Significance
The speed of neural information processing in the human central nervous system is ultimately determined by the speed of chemical transmission at synapses, because action potentials have relatively short distances to traverse. The release of synaptic vesicles containing neurotransmitters must therefore be remarkably fast as compared to other forms of membrane fusion. Six separate SNARE complexes cooperate to achieve this. But how can exactly six copies be assembled under every vesicle? Here we report that six copies of the key molecular chaperone that assembles the SNAREs can arrange themselves into a closed hexagon, providing the likely answer.
Keywords: synaptic transmission, membrane fusion, SNARE, Munc13, cryo-ET
Abstract
How can exactly six SNARE complexes be assembled under each synaptic vesicle? Here we report cryo-EM crystal structures of the core domain of Munc13, the key chaperone that initiates SNAREpin assembly. The functional core of Munc13, consisting of C1–C2B–MUN–C2C (Munc13C) spontaneously crystallizes between phosphatidylserine-rich bilayers in two distinct conformations, each in a radically different oligomeric state. In the open conformation (state 1), Munc13C forms upright trimers that link the two bilayers, separating them by ∼21 nm. In the closed conformation, six copies of Munc13C interact to form a lateral hexamer elevated ∼14 nm above the bilayer. Open and closed conformations differ only by a rigid body rotation around a flexible hinge, which when performed cooperatively assembles Munc13 into a lateral hexamer (state 2) in which the key SNARE assembly-activating site of Munc13 is autoinhibited by its neighbor. We propose that each Munc13 in the lateral hexamer ultimately assembles a single SNAREpin, explaining how only and exactly six SNARE complexes are templated. We suggest that state 1 and state 2 may represent two successive states in the synaptic vesicle supply chain leading to “primed” ready-release vesicles in which SNAREpins are clamped and ready to release (state 3).
The speed of neural information processing in the human central nervous system is ultimately determined by the speed of chemical transmission at synapses, because action potentials have relatively short distances to traverse. The release of synaptic vesicles (SVs) containing neurotransmitters must therefore be remarkably fast as compared to other forms of membrane fusion, occurring in ∼0.2 to 1 msec after the signal to secrete from the readily releasable pool (RRP) consisting of an influx of local calcium ions (1, 2). In central synapses the RRP typically consists of 5 to 10 SVs docked within 5 nm of the presynaptic plasma membrane (PM) containing clamped partially zippered SNARE complexes (1, 3, 4). It takes ∼1 s for the isolated synaptic SNARE complexes to trigger fusion after initial docking (5, 6). Evidently, cooperation among multiple SNARE complexes is needed to explain the dichotomy of more than three orders of magnitude between isolated synaptic SNAREs and the same SNAREs in situ.
One cooperative mechanism is provided by release from a common fusion “clamp” provided by the SV calcium-sensor protein Synaptotagmin (7–9), which binds to the partially zippered SNARE complex (10, 11). The clamp likely requires further assembly into Ca2+-sensitive ring-like oligomers of up to 15 to 20 copies of Synaptotagmin at the SV–PM interface (4, 12–18), although other models have been proposed (19–21). Remarkably, including just Synaptotagmin along with the SV v-SNARE VAMP2 in reconstituted vesicles results in synchronous release in <10 msec after a Ca2+ pulse in a fully defined system containing only preassembled t-SNAREs Syntaxin1–SNAP25 in the target membrane bilayer (17, 22).
Mechanical coupling is a second, independent mechanism likely to dramatically enhance cooperativity (23). Bilayers are mechanically rigid on the ∼10-nm scale of SNARE complexes. This effectively but indirectly couples the zippering of multiple SNAREpins to each other. A quantitative model (23) based on empirical parameters yields the surprising prediction that there is an optimum number of three to six SNAREpins needed to enable submillisecond synchronous release. Direct reconstruction of the SV–PM interface from cryo-electron tomograms has provided strong and independent evidence for this prediction (4, 24). Exactly six densities were observed under each SV, each being large enough to consist of one but not two single SNAREpins with associated chaperones. Moreover, the densities were organized symmetrically around the contact point on a circle of ∼29 nm diameter, which would correspond to about 15 to 20 oligomerized Synaptotagmins (12). Expressing a ring-disrupting mutant of Synaptotagmin destroys this circular organization (24) and desynchronizes evoked release (18).
Here, we seek to clarify the mechanism by which exactly six SNARE complexes can be assembled under each SV. The specialized chaperone Munc13 is required for the first committed step in this vesicle “priming” pathway (25, 26). We now report the discovery that six copies of the core functional region of Munc13 can assemble into a closed hexagon, which could serve as the needed template. We also suggest a specific mechanism by which this unexpected oligomeric arrangement could link vesicle docking to SNARE assembly.
Results
Crystallization of Munc13C between Bilayers in Distinct Open and Closed Conformations.
Munc13C consists of the portion of Munc13-1 containing its C1–C2B–MUN–C2C domains (27) and can functionally substitute to a high degree for the full-length protein (28). The C1 domain binds diacylglycerol (DAG) or a soluble analog, phorbol ester. The C2B domain binds phosphatidylinositol 4,5-bisphosphate (PIP2) via its polybasic region and the local lipid bilayer via an aliphatic surface loop (28–31). Munc13’s C2B domain is structurally very similar to the C2B domain of Synaptotagmin, which is a Ca2+ sensor. In the case of Synaptotagmin, Ca2+ binding leads to insertion of a second aliphatic loop (loop 1) of C2B that reorients the domain on the plasma membrane surface (32–35). It seems likely that this also occurs for Munc13’s C2B domain.
The C2C domain binds the SV at some stage in the priming process. While it does not bind Ca2+, C2C avidly binds and clusters phosphatidylserine (PS)-containing liposomes and is required for SV priming (36, 37). Mutation of a single surface residue (F1658) in C2C abolishes both synaptic vesicle docking and neurotransmitter release (36). The intervening MUN domain, which affords Munc13C its arc-like structure, bears binding sites for all three synaptic SNAREs (27, 38–40). However, the structural and functional roles of these various Munc13 membrane binding domains in SNARE complex initiation have remained unclear.
To investigate the behavior of membrane-bound Munc13, we performed cryo-electron tomography (cryo-ET) and subtomogram averaging studies on Munc13-decorated vesicles. Munc13C protein was expressed and purified as described in Materials and Methods, and then added (0.5 µM final concentration) along with phospholipid vesicles (∼50 µM final concentration) in buffer consisting of 20 mM 3-(N-morpholino)propanesulfonic acid (MOPS) pH 7.4, 150 mM KCl, 1 mM ethylenediaminetetraacetic acid (EDTA)*, and 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) at room temperature. The phospholipid vesicles consisted of 80 mol% PS, 14 mol% phosphatidylcholine (PC), and 6 mol% PIP2, a composition chosen because it contains abundant PS and PIP2, both known ligands of Munc13-1.†
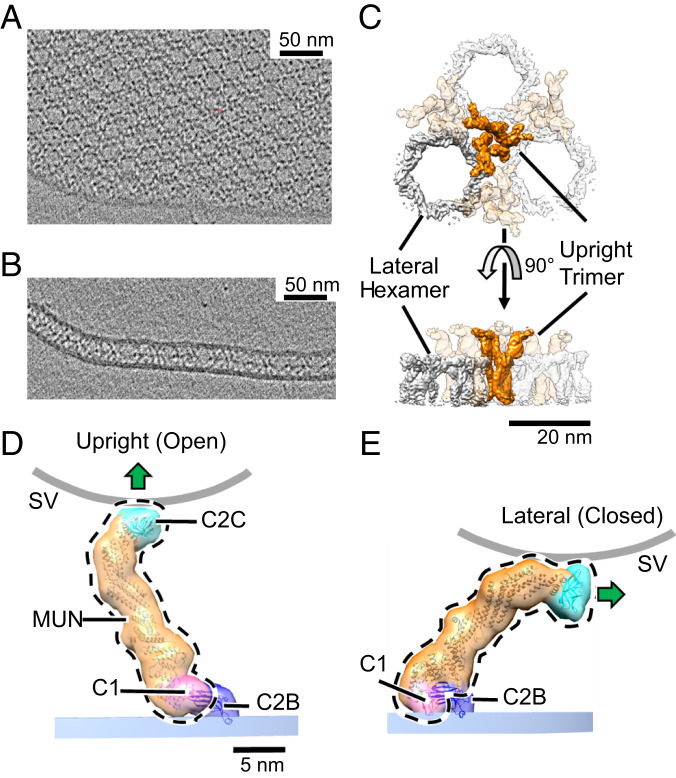
A sample was then frozen for cryo-EM after as little as ∼10 s (or with equivalent results as long as 5 min). In all cases we observed numerous lamellar crystals consisting of two extensive lipid bilayers, uniformly separated by an electron density–filled gap of ∼21 nm. The electron density was organized in a clear hexagonal pattern (Fig. 1 A, top view and B, side view). In favorable views some of the apposed bilayers can be seen to be continuous with each other, as if derived from a single vesicle that had flattened; in other cases, the two bilayers appear to derive from separate vesicles that extensively flatten as they adhere (SI Appendix, Fig. S1).
Fig. 1.
Two-dimensional crystal of membrane-bound Munc13C. (A) Representative slice of a reconstructed tomogram showing a view of the crystal perpendicular to the electron beam showing clear hexagonal packing of protein densities (black). (Scale bar 50 nm.) (B) Another slice of the reconstructed tomogram in A shows a region where the crystal plane is oriented parallel to the electron beam. This reveals a layer of protein density sandwiched between two membrane bilayers. (C) Top and side views of the reconstructed crystal 3D map, shown as an isosurface representation. The map shown is a hybrid of two separate 3D reconstructions, focused on upright trimers and lateral hexamers, respectively, to maximize the resolution of each element. The trimers (orange) are formed by Munc13C oligomerized in an open conformation (D) and hexamers (gray color) formed by Munc13C in a closed state (E). (Scale bar: 20 nm.) (D) Upright (open) and (E) lateral (closed) conformations of Munc13C. The two states are related by rigid body rotation of C1 (pink), MUN (orange), and C2C (cyan) domains of the Munc13C molecule (delineated by the black dashed line) relative to the C2B domain (dark blue). This jackknife-like motion rotates C2C and its associated membrane binding surface (shown by a green arrow) almost 90°, disrupting interactions between C2C and the synaptic vesicle membrane (curved gray line). Atomic models in D and E (ribbon diagrams) were obtained by modeling of the AlphaFold predicted Munc13C structure into the ∼10 Å 3D maps (Materials and Methods). For illustration purposes, models are enclosed by semitransparent isosurfaces of corresponding, synthetically generated 3D maps rendered at 20 Å resolution. Experimentally determined 3D maps with superposed atomic models are shown in SI Appendix, Fig. S3. The membrane bilayer region in the reconstruction is indicated by a shaded bar (PM, blue). The putative location of a synaptic vesicle membrane is shown as a thick gray line (SV), whose curvature approximates a sphere with a diameter of 45 nm.
The three dimensional (3D) density maps of the crystal were reconstructed at ∼10 Å resolution (SI Appendix, Fig. S2) via a multistage workflow that involved an iterative process including subtomogram averaging and 3D classification using the emClarity, RELION 3.1 and Warp/M software packages (42–46). To address the flexibility in the crystal we implemented particle symmetry expansion and subsequent 3D classification with a focus on areas of interest. Details are provided in Materials and Methods and in SI Appendix.
Remarkably, two distinct conformations of Munc13C were found, which we refer to as “upright” and “lateral” (Fig. 1 D and E). All the upright molecules are attached by their C2B domains to one bilayer, and all the lateral molecules are attached by their C2B domains to the other bilayer. The crystal is formed when these two, antiparallel conformations intercalate with each other, sandwiched between the two bilayers (SI Appendix, Fig. S3 A and B). We regard this aspect of the crystal as fortuitous (Discussion).
In the upright conformation (Fig. 1D and SI Appendix, Fig. S3C) Munc13C is vertically oriented to span the two bilayers, contacting one with C2B and the other with C2C, resulting in their rigid separation by ∼21 nm. As C2B binds PIP2, which is found in PMs but not SVs (47), we consider the bilayer to which C2B is bound to represent the PM in the structure. As C2C binds the SV we consider the opposing bilayer to represent the SV in the structure in its relation to the upright conformation. An upright Munc13C could therefore capture a SV and restrict it to a distance of about 21 nm from the plasma membrane. Notably, upright Munc13C places a critical Phe residue (F1658), strongly required for vesicle binding and docking at the active zone PM (36), in direct contact with the SV-representing bilayer (SI Appendix, Fig. S5B).
In its lateral conformation (Fig. 1E and SI Appendix, Fig. S3D), Munc13C now departs at an angle from the PM-representing bilayer. Together with its arc-like shape, this rotation with respect to the upright conformation brings the top of its C2C domain closer to the PM-representing bilayer (∼14 nm), while simultaneously orienting the C2C membrane-binding surface (thick arrows in Fig. 1 D and E) nearly perpendicular to the PM rather than parallel to it (as it is positioned in state 1) inconsistent with a SV binding to the same surface employed in the upright conformation. The lateral molecules give rise to the prominent band of electron density lying about halfway between the apposed bilayers in the crystal (Fig. 1B).
The lateral conformation state closely resembles the high-resolution structure of a truncated Munc13C, which lacks C2C crystalized from solution without membranes (SI Appendix, Figs. S3D, S4A and S6A) (41). While the truncated X-ray structure lacks C2C, C2C is well resolved in our structures, clearly defining its alternative relationships to both the adjoining MUN domain and the contacting bilayer. Moreover, the observed lateral structure is well modeled by AlphaFold (48), which predicts atomic features of regions not previously crystallized (SI Appendix, Figs. S3 and S5) linking C2C and the C-terminal subdomain of MUN, MUN-D (MUN consists of subdomains A–D proceeding in the carboxyl-terminal direction).
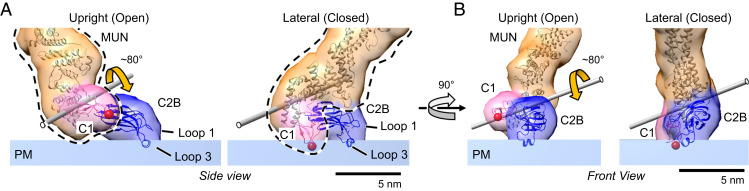
In the lateral conformation interdomain contacts assemble C1–C2B–MUN–C2C into a single rigid unit. In the upright conformation, the contacts between C2B and both C1 and MUN are broken, but the contacts between C1 and MUN still remain (see Fig. 4 and SI Appendix, Fig S4 and Fig. S6B). This allows C1 and MUN–C2C to move as a single rigid unit separately from C2B in the upright conformation. The lateral and upright orientations are therefore related to each other by a “jackknife” motion (Movie S1). The jackknife is “closed” in the lateral conformation (which we alternately refer to as closed, or state 2) but “open” in the upright conformation (which we alternately refer to as open, or state 1). When taking bilayer attachment into account, closing the jackknife results in a rigid body rotation of the C1–MUN–C2C unit (relative to the PM-representing bilayer) about a single axis (indicated in Fig. 4 and illustrated in Movie S1). C2B remains in a fixed position on the PM bilayer during this motion.
Fig. 4.
Configuration of the N-terminal membrane-binding domains in the open and closed Munc13C conformations. (A) Side view of open (Left) and closed (Right) N-terminal end of Munc13C. The difference between the configurations is based on the jackknife-like action of the C1 (pink), MUN (orange), and C2C (not shown in the figure) domains relative to C2B (blue). The C2B orientation with respect to the bilayer is the same in both conformations, with loop 3 helix inserted into the phospholipid headgroup plane and the beta-strands parallel to the membrane surface. The C1–MUN domains (highlighted by a black dashed line) acts as a rigid unit with a jackknife-like movement about the axis shown by the gray rod and a maximum rotation angle of ∼80° (rotation axis and angle between the two C1–MUN orientations was estimated using the “measure rotation” command in Chimera). During this movement, the orientation of the C1 domain changes dramatically, with the loops that form the DAG-binding pocket (red sphere) making partial contact with the bilayer in the closed conformation. (B) Corresponding front view of the open and closed configurations. Protein structures are depicted as fitted atomic models and corresponding synthetic 3D map isosurfaces as in Figs. 1–3.
Distinct Oligomeric Organizations of Open and Closed Conformers in the Crystal.
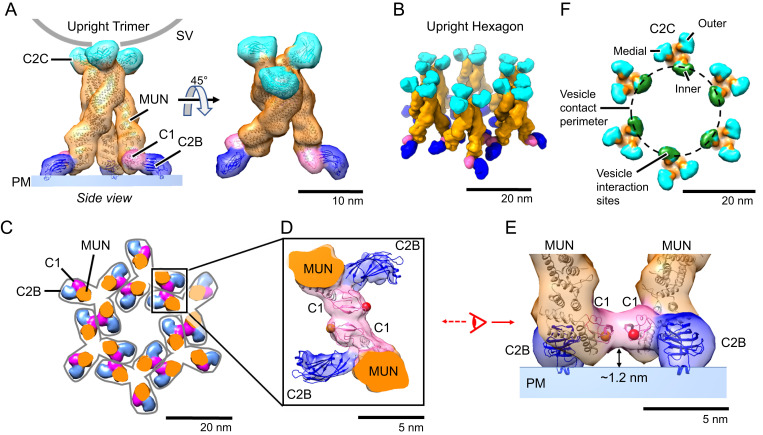
The open, upright state molecules are further organized within the crystal as symmetrical “upright trimers” (Figs. 1C and 2A). Each upright trimer is attached at the PM equivalent by an N-terminal “pedestal” consisting of the three C1–C2B units radiating outward in the plane of the PM 120° apart (Fig. 2A) and attached at the other end to the apposed bilayer by symmetrical contacts made by its three C-terminal C2C domains (Fig. 2A). SI Appendix, Fig. S8, shows a side view and opening of the extensive trimer interface among the convex surfaces of the three curved MUN domains involving primarily their MUN-D subdomains. This interaction appears to be mainly electrostatic in nature (SI Appendix, Fig. S8).
Fig. 2.
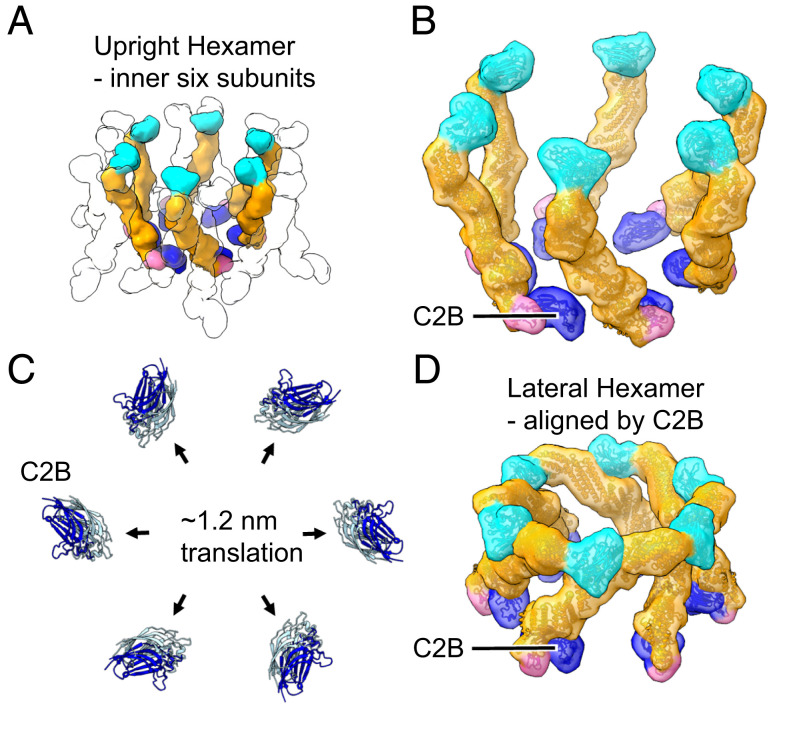
Munc13C trimer organization. (A) Upright Munc13C trimer side (Left) and oblique (Right) views (membrane is omitted for clarity). In this figure and the following, except where noted, protein structures are depicted as fitted atomic models (ribbon diagrams) and corresponding synthetic 3D map isosurfaces as in Fig. 1 D and E. For this conformer, the C2C membrane binding surface is oriented upward, allowing it to bind to a vesicle (gray line represents a vesicle with a diameter of 45 nm). The C1 domain is colored pink, C2B in dark blue, MUN domain in orange, and C2C in cyan. (B) Oblique view of the hexagonal array of upright trimers found in the Munc13C crystal (viewing orientation is the same as in A, Right). (C) View of the hexagonal array as in B but showing a horizontal slice focused on the C1/C2B domains, 5 nm above the PM shown in A. Gray outlines delineate the base of each trimer. Domain coloring is the same as in A. Each trimer is connected with neighboring trimers through antiparallel interactions of the C1 domains as shown in D. (D) Expanded view of interactions between neighboring C1 domains (pink). The reconstructed 3D map density indicates a tight contact between the C1 domains of adjacent trimers. Such interactions keep the C1 domains parallel to the membrane as shown in E. This orientation also keeps C1 domains ∼1.2 nm above the membrane with the DAG-binding pocket facing parallel to the membrane (represented with a red sphere, based on the position of the phorbol ester in the structure of PKC C1 bound to the phorbol ester [PDB: 1PTR]). (F) Top view of the trimeric hexamer, focusing on the C2C domains. The dashed line represents the edge of a 45-nm vesicle if it were parked in the middle of the trimeric hexamer, bound to the innermost ring of C2C domains (green). It is likely that medial and outer rings of C2C (cyan) of this assembly are sterically limited in interacting with the parked vesicle.
Six such upright trimers are further arranged into an “upright hexagon” (Fig. 2B shows an oblique view) linked together near the PM by virtue of antiparallel contacts between the C1-bearing tips of adjacent upright trimer pedestals. These C1–C1 “dyad” contacts (magenta) are visible in a 5 nm slice through the structure parallel to and just above the plane of the PM (Fig. 2C; C1–C1 dyad detail views are given in Fig. 2 D, top view and 2E, side view).
On the opposite side of the upright hexagon, the C2C domain of each of the 18 Munc13C monomers binds the apposed bilayer that represents the SV except with respect to its curvature (Fig. 2F). These C2Cs interact equivalently with the flat bilayer membrane in our two-dimensional (2D) crystal, but when the apposed membrane is curved, as in an actual SV, this can no longer be the case. Thus, when a hypothetical 45-nm diameter SV vesicle (broken circle in Fig. 2F) is docked to an upright hexagon, only the six “inner” C2CI domains, facing the inside of the hexagon, are favorably positioned for vesicle binding (Fig. 2F). The six “medial” C2CM domains are next-best positioned, while the “outer” C2CO would appear unable to bind the SV.
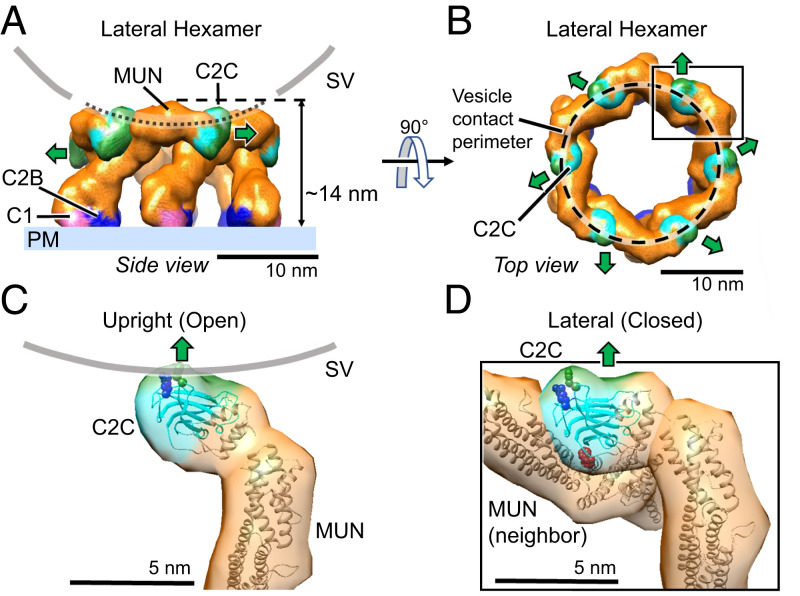
In contrast to the upright hexagon, the closed lateral state conformers are assembled into “lateral hexamer” cages, each consisting of only six Munc13C molecules (Figs. 1C and 3 A, side view and 3B, top view). Each closed conformer now uses its C2C domain to interact with its neighbor’s MUN-B and -C subdomains. When this contact occurs repeatedly the closed, lateral state conformation results in a lateral hexamer, which cannot accept additional subunits. Its top surface is elevated about 14 nm above the PM-representing bilayer. In going from open, upright state 1 to closed, lateral state 2, the C2C domain is thus repurposed from membrane binding to a previously unsuspected protein-binding mode.
Fig. 3.
Munc13C closed conformation assembles into a hexagonal cage within the crystal. (A) Side view and (B) top view of the hexagonal cage (∼14 nm tall) formed by Munc13C molecules in the closed, lateral conformation. Domain coloring is the same as in Fig. 2. In this configuration, the C2C membrane-binding surface (green color) is oriented parallel to the bilayer (green arrow). The gray line corresponds to the contacting contour of a synaptic vesicle (45 nm in diameter). The vesicle is centered on the hexagon opening and lowered to make contact with the top surface of the hexagon cage. The contacting circular contour is shown in B with a dashed line. In this configuration, the C2C membrane interaction surface is sterically excluded from interacting with the vesicle membrane. (C) The C-terminal end of Munc13C in the upright (open) configuration. The orientation and interaction with the synaptic vesicle membrane surface (gray line) is based on a 3D reconstruction of Munc13C trimer (SI Appendix, Fig. S2 D–F and S5B). Important membrane-interacting amino acids (36) R1598 and F1658 are shown as blue and green spheres, respectively. (D) Zoomed-in view of the C-terminal end of Munc13C in a lateral hexamer conformation. Here, the C2C domain interacts with the MUN domain from a neighboring Munc13C molecule. The C2C membrane-interacting surface (shown in green) oriented toward the outside of the hexagon (green arrow), interacts with the MUN domain of an upright molecule (SI Appendix, Fig. S7, not shown here). Additionally, the C2C domain binds the MUN domain of the neighboring closed (lateral) Munc13C (shown in detail in SI Appendix, Fig. S7). This occludes the NF pocket (shown by red spheres of N1128 and F1131) which is responsible for Syntaxin1A opening and activation (55). Protein structures are depicted as fitted atomic models and corresponding synthetic 3D map isosurfaces as in Figs. 1 and 2.
SNARE Assembly Is Autoinhibited in the Lateral Hexamer.
The lateral hexamer structure also reveals an additional functional role served by C2C lateral contacts in the closed state: modulating SNARE assembly. The membrane-distal SNARE motifs of the three synaptic SNAREs are assembled into a half-zippered four-helix bundle in a concerted reaction (49–51) involving a metastable “template complex” (50, 52) of VAMP2 and Syntaxin1 with the chaperone Munc18 that positions these two SNAREs to receive the third SNARE, SNAP25. Syntaxin enters this reaction prebound to Munc18 in a closed conformation (53, 54). Critically, Munc13 “opens” this closed complex by binding to an exposed linker region in Syntaxin (38, 40), thereby exposing both the SNARE motif of Syntaxin and the templating surface of Munc18 and enabling all subsequent events.
Two key amino acids (N1128 and F1131) in the MUN-B subdomain are needed to enable Munc13 to bind the Syntaxin linker and open the closed complex of Syntaxin–Munc18 to assemble SNAREpins. These residues project from the same side of helix H6 (38, 55) and, remarkably, they are both occluded by lateral contacts of the C2C domain within the lateral hexamer (Fig. 3D and SI Appendix, Fig. S7). This implies that Munc13’s catalytic activity would be autoinhibited in the lateral hexamer. The lateral hexamer will also prevent SNARE assembly by limiting the SV from approaching the PM closer than ∼12 nm (modeling suggests a 45-nm diameter sphere could sit atop the hexamer with its lowest point 12 nm from the bilayer).
C2B Is Identically Positioned in Open and Closed Conformations.
Fig. 4 compares details of the membrane attachments of the C1–C2B units in the open and closed conformations. The C2B domain is similarly if not identically positioned in the two conformations, with one of its aliphatic, Ca2+-binding loops (loop 3) inserted into the bilayer while the other (loop 1) is not. The polybasic region, which binds PIP2 (33, 56), is closely applied to the bilayer surface in both cases (SI Appendix, Fig. S4). These features are all expected from previous structural studies in the absence of Ca2+ (33, 35) and closely mirror the orientation of the Synaptotagmin C2B domain in the absence of Ca2+ and the presence of Mg2+ (32). One subtle difference is that loop 3 of Munc13 C2B inserts in the absence of Mg2+ (as noted previously, we routinely omitted Mg2+ from the crystallization buffer to maximize negative surface charge) probably because it carries a higher positive charge and more hydrophobicity than loop 3 of Synaptotagmin1 C2B, which requires Mg2+ to productively insert during binding of PS in the bilayer (29, 32).
The Structure Suggests that DAG Binding to the C1 Domain Favors the Closed Conformation.
In contrast to C2B, the positioning of C1 relative to the PM differs markedly between the two states. In closed state 2, C1 (like C2B) is oriented with its DAG binding surface (red sphere) directly in contact with the bilayer (Fig. 4 A and B, Right images). However, in the open upright state, C1 is now oriented about 80° away from the plane of the membrane (Fig. 4 A and B, Left images) incompatible with binding membrane–inserted DAG. Moreover, C1 as a whole is elevated ∼1.2 nm above the bilayer surface (Figs. 2E and 4 A and B, Left images and SI Appendix, Fig. S4B), which would independently preclude membrane-inserted DAG from binding. The elevation of C1 above the bilayer also enables the C1 dyad contact between neighboring open trimers (Fig. 2 C and D). The residues involved in this contact (569 to 575 and 590 to 592) are 100% conserved among vertebrates and differ only slightly among rat and worm and fruit fly.
We note that DAG was not included in the liposomes from which the crystals formed. Binding of DAG to C1 is therefore expected to stabilize the closed conformation (and the lateral hexagon oligomeric state) relative to the open conformation (and upright trimer). Binding of DAG to many C1 domains has been found to release tightly bound Zn++ and to concomitantly unfold portions of the protein (57). If this occurs in Munc13, one likely effect would be to destabilize the C1–C1 dimer interface. Thus, DAG may simultaneously destabilize the C1 dimer (and therefore the open conformation) while simultaneously stabilizing the alternative closed conformation by direct membrane binding.
With this in mind, we can hypothetically simulate the proposed effect of DAG (Fig. 5). For simplicity, Fig. 5A shows the only the inner six copies of Munc13C in the open hexagon arrangement (referring to Fig. 2C) whose C2CI domains can contact the SV. DAG binding would trigger a rigid body rotation of C1–MUN–C2C (Fig. 5 B and D show before and after and Movie S2 shows the transition), which brings these inner six monomers into the exact arrangement of the lateral hexamer. While the outer and medial C2C-bearing Munc13Cs of a complete open hexagon will also transition, they would rotate away from the hexamer cage and thus not contribute to it.
Fig. 5.
Comparison of hexamer rings of open and closed Munc13C found in the 2D crystal. (A) Depiction of the innermost ring of six Munc13C molecules from the trimeric hexamer of open Munc13C molecules found in the crystal (Fig. 2B). Positions of medial and outer subunits are indicated by empty silhouette outlines. (B) Close-up view of the six inner Munc13C molecules in A. (C) C2B domains (pale blue) from the six open Munc13C molecules in A and B aligned with C2B domains (dark blue) from the hexagonal cage of closed Munc13C molecules (Fig. 3). The two C2B arrangements are related by rigid-body translations of ∼1.2 nm corresponding to outward movement of the C2B domains from the center. (D) View of the hexameric cage of closed Munc13C molecules, oriented so its C2B domains are aligned with C2B domains of the six open Munc13C molecules in B. Comparison of open and closed Munc13C arrangements from B and D, respectively, reveals an economic motion in which the C2B “feet” splay outwards, while jackknife closure by the rest of the molecule brings C2C domains into contact with their counterclockwise, lateral neighbors. See Movie S2.
Discussion
Our study reveals two distinct conformations of Munc13 in the absence of calcium. In the previously unknown open conformation, Munc13 extends nearly vertically ∼21 nm to link the two membranes, holding them rigidly apart as a result of its trimeric organization with a pedestal of C1 and C2B at the plasma membrane–binding end and a triad of C2C domains at the synaptic vesicle–binding end. In the alternative closed conformation, Munc13 is now tilted toward the plane of the membrane, extending laterally at a maximum height of ∼14 nm. In the closed conformation interdomain contacts assemble C1–C2B–MUN–C2C into a single rigid unit. In the open conformation, the contacts between C2B and both C1 and MUN are broken, but the contacts between C1 and MUN still remain (Fig. 4 and SI Appendix, Fig. S4 and S6B). As a result, the membrane-binding surfaces of C2C and C2B are 90° askew in the closed conformation. This change in geometry causes the surface of C2C that contacts the vesicle in the open conformation to face away from the vesicle in the closed conformation. Notably, the positioning of C2B on the membrane is not discernibly altered during this conformational switch.
Aspects of the closed conformation have been previously captured in crystals of Munc13 lacking C2C (41) and at likely lower resolution in cryo-EM tomograms of Munc13C attached to membranes by DAG and PIP2 (58). Our results add considerable clarity both by including C2C at sufficient resolution to establish that C2C is rigidly linked to the main body of the MUN domain and by directly establishing the positioning of Munc13 on the membrane surface in the closed conformation in the absence of Ca2+. Camacho et al. have recently reported molecular dynamic simulations of the bilayer C1–C2B geometries in the absence and the presence of Ca2+ (59). The simulation utilized the closed conformation, and it predicts a nearly perpendicular orientation of MUN in the absence of Ca2+ in contrast to the tilted, lateral geometry we have observed experimentally. Simulation of DAG and Ca2+ binding predicts a very “slanted” arrangement in which the bottom of the MUN domain appears to be elevated maximally about 5 nm from the bilayer surface. Such an arrangement could correspond to an intermediate transition state in the Ca2+-dependent progression from state 2 to state 3 in the model we will detail below (Fig. 6).
Fig. 6.
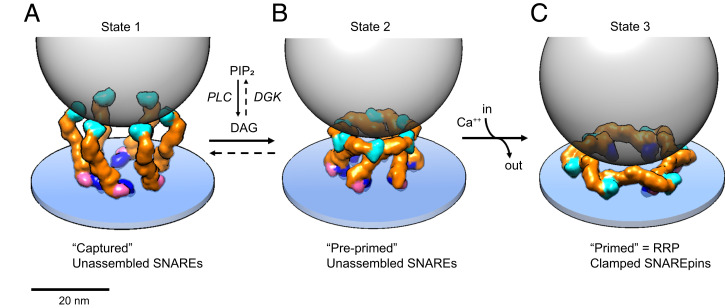
Model of how Munc13C may orchestrate vesicle capture, priming, and release. (A) In state 1, the synaptic vesicle (45 nm diameter gray transparent sphere) is captured by the six inner upright Munc13 molecules (other Munc13 molecules that could be involved in this potentially trimeric interaction are not shown for simplicity). In this configuration, each C2C domain is bound to the vesicle membrane and the bottom of the vesicle is now positioned ∼18 nm above the plasma membrane, a separation at which SNAREpins cannot assemble. (B) DAG binding effectively switches the conformation from open (upright) to closed (lateral) rearranging the Munc13C protomers into the closed, hexagon cage (state 2). The hexagon cage now separates the bottom of the synaptic vesicle from the plasma membrane by a minimum of ∼12 nm, still too far for SNAREpins to assemble. SNAREpins are also unable to assemble in state 2 due to autoinhibition. Specifically, the catalytic pocket on the surface of the MUN domain needed to open the closed Syntaxin1A–Munc18 complex is occluded by the C2C domain of the neighboring protomer. (C) State 3 is the RRP, consisting of primed vesicles with approximate half-zippered SNAREpins clamped by Synaptotagmins. State 2 is proposed to transition to state 3 as the result of an influx of Ca2+ binding to Munc13 C2B domains and causing them to rotate toward the plasma membrane when they insert Ca2+-binding loop 1 into the bilayer along with previously inserted loop 3. This is also expected to rotate MUN–C2C toward and directly against the plasma membrane. The orientation of Munc13 on the plasma membrane is unknown but is based on an illustrative model (SI Appendix, Fig. S9). The actual orientation might be more radially open. The bottom of the synaptic vesicle is now positioned ∼4 to 5 nm above the PM surface, with SNAREpins partially assembled and forming complexes with Synaptotagmin-1 C2B domains, thus priming the vesicle for ready release when the synapse is next activated.
The crystal organization of Munc13C bound to lipid bilayers further reveals entirely unsuspected and distinct oligomeric arrangements of each conformation, which we term the upright trimer and the lateral hexamer. The trimers are stabilized in the open conformation by ionic interactions between MUN domains at the trimer interface and are further assembled into a hexagonal lattice by C1–C1 interactions at their pedestals. The trimers and hexamers are oppositely oriented in the crystal. All of the trimer C2B domains are attached to one bilayer, while all the hexamer C2B domains are attached to the other. The hexamers and trimers contact each other. Specifically, C2C of each lateral hexamer contacts a MUN domain of a neighboring oppositely oriented upright trimer, as detailed in SI Appendix, Fig. S7. While this interaction is likely important for crystallization, there is no reason to suspect it is physiologically relevant. On the other hand, there are several reasons to suspect that the interactions giving rise to the upright trimer and lateral hexamer assemblies are physiologically important.
First, the crystal assembles reproducibly and spontaneously within seconds of mixing lipid vesicles with Munc13C protein. Protein crystallization is typically a slow process requiring precipitants to lower solubility. It is this aspect that often leads to nonphysiological low-affinity crystal contacts.
Second, the closed conformation (lateral hexamer state) likely represents the lowest energy state of Munc13C because it is isomorphous with that of C2C-truncated Munc13C in a pure protein crystal (41) consisting of a single rigid unit from its N-terminal C1 domain to its C-terminal C2C domain. These rigid closed conformers are themselves rigidly oriented on the bilayer surface by means of at least three points of attachment: 1) the polybasic region of C2B with PIP2; 2) bilayer insertion of an aliphatic loop of C2B (Fig. 4 A and B, Right and SI Appendix, Fig. S4); and 3) C1 to DAG in the bilayer when this lipid is present. These constraints cooperate to rigidly orient and position the C2C domains in space. It would seem to be an extraordinary coincidence that these domains just happen to be at the exact height and rotational orientations needed to form a closed hexagon (Fig. 3A), and an even greater coincidence that in doing so they just happen to be bound at the critical site that initiates SNARE assembly (Fig. 3D and SI Appendix, Fig. S7).
Third, the six innermost copies of Munc13C (whose inward-facing C2CI domains are positioned to bind the SV; Fig. 2F) of the upright hexagon are attached to the PM by six C2B domains, which lie on a circle of ∼20-nm diameter. It would be surprising if this were coincidentally similar to the positioning of the six SNAREpin-containing exocytosis modules observed by cryo-EM tomography underneath each synaptic vesicle in the RRP in situ (4).
Finally, single molecule studies show that a minimum of 3 copies of Munc13C are needed to first detect binding of PS-containing phospholipid vesicles to planar bilayers, mediated primarily by their C2C domains (60). This could be indicative of binding by a trimer of Munc13C. Efficient binding of a single vesicle requires a minimum of 6 copies of Munc13C. A second vesicle does not bind until 12 copies of Munc13C are present (60), consistent with quantal and cooperative binding involving groups of 6 copies of Munc13. Further research will be needed to more specifically relate these single-molecule/single-vesicle results to the oligomeric organizations of Munc13C in the crystal.
Additionally, the effects of mutations engineered into the numerous contact sites revealed by our data will be needed to firmly test the proposed functional relevance of the deduced trimeric and hexameric oligomeric states. These oligomers will also need to be demonstrated in biochemical experiments in which they can form separately from each other outside of the crystal and be shown to function as on-pathway intermediates in reconstituted fusion reactions.
Three-State Model for Capture and Priming of Synaptic Vesicles.
Previous studies (30, 41) are consistent with the closed conformation described here, the main differences being that our structure includes C2C, is much more precise, and reveals a rigid orientation relative to the PM. Importantly, neither the open conformation nor any oligomeric organization was reported. One major difference is that in the crystal Munc13 is attached to a flat bilayer rather than a curved vesicle (58). In this respect our conditions are much more realistic. In particular, neither the C1–C2B pedestal of the upright trimer (∼23 nm in its largest chord on the membrane surface) nor the extensive flat membrane footprint of the membrane-contacting surface of the lateral hexagon (∼27 nm in diameter) could effectively assemble on the ∼50- to 100-nm diameter vesicles employed by Gipson et al. (58).
We hypothesize that the proposed open → closed conformational switch (Fig. 5) is a key step in vesicle priming that fits naturally within an overall three-state assembly pathway (Fig. 6) that enables precise regulation of the Ca2+-dependent supply chain of RRP vesicles (61).
In state 1 we propose that the synaptic vesicle is locally captured by one or more Munc13 C2C domains presented in the open conformation, positioning the bottom of the vesicle at a fixed distance of ∼18 nm from the active zone PM bilayer (Fig. 6A). The capturing Munc13s could most simply be preorganized into upright trimers or they could accrete individually as monomers. In either case, their local concentration would eventually approximate that in the crystal, so they would be expected to spontaneously assemble into all or part of the 18-member upright hexagon observed in the crystal, depending on the number of copies available (Fig. 2B; also see below concerning other structures in the active zone) in which six upright trimers are linked at their pedestals by the tips of their radiating C1 domains. The MUN–MUN interfaces stabilizing the trimer (SI Appendix, Fig. S8) and the C1 dyad (Fig. 2 D and E) function synergistically in this model. However, it is possible that either interface alone could suffice for this purpose or that the C2C contacts with the SV also stabilize the trimer. State 1 vesicles would be difficult to distinguish from the many other SVs that are more than ∼10 nm from the PM, the bulk of which are entrapped in condensates with Synapsins (62).
In state 2 we propose that the six inner copies of Munc13 in state 1 (Fig. 6A) have now switched from open to their closed conformations in which they now self-assemble into lateral hexamers (Figs. 5 and 6B). This advances the captured SV into a more mature but still “preprimed” state in which SNAREs are still unable to assemble with each other. The very nature of a closed hexagon dictates that this transition can only occur when the vesicle has accumulated six Munc13s and that any extra copies would be excluded. If each Munc13 can assemble only one SNAREpin, then this could simply and elegantly explain the remarkable finding that there are ultimately exactly six SNAREpin-containing densities under each closely docked SVs (4, 24).
We suggest that SNAREpins cannot yet assemble at this stage for two reasons. First, the catalytic site that initiates assembly is autoinhibited in each MUN domain in the lateral hexamer by its neighbor’s C2C domain (Fig. 3D and SI Appendix, Fig. S7). Second, the height of the hexamer is expected to preclude SNARE assembly by acting as a barrier, keeping SV and PM at least ∼12 nm apart. Most importantly, SNAREpins are not stable at interbilayer separations >8.5 ± 1.5 nm as revealed by surface forces apparatus experiments (63). Additionally, when optical tweezers are used to pull apart the bead-attached VAMP–Syntaxin–Munc18 template complex (52), dissociation occurs when their separation corresponds to an interbilayer distance of ∼10.5 nm (see SNAREpin assembly distance section in SI Appendix for the basis of this conclusion). Note that while the catalytic site of MUN is sterically available in the upright conformation, the even greater intermembrane separation (∼18 nm) precludes SNAREpin assembly.
Thus, the lateral hexagon is expected to stabilize an intermediate preprimed state of assembly of the SNARE complex, “primed” being defined as a state containing partially zippered four helix bundles (64, 65). Whether the SV directly binds to the surface of the hexagon (as diagrammed in Fig. 6B) or instead is simply prevented from approaching the PM by the hexagon is unclear. The vesicles would certainly be retained locally by Synaptotagmin binding to PM PIP2, which is absolutely required for SV docking at the active zone (56, 66, 67).
In state 3, RRP vesicles are stochastically produced from state 2 by a round of Ca2+ influx and efflux (Fig. 6C). This model for the RRP corresponds closely to our previously described “buttressed ring” (15) with certain important updates. Use of the synapse depletes the RRP, which then needs to be replenished. Our model for state 2 suggests a simple mechanism by which Ca2+ entry can trigger this. Ca2+ binding to the six Munc13 C2B domains in state 2 is expected to reorient them as their aliphatic, Ca2+-binding loop 1 now inserts into the PM bilayer, simultaneously rotating their attached MUN domains toward the PM (see SI Appendix, Fig. S9 for more detail). Critically, this motion simultaneously removes the autoinhibiting C2C domains from their MUN domains, now enabling SNAREpin assembly to proceed. Assembly requires not only opening of closed Syntaxin by MUN and then templating by Munc18, but also an unobstructed approach of the SV to the PM; and lateral hexagon disassembly also now permits this. When Ca2+ is later pumped out, the now half-zippered SNAREpins, clamped by Synaptotagmins, would stabilize the proposed flattened radial or ring-like arrangement of MUN domains in state 3, preventing them from returning to state 2 when the Ca2+ is cleared out.
Proposed Regulation of the Stability of State 2 by Diacylglycerol.
While the membrane interfaces of Munc13’s PIP2-binding C2B domains in state 1 and state 2 are indistinguishable, the membrane topologies of the C1 domains in the two states are radically different. In state 1 the DAG-binding site of C1 is both elevated above and pointing away from the membrane surface, a geometry that precludes it from binding DAG in the lipid bilayer (Fig. 4 A and B, Left). But in state 2 the same site is now closely applied to the lipid bilayer surface, favoring DAG binding (Fig. 4 A and B, Right). This implies that membrane-bound DAG can bind to C1 in state 2 but not state 1. In the simplest model, the energy associated with DAG binding will lower the rate of the back reaction 2 → 1 without affecting the rate of the forward reaction 1→ 2, thereby shifting the conformational equilibrium in favor of state 2. Accordingly, the local concentration of DAG is likely to increase the flux of SVs toward state 3 (RRP). In addition, and more speculatively, DAG binding could synergistically destabilize state 1 by disrupting the dimers of C1, provided that the tightly coordinated Zn++ is released in the process, as occurs with other C1 domains (57).
The likely physiological effects of DAG on states 2 and 3 can be deduced from the pharmacological effects of phorbol esters. These drugs, which are analogs of DAG, increase the probability of release from the RRP (25, 68), suggesting that state 3 is destabilized by phorbol ester. Because consistent changes in RRP have not been reported (25, 68), it is likely the phorbol ester also increases the flux from state 2 to state 3 needed to maintain the RRP. This pharmacology suggests that both states 2 and 3 are both stabilized by DAG because phorbol esters should be regarded mechanistically as DAG antagonists, not agonists, as they relate to membrane binding. Whereas DAG is fully inserted into the lipid bilayer, the much smaller and less hydrophobic phorbol ester should be much more weakly membrane attached. Because phorbol ester competes with DAG for binding to C1, it follows that ester binding will reduce C1–DAG-mediated contacts with the bilayer. To the extent that states 2 or 3 are stabilized by the additional DAG-mediated bilayer attachment, they would then be destabilized when phorbol esters replace bound DAG.
The crystal packing requires equal numbers of Munc13 molecules in upright trimers and lateral hexagons. The efficient assembly of crystals from liposomes lacking DAG therefore suggests that the intrinsic energy difference between the two oligomers is minimal in the absence of DAG. Attempts to form similarly extensive and well-ordered crystals in the presence of DAG were unsuccessful. This can now be readily explained because lateral hexagons are expected to predominate over upright trimers in the presence of DAG. Although some extensive crystalline regions were found in samples containing DAG, they were only rarely observed. However, disorganized lateral hexagons were readily found (SI Appendix, Fig. S10). As noted, we intentionally utilized nonphysiological lipid compositions to help drive the formation of 2D crystals. Under these conditions, states 1 and 2 appear to have comparable stability (existing in comparable numbers). It is difficult to predict the relative stability of these states under more realistic lipid conditions.
Possible Relationships to Other Structures in the Active Zone.
Though the core Munc13C domain can replace full-length Munc13-1 to a very high degree (28), it contains only approximately two-thirds of the full Munc13-1 sequence. The complete protein contains an extensive, poorly conserved N-terminal portion that begins with another C2 domain, C2A, followed by a long apparently unstructured region (containing a calmodulin-binding site), which then continues into C1 and the Munc13C protein we crystallized. The C2A domain binds to proteins located in fixed cells in 40- to 50-nm diameter “dense projections” (DPs) (69), which are likely protein condensates and more fluidly organized in life (70). In fixed synapses, the similarly sized SVs and DPs are interspersed and closely packed in a hexagonal array. Clusters of Munc13 have been consistently described at docking sites (71, 72), and quantitative superresolution microscopy suggests there are at least nine copies of Munc13 associated with each such RRP vesicle (73). The lateral hexamer (Fig. 3 A and B; 6 subunits, ∼27-nm outer diameter) could be easily accommodated in the space likely to be available. The upright hexagon (Fig. 2 B, C, and F) composed of six upright trimers, being ∼50 nm in diameter, would be a tighter fit. An interesting variation on state 1 that would easily fit would be an analogous 12-subunit upright hexagon (SI Appendix, Fig. S11) produced by omitting the six medial monomers whose C1 domains are not engaged in the C1–C1 dyad interactions in the upright hexagon. This results in an upright hexagon with an ∼37-nm footprint, still held together by the original six antiparallel C1–C1 dyad contacts. With the native, full-length protein, a pair of C2A-containing unstructured domains would then emanate from each vertex of the hexagon to contact a neighboring DP condensate. This model would be in keeping with the fact that C2A homodimerizes (74), suggesting that Munc13C domains enter DP condensates already paired.
Relationship to Previous Models.
Careful physiologic, kinetic, and morphological measurements and analysis (75) have previously suggested that a “tight” RRP is replenished with each Ca2+ wave from a “loose” pool of synaptic vesicles located >5 nm from the plasma membrane (76). This loose population could correspond to our state 2. A key difference is that it was posited (76) that SNAREpins have already zippered in the loose vesicles though less so than in the tight RRP vesicles (hence the choice of the term loose). By contrast, SNAREpins cannot initiate zippering based on the structure of state 2 in our model. The evidence cited for SNARE engagement in the kinetically defined loose state (77) is indirect, and more recent structural biology makes a proposed “less than half-zippered” intermediate loose state implausible because half-zippered SNARE complexes are produced in a concerted reaction from the Munc18 template complex (50, 52). More generally, it has long been appreciated that some sort of tilted/slanted state is needed sterically to enable SNARE assembly (21). These expectations are consistent with the well-defined open and closed states we have now found, with the critical distinction that the closed state—assembled into the lateral hexagon—actually prevents rather than enables SNAREpin assembly.
SNAREpins forming during the final assembly of the RRP ultimately must be captured by Synaptotagmin and clamped before they could complete assembly to minimize spontaneous neurotransmitter release. Synaptotagmin oligomerization is required for the ring-like arrangement of clamped SNARE complexes because a targeted mutation (F349A) that prevents oligomerization disrupts the circular arrangement but not the assembly of the exocytosis modules themselves (4, 24), and at the same time, this mutation also desynchronizes evoked release and increases spontaneous release (18).
Our results concerning Munc13 oligomers can be reconciled with these previously proposed roles of Synaptotagmin oligomers in the buttressed rings model (15) with a simple modification of the model, as follows: Combining the EM structure for the packing of Synaptotagmin C2B domains in their rings (12, 14) with the X-ray crystal structures of Synaptotagmin–SNARE complexes (11) suggested that SNAREpins emanating from both neighboring and even next-nearest neighboring C2Bs in the ring will sterically clash (15). This would mean a ring of no fewer than 18 C2B domains would be needed to clamp the six SNAREpins that power fusion from an RRP SV. SI Appendix, Fig. S12 shows an 18-member Synaptotagmin C2B ring superimposed in the same plane (just above the PM) as the six Munc13 C2B domains of the lateral hexagon cage (state 2). Interestingly, they overlap closely overall and a Munc13 C2B domain occupies the position of every third Synaptotagmin C2B domain. This suggests the intriguing possibility of a hybrid C2B oligomer ring-like topology in which each Munc13-derived C2B is flanked by two Synaptotagmin-derived C2B domains. Alternatively, the six Munc13 C2Bs could be located periodically on the outside of a continuous 18-member ring of Synaptotagmin1 C2B domains as originally proposed (15).
Summary and Perspectives.
In sum, the crystal structure naturally suggests a parsimonious view of the supply chain that results in readily releasable vesicles at neuronal synapses, illuminated with unprecedented precision by high-resolution structures of proteins on membranes in reasonable approximations of their native environments. A central finding is that successive topological rearrangements of Munc13 likely orchestrate this process. Each use of a synapse results in a transient peak of local Ca2+, leading to release of some SVs from the RRP, thereby creating the need for resupply. Ca2+ influx resulting from repeated use of the synapse transitions state 2 to the RRP (state 3). Biophysical simulations suggest that as few as three or as many as six (but not many more) SNAREs will suffice for submillisecond fusion due to mechanical coupling following synchronous release from the clamped state (bound to Synaptotagmin oligomers). Having six Munc13s would always assure that a sufficient number of SNAREpins is present even when chaperoned assembly is <100% efficient.
The insights we have outlined in this and previous reports (12, 32) have resulted from structural studies of Synaptotagmin and Munc13 in the relatively natural environment of membranes. Continuing this approach to include successive states of docked and primed vesicles whose functional activity can be ascertained will inform models such as we have proposed here and holds the promise of achieving a complete physical–chemical understanding of the mechanism that sets the pace of central neurotransmission and thereby ultimately the speed with which we can process information.
Materials and Methods
Materials.
The lipids used in this study, including 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (sodium salt) (DOPS), L-α-phosphatidylinositol-4,5-bisphosphate (brain, porcine) (ammonium salt) (PIP2) were purchased from Avanti Polar lipids.
The cDNA construct with His12–Munc13C (Munc13–1 residues 529 to 1735 with residues 1408 to 1452 replaced by the sequence EF) was expressed in a modified pCMV–AN6 vector, which includes a PreScission cut site following the His12 tag.
Protein Expression and Purification.
Munc13 protein was expressed and purified as described previously (27). Munc13C was expressed in ExpiHEK-293 cell cultures using ExpiFectamine as a transfection reagent (Thermo Fisher). Briefly, thawed cells were passaged three times prior to use. Posttransfection, cells were grown for 72 h before being spun down and rinsed in ice-cold buffer (140 mM KCl, 50 mM Hepes, pH 7.4). The pellet was resuspended in buffer and lysed using a Dounce homogenizer three times and spun each time at 900 × g to separate the soluble protein from dense cell fractions. Triton X-100 (5% final, Sigma-Aldrich) was added and the lysate was clarified using at 140,000 × g or 30 min at 4 °C using a Type45Ti rotor (Beckman Coulter, AG). The supernatant was incubated overnight with His-Pur Ni-NTA beads (Thermo Fisher), in the presence of DNase 1, RNase A (AmericanBio), and Benzonase (Sigma-Aldrich) to remove nucleotide contamination. The protein was further washed in the lysis buffer (without Triton X-100) before being cleaved with PreScission protease for 2 h at room temperature. The eluted protein was further purified via gel filtration (Superdex 200, GE Healthcare).
Lipid Membrane Preparation.
Vesicles were prepared with a lipid composition consisting of DOPC/DOPS/PIP2 in a molar ratio of 14/80/6. The lipid stocks were mixed in a chloroform with addition of 20 μL methanol to dissolve PIP2 and the solvent was evaporated under N2 gas followed by vacuum drying for 1 h. The resulting dried lipid film was rehydrated for 1 h at room temperature with constant vortexing in buffer containing 20 mM MOPS pH 7.4, 150 mM KCl, 1 mM EDTA, and 0.5 mM TCEP at a final lipid concentration of 1 mM. After rehydration, the mixture was sonicated for 5 min using a bath sonicator (Branson Ultrasonics). To remove large aggregates of lipid membranes, the prepared solution was stored overnight at 4 °C. Vesicles were separated from sedimented aggregates and used for crystallization experiments.
Protein Crystallization.
The protocol resulted in unilamellar vesicles with broad range in sizes. Crystals were formed immediately before the freezing by mixing 1 µM Munc13C with 100 µM lipid membranes in 1:1 (vol/vol) ratio, total volume of 20 µL. Once mixed, samples were incubated at room temperature for 5 min before freezing.
Electron Microscopy Sample Preparation and Data Acquisition.
Samples were vitrified using a Vitrobot Mark IV (Thermo Fisher Scientific) held at 8 °C with 100% humidity. Bovine serum albumin (BSA)-coated, 10-nm Gold Tracer beads (Aurion) were added immediately prior to vitrification to the samples as fiducial markers. Samples (2.5 µL) were applied to freshly glow-discharged 200 mesh Lacey Formvar/carbon grids directly in the blotting chamber, and grids were blotted for 5 s with blot force −1 and then plunge frozen in liquid ethane cooled by liquid nitrogen. Initially, samples were screened with a Glacios Cryo TEM 200 kV (Thermo Fisher Scientific) equipped with a K2 Summit direct electron detector (Gatan). Low- and medium-resolution montages were acquired for a complete grid and selected squares overview. The K2 camera was used in counting mode, and detector dark and gain references were collected prior to each data acquisition session. Cryoelectron tomograms were acquired using SerialEM with a bidirectional scheme from 0°; −51° to +51°, 3° increment, 35 frames, and 54 e−/A2 total dose. The nominal magnification was 13,500× resulting in a pixel size of 3.019 Å. Nominal defocus was set to −4 µm. Collected tilt series were selected for further processing based on the visual appearance of the crystal, resulting in 10 total tomograms for initial 3D reconstruction. The final dataset was collected using a 300 kV Titan Krios G2 transmission electron microscope (Thermo Fisher Scientific) equipped with GIF Quantum LS energy filter mounted in front of a K3 Summit direct electron detector (Gatan). The slit width of the filter was set to 20 eV. Filter tuning was done using Digital Micrograph software (Gatan). The tilt-series images were acquired using SerialEM at a nominal magnification of 42,000× (corresponding to a calibrated physical pixel size of 2.1 Å) using dose symmetric scheme (78) with tilt range ± 51° and 3° increment. The tilt images were acquired as 11,520 × 8,184 super-resolution movies of 12 frames for the first session or 14 frames for the second one per each tilt corresponding to a tilt series total dose ∼100 e−/Å2 and ∼120 e−/Å2, respectively. Defocus nominal range was set to −3.5 µm to −5 µm.
Image Processing, Tomogram Reconstruction, Model Fitting, Data Analysis, and Availability.
These are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Shyam Krishnakumar and Dr. Frederic Pincet for their comments and critique of the manuscript. We are grateful to the staff of the Yale Cryo-EM Resource facilities and the High-Performance Computing Facility for expert support and guidance. We also wish to thank former member of the C.V.S. laboratory, Kimberley Gibson, for help with PEET and emClarity. This research was supported by grants from NIH DK 027044 and NIH GM 110530 (C.V.S.).
Footnotes
Reviewers: J.D., Weill Cornell Medicine; and T.S., Heidelberg University.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2121259119/-/DCSupplemental.
*Zn++ was not present, but is required for folding of C1. It is unlikely that already Zn++ was removed by chelation during the brief exposure to EDTA because the density of our reconstruction of C1 conforms with the known X-ray structure (41). Additionally, crystallization trials in presence of 0.1 mM EGTA were similarly efficient as in presence 1mM EDTA (data not shown). Including 1 mM of Mg2+ prevented crystallization but Mg2+ was added after crystallization without effect.
†DAG, which promotes Munc13-membrane binding, was intentionally omitted; the typical requirement for DAG (and 1 to 2 mol% PIP2) for membrane binding was bypassed by employing much higher concentrations of PS and PIP2.
Data Availability
3D density maps of the hexagon, trimer and isolated lateral, upright Munc13C as well as a composite map of six trimers and one hexagon have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-25737, EMD-25738, EMD-25739, EMD-25740, and EMD-25741, respectively. For the original reconstructions the depositions include the corresponding EM density map, both half maps and the mask used for the final FSC calculations. Atomic models have been deposited in the Protein Data Bank under accession number 7T7V for lateral Munc13C, 7T7X for upright Munc13C, 7T7C for the hexagonal arrangement of lateral and upright Munc13C, 7T7R for the upright Munc13C trimer and 7T81 for the lateral hexagon surrounded by upright trimers.
References
- 1.Südhof T. C., Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron 80, 675–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaeser P. S., Regehr W. G., Molecular mechanisms for synchronous, asynchronous, and spontaneous neurotransmitter release. Annu. Rev. Physiol. 76, 333–363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kaeser P. S., Regehr W. G., The readily releasable pool of synaptic vesicles. Curr. Opin. Neurobiol. 43, 63–70 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Radhakrishnan A., et al. , Symmetrical arrangement of proteins under release-ready vesicles in presynaptic terminals. Proc. Natl. Acad. Sci. U.S.A. 118, e2024029118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramakrishnan S., et al. , High-throughput monitoring of single vesicle fusion using freestanding membranes and automated analysis. Langmuir 34, 5849–5859 (2018). [DOI] [PubMed] [Google Scholar]
- 6.van den Bogaart G., et al. , One SNARE complex is sufficient for membrane fusion. Nat. Struct. Mol. Biol. 17, 358–364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brose N., Petrenko A. G., Südhof T. C., Jahn R., Synaptotagmin: A calcium sensor on the synaptic vesicle surface. Science 256, 1021–1025 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Geppert M., et al. , Synaptotagmin I: A major Ca2+ sensor for transmitter release at a central synapse. Cell 79, 717–727 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Littleton J. T., Stern M., Schulze K., Perin M., Bellen H. J., Mutational analysis of Drosophila synaptotagmin demonstrates its essential role in Ca(2+)-activated neurotransmitter release. Cell 74, 1125–1134 (1993). [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q., et al. , Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature 525, 62–67 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Q., et al. , The primed SNARE-complexin-synaptotagmin complex for neuronal exocytosis. Nature 548, 420–425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J., et al. , Calcium sensitive ring-like oligomers formed by synaptotagmin. Proc. Natl. Acad. Sci. U.S.A. 111, 13966–13971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J., et al. , Circular oligomerization is an intrinsic property of synaptotagmin. eLife 6, e27441 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zanetti M. N., et al. , Ring-like oligomers of synaptotagmins and related C2 domain proteins. eLife 5, e17262 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothman J. E., Krishnakumar S. S., Grushin K., Pincet F., Hypothesis - buttressed rings assemble, clamp, and release SNAREpins for synaptic transmission. FEBS Lett. 591, 3459–3480 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramakrishnan S., et al. , Synaptotagmin oligomers are necessary and can be sufficient to form a Ca2+ -sensitive fusion clamp. FEBS Lett. 593, 154–162 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramakrishnan S., Bera M., Coleman J., Rothman J. E., Krishnakumar S. S., Synergistic roles of Synaptotagmin-1 and complexin in calcium-regulated neuronal exocytosis. eLife 9, e54506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tagliatti E., et al. , Synaptotagmin 1 oligomers clamp and regulate different modes of neurotransmitter release. Proc. Natl. Acad. Sci. U.S.A. 117, 3819–3827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunger A. T., Choi U. B., Lai Y., Leitz J., Zhou Q., Molecular mechanisms of fast neurotransmitter release. Annu. Rev. Biophys. 47, 469–497 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunger A. T., Leitz J., Zhou Q., Choi U. B., Lai Y., Ca2+-triggered synaptic vesicle fusion initiated by release of inhibition. Trends Cell Biol. 28, 631–645 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizo J., Mechanism of neurotransmitter release coming into focus. Protein Sci. 27, 1364–1391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bera M., Ramakrishnan S., Coleman J., Krishnakumar S. S., Rothman J. E., Molecular determinants of complexin clamping in reconstituted single-vesicle fusion. biorxiv [Preprint] (2021). 10.1101/2021.07.05.451112. Accessed 10 September 2021. [DOI]
- 23.Manca F., et al. , SNARE machinery is optimized for ultrafast fusion. Proc. Natl. Acad. Sci. U.S.A. 116, 2435–2442 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., et al. , Symmetrical organization of proteins under docked synaptic vesicles. FEBS Lett. 593, 144–153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basu J., Betz A., Brose N., Rosenmund C., Munc13-1 C1 domain activation lowers the energy barrier for synaptic vesicle fusion. J. Neurosci. 27, 1200–1210 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betz A., Okamoto M., Benseler F., Brose N., Direct interaction of the rat unc-13 homologue Munc13-1 with the N terminus of syntaxin. J. Biol. Chem. 272, 2520–2526 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Kalyana Sundaram R. V., et al. , Munc13 binds and recruits SNAP25 to chaperone SNARE complex assembly. FEBS Lett. 595, 297–309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X., et al. , Functional synergy between the Munc13 C-terminal C1 and C2 domains. eLife 5, e13696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shin O. H., et al. , Munc13 C2B domain is an activity-dependent Ca2+ regulator of synaptic exocytosis. Nat. Struct. Mol. Biol. 17, 280–288 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michelassi F., Liu H., Hu Z., Dittman J. S., A C1-C2 Module in Munc13 Inhibits Calcium-Dependent Neurotransmitter Release. Neuron 95, 577–590.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabachinski G., Yamaga M., Kielar-Grevstad D. M., Bruinsma S., Martin T. F., CAPS and Munc13 utilize distinct PIP2-linked mechanisms to promote vesicle exocytosis. Mol. Biol. Cell 25, 508–521 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grushin K., et al. , Structural basis for the clamping and Ca2+ activation of SNARE-mediated fusion by synaptotagmin. Nat. Commun. 10, 2413 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bai J., Tucker W. C., Chapman E. R., PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat. Struct. Mol. Biol. 11, 36–44 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Hui E., Bai J., Chapman E. R., Ca2+-triggered simultaneous membrane penetration of the tandem C2-domains of synaptotagmin I. Biophys. J. 91, 1767–1777 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo W., Herrick D. Z., Ellena J. F., Cafiso D. S., The calcium-dependent and calcium-independent membrane binding of synaptotagmin 1: Two modes of C2B binding. J. Mol. Biol. 387, 284–294 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quade B., et al. , Membrane bridging by Munc13-1 is crucial for neurotransmitter release. eLife 8, e42806 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Padmanarayana M., et al. , A unique C2 domain at the C terminus of Munc13 promotes synaptic vesicle priming. Proc. Natl. Acad. Sci. U.S.A. 118, e2016276118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S., et al. , Conformational change of syntaxin linker region induced by Munc13s initiates SNARE complex formation in synaptic exocytosis. EMBO J. 36, 816–829 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang S., et al. , Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly. Nat. Commun. 10, 69 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma C., Li W., Xu Y., Rizo J., Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat. Struct. Mol. Biol. 18, 542–549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu J., et al. , Mechanistic insights into neurotransmitter release and presynaptic plasticity from the crystal structure of Munc13-1 C1C2BMUN. eLife 6, e22567 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Himes B. A., Zhang P., emClarity: Software for high-resolution cryo-electron tomography and subtomogram averaging. Nat. Methods 15, 955–961 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bharat T. A. M., Russo C. J., Löwe J., Passmore L. A., Scheres S. H. W., Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure 23, 1743–1753 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tegunov D., Xue L., Dienemann C., Cramer P., Mahamid J., Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5 Å in cells. Nat. Methods 18, 186–193 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takamori S., et al. , Molecular anatomy of a trafficking organelle. Cell 127, 831–846 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma C., Su L., Seven A. B., Xu Y., Rizo J., Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shu T., Jin H., Rothman J. E., Zhang Y., Munc13-1 MUN domain and Munc18-1 cooperatively chaperone SNARE assembly through a tetrameric complex. Proc. Natl. Acad. Sci. U.S.A. 117, 1036–1041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lai Y., et al. , Molecular mechanisms of synaptic vesicle priming by Munc13 and Munc18. Neuron 95, 591–607.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiao J., et al. , Munc18-1 catalyzes neuronal SNARE assembly by templating SNARE association. eLife 7, e41771 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Misura K. M., Scheller R. H., Weis W. I., Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature 404, 355–362 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Dulubova I., et al. , Munc18-1 binds directly to the neuronal SNARE complex. Proc. Natl. Acad. Sci. U.S.A. 104, 2697–2702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang X., et al. , Syntaxin opening by the MUN domain underlies the function of Munc13 in synaptic-vesicle priming. Nat. Struct. Mol. Biol. 22, 547–554 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park Y., et al. , Controlling synaptotagmin activity by electrostatic screening. Nat. Struct. Mol. Biol. 19, 991–997 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao F., et al. , Are zinc-finger domains of protein kinase C dynamic structures that unfold by lipid or redox activation? Antioxid. Redox Signal. 14, 757–766 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gipson P., et al. , Morphologies of synaptic protein membrane fusion interfaces. Proc. Natl. Acad. Sci. U.S.A. 114, 9110–9115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camacho M., et al. , Control of neurotransmitter release by two distinct membrane-binding faces of the Munc13-1 C1C2B region. eLife 10, e72030 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F., et al. , Vesicle capture by membrane-bound Munc13-1 requires self-assembly into discrete clusters. FEBS Lett. 595, 2185–2196 (2021). [DOI] [PubMed] [Google Scholar]
- 61.Silva M., Tran V., Marty A., Calcium-dependent docking of synaptic vesicles. Trends Neurosci. 44, 579–592 (2021). [DOI] [PubMed] [Google Scholar]
- 62.Milovanovic D., Wu Y., Bian X., De Camilli P., A liquid phase of synapsin and lipid vesicles. Science 361, 604–607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F., et al. , Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat. Struct. Mol. Biol. 14, 890–896 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Hua S. Y., Charlton M. P., Activity-dependent changes in partial VAMP complexes during neurotransmitter release. Nat. Neurosci. 2, 1078–1083 (1999). [DOI] [PubMed] [Google Scholar]
- 65.Prashad R. C., Charlton M. P., SNARE zippering and synaptic strength. PLoS One 9, e95130 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Honigmann A., et al. , Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat. Struct. Mol. Biol. 20, 679–686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pérez-Lara Á., et al. , PtdInsP2 and PtdSer cooperate to trap synaptotagmin-1 to the plasma membrane in the presence of calcium. eLife 5, e15886 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lou X., Korogod N., Brose N., Schneggenburger R., Phorbol esters modulate spontaneous and Ca2+-evoked transmitter release via acting on both Munc13 and protein kinase C. J. Neurosci. 28, 8257–8267 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Südhof T. C., The presynaptic active zone. Neuron 75, 11–25 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X., Wu X., Wu H., Zhang M., Phase separation at the synapse. Nat. Neurosci. 23, 301–310 (2020). [DOI] [PubMed] [Google Scholar]
- 71.Böhme M. A., et al. , Active zone scaffolds differentially accumulate Unc13 isoforms to tune Ca(2+) channel-vesicle coupling. Nat. Neurosci. 19, 1311–1320 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Reddy-Alla S., et al. , Stable positioning of Unc13 restricts synaptic vesicle fusion to defined release sites to promote synchronous neurotransmission. Neuron 95, 1350–1364.e12 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Sakamoto H., et al. , Synaptic weight set by Munc13-1 supramolecular assemblies. Nat. Neurosci. 21, 41–49 (2018). [DOI] [PubMed] [Google Scholar]
- 74.Lu J.et al., . Structural Basis for a Munc13–1 Homodimer to Munc13–1/RIM Heterodimer Switch. PLoS Biol. 4, e192 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miki T., Nakamura Y., Malagon G., Neher E., Marty A., Two-component latency distributions indicate two-step vesicular release at simple glutamatergic synapses. Nat. Commun. 9, 3943 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neher E., Brose N., Dynamically primed synaptic vesicle states: Key to understand synaptic short-term plasticity. Neuron 100, 1283–1291 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Xu T., et al. , Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell 99, 713–722 (1999). [DOI] [PubMed] [Google Scholar]
- 78.Hagen W. J. H., Wan W., Briggs J. A. G., Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
3D density maps of the hexagon, trimer and isolated lateral, upright Munc13C as well as a composite map of six trimers and one hexagon have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-25737, EMD-25738, EMD-25739, EMD-25740, and EMD-25741, respectively. For the original reconstructions the depositions include the corresponding EM density map, both half maps and the mask used for the final FSC calculations. Atomic models have been deposited in the Protein Data Bank under accession number 7T7V for lateral Munc13C, 7T7X for upright Munc13C, 7T7C for the hexagonal arrangement of lateral and upright Munc13C, 7T7R for the upright Munc13C trimer and 7T81 for the lateral hexagon surrounded by upright trimers.