Significance
Protein palmitoylation is one of the most highly abundant protein modifications, through which long-chain fatty acids get attached to cysteines by a thioester linkage. It plays critically important roles in growth signaling, the organization of synaptic receptors, and the regulation of ion channel function. Yet the molecular mechanism of the DHHC family of integral membrane enzymes that catalyze this modification remains poorly understood. Here, we present the structure of a precatalytic complex of human DHHC20 with palmitoyl CoA. Together with the accompanying functional data, the structure shows how a bivalent recognition of palmitoyl CoA by the DHHC enzyme, simultaneously at both the fatty acyl group and the CoA headgroup, is essential for catalytic chemistry to proceed.
Keywords: protein S-acylation, integral membrane enzyme, DHHC acyltransferase, fatty acyl-CoA, membrane protein structure
Abstract
S-acylation, also known as palmitoylation, is the most abundant form of protein lipidation in humans. This reversible posttranslational modification, which targets thousands of proteins, is catalyzed by 23 members of the DHHC family of integral membrane enzymes. DHHC enzymes use fatty acyl-CoA as the ubiquitous fatty acyl donor and become autoacylated at a catalytic cysteine; this intermediate subsequently transfers the fatty acyl group to a cysteine in the target protein. Protein S-acylation intersects with almost all areas of human physiology, and several DHHC enzymes are considered as possible therapeutic targets against diseases such as cancer. These efforts would greatly benefit from a detailed understanding of the molecular basis for this crucial enzymatic reaction. Here, we combine X-ray crystallography with all-atom molecular dynamics simulations to elucidate the structure of the precatalytic complex of human DHHC20 in complex with palmitoyl CoA. The resulting structure reveals that the fatty acyl chain inserts into a hydrophobic pocket within the transmembrane spanning region of the protein, whereas the CoA headgroup is recognized by the cytosolic domain through polar and ionic interactions. Biochemical experiments corroborate the predictions from our structural model. We show, using both computational and experimental analyses, that palmitoyl CoA acts as a bivalent ligand where the interaction of the DHHC enzyme with both the fatty acyl chain and the CoA headgroup is important for catalytic chemistry to proceed. This bivalency explains how, in the presence of high concentrations of free CoA under physiological conditions, DHHC enzymes can efficiently use palmitoyl CoA as a substrate for autoacylation.
The most pervasive form of lipid modification of eukaryotic proteins occurs in the form of covalent attachment of long-chain fatty acids to cysteines through a thioester linkage known as protein S-acylation (1, 2). Since the most common form of S-acylation involves C16 or palmitic acid, protein S-acylation is commonly referred to as protein palmitoylation. There are more than 3,500 proteins that are known to be palmitoylated (3, 4), underscoring the importance of palmitoylation in virtually all areas of cellular physiology. Protein S-acylation is catalyzed by integral membrane enzymes that belong to the DHHC family of protein acyl transferases (5, 6). The name derives from the catalytic D-H-H-C motif that resides on an intracellular loop between two transmembrane helices. Consequently, protein S-acylation is a modification that occurs on the cytosolic face of the membrane. DHHC enzymes localize to organellar as well as plasma membranes, and the substrates range from small GTPases such as Ras, neuronal scaffolding proteins such as PSD-95, and integral membrane proteins such as G-protein coupled receptors and ion channels.
DHHC enzymes catalyze substrate S-acylation in a two-step mechanism (7). In the first step, they catalyze self-acylation at the catalytic cysteine using the universal first substrate, fatty acyl-CoA, typically palmitoyl CoA. In a second step, the S-acylated DHHC transfers the acyl group onto a substrate protein in a transacylation reaction. The autoacylation reaction is thus the essential first step for substrate S-acylation and has been the biochemically best studied for DHHC palmitoyltransferases (7, 8). Recently, the structures of two DHHC palmitoyltransferases were reported, namely human DHHC20 and a zebrafish homolog of DHHC15, lending the first atomic insights into the three-dimensional architecture of the DHHC enzyme family (9). Together with the structure of an inhibited form of human DHHC20, in a covalent complex with 2-bromopalmitate (9), these structures yielded tremendous insights into the detailed structural chemistries of the catalytic site, the Zn2+-binding motifs, and the groove where the acyl chain gets inserted (10). However, in order to understand the complete structural basis of substrate S-acylation by DHHC palmitoyltransferases, it is essential to have structural descriptions of every functionally important state. Critical missing pieces from the early steps in the enzymatic mechanism are how DHHC enzymes recognize fatty acyl-CoA and how its reactive thioester group is stabilized near the catalytic site so as to foster spontaneous self-acylation. Previous experiments showed that residues in the cavity of DHHC enzymes impact fatty acyl chain selectivity (9). However, without knowledge about residues that contact the CoA headgroup, we do not currently understand how CoA headgroup contacting residues contribute to fatty acyl selectivity. These are important questions because the CoA headgroup is an important player in the recognition of fatty acyl-CoAs by other proteins that utilize this ubiquitous reagent in cellular biochemistry. Here, we use X-ray crystallography and molecular dynamics simulations (MD) to characterize the interactions of human DHHC20 with palmitoyl CoA and report the structure of the complex. We carry out follow-up biochemical experiments that corroborate the structure and lend insights into the initial step of protein palmitoylation by a human DHHC palmitoyltransferase.
Results and Discussion
A Catalytically Inactive Mutant of Human DHHC20 Binds Palmitoyl CoA.
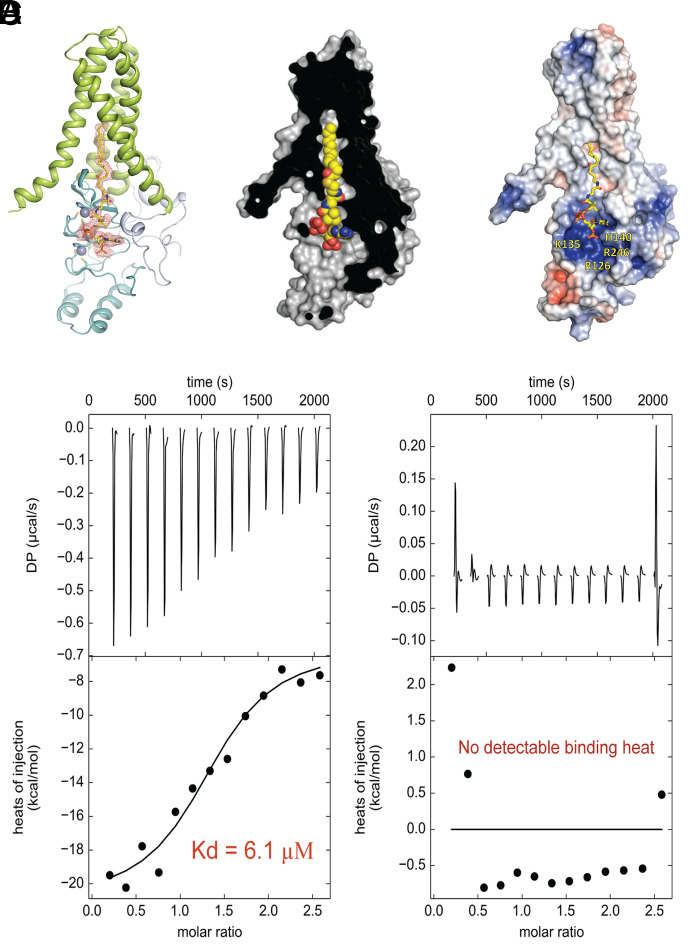
We wanted to obtain a structural snapshot of the early stages of the encounter of human DHHC20 (hereafter referred to as hDHHC20) with palmitoyl CoA (hereafter referred to as PCoA), specifically the complex before catalysis proceeds. To do so, we decided to focus on a catalytically inactive mutant to ensure the capture of an intact PCoA in complex with DHHC20. Specifically, we purified a serine mutant of the catalytic cysteine and investigated the interaction with PCoA using isothermal titration calorimetry. These experiments showed that the catalytically inactive mutant of human DHHC20 (hereafter referred to as hDHHS20) binds PCoA with 6 μM affinity (Fig. 1D), making the hDHHS20-PCoA complex a feasible structural target using X-ray crystallography at the outset, considering the high concentrations used in crystallization trials. After several rounds of careful optimizations, the hDHHS20-PCoA complex eventually gave crystals in lipidic cubic phase that diffracted to ∼2.9 Å.
Fig. 1.
Structure and affinity of the complex between hDHHS20 and palmitoyl CoA (PCoA). (A) The protein in cartoon and the PCoA in stick rendition with a 2mFo-DFc electron density map contoured at 1.0σ in orange mesh. (B) The protein in surface rendition and the PCoA in space filling rendition showing the acyl chain–binding cavity. (C) The protein in surface rendition colored by electrostatic potential with positively charged surface in blue and negative charged surface in red. The PCoA is shown in stick rendition. (D) Isothermal titration calorimetric analysis of hDHHS20 binding to PCoA (Left) and that of hDHHS20 with buffer (Right).
Structure of hDHHS20 Bound to Palmitoyl CoA.
The overall structure of the protein in the hDHHS20-PCoA complex is similar to that of apo-hDHHC20 (Fig. 1 A and B), with the tepee-shaped architecture of the transmembrane domain and two Zn2+ ions bound to the DHHC domain (9). PCoA binds to hDHHS20 in an extended configuration (Fig. 1 A and B), whereby the two points on the protein that contact the tip of the fatty acyl chain and the terminus of the CoA headgroup are ∼36 Å away. PCoA is amphipathic in nature with a hydrophobic alkyl chain and a highly charged sugar phosphate–based headgroup. Consequently, the features of the protein surfaces where it contacts each of these motifs are also very different; the acyl chain inserts into a hydrophobic pocket within the transmembrane region of the protein, while the CoA headgroup binds to a positively charged surface that is exposed to the cytosol (Fig. 1C). Although in our previously published structure of hDHHC20 we observed electron density close to this positively charged cytoplasmic surface, which we interpreted as a free CoA purely based on crystallographic considerations (9), our current structure reveals that in the complex with an intact PCoA, the orientation of the CoA headgroup is reversed relative to that earlier model. It is also worth noting that in comparison with the structure of hDHHC20 covalently modified by 2-bromopalmitate (hereafter referred to as the hDHHC20-2BP complex), the acyl chain of PCoA in the hDHHS20-PCoA complex is not inserted fully into the acyl chain–binding cavity (SI Appendix, Fig. S3). One possible explanation is that complete insertion requires a reaction of the acyl group with the catalytic cysteine (now mutated to serine). However, careful examination of the PCoA-binding mode in our structure shows that PCoA is bound at the interface between two hDHHS20 monomers in the crystal lattice, with the CoA moiety making contacts with both monomers, which are likely to influence its configuration (SI Appendix, Fig. S2). Although it has been suggested that DHHC enzymes could exist in a partial dimeric population in cellulo (11), it was unclear at the outset if dimerization is necessary for PCoA recognition or enzymatic activity of hDHHC20. Hence, we decided to resolve this question next.
hDHHC20 Is Fully Active as a Monomer.
hDHHC20 purifies as a dimer in detergents as judged by analytical ultracentrifugation (SI Appendix, Fig. S1A). However, two different crystal forms in the lipid environment of lipidic cubic phase crystallization conditions reveal starkly different dimerization interfaces—one resulting from a parallel arrangement and the other from an antiparallel configuration (9). This observation led us to conclude that these dimerization interfaces are weak and disengage relatively easily; indeed, it has been shown for other proteins that partial de-lipidation can lead to weak dimerization in detergents (12). Nonetheless, to resolve whether dimerization is crucial and ascertain whether the contacts to the second monomer in the crystal lattice are mechanistically significant, we sought to purify hDHHC20 in a monomeric state and to evaluate its functional activity. To that end, we purified hDHHC20 in a lipid-rich condition, reasoning this approach might preclude dimerization. Indeed, the addition of lipids at high concentration during the affinity chromatography and size-exclusion purification caused a distinct right shift in the size-exclusion profile of hDHHC20, supporting successful purification of monomeric hDHHC20 under these conditions (SI Appendix, Fig. S1B). The coupled enzyme assay (7, 9) demonstrated robust enzymatic activity of the monomeric enzyme. These results thus verify the hypothesis that the enzymatic activity of hDHHC20 is fully contained in the monomeric protein. To further support our conclusions, we showed that monomeric hDHHC20 was potently inhibited by 2-bromopalmitic acid (IC50 = 4.5 μM), a widely used inhibitor of DHHC enzymes (9, 13). In view of these results, we next decided to use MD simulations to refine the binding mode of palmitoyl CoA to monomeric hDHHC20 using our crystal structure as a starting point.
Refinement of PCoA-Binding Pose through MD Simulations.
As discussed in the previous section, while the crystal structure reported here features two hDHHS20 proteins in the unit cell (SI Appendix, Fig. S2A), our in vitro assays demonstrate that monomeric hDHHC20 is the functional form of the enzyme. In the crystal lattice, the second protein in the unit cell forms multiple contacts to the bound PCoA headgroup, which would not exist for monomeric hDHHC20 (SI Appendix, Fig. S2C); indeed, PCoA mediates most of the contacts between the two hDHHS20 molecules, whose structures are nearly identical (Cα-trace RMSD of 0.5 Å). Specifically, we observe that the phosphate group in PCoA that is closest to the acyl chain (P2) forms salt bridges with Lys237 and Arg239 of the second hDHHS20 molecule (SI Appendix, Fig. S2C). The adenine ring in the headgroup is also deeply buried in the protein–protein interface, forming concurrent hydrogen bonds with the backbone carbonyls of Ser244 and Phe245 of the second hDHHS20 copy. In addition to these interactions, the electron density reveals a strong signal in close proximity to the distal headgroup phosphate (P3), which appears to be a phosphate ion from the crystallographic buffer bound to the protein through salt bridges with Arg246 and Arg126 (SI Appendix, Fig. S1). Altogether, these observations suggest that the precise pose adopted by PCoA bound to monomeric hDHHC20 might differ from that observed in this crystal lattice.
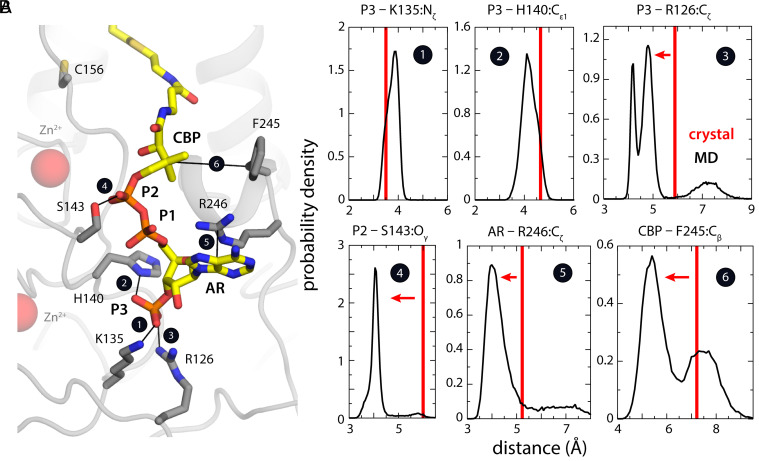
To examine which adjustments in the PCoA-binding pose might result from isolating a monomeric hDHHC20-PCoA complex, we carried out an all-atom, 500-ns MD simulation of a single copy of the enzyme–lipid complex embedded in a POPC bilayer (SI Appendix, Fig. S4); this choice of lipid had already been validated through the demonstration that hDHHC20 retains activity when reconstituted in a lipid nanodisc with PC lipids (14). In this construct, we reversed the Ser mutation of Cys156 and omitted the phosphate ion near the PCoA headgroup. Reassuringly, the simulation preserved the overall features of the complex as well as several of the key protein–ligand interactions observed in the crystal structure (Figs. 2 and 3). For example, in the PCoA headgroup, the distal phosphate maintains its contacts with His140 and Lys135 (Fig. 2 A and B), both of which are highly conserved residues (9). However, the simulation also revealed significant changes in the binding pose (Figs. 2 and 3). In the PCoA headgroup, both the adenine group and P2 phosphate reconfigure to form new contacts and compensate for the interactions lost (Fig. 2 A and B). Specifically, the P2 phosphate engages with the highly conserved Ser143 while the adenine group reorients toward the solvent and forms a π-cation interaction with Arg246, which repositions in absence of the extraneous phosphate ion (Fig. 2 A and B). The other contact for this phosphate ion in the crystal structure, Arg126, also repositions and becomes the third direct coordinating contact for distal phosphate P3 (Fig. 2 A and B). Finally, it is worth noting that the absence of the second protein in the unit cell causes the hydrophobic quaternary carbon adjacent to P2 to shift toward Phe245, seemingly so as to minimize its exposure to the polar solvent (Fig. 2 A and B).
Fig. 2.
Refinement of the PCoA-binding pose with MD simulations of a single hDHHC20-PCoA complex. (A) Network of interactions formed by the PCoA head group in the refined structure. Black lines indicate the key interactions that seem to define the binding pose. The configuration depicted represents the most probable state based on analysis of probability distributions for each of those protein–ligand distances shown in B. (B) Probability distributions (black curves) for each of the protein–ligand interactions highlighted in A, calculated from a 500-ns MD trajectory. Note the distances shown are for the central atoms in the interacting groups (e.g., P in phosphate, Cζ in arginine, etc) for conciseness. The value of that distance in the crystal structure (i.e., the starting point of the simulation) is also indicated (red lines).
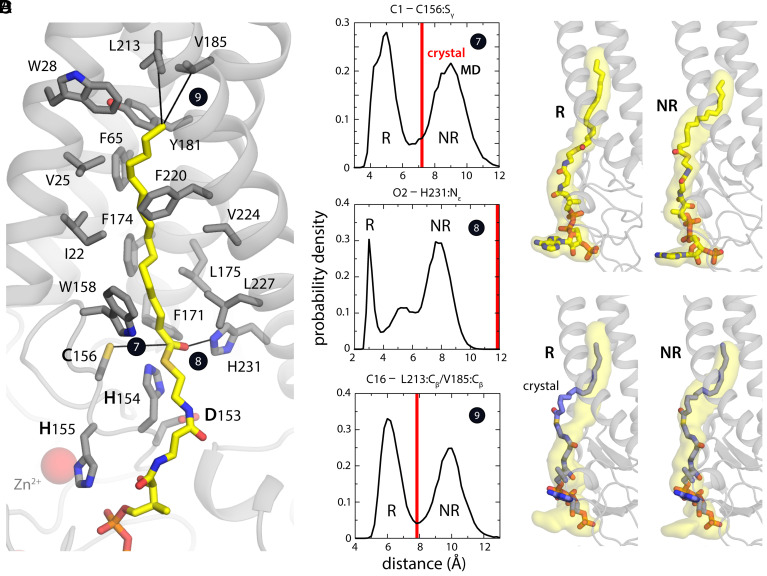
Fig. 3.
Reactive (R) and nonreactive (NR) configurations of bound PCoA. (A) Configuration of the fatty acyl chain and thioester group in the R state of the hDHHC20-PCoA complex resulting from a 0.5-µs MD trajectory. Black lines again highlight distances used to describe adjustments in the binding pose relative to the crystal structure. (B) Probability distributions derived from this trajectory (black curves) for the protein–ligand interaction distances highlighted in A compared with crystal structure values (red lines). (C) Comparison of the R and NR configurations observed in simulation. Alongside specific simulation snapshots corresponding to the peaks in the probability distributions in B, the figure shows three-dimensional (3D) density maps (yellow volume) calculated for all snapshots in each state (R: d7 ≤ 6.5 Å; NR: d > 6.5 Å). (D) The configuration of PCoA in the crystal structure is compared with the 3D density maps derived from simulation data for the R and NR states. Several configurations of this putatively R state are freely available for download at https://github.com/Faraldo-Gomez-Lab-at-NIH/Download.
In contrast to the PCoA headgroup, the crystal structure of the hDHHC20-PCoA shows no contacts for the palmitoyl chain with a second protein (SI Appendix, Fig. S2A). It is thus intriguing that the thioester group, which must react with Cys156, instead projects away from the catalytic site and that the hydrocarbon chain appears not to occupy the full volume of the binding cavity. The structure therefore appears to capture a noncatalytic intermediate. Indeed, our simulation of the (wild-type) complex reveals two distinct conformational states, comparably populated, which differ primarily in the proximity of the carbonyl in PCoA to the active site in the protein (Fig. 3 A and B). In one state, seemingly nonreactive (NR), the fatty acyl chain is configured very similarly to what is observed in the crystal structure (Fig. 3 C and D); in the other, however, the thioester group in PCoA is much closer to the catalytic site and appears to be primed for catalysis (Fig. 3 A, C, and D). Interestingly, this seemingly reactive (R) state appears to be stabilized by a specific hydrogen bond between the carbonyl group in PCoA and His231 (Fig. 3A), which is not well conserved but is often replaced by other side chains capable of H bonding, such as glutamine (9). The proximity of the thioester group to the catalytic site in the reactive state correlates with a shift in the palmitoyl chain, which inserts itself deeper into the hydrophobic pocket (Fig. 3 A, C, and D). By contrast, in the NR state, the palmitoyl chain buckles out of the binding pocket near Trp158 as is also observed in the crystal structure.
In summary, we posit that by combining X-ray crystallography and MD simulations of the hDHHC20-PCoA complex, we have identified the mode by which hDHHC20 recognizes the CoA head group and have also captured the palmitoyl chain in the two states that immediately precede the autoacylation reaction. We also posit that the R state represents the precatalytic complex of hDHHC20 with PCoA.
Functional Assays Support Observed Recognition Mode.
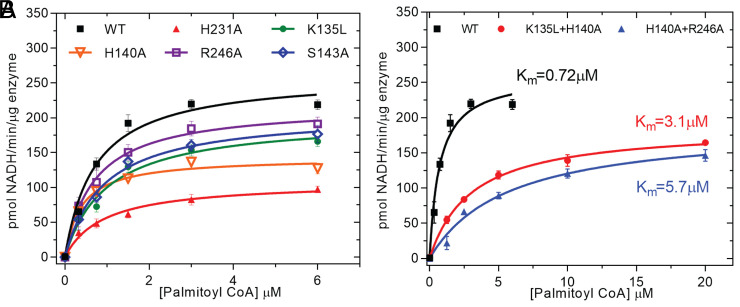
We decided to directly test the predictions made by the MD simulations about the R state being the precatalytic complex of hDHHC20 with PCoA by extensive mutagenesis and testing the activity of the mutants. It is important to point out that several key contacts between hDHHC20 and PCoA in the crystal structure were preserved in the putative R state observed in MD simulations (Fig. 2A and SI Appendix, Fig. S2B). Of these, the mutation of Lys135 and His140 led to significant erosion of enzymatic activity (Fig. 4A). This effect became even more prominent when the mutations were combined (Fig. 4B). The critical determinants for testing the predictions from MD simulations were contacts that were developed in the R state but did not exist in the crystal structure. The contact between His231 and the carbonyl group of PCoA is one example. The His231Ala mutant is one of the most striking single mutants we tested in terms of compromising enzymatic activity (Fig. 4A), consistent with the hypothesis that a residue with H-bonding potential is key to stabilize the R state. Another example is Arg246Ala, which abolishes a π-cation interaction with PCoA in the R state (Fig. 2A) and also lowers enzymatic activity (Fig. 4A). Finally, Ser143 makes a close contact with P2 phosphate in the R state only, and the mutation of this residue to alanine appreciably lowered enzymatic activity (Fig. 4A).
Fig. 4.
Mutagenesis based testing of MD simulations results. Analysis of the enzymatic activity of selected single mutants (A) and double mutants (B). The coupled-enzyme assay was used, and Michaelis–Menten fits are shown.
Mechanistic Significance and Putative Gating Mechanism of the Acyl Chain–Binding Pocket.
It is increasingly recognized that membrane receptors and transport proteins are regulated by specific lipids (15–22). Structural studies of these effects have typically revealed lipids bound on sites where their headgroups form ionic interactions, seemingly conferring specificity, while the alkyl chains pack against hydrophobic regions of the protein surface. The mode in which hDHHC20 recognizes the PCoA headgroup is thus not atypical; however, that these enzymes have evolved a binding pocket for the acyl chain is, in our view, very intriguing.
A fundamental difference between lipid regulation of membrane protein activity and lipid chemistry by membrane enzymes is that the former does not require that the same lipid molecule be bound to the protein throughout the process that is modulated. For example, in the lifetime of the open state of a PIP2-activated ion channel, a given recognition site might be occupied by many different PIP2 molecules, exchanging between the bulk membrane and the interaction site. By contrast, enzymes such as DHHC require that one substrate molecule reside at the “reaction site” long enough for catalysis to occur. To this end, a high-affinity headgroup interaction would be a counterproductive solution; after all, the CoA headgroup must come off readily after PCoA is cleaved off. Thus, while this interaction must confer specificity against other lipid types, it must also be labile. Instead, we hypothesize that it is the acyl chain–binding pocket that is key to extend the lifetime of the bimolecular complex.
We tested this hypothesis computationally and experimentally. Specifically, we designed a computer simulation protocol to assess how the stability of the hDHHC20-CoA interaction differs when in context of the complete PCoA or in absence of the palmitoyl chain (Methods). In this comparative simulation protocol (23), spontaneous dissociations of a ligand are induced by perturbing its interactions with all other elements of the system (not only the protein but also the solvent, etc.). This perturbation scales down the magnitude of the free energy barriers and minima underlying binding and unbinding, thereby accelerating the kinetics of these processes exponentially. Dissociation trajectories can be thus observed within a relatively short simulation time, without imposing a specific direction of movement, and the impact of differential factors (in this case, the presence or absence of the palmitoyl chain) can be assessed systematically.
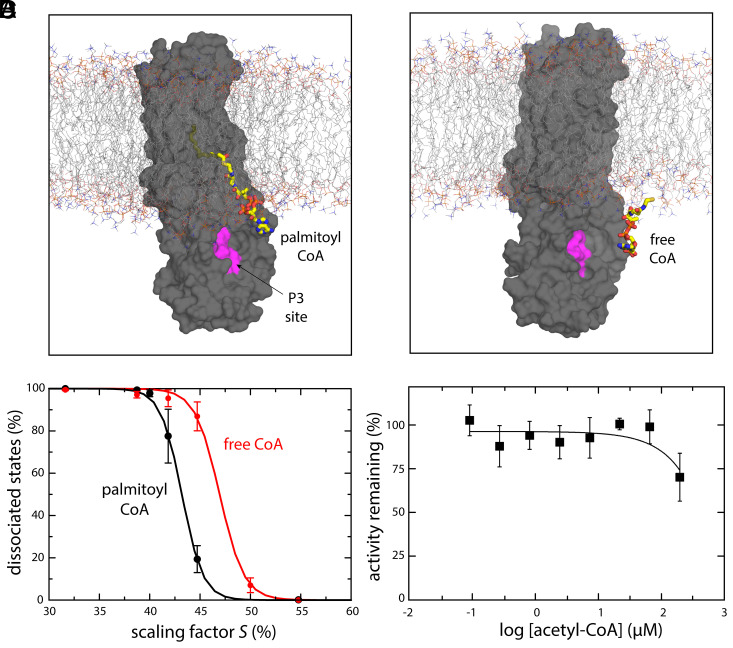
This analysis shows that the effective strength of the protein–CoA interaction is clearly enhanced by the acyl chain, relative to free CoA (Fig. 5 A and B). Specifically, the simulation data shows that in the range of simulation conditions in which headgroup dissociation occurs (within a given timeframe), the probability of dissociation of free CoA is much greater than that of CoA linked to the palmitoyl chain, everything else being equal (Fig. 5 A and B). That is, if CoA is linked to the fatty acyl chain, the sigmoidal dissociation probability curve is left-shifted (indicating dissociation requires a larger perturbation) relative to free CoA. Indeed, inspection of the dissociation trajectories in either case (Fig. 5A) shows that when free CoA dissociates, it does so readily. By contrast, in the context of the bound fatty acyl chain, the dissociated headgroup remains in close proximity to the protein and often rebinds.
Fig. 5.
Fatty acyl chain enhances hDHHC20-PCoA headgroup interaction. (A and B) Snapshots of two MD trajectories used to quantify the relative probability of dissociation of the CoA headgroup in the presence or absence of the palmitoyl tail, respectively, induced with the solute-tempering method (Methods). DHHC20 (gray surface) is shown embedded in the POPC lipid bilayer (thin lines) with the site of interaction for the distal phosphate in PCoA/CoA (P3) highlighted, that is, K135, H141, and R126 (magenta); the solvent is not shown for clarity. In both cases, the CoA group (thick bonds) is fully dissociated. (C) The probability of CoA dissociation, quantified as the percentage of dissociated configurations observed in a 50-ns solute-tempering trajectory, averaged over all calculated trajectories for a given scaling factor S (n = 3 to 6), is shown as a function of S in the presence (black) or absence (red) of the palmitoyl tail. Error bars denote the SEM. The data were fitted to a sigmoidal function f = 1/(1 + exp[m (S − n)]). The resulting parameters (C.C. > 99.9%) are PCoA, m = 95.6, n = 0.43; free CoA, m = 82.6, n = 0.47. (D) Experimental data for inhibition of hDHHC20 enzymatic activity by added CoA. Shown is the percentage activity as function of the log of acetyl CoA concentration in μM.
These computational observations can be verified experimentally. To do so, we carried out an activity assay in which we tested if increasing concentrations of acetyl CoA could inhibit the autoacylation reaction. Consistent with the computational results, we found that PCoA is clearly a much stronger binder than soluble CoA; even at concentrations larger than 100 µM, acetyl CoA has no discernable inhibitory effect (Fig. 5D), implying that it neither precludes PCoA recognition nor accelerates PCoA dissociation prior to catalysis. That soluble CoA is ineffective is in clear contrast to comparable assays of lipid modulation of ion channels, which have shown, for example, that soluble variants of PIP2 can completely recapitulate the regulatory effects of PIP2 itself (15). This contrast underscores the rather fundamental differences between lipid transfer enzymes and lipid-regulated membrane proteins in regard to the recognition of lipids.
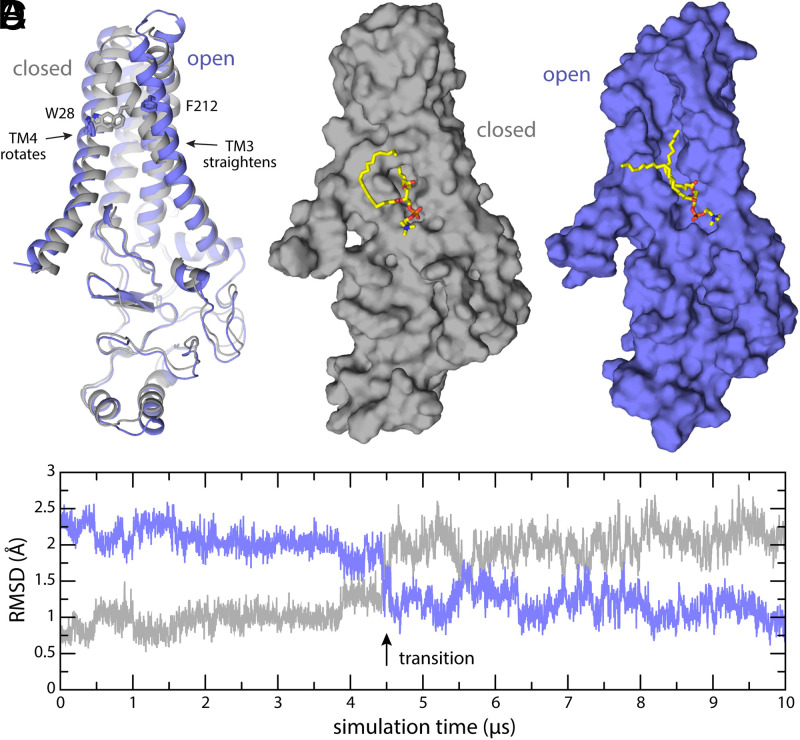
We would argue, therefore, that the acyl chain–binding pocket of hDHHC20 is a crucial mechanistic feature, evolved to meet the specific requirements of a bimolecular chemical reaction, which are arguably distinct from those of lipid regulation processes. An open question, however, is how this binding pocket becomes occupied by the acyl chain. To begin to address question, we carried out a microsecond-long MD simulation of apo-state hDHHC20, hoping that the trajectory would reveal spontaneous fluctuations in the protein structure indicative of a potential mechanism of recognition. Indeed, this long trajectory revealed what appears to be a breathing mechanism wherein a reconfiguration of helices TM3 and TM4 transforms the acyl chain–binding pocket into an open cleft exposed to the bilayer interior (Fig. 6). No changes occur elsewhere in the structure, however, including the binding site for the PCoA headgroup on the cytosolic domain. This transformation is seemingly slow and did not reverse in the timescale of the simulation, and so we must interpret these preliminary results with caution. Nonetheless, the observed structural change is consistent with the notion that the mechanism of PCoA capture begins by recognition of the headgroup, which in turn would foster binding of the acyl chain to the open cleft between TM3 and TM4; closure of this cleft would secure the PCoA chain and enhance the headgroup interaction, ultimately leading to a reactive configuration. Further studies will be required to experimentally refute or confirm this mechanism; a possible approach would be to engineer a reversible linkage between helices TM3 and TM4 so as to preclude opening of the cleft. We anticipate the enzymatic rate will be significantly diminished by this linkage.
Fig. 6.
Putative mechanism of gating of the PCoA-binding pocket. (A) The experimental structure of apo-state DHHC20 (9) (gray) is compared with the final snapshot of a 10-µs MD trajectory (purple). The trajectory reveals the transformation of the binding pocket into an open cleft through local but consequential changes in helices TM3 and TM4. For example, the side chains of Trp28 and Phe212, which are in direct contact and contribute to occlude the binding pocket in the closed state, become separated by 16 Å in the putatively open state. (B and C) Same as A on a surface representation. Membrane and solvent are omitted for clarity, except the POPC lipid molecule that partially occupies the binding pocket in absence of PCoA. (D) Time-series of the RMSD between the snapshots observed in simulation and either the experimental structure of apo-state DHHC20 (gray) or the putatively open state depicted in A and C. For clarity, the RMSD is quantified only for the backbone of the transmembrane span. The approximate time point in which the opening transition occurs is indicated.
CoA Binding and Acyl Chain Length Selectivity.
Having established the importance of both acyl chain and CoA headgroup recognition in the enzymatic reaction of hDHHC20 and, by inference, other DHHC enzymes, we next turned our attention to the contribution of CoA-binding residues to acyl chain length selectivity. Although DHHC enzymes predominantly catalyze attachment of the 16-carbon palmitic acid to cysteines, both in cellulo (24, 25) and in vitro (8, 9), they can also attach longer or shorter fatty acids, albeit with lower preference. We had shown previously that engineering mutations at the delimiting end of the acyl-binding cavity, which contacts the tip of the hydrophobic chain, can notably change the acyl chain length selectivity (9). However, the role of the CoA binding residues has not been elucidated since their identity was not known before this study. We tested the acyl selectivity profile of mutants of two residues that contact the PCoA headgroup in the R state. Interestingly, both H231A and R246A mutants displayed similar acyl chain length selectivity as the wild-type enzyme (SI Appendix, Fig. S5). This finding demonstrates that the fatty acyl–binding cavity and the CoA headgroup–binding module do not contribute equally to the acyl chain length selectivity. This is also consistent with the above hypothesis that recognition of the PCoA headgroup and of the acyl chain occur at different times in the reaction trajectory. Future computational and experimental studies should no doubt examine this hypothesis in greater detail.
Conclusions
Our crystal structure, computer simulations, and biochemical experiments reveal the structure of a precatalytic complex of hDHHC20 with PCoA. This structure specifically reveals how DHHC enzymes recognize the CoA headgroup together with the acyl chain to achieve a two-site contact recognition strategy for PCoA. PCoA is an amphipathic molecule with a hydrophobic fatty acyl tail and a charged diphosphoadenosine headgroup linked by a flexible phosphopantethiene linker. PCoA binds to hDHHC20 in an extended conformation, with the fatty acyl chain inserted in a membrane-embedded cavity enclosed by the transmembrane domain and the charged headgroup recognized through multiple contacts by a positively charged protein surface in the cytosol. The contacts to the acyl chain (hydrophobic) and those to the headgroup (polar) take place in very different milieu with no specific contacts to the phosphopantethiene linker. Our biochemical and computational results show that both interactions make important contributions to the overall hDHHC20-PCoA interaction since free CoA is not able to inhibit enzymatic activity or stably associate with the enzyme. Thus, we posit that in this context, PCoA is not a typical small molecule ligand. Rather, it has two chemical moieties, the CoA headgroup and the fatty acyl chain, each of which also occur as part of other molecules in the cell, such as acetyl CoA and phospholipids. In PCoA, the fatty acyl chain and the CoA are linked by the flexible phosphopantethiene linker, thus making PCoA a bivalent ligand/substrate. Consequently, formation of contacts at both sites, namely the acyl chain and the diphosphoadenosine headgroup, sets the stage for autoacylation to proceed, which is the first step of catalysis by hDHHC20. This bivalency is key from a physiological standpoint in that it permits PCoA to compete out the high intracellular concentrations of free CoA (∼100 to 400 μM) (26), which otherwise would saturate the CoA-binding site and inhibit the DHHC enzymes, preventing them from acting on target proteins. We envisage that the notion of bivalency will translate to other proteins whose function is to catalyze transfer reactions using specific lipids as substrates.
Interestingly, our MD simulations revealed a persistent NR state of PCoA as well, where the conformation of the acyl chain is very similar to that of PCoA in the crystal structure. Long-time simulations of apo-hDHHC20 also indicate that in the absence of PCoA, the binding pocket can become an open cleft through localized conformational changes. While these mechanistic insights no doubt require further examination, the existing results seem to imply that the association of hDHHC20 with PCoA is initiated by the recognition of the CoA headgroup at the cytosolic site, resulting in an early encounter complex. We suggest our crystal structure captures a subsequent intermediate arising from this first encounter complex following recognition of the acyl chain and closure of its hydrophobic binding cleft without complete insertion of the acyl chain into the binding cavity. As observed in simulation, the conversion of this intermediate into a final R configuration, representing the precatalytic complex, would follow, requiring full insertion of the chain into its pocket. Other intermediate NR conformations could also exist en route to the R state that are not captured by the crystal structure. In any case, it is likely that the chain-length selectivity of hDHHC20 reflects the final step in the PCoA recognition process; that is, while first-encounter complexes are likely to be similarly viable with acyl-CoA molecules of different chain length, the likelihood of the reactive configuration would depend on the degree of fitness of the acyl chain to its binding pocket within the protein. This model would explain why our experiments show that mutation of residues that contact the headgroup do not change the acyl chain length selectivity of hDHHC20; by contrast, mutations deep into the binding pocket are very influential as we have previously shown (9).
Although DHHC enzymes are critically important in a range of physiological processes and have now been connected to several human diseases including, most recently, COVID-19 (27, 28), a structure-based understanding of their mechanism has only been forthcoming recently. The structural snapshots presented here constitute an important early step in the entire catalytic cycle of hDHHC20. Elucidation of the entire enzymatic cycle at the atomic level will require continued systematic efforts along the directions presented here.
Methods
MD Simulation of hDHHC20-PCoA Complex.
The starting condition for the simulation of hDHHC20-PCoA is the crystal structure of hDHHS-PCoA, after reversing the serine mutation of Cys156. Protonation probabilities for each of the ionizable residues in the protein–ligand complex were evaluated using electrostatic energy calculations and a Monte Carlo algorithm as described elsewhere (29). The protein–ligand complex with two Zn2+ ions bound was embedded in a preequilibrated POPC bilayer using GRIFFIN (30). The internal geometry of the Zn2+-binding sites was maintained throughout all simulations using a network of ion–protein and protein–protein distance restraints involving the four coordinating residues for each Zn2+. The resulting simulation system includes 222 POPC lipids, 21,633 water molecules, 33 Na+, and 41 Cl− (100 mM NaCl plus counterions of the protein and ligand net charge) in an orthorhombic box of ∼89 × 89 × 119 Å. The preparation of the simulations entailed a multistage equilibration phase whereby a series of (primarily) internal coordinate restraints acting on different portions of the protein structure were gradually weakened and removed over 100 ns. A trajectory of 0.5 µs was then calculated to refine the binding pose of PCoA in the context of the experimental structure of the protein. To preclude an artificial relaxation of the protein, a weak harmonic restraint (k = 6 kcal/mol/Å2) acting on the RMSD of the Cα trace (relative to the crystal structure) was applied throughout the simulation. On average, the RMSD of the resulting ensemble of protein configurations is 1.5 ± 0.1 Å for the complete Cα trace and 0.85 ± 0.1 Å for the transmembrane span only. The simulations were carried out using NAMD 2.13 (31, 32) and the CHARMM36 force field (33, 34) at constant temperature (298 K) and constant pressure (1 atm) with periodic boundary conditions. Force field parameters for PCoA and CoA were those used in a previous study (14). The integration time step was 2 fs. Electrostatic interactions were calculated using the particle-mesh Ewald method, with a real-space cutoff value of 12 Å; van der Waals interactions were also cutoff at 12 Å, with a smooth switching function taking effect at 10 Å. As described above, this 0.5-µs simulation resulted in a well-defined configuration of the PCoA headgroup but two alternative configurations of the acyl chain, referred to as R and NR.
MD Simulation of apo hDHHC20.
To more precisely quantify the intrinsic conformational landscape of hDHHC20, in the absence of PCoA, a 10-µs MD trajectory was calculated with an Anton 2 supercomputer (35) using again the CHARMM36 forcefield at constant temperature (298 K) and pressure (1 atm), respectively set with the Nose–Hoover thermostat and the Martyna–Tobias–Klein barostat and with periodic boundary conditions and an integration time step of 2.5 fs. Electrostatic interactions were calculated using the Gaussian-split Ewald method; van der Waals interactions were cutoff at 10 Å. To prevent degradation of the hDHHC20 secondary structure in the microsecond time scale, we applied soft restraints on all φ and ψ dihedral angles along the protein backbone. These restraints are identical to those used in previous studies based on microsecond simulations calculated on Anton (36, 37) but with a significantly weaker force constant equal to 1 kcal/mol. Accordingly, the resulting ensemble of protein configurations shows a significant conformational heterogeneity; RMSD values relative to starting configuration range from ∼1.5 to ∼3.5 Å for the entire backbone (2.4 Å on average) and from ∼0.6 to ∼2 Å for the transmembrane span only (1.4 Å on average). This 10-µs trajectory was initiated with the final configuration of a preparatory 700-ns trajectory obtained with NAMD in a previously study (14), which had been as initiated with the crystal structure of apo-state hDHHC20 (9). This preparatory trajectory was carried out to optimize the orientation of the protein in the bilayer as well as the detailed features of the protein–lipid interface, and so it featured an RMSD restraint acting on the protein backbone of force constant 60 kcal/mol/Å2. The RMSD for the ensemble of configurations in this preparatory trajectory, relative to the crystal structure, is 1.0 ± 0.1 Å for the entire backbone and 0.5 ± 0.1 Å for the transmembrane span only.
Solute-Tempering MD Simulations of Free CoA and PCoA Headgroup Dissociation.
A complex of hDHHC20 and free CoA was constructed on the basis of a snapshot of the 500-ns simulation of the hDHHC20-PCoA complex that closely resembles the time average over this trajectory. After truncating the acyl tail, this configuration was equilibrated for 50 ns while restricting changes in the internal structure of CoA and its relative position relative to the protein using a flat-bottom RMSD restraint. To evaluate the relative persistence of the protein–CoA interaction in each system, a series of simulations was then carried out in each case using the solute-tempering method (23, 38, 39). In this scheme, the “solute” was considered to be either the PCoA headgroup or free CoA. That is, nonbonded interactions between CoA and all other components in the simulation system were scaled by a factor S = , nonbonded interactions within CoA were scaled by a factor λ, and all other interactions in the system were intact. (In PCoA, the head group was defined as above the C3P carbon.) After a series of initial tests, a total of 27 independent (nonexchanging) simulations of 50 ns each were carried out for the hDHHC20-CoA complex for λ values equal to 0.05, 0.10, 0.15, 0.16, 0.175, 0.20, 0.30, and 0.40 (three to six trajectories per λ). Analogously, for the hDHHC20-PCoA complex, 30 independent simulations of 50 ns each were carried out for λ values equal to 0.10, 0.15, 0.175, 0.20, 0.25, 0.30, and 0.40 (three to six trajectories per λ). The dissociation of CoA was monitored over time by evaluating an average of four distances that reflect direct interactions with hDHHC20 in the associated state; the characteristic value of this average is 6.5 ± 0.2 Å for the hDHHC20-PCoA complex and 6.8 ± 0.3 Å for the hDHHC20-CoA complex. Based on the visual inspection of trajectories calculated with λ = 0.1, it was estimated that full dissociation occurred when this average distance was equal to or greater than 11 Å. To determine the percentage of dissociated configurations in each simulation, this distance was evaluated throughout the last 30 ns; the first 20 ns were considered as equilibration and not included in the analysis. All solute-tempering simulations were carried out using NAMD 2.13 (31), with the same specifications as those noted above.
Protein Purification.
The human DHHC20 containing a C156S mutation (hDHHS20) was prepared following an established procedure (9). Briefly, an hDHHS20 expression construct was cloned into a modified pPICZ vector with an N-terminal 10×His tag followed by an mVenus coding sequence and a PreScission cleavage site. The hDHHS20 protein was overexpressed in Pichia pastoris strain SDM1163 grown in buffered glycerol-complex medium (0.1 M potassium phosphate, pH 6.0, 3.4 g/L yeast nitrogen base, 1% glycerol, 0.4 µg/mL biotin, and 100 µg/mL zeocin) cultures. Cells were allowed to grow for ∼24 h to an optical density at a wavelength of 600 nm of ∼20 at 30 °C. Then, cells were pelleted by centrifugation and resuspended in 1.5 L of buffered methanol-complex medium (0.1 M potassium phosphate, pH 6.0, 3.4 g/L yeast nitrogen base, 1% methanol, 0.4 µg/mL biotin, and 25 µg/mL zeocin). Cells were incubated for another ∼36 h at 23 °C. Cells were harvested by centrifugation. The pelleted cells were flash frozen in liquid nitrogen and stored at −80 °C. Frozen cells were disrupted using Retsch MM400 millers with liquid nitrogen cooling. Approximately 20 g milled cell powder was suspended in ∼100 mL lysis buffer containing 100 mM Hepes, pH 7.9, 450 mM NaCl, 5 mM βME, 1.5% (wt/vol) n-dodecyl-β-D-maltoside (DDM), protease inhibitors [benzamidine, 4-(2-aminoethyl)benzenesulfonyl fluoride, soy trypsin inhibitor, pepstatin, and leupeptin], and deoxyribonuclease. The lysate was extracted using a magnetic stirrer at 4 °C for 2 h and then centrifuged at 38,000 × g for 30 min at 4 °C. The supernatant was loaded onto a column containing 2.5 mL TALON resin (Clontech). The column was washed with 30 mL 50 mM Hepes, pH 7.5, 250 mM NaCl, 5 mM βME, 2 mM tris(2-carboxyethyl)phosphine (TCEP), 0.1 mg/mL POPC:POPG:POPA (3:1:1) lipids, and 1 mM DDM buffer containing 25 mM imidazole followed by an additional wash with 5 mL same buffer without imidazole. Prior to on-column PreScission protease cleavage, the protein-bound resin was resuspended in 6 mL wash buffer without imidazole. The resin slurry was then rotated for 12 h at 4 °C. The cleaved protein was collected by gravity flow and concentrated with a 50-kDa molecular weight cutoff 15-mL concentrator (Millipore) to ∼1.0 mL. The protein was then reductively methylated using a formaldehyde and dimethylamineborane complex following standard protocols. The next morning, the methylation reaction was quenched with 100 mL 1 M Tris HCl, pH 8.0, and 5 mL 1 M dithiothreitol (DTT) for 1 h at 4 °C. The protein was concentrated to ∼0.25 mL The concentrated protein sample was loaded onto a Superdex 200 Increase size exclusion column equilibrated with buffer that consists of 50 mM Hepes, pH 7.5, 150 mM NaCl, 2 mM TCEP, 0.05 mg/mL POPC:POPG:POPA (3:1:1) lipids, and 0.5 mM DDM buffer at 4 °C. The fractions containing the target protein were pooled and concentrated to ∼15 to 20 mg/mL.
Crystallization and Structure Determination.
The concentrated protein was incorporated into monoolein using established protocols (40), and crystallization trials were set up on 96-well Microbatch plates (Hampton Research) as described in the previous study (9). The best diffracting hDHHS20/Palmitoly-CoA complex crystals were obtained in 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer, pH 6.2, 20 mM KH2PO4, 26.5% polethylene glycol 300, 5 mM DTT, 2.2% 2,5-Hexanediol, and 500 μM Palmitoyl-CoA. Diffraction data were collected at the GM/CA-CAT 23ID-D and 23ID-B beamlines, respectively, at the Advanced Photon Source at the Argonne National Laboratory in Chicago, IL. Molecular replacement with the program PHASER (41) was used to obtain the initial phase of the hDHHS20/Palmitoly-CoA complex structure. The structure of the human DHHC20 (Protein Data Bank [PDB] entry: 6BMN) was used as a search model. The final model was obtained after iterative cycles of manual model building with COOT and refinement using PHENIX. MOLPROBITY (42) was used to evaluate the quality of the refined structure. The statistics for the structure are shown in SI Appendix, Table S1. The coordinates and structure factors of the hDHHS20/Palmitoly-CoA complex structure has been deposited to the PDB with the accession code 7KHM.
Isothermal Titration Calorimetry.
The hDHHS20 protein sample was purified in the same manner as described in Protein Purification. Data were collected during 15 3-μL injections of 200 µM hDHHS20 into 15 μM Palmitoly-CoA at 20 °C with stirring at 480 rpm and 180-s injection spacing using a MicroCal i200 microcalorimeter (GE Healthcare). The heat exchanged during each injection was integrated using NITPIC v1.2.0 (43) and fit to nonlinear regression analysis as a single-site binding using SEDPHAT v12.1b, obtaining thermodynamic parameters. GUSSI v1.1.0 (44) was used to plot the processed data.
DHHC20 Mutant Analyses.
The Venus-hDHHC20 wild type and mutants were cloned into the pEG-Bacmam vector and expressed in human embryonic kidney (HEK) 293T cells grown in Dulbecco’s Modified Eagle Medium with 2 mM glutamine, 2% fetal bovine serum, and 100 units/mL penicillin/streptomycin. A total of 30 μg DNA was complexed with 90 μg polyethyleneimine and used to transfect 150-mm plates of ∼80% confluent HEK293T cells. Cells were harvested after 48 h and washed with phosphate-buffered saline. The cell pellets were resuspended in 2 mL 40 mM Tris HCl, pH 7.2, 270 mM NaCl, 5 mM βME, and 5 mM MgCl2 containing DNase and protease inhibitors. A total of 300 μL 0.4 M stock DDM was added to the cell suspension and rotated for 2 h at 4 °C. Cell debris was removed by centrifugation at 21,000 × g for 10 min at 4 °C. The supernatant was then applied to 50 μL TALON resin in a microfuge tube and incubated on a rotator for ∼1 h at 4 °C. Resin was first washed with 3 mL 40 mM Tris HCl, pH 7.2, 270 mM NaCl, 1 mM TCEP containing 25 mM imidazole and 1 mM DDM. Protein was eluted by ∼80 μL 20 mM Tris HCl, pH 7.2, 135 mM NaCl, 1 mM TCEP, and 5 mM βME buffer containing 250 mM imidazole. Protein concentration was estimated by running a stain-free sodium dodecyl sulfate polyacrylamide gel electrophoresis (Bio-Rad) gel with previously purified Venus-hDHHC20 standards from which a linear calibration curve could be constructed. Coupled-enzyme assays to measure Michaelis–Menten constants were carried out as described in the previous study (9). For the acyl-CoA chain length selectivity experiments, enzyme and acyl-CoA concentrations for the coupled-enzyme assay were kept fixed at 10 nM and 10 μM, respectively. The normalized activity for the wild-type and mutant DHHC20 enzymes was obtained by taking the initial rate of each reaction normalized to 1 with regard to that enzyme’s activity in the presence of palmitoyl-CoA.
Sedimentation Velocity.
Sedimentation velocity experiments were conducted at 50,000 rpm and 20 °C on a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge following standard protocols (45). Pooled protein fractions in 50 mM Hepes (pH 7.5), 150 mM NaCl, 2 mM TCEP, and 0.5 mM DDM obtained from size exclusion chromatography along with a matching solvent buffer without detergent were used. Samples at concentrations of ∼10 μM were loaded into two-channel, 12-mm path length sector-shaped cells and thermally equilibrated at zero speed. Absorbance and interference velocity scans were subsequently acquired at ∼5-min intervals—absorbance data were collected in a continuous mode as single measurements at 280 nm using a radial spacing of 0.003 cm. Time-corrected (46) data were analyzed in SEDFIT 15.01c (47) in terms of a continuous c(s) distribution of sedimenting species using an s range of 0 to 20 with a linear resolution of 400 and a maximum entropy regularization CI of 0.95. In all cases, excellent fits were observed with RMSDs ranging from 0.0037 to 0.0053 absorbance units and 0.0079 to 0.013 fringes. The c(s) distributions were imported into GUSSI 1.2.1 (47), and analyzed using the membrane integration tool (48) to characterize the major species observed. The protein partial specific volume and absorbance extinction coefficient at 280 nm were calculated based on the amino acid composition in SEDNTERP (49). The interference refractive index increment was calculated based on the amino acid sequence in SEDFIT 15.01c (50). A partial specific volume of 0.820 cm3 ⋅ g−1 (51) and refractive index increment of 0.143 cm3 ⋅ g−1 (48) were used for DDM. The solvent density ρ and viscosity η were determined experimentally at 20 °C on an Anton Paar DMA 5000 density meter and Anton Paar AMVn rolling ball viscometer, respectively. Sedimentation coefficients were corrected to standard conditions in water at 20 °C, s20,w.
Monomeric hDHHC20 Purification.
We previously discovered that excessive amounts of phospholipids during purification prevents a monomer‐to‐dimer transition of hDHHC20. To prepare the monomeric form of hDHHC20, ∼5 g yeast powder containing the Venus-hDHHC20 wild type was processed as described in Protein Purification, resulting in ∼25 mL of supernatant from the centrifuged lysate. This supernatant was loaded onto a Bio-Rad Econo column containing 1 mL TALON resin (Clontech). The column was washed with 15 mL 50 mM Hepes, pH 7.5, 250 mM NaCl, 5 mM βME, 2 mM TCEP, 1.0 mg/mL POPC:POPG:POPA (3:1:1) lipids, and 2 mM DDM buffer containing 25 mM imidazole followed by elution with ∼3 mL buffer with 200 mM imidazole. The elution was diluted 10-fold with the same buffer without imidazole. Fluorescence size-exclusion chromatography was used to assess the gel filtration profile of the purified protein. Enzymatic activity of the purified sample was with the coupled-enzyme protein acyltransferase (PAT) assay. For comparison, the same sample was prepared without the phospholipid additives in the purification buffer.
PAT Activity Inhibition Assays.
Inhibition assays were carried out in 384-well low-volume plates (Thermo Fisher) at 30 °C. Plates were read in a Tecan M1000Pro fluorimeter. A total of 10 µL reaction solution A (0.25 mM oxidized nicotinamide adenine dinucleotide, 0.2 mM thiamine pyrophosphate, 2 mM 2-oxoglutarate, and 10 nM DHHC enzyme in a buffer containing 25 mM MES, pH 6.5, 50 mM NaCl, 1 mM DTT, 0.4 mM DDM, and 0.04 mg/mL POPC:POPG:POPA [3:1:1] lipids) and 10 µL reaction solution B (α-ketoglutarate dehydrogenase complex in the same buffer) were prepared. To determine the half-maximal inhibitory concentration (IC50) of 2-bromopalmitate or acetyl CoA, the compound was serially diluted in 10 μL reaction solution A. For the acetyl CoA inhibition assay, the phospholipid additives were removed from the assay buffer. Prior to the start of the assay, the reaction solution A and B were mixed and incubated at room temperature for 10 min. A total of 10 µM palmitoyl-CoA was added into the assay mixture to initiate the reaction. The data were collected and analyzed as described in the previous study (9). An average IC50 of three independent experiments was calculated based on the inhibition of NADH fluorescence signaling using nonlinear least square function in GraphPad Prism 8.
Supplementary Material
Acknowledgments
This work was supported by the Divisions of Intramural Research of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (C.-J.L., M.S.R., F.S., and A.B.); the National Heart, Lung and Blood Institute (R.S. and J.D.F.-G.); and the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (R.G.). Computational resources were provided in part by the NIH Supercomputing Facility Biowulf. The Pittsburgh Supercomputing Center provided access to an Anton2 computer donated by D. E. Shaw Research through NIH Grant R01-GM1169161. Crystals for this project were screened and datasets collected at various stages at the following beamlines: The Northeastern Collaborative Access Team (NE-CAT) and The General Medical Sciences and Cancer Institutes Structural Biology Facility (GM/CA) at the Advanced Photon Source, Argonne National Laboratory. NE-CAT is funded by the National Institute of General Medical Sciences from the NIH (P30 GM124165). The Eiger 16M detector on 24-ID-E is funded by an NIH–Office of Research Infrastructure Programs High-End Instrument Grant (S10OD021527). GM/CA has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). The Eiger 16M detector at GM/CA X-ray Science Division was funded by NIH Grant S10 OD012289. This research used resources of the Advanced Photon Source, a US Department of Energy (DOE), Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. C.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022050119/-/DCSupplemental.
Data Availability
Crystal structure data have been deposited in PDB (accession no. 7KHM).
References
- 1.Chamberlain L. H., Shipston M. J., The physiology of protein S-acylation. Physiol. Rev. 95, 341–376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jiang H., et al. , Protein lipidation: Occurrence, mechanisms, biological functions, and enabling technologies. Chem. Rev. 118, 919–988 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blanc M., David F. P. A., van der Goot F. G., SwissPalm 2: Protein S-palmitoylation database. Methods Mol. Biol. 2009, 203–214 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Zhou B., et al. , Low-background acyl-biotinyl exchange largely eliminates the coisolation of non-S-acylated proteins and enables deep S-acylproteomic analysis. Anal. Chem. 91, 9858–9866 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lobo S., Greentree W. K., Linder M. E., Deschenes R. J., Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 277, 41268–41273 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Roth A. F., Feng Y., Chen L., Davis N. G., The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 159, 23–28 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchell D. A., Mitchell G., Ling Y., Budde C., Deschenes R. J., Mutational analysis of Saccharomyces cerevisiae Erf2 reveals a two-step reaction mechanism for protein palmitoylation by DHHC enzymes. J. Biol. Chem. 285, 38104–38114 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jennings B. C., Linder M. E., DHHC protein S-acyltransferases use similar ping-pong kinetic mechanisms but display different acyl-CoA specificities. J. Biol. Chem. 287, 7236–7245 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rana M. S., et al. , Fatty acyl recognition and transfer by an integral membrane S-acyltransferase. Science 359, eaao6326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stix R., Lee C. J., Faraldo-Gómez J. D., Banerjee A., Structure and mechanism of DHHC protein acyltransferases. J. Mol. Biol. 432, 4983–4998 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai J., Linder M. E., Oligomerization of DHHC protein S-acyltransferases. J. Biol. Chem. 288, 22862–22870 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan L., Smirnova I. N., Verner G., Nagamori S., Kaback H. R., Manipulating phospholipids for crystallization of a membrane transport protein. Proc. Natl. Acad. Sci. U.S.A. 103, 1723–1726 (2006). Correction in: Proc. Natl. Acad. Sci. U.S.A. 103, 6073 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varner A. S., De Vos M. L., Creaser S. P., Peterson B. R., Smith C. D., A fluorescence-based high performance liquid chromatographic method for the characterization of palmitoyl acyl transferase activity. Anal. Biochem. 308, 160–167 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Stix R., Song J., Banerjee A., Faraldo-Gómez J. D., DHHC20 palmitoyl-transferase reshapes the membrane to foster catalysis. Biophys. J. 118, 980–988 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen S. B., Lipid agonism: The PIP2 paradigm of ligand-gated ion channels. Biochim. Biophys. Acta 1851, 620–628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ciardo M. G., Ferrer-Montiel A., Lipids as central modulators of sensory TRP channels. Biochim. Biophys. Acta Biomembr. 1859, 1615–1628 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Antollini S. S., Barrantes F. J., Fatty acid regulation of voltage- and ligand-gated ion channel function. Front. Physiol. 7, 573 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson M. J., Baenziger J. E., Structural basis for the modulation of pentameric ligand-gated ion channel function by lipids. Biochim. Biophys. Acta Biomembr. 1862, 183304 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Poveda J. A., Marcela Giudici A., Lourdes Renart M., Morales A., González-Ros J. M., Towards understanding the molecular basis of ion channel modulation by lipids: Mechanistic models and current paradigms. Biochim. Biophys. Acta Biomembr. 1859, 1507–1516 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Zaydman M. A., Cui J., PIP2 regulation of KCNQ channels: Biophysical and molecular mechanisms for lipid modulation of voltage-dependent gating. Front. Physiol. 5, 195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fürst O., Mondou B., D’Avanzo N., Phosphoinositide regulation of inward rectifier potassium (Kir) channels. Front. Physiol. 4, 404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilgemann D. W., Feng S., Nasuhoglu C., The complex and intriguing lives of PIP2 with ion channels and transporters. Sci. STKE 2001, re19 (2001). [DOI] [PubMed] [Google Scholar]
- 23.Li M., et al. , Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP. eLife 8, e47060 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang X., et al. , Heterogeneous fatty acylation of Src family kinases with polyunsaturated fatty acids regulates raft localization and signal transduction. J. Biol. Chem. 276, 30987–30994 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Greaves J., et al. , Molecular basis of fatty acid selectivity in the zDHHC family of S-acyltransferases revealed by click chemistry. Proc. Natl. Acad. Sci. U.S.A. 114, E1365–E1374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dansie L. E., et al. , Physiological roles of the pantothenate kinases. Biochem. Soc. Trans. 42, 1033–1036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puthenveetil R., et al. , S-acylation of SARS-CoV-2 spike protein: Mechanistic dissection, in vitro reconstitution and role in viral infectivity. J. Biol. Chem. 297, 101112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mesquita F. S., et al. , S-acylation controls SARS-CoV-2 membrane lipid organization and enhances infectivity. Dev. Cell 56, 2790–2807.e8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eicher T., et al. , Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife 3, e03145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Staritzbichler R., Anselmi C., Forrest L. R., Faraldo-Gómez J. D., GRIFFIN: A versatile methodology for optimization of protein-lipid interfaces for membrane protein simulations. J. Chem. Theory Comput. 7, 1167–1176 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phillips J. C., et al. , Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiorin G., Klein M. L., Hénin J., Using collective variables to drive molecular dynamics simulations. Mol. Phys. 111, 3345–3362 (2013). [Google Scholar]
- 33.Best R. B., et al. , Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klauda J. B., et al. , Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw D. E., et al. , “Anton 2: Raising the bar for performance and programmability in a special-purpose molecular dynamics supercomputer” in SC14: International Conference for High Performance Computing, Networking, Storage and Analysis (IEEE, 2014), pp. 41–53.
- 36.Pan A. C., et al. , Atomic-level characterization of protein-protein association. Proc. Natl. Acad. Sci. U.S.A. 116, 4244–4249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jensen M. O., et al. , Mechanism of voltage gating in potassium channels. Science 336, 229–233 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Jo S., Jiang W., A generic implementation of replica exchange with solute tempering (REST2) algorithm in NAMD for complex biophysical simulations. Comput. Phys. Commun. 197, 304–311 (2015). [Google Scholar]
- 39.Liu P., Kim B., Friesner R. A., Berne B. J., Replica exchange with solute tempering: A method for sampling biological systems in explicit water. Proc. Natl. Acad. Sci. U.S.A. 102, 13749–13754 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caffrey M., Cherezov V., Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 4, 706–731 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Cryst. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis I. W., et al. , MolProbity: All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scheuermann T. H., Brautigam C. A., High-precision, automated integration of multiple isothermal titration calorimetric thermograms: New features of NITPIC. Methods 76, 87–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brautigam C. A., Zhao H., Vargas C., Keller S., Schuck P., Integration and global analysis of isothermal titration calorimetry data for studying macromolecular interactions. Nat. Protoc. 11, 882–894 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao H., Brautigam C. A., Ghirlando R., Schuck P., Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr. Protoc. Protein Sci. 71, 20.12.1–20.12.49 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H., et al. , Recorded scan times can limit the accuracy of sedimentation coefficients in analytical ultracentrifugation. Anal. Biochem. 437, 104–108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuck P., Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.le Maire M., et al. , Gel chromatography and analytical ultracentrifugation to determine the extent of detergent binding and aggregation, and Stokes radius of membrane proteins using sarcoplasmic reticulum Ca2+-ATPase as an example. Nat. Protoc. 3, 1782–1795 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Cole J. L., Lary J. W., Moody T. P., Laue T. M., Analytical ultracentrifugation: Sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 84, 143–179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao H., Brown P. H., Schuck P., On the distribution of protein refractive index increments. Biophys. J. 100, 2309–2317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.le Maire M., Champeil P., Moller J. V., Interaction of membrane proteins and lipids with solubilizing detergents. Biochim. Biophys. Acta 1508, 86–111 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure data have been deposited in PDB (accession no. 7KHM).