Significance
Biofilms are communities of surface-attached bacteria exhibiting biofilm-specific properties. Although anaerobic biofilms impact health, industry, and environment, they are mostly studied in aerobic bacterial species. Here, we studied biofilm formation in Bacteroides thetaiotaomicron, an anaerobic gut symbiont degrading diet sugars and contributing to gut maturation. Although B. thetaiotaomicron adhesion contributes to intestinal colonization, little is known about the determinants of its biofilm capacities. We identified that bile is a physiologically relevant gut signal inducing biofilm formation in B. thetaiotaomicron and other gut Bacteroidales. Moreover, we showed that, in contrast to the known scaffolding role of extracellular DNA, bile-dependent biofilm requires a DNase degrading matrix DNA, thus revealing a previously unrecognized factor contributing to the adhesion capacity of major gut symbionts.
Keywords: biofilm, Bacteroides thetaiotaomicron, DNase, gut microbiota
Abstract
Bacteroides thetaiotaomicron is a gut symbiont that inhabits the mucus layer and adheres to and metabolizes food particles, contributing to gut physiology and maturation. Although adhesion and biofilm formation could be key features for B. thetaiotaomicron stress resistance and gut colonization, little is known about the determinants of B. thetaiotaomicron biofilm formation. We previously showed that the B. thetaiotaomicron reference strain VPI-5482 is a poor in vitro biofilm former. Here, we demonstrated that bile, a gut-relevant environmental cue, triggers the formation of biofilm in many B. thetaiotaomicron isolates and common gut Bacteroidales species. We determined that bile-dependent biofilm formation involves the production of the DNase BT3563 or its homologs, degrading extracellular DNA (eDNA) in several B. thetaiotaomicron strains. Our study therefore shows that, although biofilm matrix eDNA provides a biofilm-promoting scaffold in many studied Firmicutes and Proteobacteria, BT3563-mediated eDNA degradation is required to form B. thetaiotaomicron biofilm in the presence of bile.
Bacteroides thetaiotaomicron, one of the most abundant bacterial gut symbionts, is involved in the degradation of complex polysaccharides and the maturation of the host immune system (1–3). B. thetaiotaomicron colonizes the gel-like mucus layer of intestinal epithelia cells (IECs) in both healthy and diseased conditions, and also adheres to food particles (4–6), suggesting that adhesion to host surfaces and the formation of bacterial biofilm aggregates could play an important biological role. Consistently, the transcriptional profile of B. thetaiotaomicron colonizing the mouse gut is more similar to B. thetaiotaomicron in vitro biofilm than to in vitro planktonic culture (7). Whereas the reference strain VPI-5482 only forms poor biofilms in vitro, several mutations leading to increased aggregation and biofilm capacity in vitro were recently identified, revealing the importance of capsular polysaccharides and type V pilus in mediating adhesion (8, 9). Moreover, polysaccharide utilization loci (PUL) have been proposed to mediate Bacteroides adhesion to substrates. PULs are surface structures involved in sugar degradation that are composed of at least one TonB-dependent receptor and one cell surface lipoprotein binding a glycan substrate (3, 10). Mucin-O-glycan–degrading PUL might mediate adhesion to mucin and therefore favor mucus colonization in the gut upon bile stimulation (3, 7). Although several gut environmental cues and compounds, including antibiotics, oxygen, metal concentration, mucins, immunoglobulin A, and bile (11–17), were shown to modulate bacterial biofilm capacity in various bacterial species, gut-relevant conditions promoting B. thetaiotaomicron biofilm formation are still poorly understood.
In the closely related Bacteroides fragilis species, both subinhibitory concentrations of the antibiotic enrofloxacin and bile were independently shown to increase in vitro biofilm formation, with consequences on B. fragilis physiology (15, 18). In this study, we showed that exposure to subinhibitory concentrations of bile extract induced biofilm formation in B. thetaiotaomicron strain VPI-5482 and in the majority of tested B. thetaiotaomicron clinical isolates and several common gut Bacteroidales species. Using transposon mutagenesis, we identified a B. thetaiotaomicron VPI-5482 operon, BT3560-3563, involved in bile-dependent biofilm formation. We showed that BT3563 is an extracellular DNase that is well-represented in B. thetaiotaomicron species and that it degrades extracellular DNA (eDNA) in biofilms formed in the presence of bile. Our study therefore shows that, in contrast to the biofilm-promoting role of bacterial extracellular DNA in many species (19), degradation of eDNA by BT3563 or by other extracellular nucleases may be a widespread requirement to achieve biofilm formation in the presence of bile, as shown with B. thetaiotaomicron.
Results
Bile Induces Biofilm Formation in B. thetaiotaomicron and Several Bacteroidales.
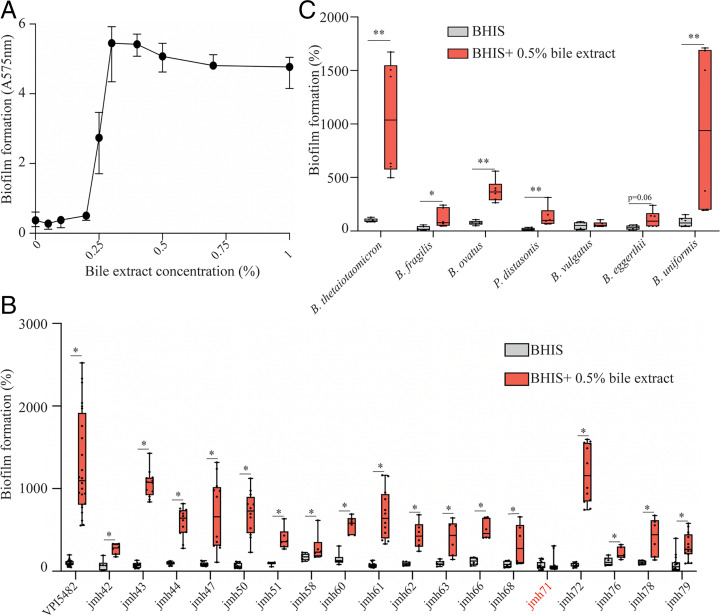
To identify gut-relevant conditions inducing biofilm formation in B. thetaiotaomicron VPI-5482, we supplemented brain heart infusion supplemented (BHIS) growth medium with various components of the normal intestinal environment, including sugars (0.5% D-glucose, D-mannose, D-rhamnose, D-cellobiose, and D-maltose), hemin (25 mg/L and 50 mg/L), mucin (0.1% and 0.5%), or a mix of bovine– ovine bile extract (0.1%, 0.5%, and 1%) (hereafter referred to as bile) for 24 h (SI Appendix, Fig. S1). We showed that addition of bile increased biofilm formation at 24 h without impacting viable cell count (minimal inhibitory concentration for bile is 4%) (SI Appendix, Figs. S1 and S2). This increase was maximal at 48 h in the presence of 0.3 to 1% bile (Fig. 1A). We then tested biofilm formation at 48 h in the presence of 0.5% bile in a collection of B. thetaiotaomicron clinical isolates (8) (SI Appendix, Table S1) and a representative strain of the most common gut Bacteroides and Parabacteroides species (Fig. 1 B and C). We showed that, except for B. thetaiotaomicron strain jmh71 and the tested Bacteroides vulgatus strain, all strains displayed an increased biofilm phenotype (Fig. 1 B and C), indicating that bile is a widespread inducer of biofilm formation among gut Bacteroidales species.
Fig. 1.
Bile extract promotes biofilm formation of several Bacteroidales isolates. (A) B. thetaiotaomicron VPI-5482 48-h biofilm formation as a function of bile extract concentration. Mean of six biological replicates, error bars represent SD. (B and C) A 96-well plate crystal violet 48-h biofilm assay. Mean of B. thetaiotaomicron VPI-5482 grown in BHIS is adjusted to 100%. Minimum-maximum (min-max) boxplot of 6 to 12 biological replicates for each strain. *P value <0.05, **P value <0.005, Mann–Whitney U test.
Exposure to Bile Induced a Major Transcriptional Shift but Did Not Reveal Biofilm Functions.
To understand the impact of bile on B. thetaiotaomicron physiology, we performed a RNA-sequencing (RNA-seq) analysis comparing B. thetaiotaomicron grown overnight in the presence or absence of 0.5% bile. We found that bile induces a widespread transcriptional shift, with 17% genes showing a significant change (Dataset S1A). In the presence of bile, out of 4,816 genes, 378 and 429 genes were significantly up- and down-regulated, respectively (Dataset S1 B and C). About half of these genes could be assigned a functional Clusters of Orthologous Groups (COG) category, spanning almost all COG categories (Dataset S1D). The 5 genes showing the highest fold change (>50) are organized in two operons, BT2793-2795, encoding a bile-specific tripartite multidrug efflux system , and BT0691-0692, predicted to encode an outer membrane protein and a calcineurin superfamily phosphohydrolase, respectively. Consistently, both of these loci were previously shown to be important for fitness under bile salt stress (20) and are probably involved in bile tolerance. Although we did not observe any induction of genes encoding capsular polysaccharides (21, 22) or type V pilin homologs (23), that were previously shown to contribute to B. thetaiotaomicron biofilm formation (8, 9), several PUL involved in mucin-O-glycan and host-derived sugar degradation (3) that could contribute to adhesion to the mucus layer were up-regulated, suggesting that bile exposure might increase B. thetaiotaomicron adhesion capacity also in vivo. These results showed that exposure to bile leads to important physiological adaptations in B. thetaiotaomicron that might increase its colonization capacity. This transcriptomic approach did not, however, reveal any regulation of genes encoding known B. thetaiotaomicron determinants of biofilm formation in vitro.
Identification of an Operon Involved in Bile-Dependent Biofilm Formation.
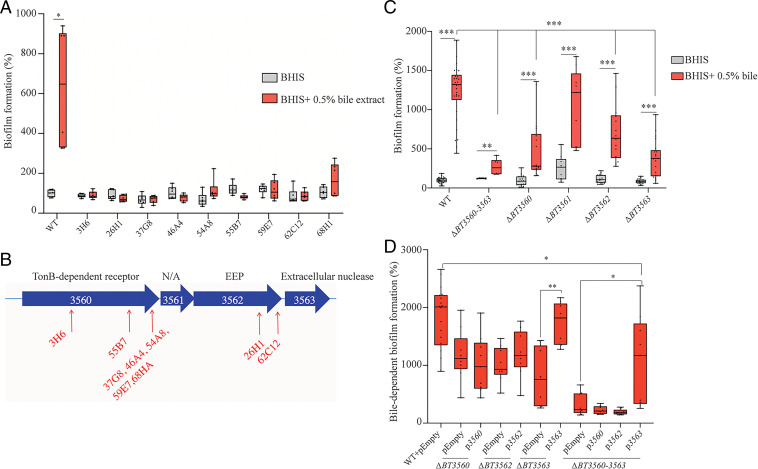
To identify the genetic determinants involved in bile-dependent biofilm formation, we performed a random transposon mutagenesis in B. thetaiotaomicron VPI-5482 and screened for mutants failing to produce biofilm in the presence of 0.5% bile. We identified 9 biofilm-negative transposon mutants out of 6,510 mutants screened (Fig. 2A), all corresponding to transposon insertions in a potential operon (SI Appendix, Fig. S3) encoding a TonB-dependent receptor (BT3560, 7 insertions), a hypothetical protein (BT3561), and two putative membrane-associated lipoproteins with nuclease activity (BT3562, 2 insertions and BT3563) (Fig. 2B). Whereas deletion of BT3561 had no impact on biofilm formation, the deletion of BT3560, BT3562, BT3563, or of the whole BT3560-3563 region significantly reduced bile-dependent biofilm formation compared to WT without an associated growth defect (Fig. 2C and SI Appendix, Fig. S4). Since crystal violet stains both cells and extracellular matrix (ECM), we resuspended biofilms and numbered colony forming units (CFUs) and showed that ΔBT3560-3563 reduction in biofilm formation was due to a reduction of cell number and not ECM production (SI Appendix, Fig. S5). Complementation of ΔBT3563 and ΔBT3560-3563 mutants by pBT3563, a plasmid constitutively expressing BT3563, restored bile-dependent biofilm formation (Fig. 2D), whereas ΔBT3560 or ΔBT3562 strains were not complemented upon addition of pBT3560 or pBT3562, suggesting that deletion of BT3560 and BT3562 might have a polar effect on the downstream BT3563 gene (Fig. 2D). However, since a ΔBT3560-3563 mutant formed slightly but reproducibly less biofilm than a ΔBT3563 simple mutant, BT3560 and BT3562 might also contribute to biofilm formation in the presence of bile. Taken together, these results showed that BT3563 is necessary for proper bile-dependent biofilm formation of B. thetaiotaomicron VPI-5482.
Fig. 2.
BT3560-3563 is required for bile-dependent biofilm formation. (A and C) A 96-well plate crystal violet biofilm assay after 48-h growth in BHIS or BHIS + 0.5% bile extract. (B) Schema of the BT3560-3563 genetic locus. Red arrows indicate transposon insertion points. N/A, not annotated; EEP, exonuclease/endonuclease/phosphatase family. (D) A 96-well plate crystal violet biofilm assay after 48-h growth in BHIS + 0.5% bile extract. WT in BHIS is not represented. (A, C, and D) Mean of WT in BHIS is adjusted to 100%. Min-max boxplot of 6 to 18 biological replicates for each strain, each replicate is the mean of two technical replicates. *P value <0.05, **P value <0.005, ***P value <0.0005, Mann–Whitney U test.
BT3563 is an Extracellular DNase Degrading eDNA in Biofilms Formed in the Presence of Bile.
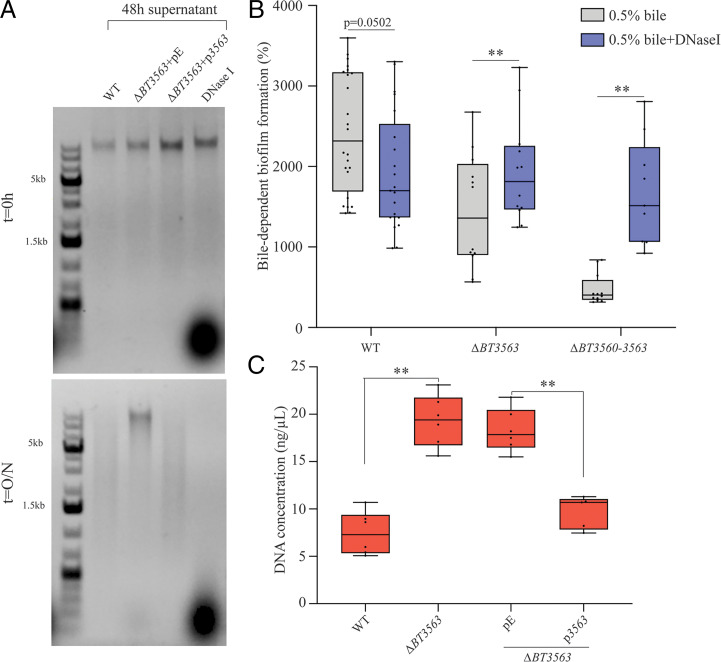
To investigate the potential nuclease activity of BT3563, we exposed genomic DNA to B. thetaiotaomicron 24-h and 48-h bile-free culture supernatant. We showed that DNA was degraded after an overnight incubation with a wild-type (WT) supernatant. This degradation was abolished when we used a ΔBT3563 strain carrying the empty vector (Fig. 3A and SI Appendix, Fig. S6). Complementation with pBT3563 plasmid, constitutively expressing BT3563, restored DNA degradation, confirming the DNase activity of BT3563 (Fig. 3A). To confirm the contribution of BT3563 extracellular DNase to bile-dependent biofilm formation, we complemented the absence of BT3563 in ΔBT3560-3563 and ΔBT3563 strains by adding purified DNase I to the growth medium. We observed an increase of bile-dependent biofilm formation in ΔBT3560-3563 and ΔBT3563 strains in the presence of DNase I, but a decrease in WT biofilm formation (Fig. 3B). This prompted us to measure the concentration of eDNA in the ECM of B. thetaiotaomicron biofilms formed in the presence of bile. We showed that, whereas deletion of BT3563 did not significantly impact ECM protein concentration (SI Appendix, Fig. S7), it increased eDNA concentration compared to WT. Upon addition of pBT3563 in trans, the eDNA concentration was reduced back to WT level, which is consistent with BT3563-dependent degradation of DNA in the ECM (Fig. 3C). Addition of bile was not necessary to observe BT3563-mediated DNA degradation (Fig. 3A), and our transcriptomic analysis showed no increase of BT3563 transcription in the presence of bile (Dataset S1A), suggesting that bile might not directly impact BT3563 production or activity. We then assessed the impact of bile on eDNA concentration itself and observed an increase of eDNA concentration in B. thetaiotaomicron supernatant of cultures grown in the presence of bile, indicating that bile could increase eDNA release (SI Appendix, Fig. S8). These results showed that BT3563 is an extracellular DNase that degrades eDNA in the biofilm matrix and that bile exposure increases the release of eDNA, which might be one of the mechanisms by which bile exposure favors biofilm formation.
Fig. 3.
BT3563 degrades nucleic acids in the extracellular matrix of bile-dependent biofilms. (A) Incubation of B. thetaiotaomicron genomic DNA (23 ng/µL final concentration) with the supernatant of 48-h bile-free cultures, as indicated, and loaded immediately on a 1% agarose gel (t = 0 h) and after an overnight incubation at 37 °C (t = O/N). A control using DNase I rather than supernatant is shown in the last lane. pE, empty vector; p3563, vector constitutively expressing BT3563. (B) A 96-well plate crystal violet biofilm assay after 48-h growth in BHIS + 0.5% bile extract, with or without DNase I. Mean of WT in BHIS is adjusted to 100% (not represented). Min-max boxplot of 9 to 21 biological replicates for each strain, each replicate is the mean of two technical replicates. **P value <0.005, Wilcoxon matched-pairs signed rank test. (C) Extracellular DNA concentration (ng/µL) in the purified extracellular matrix of bile-dependent biofilms grown in BHIS + 0.5% bile extract for 48 h. Min-max boxplot of 6 biological replicates for each strain. **P value <0.005, Mann–Whitney U test. pE, empty vector; p3563, vector constitutively expressing BT3563.
Extracellular DNase Activity Is Necessary to Observe Maximum Bile-Dependent Biofilm Formation.
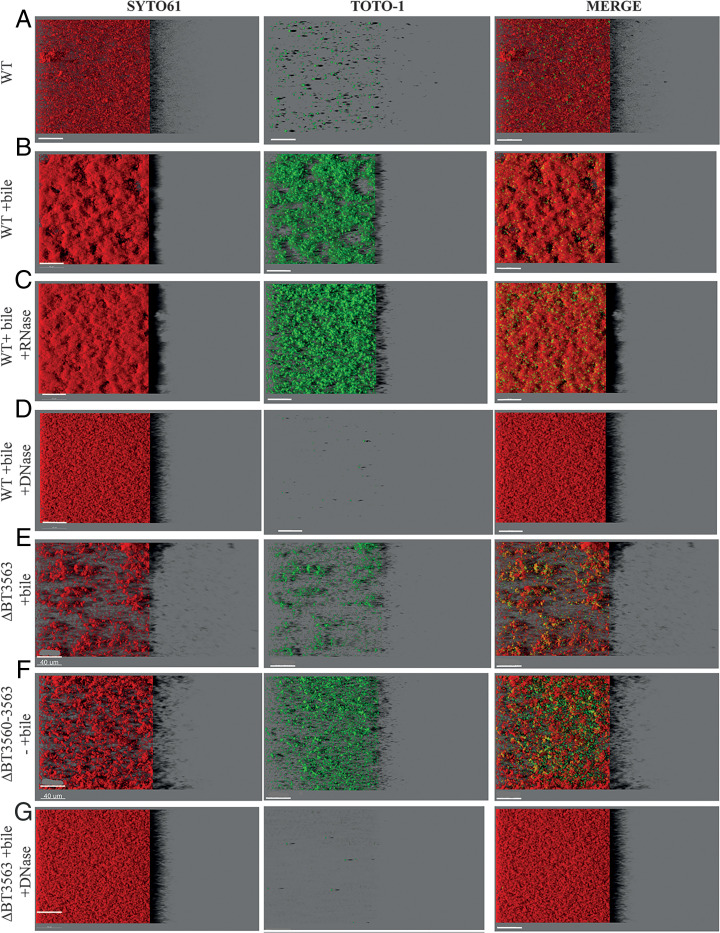
To analyze the distribution and abundance of eDNA in the ECM of B. thetaiotaomicron bile-dependent biofilms, we labeled eDNA with TOTO-1 dye and imaged 48-h 96-well plate biofilms by confocal laser scanning microscopy (CLSM). For each condition, a representative well is shown in Fig. 4; the five others are shown in SI Appendix, Fig. S9; a three-dimensional (3D) reconstruction of the biofilm for one well is shown in SI Appendix, Fig. S10 and quantification of TOTO-1 and SYTO61 fluorescence is shown in SI Appendix, Fig. S11. In the presence of bile, B. thetaiotaomicron biofilm was expectedly denser and more structured than in the absence of bile (Fig. 4 A and B and SI Appendix, Figs. S9 and S10). Extracellular nucleic acids were 20 times more abundant in the presence of bile, when correcting for cell abundance (SI Appendix, Fig. S11C) and they were distributed diffusely in the entire biofilm, with spots potentially corresponding to dead cells (Fig. 4 A and B and SI Appendix, Figs. S9 and S10). To determine the nature of these nucleic acids, we grew B. thetaiotaomicron biofilms in the presence of bile and DNase I or RNase I. Whereas RNase I treatment had little impact on B. thetaiotaomicron biofilms (Fig. 4C and SI Appendix, Figs. S9–S11), addition of DNase I decreased by 20-fold the amount of extracellular nucleic acids detected (Fig. 4D and SI Appendix, Fig. S11C), close to the levels observed for biofilms formed in the absence of bile. This confirms that DNA was the most abundant nucleic acid in the ECM upon bile induction. Addition of DNase I also led to a loss of B. thetaiotaomicron biofilm 3D structure, suggesting eDNA might be an important structuring component of the ECM (Fig. 4D and SI Appendix, Figs. S9 and S10). We then showed that, whereas the ΔBT3563 mutant formed scattered biofilms in the presence of bile, with dispersed or small clumps of cells (Fig. 4 E and F and SI Appendix, Figs. S9 and S10), addition of DNase I during growth of ΔBT3563 restored a WT homogenous and dense biofilm phenotype (Fig. 4 D and G and SI Appendix, Figs. S9 and S10). One hypothesis would be that DNase activity might be required to prevent repulsion between clumps of cells, or between cells and some components of the extracellular matrix, allowing the formation of denser structures. By contrast to eDNA quantification in the extracted ECM, we saw no increase in TOTO-1 labeling of ΔBT3563 or ΔBT3560-3563 mutants compared to WT in the presence of bile (SI Appendix, Fig. S11C), which could be due to a lack of sensitivity of eDNA quantification by CLSM. These results demonstrated that DNA, rather than RNA, contributes to biofilm structure in the presence of bile and that the presence of either BT3563 or the addition in culture of DNase I lead to the formation of denser bacterial structures, allowing maximum biofilm formation.
Fig. 4.
(A–G) Visualization of B. thetaiotaomicron biofilms. IMARIS easy 3D projections from CLSM images of B. thetaiotaomicron biofilms grown in plates in the absence or presence of 0.5% bile extract (“+bile”), RNase I (“+RNase”), or DNase I (“+DNase”). Cells are labeled with SYTO61 dye (Left, in red). eDNA and dead cells are labeled with TOTO-1 (Middle, in green). The merged image is shown on the Right. For each image, the virtual shadow projection of the biofilm is shown on the Right. (Scale bars, 40 µm.)
BT3563 Homologs Are Involved in Bile-Dependent Biofilm Formation of B. thetaiotaomicron Clinical Isolates.
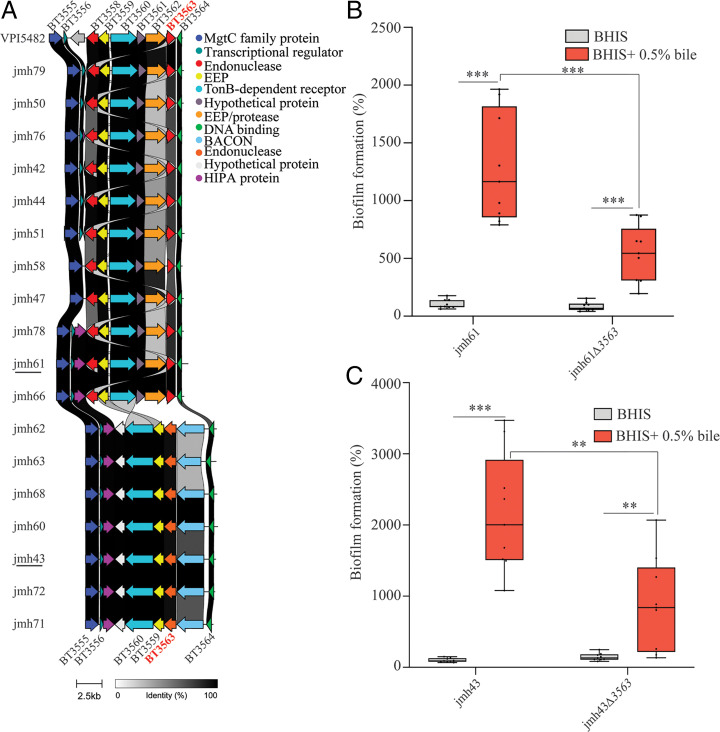
To further investigate the correlation between BT3563 and bile-dependent biofilm, we sequenced the whole genome of the 18 B. thetaiotaomicron jmh isolates (24). We found a BT3563 homolog in all tested strains, even in B. thetaiotaomicron jmh71, the only strain that did not form biofilms in the presence of bile (SI Appendix, Table S2), suggesting jmh71 could be lacking additional factors needed for bile-dependent biofilm formation apart from BT3563. In 11 out of the 18 jmh strains (jmh42, 44, 47, 50, 51, 58, 61, 66, 76, 78, and 79), the homology with VPI-5482 BT3563 exceeded 80% homology (SI Appendix, Table S2) and the genomic region around the detected BT3563 homolog gene was similar to that of VPI-5482 and included homologs of genes BT3558 to BT3564 (as depicted in Fig. 5A). In the 7 other strains (jmh43, 60, 62, 63, 68, 71, and 72), the homology with VPI-5482 BT3563 was weaker (34% identity, SI Appendix, Table S2), and the genetic environment of their BT3563 homologs differed, including a homolog of BT3559, BT3560, BT3564, as well as a gene coding for a protein carrying a Bacteroides-associated carbohydrate binding often N-terminal (BACON) domain Fig. 5A. We also showed that Bacteroides ovatus and Bacteroides uniformis are the only Bacteroidales strains tested with a BT3555-BT3564 region similar to B. thetaiotaomicron VPI-5482, including a homolog of BT3560, BT3562, and BT3563 (SI Appendix, Fig. S12). In the other strains tested, this region is either completely missing (Parabacteroides distasonis) or present without a BT3563 homolog (B. fragilis, B. vulgatus, Bacteroides eggerthii) (SI Appendix, Fig. S12). A more comprehensive search revealed that BT3563 homologs were present in many diverse Bacteroidales, and at least in one example of Flavobacterium columnare and Salinivirga cyanobacteriivorans, non-Bacteroidales Bacteroidetes. However, the synteny of the BT3560-3564 region was conserved only in strains from the Bacteroides, Prevotella, Alloprevotella, Porphyromonas, and Alistipes genera (SI Appendix, Fig. S13). We tested the contribution of BT3563 homologs to B. thetaiotaomicron bile-dependent biofilm formation in two randomly selected strains, jmh61 and jmh43. In these strains, the BT3563 proteins share, respectively, 87% and 34% of protein identity with VPI-5482 BT3563, and a different genetic organization around the BT3563 homolog is observed (SI Appendix, Table S2 and Fig. 5A). We showed that deletion of the BT3563 homolog in both jmh61 and jmh43 led to a reduction of bile-dependent biofilm formation (Fig. 5 B and C), indicating that the contribution of VPI-5482 BT3563 extracellular DNase to bile-dependent biofilm formation is a widespread property of B. thetaiotaomicron species.
Fig. 5.
BT3563 homologs are involved in bile-dependent biofilm formation of B. thetaiotaomicron clinical isolates. (A) Comparison of the genetic organization around BT3563 homologs in B. thetaiotaomicron isolates. EEP, endonuclease/exonuclease/phosphatase superfamily; BACON, Bacteroides-associated carbohydrate binding often N-terminal; HIPA, serine/threonine-protein kinase toxin HipA. BT3563 homologs sharing more than 80% sequence identity with VPI-5482 BT3563 are shown in red (for jmh42, 44, 47, 50, 51, 58, 61, 66, 76, 78, and 79) and BT3563 homologs sharing 34% sequence identity with VPI-5482 BT3563 are shown in orange (for jmh43, 60, 62, 63, 68, 71, and 72). (B and C) A 96-well plate crystal violet biofilm assay after 48-h growth in BHIS or BHIS + 0.5% bile extract. Mean of jmh61 or jmh43 in BHIS is adjusted to 100%. Min-max boxplot of nine biological replicates for each strain; each replicate is the mean of two technical replicates. **P value <0.005, ***P value <0.0005, Mann–Whitney U test.
Discussion
In this report, we showed that physiological concentrations of bile extract induce the formation of biofilm in almost all tested B. thetaiotaomicron strains, as well as in representative Bacteroidales species. This widespread induction by a compound exclusive to the intestinal environment suggests that biofilm formation might be an important aspect of Bacteroidales biology in the gut. Bile concentration in the gut can range between 0.2 and 2% (25), which is consistent with the concentrations required to induce maximum B. thetaiotaomicron biofilm formation (0.3 to 1%). However, the concentration of intestinal bile is highly variable, depending on the individual, time of the day, diet, and the precise localization within the gut. Moreover, bile composition changes along the intestine and also depends on environmental parameters such as diet and gut microbiota composition. Thus, we can expect that the bile concentration and composition necessary to induce biofilm formation are only reached at certain times or in specific locations in the gut.
Although most studies have been performed with pathogenic bacteria, rather than with symbiotic members of the community, bile was previously shown to induce biofilm formation of at least one representative of all four major bacterial phylum of the gut microbiota: Firmicutes (Clostridioides difficile and Listeria monocytogenes), Bacteroidetes (B. fragilis), Proteobacteria (Vibrio cholerae), and Actinobacteria (Bifidobacterium) (15, 17, 26–28), suggesting that bile is an important gut signal for biofilm formation. Consistently, we observed that bile extract induced biofilm formation in almost all tested B. thetaiotaomicron strains, as well as in representative strains of most of the common gut Bacteroidales species. Depending on the model considered in each of these studies, the concentration, incubation time, and type of bile component necessary to trigger biofilm formation varied greatly. Bile-dependent biofilm formation also involved different ECM components and different determinants. This suggests that although bile components might be a conserved signal for biofilm formation, the mechanism of biofilm induction differs. Bile could act as a signal triggering biofilm formation to stably colonize the gut. Alternatively, biofilm formation could be a conserved bacterial protection mechanism against the cytotoxic effects of bile, such as lipid membranes and DNA damage and protein denaturation (29). Indeed, bacteria within biofilms are known to be more tolerant to various antimicrobials, including bile acids, than planktonic bacteria, and bile-dependent biofilm formation has been shown to increase tolerance to bile-mediated killing in different species, including B. fragilis (15, 17, 26). Although, the production of a bile salt hydrolase (BSH) (30) and a tripartite multidrug resistance efflux pump, BT2793-2795 , confer a high resistance to bile to B. thetaiotaomicron, bile-induced biofilm formation in the gut probably constitutes an additional strategy by which gut bacteria protect themselves from bile toxicity (31). Exposure of B. thetaiotaomicron to bile also induces various stress responses and increases the production of efflux pumps (25), suggesting the activation of stress tolerance mechanisms that could provide cross-protection against other damaging agents, such as antibiotics (15, 17). Interestingly, in Klebsiella pneumoniae, an absence of poly-N-acetylglucosamine production significantly reduced bile-dependent biofilm formation in vitro and also impaired its ability to colonize the mouse gut, showing that bile-dependent biofilm formation could impact colonization efficiency (32). Moreover, bile-mediated biofilm formation might be an important aspect of gut microbiota competition and cooperation. Bile is a complex mix of cholesterol, different bile acids, and proteins. Some studies tested purified bile acids, rather than bile extract, and showed that all bacteria did not induce biofilm in response to the same bile acid. For instance, C. difficile reacted to chenodeoxycholate and deoxycholate (17), Bacteroides breve to conjugated bile acids (taurocholate, glycocholate, taurodeoxycholate, and glycodeoxycholate) (33), and Lactobacillus to taurocholate (34). Conversely, taurine-conjugated bile acids disperse biofilms of V. cholerae or Pseudomonas aeruginosa (35, 36). Interestingly, members of the gut microbiota can transform bile acids, with potential consequences on biofilm formation. In particular, several Clostridia can transform primary bile acids, such as cholate, into secondary bile acids, such as deoxycholate, by 7α-dehydroxylation, and various bacteria possess a BSH enzyme that can remove the conjugated taurine or glycine residue of specific bile acids (37). These bile modifications by members of the gut microbiota render bile acids less soluble and more cytotoxic, but they could also impact gut microbiota physiology (38, 39) and the biofilm formation capacity of other bacteria, and therefore their colonization efficiency. Consistently, Clostridium scindens was shown to increase C. difficile biofilm formation in vitro by converting cholate to deoxycholate (17).
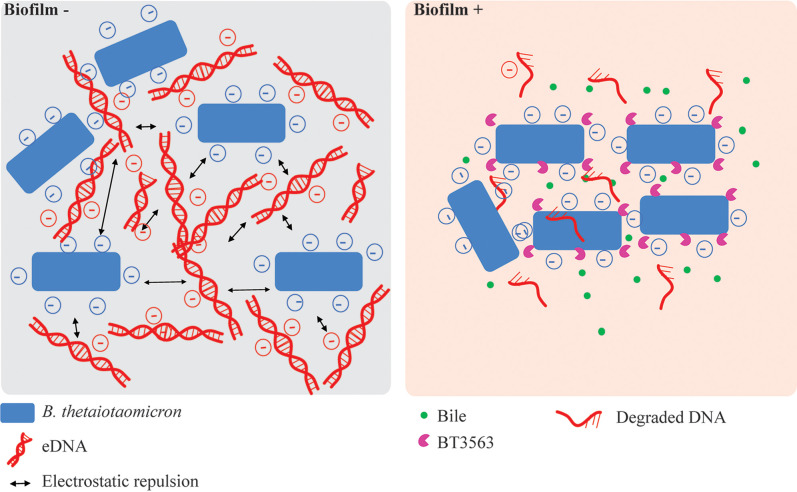
Our results indicate that the simultaneous presence of bile, eDNA, and the extracellular DNase BT3563 are required to observe dense, structured B. thetaiotaomicron biofilms. Presence of bile in the growth medium led to important physiological adaptations, as 17% of B. thetaiotaomicron genes were differentially expressed, but the precise contribution of bile to biofilm formation is still unclear. Although the mechanism of bile-dependent biofilm formation has not been investigated for all bacteria, in some cases, bile was shown to increase the production of exopolysaccharide (32, 40, 41), cyclic-di-GMP (42), and various adhesins such as autotransporters (43) and curli (44). However, very little is known about B. thetaiotaomicron biofilm determinants. Capsular polysaccharide 8 and type V pili production were previously shown to increase B. thetaiotaomicron adhesion capacity, but these genes were not induced in the presence of bile. However, B. thetaiotaomicron mucin-O-glycan–degrading PUL were up-regulated in biofilms grown in chemostats compared to planktonic cultures and in the mucus compared to the lumen (7, 45), and they were also induced in the presence of bile, suggesting bile exposure might prime bacteria for adhesion and colonization to the mucus layer in vivo. Bile also increased the release of eDNA, which could modulate biofilm formation. eDNA is a well-known component of the biofilm ECM and numerous studies have shown that addition of DNase to the growth media prevented biofilm formation or dispersed preformed biofilm in both gram-positive and gram-negative bacteria (19). Consistently, addition of DNase I led to a small reduction in the bile-dependent biofilm formation of B. thetaiotaomicron, suggesting that eDNA is an important ECM component (Fig. 3B). However, unexpectedly, we show that the extracellular DNase BT3563 is also necessary for bile-dependent biofilm formation, suggesting that partial degradation of eDNA might be necessary for biofilm formation in the presence of bile. Neither BT3563 transcription nor BT3563 DNase activity was increased by bile, so the link between bile exposure and BT3563 function remains unclear. BT3563 carries a lipoprotein signal commonly found in surface-exposed outer membrane proteins in Bacteroides. Consistently, BT3563 has been detected in the B. thetaiotaomicron outer membrane and was enriched in outer-membrane vesicles (OMVs) (46), suggesting BT3563 might be secreted to the supernatant through OMV production. We hypothesize that cell- or OMV-associated BT3563 could degrade eDNA during biofilm formation, maintaining a specific eDNA concentration or organization within the ECM to form structured bile-dependent biofilms. Deletion of BT3563 led to the formation of scattered biofilms in the presence of bile, composed of clumps of cells rather than a dense layer at the bottom of the well. To allow the formation of denser structures, BT3563 might be required to remove eDNA from cell vicinity and prevent electrostatic repulsion between the negatively charged eDNA and bacterial surface or some component of the ECM such as exopolysaccharides (Fig. 6). For instance, the production of positively charged proteins was necessary to prevent electrostatic repulsion between eDNA and Staphylococcus aureus cells in biofilms, and perturbation of these proteins led to an increasingly porous biofilm, reminiscent of our ΔBT3563 biofilms (47, 48). It is also to be noted that a ΔBT3560-3563 mutant formed less bile-dependent biofilm than a ΔBT3563 simple mutant, suggesting the TonB-dependent receptor BT3560 and/or the nuclease BT3562 could also contribute to biofilm formation. Moreover, ΔBT3560-3563 still formed biofilm in the presence of bile, demonstrating that additional factors could be involved.
Fig. 6.
Model of DNase-mediated biofilm formation. In the absence of BT3563, the electrostatic repulsion between B. thetaiotaomicron cells and eDNA prevents close association of cells and biofilm formation. In presence of bile and BT3563, degradation of eDNA surrounding the bacterial cells by BT3563 allows the formation of tight aggregates of cells and biofilm formation.
Our study therefore demonstrates that gut-relevant environmental cues such as bile can strongly stimulate B. thetaiotaomicron biofilm formation. This induction relied on the production of an extracellular DNase, BT3563, that is present in all B. thetaiotaomicron isolates, and in many diverse Bacteroidetes. Although our study focused exclusively on in vitro experiments and will therefore need to be confirmed in animal models, our results suggest that, in addition to the well-established structural role of eDNA in the biofilm matrix, partial eDNA degradation might also represent an important aspect of biofilm formation in B. thetaiotaomicron species, and possibly in other Bacteroidales.
Materials and Methods
Bacterial Strains and Growth Conditions.
Bacterial strains used in this study are listed in SI Appendix, Table S3. B. thetaiotaomicron and other Bacteroidales strains were grown in BHIS broth supplemented with erythromycin 15 µg/mL (erm), tetracycline 2.5 µg/mL (tet), gentamycin 200 µg/mL (genta), 5′-fluoro-2′-deoxyruidin 200 µg/mL (FdUR), anhydrotetracycline (0.1 µg/mL), D-glucose, D-mannose, D-rhamnose, D-cellobiose, D-maltose (0.5% wt/vol), hemin from bovine (25 mg/L and 50 mg/L), porcine mucin extract (0.1% and 0.5% wt/vol), bile extract from bovine and ovine (Sigma, B8381) at 0.5% unless indicated otherwise, DNase I (Thermo Scientific, VF304452) 98 U/mL, or RNase1 (Thermo Scientific, EN0601) 0.06 U/mL when required. Cultures were incubated at 37 °C in anaerobic conditions using jars with anaerobic atmosphere generators (GENbag anaero, Biomerieux, No. 45534) or in a C400M Ruskinn anaerobic-microaerophilic station. Escherichia coli S17λpir was grown in Miller’s lysogeny broth (LB) (Corning) supplemented with ampicillin (100 µg/mL) when required and incubated at 37 °C with 180 rpm shaking. Cultures on solid media were done in BHIS with 1.5% agar, and antibiotics were added when needed. Bacteria were always streaked from glycerol stock on BHIS-agar before being grown in liquid cultures. All media and chemicals were purchased from Sigma-Aldrich unless indicated otherwise.
All experiments and genetic constructions of B. thetaiotaomicron were made in VPI-5482Δtdk background (49) unless indicated otherwise, which was developed for a two-step selection procedure of unmarked gene deletion by allelic exchange, as previously described. Therefore, the VPI-5482Δtdk is referred to as wild type or VPI-5482 in this study.
Genome Analysis.
Genomic DNA of clinical strains was prepared from overnight cultures using the DNeasy blood and tissue kit (Qiagen). Illumina whole-genome sequencing was performed by the Plateforme de Microbiologie Mutualisée (P2M) of Institut Pasteur and the genomes were assembled using SPAdes v3.13.0 (50) and when necessary were reassembled using Unicycler (51). The obtained genomes were annotated using RASTtk (52, 53) on the patricbrc.org database (54) and we searched BT3563 nucleic acid and amino acid sequence in these genomes using the BLASTp tool of patricbrc.org (55, 56). All of these genomes were made available publicly on the patricbrc.org database (see SI Appendix, Table S1 for accession Nos.). Moreover, this whole-genome shotgun project has been deposited at DNA Data Bank of Japan (DDBJ)/European Nucleotide Archive (ENA)/GenBank under the Bioproject accession PRJNA725886 (24). The accession Nos. for each strain are shown in SI Appendix, Table S1. Sequences of the region surrounding BT3563 homologs were extracted and the gene cluster comparison was made using the clinker and clustermap.js pipeline (57). Synteny analysis was performed using either the web-based tool Synttax (58) searching for homologs of BT3563 using a custom selection of diverse Bacteroidetes or the genome browser tool of the MicroScope plateforme (59) looking for B. thetaiotaomicron BT3555-3564 region homology in the seven Bacteroidales strains tested.
A 96-Well Crystal Violet Biofilm Formation Assay.
Overnight culture was diluted to OD600 = 0.05 in 100 µL BHIS and inoculated in technical duplicates in polystyrene Greiner round-bottom 96-well plates. The wells at the border of the plates were filled with 200 µL of water to prevent evaporation. Incubation was done at 37 °C in anaerobic conditions for 48 h. The supernatant was removed by careful pipetting and the biofilms were fixed using 100 µL of Bouin’s solution (picric acid 0.9%, formaldehyde 9%, and acetic acid 5%; HT10132, Sigma-Aldrich) for 10 min. Then the wells were washed once with water by immersion and flicking, and the biofilm was stained with 125 µL 1% crystal violet (V5265, Sigma-Aldrich) for 10 min. Crystal violet solution was removed by flicking and biofilms were washed twice with water. Stained biofilms were resuspended in 1:4 acetone:ethanol mix and absorbance at 575 nm was measured using a TECAN Infinite M200 PRO plate reader.
Targeted Mutagenesis.
A list of all the primers used in this study can be found in SI Appendix, Table S4. Deletion mutants in B. thetaiotaomicron VPI 5482 were constructed using the previously described vector for allelic exchange pExchange-tdk (49). Deletion mutants in B. thetaiotaomicron clinical isolates were constructed using the recently described suicide vector pLGB13 for allelic exchange in Bacteroides natural isolates (Addgene 126618) (60). Briefly, 1-kb regions upstream and downstream of the target sequence and the vector (pExchange-tdk or pLGB13) were amplified by PCR using Phusion Flash High-Fidelity PCR Master Mix (Thermo Fisher Scientific, F548). All three fragments were ligated using Gibson assembly: the inserts and the plasmids were mixed with Gibson master mix 2× (100 µL 5× ISO buffer, 0.2 µL 10,000 U/mL T5 exonuclease (NEB No. M0363S), 6.25 µL 2,000 U/mL Phusion HF polymerase (NEB No. M0530S), 50 µL 40,000 U/mL Taq DNA ligase (NEB No. M0208S), 87 µL dH2O for 24 reactions) and incubated at 50 °C for 35 min. The resulting mix was transformed in E. coli S17λpir that was used to deliver the vector to B. thetaiotaomicron by conjugation. Conjugation was carried out by mixing exponentially grown cultures (OD600 = 0.6) of the donor and the recipient strain in a 2:1 ratio. The mixture was spotted on BHIS-agar plates and incubated at 37 °C in aerobic conditions overnight. The mix was then streaked on BHIS agar supplemented with antibiotic—for selection of B. thetaiotaomicron transconjugants that had undergone the first recombination event—and gentamicin to ensure exclusion of any E. coli growth. Eight of the resulting colonies were grown overnight in BHIS with no antibiotic to allow a second recombination event, and the culture was plated on BHIS-agar plates supplemented with either FdUR (to counterselect pEchange-tdk) or anhydrotetracycline (to counterselect pLGB13) to select for loss of plasmid. The resulting deletion mutants were confirmed by PCR and sequencing.
We used the pNBU2-bla-erm vector (61) or pNBU2-bla-tet (9) vector for complementation, which inserts in the 5′ untranslated region of the tRNA-Ser, in which we previously cloned the constitutive promoter of BT1311 encoding the sigma factor RpoD (8). Target genes were amplified by PCR using Phusion Flash High-Fidelity PCR Master Mix from start codon to stop codon and they were cloned after BT1311 promoter by Gibson assembly. The Gibson mix was transformed in E. coli S17λpir and the resulting E. coli was used to transfer the plasmid to B. thetaiotaomicron by conjugation (see above).
Random Transposon Mutagenesis.
pSAMbt, the previously published tool for random mariner-based transposon mutagenesis in B. thetaiotaomicron (62) was conjugated in B. thetaiotaomicron as described above. After streaking on BHIS-erm-genta agar plates, isolated colonies were resuspended in 100 µL BHIS in 96-well plates, grown overnight, and tested for biofilm formation as described above. The selected clones were then streaked on a fresh BHIS-erm-genta agar plate and three isolated colonies were tested for biofilm formation to ensure no mix of transposon mutants had occurred during preparation of the library. The genomic DNA of the validated clones was extracted using the DNeasy blood and tissue kit (Qiagen) and sent for whole-genome sequencing at the Mutualized Platform for Microbiology of Institut Pasteur.
Growth Curve.
A total of 2.5 µL of overnight cultures was added to 200 µL BHIS that had previously been incubated in anaerobic condition overnight to remove dissolved oxygen, in Greiner flat-bottom 96-well plates. A plastic adhesive film (adhesive sealing sheet; Thermo Scientific, AB0558) was added on top of the plate inside the anaerobic station, and the plates were then incubated in a TECAN Infinite M200 Pro spectrophotometer for 24 h at 37 °C. OD600 was measured every 30 min, after a 900-s orbital shaking of 2 mm amplitude.
Viable Cells Count.
Overnight cultures were diluted to OD600 = 0.5 in a final volume of 1 mL of NaCl 0.85%. Aliquots were centrifugated at 6,000 × g (7,000 rpm) for 7 min and the supernatant was discarded. After washing twice with NaCl 0.85%, 2 µL of propidium iodide was added and the samples were incubated in anaerobic condition and protected from light at 37 °C, for 30 min. The number of viable bacteria was then quantified using a MACSQuant VYB flow cytometer and MACSQuantify Software V2.11.
Extracellular Matrix Extraction and Quantification.
ECMs were extracted based on previously described methods (63). A total of 2 mL of 48-h biofilm cells grown in the presence of bile were harvested and centrifugated at 5,000 × g for 10 min. The pellet was washed twice with NaCl 0.85% and weighed, then resuspended in an extraction buffer (Tris⋅HCl pH 8.0; 1.5 M NaCl) at a 1:10 mass-volume ratio and incubated at 25 °C for 30 min with agitation. Then, cells were removed by centrifugation at 15,000 × g and 25 °C for 10 min and the supernatant containing the extracted ECMs was stored at −20 °C until use. The amount of DNA, and proteins in the ECM were measured using a Qubit 3.0 fluorometer (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Nuclease Activity of the Supernatant.
Overnight or 48-h cultures were centrifugated for 6.5 min at 6,000 × g. When applicable, we measured eDNA concentration using the QuBit HS double-stranded DNA kit (Invitrogen). A total of 50 µL of supernatant was mixed with B. thetaiotaomicron VPI-5482 genomic DNA. 10 µL was used to run on a 1% agarose gel and colored with ethidium bromide. The remaining 40 µL was incubated at 37 °C overnight for approximatively 18 h, and then 10 µL was used to run a 1% agarose gel and colored with ethidium bromide. DNase I was used as a positive control of DNase activity.
CLSM.
For confocal laser scanning microscopy, biofilms were grown in 96-well plates (µclear, Greiner Bio-One). A total of 100 µL of BHIS, supplemented with 0.5% bile extract, DNase I (Thermo Scientific, VF304452) 98 U/mL, or RNase1 (Thermo Scientific, EN0601) 0.06 U/mL when required, was added to each well and the plates were incubated at 37 °C, in static condition 48 h under anaerobic conditions. The unwashed biofilms were then directly stained in red with 20 μM of SYTO61 (Life Technologies; cell permeant nucleic acid dye to contrast all the bacteria) and in green with 0.4 µM of TOTO-1 (Thermo Scientific; cell impermeant DNA dye to contrast eDNA). After 15 min of incubation, Z stacks of horizontal plane images were acquired in 1-μm steps using CLSM (Leica TCS SP8, INRAE MIMA2 microscopy platform) with a water 63× immersion lens (numerical aperture [NA] = 1.2). Two stacks of images were acquired randomly on three independent samples at 800 Hz. Fluorophores were excited and emissions were captured as prescribed by the manufacturer. Simulated 3D fluorescence projections were generated using IMARIS 9.3 software (Bitplane).
RNA-Seq Analysis.
Overnight cultures were mixed with RNAprotect (Qiagen) to prevent RNA degradation, and the pellet was kept at −80 °C. Total RNA was extracted using MP Biomedicals FastRNA Pro Blue kit by the provider’s manual and treated with an Ambion Turbo DNA-free kit to remove possible DNA contamination. Total RNA from four independent replicates was checked on RNA6000 Nano chips on a Bioanalyzer (Agilent) for its quality and integrity. Ribosomal RNA depletion was performed using the Bacteria RiboZero kit (Illumina). From rRNA-depleted RNA, directional libraries were prepared using the TruSeq Stranded mRNA Sample preparation kit following the manufacturer’s instructions (Illumina). Libraries were checked for quality on Bioanalyzer DNA 1000 chips (Agilent). Quantification was performed with the fluorescent-based quantitation Qubit dsDNA HS Assay kit (Thermo Fisher Scientific). Sequencing was performed as a single read run for 65-bp sequences on a HiSEq 2500 Illumina sequencer (65 cycles). The multiplexing level was 16 samples per lane. Bioinformatics analysis was performed using the RNA-seq pipeline from Sequana.
Supplementary Material
Acknowledgments
We thank Rebecca Stevick, Christophe Beloin, Susan Joyce, and David Clarke for critical reading of the manuscript. We are grateful to Andy Goodman, Justin Sonnenburg, and Laurie Comstock for providing the genetic tools used in this study. This work was supported by an Institut Pasteur grant and by the French government’s Investissement d’Avenir Program, Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (Grant ANR-10-LABX-62-IBEID) and the Fondation pour la Recherche Médicale (Grant DEQ20180339185). N.B. was supported by a Ministère Français de l’Education Nationale, de l’Enseignement Supérieur et de la Recherche fellowship. J.M. was supported by the Pasteur Paris University International Doctoral Program and the Fondation pour la Recherche Médicale (Grant FDT20160435523). The RNA-seq experiments were performed by the Biomics Platform, C2RT, Institut Pasteur, Paris, France, supported by France Génomique (Grant ANR-10-INBS-09-09) and Infrastructures en Biologie Santé et Agronomie (IBISA).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. E.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2111228119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information. The genomes of strains sequenced in this article are available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA725886 (24).
References
- 1.Eckburg P. B., et al. , Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wexler H. M., Bacteroides: The good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens E. C., Chiang H. C., Gordon J. I., Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Macfarlane S., Macfarlane G. T., Composition and metabolic activities of bacterial biofilms colonizing food residues in the human gut. Appl. Environ. Microbiol. 72, 6204–6211 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenburg J. L., et al. , Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Mark Welch J. L., Hasegawa Y., McNulty N. P., Gordon J. I., Borisy G. G., Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl. Acad. Sci. U.S.A. 114, E9105–E9114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.TerAvest M. A., et al. , Regulated expression of polysaccharide utilization and capsular biosynthesis loci in biofilm and planktonic Bacteroides thetaiotaomicron during growth in chemostats. Biotechnol. Bioeng. 111, 165–173 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mihajlovic J., et al. , A putative type v pilus contributes to Bacteroides thetaiotaomicron biofilm formation capacity. J. Bacteriol. 201, e00650-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Béchon N., et al. , Capsular polysaccharide cross-regulation modulates Bacteroides thetaiotaomicron biofilm formation. MBio 11, e00729-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grondin J. M., Tamura K., Djean G., Abbott D. W., Brumer H., Polysaccharide utilization loci: Fueling microbial communities. J. Bacteriol. 199, e00860-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bollinger N., Hassett D. J., Iglewski B. H., Costerton J. W., McDermott T. R., Gene expression in Pseudomonas aeruginosa: Evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183, 1990–1996 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollinger R. R., et al. , Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: Role of type 1 pili. Mol. Immunol. 43, 378–387 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Ahn S. J., Burne R. A., Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans. J. Bacteriol. 189, 6293–6302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman L. R., et al. , Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Pumbwe L., et al. , Bile salts enhance bacterial co-aggregation, bacterial-intestinal epithelial cell adhesion, biofilm formation and antimicrobial resistance of Bacteroides fragilis. Microb. Pathog. 43, 78–87 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Sicard J. F., Bihan G. L., Vogeleer P., Jacques M., Harel J., Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect. Microbiol. 7, 387 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubois T., et al. , A microbiota-generated bile salt induces biofilm formation in Clostridium difficile. npj Biofilms Microbiomes 5, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva J. O., et al. , In vitro effect of antibiotics on biofilm formation by Bacteroides fragilis group strains isolated from intestinal microbiota of dogs and their antimicrobial susceptibility. Anaerobe 28, 24–28 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Okshevsky M., Meyer R. L., The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 41, 341–352 (2015). [DOI] [PubMed] [Google Scholar]
- 20.H. Liuet al., . Functional genetics of human gut commensal Bacteroides thetaiotaomicron reveals metabolic requirements for growth across environments. Cell Reports 34, 108789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J., et al. , A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Coyne M. J., Comstock L. E., Niche-specific features of the intestinal bacteroidales. J. Bacteriol. 190, 736–742 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Q., et al. , A distinct type of pilus from the human microbiome. Cell 165, 690–703 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.N. Béchon, et al., DNA Data Bank of Japan (DDBJ)/European Nucleotide Archive (ENA)/GenBank. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA725886. Deposited 28 April 2021. [Google Scholar]
- 25.Kristoffersen S. M., et al. , Low concentrations of bile salts induce stress responses and reduce motility in Bacillus cereus ATCC 14570. J. Bacteriol. 189, 5302–5313 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hung D. T., Zhu J., Sturtevant D., Mekalanos J. J., Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 59, 193–201 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Begley M., Kerr C., Hill C., Exposure to bile influences biofilm formation by Listeria monocytogenes. Gut Pathog. 1, 11 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambalam P., Kondepudi K. K., Nilsson I., Wadström T., Ljungh A., Bile enhances cell surface hydrophobicity and biofilm formation of bifidobacteria. Appl. Biochem. Biotechnol. 172, 1970–1981 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Urdaneta V., Casadesús J., Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. (Lausanne) 4, 163 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao L., et al. , A selective gut bacterial bile salt hydrolase alters host metabolism. eLife 7, e37182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flemming H. C., et al. , Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Chen K. M., et al. , The role of pgaC in Klebsiella pneumoniae virulence and biofilm formation. Microb. Pathog. 77, 89–99 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Kelly S. M., et al. , Bifidobacterial biofilm formation is a multifactorial adaptive phenomenon in response to bile exposure. Sci. Rep. 10, 11598 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ambalam P., Kondepudi K. K., Nilsson I., Wadström T., Ljungh Å., Bile stimulates cell surface hydrophobicity, Congo red binding and biofilm formation of Lactobacillus strains. FEMS Microbiol. Lett. 333, 10–19 (2012). [DOI] [PubMed] [Google Scholar]
- 35.Hay A. J., Zhu J., Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infect. Immun. 83, 317–323 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanchez L. M., et al. , Biofilm formation and detachment in gram-negative pathogens is modulated by select bile acids. PLoS One 11, e0149603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molinero N., Ruiz L., Sánchez B., Margolles A., Delgado S., Intestinal bacteria interplay with bile and cholesterol metabolism: Implications on host physiology. Front. Physiol. 10, 185 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu D., Sorg J. A., Sun X., Clostridioides difficile biology: Sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect. Microbiol. 8, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng S., Zhu L., Faden H. S., Interactions of bile acids and the gut microbiota: Learning from the differences in Clostridium difficile infection between children and adults. Physiol. Genomics 51, 218–223 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Crawford R. W., Gibson D. L., Kay W. W., Gunn J. S., Identification of a bile-induced exopolysaccharide required for Salmonella biofilm formation on gallstone surfaces. Infect. Immun. 76, 5341–5349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickerson K. P., et al. , Analysis of Shigella flexneri resistance, biofilm formation, and transcriptional profile in response to bile salts. Infect. Immun. 85, e01067-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koestler B. J., Waters C. M., Bile acids and bicarbonate inversely regulate intracellular cyclic di-GMP in Vibrio cholerae. Infect. Immun. 82, 3002–3014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Köseoğlu V. K., Hall C. P., Rodríguez-López E. M., Agaisse H., The autotransporter IcsA promotes shigella flexneri biofilm formation in the presence of bile salts. Infect. Immun. 87, e00861-18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.González J. F., et al. , Human bile-mediated regulation of Salmonella curli fimbriae. J. Bacteriol. 201, e00055-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., et al. , The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 6, 8292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valguarnera E., Scott N. E., Azimzadeh P., Feldman M. F., Surface exposure and packing of lipoproteins into outer membrane vesicles are coupled processes in Bacteroides. MSphere 3, e00559-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dengler V., Foulston L., DeFrancesco A. S., Losick R., An electrostatic net model for the role of extracellular DNA in biofilm formation by Staphylococcus aureus. J. Bacteriol. 197, 3779–3787 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kavanaugh J. S., et al. , Identification of extracellular DNA-binding proteins in the biofilm matrix. MBio 10, e01137-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koropatkin N. M., Martens E. C., Gordon J. I., Smith T. J., Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–1115 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bankevich A., et al. , SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wick R. R., Judd L. M., Gorrie C. L., Holt K. E., Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput. Biol. 13, e1005595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brettin T., et al. , RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wattam A. R., et al. , Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45 (D1), D535–D542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis J. J., et al. , The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 48 (D1), D606–D612 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boratyn G. M., et al. , BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Leary N. A., et al. , Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44 (D1), D733–D745 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilchrist C. L. M., Chooi Y.-H., Clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475 (2021). 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 58.Oberto J., SyntTax: A web server linking synteny to prokaryotic taxonomy. BMC Bioinformatics 14, 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vallenet D., et al. , MicroScope: An integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 48 (D1), D579–D589 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.García-Bayona L., Comstock L. E., Streamlined genetic manipulation of diverse bacteroides and parabacteroides isolates from the human gut microbiota. MBio 10, e01762-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang J., Shoemaker N. B., Wang G. R., Salyers A. A., Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene. J. Bacteriol. 182, 3559–3571 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goodman A. L., et al. , Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiba A., Sugimoto S., Sato F., Hori S., Mizunoe Y., A refined technique for extraction of extracellular matrices from bacterial biofilms and its applicability. Microb. Biotechnol. 8, 392–403 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information. The genomes of strains sequenced in this article are available at https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA725886 (24).