Significance
The extracellular concentration of adenosine triphosphate (ATP) reaches several hundred micromoles in the inflamed tissues or tumor environment. A high concentration of ATP activates P2X7, a purinergic receptor, and induces the formation of a nonselective cation channel, accompanied by reversible phosphatidylserine (PtdSer) exposure, leading to cell lysis. Here, we found that Xk and Vps13a complexed at plasma membranes are indispensable for ATP-induced PtdSer exposure and cell lysis. Patients with McLeod syndrome and chorea-acanthocytosis are known to carry the mutation in XK and VPS13A genes, respectively, suggesting that a defect of XK-VPS13A–mediated phospholipid scrambling is responsible for neuroacanthocytosis, a progressive movement disorder, and cognitive and behavior changes.
Keywords: phosphatidylserine, XKR scramblase, P2X7 receptor, cell death, regulatory T cells
Abstract
A high extracellular adenosine triphosphate (ATP) concentration rapidly and reversibly exposes phosphatidylserine (PtdSer) in T cells by binding to the P2X7 receptor, which ultimately leads to necrosis. Using mouse T cell transformants expressing P2X7, we herein performed CRISPR/Cas9 screening for the molecules responsible for P2X7-mediated PtdSer exposure. In addition to Eros, which is required for the localization of P2X7 to the plasma membrane, this screening identified Xk and Vps13a as essential components for this process. Xk is present at the plasma membrane, and its paralogue, Xkr8, functions as a phospholipid scramblase. Vps13a is a lipid transporter in the cytoplasm. Blue-native polyacrylamide gel electrophoresis indicated that Xk and Vps13a interacted at the membrane. A null mutation in Xk or Vps13a blocked P2X7-mediated PtdSer exposure, the internalization of phosphatidylcholine, and cytolysis. Xk and Vps13a formed a complex in mouse splenic T cells, and Xk was crucial for ATP-induced PtdSer exposure and cytolysis in CD25+CD4+ T cells. XK and VPS13A are responsible for McLeod syndrome and chorea-acanthocytosis, both characterized by a progressive movement disorder and cognitive and behavior changes. Our results suggest that the phospholipid scrambling activity mediated by XK and VPS13A is essential for maintaining homeostasis in the immune and nerve systems.
Plasma membranes are composed of a lipid bilayer of phospholipids. Phospholipids with different head groups and acyl residues are asymmetrically distributed between the outer and inner leaflets of the plasma membrane (1, 2). Phosphatidylserine (PtdSer), phosphatidylethanolamine (PtdEtn), and phosphatidylinositol are restricted to the inner leaflet of plasma membranes, while the majority of phosphatidylcholine (PtdCho) and sphingomyelin localize to the outer leaflet. The asymmetrical distribution of phospholipids contributes to signal transduction, the partitioning of proteins, and the maintenance of membrane integrity (3). ATP11A and ATP11C, which are P4-type ATPases present in plasma membranes together with CDC50A, function as flippases that maintain PtdSer and PtdEtn in the inner leaflet (4, 5).
The asymmetrical distribution of phospholipids at the plasma membrane is broken down in various biological processes (1, 6). When exposed on the cell surface, PtdSer functions as a signaling molecule or activates enzymes. We recently elucidated the molecular mechanisms by which apoptotic cells and activated platelets expose PtdSer on cell surfaces (6). When cells undergo apoptosis, caspases are activated and destroy the flippase activity of ATP11A and ATP11C, while increases in intracellular Ca2+ in activated platelets inactivate them. PtdSer is exposed in apoptotic cells within hours and within minutes in activated platelets (1, 6). The blockade of flippases is not sufficient to quickly expose PtdSer because of the difficulties associated with the movement of its hydrophilic head groups in hydrophobic lipid bilayers (7). Caspases or Ca2+ activate Xkr8 or TMEM16F, respectively, which elicit phospholipid scramblase activity that nonspecifically translocates phospholipids in both inward and outward directions (8, 9). Scramblases at plasma membranes provide a path or cleft for the hydrophilic head groups of phospholipids to translocate between two lipid layers (10, 11). Therefore, PtdSer is transferred to the extracellular surface, serves as an “eat me” signal for apoptotic cells, or provides the scaffold on platelets for blood-clotting factors (6).
Another process that exposes PtdSer to the cell surface is the stimulation of T lymphocytes with a high concentration of adenosine triphosphate (ATP) (12–15). ATP is the intracellular energy currency present in the cytosol at a concentration of ∼4 mM (16). Its extracellular concentration is low or in the nanomolar range under physiological conditions, but reaches several hundred micromoles in inflamed tissues or the tumor environment (17, 18). When released from cells, ATP and its metabolites bind to purinergic receptors (P1, P2X, and P2Y), which are G-protein-coupled receptors or ion channels (18). P2X7, encoded by P2rx7, is an ATP-gated cation channel composed of the homotrimer of a protein that possesses two transmembrane helices with short N- and long C-terminal cytoplasmic domains (19). It is expressed in various cells, including immune cells and some tumor cells (20–22). A previous study demonstrated that the short exposure of P2X7 to a high dose of ATP induced the formation of a nonselective cation channel, leading to potassium efflux and sodium and calcium influx (23), and this was accompanied by the reversible exposure of PtdSer (12–15, 24). On the other hand, long-term exposure to a high concentration of ATP kills cells by rupturing their membranes. This type of cell death in macrophages, called pyroptosis, is mediated by NLRP3 inflammasomes that activate caspase 1, followed by the pore-forming protein Gasdermin D (25) and Ninjurin1 (26). On the other hand, temperature-independent P2X7-mediated rapid cell shrinkage and PtdSer exposure in mouse T cells (14) suggest the existence of an inflammasome-independent cell-death pathway.
We previously reported that mouse T cell transformants expressing P2X7 responded to a high concentration of ATP for PtdSer exposure (27). CRISPR/Cas9 screening identified Eros as an essential molecule for the expression of P2X7 at the plasma membrane. In the present study, we further characterized the molecules responsible for P2X7-mediated PtdSer exposure in a T cell line and found that Xk and Vps13a were indispensable for this process. Xk and Vps13a formed a complex at plasma membranes and scrambled phospholipids, leading to membrane rupture. We also showed that the Xk–Vps13a pathway played an important role in the ATP-induced killing of mouse CD25+CD4+ T cells. XK and VPS13A are the genes responsible for McLeod syndrome (28, 29) and chorea-acanthocytosis, respectively (30–32), which are both characterized by a progressive movement disorder and cognitive and behavior changes. These results suggest that the Xk-Vps13a–mediated scrambling of phospholipids (and cell killing) plays an essential role in the immune and nervous systems.
Results
Genome-Scale CRISPR/Cas9 Screening for Molecules Involved in P2X7-Mediated PtdSer Exposure.
We performed CRISPR/Cas9 screening to identify the molecules involved in P2X7-mediated PtdSer exposure (27). The mouse T cell lymphoma cell line WR19L expresses TMEM16F at the plasma membrane, which is responsible for Ca2+-dependent scrambling or PtdSer exposure (33). The mouse P2rx gene family comprises seven members that function as ATP-induced, cation-specific ion channels (34). Some P2rx family members are ubiquitously expressed, while others are expressed in a tissue-specific manner. The ATP analog 2′(3′)-O-(4-benzoylbenzoyl)ATP (BzATP) activates P2X7, but not other P2rx family members (35). Therefore, WR19L cells deficient in Tmem16f and P2rx7 (DKO) do not respond to Ca2+ ionophores or BzATP for PtdSer exposure.
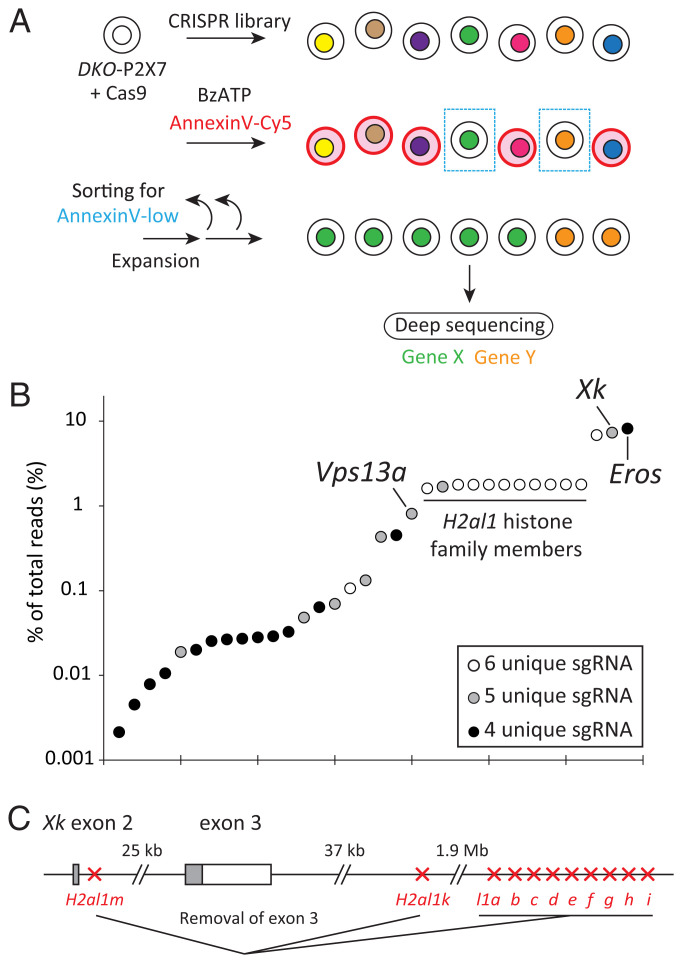
When DKO cells were transformed with a high expression level of mouse P2X7, the transformants (DKO-P2X7) reversibly exposed PtdSer in response to BzATP. DKO-P2X7 cells treated at 4 °C with BzATP exposed PtdSer within 5 min. Following the removal of BzATP from the buffer, exposed PtdSer remained on the cell surface at 4 °C for at least 30 min, but was quickly internalized at 37 °C. We utilized this characteristic in CRISPR/Cas9 screening for the molecules responsible for P2X7-mediated PtdSer exposure. DKO-P2X7 cells were transformed with Cas9 and subsequently with a Genome-Scale CRISPR/Cas9 Knockout (GeCKO) library (36) at a multiplicity of infection of 0.3 (Fig. 1A). Cells were then stimulated with BzATP, and the population that was stained at a low level with Annexin V was sorted. This procedure of the stimulation with BzATP, sorting, and expansion was repeated three times. The single-guide RNA (sgRNA) sequences integrated into the genomic DNA of the sorted cells were analyzed by next-generation sequencing (NGS).
Fig. 1.
Genome-wide CRISPR/Cas9 screening for genes responsible for P2X7-mediated PtdSer exposure. (A) Scheme of genome-wide CRISPR/Cas9 screening. Cas9 was transduced to Tmem16f−/−P2rx7−/− (DKO) WR19L cell transformants expressing mouse P2X7. The CRISPR library was then introduced by a lentivirus, and stable transformants were stimulated with BzATP for PtdSer exposure. The cell population with a reduced ability to respond to BzATP for PtdSer exposure was sorted by using a cell sorter and expanded. This procedure of the stimulation with BzATP, sorting, and expansion was repeated three times. The DNA fragment coding for sgRNA was amplified by PCR from the genomic DNA of sorted cells and subjected to deep sequencing. (B) Genes targeted by enriched sgRNAs. The genes in which four, five, or six unique sgRNAs were detected in deep sequencing were plotted with their abundance (percentage of the total number of reads). (C) Deletion of exon 3 of the Xk gene by sgRNA for H2al1 histone family members. Eleven well-conserved histone H2al1 family genes are present in the flanking region of exon 3 of the Xk gene. The sgRNAs recognizing these H2al1 histone genes may remove exon 3 of the Xk gene.
We previously reported (27) that the sgRNAs targeting Eros (CYBC1) were the most enriched, occupying 8.2% of all reads (Fig. 1B). Eros, as a chaperone, was necessary for P2X7 to be stably expressed in the plasma membrane. Further analyses of NGS reads identified 34 genes for which four to six unique sgRNA sequences were present (Fig. 1B). Among them, 7.3% of sgRNAs were against Xk (28). We also noted the enrichment of sgRNAs for 11 H2al1 histone family members (37), the genes of which were located at intron 2 and downstream of exon 3 of the Xk gene (Fig. 1C) in the X chromosome. Although H2al1 may have played some role in P2X7-mediated PtdSer exposure, the most likely explanation is that the Cas9–sgRNA complex deleted exon 3 of the Xk gene by simultaneously cleaving H2al1 variant genes because all H2al1 variants possess a 99% identical sequence in the coding region (SI Appendix, Fig. S1). These results suggested the involvement of Xk in P2X7-mediated PtdSer exposure.
Xk is a membrane protein with unknown functions; however, its mutation is responsible for McLeod syndrome, a neuroacanthocytosis syndrome (28, 29). Another syndrome, chorea-acanthocytosis, with a similar phenotype (neuroacanthocytosis), is caused by loss-of-function mutations in the Vps13a gene (30–32). Five different sgRNAs for Vps13a were enriched in a cell population showing a reduced ability to expose PtdSer in response to BzATP. The Vps13a gene was ranked next to H2al1 or Xk, occupying ∼1% of all reads (Fig. 1B).
Requirement of Xk and Vps13a for P2X7-Mediated PtdSer Exposure.
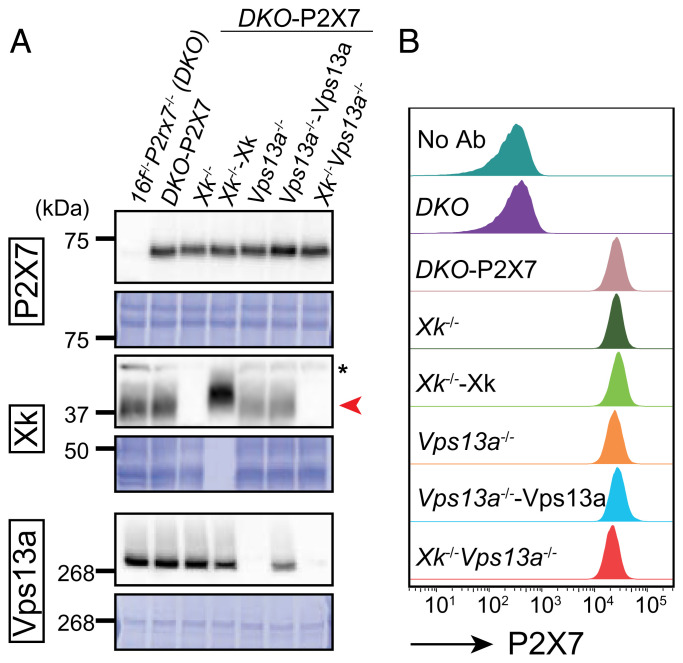
To examine whether Xk and Vps13A are required for P2X7-mediated PtdSer exposure, the genes for Xk, Vps13a, or both were knocked out in DKO-P2X7 cells by using the CRISPR/Cas9 system (SI Appendix, Fig. S2). The null mutation of neither Xk nor Vps13a affected the expression level of P2X7 in total cell lysates (Fig. 2A) or on the cell surface (Fig. 2B). The transformation of Xk−/− or Vps13a−/− DKO-P2X7 with the respective gene (Xk or Vps13a) did not affect the total or cell-surface expression level of P2X7. These results suggested that, in contrast to Eros (27), Xk and Vps13a are not essential for P2X7 to be expressed on the cell surface. The results obtained also demonstrated that the null mutation in or external expression of Xk did not affect the expression level of Vps13a and vice versa (Fig. 2A).
Fig. 2.
Establishment of P2X7-, Xk-, or Vps13a-deficient cell lines. (A) Western blot analysis. The P2rx7 gene in Tmem16f−/− WR19L cells was knocked out to generate 16f−/−P2rx7−/− cells (DKO). DKO was transformed with mouse P2X7 to establish DKO-P2X7. The Xk gene in DKO-P2X7 was knocked out in Xk−/−, and Xk−/−DKO-P2X7 was transformed with FLAG-tagged mouse Xk to generate Xk−/−DKO–P2X7–Xk (Xk−/−-Xk). In Vps13a−/−, the Vps13a gene was knocked out in DKO-P2X7, and Vps13a−/−DKO-P2X7 was transformed with mouse Vps13a in Vps13a−/−–Vps13a. In Xk−/−Vps13a−/−, Xk, and Vps13a genes were knocked out in DKO-P2X7. Approximately 4.7 μg of protein per lane (except for Xk−/−–Xk containing 94 ng of protein) of whole-cell lysates was separated by SDS-PAGE. Western blotting was performed with Abs against P2X7, Xk, and Vps13a, and membranes were stained with CBB. The red arrowhead shows Xk. *, non-specific band. (B) The cell-surface expression of P2X7. DKO, DKO-P2X7, Xk−/−, Xk−/−–Xk, Vps13a−/−, Vps13a−/−–Vps13a, and Xk−/−Vps13a−/− cells were stained with Alexa 647–anti-P2X7 Ab. The Alexa 647-staining profiles in the SYTOX Blue-negative population are shown.
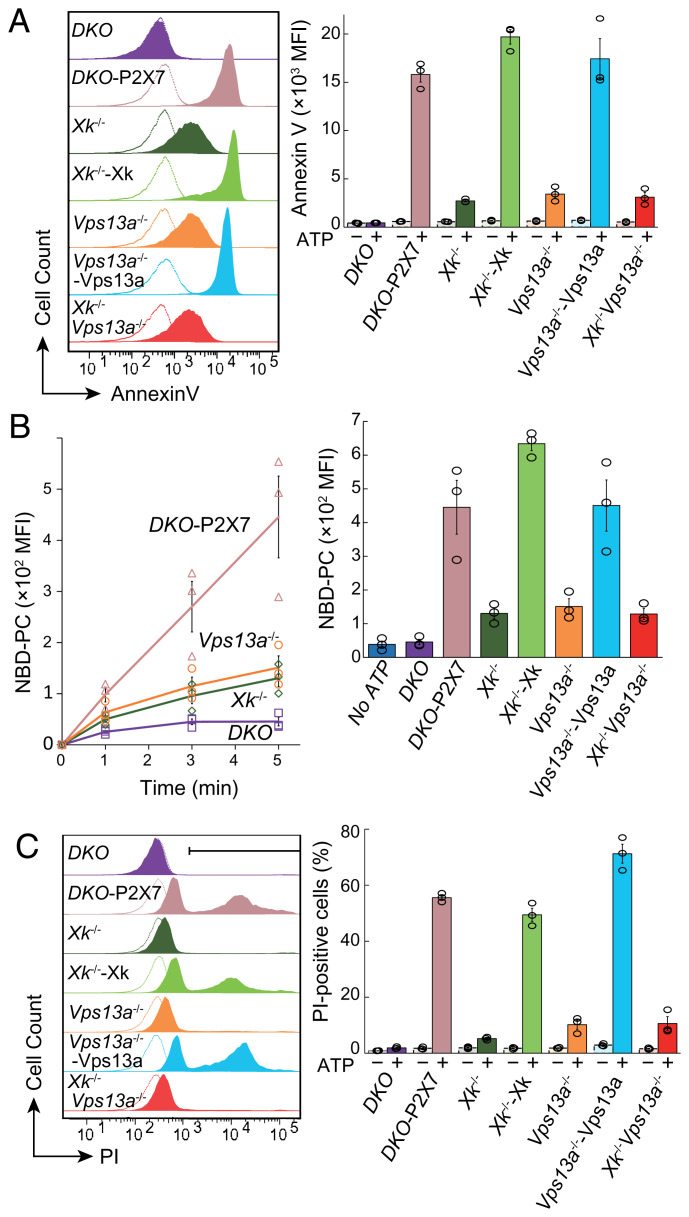
DKO-P2X7 cells stimulated at 4 °C with a high concentration of ATP (500 μM) robustly exposed PtdSer within 5 min (Fig. 3A). DKO cells did not respond to ATP for PtdSer exposure, confirming that P2X7 mediated ATP-induced PtdSer exposure. A deficiency of either Xk or Vps13a severely reduced ATP-induced PtdSer exposure to a similar level (Fig. 3A). The transformation of Xk−/− or Vps13a−/− cells with the respective gene restored the ability of these cells to expose PtdSer when stimulated with ATP, confirming that the reduction in PtdSer exposure was due to the lack of Xk or Vps13a. The double deficiency of Xk and Vps13a had no additional effects on PtdSer exposure, suggesting that Xk and Vps13a functioned in the same pathway.
Fig. 3.
Requirement of Xk and Vps13a for ATP-induced phospholipid scrambling and cytolysis. (A) DKO, DKO-P2X7, Xk−/−DKO-P2X7 (Xk−/−), Xk−/−DKO-P2X7/Xk (Xk−/−–Xk), Vps13a−/−DKO-P2X7 (Vps13a−/−), Vps13a−/−DKO-P2X7/Vps13a (Vps13a−/−–Vps13a), and Xk−/−Vps13a−/−DKO-P2X7 (Xk−/−Vps13a−/−) cells were unstimulated or stimulated with 500 µM ATP at 4 °C for 5 min, stained with Annexin V in the presence of 2.5 µg/mL PI, and analyzed by flow cytometry. The Annexin V staining profile in the PI-negative population is shown on the left. (B) Cells were incubated at 4 °C with 500 µM ATP in the presence of 250 nM NBD-PC for the indicated period, and BSA-nonextractable NBD-PC was detected by flow cytometry. (C) Cells were successively incubated at 4 °C for 5 min and at room temperature for 10 min in Annexin buffer containing 500 µM ATP and 2.5 µg/mL PI and then analyzed by flow cytometry. The PI-positive cell population is indicated by a horizontal bar in the top panel. Experiments in A–C were performed three times. The average values of mean fluorescent intensity (MFI) at 5 min are shown with SE (bar) on the right for A and B. The average percentage of PI-positive cells is plotted with SE in C.
Requirement of Xk and Vps13a for Phospholipid Scrambling and Cytolysis.
Xk is a member of the Xkr family, to which the caspase-dependent scramblases Xkr4, Xkr8, and Xkr9 belong (8, 38). Since scramblases transport phospholipids bidirectionally and nonspecifically between the lipid bilayer (1), we considered the stimulation of P2X7 to induce the bidirectional nonspecific transport of phospholipids. Accordingly, ATP-stimulated DKO-P2X7 cells incorporated fluorescence-labeled PtdCho {1-oleoyl-2-{6-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]hexanoyl}-sn-glycero-3-phosphocholine (NBD-PC)} at 4 °C in a P2X7-dependent manner (Fig. 3B). The null mutation in Xk or Vps13a in DKO-P2X7 cells markedly reduced the incorporation of NBD-PC. These results indicated that the binding of ATP to P2X7 activated phospholipid scrambling requiring Xk and Vps13a.
The sustained stimulation of P2X7 with ATP has been shown to induce lytic cell death (39, 40). When DKO-P2X7 cells were incubated at room temperature for 10 min following an incubation at 4 °C for 5 min, more than 50% of cells were stained by propidium iodide (PI), indicating that they underwent necrosis (Fig. 3C). A deficiency of either Xk or Vps13a prevented this process, and the transformation of Xk−/− or Vps13a−/− cells with the corresponding gene rescued the process. These results indicated that P2X7-mediated ATP-induced necrosis was also dependent on Xk and Vps13a and suggested that phospholipid scrambling executed by Xk and Vps13a is a prerequisite for P2X7-mediated cytolysis.
Complex Formation between Xk and Vps13a.
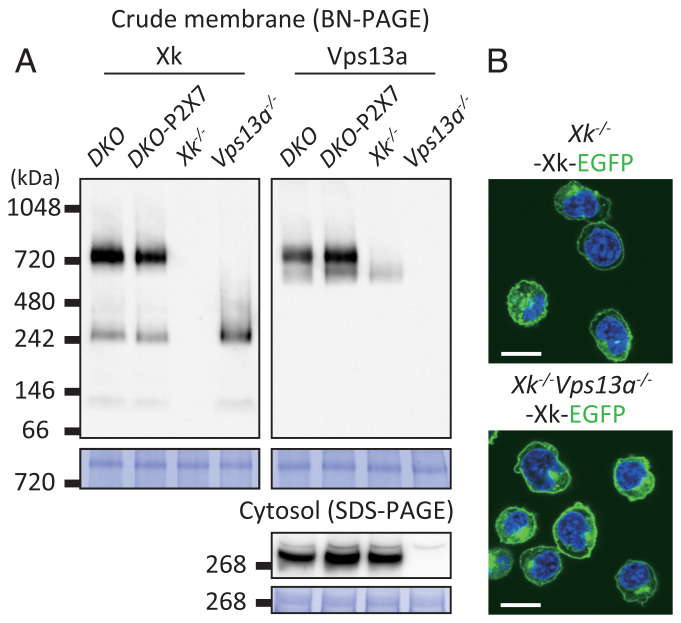
Mammalian Vps13a, a protein of ∼360 kDa, belongs to the VPS13 family (41). Some human VPS13 family members and yeast Vps13 have been shown to transport phospholipids from the endoplasmic reticulum (ER) to intracellular organelles, particularly mitochondria (42, 43). Leonzino et al. (41) postulated that VPS13 interacts with “scramblases” at the ER and target membranes. Since Xk is a relative of Xkr8 scramblase (38), we speculated that Xk and Vps13a form a complex in membranes. Membrane fractions were prepared from DKO, DKO-P2X7, Xk−/−DKO-P2X7, and Vps13a−/−DKO-P2X7, solubilized with 1.0% n-dodecyl-β-d-maltopyranoside (DDM) and 0.1% cholesteryl hemisuccinate (CHS), and separated by using Blue-native polyacrylamide gel electrophoresis (BN-PAGE) (Fig. 4A). Western blotting of the separated proteins with anti-Xk antibody (Ab) detected bands with ∼760 and 250 kDa in DKO and DKO-P2X7 cells. Neither band was detected in Xk−/− cells, indicating that they contained Xk.
Fig. 4.
Complex formation between Xk and Vps13a in WR19L cells. (A) BN-PAGE analysis of Xk and Vps13a in WR19L cells. The solubilized crude membrane fractions (4.2 µg of protein) and the cytosolic fractions (21 µg of protein) from DKO, DKO-P2X7, Xk−/−DKO-P2X7 (Xk−/−), and Vps13a−/−DKO-P2X7 (Vps13a−/−) were separated by BN-PAGE or SDS-PAGE and analyzed by Western blotting with anti-Xk (Left) or anti-Vps13a Ab (Right). Each membrane was stained with CBB and shown in the Lower panels. The positions of Mr. standard proteins are shown with their Mr. (B) The cellular localization of Xk. Xk−/−DKO-P2X7 (Xk−/−) and Xk−/−Vps13a−/−DKO-P2X7 (Xk−/−Vps13a−/−) cells were transformed with Xk–EGFP and observed by confocal microscopy in the presence of 5 µg/mL Hoechst 33342. EGFP and Hoechst signals are shown in green and blue. (Scale bars, 10 µm.)
The 760-kDa band was absent in Vps13a−/− cells, while the intensity of the 250-kDa band was stronger than those in DKO and DKO-P2X7 cells. Since membrane proteins often behave peculiarly in BN-PAGE or migrate like molecules twice larger than expected from its calculated molecular mass (Mr.) (44), the 250-kDa band may be a homodimer of Xk with a calculated Mr. of 51,114, while the 760-kDa band may be a complex of Xk and Vps13a with a calculated Mr. of 359,394. In support of this assumption, Western blotting with anti-Vps13a Ab detected the 760-kDa band in DKO and DKO-P2X7 cells. The null mutation in Xk resulted in the 760-kDa band being undetectable by anti-Vps13a Ab (Fig. 4A), suggesting that the majority of Vps13a in the membrane is associated with Xk. A faint band of ∼670 kDa detected by anti-Vps13a Ab in DKO, DKO-P2X7, and Xk−/− DKO-P2X7 cells suggested that a minor population of Vps13a interacted with molecules other than Xk in membranes.
In contrast to the results obtained from membrane fractions, the amount of the Vps13a protein in total cell lysates (Fig. 2A) or in the cytosol (Fig. 4A) was not affected by the Xk null mutation, indicating that a small portion of Vps13a was tethered or recruited to membranes via Xk. When the green fluorescent protein (GFP)-tagged Xk protein was expressed in Xk−/− DKO-P2X7 cells, the majority of GFP was present at the plasma membrane (Fig. 4B), suggesting that Xk and Vps13a function at the plasma membrane. The Vps13a null mutation did not exert adverse effects on the localization of Xk at the plasma membrane (Fig. 4B), indicating that Vps13a was not necessary for the localization of Xk at the plasma membrane.
Effects of Xk on ATP-Induced Cytolysis of CD25+CD4+ T Cells.
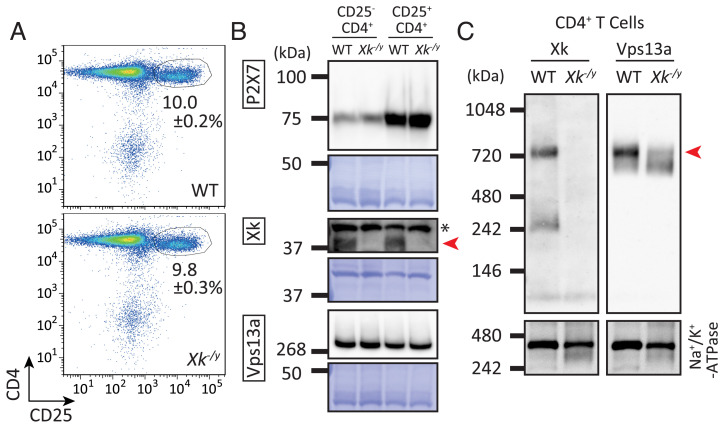
CD25+CD4+ T cells are susceptible to a high concentration of ATP for PtdSer exposure and cell death (12, 15). To examine the involvement of Xk and Vps13a in this process, we prepared CD25+ or CD25−CD4+ T cells from the spleens of wild-type and Xk−/y mice (Fig. 5A) and examined the expression of P2X7, Xk, and Vps13a. Consistent with previous findings (12, 15), Western blotting with anti-P2X7 Ab indicated that the expression of P2X7 was approximately threefold higher in CD25+CD4+ T cells than in CD25−CD4+ T cells (Fig. 5B). On the other hand, Xk and Vps13a were expressed at similar levels in CD25+ and CD25−CD4+ T cells. An analysis of the membrane proteins of CD4+ T cells by BN-PAGE indicated that Xk was present in the 760- and 250-kDa bands, similar to WR19L cells (Fig. 5C). Anti-Vps13a Ab recognized the 760-kDa band, and its intensity was markedly, but not completely, reduced in Xk-null CD4+ T cells, indicating that Vps13a in the membrane mainly complexed with Xk to form the 760-kDa complex. The minor band around 670 kDa may represent the complex that Vps13a forms with other proteins in membranes, as discussed above in WR19L cells.
Fig. 5.
Expression of P2X7, Xk, and Vps13a in mouse splenic T cells. (A) The CD25+ population in CD4+ T cells. CD4+ T cells from Xk+/y and Xk−/y mice were stained with PE–anti-CD4 mAb and FITC–anti-CD25 mAb. Staining profiles in the SYTOX Blue-negative population are shown. The percentage of the CD25+ population in CD4+ splenic T cells from three different mice are indicated as the mean ± SE. (B) Expression of P2X7, Xk, and Vps13a in CD4+ T cells. CD25−CD4+ and CD25+CD4+ populations were collected from the splenic CD4+ T cells of Xk+/y and Xk−/y mice by FACSAria II. Whole-cell lysates were prepared from cells, and 2.2 μg of protein per lane was separated by SDS-PAGE. Western blotting was performed with Abs against P2X7, Xk, or Vps13a and membranes stained with CBB. The red arrowhead shows Xk. *, non-specific band. (C) Complex formation between Xk and Vps13a in CD4+ T cells. The solubilized membrane fractions of CD4+ T cells (850 ng of protein) from Xk+/y (lane 1) and Xk−/y (lane 2) littermate male mice at 11 wk were separated by BN-PAGE, followed by Western blotting with anti-Xk (C, Left) or anti-Vps13a Ab (C, Right). Membranes were reprobed with anti-Na+/K+-ATPase mAb in the bottom panels. WT, wild-type. The red arrowhead shows the Xk-Vps13a complex.
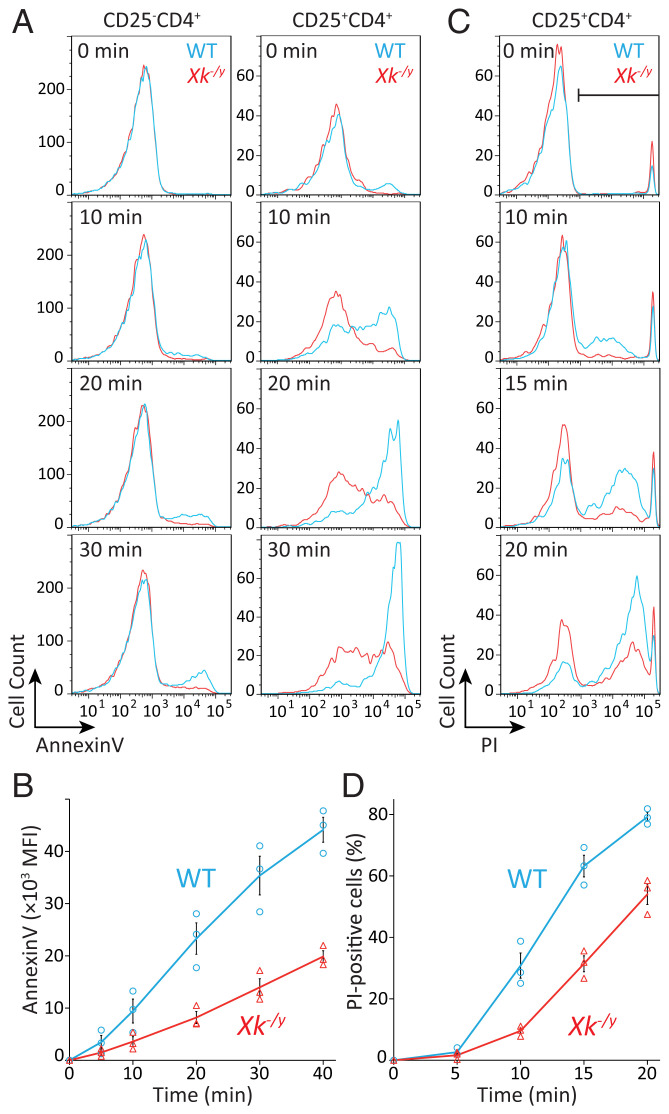
PtdSer exposure and necrotic cell death (PI positivity) were followed by the treatment of CD4+ T cells with 500 μM ATP at 10 or 37 °C, respectively. As shown in Fig. 6 A and B, consistent with their P2X7 expression levels, CD25+CD4+ T cells responded to ATP and gradually exposed PtdSer, whereas CD25−CD4+ T cells failed to respond to ATP. The deficiency of Xk in CD25+CD4+ T cells delayed the kinetics of PtdSer exposure to ∼40% of that in wild-type cells. The generation of PI-positive necrotic cells was also retarded without Xk, particularly at the early stage or within 15 min of the stimulation by ATP (Fig. 6 C and D). These results indicated that the Xk–Vps13a complex was involved in ATP-induced, P2X7-mediated PtdSer exposure and cell death in CD25+CD4+ T cells.
Fig. 6.
Involvement of Xk in ATP-induced PtdSer exposure and cytolysis in CD25+CD4+ T cells. (A) Effects of Xk on ATP-induced PtdSer exposure in CD4+ T cells. CD4+ T cells from Xk+/y and Xk−/y mice were mixed with PE–anti-CD4 mAb and FITC–anti-CD25 mAb, stimulated with 500 µM ATP at 10 °C for the indicated period in the presence of Annexin V and SYTOX Blue, and analyzed by flow cytometry. The Annexin V staining profile in SYTOX Blue-negative CD25−CD4+ (A, Left) and CD25+CD4+ (A, Right) populations is shown. (B) Experiments were performed with three different mice, and the average MFI of Annexin V staining in the CD25+CD4+ population is plotted with SE (bar). (C) Effects of Xk on ATP-induced cytolysis in CD25+CD4+ T cells. CD4+ T cells from wild-type (WT) and Xk-deficient mice were sequentially stained with LIVE/DEAD violet fluorescent dye and with APC–anti-CD4 mAb and APC–Cy7–anti-CD25 mAb. Cells were then stimulated with 500 µM ATP at 37 °C for the indicated period in the presence of 2.5 µg/mL PI and analyzed by flow cytometry. PI staining profiles in the LIVE/DEAD-negative CD25+CD4+ population are shown. Experiments were performed with three different mice. (D) The average percentage of PI-positive cells at the indicated time is plotted with SE (bars). The cell population indicated by the horizontal bar was regarded as PI-positive.
Discussion
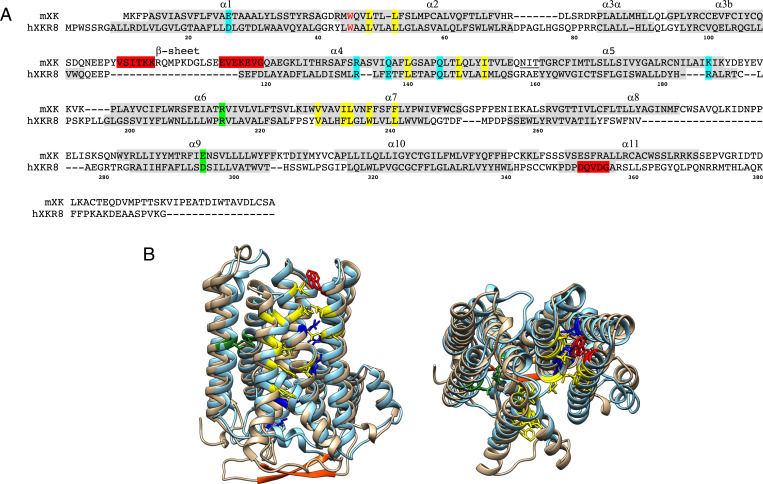
We herein demonstrated that P2X7-mediated PtdSer exposure and cytolysis (14, 39) required Xk and Vps13a, human genes of which were responsible for McLeod syndrome (28, 29) and chorea-acanthocytosis (30–32), respectively. Xk and Vps13a formed a complex at the plasma membrane, which is consistent with recent findings showing the coimmunoprecipitation of XK and VPS13A (45, 46). Xk is associated with a glycoprotein Kell with apparent Mr. of 100 kDa in red blood cells (47). However, the Kel-null mutation did not affect the migration in Xk in BN-PAGE (SI Appendix, Figs. S2 and S3), which agrees with the report that the expression of Kell is restricted to erythroid cells (48). Xk is a paralogue of Xkr8 that bidirectionally and nonspecifically scrambles phospholipids at plasma membranes in a caspase- or kinase-dependent manner (38, 49). We recently reported that human XKR8 formed a cuboid-like structure complexed with Basigin (10). Although the overall identity of the amino acid sequence of mouse Xk is less than 20% that of Xkr8 (Fig. 7A), its tertiary structure predicted by AlphaFold2 (50) is very similar to that of human XKR8 (Fig. 7B). The characteristic features of XKR8 (the presence of a hydrophobic cleft on the upper part of the molecule and charged residues in the transmembrane helices) were well conserved in mouse Xk. The amino acids (Arg-222 and Glu-327) in XK mutated in human patients with McLeod syndrome (29) formed a salt bridge in human XKR8 (Arg-214 and Asp-295) to stabilize the protein (10) (Fig. 7A). Asp-26 and Lys-134, which are indispensable for the scrambling activity of mouse Xkr8, were replaced by homologous amino acids (Glu and Arg, respectively) in mouse Xk (Fig. 7A), supporting the function of Xk as a scramblase. Caspase 3 during apoptosis cleaves off the C-terminal region of Xkr8 to activate the scramblase. Xk is not activated during apoptosis (38) and, accordingly, does not contain a caspase-recognition site. Xk, but not Xkr8, carries a β-hairpin between transmembrane α3 and α4 in the cytoplasmic region (Fig. 7). A β-hairpin is often involved in protein–protein interaction (42, 51, 52), and human VPS13A binds to cellular organelles via its C-terminal region (42), suggesting that the β-hairpin of Xk binds to β-strands in the C-terminal region of Vps13a (SI Appendix, Fig. S4).
Fig. 7.
Conserved structure between Xkr8 and Xk. (A) The amino acid sequences of mouse Xk and human XKR8 are aligned with α-helices numbered. Helices α1 to α10 are in the membrane. The residues (R214 and D295) highlighted in green form a salt bridge in XKR8 and are mutated in XK of McLeod syndrome patients. The conserved charged residues in the transmembrane are highlighted in blue, while the hydrophobic residues forming the cleft for a phospholipid in XKR8 are highlighted in yellow. The tryptophan residue (W45) that serves as a gate keeper (10) is shown in red. A caspase-recognition site in XKR8 and the β-hairpin in Xk are highlighted in red. Numbers at the bottom indicate the position of amino acid of XKR8. A putative N-glycosylation site at the extracellular loop between α4 and α5 of Xk is underlined. (B) Side (left) and top (right) views of the structure of mouse Xk (tan), predicted by AlphaFold, was superimposed on human XKR8 structure (cyan) (10). The residues were colored as above. The C-terminal tail regions of XKR8 and Xk were not included since the cryo-electron microscopy structure of hXKR8 did not reveal it (10).
The binding of ATP to P2X7 quickly activates the Xk–Vps13a complex to scramble phospholipids at 4 °C. ATP-engaged P2X7 has been shown to induce a number of biological processes that include PtdSer exposure, the formation of nonspecific cation channels, large molecule permeabilization, ATP release, membrane blebbing and actin reorganization, cytokine secretion, cell death, and cell growth (21, 23, 53, 54). The downstream molecules responsible for some of these processes have been identified, such as pannexin for ATP release (55), ROCK for membrane blebbing (56), and NLRP3 inflammasomes for interleukin-1β secretion (57). However, the molecular mechanisms by which the stimulation of P2X7 leads to various biological processes have not yet been elucidated in most cases. Similarly, the mechanism by which P2X7 activates the Xk–Vps13a complex remains unclear (SI Appendix, Fig. S4). Although the activation of P2X7 causes a Ca2+ influx, an intracellular Ca2+ chelator [1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid tetrakis(acetoxymethyl ester)] did not inhibit P2X7-mediated PtdSer exposure (27), indicating that a change in the intracellular concentration of Ca2+ is not responsible for the activation of Xk–Vps13a. In contrast to other P2X family members, P2X7 carries an extended C-terminal region that is required for ATP-stimulated cytolytic pore formation (58). The tertiary structure of P2X7 showed that this domain binds to a guanosine nucleotide with high affinity (19). Similar to other guanosine nucleotide-binding proteins, P2X7 may transiently interact with the Xk–Vps13a complex to activate its scramblase activity.
The Xkr8–Basigin complex is present at plasma membranes, and no other proteins appear to interact with this complex (59). The structure of Xkr8 also supports its intrinsic ability to scramble phospholipids (10). Mouse Xk is localized to the plasma membrane (38) (Fig. 4), and the Xk–Vps13a complex is likely to be present at the plasma membrane. Xk in Vps13a−/− cells remained in the membrane fraction, whereas Vps13a in Xk−/− cells was present at negligible levels in the membrane fraction, indicating that the Xk protein recruited Vps13a to the plasma membrane. These results also suggest that Xk is the major partner of Vps13a at plasma membranes in T lymphocytes. The strong reduction of PtdSer exposure in Vps13a-deficient cells indicated that the signal from P2X7 activated Vps13a-engaged Xk, but not its unengaged form, to scramble phospholipids. Further studies are warranted to compare the structures of Xk, Xk–Vps13a, and the activated Xk–Vps13a complex.
Xk-Vps13a–mediated PtdSer externalization was reversible at a low temperature or 4 °C. However, following a shift to room temperature, cells were quickly lysed. We previously reported that CDC50A-deficient (flippase-null) WR19L cells or those expressing the constitutively active form of TMEM16F scramblase continuously exposed PtdSer (4, 60). These cells grew at a slightly lower rate than wild-type cells, but were not lysed in the steady-state conditions. As described above, P2X7 mediates a number of biological processes (21, 23, 53, 54). In addition to the breakdown of the asymmetrical distribution of phospholipids at plasma membranes, a marked change in cell permeability (61) or the activation of the Ninjurin1-like molecule (26) may be needed for cell lysis.
Since CD25+CD4+ regulatory T cells, but not CD25−CD4+ T cells or CD8+ T cells, express P2X7 at a high level, the elevated concentration of ATP in inflamed tissues and the tumor environment has been proposed to enhance the immune system by down-regulating regulatory T cells in order to attack infected cells or tumor cells (18, 54, 62). However, when this system is excessively activated, it may lead to autoimmune disease (63, 64). The involvement of the Xk–Vps13a complex in the ATP-induced cytolysis of CD25+CD4+ T cells suggested that this process can be controlled by the up- or down-regulation of Xk-Vps13a–mediated phospholipid scrambling. In contrast to CD25+CD4+ T cells, CD25−CD4+ T cells only expressed P2X7 at negligible levels, whereas the expression levels of Vps13a and Xk were similar to those in CD25+CD4+ T cells, suggesting that a signal other than P2X7 activates Xk–Vps13a to scramble phospholipids. Under various conditions, such as adhesion to endothelial cells (65) and the activation by antigen recognition (66), T cells expose PtdSer. Further studies are needed to establish whether patients with McLeod syndrome and chorea-acanthocytosis have abnormalities in their immune system. In addition to the immune system, the development of neuroacanthocytosis in patients deficient in XK or VPS13A (29, 32) suggests that this system is involved in the homeostasis of neurons, red blood cells, and muscle cells. Since P2X7 is mainly expressed in the immune system (67), it is unlikely that P2X7 triggers the Xk-Vps13a–mediated scrambling in these cells. It will be interesting to identify the biological processes that activate the Xk–Vps13a system in these cells.
Materials and Methods
Cell Lines, Plasmids, Abs, and Reagents.
Tmem16f−/−P2rx7−/− WR19L cells (DKO), a derivative of mouse WR19L cells (ATCC; TIB-52), were described previously (27) and grown in Roswell Park Memorial Institute (RPMI) 1640 supplemented with 10% fetal calf serum (FCS). HEK293T cells (ATCC; CRL-3216) were grown in Dulbecco’s modified Eagle medium–10% FCS.
pPEF-BOS was derived from pEF-BOS, into which a puromycin resistance gene was introduced from pPUR (Clontech). pX459v2 was from Addgene. pCMV-VSV-G was provided by H. Miyoshi, RIKEN BioResource Center (Tsukuba, Japan). The pMXs-puro retroviral vector and pGag-pol-IRES-bsr packaging plasmid were from T. Kitamura, Institute of Medical Science, University of Tokyo, Tokyo. pAdVAntage was purchased from Promega.
Alexa 647-conjugated rat anti-mouse P2X7 monoclonal Ab (mAb) (clone Hano43) was from Bio-Rad Laboratories. Phycoerythrin (PE)-conjugated or allophycocyanin (APC)-conjugated rat anti-mouse CD4 mAb (clone RM4-5) was from BD Biosciences. Fluorescein isothiocyanate (FITC)- or APC–Cy7-rat anti-mouse CD25 mAb (clone PC61) was from BioLegend. Rabbit anti-P2X7 Ab was from Alomone Labs (APR-004). Rabbit anti-XK Ab (HPA019036) and rabbit anti-Chorein Ab (NBP1-85641) were from Atlas Antibodies and Novus Biologicals, respectively. Mouse anti-Na+/K+–ATPase mAb (clone 464.6) was from Abcam. Horseradish peroxidase (HRP)–goat Abs against rabbit immunoglobulins (Igs) (P0448) or mouse Igs (P0447) were from Agilent Technologies. Cy5–Annexin V was from BD Biosciences. SYTOX Blue and Hoechst 33342 were from Thermo Fisher Scientific. NBD-PC was from Avanti Polar Lipids. DDM and CHS were from Dojindo Molecular Technologies and Merck, respectively.
Genome-Wide CRISPR/Cas9 Screening.
Screening with the mouse CRISPR Knockout Pooled Library (GeCKO version [v] 2) was as described previously (27). In brief, 2 × 107 DKO transformants expressing mouse P2X7 and Cas9 were infected with a lentivirus carrying the GeCKO v2 sgRNA library at a multiplicity of infection of 0.3 and cultured in the presence of 1 µg/mL puromycin. The resultant cells (3 × 107) were chilled at 4 °C and stimulated with 60 µM BzATP for 5 min in 3 mL of Annexin buffer (10 mM Hepes–NaOH [pH 7.5], 140 mM NaCl, and 2.5 mM CaCl2). Cells were diluted 15-fold with chilled buffer, spun at 300 × g for 3 min, and suspended in 3 mL of Annexin buffer containing Cy5–Annexin V. Approximately 0.8% of cells with the lowest ability to bind Annexin V were collected by FACSAria II (BD Biosciences) and expanded. This procedure (stimulation with BzATP, sorting for reduced Annexin binding, and expansion) was repeated two more times. With genomic DNA from the sorted cells, DNA fragments containing the sgRNA sequence were amplified by PCR using PrimeSTAR GXL DNA Polymerase (Takara Bio) and primers carrying the sequence on the vector for the sgRNA library. The PCR product was subjected to PCR using PrimeSTAR HS DNA polymerase (Takara Bio) for NGS with a mixture of forward primers (NGS-Lib-Fwd-1–10) and a common reverse primer as described previously (27). The PCR product was purified and analyzed with MiSeq (Illumina) using the MiSeq Reagent Kit v3 (Illumina). NGS reads were related to reference sgRNA sequences in the library, and the number of each sgRNA was counted by using custom-made software from Amelieff.
Gene Editing and Transformation of Cell Lines.
Xk and Vps13a genes in WR19L cells were knocked out by using the CRISPR/Cas9 system as described previously (4). Complementary oligonucleotides carrying the sgRNA target sequence (Xk, 5′-CTTTCTCCACCTCCTCTGAA-3′; Vps13a, 5′-TGACCAACTTTAACTTTGAA-3′; and Kel, 5′-GACAGCTAGCAGCACCCATC-3′) were annealed, ligated into pX459v2, and introduced into WR19L cells by electroporation using a NEPA21 Super Electroporator (Nepa Gene). In some cases, transfection was performed twice at a 3-d interval. At 20 to 30 h later, cells were treated with 1 μg/mL puromycin for 34 h. Single clones isolated by limiting dilutions were screened by sequencing the target regions of the Xk, Vps13a, and Kel genes. Cells containing the out-of-frame mutation were identified as Xk−/−, Vps13a−/−, Xk−/−Vps13a−/−, and Kel−/− cells.
Mouse Xk complementary DNA (cDNA) (NM_023500) was as described previously (38), tagged with FLAG or enhanced GFP (EGFP) at the C terminus, and inserted into the pMXs-puro vector. Mouse Vps13a cDNA (NM_173028) was prepared by RT-PCR from the bone marrow of C57BL/6J mice and inserted into pPEF-BOS. The authenticity of cDNAs was confirmed by DNA sequencing. Regarding transformation, HEK293T cells were transfected with pMXs-puro carrying Xk cDNA with pGag-pol-IRES-bsr, pCMV-VSV-G, and pAdVAntage using Fugene 6 (Promega) and cultured for 48 h. Retroviruses in the supernatant were collected by centrifugation at 6,000 × g at 4 °C for 16 h and used to infect WR19L cells. pPEF-BOS carrying Vps13a cDNA was introduced into WR19L cells by electroporation using NEPA21. Stable transformants were selected by culturing in the presence of 1 µg/mL puromycin.
Mice.
Xk-targeted mice (C57BL/6N-Atm1Brd Xktm1a(KOMP)Mbp/MbpMmucd) were obtained from Mutant Mouse Resource and Research Centers and crossed with CAG-Cre transgenic mice [B6.Cg-Tg(CAG-Cre)CZ-MO2Osb] (RIKEN) to generate mice that lost a part of exon 3 in the Xk gene (SI Appendix, Fig. S5). Knockout mice were maintained on a C57BL/6N genetic background.
All mice were housed in a specific pathogen-free facility at the Research Institute for Microbial Diseases, Osaka University. All mouse studies were approved by the Ethics Review Committee for Animal Experimentation of the Research Institute for Microbial Diseases, Osaka University.
Preparation of Splenic CD4+ T Cells.
To prepare T cells from the mouse spleen, the spleens of male mice aged 8 to 11 wk were mashed in phosphate-buffered saline (PBS) containing 2% FCS (PBS/FCS) with frosted glass slides, and dispersed cells were passed through a 70-µm cell strainer. CD4+ T cells were selected by using an EasySep Mouse CD4+ T Cell Isolation Kit (STEMCELL Technologies), according to the manufacturer’s instructions. In brief, cells from the spleen were suspended in PBS/FCS containing 1 mM ethylenediaminetetraacetic acid (EDTA) at a density of 1 × 108 cells/mL and mixed with 1/20 volume of rat serum and 1/20 volume of a mixture of biotinylated Abs against mouse splenocytes other than CD4+ T cells. The mixture was incubated at room temperature for 10 min, and Ab-labeled cells were removed by using streptavidin magnetic beads. Isolated CD4+ T cells were stained on ice for 30 min with 1 µg/mL PE- or APC-labeled anti-CD4 mAb and 1 µg/mL FITC–anti-CD25 mAb in PBS/FCS, washed with PBS, and analyzed by flow cytometry using a FACSCanto II (BD Biosciences) or collected by FACSAria II (BD Biosciences).
Flow Cytometry for P2X7.
P2X7 on the cell surface was detected by flow cytometry as described previously (27). In brief, 5 × 105 cells were washed with PBS/FCS and incubated on ice for 30 min in 100 µL of PBS/FCS containing 40-fold diluted Alexa 647–anti-P2X7 mAb. Cells were washed with PBS/FCS, suspended in 250 µL of PBS/FCS containing 250 nM SYTOX Blue, and analyzed by flow cytometry using FACSCanto II. Data were analyzed by FlowJo (BD Biosciences).
ATP-Induced PtdSer Exposure and Cell Death.
ATP-induced PtdSer exposure was assayed by Annexin V binding, while dead cells were detected by PI staining as described previously (9, 27), with slight modifications. In brief, DKO-derived cells in RPMI 1640–10% FCS were preincubated at 4 °C for 10 min, collected by centrifugation, and resuspended in chilled Annexin buffer containing 300- to 400-fold diluted Cy5–Annexin V and 2.5 µg/mL PI. Cells were stimulated at 4 °C with 500 µM ATP for 5 min, and an aliquot of cells was analyzed by flow cytometry using FACSCanto II. The remaining cells were kept at room temperature for an additional 10 min and analyzed by flow cytometry.
To detect PtdSer exposure in splenic T cells, 1 × 106 CD4+ T cells were initially stained with 1 µg/mL PE–anti-CD4 mAb and 1 µg/mL FITC–anti-CD25 mAb on ice for 30 min, preincubated in PBS at 10 °C for 10 min, and then incubated with ATP in Annexin buffer containing Cy5–Annexin V and 250 nM SYTOX Blue at 10 °C. To assay ATP-dependent cell death, 1 × 106 CD4+ T cells were incubated on ice for 30 min in PBS with 1,000-fold-diluted Violet Dead Cell Stain (LIVE/DEAD Fixable Dead Cell Stain Kit, Thermo Fisher Scientific), stained for 30 min with 1 µg/mL APC–anti-CD4 mAb and 1 µg/mL APC–Cy7–anti-CD25 mAb in chilled PBS/FCS, and washed with PBS. Cells were then preincubated at 37 °C for 10 min in 1 mL of Annexin buffer containing 2.5 µg/mL PI. ATP was added to the mixture at a final concentration of 500 µM and incubated at 37 °C. An aliquot of the cell suspension was analyzed by flow cytometry for PI+ in the LIVE/DEAD-negative population.
ATP-Induced Internalization of PtdCho.
The internalization of PtdCho to the inner leaflet of the plasma membrane was assayed as described (9), with modifications. In brief, 3 × 105 cells in RPMI 1640–10% FCS were preincubated at 4 °C for 10 min, collected by centrifugation, and incubated at 4 °C in 300 µL of chilled Annexin buffer containing 250 nM NBD-PC and 500 µM ATP. After the incubation, a 90-µL aliquot was mixed with 150 µL of Annexin buffer containing 5 mg/mL fatty acid-free bovine serum albumin (BSA), incubated on ice for 1 min, and analyzed by FACSCanto II.
Confocal Microscopy.
WR19L cell transformants expressing Xk–EGFP were washed with Hanks’ Balanced Salt Solution (HBSS) supplemented with 2% FCS (HBSS/FCS) and suspended in HBSS/FCS containing 5 µg/mL Hoechst 33342. Cells were then seeded on a glass-bottomed dishes (Matsunami) and observed under a confocal fluorescence microscope (FV1000-D, Olympus).
Preparation of Whole-Cell Lysates, Membrane Fractions, and Cytosolic Fractions.
To prepare whole-cell lysates, cells were incubated at 4 °C for 1 h in lysis buffer [20 mM Tris⋅HCl (pH 7.5), 1% DDM, 0.1% CHS, 50 mM KCl, 1 mM MgCl2, 10% glycerol, 1 mM (p-amidinophenyl)methanesulfonyl fluoride hydrochloride (p-APMSF), and a protease inhibitor mixture (cOmplete, EDTA-free; Roche Diagnostics)]. After the removal of insoluble materials by centrifugation at 20,000 × g at 4 °C for 20 min, the supernatants obtained were used as whole-cell lysates.
Regarding membrane and cytosolic fractions, 1.8 to 8.8 × 107 cells were washed with PBS and homogenized at 4 °C using a Dounce homogenizer in 2.5 mL of solution A (10 mM Tris⋅HCl [pH 7.5] and 1 mM p-APMSF). The homogenate was diluted with 2.5 mL of solution B [10 mM Tris⋅HCl (pH 7.5), 500 mM sucrose, 100 mM KCl, 10 mM MgCl2, and 1 mM p-APMSF] and sequentially spun at 800 × g at 4 °C for 10 min and then at 8,000 × g for 10 min to remove nuclei and mitochondria. Samples were then spun at 100,000 × g for 60 min. The supernatants were concentrated by ultrafiltration membranes (Amicon Ultra 10K, Merck) and used as the cytosolic fractions. The precipitates were suspended in 120 to 200 µL of the lysis buffer, passed through a 29G needle, and then incubated with rotation at 4 °C for 2 h. Insoluble materials were removed as described above, and the supernatant was used as the crude membrane fraction.
SDS-PAGE, BN-PAGE, and Western Blotting.
Samples for sodium dodecyl sulfate (SDS)-PAGE were incubated at room temperature for 2 h in SDS sample buffer (62.5 mM Tris⋅HCl [pH 6.8], 2% SDS, 10% glycerol, 2.5% 2-mercaptoethanol, and 0.005% bromophenol blue) and separated by 7.5% or 10% SDS-PAGE (Nacalai Tesque). Precision Plus Protein Dual Color Standards (Bio-Rad Laboratories) and HiMark Prestained Protein Standard (Thermo Fisher Scientific) were used as molecular weight (m.w.) markers. Samples for BN-PAGE were mixed with 1/4 volume of NativePAGE Sample Buffer (4×) and 1/20 volume of NativePAGE 5% Coomassie Brilliant Blue (CBB) G-250 Sample Additive (20×) and loaded onto a NativePAGE Novex 4 to 16% BisTris gel (Thermo Fisher Scientific). After electrophoresis at 150 V at 4 °C for 35 min, the concentration of CBB G-250 in the running buffer was changed from 0.02 to 0.002%, and samples were subjected to further successive electrophoresis at 150 V for 25 min, 250 V for 30 min, and 350 V for 20 min. The NativeMark Unstained Protein Standard (Thermo Fisher Scientific) was used as m.w. markers.
Regarding Western blotting, proteins in gels were transferred to polyvinylidene difluoride membranes (Immobilon-P, Merck) immediately after SDS-PAGE or after the incubation of BN-PAGE gels in SDS-PAGE running buffer (25 mM Tris⋅HCl [pH 8.3], 192 mM glycine, and 0.1% SDS) at room temperature for 15 min. Nonspecific binding sites on the membranes were blocked by incubating with 5% skim milk and probed with HRP-labeled Abs, followed by detection with the Immobilon Western Chemiluminescent HRP substrate (Merck) or Western Lightning Plus-ECL (PerkinElmer). As a loading control, proteins on the membrane were stained with CBB R-250 or reprobed with anti-Na+/K+–ATPase mAb after quenching HRP with 15% H2O2 in PBS (68).
Supplementary Material
Acknowledgments
We thank M. Kamada for secretarial assistance. This work was supported in part by Japan Society for the Promotion of Science Grant-in-Aid 21H04770 (to S.N.).
Footnotes
Reviewers: M.H., Friedrich-Alexander-Universitat Erlangen-Nurnberg Naturwissenschaftliche Fakultat; A.L., Technische Universitat Dresden; and J.S., Walter and Eliza Hall Institute of Medical Research.
Competing interest statement: Y.R. is on a leave of absence from Otsuka Pharmaceutical Co. S.N. is one of the co-authors of a review article on cell death published in January 2018 [L. Galluzzi et al., Cell Death Differ. 25, 486–541 (2018)], of which two of the referees were co-authors.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2119286119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix. All reagents developed in this report are available on request (to S.N.).
References
- 1.Bevers E. M., Williamson P. L., Getting to the outer leaflet: Physiology of phosphatidylserine exposure at the plasma membrane. Physiol. Rev. 96, 605–645 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lorent J. H., et al. , Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 16, 644–652 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doktorova M., Symons J. L., Levental I., Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat. Chem. Biol. 16, 1321–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Segawa K., et al. , Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science 344, 1164–1168 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Andersen J. P., et al. , P4-ATPases as phospholipid flippases—Structure, function, and enigmas. Front. Physiol. 7, 275 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagata S., Suzuki J., Segawa K., Fujii T., Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 23, 952–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kornberg R. D., McConnell H. M., Lateral diffusion of phospholipids in a vesicle membrane. Proc. Natl. Acad. Sci. U.S.A. 68, 2564–2568 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki J., Denning D. P., Imanishi E., Horvitz H. R., Nagata S., Xk-related protein 8 and CED-8 promote phosphatidylserine exposure in apoptotic cells. Science 341, 403–406 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Suzuki J., Umeda M., Sims P. J., Nagata S., Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Sakuragi T., et al. , The tertiary structure of the human Xkr8-Basigin complex that scrambles phospholipids at plasma membranes. Nat. Struct. Mol. Biol. 28, 825–834 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunner J. D., Lim N. K., Schenck S., Duerst A., Dutzler R., X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Aswad F., Kawamura H., Dennert G., High sensitivity of CD4+CD25+ regulatory T cells to extracellular metabolites nicotinamide adenine dinucleotide and ATP: A role for P2X7 receptors. J. Immunol. 175, 3075–3083 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Courageot M.-P., Lépine S., Hours M., Giraud F., Sulpice J.-C., Involvement of sodium in early phosphatidylserine exposure and phospholipid scrambling induced by P2X7 purinoceptor activation in thymocytes. J. Biol. Chem. 279, 21815–21823 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Taylor S. R. J., et al. , Sequential shrinkage and swelling underlie P2X7-stimulated lymphocyte phosphatidylserine exposure and death. J. Immunol. 180, 300–308 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Scheuplein F., et al. , NAD+ and ATP released from injured cells induce P2X7-dependent shedding of CD62L and externalization of phosphatidylserine by murine T cells. J. Immunol. 182, 2898–2908 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Yoshida T., Kakizuka A., Imamura H., BTeam, a novel BRET-based biosensor for the accurate quantification of ATP concentration within living cells. Sci. Rep. 6, 39618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pellegatti P., et al. , Increased level of extracellular ATP at tumor sites: In vivo imaging with plasma membrane luciferase. PLoS One 3, e2599 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linden J., Koch-Nolte F., Dahl G., Purine release, metabolism, and signaling in the inflammatory response. Annu. Rev. Immunol. 37, 325–347 (2019). [DOI] [PubMed] [Google Scholar]
- 19.McCarthy A. E., Yoshioka C., Mansoor S. E., Full-length P2X7 structures reveal how palmitoylation prevents channel desensitization. Cell 179, 659–670.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benzaquen J., et al. , Alternative splicing of P2RX7 pre-messenger RNA in health and diseases: Myth or reality? Biomed. J. 42, 141–154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Di Virgilio F., Dal Ben D., Sarti A. C., Giuliani A. L., Falzoni S., The P2X7 receptor in infection and inflammation. Immunity 47, 15–31 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Grassi F., The P2X7 receptor as regulator of T cell development and function. Front. Immunol. 11, 1179 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.North R. A., Molecular physiology of P2X receptors. Physiol. Rev. 82, 1013–1067 (2002). [DOI] [PubMed] [Google Scholar]
- 24.MacKenzie A., et al. , Rapid secretion of interleukin-1beta by microvesicle shedding. Immunity 15, 825–835 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Galluzzi L., et al. , Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kayagaki N., et al. , NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 591, 131–136 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Ryoden Y., Fujii T., Segawa K., Nagata S., Functional expression of the P2X7 ATP receptor requires Eros. J. Immunol. 204, 559–568 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Ho M., et al. , Isolation of the gene for McLeod syndrome that encodes a novel membrane transport protein. Cell 77, 869–880 (1994). [DOI] [PubMed] [Google Scholar]
- 29.Jung H. H., Danek A., Walker R. H., Frey B. M., Gassner C., “McLeod neuroacanthocytosis syndrome” in GeneReviews, Adam M. P., et al., Eds. (U.S. National Library of Medicine, Seattle, 2019), pp. 1–23. [Google Scholar]
- 30.Ueno S., et al. , The gene encoding a newly discovered protein, chorein, is mutated in chorea-acanthocytosis. Nat. Genet. 28, 121–122 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Rampoldi L., et al. , A conserved sorting-associated protein is mutant in chorea-acanthocytosis. Nat. Genet. 28, 119–120 (2001). [DOI] [PubMed] [Google Scholar]
- 32.Velayos Baeza A., et al. , “Chorea-acanthocytosis” in GeneReviews, Adam M. P. et al., Eds. (U.S. National Library of Medicine, Seattle, 2019), pp. 1–24. [Google Scholar]
- 33.Fujii T., Sakata A., Nishimura S., Eto K., Nagata S., TMEM16F is required for phosphatidylserine exposure and microparticle release in activated mouse platelets. Proc. Natl. Acad. Sci. U.S.A. 112, 12800–12805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmid R., Evans R. J., ATP-gated P2X receptor channels: Molecular insights into functional roles. Annu. Rev. Physiol. 81, 43–62 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Constantinescu P., et al. , P2X7 receptor activation induces cell death and microparticle release in murine erythroleukemia cells. Biochim. Biophys. Acta 1798, 1797–1804 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Shalem O., et al. , Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Govin J., et al. , Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J. Cell Biol. 176, 283–294 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki J., Imanishi E., Nagata S., Exposure of phosphatidylserine by Xk-related protein family members during apoptosis. J. Biol. Chem. 289, 30257–30267 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Di Virgilio F., et al. , Cytolytic P2X purinoceptors. Cell Death Differ. 5, 191–199 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Mackenzie A. B., Young M. T., Adinolfi E., Surprenant A., Pseudoapoptosis induced by brief activation of ATP-gated P2X7 receptors. J. Biol. Chem. 280, 33968–33976 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Leonzino M., Reinisch K. M., De Camilli P., Insights into VPS13 properties and function reveal a new mechanism of eukaryotic lipid transport. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1866, 159003 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar N., et al. , VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 217, 3625–3639 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeshaw W. M., et al. , Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. eLife 8, e43561 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heuberger E. H., Veenhoff L. M., Duurkens R. H., Friesen R. H. E., Poolman B., Oligomeric state of membrane transport proteins analyzed with blue native electrophoresis and analytical ultracentrifugation. J. Mol. Biol. 317, 591–600 (2002). [DOI] [PubMed] [Google Scholar]
- 45.Park J.-S., Neiman A. M., XK is a partner for VPS13A: A molecular link between chorea-acanthocytosis and McLeod syndrome. Mol. Biol. Cell 31, 2425–2436 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urata Y., et al. , Novel pathogenic XK mutations in McLeod syndrome and interaction between XK protein and chorein. Neurol. Genet. 5, e328 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russo D., Redman C., Lee S., Association of XK and Kell blood group proteins. J. Biol. Chem. 273, 13950–13956 (1998). [DOI] [PubMed] [Google Scholar]
- 48.Lee S., Zambas E. D., Marsh W. L., Redman C. M., The human Kell blood group gene maps to chromosome 7q33 and its expression is restricted to erythroid cells. Blood 81, 2804–2809 (1993). [PubMed] [Google Scholar]
- 49.Sakuragi T., Kosako H., Nagata S., Phosphorylation-mediated activation of mouse Xkr8 scramblase for phosphatidylserine exposure. Proc. Natl. Acad. Sci. U.S.A. 116, 2907–2912 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham B. C., Wells J. A., Minimized proteins. Curr. Opin. Struc. Biol. 7, 457–462 (1997). [DOI] [PubMed] [Google Scholar]
- 52.Chothia C., et al. , Conformations of immunoglobulin hypervariable regions. Nature 342, 877–883 (1989). [DOI] [PubMed] [Google Scholar]
- 53.Choi J. H., Ji Y. G., Ko J. J., Cho H. J., Lee D. H., Activating P2X7 receptors increases proliferation of human pancreatic cancer cells via ERK1/2 and JNK. Pancreas 47, 643–651 (2018). [DOI] [PubMed] [Google Scholar]
- 54.Virgilio F. D., Sarti A. C., Falzoni S., Marchi E. D., Adinolfi E., Extracellular ATP and P2 purinergic signalling in the tumour microenvironment. Nat. Rev. Cancer 18, 601–618 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Pelegrin P., Surprenant A., Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 25, 5071–5082 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morelli A., et al. , Extracellular ATP causes ROCK I-dependent bleb formation in P2X7-transfected HEK293 cells. Mol. Biol. Cell 14, 2655–2664 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qu Y., Franchi L., Núñez G., Dubyak G. R., Nonclassical IL-1 β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 179, 1913–1925 (2007). [DOI] [PubMed] [Google Scholar]
- 58.Smart M. L., et al. , P2X7 receptor cell surface expression and cytolytic pore formation are regulated by a distal C-terminal region. J. Biol. Chem. 278, 8853–8860 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Suzuki J., Imanishi E., Nagata S., Xkr8 phospholipid scrambling complex in apoptotic phosphatidylserine exposure. Proc. Natl. Acad. Sci. U.S.A. 113, 9509–9514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Segawa K., Suzuki J., Nagata S., Constitutive exposure of phosphatidylserine on viable cells. Proc. Natl. Acad. Sci. U.S.A. 108, 19246–19251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Virginio C., MacKenzie A., North R. A., Surprenant A., Kinetics of cell lysis, dye uptake and permeability changes in cells expressing the rat P2X7 receptor. J. Physiol. 519, 335–346 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Belkaid Y., Regulatory T cells and infection: A dangerous necessity. Nat. Rev. Immunol. 7, 875–888 (2007). [DOI] [PubMed] [Google Scholar]
- 63.Figliuolo V. R., et al. , P2X7 receptor promotes intestinal inflammation in chemically induced colitis and triggers death of mucosal regulatory T cells. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1183–1194 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Schenk U., et al. , ATP inhibits the generation and function of regulatory T cells through the activation of purinergic P2X receptors. Sci. Signal. 4, ra12 (2011). [DOI] [PubMed] [Google Scholar]
- 65.Qu J., et al. , Phosphatidylserine-dependent adhesion of T cells to endothelial cells. Biochim. Biophys. Acta 1501, 99–115 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Fischer K., et al. , Antigen recognition induces phosphatidylserine exposure on the cell surface of human CD8+ T cells. Blood 108, 4094–4101 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Jacob F., Pérez Novo C., Bachert C., Van Crombruggen K., Purinergic signaling in inflammatory cells: P2 receptor expression, functional effects, and modulation of inflammatory responses. Purinergic Signal. 9, 285–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sennepin A. D., et al. , Multiple reprobing of Western blots after inactivation of peroxidase activity by its substrate, hydrogen peroxide. Anal. Biochem. 393, 129–131 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. All reagents developed in this report are available on request (to S.N.).