Significance
The targeting of mammalian ErbB receptor signaling by a venom toxin to cause hypersensitivity is a mode of action that has not previously been described. Natural selection of a defensive toxin to target ErbB signaling provides compelling independent evidence for a fundamental role of this receptor and its ligands in mammalian pain. The evolution of a toxin in ant venom to mimic a vertebrate nociceptive hormone serves as an example of both convergent evolution and molecular mimicry, illustrating how natural selection can shape the gene product of one organism to resemble that of another.
Keywords: mimicry, Gilbertian mimicry, EGFR, ErbB, convergent evolution
Abstract
Venoms are excellent model systems for studying evolutionary processes associated with predator–prey interactions. Here, we present the discovery of a peptide toxin, MIITX2-Mg1a, which is a major component of the venom of the Australian giant red bull ant Myrmecia gulosa and has evolved to mimic, both structurally and functionally, vertebrate epidermal growth factor (EGF) peptide hormones. We show that Mg1a is a potent agonist of the mammalian EGF receptor ErbB1, and that intraplantar injection in mice causes long-lasting hypersensitivity of the injected paw. These data reveal a previously undescribed venom mode of action, highlight a role for ErbB receptors in mammalian pain signaling, and provide an example of molecular mimicry driven by defensive selection pressure.
Venom has evolved on more than 100 independent occasions in the animal kingdom, primarily for use in either predation, defense, or both (1). Most venoms are complex mixtures of peptide toxins, and several lines of evidence (for a review, see ref. 2) suggest that these toxins may be “tuned” by natural selection for maximum efficacy in targeted prey, predators, and competitors. Because of the direct link between genotype and functional phenotype, venoms represent excellent model systems for studying evolutionary processes associated with ecological interactions, particularly between predator and prey (3).
Most ant species (Hymenoptera: Formicidae) are venomous and use their venom for both predation and defense. Only recently has the composition and mode of action of ant venoms begun to be investigated in detail. Most peptide toxins in ant venoms appear to be derived from a single gene family known as the aculeatoxins—amphipathic peptides that target cell membranes, a mode of action that engenders a dual function in predation (paralysis of invertebrate prey) and defense (immediate nociception) (4, 5). However, other structural classes of peptide toxins exist in ant venoms and their functions remain enigmatic (4, 6, 7).
Here, we present the discovery of a peptide toxin (Mg1a) from venom of the Australian giant red bull ant Myrmecia gulosa (Fig. 1A). Mg1a has convergently evolved to structurally and functionally mimic vertebrate epidermal growth factor (EGF)–like peptide hormones. This mimicry enables Mg1a to serve a defensive function for the ant by inducing long-lasting hypersensitivity in mammalian predators.
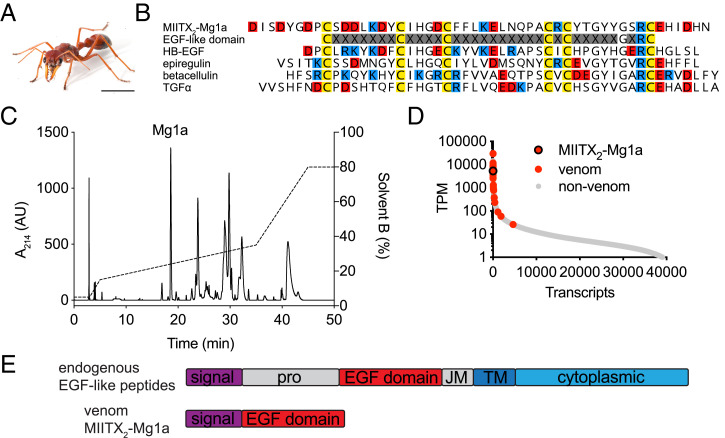
Fig. 1.
Mg1a is a major component of M. gulosa venom. (A) The Australian giant red bull ant M. gulosa. (Scale bar, 5 mm.) (B) Alignment of the sequence of Mg1a with a consensus EGF-like domain and several EGF-like peptide hormones: human HBEGF (UniProt accession no. Q99075), epiregulin (UniProt accession no. O14944), betacellulin (UniProt accession no. P35070), and TGFα (UniProt accession no. P01135). Cys, Glu/Asp, and Arg/Lys residues are colored yellow, red, and blue, respectively. (C) An RP-HPLC chromatogram of M. gulosa venom. The peak corresponding to Mg1a (as determined by MS) is labeled. AU, arbitrary units. (D) The Mg1a-encoding transcript is found in the highly expressed portion of the venom-apparatus transcriptome with an estimated expression level like that of other venom peptides (highlighted in red). (E) The prepropeptide structures of endogenous EGF-like peptide hormones and Mg1a. JM, juxtamembrane domain; pro, propeptide; TM, transmembrane domain.
Results
MIITX2-Mg1a Is a Major Component of M. gulosa Venom.
In a previous study (4), we showed that the venom of M. gulosa was composed primarily of amphipathic peptides derived from the aculeatoxin gene family, but with one exception: a unique cysteine-rich peptide, MIITX2-Mg1a (hereafter Mg1a), which is unrelated to the aculeatoxins. The cysteine spacing and conservation of Gly39 and Arg41 residues (8) in Mg1a suggested that it might adopt an EGF-like fold (Fig. 1B). The EGF-like fold is characteristic of an ancient superfamily of peptide hormones that have diverged across Metazoa (9); in insects, 4 different EGF-like peptide hormones (Spitz, Vein, Gurken, and Keren) have been reported, while mammals have 11 EGF-like peptide hormones: EGF, amphiregulin, epigen, epiregulin, betacellulin, heparin-binding EGF-like growth factor (HBEGF), transforming growth factor-α (TGFα), and 4 neuregulins (10). While all EGF-like peptides retain the same overall fold, divergence of amino acid sequence and intercysteine spacing has resulted in distinct pharmacological profiles. For example, the insect EGF-like peptide hormone Spitz is a ligand of the insect receptor (ErbB), while mammalian EGF-like peptide hormones are ligands of one or more mammalian receptors (ErbB1 to 4, also known as HER1 to 4, including heterodimers thereof) (10). A comparison of the amino acid sequence of Mg1a with that of several of the human EGF (hEGF)–like peptide hormones (Fig. 1B) illustrates the shared sequence features.
In order to assess Mg1a in the context of the whole venom, we collected “stung” venom from several individuals of M. gulosa (by inciting the ants to sting a thin layer of parafilm from which venom droplets were collected) and analyzed the venom by reversed-phase high-performance liquid chromatography (RP-HPLC) (Fig. 1C), using tandem mass spectrometry (MS/MS) to identify the fraction corresponding to Mg1a (SI Appendix, Fig. S1). In the HPLC chromatogram of venom collected from eight ants, one of the major peaks corresponded to Mg1a (Fig. 1C). Analysis of the expression of Mg1a in the venom-gland transcriptome of M. gulosa was also consistent with Mg1a being a major venom component: The transcript encoding Mg1a had an estimated expression level of 3,854 transcripts per million (TPMs), making it one of the most highly expressed transcripts (Fig. 1D). For comparison, the estimated expression level of the endogenous Spitz peptide of M. gulosa in the venom-gland transcriptome was 18 TPMs (and the mature peptide was not detected in the venom).
One other factor supported the designation of Mg1a as a venom component (as opposed to an endogenous hormone): The precursor architecture of almost all invertebrate and vertebrate endogenous EGF-like peptide hormones consists of a signal peptide, a propeptide, and the mature EGF-like peptide domain (most often a single copy), followed by short juxtamembrane and transmembrane domains and finally a cytoplasmic domain (Fig. 1E). The endogenous EGF-like peptide hormones are tethered to the cell membrane after their translation, requiring enzymatic cleavage to release the active peptide (11). By contrast, the precursor of Mg1a consists solely of the secretory signal peptide followed directly by the mature EGF-like peptide—a precursor architecture that resembles that of other directly secreted peptides, including almost all venom peptides.
Mg1a Is an Analog of Mammalian EGF Peptide Hormones.
We used the mature peptide sequence of Mg1a as a query to search (using blastp) the National Center for Biotechnology Information nonredundant (NCBI nr) protein sequence database for related sequences. All of the top 100 alignments were vertebrate EGF-like peptide hormones, predominantly sequences annotated as HBEGF and betacellulin (79/100). When sorted by sequence identity, 5 of the top 10 hits (with sequence identity of 59% and identical intercysteine loop lengths) were to EGF-like peptide hormone sequences of marsupials (Mammalia: Marsupialia), for example the Tammar wallaby (Notamacropus eugenii), koala (Phascolarctos cinereus), and common wombat (Vombatus ursinus) (Fig. 2A). We repeated this analysis using the mature peptide sequence of Mg1a as a query to search (using tblastn) the NCBI Transcriptome Shotgun Assembly (TSA) Database. Again, the top 100 hits were vertebrate EGF-like peptide hormones, and of these the top 2 (with sequence identity of 59% and identical intercysteine loop lengths) were to EGF-like peptide hormone sequences of marsupials: the fat-tailed dunnart (Sminthopsis crassicaudata) and long-nosed bandicoot (Perameles nasuta). For comparison, the Spitz peptide hormones of M. gulosa, the Indian jumping ant (Harpegnathos saltator), western honey bee (Apis mellifera), and Drosophila melanogaster have sequence identities of 26 to 33% with Mg1a and a distinct intercysteine loop length (Fig. 2A). While these analyses were subject to limitations (e.g., the dynamic nature of the search databases and the relatively short length of Mg1a), it was clear that, despite its insect origin, the amino acid sequence and intercysteine loop length of the Mg1a mature peptide resembled vertebrate rather than insect EGF-like hormones, with a consistent signal emerging for similarity to those of marsupials.
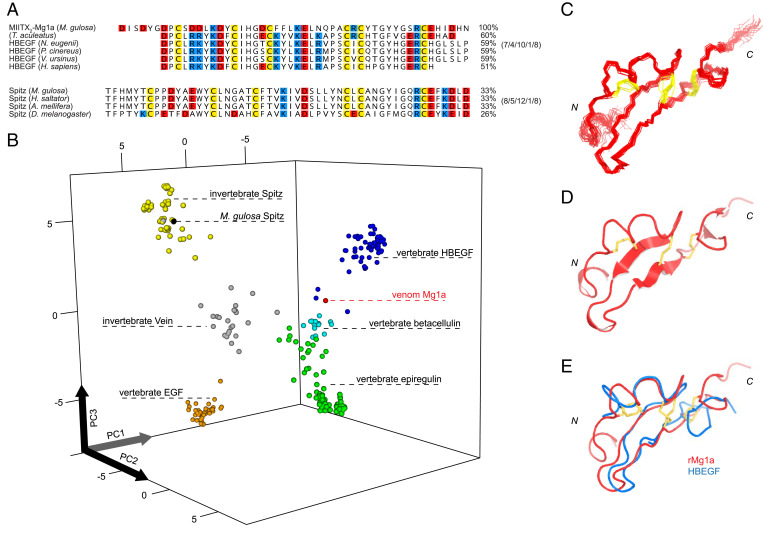
Fig. 2.
Mg1a from the venom of M. gulosa is an analog of the vertebrate EGF-like peptide hormones. (A, Top) Sequence alignment of Mg1a (mature peptide) with the EGF-like domain of the short-beaked echidna (T. aculeatus) peptide, Tammar wallaby (N. eugenii) HBEGF (ARH02599.1), koala (P. cinereus) HBEGF (XP_020860891.1), common wombat (V. ursinus) HBEGF (XP_027712301.1), and Homo sapiens HBEGF (UniProt accession no. Q99075). (A, Bottom) Alignment of Spitz sequences from M. gulosa, the Indian jumping ant (H. saltator), western honey bee (A. mellifera), and D. melanogaster for comparison. Percentage sequence identity with Mg1a is indicated (Right). The intercysteine loop length of each group is indicated (Far Right). (B) Quantitative map of sequence space for the EGF-like peptide hormone superfamily. Each point represents a nonredundant sequence. Points are colored as follows: EGF, orange; HBEGF, blue; betacellulin, cyan; epiregulin, green; Spitz, yellow; Vein, gray; Mg1a, red; M. gulosa Spitz, black. (C) Ensemble of the final 20 structures of rMg1a superimposed over the backbone heavy atoms of residues 7 to 47 (showing backbone heavy atoms). N and C termini are labeled. (D) Ribbon representation of the closest-to-average structure of rMg1a. (E) Alignment of the closest-to-average structure of rMg1a (red) with that of HBEGF (blue; PDB ID code 1XDT), superimposed over all backbone heavy atoms. Disulfide bonds are shown in yellow.
Using principal-component analysis (PCA), we generated a quantitative map of protein sequence space for Mg1a and the vertebrate and invertebrate EGF-like peptide hormones (EGF, HBEGF, betacellulin, epiregulin, Spitz, and Vein) based on the biophysical properties of each residue in the EGF-like domain of each mature peptide sequence (Fig. 2B) (12). The map produced well-defined groupings for each of the EGF-like peptide hormone families. Consistent with the results of our blast analyses, Mg1a grouped near the vertebrate families (betacellulin, HBEGF, and epiregulin), in contrast to endogenous M. gulosa Spitz, which grouped with other Spitz sequences.
We then tested whether the observed relationships in primary structure were mirrored by the tertiary structure. For these experiments, we produced uniformly 15N, 13C–labeled Mg1a by recombinant expression in bacteria (SI Appendix, Fig. S2). The recombinant peptide was produced with an additional Gly residue at the N terminus, and we refer to it as rMg1a. One-dimensional (1D) 1H and 2D [1H-15N]-HSQC (heteronuclear single quantum coherence) NMR spectra indicated that rMg1a adopted a single major conformation and was highly structured in solution (SI Appendix, Fig. S3). Structures were generated in CYANA using 671 nuclear Overhauser effect (NOE)–derived distance constraints and 75 dihedral-angle constraints. Structural constraints and statistics are summarized in SI Appendix, Table S1. Determination of the solution structure of rMg1a confirmed that it had an EGF-like fold (Fig. 2C) with disulfide connectivity of Cys I–III, II–IV, and V–VI and two small antiparallel β-sheets: The longer sheet (Gly21–Phe24 and Cys34–Tyr37) makes up the central core of the fold, while the shorter sheet (Tyr39–Tyr40 and His46–Ile47) is near the C terminus. Alignment of the closest-to-average structure of rMg1a with those of betacellulin (Protein Data Bank [PDB] ID code 1IP0) (Fig. 2D), HBEGF (PDB ID code 1XDT), epiregulin (PDB ID code 1K36), and Spitz (PDB ID code 3CA7) yielded backbone heavy-atom rmsd values of 1.37, 1.84, 1.96, and 2.87 Å, respectively. Thus, at the level of both primary (sequence identity, intercysteine loop lengths, biophysical properties) and tertiary structure, the venom peptide Mg1a more closely resembles vertebrate EGF-like peptide hormones than insect Spitz.
Together, these results led us to hypothesize that the natural target of Mg1a was not the endogenous insect ErbB receptor but a vertebrate ErbB receptor, and that its presence in the venom of M. gulosa reflects its evolution as a defensive weapon against vertebrate predators. The fact that several of the top alignments in the blast search were with EGF-like peptide hormones from Australian mammals implicated predation pressure by the insectivores among these as a potential driver of this evolutionary process. Several Australian mammals are known to feed opportunistically on solitary Myrmecia workers, but only one, the short-beaked echidna (Tachyglossus aculeatus), a monotreme, will attack colonies to feed on the brood (eggs, larvae, and pupae) (13). Because the latter likely represents a much stronger selection pressure on Myrmecia than occasional predation on solitary workers, we predicted that Mg1a may have evolved under selection pressure from—and therefore resemble an endogenous EGF-like peptide of—the echidna or other now-extinct mammals with similar foraging behavior. To further examine this potential evolutionary scenario, we searched the recently published genome of the echidna (14) using Mg1a as the query. Consistent with our hypothesis, we identified a peptide sequence in the echidna genome which, when aligned with Mg1a, was 60% identical (Fig. 2A). Thus, postdivergence of insect and vertebrate EGF-like peptide hormones, the ant venom peptide Mg1a appears to have convergently evolved to more closely resemble the EGF-like hormone(s) of its vertebrate predator(s).
Mg1a Is a Potent Ligand of Mammalian ErbB1.
If Mg1a is the product of selection pressure to target a mammalian ErbB receptor, potent activity at this receptor would be expected. We determined the potency of rMg1a at human ErbB1 using a bioluminescence resonance energy transfer (BRET) assay that quantifies the dimerization (required for activation) of the human ErbB1 receptor (hErbB1) transiently expressed in HEK293T cells (SI Appendix, Fig. S4). rMg1a was active at hErbB1 (Fig. 3A) with a half-maximal effective concentration (EC50) of 6.3 nM (95% CI, 2.0 to 16.6 nM, n = 3) (Fig. 3B). For comparison, hEGF had an EC50 of 10.5 nM (95% CI, 6.4 to 16.6 nM, n = 3) in the same assay.
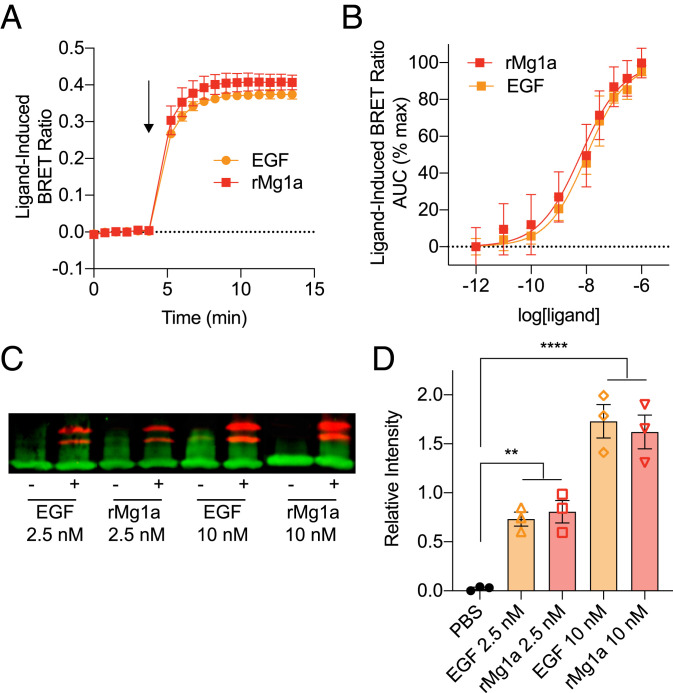
Fig. 3.
Mg1a is a potent ligand of mammalian ErbB1. (A) Ligand-induced BRET ratio of HEK293T cells transiently expressing hErbB1/Rluc8 and Grb2/Venus following stimulation with 100 nM rMg1a or hEGF. The arrow indicates the time of ligand addition. Data are mean ± SEM (n = 3). (B) Concentration–response curves of ligand-induced BRET ratio for HEK293T cells transiently expressing hErbB1/Rluc8 and Grb2/Venus following stimulation with rMg1a or hEGF. Data are mean ± SEM (n = 3). AUC, area under the curve. (C) Representative Western blot showing phosphorylated Erk1 and Erk2 (red) and the control housekeeping protein GAPDH (green) after stimulation of HEK293 cells transiently expressing hErbB1 with 2.5 or 10 nM rMg1a or hEGF. (D) Quantitative Western blot data showing relative intensity over the GAPDH loading control. Data are expressed as mean ± SEM (n = 3). **P < 0.01, ****P < 0.0001.
Western blot analysis was used to investigate the ability of rMg1a to induce phosphorylation of extracellular signal-regulated kinases (ERKs) 1 and 2, downstream signaling molecules of ErbB receptors (15). Mg1a caused a concentration-dependent increase in phosphorylation of Erk1/2 (P < 0.01 for 2.5 nM and P < 0.0001 for 10 nM, Šídák’s multiple comparisons), equivalent to that caused by hEGF (Fig. 3 C and D).
These data indicate that, consistent with its vertebrate-like primary and tertiary structure, rMg1a is a potent ligand of mammalian ErbB1 with activity comparable to that of hEGF, an endogenous ligand of the receptor.
Intraplantar Injection of Mg1a in Mice Causes Long-Lasting Hypersensitivity.
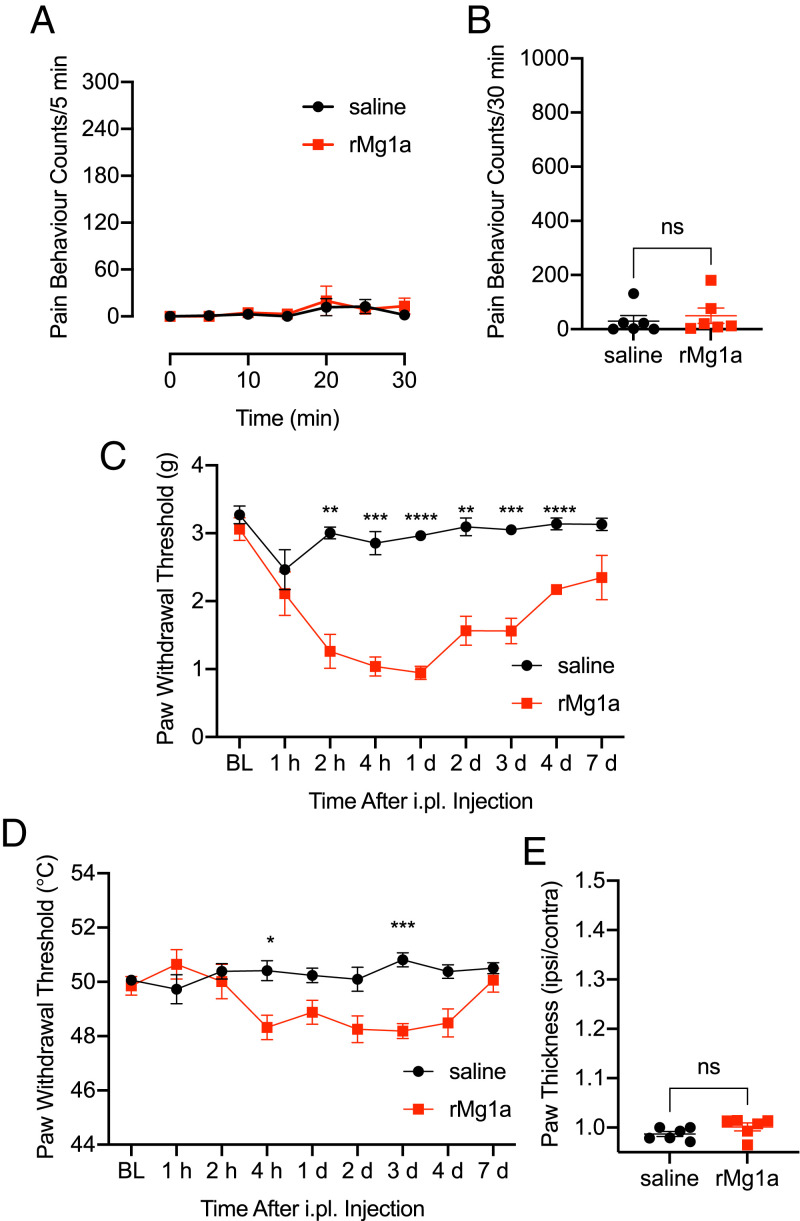
Myrmecia use their sting for both predation (to incapacitate other arthropods, which they feed to their larvae) and defense (to induce pain in, and thus deter, potential predators). Our results suggested that Mg1a has evolved as a defensive weapon against vertebrate predators. Accordingly, we observed no effect on crickets (Acheta domesticus), blowflies (Lucilia cuprina), or fruit flies (D. melanogaster) after intraabdominal injection of rMg1a up to a dose of 41 nmol/g (SI Appendix, Fig. S5). Shallow intraplantar (i.pl.) injection of rMg1a (200 pmol) in mice did not cause spontaneous nocifensive behavior (sum of pain-behavior counts at 30 min for saline versus rMg1a, P = 0.58, t test, t = 0.5712, degrees of freedom [df] 10) (Fig. 4 A and B). However, it caused a significant decrease in paw-withdrawal threshold to both mechanical (P < 0.01 to P < 0.0001, two-way ANOVA, Šídák’s multiple comparisons) and thermal (P < 0.05 to P < 0.001, two-way ANOVA, Šídák’s multiple comparisons) stimuli (Fig. 4 C and D). This hypersensitivity was delayed in onset (beginning at 2 to 4 h after injection) and long-lasting (3 to 4 d after injection). Over the time course of this experiment, we observed no visual signs of inflammation (e.g., redness or swelling), and paw thickness measured at 3 d following injection was not significantly different between saline- and rMg1a-treated mice (P = 0.16, t test, t = 1.501, df 10), indicating that the hypersensitivity elicited by injection of rMg1a was independent of overt inflammation.
Fig. 4.
Intraplantar injection of Mg1a in mice caused long-lasting hypersensitivity. (A) Spontaneous pain-behavior counts for 30 min following i.pl. injection of rMg1a (200 pmol) or saline. Data are mean ± SEM (n = 6 per group). (B) Sum of pain-behavior counts at 30 min after i.pl. injection of 200 pmol rMg1a. Data are mean ± SEM (n = 6 per group). ns, not significant. (C) Paw-withdrawal threshold to mechanical stimulus. Data are mean ± SEM (n = 6 per group). **P < 0.01, ***P < 0.001, ****P < 0.0001, two-way ANOVA, Šídák’s multiple comparisons. Note: The x axis is nonlinear. (D) Paw-withdrawal threshold to thermal stimulus. Data are mean ± SEM (n = 6 per group). *P < 0.05, ***P < 0.001, two-way ANOVA, Šídák’s multiple comparisons. Note: The x axis is nonlinear. (E) Paw thickness (ipsilateral/contralateral) measured 3 d after i.pl. injection of 200 pmol rMg1a. Data are mean ± SEM (n = 6 per group).
These data are consistent with our hypothesis that Mg1a has evolved specifically as a defensive weapon against mammalian predators and reveal that its likely function is to cause long-lasting hypersensitivity at the injection site.
Origin of Weaponized EGF-like Peptides in Ant Venoms.
We next investigated how broadly EGF-like peptides were found in the venoms of ants and other taxa. We began by exploring other Myrmecia species (subfamily Myrmeciinae). To this end, we generated a venom-gland transcriptome of Myrmecia chrysogaster, the gold-tailed bull ant, which is also endemic to Eastern Australia. We identified a single, highly expressed (7,303 TPMs), ortholog of Mg1a that we named MIITX2-Mc1a. This ortholog shared the same precursor architecture as Mg1a and the mature peptide was also analogous to vertebrate EGF-like peptide hormones (SI Appendix, Fig. S6 A and B). As was the case with Mg1a, several EGF-like hormones of marsupials were among the top hits to Mc1a (the mature peptide) in a blastp search of the NCBI nr database.
We then explored ant venoms outside of the subfamily Myrmeciinae. In a venom-gland transcriptome of Rhytidoponera metallica (subfamily Ectatomminae), the greenhead ant, which is widely distributed in Australia, we identified a suite of precursors encoding EGF-like peptides with similar signal peptide sequences and the same precursor architecture as Mg1a (SI Appendix, Fig. S6A), indicative of homology and consistent with a role in venom. A search (using blastp) of the NCBI nr protein database suggested that the R. metallica peptides were most closely related to vertebrate betacellulin (SI Appendix, Fig. S6A), while PCA suggested an intermediate positioning between the invertebrate EGF-like domains Vein and Spitz and the vertebrate EGF-like domains (SI Appendix, Fig. S6C). Another EGF-like peptide, Mri1a (UniProt accession no. A0A6G9KJM3; European Molecular Biology Laboratory accession no. QIQ51456.1), has been reported in the venom of the Eurasian ant Manica rubida (subfamily Myrmicinae) (6). Analysis of its precursor sequence and architecture revealed that it too belongs to the same gene family as Mg1a (SI Appendix, Fig. S6A). In the case of Mri1a, however, according to both blastp and PCA, the mature peptide sequence was clearly most similar to that of insect Spitz, as was the peptide’s intercysteine spacing (SI Appendix, Fig. S6 A and C). Thus, multiple species of ant have EGF-like peptide toxins in their venoms—in Myrmecia these venom peptides have evolved to resemble the EGF-like hormones of these ants’ vertebrate predator(s), while in M. rubida the venom peptide resembles the insect hormone Spitz and presumably reflects selection for efficacy in this ant’s insect predators/prey.
To further investigate the origin of EGF-like peptide toxins in ant venoms, we used tblastn to search (using the Mg1a, Mc1a, and Mri1a precursor protein sequences as queries) the NCBI TSA Database and the Hymenoptera Genome Database (16), which included genomic datasets from 58 species of Hymenoptera including 24 ant species. We identified additional homologous precursor sequences from genomes or whole-body transcriptomes of five other ant species (Myrmica sulcinodus, Myrmica rubra, Formica aquilonia, Pogonomyrmex californicus, and Pogonomyrmex barbatus). No homologous ant venom EGF-like toxin sequences were detected in hymenopterans outside of the formicoid clade of the Formicidae, suggesting that within Hymenoptera, the EGF-like toxin gene family is a unique innovation of the formicoid ant lineage. Some of the detected sequences encoded mature peptides which, like Mri1a, were insect-like, while other sequences encoded mature peptides which, like Mg1a and Mc1a, were vertebrate-like (according to blastp searches of the NCBI nr database). However, of the predicted vertebrate-like mature peptides, none exhibited the similarity to marsupial/monotreme EGF-like hormones that we observed for the Myrmecia venom peptides.
The similarity of the venom peptide Mri1a to the insect hormone Spitz was suggestive of an evolutionary link between the ant EGF-like venom peptide toxins and the endogenous insect hormone; that is, the ant venom EGF-like gene family appears to be derived from the spitz gene. We were unable to resolve the phylogeny of the ant venom EGF-like toxins, presumably due to saturation effects (SI Appendix, Fig. S7). However, endogenous Spitz hormones from diverse hymenopterans grouped closely together on short branches indicative of minimal sequence divergence. By contrast, the ant venom EGF-like toxins, which were derived from a much narrower range of species, exhibited high levels of sequence divergence—a signature of many gene families that have been co-opted for use in venoms (17, 18), and suggestive of functional divergence. The high level of sequence divergence in the ant venom EGF-like peptides may have been driven by selection for efficacy at the receptor(s) of different predators or prey. For example, the similarity of Mri1a from M. rubida to insect Spitz suggests that this peptide has retained its ancestral activity, to target the insect ErbB receptor of this species’ insect predators/prey, either as a defensive or predatory weapon. On the other hand, the structural and functional similarity of Mg1a and Mc1a from Myrmecia spp. to vertebrate EGF-like hormones suggests that they evolved as a defensive weapon to target the ErbB receptor(s) of the ant’s vertebrate predator(s).
Outside Formicidae, there have been reports of EGF-like peptides in the venoms of sea anemones of the genus Stichodactyla (19–21). These venom peptides share a similar “refined” precursor architecture to the ant venom EGF-like peptides. Analysis of the mature peptide sequences revealed highest similarity to the arthropod EGF-like peptide Vein (SI Appendix, Fig. S6B), suggesting that the receptor for these venom peptides is the arthropod ErbB homolog and that their function is for predation or defense against arthropods. This represents an independent convergent incorporation of EGF-like peptides into venom, the origins of which are unclear given the reported absence of an endogenous EGF signaling system in Cnidaria (9).
Discussion
Here, we present the discovery and characterization of an EGF-like peptide from venom of the ant M. gulosa, which is a potent ligand of the mammalian ErbB1 receptor and causes long-lasting hypersensitivity in mammals. Most algesic venom toxins characterized to date target ion channels or receptors in the nervous or cardiovascular system, or target cell membranes. The exploitation of ErbB signaling by a toxin in order to cause pain extends the known molecular mechanisms of envenomation beyond conventional neurotoxins, cardiotoxins, and membrane perturbants. Moreover, while most described venom toxins have an immediate and direct effect, the delayed and long-lasting hypersensitivity caused by Mg1a is, to our knowledge, unprecedented.
In the context of defense, the advantage of causing immediate nociception is easily understood. Indeed, stings of Myrmecia do cause immediate intense pain, which results from the action of one or more membrane-targeting peptides (4). However, the advantages of a delayed and indirect, albeit long-lasting, effect are less obvious. One explanation for this effect is that an initial dose of Mg1a, as might occur in an attack by a predator on a Myrmecia nest, would cause long-lasting hypersensitivity and perhaps reduce the duration of the attack, discourage future attacks, or enhance the algesic actions of subsequent exposure to other pain-causing venom peptides. Further direct observations of Myrmecia–predator interactions are necessary to test this hypothesis.
One important outcome of this study is the implication of ErbB signaling in mammalian pain—more specifically long-lasting hypersensitivity. The fact that the exploitation of this signaling pathway for defensive purposes in Myrmecia has been naturally selected for implies that it may play a fundamental role in nociception in mammals, one that until recently has gone largely unnoticed. Mammalian EGF-like peptide hormones have mostly been studied in the context of development, although they also play important physiological roles in adults. For example, some are major components of wound fluid (22) and are involved in epithelialization in skin wound healing (23). ErbB receptor inhibitors are used for the treatment of a range of cancers, and several case reports have indicated that ErbB receptor inhibition may relieve certain cancer pain (for a review, see ref. 24). Recent independent studies have reported roles for epiregulin-, EGF-, and HBEGF-ErbB signaling in pain (25–27) and, as in the present study, each reported a specific role in hypersensitivity.
From these studies, we can begin to formulate a hypothesis for the role played by ErbB signaling in pain. Following an injury, high concentrations of EGF-like peptide hormones may be released at the wound site where they serve a critical role in wound healing and, independent of inflammation, sensory neurons innervating the site of injury may become sensitized via activation of ErbB receptors expressed on nerve terminals or cells interacting with these. While long-term pain is often viewed as pathological, hypersensitivity of a wound site may be adaptive (i.e., for protection of a wound during healing). Mg1a in the venom of M. gulosa may have evolved to exploit this pain pathway in the absence of a genuine injury. Thus, activation of ErbB by certain EGF-like peptide hormones may represent an important noninflammatory mediator of wound hypersensitivity, and inhibition of these pathways may be an effective treatment for certain pain pathologies.
Our results allow us to build a hypothesis of the evolution of EGF-like peptide toxins in the venoms of ants. We can deduce that early in the formicoid lineage, a duplication of a gene, probably that encoding the peptide hormone Spitz, allowed its modification for direct secretion (i.e., loss of domains other than the signal and mature peptide or recombination of the mature peptide with another signal peptide) and recruitment to the venom. Supporting this interpretation, we found that the venom of M. rubida contains a peptide that closely resembles endogenous Spitz, suggesting retention of its ancestral function targeting the insect ErbB receptor but which is now directed exogenously for predation and/or defense against other insects. In Myrmecia, and possibly other lineages of ants, selection pressure for defense against one or more vertebrate predators resulted in the evolution of the venom peptide to resemble, both structurally and functionally, a nociceptive peptide hormone of the predator(s).
This scenario draws strong parallels with the recent discovery of an insulin-like toxin in the venom of the fish-hunting cone snail Conus geographus, that has convergently evolved to mimic, structurally and functionally, the endogenous insulin hormone of its fish prey (28). When hunting, the snail releases the venom insulin into the water, where it acts on the fish’s endocrine system to cause sedation, thus facilitating prey capture.
Both of these toxins are examples of convergent evolution and, more specifically, of biological mimicry—where one organism (the mimic) gains a selective advantage by mimicking an aspect (or multiple aspects) of another organism (the model) in order to deceive it or another (the dupe). The scenario involving the cone snail and the fish is a form of aggressive molecular mimicry, known as Batesian–Wallacian mimicry (a bipolar aggressive system in which the model belongs to the dupe’s species and the dupe is deceived by the model). The scenario of the bull ant and its predator(s) is different. It is a rare form of mimicry known as Gilbertian mimicry, a bipolar protective system in which the model belongs to the dupe’s species and the dupe is deceived by the model (29). Gilbertian mimicry has been described in certain species of the plant genus Passiflora, where parts of the plants have evolved to visually mimic Heliconius butterfly eggs that are near to hatching (30). To avoid intraspecific competition, Heliconius females avoid ovipositing on plants already bearing such eggs (or egg mimics) and thus the plant is afforded protection (31). In this study, the scenario presented is analogous to the Gilbertian mimicry described in Passiflora and Heliconius, yet it occurs not at the organismic level but at the molecular level. Here, a molecule (Mg1a) produced by the ant M. gulosa mimics an endogenous nociceptive molecule of its vertebrate predator(s), and its injection causes long-lasting hypersensitivity and presumably a reduction in predation, thus protecting the ant. While Gilbertian mimicry appears to be a rare phenomenon (until now it had only been described in the Passiflora–Heliconius interaction), we predict that more examples of this type of mimicry may have evolved, particularly in cases where nociceptive molecules are concerned.
This study illustrates how natural selection can mold the gene product of one organism to resemble that of another (in this case, the gene product of the prey resembles that of the predator). It is an example of both convergent evolution and an unusual form of molecular mimicry, one that brings to mind the ancient proverb “To know your enemy, you must become your enemy.”
Materials and Methods
Collection of Venom and Purification of Mg1a.
Eight adult female M. gulosa were collected from a single site near Brisbane, QLD, Australia. Venom was acquired by inciting ants to sting a thin layer of parafilm, which was rinsed with purified water.
Approximately 500 μg of venom was separated on a Phenomenex Gemini NX-C18 column (250 × 4.6 mm; particle size 3 μm; pore size 110 Å) using a gradient of 5 to 80% solvent B (90% acetonitrile [ACN], 0.05% trifluoroacetic acid) over 60 min at a flow rate of 1 mL⋅min−1. Fractions were collected on the basis of absorbance at 214 nm and matrix-assisted laser/desorption ionization MS was used to confirm the identity of the peptide(s) present in each fraction. The fraction corresponding to Mg1a was quantified via absorbance at 280 nm (NanoDrop 2000C spectrophotometer; Thermo Fisher Scientific), divided into aliquots, and dried by vacuum centrifugation.
The complete primary structure of Mg1a was confirmed by liquid chromatography MS/MS analysis of native, reduced, and alkylated peptide. A 2-µg aliquot of Mg1a was analyzed on an AB Sciex TripleTOF 5600 mass spectrometer equipped with a Turbo-V source heated to 550 °C. A second 2-µg aliquot was first reduced and alkylated (incubated in 50 mM ammonium carbonate, pH 11, 48.75% ACN, 1% 2-iodoethanol, 0.25% triethylphosphine for 1 h at 37 °C) and analyzed on the same instrument. Experimental MS/MS spectra of native, reduced, and alkylated Mg1a were compared against theoretical peak lists for each peptide generated using MS-Product in ProteinProspector v5.22.1 (https://prospector.ucsf.edu/prospector/).
Principal-Component Analysis.
Sequences of EGF, HBEGF, betacellulin, epiregulin, Spitz, and Vein were downloaded from UniProtKB, and were truncated to the EGF-like domain of each. Misannotated and fragmented sequences were removed and identical sequences were clustered using Cd-hit v4.6 (32). The remaining 313 unique sequences were aligned using local pairwise alignment (L-INS-i) with the regional alignment tool (v0.2) in MAFFT v7.304b (33). After adjusting the alignment of structurally conserved cysteine residues, we then clustered and projected the biophysical properties of each sequence in a multidimensional sequence space as described previously (12). Briefly, the alignment was converted into a numerical matrix and analyzed using PCA to summarize the main covarying sets of properties using sequence space (R) codes obtained from GitHub, https://github.com/TS404/SeqSpace. The variables used were molecular mass (Da), net charge (coulombs), hydrophobicity (Doolittle index), disorder propensity (TOP-IDP), disulfide potential (binary descriptor), and occupancy (binary descriptor).
Recombinant Expression of Mg1a.
A synthetic gene encoding Mg1a, with codons optimized for high-level expression in Escherichia coli, was produced by GeneArt (Life Technologies). The synthetic gene also incorporated a tobacco etch virus (TEV) protease recognition site immediately N-terminal to the Mg1a gene to minimize the presence of additional nonnative residues after protease cleavage. The synthetic gene was inserted into a pLicC expression plasmid (derived from pMCSG9) (34), which enables expression of Mg1a as a His6-tagged maltose-binding protein (MBP) fusion protein, with the incorporated TEV protease recognition site between the MBP- and Mg1a-coding regions. E. coli BL21(λDE3) were transfected with 10 ng/μL plasmid, incubated on ice, and then heat-shocked. Cells were plated on Luria–Bertani (LB) agar containing 100 μg/mL ampicillin (Astral Scientific) and grown for 18 h. Suitable single colonies were used to spike LB broth containing 100 μg/mL ampicillin, and grown for 18 h at 37 °C before being transferred to media for autoinduction. A modified autoinduction medium was used based on ZYP5052 (35), where lactose was replaced by a low concentration of isopropyl β-d-1-thiogalactopyranoside (IPTG) (36), resulting in increased expression yields. The medium, referred to as ZYP5050I (due to the presence of 0.5% [weight/volume; wt/vol] glycerol, 0.05% [wt/vol] glucose, 0.0% lactose, and the presence of IPTG), contained 10 g/L tryptone, 5 g/L yeast extract, 54.3 mM glycerol, 2.8 mM glucose, 25 mM (NH4)2SO4, 50 mM KH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 5 μM FeCl3, 2 μM CaCl2, 1 μM MnCl2, 1 μM ZnSO4, 0.2 μM CoCl2, 0.2 μM CuCl2, 0.2 μM Na2MoO4, 0.2 μM Na2SeO3, 0.2 μM H3BO3, 40 μM IPTG, 100 μL/L Antifoam SE-15 (Sigma), and 100 μg/mL ampicillin. The culture was incubated at 30 °C, with shaking at 300 rpm, until the optical density at 600 nm (OD600) reached 0.8 to 1.2, as determined via spectrophotometry (SmartSpec 3000; Bio-Rad). The temperature was then lowered to 17 °C, and the culture was grown until the OD600 was >5 (∼21 h later). The cells were then harvested via centrifugation (6,200 × g, 20 min, 4 °C), the supernatant was discarded, and the pellet was dissolved in Tris⋅HCl NaCl (TN) buffer (20 mM Tris, 200 mM NaCl, pH 8.0).
The cell pellet was homogenized and lysed using a constant-pressure cell disruptor (25 to 32 kpsi; TS Series; Constant Systems; 6 to 14 °C). The cell lysate was then centrifuged (39,000 × g, 30 min, 4 °C) and the soluble lysate was collected. The soluble lysate was then incubated with 50% Ni-NTA Superflow resin (Qiagen) equilibrated in TN buffer for 30 min at 4 °C with gentle mixing. The mixture was then added to an empty gravity-flow column and washed with TN buffer containing 15 mM imidazole to remove weakly bound bacterial proteins. Next, the His-tagged protein was eluted in TN buffer containing 250 mM imidazole. Finally, the sample was concentrated and exchanged into buffer without imidazole via a repeated concentration–dilution process through a Millipore Amicon Ultra-15 30K centrifugal filter device (3,000 rpm, 10 min). TEV protease (50 μg/mL), produced in-house using the protocol described by ref. 37, and dithiothreitol (0.5 μM) were added to the protein solution to cleave the recombinant Mg1a from the His6-MBP fusion protein. The cleavage sample was incubated overnight (4 °C, with gentle rolling) and the final product was purified by RP-HPLC as described above.
To produce isotopically labeled rMg1a for NMR studies, E. coli were grown in a minimal medium containing [13C]glucose, [13C]glycerol, and 15NH4Cl as the sole carbon and nitrogen sources, respectively. Again, an autoinduction method was used (35). After depletion of the [13C]glucose, at which point induction of protein expression begins, [13C]glycerol is utilized by the cells for continued growth to high density. In order to minimize the cost of expensive [13C]reagents, cells were precultured in rich media prior to further culturing and expression at a high cell density in minimal media (38), and [13C]glycerol concentration was reduced from 0.5 to 0.2% in the final media. Cells were precultured in 2 L buffered Terrific Broth containing 100 μg/mL ampicillin at 37 °C (180 rpm) until OD600 reached 1.2 to 1.5, and then the cells were pelleted and resuspended in 250 mL phosphate buffer (5 mM Na2SO4, 50 mM KH2PO4, 60 mM Na2HPO4) to remove any residual rich media. Cells were pelleted again via centrifugation, the supernatant was discarded, and pellets were resuspended in 250 mL autoinduction medium. The minimal autoinduction medium, which we called P-2050I (due to the presence of 0.2% [wt/vol] glycerol, 0.05% [wt/vol] glucose, 0.0% lactose, and the presence of IPTG), contained 1 mM MgSO4, 10 μM FeCl3, 4 μM CaCl2, 21.7 mM [13C]glycerol, 2.8 mM [13C]glucose, 9.2 mM 15NH4Cl, 5 mM Na2SO4, 50 mM KH2PO4, 60 mM Na2HPO4, 1 mM MgSO4, 5 μM FeCl3, 2 μM CaCl2, 1 μM MnCl2, 1 μM ZnSO4, 0.2 μM CoCl2, 0.2 μM CuCl2, 0.2 μM Na2MoO4, 0.2 μM Na2SeO3, 0.2 μM H3BO3, 0.2 µM vitamin B12, 0.2 µM nicotinic acid, 0.2 µM pyridoxine⋅HCl, 3 µM thiamine⋅HCl, 0.2 µM p-aminobenzoic acid, 5 nM folic acid, 5 nM riboflavin, 40 µM IPTG, 100 μg/mL ampicillin, and 100 μL/L Antifoam SE-15. Cells were grown in darkness at 30 °C for 2.5 h before the temperature was switched to 17 °C and cells were cultured for a further 22 h with shaking at 300 rpm. After incubation, cells were harvested and lysed, and Mg1a was purified as outlined above. The extent of 13C and 15N incorporation was determined via electrospray ionization MS/MS.
NMR Spectroscopy and Structure Determination.
NMR samples containing 15N/13C-labeled rMg1a were prepared by dissolving lyophilized peptide in 300 μL of NMR buffer (20 mM sodium phosphate, pH 6.0, 5% 2H2O) at a concentration of 350 µM. For resonance assignments, spectra were acquired at 293 K using a Bruker Avance III 600-MHz spectrometer equipped with a cryogenically cooled probe. Two-dimensional [15N-1H]-HSQC, 3D HNCO, 3D HNCA, 3D CBCA(CO)NH, 3D HBHA(CO)NH, and 3D HNCACB spectra were used to generate backbone-resonance assignments while 3D HCCH-correlation spectroscopy and 2D [13C-1H]-HSQC spectra were used for side-chain carbon and proton assignments. Nonuniform sampling data were reconstructed with compressed sensing using qMDD, uniform sampling data were processed in TOPSPIN (v3.2), and chemical shift analysis was performed using CcpNmr Analysis (v2.1.5) (39). Interproton distance restraints were obtained from 3D 13C-aliphatic, 13C-aromatic, and 15N-NOE spectroscopy (NOESY)-HSQC spectra (250-ms mixing time). The uniformly sampled NOESY spectra were acquired at 293 K on a Bruker Avance II+ 900-MHz spectrometer equipped with a cryoprobe and processed using the Rowland NMR Toolkit; rnmrtk.uchc.edu/rnmrtk/RNMRTK.html). They were manually peak-picked and automatically assigned using CYANA (v3.98.13) (40). Dihedral-angle restraints were obtained from the TALOS-N prediction program (41) with the constraint range for structural calculation set to twice the SD. CYANA was used to calculate 200 structures of rMg1a, and then the top 30 were selected based on the final CYANA target function. With ranking based on their stereochemical properties as judged by MolProbity (42), an ensemble of 20 structures was chosen to represent the final solution structure of rMg1a.
Bioluminescence Resonance Energy Transfer Assay.
The plasmids pcDNA3-ErbB1/Rluc8 and pcDNA3-Grb2/Venus have been described previously (43) and were gifts of Kevin Pfleger, Harry Perkins Institute of Medical Research, University of Western Australia, Perth, WA, Australia.
HEK293T cells were seeded at 25,000 cells per well in poly-d-lysine (0.1 mg/mL; Sigma-Aldrich; P6407) –coated white 96-well plates 24 h before transfection. FuGene (Promega; E2311) was used, as per the manufacturer’s instructions, to transiently transfect 5 ng pcDNA3-ErbB1/Rluc8 and 7 ng pcDNA3-Grb2/Venus per well. At 24 h posttransfection, cells were serum-starved in FluoroBRITE (Gibco; A18967) supplemented with 25 mM Hepes (Gibco; 15630), 2 mM l-glutamine (Gibco; 25030), penicillin (100 U/mL), streptomycin (100 µg/mL), and amphotericin B (250 ng/mL; Gibco; 15240). BRET-based assays were performed 6 h later at 37 °C using a Tecan Spark microplate reader. The medium in each well was replaced with Dulbecco’s phosphate-buffered saline (DPBS; Gibco; 2160) containing 5 µM coelenterazine-h (Promega; S2011). Light emissions at 430 to 485 nm (Rluc8) and 520 to 560 nm (Venus) were measured at 45-s intervals before and after stimulation with vehicle or ligand. The BRET ratio was calculated as the light emitted at 520 to 560 nm divided by the light emitted at 430 to 485 nm. The average of triplicate vehicle-treated wells was subtracted from each ligand-treated condition to give the ligand-induced BRET ratio, correcting for background emissions.
EGF Receptor Transfection and Western Blot.
HEK293 cells were seeded on 12-well plates and grown at 37 °C, 5% CO2 until they reached ∼80% confluence. Transient transfection was achieved through incubation with 15 ng of plasmid containing human ErbB1 and Lipofectamine LTX reagent diluted in Opti-MEM. At 5 h posttransfection, the medium was aspirated off and replaced with Dulbecco’s modified Eagle’s medium (DMEM) supplemented with l-glutamine and 10% fetal bovine serum. Cells were grown for a further 24 h, and then the medium was aspirated off and replaced with serum-free DMEM to remove growth factors present in the medium. After 12 h, cells were stimulated for 5 min with either hEGF (R&D Systems; 236-EG), rMg1a, or PBS (negative control). Following stimulation, cells were placed on ice, washed twice with cold PBS, and lysed using RIPA lysis buffer (50 mM Tris⋅HCl, pH 7.5, 100 mM NaCl, 2 mM ethylenediaminetetraacetate [EDTA], 50 mM NaF, 0.1% [wt/vol] sodium dodecyl sulfate, 0.5% [wt/vol] sodium deoxycholate, 1% [vol/vol] Triton X-100, 20 mM sodium pyrophosphate, 10 mM sodium orthovanadate) as well as cOmplete EDTA-free protease inhibitor mixture (Roche) and a PhosSTOP phosphatase inhibitor tablet (Roche). The cell lysates were gently rocked for 1 h at 4 °C and centrifuged (15,000 × g for 15 min), and the supernatant was collected and frozen until analysis.
The supernatants were thawed, electrophoresed on a sodium dodecyl sulfate–polyacrylamide gel (12.5% resolving gel), and then transferred to a polyvinylidene difluoride membrane via electrophoretic transfer. Membranes were washed with PBS containing 0.1% Tween 20 (PBS/Tween) and then blocked with 1:1 PBS/Tween and Intercept (PBS) Blocking Buffer (LI-COR Biosciences; 927-70001) to prevent nonspecific binding. Membranes were then incubated overnight at 4 °C with primary antibodies against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and phosphorylated Erk1 and Erk2. The following morning, membranes were washed three times and then incubated with secondary anti-mouse and anti-goat fluorescent antibodies for 2 h before once again washing three times and visualizing using an Odyssey CLx near-infrared imaging system.
Venom-Gland Transcriptome of M. chrysogaster.
A single M. chrysogaster worker was collected near Logan, QLD, Australia. The venom duct, venom reservoir, and venom glands (together referred to as the venom apparatus) were dissected out and placed in RNAlater. Total RNA was extracted from the venom apparatus using TRIzol reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions. Complementary DNA library preparation and sequencing were performed by the Institute for Molecular Bioscience Sequencing Facility at The University of Queensland. A dual-indexed library was constructed using the TruSeq-3 Stranded mRNA Sample Prep Kit (Illumina) with oligo(dT) selection and an average insert size of 180 bp; 150-cycle paired-end sequencing was performed on an Illumina NextSeq 500 instrument. Adapter trimming of demultiplexed raw reads was performed using fqtrim (44), followed by quality trimming and filtering using PRINSEQ-lite (45) and error correction using BBnorm tadpole, a part of the BBtools package. Trimmed and error-corrected reads were assembled using Trinity v2.4.0 (46) with a k-mer length of 31 and a minimum k-mer coverage of 2. Assembled transcripts were annotated using a blastx search (47) with an E value of 1e−3 against a UniRef90 database. Estimates of transcript abundance were made using the RSEM (48) plugin of Trinity (align_and_estimate_abundance).
Insect Assays.
House crickets (A. domesticus; average mass 50 mg; Pisces Live Food) were injected intraabdominally with 2 μL water or rMg1a (doses ranging from 1.4 to 41.5 nmol/g). Following injection, crickets were placed on their back and the time taken to right themselves was recorded.
Sheep blowflies (L. cuprina; average mass 25.2 mg) were cooled on dry ice for 1 min and then injected into the lateral thorax with 2 μL water or rMg1a (41.5 nmol/g) using a Terumo insulin syringe (29-gauge needle) attached to an Arnold hand microapplicator (Burkard Manufacturing). Survival of injected flies was monitored at 10 min, 30 min, 1 h, and 24 h postinjection, with paralysis (twitching, but inability to walk or fly) and lethality recorded.
Fruit flies (D. melanogaster; 7 d old; average mass 0.78 mg) were cooled on ice for 3 min and then injected into the lateral thorax with 50 nL water or rMg1a (41 nmol/g) using a nanoinjector (Nanoject II autoinjector; Drummond Scientific) fitted with pulled borosilicate glass microinjection needles. Survival of injected flies was monitored at 10 min, 30 min, 1 h, and 24 h postinjection, with paralysis (inability to walk or fly) and lethality recorded.
Pain-Behavior Experiments.
Male 5-wk-old C57BL/6J mice used for behavioral experiments were purchased from the Animal Resources Centre (WA, Australia). Mice were housed in groups of up to four per cage, maintained on a 12/12-h light–dark cycle, and fed standard rodent chow and water ad libitum. rMg1a (200 pmol) diluted in saline containing 0.1% bovine serum albumin (BSA) was administered in a volume of 20 µL to the hind paw by shallow i.pl. injection. Negative-control animals were injected with saline containing 0.1% BSA. Following injection, spontaneous pain-behavior events were counted from video recordings by a researcher blinded to the treatment.
Prior to mechanical and thermal threshold testing, animals were acclimatized (30 min) to the MouseMet test enclosures containing a bar bottom to permit access to the plantar hind paw. Measurements were performed at 1 h, 2 h, 4 h, 1 d, 2 d, 3 d, 4 d, and 7 d after i.pl. injection of rMg1a (dose) or saline containing 0.1% BSA. Mechanical paw-withdrawal thresholds were measured using an automatic von Frey apparatus (MouseMet; Topcat Metrology) as previously described (49). Thermal threshold measurements were performed using an automated MouseMet thermal apparatus (Topcat Metrology) as previously described (50). All measurements were performed by a researcher blinded to the treatment. Baseline paw-withdrawal thresholds were determined 3 d prior to i.pl. injection.
At each time point, animals were visually inspected for inflammation of the injected paw (e.g., swelling and reddening) and, at 3 d after injection, the thickness of both hind paws was measured using electronic calipers.
Animal experiments were approved by The University of Queensland Animal Ethics Committee (Approval no. 2021/AE000448).
Phylogenetic Analysis.
Sequences of ant EGF-like toxin precursors and Spitz (truncated to signal and mature peptides only) were aligned using the L-INS-i algorithm of MAFFT v7.309 (33). We selected the most appropriate evolutionary model (JTTDCMut+G4) using ModelFinder (51) before we used IQ-TREE v2.0.6 (52) to reconstruct the molecular phylogeny by maximum likelihood, estimating branch support values by ultrafast bootstrap using 10,000 replicates (53). Trees were visualized in Archaeopteryx v0.9921 (54).
Statistics.
All statistical analyses were done using GraphPad Prism (v9.0.0). For concentration–response curves of the ligand-induced BRET ratio, data were fitted to a nonlinear regression with variable slope. For quantitative Western blot data, an ordinary one-way ANOVA with Šídák’s multiple-comparisons test was used to test significance between EGF- or rMg1a-treated and negative-control cells. For the insect paralysis assay, an ordinary one-way ANOVA with Šídák’s multiple-comparisons test was used to test significance between rMg1a-treated and negative-control animals. For the insecticidal assays, data were plotted using survival curves. For analysis of spontaneous pain, a two-way ANOVA with Šídák’s multiple-comparisons test was used to test significance over the time course of the experiment and an unpaired t test was used to test for significance of the sum of pain-behavior counts at 30 min between treated and negative-control animals. For measurements of both mechanical and thermal hypersensitivity, a two-way ANOVA with Šídák’s multiple-comparisons test was used to test for significance between treated and negative-control animals. For paw-thickness measurements, an unpaired t test was used to test for significance between treated and negative-control animals.
Supplementary Material
Acknowledgments
We acknowledge funding from the Australian Research Council (Discovery Grant DP190103787 to I.V., G.F.K., and S.D.R.), the Australian National Health & Medical Research Council (Principal Research Fellowship APP1136889 to G.F.K. and Career Development Fellowship APP1162503 to I.V.), and the Norwegian Research Council (FRIPRO-YRT Fellowship 287462 to E.A.B.U.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. G.N. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112630119/-/DCSupplemental.
Data Availability
The precursor sequence of Mg1a reported in this article has been deposited in UniProt (accession no. P0DSL4). The solution structure and chemical shifts of Mg1a have been deposited in the PDB (ID code 7R6P) and Biological Magnetic Resonance Data Bank (ID code 30928), respectively.
All study data are included in the article and/or SI Appendix.
References
- 1.Schendel V., Rash L. D., Jenner R. A., Undheim E. A. B., The diversity of venom: The importance of behavior and venom system morphology in understanding its ecology and evolution. Toxins (Basel) 11, 666 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casewell N. R., Wüster W., Vonk F. J., Harrison R. A., Fry B. G., Complex cocktails: The evolutionary novelty of venoms. Trends Ecol. Evol. 28, 219–229 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Zancolli G., Casewell N. R., Venom systems as models for studying the origin and regulation of evolutionary novelties. Mol. Biol. Evol. 37, 2777–2790 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Robinson S. D., et al. , A comprehensive portrait of the venom of the giant red bull ant, Myrmecia gulosa, reveals a hyperdiverse hymenopteran toxin gene family. Sci. Adv. 4, eaau4640 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson S. D., et al. , A pain-causing and paralytic ant venom glycopeptide. iScience 24, 103175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Touchard A., et al. , Venom peptide repertoire of the European myrmicine ant Manica rubida: Identification of insecticidal toxins. J. Proteome Res. 19, 1800–1811 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Touchard A., et al. , Isolation and characterization of a structurally unique β-hairpin venom peptide from the predatory ant Anochetus emarginatus. Biochim. Biophys. Acta 1860 (11 Pt A), 2553–2562 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Montelione G. T., et al. , Solution structure of murine epidermal growth factor determined by NMR spectroscopy and refined by energy minimization with restraints. Biochemistry 31, 236–249 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Barberán S., Martín-Durán J. M., Cebrià F., Evolution of the EGFR pathway in Metazoa and its diversification in the planarian Schmidtea mediterranea. Sci. Rep. 6, 28071 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Citri A., Yarden Y., EGF-ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Massagué J., Pandiella A., Membrane-anchored growth factors. Annu. Rev. Biochem. 62, 515–541 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Shafee T., Anderson M. A., A quantitative map of protein sequence space for the cis-defensin superfamily. Bioinformatics 35, 743–752 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Spencer C. P., Richards K., Observations on the diet and feeding habits of the short-beaked echidna (Tachyglossus aculeatus) in Tasmania. Tasman. Nat. 131, 36–41 (2009). [Google Scholar]
- 14.Zhou Y., et al. , Platypus and echidna genomes reveal mammalian biology and evolution. Nature 592, 756–762 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahn N. G., et al. , Multiple components in an epidermal growth factor-stimulated protein kinase cascade. In vitro activation of a myelin basic protein/microtubule-associated protein 2 kinase. J. Biol. Chem. 266, 4220–4227 (1991). [PubMed] [Google Scholar]
- 16.Elsik C. G., et al. , Hymenoptera Genome Database: Integrating genome annotations in HymenopteraMine. Nucleic Acids Res. 44, D793–D800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodward S. R., Cruz L. J., Olivera B. M., Hillyard D. R., Constant and hypervariable regions in conotoxin propeptides. EMBO J. 9, 1015–1020 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Safavi-Hemami H., et al. , Venom insulins of cone snails diversify rapidly and track prey taxa. Mol. Biol. Evol. 33, 2924–2934 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiomi K., et al. , An epidermal growth factor-like toxin and two sodium channel toxins from the sea anemone Stichodactyla gigantea. Toxicon 41, 229–236 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Honma T., et al. , Novel peptide toxins from the sea anemone Stichodactyla haddoni. Peptides 29, 536–544 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Madio B., Undheim E. A. B., King G. F., Revisiting venom of the sea anemone Stichodactyla haddoni: Omics techniques reveal the complete toxin arsenal of a well-studied sea anemone genus. J. Proteomics 166, 83–92 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Marikovsky M., et al. , Appearance of heparin-binding EGF-like growth factor in wound fluid as a response to injury. Proc. Natl. Acad. Sci. U.S.A. 90, 3889–3893 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shirakata Y., et al. , Heparin-binding EGF-like growth factor accelerates keratinocyte migration and skin wound healing. J. Cell Sci. 118, 2363–2370 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Borges J. P., Mekhail K., Fairn G. D., Antonescu C. N., Steinberg B. E., Modulation of pathological pain by epidermal growth factor receptor. Front. Pharmacol. 12, 642820 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin L. J., et al. , Epiregulin and EGFR interactions are involved in pain processing. J. Clin. Invest. 127, 3353–3366 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitchell R., Mikolajczak M., Kersten C., Fleetwood-Walker S., ErbB1-dependent signalling and vesicular trafficking in primary afferent nociceptors associated with hypersensitivity in neuropathic pain. Neurobiol. Dis. 142, 104961 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Wangzhou A., et al. , A ligand-receptor interactome platform for discovery of pain mechanisms and therapeutic targets. Sci. Signal. 14, eabe1648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Safavi-Hemami H., et al. , Specialized insulin is used for chemical warfare by fish-hunting cone snails. Proc. Natl. Acad. Sci. U.S.A. 112, 1743–1748 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasteur G., A classificatory review of mimicry systems. Annu. Rev. Ecol. Syst. 13, 169–199 (1982). [Google Scholar]
- 30.Gilbert L. E., “Ecological consequences of a coevolved mutualism between butterflies and plants” in Coevolution of Animals and Plants, Gilbert L. E., Raven P. H., Eds. (University of Texas Press, Austin, TX, 1975), pp. 210–240. [Google Scholar]
- 31.Williams K. S., Gilbert L. E., Insects as selective agents on plant vegetative morphology: Egg mimicry reduces egg laying by butterflies. Science 212, 467–469 (1981). [DOI] [PubMed] [Google Scholar]
- 32.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 33.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., Mark I. D., A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Studier F. W., Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Faust G., Stand A., Weuster-Botz D., IPTG can replace lactose in auto-induction media to enhance protein expression in batch-cultured Escherichia coli. Eng. Life Sci. 15, 824–829 (2015). [Google Scholar]
- 37.Tropea J. E., Cherry S., Waugh D. S., Expression and purification of soluble His(6)-tagged TEV protease. Methods Mol. Biol. 498, 297–307 (2009). [DOI] [PubMed] [Google Scholar]
- 38.Marley J., Lu M., Bracken C., A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR 20, 71–75 (2001). [DOI] [PubMed] [Google Scholar]
- 39.Vranken W. F., et al. , The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins 59, 687–696 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Güntert P., Buchner L., Combined automated NOE assignment and structure calculation with CYANA. J. Biomol. NMR 62, 453–471 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Shen Y., Bax A., Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol. Biol. 1260, 17–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Williams C. J., et al. , MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ayoub M. A., See H. B., Seeber R. M., Armstrong S. P., Pfleger K. D., Profiling epidermal growth factor receptor and heregulin receptor 3 heteromerization using receptor tyrosine kinase heteromer investigation technology. PLoS One 8, e64672 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.G. Pertea, Fqtrim: v0. 9.4 release. (2015). 10.5281/zenodo.20552. Accessed 1 February 2022. [Google Scholar]
- 45.Schmieder R., Edwards R., Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deuis J. R., et al. , An animal model of oxaliplatin-induced cold allodynia reveals a crucial role for Nav1.6 in peripheral pain pathways. Pain 154, 1749–1757 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deuis J. R., Vetter I., The thermal probe test: A novel behavioral assay to quantify thermal paw withdrawal thresholds in mice. Temperature 3, 199–207 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han M. V., Zmasek C. M., phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics 10, 356 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The precursor sequence of Mg1a reported in this article has been deposited in UniProt (accession no. P0DSL4). The solution structure and chemical shifts of Mg1a have been deposited in the PDB (ID code 7R6P) and Biological Magnetic Resonance Data Bank (ID code 30928), respectively.
All study data are included in the article and/or SI Appendix.