [34] is integrated with our proposed CapsCovNet. As depicted in Fig. 7, our model completely ignores the presence of medical equipment and embedded letters in the X-ray images. So, it can be firmly stated that our model is robust against these sort of biases. Tissue intensity and characteristics in medical images vary significantly from dissimilar acquisition techniques and for different sources of acquisition as well [55]. Hence, intensity standardization is firmly in practice for medical image segmentation and disease classification. We have used N-CLAHE histogram standardization in this article, which, apart from aiding to improve our classification accuracy, helped to extenuate the bias from different sources of acquisition.
Fig. 7.
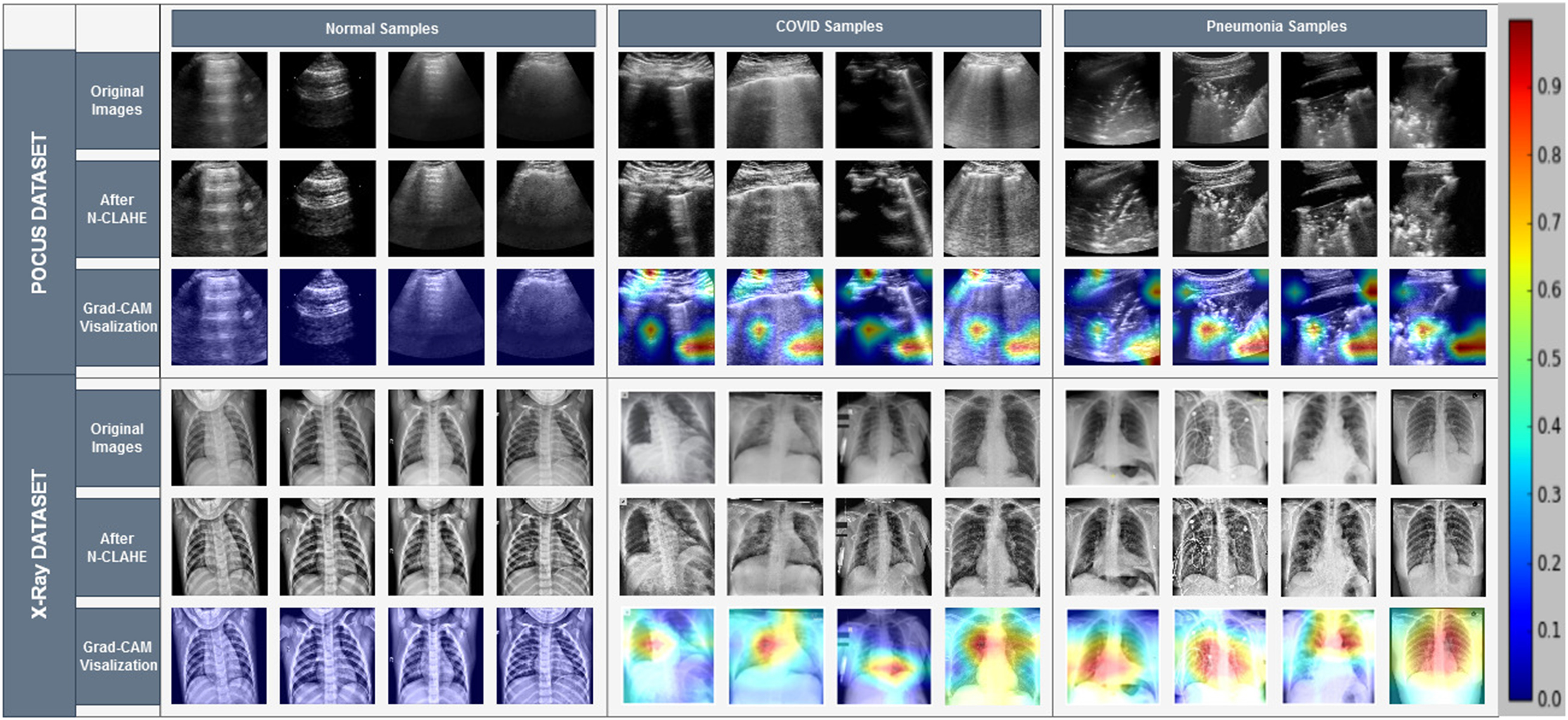
Grad-CAM visualization of US and X-ray samples.
Another important concern while working with different small-sized datasets is the relevancy of information learned by a deep learning model to actual clinical characteristics prevalent in respective diseases. As shown in [56], even without the information present in most of the lung regions, good classification results can be obtained for different proposed models, which are biased toward different factors and emphasize on the information relevant to source or method of acquisition. From Fig. 4, it is evident that the information present in the lung region is of highest importance, and therefore, the hypothesis presented in [56] is not appropriate regarding our model.
In normal patient’s chest X-ray, generally, no form of opacities is present [57], [58], and from Fig. 7, our model has not marked any form of opacity also, and it can effectively identify healthy lungs. For distinguishing COVID-19 and viral pneumonia, our model is marking central and peripheral distribution in viral pneumonia, but it is not marking both sorts of distribution in COVID-19 pneumonia. The findings from the clinical experiment presented in [50] perfectly match our model’s activation heatmap. Therefore, it can be fairly assumed that our model is learning distinguishing features, which are relevant to actual clinical features for COVID-19 detection task.
The issue of generalization of successfully identifying a new case of a normal patient from the COVID-19-affected one can be safely claimed as for all test cases in different folds, our model has detected abnormalities in lungs. Millions of patients are diagnosed with COVID-19 and pneumonia, which have many common characteristics between them, and it is very difficult to claim that our model can distinguish each and every one of those cases successfully without training it with a huge volume of data. Even there is always significant possibility of inherent biases among expert radiologists in interpreting and diagnosing disease from medical images [59]. Nevertheless, our proposed model is learning underlying complex insights from data found from the study of expert radiologists [58], which strengthens the possibility of clinical application of our work in prescreening and aiding in diagnosis of patient successfully.
VII. Conclusion
In this article, a parallelly connected convolutional block-based capsule network was proposed to diagnose COVID-19 from chest X-ray and US images. The parallel connection of convolutional block and concatenation of multiple capsule units, which are introduced in this article, bolstered the model to learn more competent features and improved the COVID-19 detection performance substantially. The proposed architecture provided significant results using a very small training dataset, and further improvement was achieved by pretraining the convolutional blocks. Moreover, the localization of the affected region found from this model can help the medical practitioners as a clinical tool. Nevertheless, further study should be required to inspect the complex pattern of COVID-19 by incorporating more patients’ data from diverse geographic locations of the world.
Funding Statement
This work was supported in part by the Natural Sciences and Engineering Research Council of Canada and in part by the Regroupment Strategique en Microelectronique du Quebec.
Footnotes
Contributor Information
A. F. M. Saif, Email: afmsaif927@gmail.com.
Tamjid Imtiaz, Email: tamjidimtiaz1995@ gmail.com.
Shahriar Rifat, Email: rifatshahriar0231@gmail.com.
Celia Shahnaz, Email: celia@eee.buet.ac.bd.
Wei-Ping Zhu, Email: weiping@ece.concordia.ca.
M. Omair Ahmad, Email: omair@ece.concordia.ca.
References
- [1].Wu F. et al. , “A new coronavirus associated with human respiratory disease in China,” Nature, vol. 579, no. 7798, pp. 265–269, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Huang C. et al. , “Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China,” Lancet, vol. 395, no. 10223, pp. 497–506, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].World Health Organization, Pneumonia of Unknown Cause—China. Emergencies Preparedness, Response, Disease Outbreak News, World Health Organization, Geneva, Switzerland, 2020. [Google Scholar]
- [4].Xu X. et al. , “A deep learning system to screen novel coronavirus disease 2019 pneumonia,” Engineering, vol. 6, no. 10, pp. 1122–1129, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang W. et al. , “Detection of SARS-CoV-2 in different types of clinical specimens,” JAMA, vol. 323, no. 18, pp. 1843–1844, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen C. M., Chou Y. H., Tagawa N., and Do Y., “Computer-aided detection and diagnosis in medical imaging,” Comput. Math. Methods Med., vol. 2013, 2013, Art. no. 790608, doi: 10.1155/2013/790608. [DOI] [Google Scholar]
- [7].Sloun R. J. G. van, Cohen R., and Eldar Y. C., “Deep learning in ultrasound imaging,” Proc. IEEE, vol. 108, no. 1, pp. 11–29, Jan. 2020, doi: 10.1109/JPROC.2019.2932116. [DOI] [Google Scholar]
- [8].Hu M. et al. , “Learning to recognize Chest-XRay images faster and more efficiently based on multi-kernel depthwise convolution,” IEEE Access vol. 8, pp. 37265–37274, 2020. [Google Scholar]
- [9].Domingues I., Pereira G., Martins P., Duarte H., Santos J., and Abreu P. H., “Using deep learning techniques in medical imaging: A systematic review of applications on CT and PET,” Artif. Intell. Rev., vol. 53, pp. 4093–4160, 2020, doi: 10.1007/s10462-019-09788-3. [DOI] [Google Scholar]
- [10].Ozturk T., Talo M., Yildirim E. A., Baloglu U. B., Yildirim O., and Acharya U. R., “Automated detection of COVID-19 cases using deep neural networks with X-ray images,” Comput. Biol. Med., vol. 121, 2020, Art. no. 103792, doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Das D., Santosh K. C., and Pal U., “Truncated inception net: COVID-19 outbreak screening using chest X-rays,” Phys. Eng. Sci. Med., vol. 43, no. 3, pp. 915–925, 2020, doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Harmon S. A. et al. , “Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets,” Nature Commun., vol. 11, no. 1, 2020, Art. no. 4080, doi: 10.1038/s41467-020-17971-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhang Q. et al. , “A GPU-based residual network for medical image classification in smart medicine,” Inf. Sci., vol. 536, pp. 91–100, 2020. [Google Scholar]
- [14].Huang Z., Zhu X., Ding M., and Zhang X., “Medical image classification using a light-weighted hybrid neural network based on PCANet and DenseNet,” IEEE Access, vol. 8, pp. 24697–24712, 2020, doi: 10.1109/ACCESS.2020.2971225. [DOI] [Google Scholar]
- [15].Tseng K., Zhang R., Chen C. M., and Hassan M. M., “DNetUnet: A semi-supervised CNN of medical image segmentation for super-computing AI service,” J. Supercomput., vol. 77, pp. 3594–3615, 2020, doi: 10.1007/s11227-020-03407-7. [DOI] [Google Scholar]
- [16].LeCun Y., Bengio Y., and Hinton G., “Deep learning,” Nature, vol. 521, pp. 436–444, 2015, doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- [17].Krizhevsky A., Sutskever I., and Hinton G. E., “ImageNet classification with deep convolutional neural networks,” Int. Conf. Neural Inf. Process. Syst., 2012, pp. 1097–1105. [Google Scholar]
- [18].Fang Y. et al. , “Sensitivity of chest CT for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. E115–E117, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ai T. et al. , “Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases,” Radiology, vol. 296, no. 2, pp. E32–E40, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Brunese L., Mercaldo F., Reginelli A., and Santone A., “Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-Rays,” Comput. Methods Programs Biomed., vol. 196, 2020, Art. no. 105608, doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Apostolopoulos I. D. and Mpesiana T. A., “COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks,” Phys. Eng. Sci. Med., vol. 43, no. 2, pp. 635–640, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang L., Lin Z. Q., and Wong A., “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest radiography images,” Sci. Rep., vol. 10, 2020, Art. no. 19549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Song Y. et al. , “Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images,” IEEE/ACM Trans. Comput. Biol. Bioinform., to be published, doi: 10.1109/TCBB.2021.3065361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ardakani A. A., Kanafi A. R., Acharya U. R., Khadem N., and Mohammadi A., “Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks,” Comput. Biol. Med., vol. 121, 2020, Art. no. 103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hinton G., Sabour S., and Frosst N., “Matrix capsules with EM routing,” in Proc. Int. Conf. Learn. Represent., 2018, pp. 1–15. [Google Scholar]
- [26].Saif A. F. M., Shahnaz C., Zhu W., and Ahmad M. O., “Abnormality detection in musculoskeletal radiographs using capsule network,” IEEE Access, vol. 7, pp. 81494–81503, 2019, doi: 10.1109/ACCESS.2019.2923008. [DOI] [Google Scholar]
- [27].Reddy U. M., Filly R. A., and Copel J. A., “Prenatal imaging: Ultrasonography and magnetic resonance imaging,” Obstet. gynecol., vol. 112, no. 1, pp. 145–157, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liu S. et al. , “Deep learning in medical ultrasound analysis: A review,” Engineering, vol. 5, no. 2, pp. 261–275, 2019, doi: 10.1016/j.eng.2018.11.020. [DOI] [Google Scholar]
- [29].Becker A. S., Mueller M., Stoffel E., Marcon M., Ghafoor S., and Boss A., “Classification of breast cancer in ultrasound imaging using a generic deep learning analysis software: A pilot study,” Brit. J. Radiol., vol. 91, 2018, Art. no. 20170576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Han S. et al. , “A deep learning framework for supporting the classification of breast lesions in ultrasound images,” Phys. Med. Biol., vol. 62, no. 19, 2017, Art. no. 7714. [DOI] [PubMed] [Google Scholar]
- [31].Savaş S., Topaloğlu N., Kazcı Ö., and Koşar P. N., “Classification of carotid artery intima media thickness ultrasound images with deep learning,” J. Med. Syst., vol. 43, no. 8, pp. 1–12, 2019. [DOI] [PubMed] [Google Scholar]
- [32].Afshar P., Mohammadi A., and Plataniotis K. N., “Brain tumor type classification via capsule networks,” in Proc. IEEE Int. Conf. Image Process., 2018, pp. 3129–3133. [Google Scholar]
- [33].Toraman S., Alakus T. B., and Turkoglu I., “Convolutional CapsNet: A novel artificial neural network approach to detect COVID-19 disease from X-ray images using capsule networks,” Chaos, Solitons Fractals, vol. 140, 2020, Art. no. 110122, doi: 10.1016/j.chaos.2020.110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Selvaraju R. R., Cogswell M., Das A., Vedantam R., Parikh D., and Batra D., “GRAD-CAM: Visual explanations from deep networks via gradient-based localization,” in Proc. IEEE Int. Conf. Comput. Vis., 2017, pp. 618–626. [Google Scholar]
- [35].Born J. et al. , “POCOVID-Net: Automatic detection of COVID-19 from a new lung ultrasound imaging dataset (POCUS),” 2020, arXiv:2004.12084.
- [36].Koonsanit K., Thongvigitmanee S., Pongnapang N., and Thajchayapong P., “Image enhancement on digital X-ray images using N-CLAHE,” in Proc. 10th Biomed. Eng. Int. Conf., Aug. 2017, pp. 1–4. [Google Scholar]
- [37].Zuiderveld K., “Contrast limited adaptive histogram equalization,” in Graphics Gems IV. New York, NY, USA: Academic, 1994. [Google Scholar]
- [38].Horry M. J. et al. , “COVID-19 detection through transfer learning using multimodal imaging data,” IEEE Access, vol. 8, pp. 149808–149824, 2020, doi: 10.1109/ACCESS.2020.3016780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Chowdhury M. E. et al. , “Can AI help in screening viral and COVID-19 pneumonia?,” IEEE Access, vol. 8, pp. 132665–132676, 2020, doi: 10.1109/ACCESS.2020.3010287. [DOI] [Google Scholar]
- [40].Cohen J. P., “COVID Chest X-Ray dataset,” 2020. [Online]. Available: https://github.com/ieee8023/covid-chestxray-dataset
- [41].Mooney P., “Kaggle Chest X-Ray images (pneumonia) dataset,” 2020. [Online]. Available: https://github.com/ieee8023/covid-chestxray-datase
- [42].Afshar P., Heidarian S., Naderkhani F., Oikonomou A., Plataniotis K. N., and Mohammadi A., “COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images,” Pattern Recognit. Lett., vol. 138, pp. 638–643, 2020, doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].S.-I. S. O. M. A. I. Radiology, “COVID-19 database,” 2020. [Online]. Available: https://www.sirm.org/category/senza-categoria/covid-19/
- [44].Monteral J. C., “COVID-Chestxray database,” 2020. [Online]. Available: https://github.com/ieee8023/covid-chestxray-dataset
- [45].Radiopedia, 2020. [Online]. Available: https://radiopaedia.org/search?lang=us&page=4&q=covid+19& scope=all&utf8=%E2%9C%93
- [46].Imaging C., “This is a thread of COVID-19 CXR (All SARS-CoV-2 PCR) from my Hospital (Spain). I hope it could help,” 2020. [Online] Available: https://threadreaderapp.com/thread/1243928581983670272.html
- [47].Wang X., Peng Y., Lu L., Lu Z., Bagheri M., and Summers R. M., “ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit., Jul. 2017, pp. 2097–2106. [Google Scholar]
- [48].Mooney P., “Chest X-Ray images (Pneumonia),” 2018. [Online] Available: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- [49].Windrim L., Melkumyan A., Murphy R. J., Chlingaryan A., and Ramakrishnan R., “Pretraining for hyperspectral convolutional neural network classification,” IEEE Trans. Geosci. Remote Sens., vol. 56, no. 5, pp. 2798–2810, May 2018, doi: 10.1109/TGRS.2017.2783886. [DOI] [Google Scholar]
- [50].Bai H. X., et al. , “Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT,” Radiology, vol. 29, no. 2, pp. E46–E54, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang Y. D. et al. , “COVID-19 diagnosis via DenseNet and optimization of transfer learning setting,” Cogn. Comput., pp. 1–17, 2021. [DOI] [PMC free article] [PubMed]
- [52].Wang S. H., Nayak D. R., Guttery D. S., Zhang X., and Zhang Y. D., “COVID-19 classification by CCSHNet with deep fusion using transfer learning and discriminant correlation analysis,” Inf. Fusion, vol. 68, pp. 131–148, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nhamo G., Chikodzi D., Kunene H. P., and Mashula N., “COVID-19 vaccines and treatments nationalism: Challenges for low-income countries and the attainment of the SDGs,” Glob. Public Health, vol. 16, no. 3, pp. 319–339, 2021. [DOI] [PubMed] [Google Scholar]
- [54].Kusec R., “Reverse transcriptase-polymerase chain reaction (RT-PCR),” Clin. Appl. PCR, vol. 16, pp. 149–158, 1998. [DOI] [PubMed] [Google Scholar]
- [55].Bağci U., Udupa J. K., and Bai L., “The role of intensity standardization in medical image registration,” Pattern Recognit. Lett., vol. 31, no. 4, pp. 315–323, 2010. [Google Scholar]
- [56].Maguolo G. and Nanni L., “A critic evaluation of methods for COVID-19 automatic detection from X-ray images,” Inf. Fusion, vol. 76, pp. 1–7, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Franquet T., “Imaging of pneumonia: Trends and algorithms,” Eur. Respir. J., vol. 18, no. 1, pp. 196–208, 2001. [DOI] [PubMed] [Google Scholar]
- [58].Vilar J., Domingo M. L., Soto C., and Cogollos J., “Radiology of bacterial pneumonia,” Eur. J. Radiol., vol. 51, no. 2, pp. 102–113, 2004. [DOI] [PubMed] [Google Scholar]
- [59].Busby L. P., Jesse L. C., and Christine G. M., “Bias in radiology: The how and why of misses and misinterpretations,” Radiographics, vol. 38, no. 1, pp. 236–247, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Xi E., Bing S., and Jin Y., “Capsule network performance on complex data,” 2017, pp. 1–7, arXiv:1712.03480.
- [61].Hollósi J. and Pozna C. R., “Improve the accuracy of neural networks using capsule layers,” in Proc. IEEE 18th Int. Symp. Comput. Intell. Inform., 2018, pp. 15–18. [Google Scholar]
- [62].Tartaglione E., Barbano C. A., Berzovini C., Calandri M., and Grangetto M., “Unveiling COVID-19 from chest X-ray with deep learning: A hurdles race with small data,” Int. J. Environ. Res. Public Health, vol. 17, no. 18, Sep. 2020, Art. no. 6933. [DOI] [PMC free article] [PubMed] [Google Scholar]