Significance
HBV infections are the leading cause of chronic hepatitis and carry the risk of liver cirrhosis and cancer. The HBV life cycle is perpetuated by an RNA intermediate termed pregenomic RNA (pgRNA), which is encapsidated by the viral core protein. The pgRNA packaging process is an essential step in viral replication. Here, we investigated the role of N6-methyladenosine (m6A) modification in the recognition of pgRNA by the core protein during encapsidation. m6A modification of 5′ epsilon structural motifs serves as the recognition signal for the core protein interaction, as evidenced by the failure of 5′ epsilon m6A mutant to encapsidate pgRNA. This study identifies the structural role of m6A modification in pgRNA encapsidation and provides an avenue in RNA–protein complex interactions.
Keywords: hepatitis B virus, N6-methyladenosine, HBV pgRNA encapsidation, RNA–protein interaction
Abstract
Hepatitis B virus (HBV) contains a partially double-stranded DNA genome. During infection, its replication is mediated by reverse transcription (RT) of an RNA intermediate termed pregenomic RNA (pgRNA) within core particles in the cytoplasm. An epsilon structural element located in the 5′ end of the pgRNA primes the RT activity. We have previously identified the N6-methyladenosine (m6A)–modified DRACH motif at 1905 to 1909 nucleotides in the epsilon structure that affects myriad functions of the viral life cycle. In this study, we investigated the functional role of m6A modification of the 5′ ε (epsilon) structural element of the HBV pgRNA in the nucleocapsid assembly. Using the m6A site mutant in the HBV 5′ epsilon, we present evidence that m6A methylation of 5′ epsilon is necessary for its encapsidation. The m6A modification of 5′ epsilon increased the efficiency of viral RNA packaging, whereas the m6A of 3′ epsilon is dispensable for encapsidation. Similarly, depletion of methyltransferases (METTL3/14) decreased pgRNA and viral DNA levels within the core particles. Furthermore, the m6A modification at 5′ epsilon of HBV pgRNA promoted the interaction with core proteins, whereas the 5′ epsilon m6A site–mutated pgRNA failed to interact. HBV polymerase interaction with 5′ epsilon was independent of m6A modification of 5′ epsilon. This study highlights yet another pivotal role of m6A modification in dictating the key events of the HBV life cycle and provides avenues for investigating RNA–protein interactions in various biological processes, including viral RNA genome encapsidation in the context of m6A modification.
Hepatitis B virus (HBV) infections are one of the leading causes of chronic hepatitis, which carry the risk of severe liver disease including the development of hepatocellular carcinoma (1). About 300 million people are chronically infected with HBV worldwide (2). Current HBV therapies using nucleoside analogs directed against the viral polymerase (pol) have limited efficacy and do not eliminate the HBV DNA, which maintains its presence and resumes the infectious process upon withdrawal of antivirals (2). A better understanding of the HBV life cycle is required to identify alternative drug targets to interfere with the infectious process.
HBV is a member of the Hepadnaviridae family and contains a 3.2-kb relaxed circular DNA (rcDNA) genome (1). After viral infection, core particles disassemble in the nuclear pore and deliver rcDNA into the nucleus, where it is transformed to a covalently closed circular DNA (cccDNA) by several host enzymes (3). The HBV cccDNA is a template for viral RNA transcription. HBV DNA encodes four overlapping reading frames, which include surface (HBs), precore or “e” (HBe), and core (HBc) antigen proteins; pol, a reverse transcriptase; and X (HBx) proteins. Although HBV is a DNA virus, it replicates by the reverse transcription (RT) activity of viral pol from an RNA intermediate termed pregenomic RNA (pgRNA) to produce an rcDNA inside the core particles in the cytoplasm. HBV pol protein consists of terminal protein (TP), reverse transcriptase, and carboxyl-terminal RNase H (RH) domains. Both reverse transcriptase and RH domains are necessary structural components for HBV encapsidation (4). The TP domain is used as a protein primer to initiate RT catalyzed by the reverse transcriptase domain. The assembly of nucleocapsid is triggered by the direct interaction of pol with the 5′ epsilon structure of pgRNA (4, 5). The pol recognizes the functional stem loop of the 5′ epsilon of pgRNA and primes the synthesis of the minus-strand DNA (6). The HBV core protein consists of an N-terminal assembly domain and an arginine-rich carboxyl-terminal domain, which is required for pgRNA encapsidation (7, 8). The HBV nucleocapsid is composed of dimers of core protein, and 120 (or 90) copies of core assemble into capsids with T = 4 (∼95%) or T = 3 (5%) icosahedral symmetry (9). The limited phosphorylation of core proteins is associated with pgRNA encapsidation (10, 11). These mature capsids are enveloped by a host-derived membrane containing embedded HBs protein and egress from infected hepatocytes (12). Previous studies have highlighted specific sequences and structural requirements of the 5′ epsilon element of pgRNA in the encapsidation process (Fig. 1A) (6, 13–16). First, the lower stem and the internal bulge appear to play mainly a structural role, the sequence of which can be altered without significant effect on pgRNA packaging. Second, the specific sequences on the lower right side of the upper stem are required for encapsidation. Third, the sequences of the apical loop, in contrast to the internal bulge, contribute critically, in a sequence-specific manner, to pgRNA encapsidation.
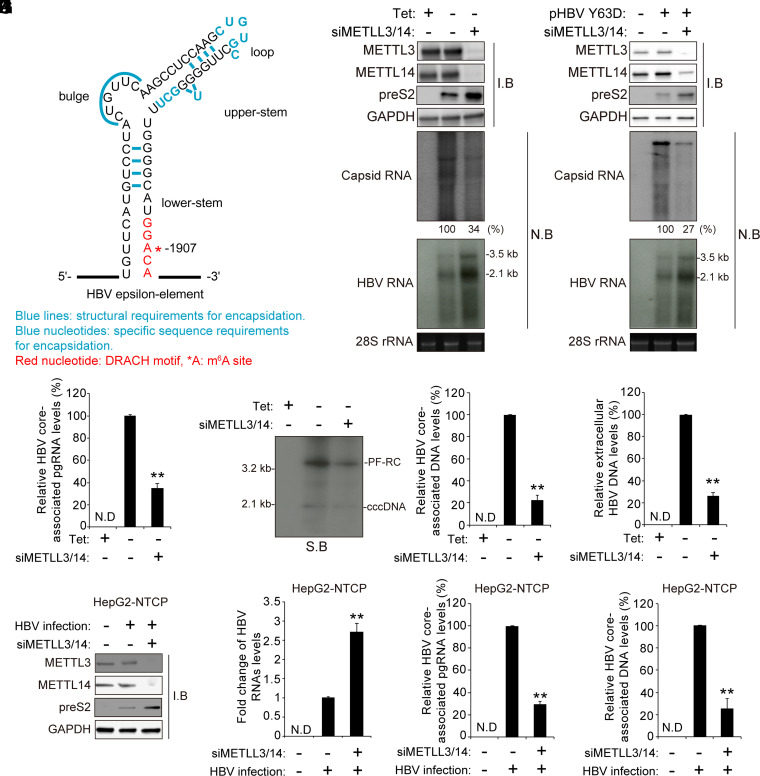
Fig. 1.
The cellular m6A methyltransferases affect the HBV pgRNA encapsidation. (A) The secondary structure of the epsilon elements is shown with the functional requirements for RNA packaging. They are represented as follows: Blue lines indicate structural requirements for encapsidation, blue nucleotides indicate specific sequence requirements for encapsidation, red nucleotides indicate DRACH motif, and *A is the m6A site. (B) HepAD38 cells stably expressing HBV were grown in the absence or presence of tetracycline for 72 h, and the cells were then transfected with the METTL3/14-specific siRNAs. After 48 h, cellular lysates, core-associated pgRNA, and total RNA were extracted from these cells for Western blotting and Northern blotting, respectively. (C) Huh7 cells were transfected with pHBV Y63D plasmid (RT-defective mutant) for 24 h, and then the siRNAs of METTL3/14 were transfected into Huh7 cells expressing pHBV Y63D plasmid. After 48 h, cellular lysates, core-associated pgRNA, and total RNA were extracted from these cells. The indicated proteins were analyzed by Western blotting. The encapsidated and cellular HBV RNA were analyzed by Northern blotting. (D–G) The siRNAs of METTL3/14 were transfected into stably expressing HBV HepAD38 cells grown in the absence or presence of tetracycline for 72 h. After 48 h, cells and supernatant were harvested. The core-associated pgRNA was analyzed by RT-qPCR (D). Hirt’s extract was prepared and subjected to Southern blot assays (E). The core-associated DNA and extracellular DNA levels were analyzed by qPCR (F and G). (H–K) HepG2-NTCP cells were infected with 2.5 × 103 genome equivalents per cell of HBV particles. After 10 d, cellular lysates, total RNA, and core-associated pgRNA and DNA were extracted from these cells. The indicated proteins were analyzed by Western blotting (H). The cellular HBV RNA and core-associated pgRNA were analyzed by RT-qPCR (I and J). The HBV core-associated DNA was assayed by qPCR (K). In E, F to G, and I to K, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. **P < 0.01. siMETTL3/14, siRNAs of METTL3/14; I.B, immunoblotting; N.B, Northern blotting; N.D, not detected.
Cellular RNAs (transfer RNAs [tRNAs], ribosomal RNAs [rRNAs], and messenger RNAs [mRNAs]) are modified by diverse chemical modifications, including N6-methyladenosine (m6A), 5-methylcytidine (m5C), and inosine in addition to N7-methylguanosine (m7G) (17). Of these, the m6A RNA methylation of the adenosine base at the nitrogen 6 position is the most prevalent and well-characterized RNA modification. m6A modification is functionally implicated in a wide range of biological processes, which include innate immune response, sex determination, stem cell differentiation, circadian clock, meiosis, stress response, and cancer development (18). The cellular methyltransferase complex, composed of methyltransferase like 3 (METTL3), METTL14, and WTAP, places m6A methylation on RNA cotranscriptionally (19). This modification is typically enriched near the stop codons and 3′-untranslated region of cellular mRNAs. The m6A-modified mRNA is recognized by YT521-B homology (YTH) domain-containing family proteins (YTHDF1-3 and YTHDC1-2), which bind m6A-containing RNAs and regulate their stability, translation, and subcellular localization (20). m6A methylation is eliminated by m6A demethylases such as fat mass and obesity-associated protein and ALKBH5, suggesting that m6A modification is reversibly catalyzed by methyltransferases and demethylases (19). In addition to cellular RNAs, the viral transcripts of DNA viruses, as well as the RNA viral genomes, are methylated. These modifications play key roles at various levels in the viral life cycle and disease pathogenesis associated with them (21–27).
Previously, we identified a single m6A-consensus motif (DRACH) at nucleotide (nt) position 1907, which is localized in the lower stem loop of the epsilon structure of HBV RNAs (Fig. 1A) (25). All HBV transcripts bear this consensus motif at the 3′ end epsilon structure, but pgRNA carries this motif twice, at 5′ and 3′ epsilon structures, owing to the terminal redundancy of sequences at its 5′ and 3′ end. m6A modification in the HBV epsilon element plays a differential dual role in the viral life cycle depending on its position in the viral RNA (25). m6A at 3′ epsilon reduces RNA stability, whereas m6A at 5′ epsilon increases HBV core-associated DNA levels. In the case of m6A at the 3′ end of viral RNAs, YTHDF2 and 3 proteins recognize this m6A site to promote the degradations of viral transcripts. However, the role of m6A in the pgRNA 5′ epsilon element, if any, in the core-associated DNA synthesis was not investigated. Here, we reasoned that the m6A modification in the 5′ epsilon structure could influence the pgRNA encapsidation followed by reverse transcriptase-mediated viral DNA synthesis because the structural requirements of 5′ epsilon are important in pgRNA encapsidation and m6A modification has been described to alter RNA structure (6, 14, 16, 28, 29). Thus, we investigated the functional role of m6A methylation at 1907 nt of 5′ epsilon in pgRNA packaging. Our results demonstrate that m6A methylation at 5′ epsilon induced pgRNA encapsidation and thus affected rcDNA synthesis and core particles production. The mutation of the m6A site in 5′ epsilon abrogates the interaction with core protein resulting in the decrease in the viral DNA genome synthesis within the core particles. The preferential core–pgRNA interaction is likely promoted by the distortion of the lower stem of 5′ epsilon caused by m6A methylation of 1907 nt. The results of our study highlight a key role of m6A modification in 5′ epsilon in the HBV pgRNA encapsidation. Its (m6A) role in the nucleocapsid assembly adds to the list of myriad functions associated with m6A RNA methylation of HBV RNAs. The global effect of the chemical modification of RNA is understood to maintain the homeostasis of the infectious process during chronic hepatitis B.
Results
Cellular m6A Methyltransferases Affect HBV pgRNA Encapsidation.
To investigate whether cellular m6A methyltransferases (METTL3/14) affect HBV pgRNA encapsidation, HepAD38 cells, which stably express the HBV genome (30), were transfected with small interfering RNAs (siRNAs) specific to METTL3/14. Total HBV RNA and core-associated pgRNA were extracted from these cells and subjected to RT-qPCR and Northern blot assay, respectively. The silencing of METTL3/14 complex increased HBV transcripts and viral protein levels (Fig. 1B; SI Appendix, Fig. 1 A and B), consistent with previous results (25). Interestingly, the core-associated pgRNA levels were decreased by the depletion of METTL3/14 despite increased viral RNA levels (Fig. 1B). We next used the HBV Y63D polymerase mutant, which upon transfection, accumulates pgRNA in the core particles due to its defect in priming activity (31). As observed in Fig. 1B, there was an increase in HBV RNA and protein levels, but the packaged pgRNA levels in core particles were dramatically reduced in METTL3/14-depleted cells (Fig. 1C). Depletion of METTL3/14 decreased the levels of rcDNA, core-associated DNA, and extracellular HBV DNA (Fig. 1 D–G), suggesting that the viral encapsidation requires m6A modification of pgRNA. We verified these observations using the HBV infection system (Fig. 1 H–K; SI Appendix, Fig. 1C). HBV infection results also showed that the silencing of METTL3/14 increased HBV protein and RNA levels, while it decreased the core-associated pgRNA and viral DNA levels. Collectively, these results suggest that the cellular m6A methyltransferases are required for HBV RNA packaging to promote viral DNA genome synthesis.
The m6A Methylation of the 5′ Epsilon Element Is Essential for HBV pgRNA Packaging.
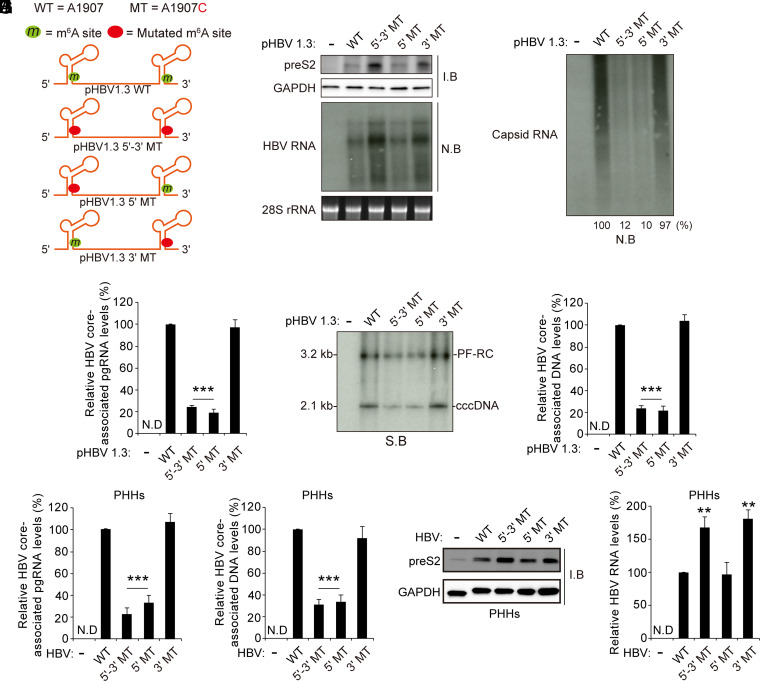
Previously, we identified a single m6A site at 1907 nt of HBV transcripts (25). The adenosine at 1907 nt is located in the lower stem of the epsilon element. To determine whether m6A modification of HBV RNA affects viral pgRNA packaging, we generated m6A site mutation in HBV 5′ and 3′, 5′, or 3′ epsilon structure (Fig. 2A). We analyzed the levels of HBV RNA and core-associated pgRNA in the indicated plasmids transfected cells (Fig. 2 B and C; SI Appendix, Fig. 2 A and B). Compared to the wild-type (WT) HBV-transfected cells, viral protein and RNA levels were increased only in HBV 5′-3′ mutant (MT) and 3′ MT–transfected cells, but not in HBV 5′ MT as analyzed by Western and Northern blot assays, respectively (Fig. 2B). These results are consistent with the previous report, which showed that only the m6A modification of 3′ epsilon regulates viral RNA stability and protein expression (25). However, the mutation of the m6A site in the 5′ epsilon element dramatically reduced the capsid pgRNA level without affecting viral RNA stability (Fig. 2 C and D). Although viral RNA levels were increased due to the mutation of the m6A site of 3′ epsilon in 5′ and 3′ MT–transfected cells, the core-associated pgRNA levels were substantially decreased by the deficient of m6A methylation of 5′ epsilon. The decreased core-associated pgRNA levels by the mutation of the m6A site of 5′ epsilon resulted in the decreases in rcDNA and core-associated DNA levels (Fig. 2 E and F). We further confirmed these results using the primary human hepatocytes (PHHs) infection system (Fig. 2 G–J). Similarly, the mutation of the m6A site of 5′ epsilon reduced HBV core-associated DNA and pgRNA levels in HBV-infected PHHs (Fig. 2 G and H). The mutation of the m6A site of 3′ epsilon did not affect pgRNA encapsidation but increased viral protein and RNA levels (Fig. 2 I and J). These results suggest that the m6A modification of the 5′ epsilon structure plays a critical role in the pgRNA packaging process.
Fig. 2.
The m6A modification of the 5′ epsilon elements promotes HBV pgRNA encapsidation. (A) Schematics indicate the 5′ and 3′ m6A sites in the epsilon structures of the HBV pgRNA. Green circles indicate the m6A site, and red circles indicate A1907C mutation in the HBV pgRNA. pHBV 1.3-mer 5′-3′ MT contains the A1907C mutation at the 5′ and 3′ ends, pHBV 1.3-mer 5′ MT within the epsilon structure contains the A1907C mutation at the 5′ end, and the pHBV 1.3-mer contains the 3′ MT at the 3′ end. (B–F) The indicated pHBV 1.3 plasmids were transfected into Huh7 cells. After 72 h, cellular lysates, total RNA, core-associated pgRNA, HBV DNA, and core-associated DNA were extracted. The indicated proteins or HBV RNAs were each analyzed by Western or Northern blotting, respectively (B). Encapsidated pgRNA was detected by Northern blotting (C). The core-associated pgRNA was analyzed by RT-qPCR (D). Hirt’s DNA extracts were prepared, and the protein-free (PF) rcDNA and cccDNA were analyzed by Southern blotting (E). The core-associated DNA was analyzed by qPCR (F). (G–J) PHHs were infected with 2.5 × 103 genome equivalents per cell of each HBV WT, 5′-3′ MT, 5′ MT, or 3′ MT infectious particles. After 10 d, PHHs were harvested to assess the expression of viral protein and RNA, core-associated pgRNA, and DNA, respectively. In D, F, G, H, and J, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. ***P < 0.001. I.B, immunoblotting; N.B, Northern blotting; N.D, not detected; S.B, Southern blotting.
To determine whether m6A sites other than the consensus DRACH motif at 1907 nt present in the HBV DNA genome affect pgRNA packaging, we depleted cellular m6A methyltransferases in HBV WT or 5′ and 3′, 5′, or 3′ MT-transfected cells and analyzed the core-associated pgRNA levels and viral DNA synthesis (SI Appendix, Fig. 2 C–F). The silencing of METTL3/14 reduced the core-associated pgRNA in HBV WT and 3′ MT plasmid–transfected cells, whereas in the case of the 5′ MT, depletion of METTL3/14 did not affect the capsid pgRNA levels. Similarly, the absence of METTL3/14 reduced the core-associated DNA and rcDNA levels in HBV WT and 3′ MT plasmid–transfected cells. But HBV core-associated DNA and rcDNA levels in 5′ MT plasmid–transfected cells were not affected by METTL3/14 expression. These results further support the view that m6A modification of the 5′ epsilon structure is relevant for pgRNA encapsidation.
The m6A Methylation of HBV 5′ Epsilon Regulates pgRNA Encapsidation Independently of m6A Reader Proteins.
Since cellular m6A reader proteins (YTHDF1-3 and YTHDC2) bind m6A-bearing RNAs to regulate their stability and translation (20), we analyzed the effect of m6A reader proteins on HBV pgRNA packaging. Because m6A modification of the 3′ epsilon structure negatively regulates HBV RNA stability by recruitment of YTHDF2/3 proteins (25), we used HBV 3′ MT plasmid to exclude the effect of YTHDF proteins on m6A of the 3′ epsilon structure. We depleted YTHDF1-3 proteins using specific siRNAs in HBV 3′ MT plasmid–transfected cells. Interestingly, we observed that any YTHDF proteins did not affect HBV core-associated pgRNA and DNA levels (SI Appendix, Fig. 3 A–C). The silencing of YTHDC2, an RNA helicase known to affect m6A methylated RNA translation (32), also did not affect the pgRNA packaging process (SI Appendix, Fig. 3 E–F). These results together suggest that the m6A-mediated pgRNA encapsidation process is independent of m6A reader proteins.
The m6A Methylation of HBV 5′ Epsilon Does Not Affect the Interaction with HBV Pol Protein.
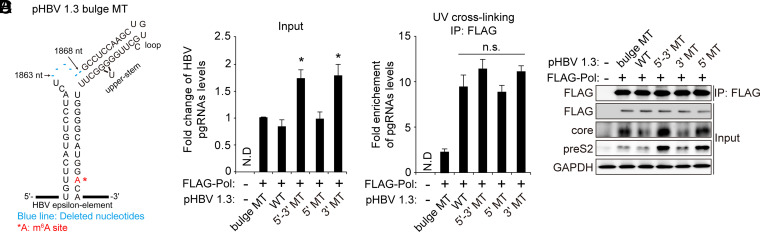
The HBV viral packaging process is triggered by the interaction of pol with the 5′ epsilon structure of pgRNA, which leads to the recognition of pgRNA structural motifs by the core protein to initiate encapsidation (4–7). Hence, we tested whether m6A modification affects the interaction between pol and pgRNA to regulate encapsidation. We used the ultraviolet (UV)-mediated cross-linking method to capture RNA–protein complexes (33) and conducted immunoprecipitation experiments with cell lysates from the Huh7 cells in which FLAG–pol and pHBV 1.3 bulge MT, WT, or MT (5′-3′, 5′, or 3′) plasmids were cotransfected (Fig. 3). We generated pHBV 1.3 bulge MT as a negative control (Fig. 3A), which contains the deletion of the bulge and upper-stem nucleotides corresponding to 1863 to 1868 nt to disrupt the interaction between pol and the 5′ epsilon of pgRNA (34). As observed before, HBV RNA and proteins levels were increased by the mutation of the m6A site of 3′ epsilon, but not in the HBV 5′ MT-expressing cells (Fig. 3 B and D). However, there were no appreciable differences in the levels of the UV cross-linked pgRNAs and FLAG–pol complexes from the cellular lysates expressing HBV WT and m6A site–mutated HBV genomes (Fig. 3C). The bulge structure mutated pgRNA transcribed from pHBV 1.3 bulge MT was not enriched by FLAG-tagged Pol. These results indicate that m6A modification of 5′ epsilon is not required for the interaction with pol.
Fig. 3.
The m6A modification of the 5′ epsilon of the HBV pgRNA does not affect viral pol–5′ epsilon interaction. (A) pHBV 1.3 Bulge MT was generated by the deletion of nucleotides from 1863 to 1868 nt of pHBV 1.3-mer. They are represented as follows: Blue lines indicate the position of deleted nucleotides, and *A is the m6A site. (B–D) Huh7 cells were cotransfected with FLAG–pol and pHBV 1.3 WT or 5′-3′ MT, 5′ MT, or 3′ MT plasmids. After 48 h, cells were washed with PBS, and then cells were irradiated with UV for RNA–protein cross-linking. Total RNA and cell lysates were extracted from these cells. FLAG-tagged HBV Pol proteins were immunoprecipitated using anti-FLAG M2 magnetic beads from UV-irradiated cell lysates. Immunoprecipitated RNAs were extracted by TRIzol. The input HBV pgRNA was analyzed by RT-qPCR (B). Immunoprecipitated HBV pgRNA levels were normalized by input HBV pgRNA levels by RT-qPCR (C). Immunoprecipitated FLAG–Pol and the indicated input proteins were analyzed by Western blotting (D). In B and C, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. *P < 0.05. IP, immunoprecipitation; N.D, not detected; n.s., nonsignificant.
The m6A Modification of HBV 5′ Epsilon Is Required for the Interaction with HBV Core Protein.
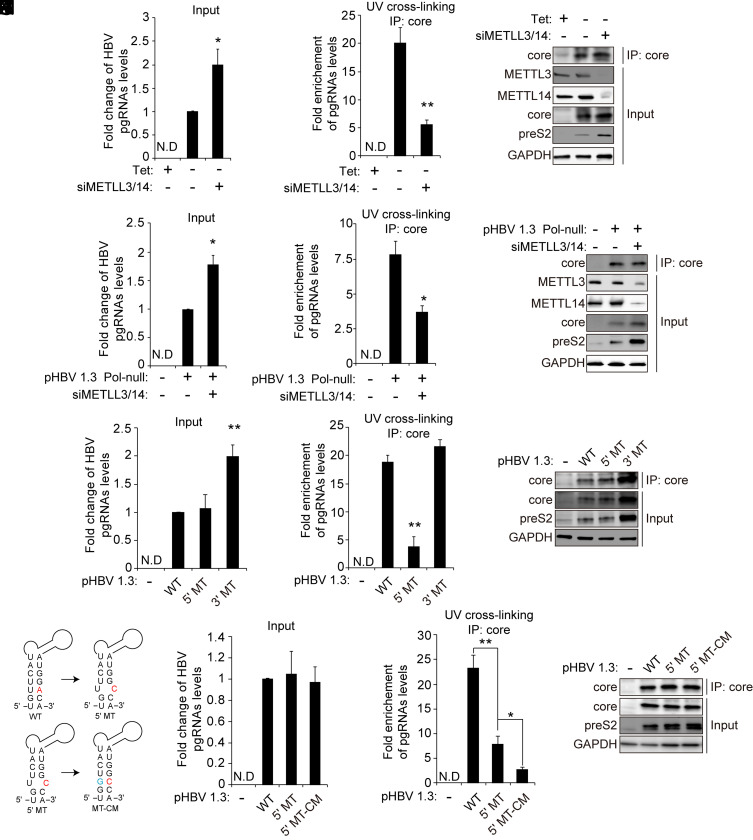
After establishing that the m6A modification of 5′ epsilon induces pgRNA encapsidation without affecting its interaction with pol, we investigated whether the m6A modification of 5′ epsilon affects the binding affinity between pgRNA and core proteins to regulate pgRNA encapsidation. The HBV expressing cells were irradiated with UV to capture the pgRNA–core protein complex, followed by immunoprecipitation of the lysates with anti-core antibodies. In METTL3/14-depleted cells, the interaction between HBV pgRNA and core proteins was decreased despite the increase in overall pgRNA expression levels (Fig. 4 A–C), indicating that the m6A modification of pgRNA is the target for core protein recognition. To further determine whether the m6A modification directly affects the recognition of 5′ epsilon by core proteins, we carried out experiments using Huh7 cells cotransfected with HBV pol-minus (null) genome and METTL3/14 siRNAs. These cells were irradiated with UV to capture the pgRNA–core protein complex, followed by immunoprecipitation of the lysates with anti-core antibodies. We observed that the silencing of METTL3/14 enzymes reduced the interaction between pgRNA and core protein (Fig. 4 D–F). These results indicate that the m6A modification of pgRNA is clearly required for the interaction with core proteins, although the pgRNA encapsidation event is not occurring because of the absence of pol protein. We next carried out immunoprecipitation of the pgRNA–core complex using the cell lysates from Huh7 cells transfected with HBV WT, 5′ MT, or 3′ MT plasmid to determine whether the m6A modification in 5′ epsilon affects the interaction with core proteins. Importantly, the m6A site mutation in the 5′ epsilon element dramatically reduced the interaction of pgRNA with core proteins compared to the HBV WT and 3′ MT-expressing cells (Fig. 4 D–F). These results suggest that the m6A methylation of HBV 5′ epsilon element promotes pgRNA encapsidation via its m6A-modified motif by inducing the interaction between pgRNA and core proteins. Because the mutation of the m6A site (A1907C) in the epsilon structure leads to a base pair mismatch in the lower stem loop, the alteration of the lower stem-loop secondary structure by A1907C mutation could affect the interaction of pgRNA with core proteins (Fig. 4G). To test this possibility, we generated compensatory mutation (CM) in the HBV 5′ MT plasmid, in which U is mutated to G to restore base pairing. Interestingly, the HBV 5′-MT-CM mutant–expressing cells displayed far more reduced binding to the core proteins compared to 5′-MT (Fig. 4 H–J). The core-associated pgRNA and DNA levels were also similarly decreased in 5′-MT-CM transfected cells compared to 5′-MT (SI Appendix, Fig. 4 A–C). These results imply that the restoration of the 5′ epsilon secondary structure by CM is not sufficient to recover core protein interaction but that the m6A modification of the target sequences is needed for recognition by core proteins. The m6A methylation is known to alter the local RNA structure and enhance the accessibility/affinity of protein binding (28, 29). In addition, the core protein binds to RNA in a nonspecific manner in vitro system, but the specific motifs encoded from pgRNA are more favored to interaction with core protein to promote nucleocapsid assembly (35–37). Thus, in this context, these results further suggest that the distorted lower stem of the 5′ epsilon structure produced by m6A modification presents a favorable conformation of pgRNA for the recognition by core protein to initiate encapsidation.
Fig. 4.
The m6A modification of the 5′ epsilon element promotes the interaction with core protein. (A–C) HepAD38 cells stably expressing HBV were grown in the absence or presence of tetracycline for 72 h, and thecells were then transfected with the METTL3/14-specific siRNAs. After 48 h, cells were irradiated with UV for RNA–protein cross-linking. Cells were harvested to extract total RNA and cellular lysates. Cellular lysates were immunoprecipitated using an anti-core antibody. The input HBV pgRNA was analyzed by RT-qPCR (A). Enriched HBV pgRNA levels were normalized by input HBV pgRNA levels by RT-qPCR (B). The indicated proteins were analyzed by Western blotting (C). (D–F) pHBV 1.3 Pol-null plasmids were transfected into Huh7 cells for 24 h, and the cells were then transfected with the METTL3/14-specific siRNAs. After 48 h, cells were irradiated with UV for RNA–protein cross-linking and harvested to extract total RNAs and cellular lysates, respectively. Cellular lysates were immunoprecipitated using an anti-core antibody. The input HBV pgRNA was analyzed by RT-qPCR (D). Enriched HBV pgRNA levels were normalized by input HBV pgRNA levels by RT-qPCR (E). The indicated proteins were analyzed by Western blotting (F). (G–I) The indicated plasmids were transfected into Huh7 cells. After 72 h, cells were UV irradiated, and total RNA and cellular lysates were extracted. The UV cross-linked RNA–protein complexes were immunoprecipitated using anti-core antibodies. The HBV pgRNA levels were assayed by qRT-PCR (G). Immunoprecipitated pgRNA levels were normalized by input pgRNA using RT-qPCR (H). The indicated proteins were analyzed by Western blotting (I). (J) The A1907C mutation in the epsilon structure is predicted to create a bubble. The compensatory U1851G mutation (blue) was generated in the 5′ epsilon (pHBV-5′-MT-CM). (K–M) Huh7 cells were transfected with the indicated plasmids. After 72 h, RNA–protein complexes from these cells were cross-linked by UV irradiation, and total RNA and cellular lysates were extracted, respectively. RNA–protein complexes were immunoprecipitated using anti-core antibodies. The input pgRNA levels were analyzed by RT-qPCR (H). Immunoprecipitated pgRNA levels were normalized by input pgRNA levels using RT-qPCR (I). The indicated proteins were analyzed by Western blotting (J). In A and B, D and E, and H and I, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. *P < 0.05, **P < 0.01. IP, immunoprecipitation.
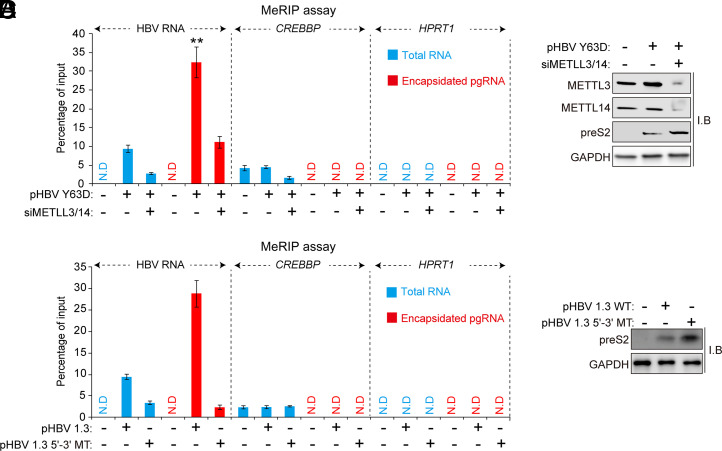
The m6A Methylated HBV pgRNA Is Enriched Inside Core Particles.
Next, we analyzed the m6A methylated or unmethylated pgRNA ratio in core particles by the methylated RNA immunoprecipitation (MeRIP) assay. For the MeRIP assay, we used the HBV Y63D mutant, which accumulates pgRNA in the core particle due to the mutation of pol activity (31). The results clearly show that m6A methylated pgRNA accumulated inside core proteins compared to total RNA fraction (Fig. 5 A and B). In the MeRIP assay, cellular CREBBP and HPRT1 were used as the positive or negative control, respectively. The silencing of METTL3/14 reduced the levels of m6A-methylated HBV RNA both from total RNA and encapsidated pgRNA fractions. We also observed similar results in the HBV 5′-3′ MT–transfected cells (Fig. 5 C and D). The presence of m6A-methylated pgRNA levels in the METTL314-depleted cells could result from the activities of methyltransferases other than METTL3/14 or the incomplete silencing of the enzymes. These results suggest that the m6A modification of the 5′ epsilon element increases the pgRNA packaging efficiency, which enriches the m6A-methylated HBV RNA inside the core particles.
Fig. 5.
The m6A methylated pgRNA is enriched in the core particles. (A and B) Huh7 cells were transfected with pHBV Y63D (RT-defective pol mutant) plasmid for 24 h, and the cells were further transfected with the siRNAs of METTL3/14 for 48 h. Total RNA and encapsidated pgRNA, and cellular lysates were isolated from these cells. Methylated RNAs from total RNA and encapsidated pgRNA fraction were immunoprecipitated from using anti-m6A antibodies. Immunoprecipitated (m6A-methylated) RNA levels were normalized by input RNA levels by RT-qPCR (A). CREBBP and HPRT1 were used for either positive or negative control. The indicated proteins were analyzed by Western blotting (B). (C and D) Huh7 cells were transfected with pHBV 1.3-mer or pHBV 1.3-mer 5′-3′ MT plasmid. After 72 h, cellular lysates, total RNA, and encapsidated pgRNA were extracted. Methylated RNAs from total RNA and encapsidated pgRNA fractions were immunoprecipitated using anti-m6A antibodies. Immunoprecipitated RNAs were analyzed by RT-qPCR (C). The indicated proteins were analyzed by Western blotting (D). In A and C, the error bars represent the SDs of three independent experiments. The P values are calculated via an unpaired Student’s t test. **P < 0.01. I.B, immunoblotting.
Discussion
The pgRNA encapsidation is the key event of the HBV life cycle (1). This step allows the continuation of viral replication. Core particles are assembled in the cytoplasm containing pgRNA–viral pol complex, wherein pgRNA is converted into DNA by a pathway involving RT to produce a partially double-stranded rcDNA. The 5′ epsilon element of pgRNA harbors cis-acting RNA encapsidation signals (14). Epsilon elements consist of a lower stem, a bulge region, an upper stem, and a tri-loop. The secondary structures of the lower stem and bulge are required for viral encapsidation (6, 13–15). The specific sequences on the lower right side of the upper stem and the apical loop play a critical role in pgRNA packaging (6, 13–15). Results presented in this study clearly demonstrate the pivotal role of m6A methylation of the 5′ epsilon elements in pgRNA encapsidation. By the extensive use of m6A mutants of HBV and the silencing strategy of METLL3/14, we show that m6A modification at 1907 nt, located in the lower stem of 5′ epsilon, promotes HBV RNA encapsidation by regulating the interaction with core proteins.
The recruitment of m6A YTHDF reader proteins regulates RNA translation and RNA stability (20). However, these m6A-binding proteins did not affect the HBV encapsidation pathway (SI Appendix, Fig. 3). Our previous experiments are consistent with the scheme in which YTHDF proteins bind m6A-modified RNAs and degrade them and only those that escape m6A-mediated RNA degradation perhaps enter the translation machinery (25, 38). In this context, this study supports the model in which m6A-mediated RNA degradation by YTHDF proteins and m6A-mediated HBV encapsidation represent distinct pathways.
Strikingly, the restoration of base pairing by the CM (5′ MT-CM) further reduced the interaction between pgRNA and core proteins, as well as the encapsidated pgRNA and core-associated DNA levels, compared to HBV 5′ MT (Fig. 4 H–J; SI Appendix, Fig. 4). Based on these results, we surmised that m6A modification may cause local structural distortion in the lower stem of the epsilon structure, and this helical distortion presents a favorable conformation for the core protein to bind and initiate viral capsid assembly. m6A modification has been described as altering RNA structure (28, 29). m6A modification is known to destabilize canonical double-stranded RNA and alter the local structures to facilitate the binding of proteins (28, 29). The selectivity of viral genomic RNAs for encapsidation from a large milieu of cellular RNAs and genomic/subgenomic viral RNAs is regulated by the biophysical properties of RNA. Viral RNA packaging is a dynamic process that occurs with high fidelity, dictated by secondary and tertiary structures and intramolecular interactions of viral RNA. In this respect, NMR-based analysis of m6A methylated epsilon is clearly needed to decipher the molecular structure that favors pgRNA encapsidation.
Despite the fact core protein has been shown to bind both RNA and DNA, indicating a nonsequence specific affinity for nucleic acids (35, 36), our results clearly suggest that the m6A modification of pgRNA induces nucleocapsid assembly by increasing the interaction with core protein without affecting the pol interaction (Figs. 3 and 4). This interaction seemingly occurs in the absence of pol, as evidenced by our results in which the pol bulge mutant and pol-null HBV genome transfections sustained m6A-modified pgRNA–core interactions (Figs. 3 and 4). Thus, the m6A-modified pgRNA 5′ epsilon might present a favorable recognition site for core protein interaction, adding a layer of the requirement of m6A modification of the pgRNA epsilon structure for nucleocapsid assembly.
Our previous studies have described a wider range of functional roles of m6A modification during HBV infection (25). These include the interferon-induced HBV RNA decay occurring in the context of m6A modification of 1907 nt (38), the requirement of HBx in methylation of viral transcripts at the sites of transcription initiation from the cccDNA template in the nucleus (39), the reduced sensitivity of RIG-I to m6A-modified RNA (40), and HBV-induced enhancement of a phosphatase and tensin homolog (PTEN) host RNA methylation and causing its degradation (41), the additional m6A modification at 1616 nt regulating HBx mRNA stability (42), and the intriguing role of fragile X mental retardation protein (FMRP) and YTHDC1 in the nuclear transport of m6A-modified viral transcripts to the cytoplasm (43). This study adds the role of m6A in the nucleocapsid assembly to the long list of myriad functions associated with m6A RNA methylation of HBV RNAs.
Our work opens a direction to investigate the role of m6A modification in RNA–protein complex interactions and particularly those of RNA viral genome packaging. The functional roles of m6A modification in various diseases including cancer and various metabolic diseases are continuously being revealed (18). Of interest in this respect is the consideration of inhibitors for m6A methyltransferases as anticancer drugs (44, 45). In this context, the pivotal role of m6A in HBV RNA encapsidation offers avenues for possible therapeutic intervention for cccDNA clearance from infected cells.
Materials and Methods
Plasmids, Antibodies, and Reagents.
The pHBV 1.3-mer plasmid was a kind gift from Dr. Wang-Shick Ryu (Yonsei University) and obtained from Addgene (65459). The FLAG–Pol plasmid was obtained from Addgene (65520). The pHBV 1.3-mer 5′-3′ MT, 5′ MT, 3′ MT, and 5′ MT-CM plasmids were previously constructed (25). The pHBV 1.3-mer Pol-null and bulge MT plasmids were generated by a QuikChange II Site-Directed Mutagenesis kit (Agilent). The ATG (start codon) in the pol open reading frame (ORF) was changed to ACG to create a pHBV 1.3-mer Pol-null plasmid. The nucleotides from 1863 to 1868 nt were deleted to generate a pHBV 1.3-mer bulge MT plasmid. The pHBV Y63D was obtained from Dr. Jianming Hu (Penn State Hershey Medical Cente) (31). Antibodies were obtained as follows: anti-YTHDF3 (#SC-379119), anti-preS2 (#SC-23944), and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; #SC-47724) antibodies from Santa Cruz Biotechnology; anti-METTL3 (#15073–1-AP) antibody from Proteintech Group; anti-METTL14 (#HPA038002) antibody from Sigma-Aldrich; anti-YTHDF1 (#86463), anti-YTHDF2 (#80014), anti-YTHDC2 (#35440), and anti-FLAG (#14793) antibodies from Cell Signaling Technology; anti-m6A antibody from Synaptic Systems; and anti-core and anti-precore antibodies were kind gifts from Dr. Jing-Hsiung James Ou (University of Southern California). Anti-preS2 antibody was diluted with a 1:200 ratio in a 5% bovine serum albumin (BSA) buffer for immunoblotting. The other antibodies were used with a 1:1,000 ratio in a 5% BSA buffer for immunoblotting. The ON-TARGET plus siRNAs of METTL3 (#L-005170-02-0005), METTL14 (#L-014169-02-0005), YTHDF1 (#L-018095-02-0005), YTHDF2 (#L-021009-02-0005), YTHDF3 (#L-017080-01-0005), and YTHDC2 (#L-014220-01-0005) were obtained from Dharmacon.
Cell Culture and Transfection.
HepAD38 cells were maintained in Roswell Park Memorial Institute medium (RPMI)-1640 with 20% fetal bovine serum and Huh7 and HepG2-NTCP cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. The HepG2-NTCP cells were provided by Dr. Wenhui Li (National Institute of Biological Sciences). The media was supplemented with 2 mM L-glutamine, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.1 mM nonessential amino acid under standard culture conditions (5% CO2, 37 °C). PHHs were purchased from Gibco and cultured according to the manufacturer’s protocol. Plasmids were transfected into cells using Mirus TransIT-LT1 reagent (Mirus) according to the manufacturer’s protocol. Lipofectamine RNAiMAX reagent was used for siRNA transfection (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Virus Production and Cell Infection.
HBV particles were harvested from the supernatants of pHBV 1.3 WT or m6A site–mutated plasmid-transfected Huh7 cells. The culture medium was centrifuged at 4 °C, 10,000 × g for 15 min. The clarified supernatants were incubated with 5% polyethylene glycol (PEG) 8000 overnight at 4 °C and then centrifuged at 4,000 rpm for 30 min at 4 °C. Pellet was redissolved in a serum-free culture medium at 1% volume of the original supernatant. For infection, the PHHs and HepG2-NTCP cells were split in collagen-coated plates and incubated for 24 h with HBV particles, which are diluted in a serum-free culture medium with 4% PEG 8000 and 2% dimethyl sulfoxide (DMSO). After being incubated with HBV particles, the cells were washed with a culture medium. Cells were incubated for 10 d with a medium changed every two days in a medium containing 2% DMSO.
Real-Time RT-qPCR.
Total RNA was isolated using the RNeasy mini kit (Qiagen). The cDNAs were synthesis from extracted total RNA using iScript Reverse Transcription Supermix (Bio-Rad). The qPCR was assessed with Ssoadvanced Universal SYBR Green supermix (Bio-Rad). Each viral RNA and mRNA expression level, normalized to GAPDH, was analyzed using the ΔΔCt method. The primers used for RT-qPCR were as follows: HBV RNA (forward primer: 5′-CTCCCCGTCTGTGCCTTCT-3′; reverse primer 5′-GCCCCAAAGCCACCCAAG-3′), HBV pgRNA (5′-CTCAATCTCGGGAATCTCAATGT-3′; reverse primer 5′-TGGATAAAACCTAGGAGGCATAAT-3′), GAPDH (forward primer: 5′-CCTGCACCACCAACTGCTTA-3′; reverse primer 5′-CATGAGTCCTTCCACGATACCA-3′).
Isolation of Encapsidated HBV pgRNA.
Cells from one well of a six-well plate were washed with PBS and incubated with 500 μL lysis buffer [50 mM Tris⋅HCl (pH7.5), 1 mM ethylenediaminetetraacetic acid (EDTA), 150 mM NaCl, and 1% Nonidet P-40 with Protease inhibitor mixture] in 37 °C for 10 min. The lysates were centrifuged for 2 min at 14,000 rpm to remove the cell debris and nuclei. The supernatant was transferred to a new centrifuge tube and then incubated with 6 U micrococcal nuclease (New England Biolabs) and 30 μL of 100 mM CaCl2 for 15 min at 37 °C for to remove unprotected free nucleic acids. The encapsidated pgRNA was extracted by the RNeasy min kit (Qiagen). The extracted encapsidated pgRNA was analyzed with RT-qPCR and Northern blot assay.
Isolation of Core-Associated HBV DNA.
Cells from one well of a six-well plate were washed with phosphate-buffered saline (PBS) and then incubated with freshly prepared 500 μL of transfection lysis buffer [50 mM Tris⋅HCl (pH 8.0), 1 mM EDTA, and 1% Nonidet P-40 with Protease inhibitor mixture] in 37 °C for 10 min. The lysates were centrifuged for 1 min at 14,000 rpm, and then the supernatants were transferred to a new centrifuge tube. The supernatant was added with 30 μL CaCl2 and 75 U micrococcal nuclease (New England Biolabs) for 45 min at 37 °C. After mixing briefly, the lysates were centrifuged for 1 min at 14,000 rpm. After brief centrifugation, 75 U micrococcal nuclease was added to the supernatant again and incubated for 45 min in a 37 °C rotator. After centrifugation for 1 min at 14,000 rpm, supernatant was transferred to a new microcentrifuge tube, and 32 μl of 0.5 M EDTA and 260 μl of 35% PEG in 1.75 M NaCl was added and kept in 4 °C for 1 h. After centrifugation at 13,000 rpm for 5 min at 4 °C, the supernatant was discarded, and the pellet was resuspended in 300 μL tris-NaCl-EDTA (TNE) buffer.
MeRIP Assay.
The extracted RNA was incubated with anti-m6A antibody (Synaptic Systems) conjugated to Protein G Dynabeads (Thermo Fisher Scientific) in MeRIP buffer [50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 0.1% Nonidet P-40] overnight at 4 °C. The immunoprecipitated RNA–bead complexes were washed with MeRIP buffer five times, and bound RNA was extracted with TRIzol (Thermo Fisher Scientific). Eluted RNA was reverse transcribed into complementary DNA (cDNA) and subjected to RT-qPCR.
Western Blotting and Immunoprecipitation.
Cells were incubated with a Nonidet P-40 lysis buffer [1% Nonidet P-40, 50 mM Tris⋅HCl (pH 8.0), 150 mM NaCl] supplemented with a protease inhibitor (Thermo Fisher Scientific) for 15 min at 4 °C. After centrifugation for 20 min at 14,000 rpm at 4 °C, cell lysates were transferred to a new microcentrifuge tube. Lysates were resolved by sodium dodecyl sulphate– polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% BSA for 1.5 h, following the by then overnight incubation with primary antibodies at 4 °C. After washing, the membranes were incubated with secondary antibody–conjugated horseradish peroxidase (Cell Signaling Technology) for 1 h. Antibody complexes were detected using a chemiluminescence substrate (Millipore). Chemiluminescence signals were detected using the ChemiDoc MP Imaging Systems (Bio-Rad).
RNA–Protein UV Cross-linking Assay.
Before harvesting, plated cells were washed with cold PBS, and then cells were exposed to 245-nm UV for 250 mJ/cm. Cell pellets were resuspended with SDS-lysis buffer [0.5% SDS, 50 mM Tris⋅HCl (pH 6.8), 1 mM EDTA, 1 mM dithiothreitol (DTT), 150 mM NaCl] supplemented with a protease inhibitor and RNase inhibitor (Thermo Fisher Scientific). Lysates were immunoprecipitated with anti-FLAG M2 magnetic beads (Sigma-Aldrich) or anti-core antibodies conjugated to Protein G Dynabeads (Thermo Fisher Scientific) for 2 h on the rotator at 4 °C. The bead complex was washed with Nonidet P-40 lysis buffer for five times. After the wash step, beads were resuspended with Nonidet P-40 lysis buffer. The half of the bead was incubated with proteinase K for 90 min at 37 °C, and then immunoprecipitated RNA was extracted using the RNeasy mini kit (Qiagen). The other half of the bead was added to protein sample buffer to be analyzed with Western blot assay.
Northern Blot Assay for HBV RNA.
The 20 μg total RNA or encapsidated pgRNA was resolved by electrophoresis on 1.2% formaldehyde agarose gel and transferred to a positively charged nylon membrane (Thermo Fisher Scientific). The membrane was fixed by UV cross-linking. The fixed membrane was prehybridized for 30 min and then incubated by ULTRAhyb hybridization buffer (Invitrogen) for 30 min and then hybridized with an internally radiolabeled double-stranded DNAprobe in ULTRAhyb hybridization buffer (Thermo Fisher Scientific) overnight. The probe was generated using [α-32P] dCTP and DECAprimeII kit (Thermo Fisher Scientific). After, washing, the membrane was exposed with BIOMAX XAR film (Sigma-Aldrich). The probe template was amplified by PCR using forward (5′- ATGGCTGCTAGGCTGTGCTGCC-3′) and reverse (5′-ATAAGGGTCGATGTCCATGCCCCAAAG-3′) primer from the pHBV1.3-mer plasmid.
Southern Blot Analysis for HBV DNA.
Modified Hirt’s extraction protocol was used to isolate protein-free viral DNA (cccDNA and protein-free rcDNA). Cells were incubated with 1.5 mL tris-EDTA (TE) buffer [10 mM Tris⋅HCl (pH 7.5), 10 mM EDTA] and 0.1 mL 10% SDS for 30 min at room temperature and then added to 0.4 mL of 5 M NaCl. After mixing briefly, the lysates were kept at 4 °C overnight. After centrifugation at 12,000 rpm for 30 min, the supernatant was transferred to a new tube and extracted two times with phenol and one time with phenol/chloroform/isopropanol. Then, the supernatant was transferred to a new tube, and two volumes of ethanol were added and kept at room temperature overnight. After centrifugation at 12,000 rpm for 15 min at 4 °C, the supernatant was discarded and the pellet was dissolved with elution buffer. This is the total HBV Hirt DNA preparation, which is a mixture of cccDNA and deproteinized (DP)-rcDNA. The extracted samples were treated with plasmid-safe ATP-dependent DNase (Lucigen). The extracted viral DNA was resolved by electrophoresis on 1.2% agarose gel, then transferred onto a nylon membrane (Thermo Fisher Scientific). The membrane was incubated with an internally radiolabeled dsDNA probe overnight.
Statistical Analysis.
All results are representative of three independent experiments. For each result, error bars represent the ±SD from at least three independent experiments. The P value was calculated using a one-tailed unpaired Student’s t test.
Supplementary Material
Acknowledgments
We thank Dr. Jianming Hu (Penn State Hershey Medical Center) for the gift of pHBV Y63D plasmid and Dr. Jing-Hsiung James Ou (University of Southern California) for the gift of anti-core/precore antibodies. This study was supported by NIH grants AI125350 and AI139234 to A.S.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120485119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Seeger C., Mason W. S., Molecular biology of hepatitis B virus infection. Virology 479-480, 672–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu J., Protzer U., Siddiqui A., Revisiting hepatitis B virus: Challenges of curative therapies. J. Virol. 93, e01032-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nassal M., HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R., Junker-Niepmann M., Schaller H., The P gene product of hepatitis B virus is required as a structural component for genomic RNA encapsidation. J. Virol. 64, 5324–5332 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch R. C., Lavine J. E., Chang L. J., Varmus H. E., Ganem D., Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as wel as for reverse transcription. Nature 344, 552–555 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Knaus T., Nassal M., The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 21, 3967–3975 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birnbaum F., Nassal M., Hepatitis B virus nucleocapsid assembly: Primary structure requirements in the core protein. J. Virol. 64, 3319–3330 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nassal M., The arginine-rich domain of the hepatitis B virus core protein is required for pregenome encapsidation and productive viral positive-strand DNA synthesis but not for virus assembly. J. Virol. 66, 4107–4116 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowther R. A., et al. , Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell 77, 943–950 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Lan Y. T., Li J., Liao W., Ou J., Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259, 342–348 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Heger-Stevic J., Zimmermann P., Lecoq L., Böttcher B., Nassal M., Hepatitis B virus core protein phosphorylation: Identification of the SRPK1 target sites and impact of their occupancy on RNA binding and capsid structure. PLoS Pathog. 14, e1007488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stannard L. M., Hodgkiss M., Morphological irregularities in Dane particle cores. J. Gen. Virol. 45, 509–514 (1979). [DOI] [PubMed] [Google Scholar]
- 13.Beck J., Nassal M., Hepatitis B virus replication. World J. Gastroenterol. 13, 48–64 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen A., Brown C., Distinct families of cis-acting RNA replication elements epsilon from hepatitis B viruses. RNA Biol. 9, 130–136 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flodell S., et al. , The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidine bulges. Nucleic Acids Res. 30, 4803–4811 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeong J. K., Yoon G. S., Ryu W. S., Evidence that the 5′-end cap structure is essential for encapsidation of hepatitis B virus pregenomic RNA. J. Virol. 74, 5502–5508 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roundtree I. A., Evans M. E., Pan T., He C., Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yue Y., Liu J., He C., RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev. 29, 1343–1355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H., Wei J., He C., Where, when, and how: Context-dependent functions of RNA methylation writers, readers, and erasers. Mol. Cell 74, 640–650 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer K. D., Jaffrey S. R., Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 33, 319–342 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzales-van Horn S. R., Sarnow P., Making the mark: The role of adenosine modifications in the life cycle of RNA viruses. Cell Host Microbe 21, 661–669 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gokhale N. S., et al. , N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20, 654–665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lichinchi G., et al. , Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 1, 16011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim G. W., Siddiqui A., N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc. Natl. Acad. Sci. U.S.A. 118, e2022024118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imam H., et al. , N6-methyladenosine modification of hepatitis B virus RNA differentially regulates the viral life cycle. Proc. Natl. Acad. Sci. U.S.A. 115, 8829–8834 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imam H., Kim G. W., Siddiqui A., Epitranscriptomic(N6-methyladenosine) Modification of Viral RNA and Virus-Host Interactions. Front. Cell. Infect. Microbiol. 10, 584283 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim G. W., Siddiqui A., The role of N6-methyladenosine modification in the life cycle and disease pathogenesis of hepatitis B and C viruses. Exp. Mol. Med. 53, 339–345 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu N., et al. , N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu N., et al. , N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 45, 6051–6063 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ladner S. K., et al. , Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 41, 1715–1720 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen D. H., Gummuluru S., Hu J., Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol. 81, 4465–4472 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao Y., et al. , m6A in mRNA coding regions promotes translation via the RNA helicase-containing YTHDC2. Nat. Commun. 10, 5332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poria D. K., Ray P. S., RNA-protein UV-crosslinking assay. Bio Protoc. 7, e2193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J., Boyer M., Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 80, 2141–2150 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatton T., Zhou S., Standring D. N., RNA- and DNA-binding activities in hepatitis B virus capsid protein: A model for their roles in viral replication. J. Virol. 66, 5232–5241 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Porterfield J. Z., et al. , Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J. Virol. 84, 7174–7184 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel N., et al. , HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly. Nat. Microbiol. 2, 17098 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imam H., Kim G. W., Mir S. A., Khan M., Siddiqui A., Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 16, e1008338 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim G. W., Siddiqui A., Hepatitis B virus X protein recruits methyltransferases to affect cotranscriptional N6-methyladenosine modification of viral/host RNAs. Proc. Natl. Acad. Sci. U.S.A. 118, e2019455118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim G. W., Imam H., Khan M., Siddiqui A., N 6-Methyladenosine modification of hepatitis B and C viral RNAs attenuates host innate immunity via RIG-I signaling. J. Biol. Chem. 295, 13123–13133 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim G. W., et al. , HBV-induced increased N6 methyladenosine modification of PTEN RNA affects innate immunity and contributes to HCC. Hepatology 73, 533–547 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim G.-W., Siddiqui A., Hepatitis B virus X (HBx) Protein expression is tightly regulated by N6-methyladenosine modification of its mRNA. J. Virol., doi: 10.1128/JVI.01655-21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim G. W., Imam H., Siddiqui A., The RNA binding proteins YTHDC1 and FMRP regulate the nuclear export of N6-methyladenosine-modified hepatitis B virus transcripts and affect the viral life cycle. J. Virol. 95, e0009721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yankova E., et al. , Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 593, 597–601 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shriwas O., Mohapatra P., Mohanty S., Dash R., The impact of m6A RNA modification in therapy resistance of cancer: Implication in chemotherapy, radiotherapy, and immunotherapy. Front. Oncol. 10, 612337 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.