Abstract
Acinetobacter baumannii is a Gram-negative bacterium responsible for many hospital-acquired infections including ventilator-associated pneumonia and sepsis. We have previously identified A. baumannii thioredoxin A protein (TrxA) as a virulence factor with a multitude of functions including reduction of protein disulfides. TrxA plays an important role in resistance to oxidative stress facilitating host immune evasion in part by alteration of type IV pili and cell surface hydrophobicity. Other virulence factors such as outer membrane vesicles (OMV) shed by bacteria have been shown to mediate bacterial intercellular communication and modulate host immune response. To investigate whether OMVs can be modulated by TrxA, we isolated OMVs from wild type (WT) and TrxA-deficient (ΔtrxA) A. baumannii clinical isolate Ci79 and carried out a functional and proteomic comparison. Despite attenuation of ΔtrxA in a mouse challenge model, pulmonary inoculation of ΔtrxA OMVs resulted in increased lung permeability compared to WT OMVs. Furthermore, ΔtrxA OMVs induced more J774 macrophage-like cell death than WT OMVs. This ΔtrxA OMV-mediated cell death was abrogated when cells were incubated with protease-K-treated OMVs suggesting OMV proteins were responsible for cytotoxicity. We therefore compared WT and mutant OMV proteins using proteomic analysis. We observed that up-regulated and unique ΔtrxA OMV proteins consisted of many membrane bound proteins involved in small molecule transport as well as proteolytic activity. Bacterial OmpA, metalloprotease, and fimbrial protein have been shown to enhance mammalian cell apoptosis through various mechanisms. Differential packaging of these proteins in ΔtrxA OMVs may contribute to the increased cytotoxicity observed in this study.
Keywords: Acinetobacter, bacteria, cytotoxicity, outer membrane vesicle, proteomic, thioredoxin
Impact statement
While most studies of thioredoxin have focused on its role in maintenance of redox homeostasis and oxidative stress defense via its antioxidant property, attempts to decipher thioredoxin-mediated functions via protein regulation is lagging. Previously, we established that deletion of TrxA in Acinetobacter baumannii impaired its ability to colonize mouse mucosal surface. Biophysical and biochemical characterization of the TrxA-null mutant (ΔtrxA) revealed an increase in surface hydrophobicity and differential membrane protein expression compared to the parental strain. In this study, we extend our knowledge of TrxA-mediated virulence regulation by examining bacterial outer membrane vesicles (OMV). Although ΔtrxA bacteria are attenuated, we observed ΔtrxA OMVs are more cytotoxic than wild type OMVs. Treatment of OMVs with proteases reduced significantly macrophage cell death compared to untreated OMVs suggesting that surface-exposed membrane proteins are responsible for the observed cytotoxic effects. Proteomic analysis suggests differential packaging of OMV contents in ΔtrxA may contribute to cytotoxicity.
Introduction
Emergent multi-drug resistant Acinetobacter baumannii is one of the major causes of hospital-acquired infections. 1 This bacterium is extremely resistant to desiccation which promotes persistence and spread in healthcare facilities.2,3 A. baumannii colonizes in various tissues and organs and frequently leads to probe-related urinary system infections, surgical site infections, catheter-related blood circulatory septicemia, and ventilation-associated pneumonia. 4 Furthermore, due to its natural competence for acquiring antimicrobial-resistance genes,5,6 A. baumannii has been highlighted by CDC as one of the top drug-resistant threats in the United States and the World Health Organization listed A. baumannii as a global “Priority 1: Critical” pathogen for new antibiotic R&D. One approach to develop new therapeutics against Acinetobacter infection is based on identification and characterization of bacterial virulence factors as novel drug targets.
Combinatorial approaches involving genomic, phenotypic, and infection model analyses have led to identification of important virulence factors, such as LPS, outer membrane vesicles (OMV), porins, and protein secretion systems in A. baumannii. 7 Typical Gram-negative bacterial OMVs are small (50–250 nm), bilayered vesicles released from the bacterial cell wall during cell growth. 8 A. baumannii OMVs contain multiple virulence factors, including outer membrane protein A (OmpA), proteases, and phospholipases. 9 A. baumannii OMVs also serve as a DNA carrier for horizontal antibiotic-resistant gene transfer. 10 Furthermore, upon interaction with mammalian hosts, A. baumannii OMVs elicit a pro-inflammatory response via surface-exposed membrane proteins in vitro and trigger a potent innate immune response in vivo. 11 These findings collectively underscore the multifaceted functions of A. baumannii OMVs and their important role in bacterial pathogenesis. During growth adaptation of A. baumannii, biogenesis and protein composition of OMVs can be modulated by in vitro stress (e.g. imipenem treatment) and differential expression of endogenous proteins (e.g. OmpA).12,13
Recently, we identified thioredoxin as a virulence factor in A. baumannii via its dissociation of the secretory component from sIgA thus evading the host immune response and facilitating bacterial colonization in the gastrointestinal tract. 14 Thioredoxin consists of thioredoxin (TrxA), thioreductase (TrxR), and NADPH. TrxA is a small ubiquitous redox protein of approximately 12 kDa which contributes to a number of cellular processes including transcription regulation, cell division, energy transduction, and several biosynthetic pathways. 15 Due to its essential role in protein disulfide bond reduction and maintenance of protein function, we investigated if deletion of TrxA gene in A. baumannii Ci79 clinical isolate affected the characteristics of OMVs by comparative proteomic and host response assays.
Materials and methods
Isolation of OMV
OMVs were isolated from the early stationary phase A. baumannii Ci79 wild type 16 and TrxA-null mutant (ΔtrxA) 14 cultures using a modified ultracentrifugation method reported by Kwon et al. 9 Luria-Bertani (LB) broth (500 ml) was inoculated with an overnight bacterial culture (5 ml), and incubated at 37°C with agitation (225 rpm) until the culture OD600 reached 0.9. The bacterial culture was centrifuged at 4000 × g for 15 min to remove intact bacteria. Supernatant was then filtered through a 0.22-μm vacuum filter to remove cell debris. The filtrate was then passed through a 100 KDa cut-off cellulose filter (Ultracel-100K, Millipore, Burlington, MA) to remove soluble proteins. The retentate was centrifuged at 150,000 × g for 2 h to pellet OMVs. The OMV pellet was resuspended in Hanks' Balanced Salt Solution (HBSS, Lonza Bioscience, Alpharetta, GA), aliquoted, and stored at –20°C until use.
Animals
All animal experiments were performed using four to six-week-old specific pathogen free C57BL/6 mice. Animals were housed in the animal facility of the University of Texas at San Antonio, and all experiments were conducted in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee (IACUC).
Transmission electron microscope
OMVs were placed onto Formvar/carbon coated grids and negatively stained with 2% (w/v) uranyl acetate for visualization using a JEM-1400(Plus) Transmission Electron Microscope (Electron Microscopy Lab, Department of Pathology, The University of Texas Health Science Center at San Antonio, San Antonio, TX).
FITC-Dextran permeability assay
Mice were intranasally inoculated with Ci79 OMV (50 μg/50 μl), ΔtrxA OMV (50 μg/50 μl), or Hanks’ Balanced Salt Solution (HBSS, Corning, Manassas, VA) alone (50 μl) as control. After 3 h, 50 µl of FITC-Dextran (MW 3000, Invitrogen, Carlsbad, CA) was introduced into the airways (10 mg/kg), and alteration of lung permeability was measured by FITC-Dextran leakage from the airways into the blood 1 h after OMV administration. Blood was obtained by cardiac puncture, transferred to a tube containing 10 μl EDTA (60 mg/ml), and centrifuged at 7000 rpm for 10 min. The Fluorescence Index of plasma FITC-Dextran was determined at 495 nm (excitation) and 528 nm (emission).
Live/dead staining by acridine orange/ethidium bromide dye
J774 cells were seeded (2.5 × 105 cells/well) for 2 h in a 96-well plate. Cells were then washed once with culture medium (DMEM + 10% FBS; D10) and incubated with OMVs at two different concentrations (1 µg or 10 µg protein in 100 µl D10 medium per well) for up to 4 h. Cells cultured with D10 alone were used as control and exhibited no more than 1% cell death at the conclusion of each experiment. Protease K at a concentration of 100 μg/ml was used to degrade OMV surface proteins in some samples. After 1 and 4 h incubations, cells were washed once with cell medium, and 75 μl acridine orange/ethidium bromide (AO/EB) dye was added, and cell viability determined using a fluorescence microscope.
OMV proteomics
The OMV-associated proteins were subjected to trypsin digestion for capillary LC/MS/MS analysis. WT and ΔtrxA OMV proteins were identified by mapping mass spectra to translated protein sequences from the Ci79 genome (Genbank accession AVOD00000000). 16 Analysis of raw mass spectrometric data was performed using the MaxQuant software package (version 1.6.3.3). 17 Tandem mass spectra (MS/MS) were searched by the Andromeda search engine 18 against Ci79 protein sequences using the following parameter settings: (a) enzyme specificity: trypsin, (b) mass tolerance: 0.5 Da, (c) maximum of two missed cleavages. Oxidations (M), acetylation (N), pyro-Glu (E), and Glnpyro (Q) were specified as variable modifications. Carbamidomethyl (C) was checked as fixed modification. The maximum false peptide and protein discovery rate were specified as 0.01. Minimum peptide length was set at seven amino acids. Proteins matched with two or more peptides (with at least one unique peptide) were considered as reliably identified. Database searches were performed with a mass tolerance of 20 ppm for precursor ion for mass calibration, and with a 6 ppm tolerance after calibration. The other parameters were set at default. 19 We used the “protein group” output from MaxQuant for all subsequent proteomic analysis. To assess differences in the abundance of proteins, intensity based absolute label-free quantification (LFQ) values were used for three biological replicates of Ci79 wild type and ΔtrxA OMVs. Two-tailed Student t test was used for statistical determination of protein abundance between WT and ΔtrxA OMV proteins.
Results
ΔtrxA OMVs enhanced lung permeability
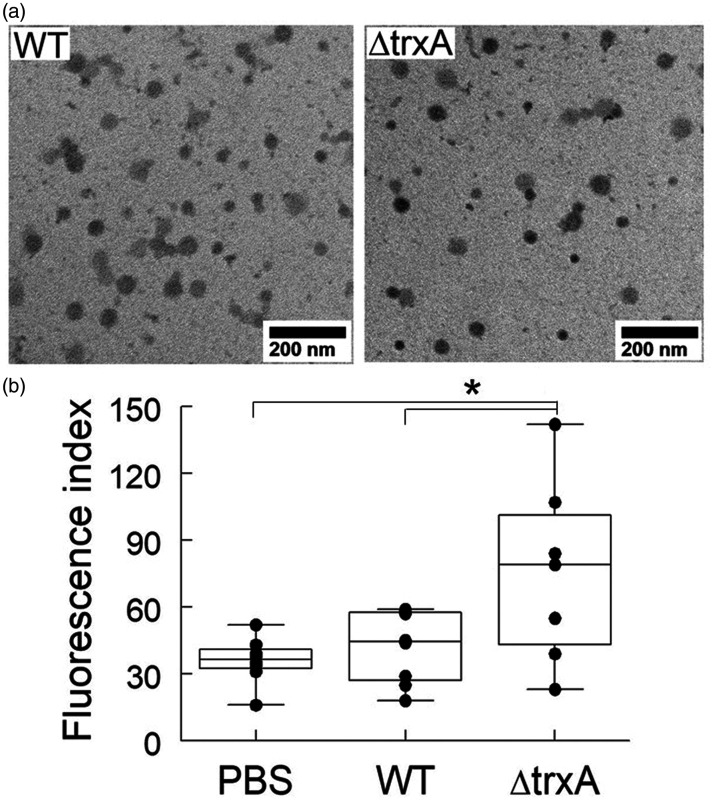
OMVs were isolated by ultracentrifugation from culture media of A. baumannii Ci79 and ΔtrxA strains grown at early stationary phase. As shown in Figure 1(a), isolated OMVs from Ci79 WT and ΔtrxA strains exhibited comparable size when viewed under the transmission electron microscope (38–80 nm and 20–95 nm, respectively). It has been reported that pulmonary inoculation of A. baumannii OMVs induced inflammation in the lungs including neutrophilic infiltration, hemorrhage, and detachment of bronchiolar epithelial cells. 11 In addition, dissemination of A. baumannii from lungs to other organs following pulmonary bacterial challenge is common. Thus, we investigated whether TrxA-deficiency altered OMV-mediated inflammation by measuring lung permeability via leakage of FITC-labeled Dextran. As shown in Figure 1(b) while no significant difference between the WT Ci79 OMVs and the PBS (control) was observed, intranasal injection of ΔtrxA OMVs (50 µg) enhanced lung permeability in mice as evidenced by the higher amount of FITC-Dextran translocated from the lungs to the blood.
Figure 1.
Treatment with ΔtrxA OMVs enhances lung permeability. (a) Transmission electron microscopy of OMVs prepared from Acinetobacter baumannii Ci79 wild type and ΔtrxA strains. (b) Mice were intranasally inoculated with Ci79 OMV (50 μg/50 μl), ΔtrxA OMV (50 μg/50 μl), or PBS alone (50 μl) as control. After 3 h, 50 µl FITC-Dextran was introduced into the airways (10 mg/kg). Alteration of lung permeability was measured by FITC-Dextran leakage (presented as fluorescence index) from the airways into the blood 1 h after OMV administration. *P < 0.05, one-way ANOVA with Tukey’s multiple comparisons.
ΔtrxA OMVs enhanced cytotoxicity is protein-dependent
The nature of OMV-associated cytotoxicity was assessed in a murine macrophage-like cell line J774. J774 cells were incubated with WT or ΔtrxA OMVs (containing 1 or 10 µg protein) for 1 or 4 h, and stained with AO/EB for death cell enumeration. We observed that both OMV preparations induced J774 cell death in a dose-dependent manner, and ΔtrxA OMVs are more cytotoxic than WT OMVs (Figure 2(a)). However, increased cell death by ΔtrxA OMVs was abrogated when OMVs were treated with protease K suggesting that the cytotoxicity is associated with OMV proteins (Figure 2(b)).
Figure 2.
ΔtrxA OMVs associated cytotoxicity is protein-dependent. (a) J774 macrophage-like cells were incubated with WT or ΔtrxA OMVs (containing 1 or 10 µg protein) for 1 or 4 h, and stained with AO/EB to determine cell death (percentage). (b) J774 cells were incubated with WT or ΔtrxA OMVs (10 μg/well) in the presence or absence of protease K (PK, 10 μg/well) for 4 h, and cell viability was determined by AO/EB staining. Five samples per group were analyzed. *P < 0.05, t-test.
Proteomic analysis of OMVs
The difference in the respective protein profiles of WT and ΔtrxA OMVs were further analyzed. Specifically, three sets of OMV preparations were subjected to trypsin digestion followed by liquid chromatography tandem mass spectrometry (LC-MS/MS). Spectra were analyzed using MaxQuant software for LFQ proteomic comparison amongst tested samples. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE 20 partner repository with dataset identifier PXD027181 and 10.6019/PXD027181. For comparative analysis, protein groups with at least two valid LFQ values out of three replicates were considered. A total of 129 and 159 proteins were identified in Ci79 and ΔtrxA OMVs, respectively (Supplementary Table S1). Among these identified proteins, 112 proteins were found in both strains, 17 unique proteins were present in the Ci79 strain, and 47 proteins were found only in the ΔtrxA strain. Common proteins included some outer membrane proteins (e.g. Omp38), metabolic proteins, periplasmic proteins (e.g. efflux proteins), and transporter proteins. For comparative protein abundance analysis, the sum of LFQ values for WT OMV proteins is comparable to that of ΔtrxA OMVs (1:1.03) indicating overall OMV protein quantity is similar in WT and ΔtrxA strains. Eighteen proteins were found to be significantly different in abundance (t test P < 0.05) in WT and ΔtrxA OMVs with a minimal 1.9-fold difference (Supplementary Table S1). These proteins as well as other top 10 “most abundant” unique WT and ΔtrxA OMV proteins are listed in Table 1. Abundant WT OMV unique proteins consisted of many ribosomal complex proteins while noticeable proteins unique to ΔtrxA include superoxide dismutase, amino acid transporter, pilus biogenesis proteins, and a putative metalloprotease (gene family of astacin-like toxins). Common OMV proteins with greater expression in ΔtrxA include aspartate-glutamate transporter, OmpA, and phospholipase.
Table 1.
List of abundant unique and differentially packing proteins identified from Acinetobacter baumannii Ci79 wild type and ΔtrxA OMVs.
| Protein ID | Protein name | LFQ WT | LFQ ΔtrxA | Unique or Δ/WT ratio |
|---|---|---|---|---|
| ETR88480.1 | substrate-binding domain of ABC aspartate-glutamate transporter | 2.96E + 07 | 1.58E + 08 | 5.35 |
| ETR85040.1 | peptidoglycan-associated lipoprotein | 2.60E + 08 | 8.44E + 08 | 3.25 |
| ETR83561.1 | ompA family protein | 3.79E + 08 | 1.12E + 09 | 2.95 |
| ETR87168.1 | phospholipase A1 family protein | 1.26E + 08 | 3.46E + 08 | 2.74 |
| ETR91147.1 | outer membrane assembly lipoYfiO family protein | 4.17E + 08 | 1.11E + 09 | 2.66 |
| ETR92825.1 | tonB-dependent copper receptor | 9.86E + 07 | 1.84E + 08 | 1.87 |
| ETR90044.1 | tannase and feruloyl esterase family protein | 6.61E + 08 | 2.08E + 08 | 0.31 |
| ETR83252.1 | toluene tolerance, Ttg2 family protein | 5.92E + 08 | ΔTrxA | |
| ETR83090.1 | superoxide dismutase domain protein | 5.07E + 08 | ΔTrxA | |
| ETR87072.1 | hypothetical protein M212_2416, partial | 4.20E + 08 | ΔTrxA | |
| ETR84903.1 | hypothetical protein M212_3000 | 3.51E + 08 | ΔTrxA | |
| ETR88966.1 | putative amino-acid transporter periplasmic solute-binding protein | 3.33E + 08 | ΔTrxA | |
| ETR92692.1 | transglycosylase SLT domain protein | 2.06E + 08 | ΔTrxA | |
| ETR92784.1 | hypothetical protein M212_0134 | 1.78E + 08 | ΔTrxA | |
| ETR83205.1 | ribosomal protein L17 | 1.63E + 08 | ΔTrxA | |
| ETR87690.1 | fimbrial family protein | 1.25E + 08 | ΔTrxA | |
| ETR84904.1 | D-alanyl-D-alanine carboxypeptidase family protein | 1.13E + 08 | ΔTrxA | |
| ETR84600.1 | putative metalloprotease | 1.12E + 08 | ΔTrxA | |
| ETR87202.1 | quinoprotein glucose dehydrogenase B | 1.08E + 08 | ΔTrxA | |
| ETR85612.1 | ompW family protein | 1.02E + 08 | ΔTrxA | |
| ETR89190.1 | peptidase M15 family protein | 9.51E + 07 | ΔTrxA | |
| ETR90544.1 | aldose 1-epimerase | 6.42E + 07 | ΔTrxA | |
| ETR83824.1 | putative rND type efflux pump | 6.27E + 07 | ΔTrxA | |
| ETR82276.1 | putative lipoprotein-34 | 6.26E + 07 | ΔTrxA | |
| ETR85745.1 | Gram-negative pili assembly chaperone | 4.50E + 07 | ΔTrxA | |
| ETR81498.1 | tonB-dependent hemoglobin/transferrin/lactoferrin receptor family protein | 3.78E + 09 | WT | |
| ETR92110.1 | ribosomal protein L20 | 4.53E + 08 | WT | |
| ETR83226.1 | ribosomal protein L22 | 2.39E + 08 | WT | |
| ETR83209.1 | 30S ribosomal protein S13 | 1.64E + 08 | WT | |
| ETR92613.1 | DNA-directed RNA polymerase, beta subunit | 1.64E + 08 | WT | |
| ETR82953.1 | hypothetical protein M212_3929 | 1.63E + 08 | WT | |
| ETR92614.1 | DNA-directed RNA polymerase, beta subunit | 1.22E + 08 | WT | |
| ETR82979.1 | tonB-dependent siderophore receptor family protein | 1.06E + 08 | WT | |
| ETR83206.1 | DNA-directed RNA polymerase, alpha subunit | 8.09E + 07 | WT | |
| ETR81473.1 | transglycosylase SLT domain protein | 7.38E + 07 | WT | |
| ETR92118.1 | transcription termination factor Rho | 5.64E + 07 | WT | |
| ETR93279.1 | polysaccharide biosynthesis/export family protein | 4.79E + 07 | WT | |
| ETR86123.1 | aconitate hydratase 2 | 2.88E + 07 | WT |
Discussion
Bacteria may secrete OMVs to promote bacterial growth and enhance pathogenesis. Bacterial OMVs can facilitate nutrient acquisition by preferential packing of glycosidases and proteases to specifically impede growth of competitors within mixed bacterial communities.21,22 Several peptidases, metalloprotease, and phospholipases were identified from our proteomic analysis. Bacterial OMVs also contribute to antibiotic resistance by carrying enzymes that mediate antibiotic protection or acting as decoys that bind to or absorb antibiotics and toxins. 23 We have identified several antibiotic resistance proteins (e.g. penicillin-binding proteins, beta-lactamases, and a putative carbapenem-associated resistance protein) from A. baumannii OMVs. Additionally, virulence determinants can be concentrated in OMVs and thus protected from host enzymatic degradation and delivered to target tissues via vesicle-associated adhesins. 23 It has been shown that OMV associated virulence factors induce host cell apoptosis and manipulate the immune response. 24 Due to their nanoparticle size, OMVs can migrate through host tissues, disrupt epithelial tight junctions, and deliver bacterial virulence factors to immune cells in underlying tissues. 25 We have observed that treatment with ΔtrxA OMVs induces J774 cell death, and increases lung permeability. Administration of ΔtrxA OMVs in lungs may injure endothelial and epithelial barriers, and lead to leukocyte infiltration which results in extravasation of the vascular fluid.
OMVs from ΔtrxA were more cytotoxic for macrophages compared to WT OMVs within a 4-h incubation period. Proteinase K-treated A. baumannii OMVs significantly reduced macrophage cell death compared to untreated OMVs suggesting that the surface-exposed membrane proteins are responsible for the observed cytotoxic effect. Among the identified 28 differentially expressed proteins, outer protein A (OmpA), putative metalloprotease, and fimbrial protein may be responsible for the induced macrophage cell death. It has been shown that A. baumannii OmpA induces early-onset apoptosis in DCs by targeting mitochondria resulting in production of reactive oxygen species (ROS) which enhanced apoptosis of AbOmpA-treated DCs. 26 In addition, A. baumannii OmpA deletion mutant strains unlike wild type did not induce apoptosis of A549 human alveolar epithelial cells. 27 Furthermore, OMVs isolated from ΔOmpA did not induce death of U937 human macrophages. 24 Collectively, this evidence supports the cytotoxic nature of A. baumannii OmpA which is 3-fold more abundant in ΔOMVs. Metalloproteases are a highly diverse set of proteolytic enzymes. Metalloprotease activity is often mediated by zinc which activates the water molecule’s nucleophilic attack on substrate peptide bonds.28,29 Metalloprotease secreted from Vibrio vulnificus activates procaspase-3-mediated apoptosis pathway. 29 The OMVs from the Porphyromonas gingivalis fimA mutant has shown to cause less HeLa apoptosis than WT. 30 The role of metalloproteases and fimbriae in A. baumannii OMV mediated cytotoxicity remains to be elucidated.
Meningococcal group B outer membrane vesicle vaccines have been used widely in Cuba, New Zealand, and Brazil to prevent invasive meningococcal disease. 31 Other OMV-based vaccines have also been developed and evaluated against bacterial infections including Acinetobacter.32–34 Results of this study provide additional insight into potential cytotoxicity factors that might need to be considered when developing OMV-based vaccines. OMVs isolated from the attenuated bacterial strain (e.g. ΔtrxA) might not be necessarily less virulent than wild type OMVs. The nature of the preferential packaging of OMV contents differ from one strain to another making it unpredictable for vaccine efficacy and cytotoxicity. We have reported that TrxA is an important virulence factor for A. baumannii colonization in the GI tract, and subsequent pneumonic infection leading to lethal sepsis.14,35 Furthermore, TrxA-mediated modulation of bacterial surface hydrophobicity via disulfide bond reduction and regulation of type IV pili is a contributing factor to A. baumannii virulence. 36 The mechanism(s) for TrxA-mediated differential OMV protein packaging remains to be determined; however, the TrxA-mediated alteration of cell hydrophobicity may play a role.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211052952 for Thioredoxin-mediated alteration of protein content and cytotoxicity of Acinetobacter baumannii outer membrane vesicles by Swathi Shrihari, Holly C May, Jieh-Juen Yu, Sara B Papp, James P Chambers, M Neal Guentzel and Bernard P Arulanandam in Experimental Biology and Medicine
ACKNOWLEDGMENTS
We gratefully acknowledge Dr. Wendell P. Griffith (Mass Spectrometry and Proteomics Core) and Mr. Mando Rodriguez (Research Computing Support Group) of the University of Texas at San Antonio for their technical assistance in proteomic analysis.
AUTHORS’ CONTRIBUTIONS: All authors participated in the design, interpretation of the studies and analysis of the data. SS, JJY, and BPA conceived and designed the experiments. SS, HJM, and SBP carried out the experiments. SS, JJY, JPC, MNG, and BPA analyzed the data. SS, JJY, JPC, and BPA wrote and edited the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship and/or publication of this article: This work was supported by The Jane and Roland Blumberg Endowment fund to BPA.
ORCID iDs: Jieh-Juen Yu https://orcid.org/0000-0001-8901-3366
Bernard P Arulanandam https://orcid.org/0000-0002-4815-5306
SupplementAL MATERIAL: Supplemental material for this article is available online.
References
- 1.Moghadam SS, Aghmiyuni ZF, Zaheri H, Arianpour N, Danaeifard MR, Roham M, Momeni M. Comparative effects of granulocyte-colony stimulating factor and colistin-alone or in combination on burn wound healing in Acinetobacter baumannii infected mice. Iran J Microbiol 2018; 10:371–7 [PMC free article] [PubMed] [Google Scholar]
- 2.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 1998; 36:1938–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wendt C, Dietze B, Dietz E, Ruden H. Survival of Acinetobacter baumannii on dry surfaces. J Clin Microbiol 1997; 35:1394–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B. Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 2017; 30:409–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramirez MS, Don M, Merkier AK, Bistue AJ, Zorreguieta A, Centron D, Tolmasky ME. Naturally competent Acinetobacter baumannii clinical isolate as a convenient model for genetic studies. J Clin Microbiol 2010; 48:1488–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Da Silva GJ, Domingues S. Insights on the horizontal gene transfer of carbapenemase determinants in the opportunistic pathogen Acinetobacter baumannii. Microorganisms 2016; 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antunes LC, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathog Dis 2014; 71:292–301 [DOI] [PubMed] [Google Scholar]
- 8.Beveridge TJ. Structures of Gram-negative cell walls and their derived membrane vesicles. J Bacteriol 1999; 181:4725–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwon SO, Gho YS, Lee JC, Kim SI. Proteome analysis of outer membrane vesicles from a clinical Acinetobacter baumannii isolate. FEMS Microbiol Lett 2009; 297:150–6 [DOI] [PubMed] [Google Scholar]
- 10.Rumbo C, Fernandez-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, Mosquera A, Chaves F, Bou G. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: a new mechanism of dissemination of carbapenem resistance genes in Acinetobacter baumannii. Antimicrob Agents Chemother 2011; 55:3084–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun SH, Lee JH, Kim BR, Kim SI, Park TI, Lee JC, Lee YC. Acinetobacter baumannii outer membrane vesicles elicit a potent innate immune response via membrane proteins. PLoS One 2013; 8:e71751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moon DC, Choi CH, Lee JH, Choi CW, Kim HY, Park JS, Kim SI, Lee JC. Acinetobacter baumannii outer membrane protein a modulates the biogenesis of outer membrane vesicles. J Microbiol 2012; 50:155–60 [DOI] [PubMed] [Google Scholar]
- 13.Yun SH, Park EC, Lee SY, Lee H, Choi CW, Yi YS, Ro HJ, Lee JC, Jun S, Kim HY, Kim GH, Kim SI. Antibiotic treatment modulates protein components of cytotoxic outer membrane vesicles of multidrug-resistant clinical strain, Acinetobacter baumannii DU202. Clin Proteomics 2018; 15:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ketter PM, Yu JJ, Guentzel MN, May HC, Gupta R, Eppinger M, Klose KE, Seshu J, Chambers JP, Cap AP, Arulanandam BP. Acinetobacter baumannii gastrointestinal colonization is facilitated by secretory IgA which is reductively dissociated by bacterial thioredoxin A. MBio 2018; 9:e01298-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar JK, Tabor S, Richardson CC. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc Natl Acad Sci U S A 2004; 101:3759–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketter P, Guentzel MN, Chambers JP, Jorgensen J, Murray CK, Cap AP, Yu JJ, Eppinger M, Arulanandam BP. Genome sequences of four Acinetobacter baumannii-A. calcoaceticus complex isolates from combat-related infections sustained in the Middle East. Genome Announc 2014; 2: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008; 26:1367–72 [DOI] [PubMed] [Google Scholar]
- 18.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 2011; 10:1794–805 [DOI] [PubMed] [Google Scholar]
- 19.Beyene GT, Kalayou S, Riaz T, Tonjum T. Comparative proteomic analysis of Neisseria meningitidis wildtype and dprA null mutant strains links DNA processing to pilus biogenesis. BMC Microbiol 2017; 17:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Perez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yilmaz S, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaino JA. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019; 47:D442–D50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elhenawy W, Debelyy MO, Feldman MF. Preferential packing of acidic glycosidases and proteases into Bacteroides outer membrane vesicles. mBio 2014; 5:e00909-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans AGL, Davey HM, Cookson A, Currinn H, Cooke-Fox G, Stanczyk PJ, Whitworth DE. Predatory activity of Myxococcus xanthus outer-membrane vesicles and properties of their hydrolase cargo. Microbiology (Reading) 2012; 158:2742–52 [DOI] [PubMed] [Google Scholar]
- 23.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat Rev Microbiol 2015; 13:605–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin JS, Kwon SO, Moon DC, Gurung M, Lee JH, Kim SI, Lee JC. Acinetobacter baumannii secretes cytotoxic outer membrane protein a via outer membrane vesicles. PLoS One 2011; 6:e17027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cecil JD, O'Brien-Simpson NM, Lenzo JC, Holden JA, Singleton W, Perez-Gonzalez A, Mansell A, Reynolds EC. Outer membrane vesicles prime and activate macrophage inflammasomes and cytokine secretion in vitro and in vivo. Front Immunol 2017; 8:1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JS, Choi CH, Kim JW, Lee JC. Acinetobacter baumannii outer membrane protein a induces dendritic cell death through mitochondrial targeting. J Microbiol 2010; 48:387–92 [DOI] [PubMed] [Google Scholar]
- 27.Gaddy JA, Tomaras AP, Actis LA. The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 2009; 77:3150–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wolfe MS. Intramembrane proteolysis. Chem Rev 2009; 109:1599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HY, Chang AK, Park JE, Park IS, Yoon SM, Lee JS. Procaspase-3 activation by a metalloprotease secreted from Vibrio vulnificus. Int J Mol Med 2007; 20:591–5 [PubMed] [Google Scholar]
- 30.Furuta N, Tsuda K, Omori H, Yoshimori T, Yoshimura F, Amano A. Porphyromonas gingivalis outer membrane vesicles enter human epithelial cells via an endocytic pathway and are sorted to lysosomal compartments. Infect Immun 2009; 77:4187–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petousis-Harris H. Impact of meningococcal group B OMV vaccines, beyond their brief. Hum Vaccin Immunother 2018; 14:1058–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li S, Chen DQ, Ji L, Sun S, Jin Z, Jin ZL, Sun HW, Zeng H, Zhang WJ, Lu DS, Luo P, Zhao AN, Luo J, Zou QM, Li HB. Development of different methods for preparing Acinetobacter baumannii outer membrane vesicles vaccine: impact of preparation method on protective efficacy. Front Immunol 2020; 11:1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pulido MR, Garcia-Quintanilla M, Pachon J, McConnell MJ. A lipopolysaccharide-free outer membrane vesicle vaccine protects against Acinetobacter baumannii infection. Vaccine 2020; 38:719–24 [DOI] [PubMed] [Google Scholar]
- 34.Huang W, Yao Y, Long Q, Yang X, Sun W, Liu C, Jin X, Li Y, Chu X, Chen B, Ma Y. Immunization against multidrug-resistant Acinetobacter baumannii effectively protects mice in both pneumonia and sepsis models. PLoS One 2014; 9:e100727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.May HC, Yu JJ, Zhang H, Wang Y, Cap AP, Chambers JP, Guentzel MN, Arulanandam BP. Thioredoxin-A is a virulence factor and mediator of the type IV pilus system in Acinetobacter baumannii. PLoS One 2019; 14:e0218505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.May HC, Yu JJ, Shrihari S, Seshu J, Klose KE, Cap AP, Chambers JP, Guentzel MN, Arulanandam BP. Thioredoxin modulates cell surface hydrophobicity in Acinetobacter baumannii. Front Microbiol 2019; 10:2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702211052952 for Thioredoxin-mediated alteration of protein content and cytotoxicity of Acinetobacter baumannii outer membrane vesicles by Swathi Shrihari, Holly C May, Jieh-Juen Yu, Sara B Papp, James P Chambers, M Neal Guentzel and Bernard P Arulanandam in Experimental Biology and Medicine