Significance
This study provides proof-of-principle evidence for intravesical delivery of messenger RNA (mRNA) via a mucoadhesive nanoparticle (NP) strategy and reveals the therapeutic potential of KDM6A in treating bladder cancer metastasis, which remains difficult due to the physiological bladder barriers. The mucoadhesive NPs could protect loaded mRNA, prolong exposure of mRNA in disease sites, and benefit the penetration and effective expression, which all represent challenging hurdles for intravesical delivery of mRNA therapeutics. mRNA local delivery can also avoid potential toxicity issues via systemic delivery and unwanted protein expression throughout the body. We expect this mucoadhesive mRNA nanotechnology can be useful for the effective up-regulation of targeted proteins in bladder tissues in situ for both mechanistic understanding and translational study of bladder-related diseases.
Keywords: KDM6A, mRNA nanoparticles, bladder cancer, intravesical delivery, elemene
Abstract
Lysine-specific demethylase 6A (KDM6A), also named UTX, is frequently mutated in bladder cancer (BCa). Although known as a tumor suppressor, KDM6A’s therapeutic potential in the metastasis of BCa remains elusive. It also remains difficult to fulfill the effective up-regulation of KDM6A levels in bladder tumor tissues in situ to verify its potential in treating BCa metastasis. Here, we report a mucoadhesive messenger RNA (mRNA) nanoparticle (NP) strategy for the intravesical delivery of KDM6A-mRNA in mice bearing orthotopic Kdm6a-null BCa and show evidence of KDM6A’s therapeutic potential in inhibiting the metastasis of BCa. Through this mucoadhesive mRNA NP strategy, the exposure of KDM6A-mRNA to the in situ BCa tumors can be greatly prolonged for effective expression, and the penetration can be also enhanced by adhering to the bladder for sustained delivery. This mRNA NP strategy is also demonstrated to be effective for combination cancer therapy with other clinically approved drugs (e.g., elemene), which could further enhance therapeutic outcomes. Our findings not only report intravesical delivery of mRNA via a mucoadhesive mRNA NP strategy but also provide the proof-of-concept for the usefulness of these mRNA NPs as tools in both mechanistic understanding and translational study of bladder-related diseases.
The Kdm6a (also known as UTX) gene encodes a histone demethylase named lysine-specific demethylase 6A (KDM6A), which is an important epigenetic regulator that functions as a histone H3K27 demethylase. The role of KDM6A as a tumor-suppressor in bladder cancer (BCa) has been well recognized (1–3). Because loss of KDM6A has been found to increase BCa risk and mutations, reduced expression of KDM6A could not only be a predictor of poor prognosis in BCa patients but may also indicate that KDM6A plays a critical role in the tumorigenesis of BCa (4). Re-expression of exogenous KDM6A via Kdm6a-DNA plasmid in different Kdm6a-null cancer cells resulted in reduced rates of cell growth and proliferation (5–7). However, the functional consequences of its inactivation and reactivation in the metastasis of BCa remain largely unexplored. Considering that muscle-invasive (MI) BCas (i.e., stages T2 to T4) have a higher chance of metastasizing to the lymph nodes or other organs (8), a clear understanding of the functional consequences of the relationship between KDM6A status and BCa metastasis may guide future mechanistic and translational studies in BCa. Nevertheless, it is still difficult to effectively up-regulate the KDM6A levels in Kdm6a-null BCa in situ to directly verify its therapeutic potential in inhibiting BCa metastasis due to various challenging hurdles.
Currently, to the best of our knowledge, all attempts to introduce exogenous KDM6A have utilized DNA transfection methods (1, 5–7). Nonviral vector lipofectamine reagents and viral vectors are currently the most commonly used tools for DNA transfection. Nevertheless, 1) nonviral lipofectamine reagents can be used only in vitro (due to poor in vivo stability) and 2) viral vector–based technologies may raise concerns such as low packaging capacity, high production cost, high immunogenicity, and potential transmission into the environment. Moreover, DNA transfection methods could increase the risk of genomic integration and mutagenesis. Keeping in mind the future transition from an improved mechanistic understanding to translational cancer research, it will not be easy to design alternative approaches that effectively introduce KDM6A functions while avoiding the above-mentioned problems. The emerging messenger RNA (mRNA) nanotechnology provides a promising alternative to address these issues (9–12). Because chemically modified mRNA is primarily active in the cytoplasm, mRNA transfection efficacy is high and genomic integration is avoided (13, 14). This excludes the potential risk of deleterious integration into the host genome (15), which is caused by viral vectors or plasmid DNAs that bring about insertional mutagenesis. Even in hard-to-transfect or nondividing cells, mRNA can still enable rapid and reliable protein expression. Additionally, in contrast to the random-onset expression kinetics offered by DNA therapeutics, mRNA enables more predictable and consistent expression kinetics both in vitro and in vivo (9, 10, 16–21). However, local delivery of mRNA drugs via a nanoparticle (NP) strategy to specific organs, especially bladder tissues, still remains challenging.
Due to the unique structure of the bladder, systemic delivery of drugs may hardly distribute to the bladder sites (i.e., blocked by the physical barriers). Instead, intravesical therapy (i.e., the instillation of drugs via a catheter into the bladder) is a common route of administration for the treatment of BCa in the clinic. However, 1) in the bladder, almost all drugs are usually rapidly washed out on first voiding of urine and 2) the exposure of the BCa tumors to the drugs is low, greatly limiting treatment efficacy (22). To prolong the dwell time of therapeutic NPs in the bladder and improve their uptake and penetration into BCa tissue (23, 24), an mRNA NP strategy that can adhere to the bladder for sustained and effective delivery of mRNA in BCa tumors is therefore urgently needed. To date, the use of mRNA NPs for intravesical delivery of mRNA to specific in situ bladder tissues remains unreported.
Herein, we provide proof of principle for the use of a mucoadhesive mRNA NP–mediated strategy to effectively up-regulated KDM6A levels in Kdm6a-null BCa in situ and explore the therapeutic potential role of KDM6A in BCa metastasis. The NPs were constructed from a Food and Drug Administration (FDA)-approved polymer, poly(lactic-co-glycolic) acid (PLGA), and used to protect and load mRNA/lipid complexes (9, 10). Because abundant functional groups (e.g., amine and sulfydryl) can effectively promote adhesion to the bladder mucosal surface for intravesical delivery (23–25), we used terminal functionalized lipid-polyethylene glycol (PEG) (i.e., DSPE-PEG-NH2, DSPE-PEG-SH, or their mixture at a fixed ratio) to coat the mRNA NP core and endow the surface of our mRNA NPs with optimized mucoadhesive properties. Our results demonstrated that the surface-engineered, mucoadhesive mRNA NPs could effectively adhere to the bladder tissue through intravesical administration, prolong the exposure of KDM6A-mRNA to the in situ BCa tumors, enhance the NP uptake/penetration, and improve the expression of KDM6A in Kdm6a-null BCa in situ. The local mRNA delivery strategy via intravesical administration could avoid unwanted protein throughout the body and potential toxicity issues via systemic delivery. We also provide evidence for the close relation between KDM6A levels and the BCa metastasis and, more importantly, showed that the therapeutic potential of KDM6A in treating Kdm6a-null BCa metastasis. The therapeutic effects could also be enhanced by combining elemene, which may potentially expand this mRNA NP strategy's application for combination cancer therapy with other therapeutics.
Results
On-Demand Introduction of Exogenous and Functional Proteins in Kdm6a-null BCa Cells via a Universal Strategy Based on mRNA NPs.
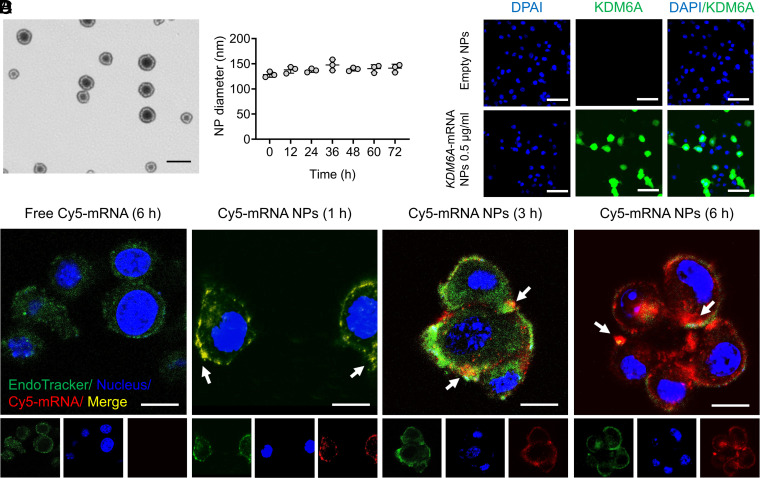
To our best knowledge, no mRNA NPs have been applied in BCa cells to date, and different cell lines may possess different transfection efficacies. Therefore, before testing the functional consequences of restoring KDM6A in Kdm6a-null BCa cells, it was necessary to first determine whether our core mRNA NP technology could be used as a universal strategy for “on-demand” introduction of exogenous and functional proteins in BCa cells. Engineered by an optimized nanoprecipitation method, negatively charged mRNA was first absorbed by cationic lipids G0-C14 via electrostatic adsorption (9, 26), and the complexes were then effectively loaded into an NP core formed of clinically approved PLGA polymers (10). 1,2-distearoyl-sn-glycero-3-phosphoethanolamine–conjugated PEG (DSPE-PEG) was used to coat the NP core to increase stability in physiological conditions (27, 28). As can be observed from the transmission electron microscopy (TEM) image, the obtained mRNA NPs were relatively uniform and spherical in structure (Fig. 1A). The average diameter of our NPs was ∼130 nm and can be maintained in phosphate-buffered saline (PBS) (containing 10% serum) at 37 °C for 72 h (Fig. 1B), indicating acceptable stability of these engineered NPs. The intracellular delivery of mRNA in Kdm6a-null KU19-19 cells (i.e., model BCa cell line without KDM6A expression in this study) was further assessed with Cy5-labeled control mRNA (Cy5-mRNA) NPs through confocal laser scanning microscopy (CLSM). Our NPs effectively delivered mRNA (red) to KU19-19 cells, while naked mRNA is hardly able to enter cells (Fig. 1C). With an increase in incubation time, the mRNA carried by the NPs effectively escaped the endosomes (green) and entered the cytoplasm.
Fig. 1.
On-demand introduction of exogenous and functional proteins in Kdm6a-null BCa cells via mRNA NPs. (A) mRNA NP morphology was observed through TEM (Scale bar, 200 nm). (B) Stability of the mRNA NPs in PBS containing 10% serum throughout 72 h. The NP diameters were confirmed through dynamic light scattering measurements. (C) CLSM imaging of Kdm6a-null KU19-19 cells after different duration of incubation (1, 3, and 6 h) with Cy5-mRNA NPs (red). CellLight Late Endosomes-GFP was used to stain endosomes (green), and Hoechst 33342 was utilized to stain nuclei (blue). Cells after 6 h of incubation with free Cy5-mRNA were assigned to the control group (Scale bars, 10 µm). (D) IF staining of KDM6A in Kdm6a-null KU19-19 cells after treatment with empty NPs or KDM6A-mRNA NPs (Scale bars, 50 µm).
To further confirm the efficacy of introducing exogenous and functional proteins in KU19-19 cells via mRNA NPs, EGFP-mRNA expressing enhanced green fluorescent proteins (EGFPs) and Luc-mRNA expressing luciferase proteins were used as model mRNAs. Both flow cytometry analysis, showing the expression of EGFPs (SI Appendix, Fig. S1A), and bioluminescence imaging, showing the expression of luciferase proteins (SI Appendix, Fig. S1 B and C), demonstrated that our mRNA NPs effectively introduced exogenous and functional proteins in KU19-19 cells. In addition, the amount of expressed proteins can be up-regulated “on-demand” by simply increasing the mRNA NP concentrations. Finally, we measured KDM6A expression in the Kdm6a-null KU19-19 BCa cells after treatment with KDM6A-mRNA NPs or empty NPs (control group) through immunofluorescence (IF) staining assays. KDM6A was effectively introduced in Kdm6a-null KU19-19 cells after the treatment of KDM6A-mRNA NPs, while the control group hardly showed any expression of KDM6A (Fig. 1D). Therefore, we were able to confirm that our core mRNA NP technology holds the potential to be a universal strategy for the “on-demand” introduction of exogenous proteins including KDM6A (Fig. 1D and SI Appendix, Fig. S2) in Kdm6a-null BCa cells.
Introduction of KDM6A via mRNA NPs in Kdm6a-null BCa Cells Prevents the Invasion and Migration of BCa Cells In Vitro.
After verifying that our mRNA NPs successfully introduced exogenous KDM6A, we focused on checking the functional consequence after KDM6A introduction in Kdm6a-null BCa cells and exploring the role of KDM6A in BCa metastasis from a cellular level. In the first part of this section’s study, it was interesting to find that Kdm6a-null KU19-19 cells exhibited higher rates of cell invasion and migration than Kdm6a-wild-type RT-4 cells (i.e., model BCa cell line with KDM6A expression in this study), via the real-time cell analyzer (RTCA) invasion and migration monitoring assays (29) (SI Appendix, Fig. S3A). In vitro wound healing assays also showed that Kdm6a-null KU19-19 cells migrated obviously after 24 h of incubation, while Kdm6a-wild-type RT-4 cells seldom migrated during the same incubation period (SI Appendix, Fig. S3B). In the transwell assays, we also observed that invasive Kdm6a-null KU19-19 cells were significantly more numerous than invasive Kdm6a-wild-type RT-4 cells (SI Appendix, Fig. S4). Because previous studies have demonstrated that increased filopodia formation is an essential precondition for the invasion of cancer cells (30), we then performed cell morphology and cytoskeleton analyses using rhodamine-conjugated phalloidin staining by IF microscopy. Consistently, most Kdm6a-null KU19-19 cells exhibited long and extended filopodia, while Kdm6a-wild-type RT-4 cells seldom exhibited any filopodia (SI Appendix, Fig. S5). Judging from all these interesting phenomena between the two cell lines, it might be possible that the loss of KDM6A in BCa cells may contribute to invasive cellular migration.
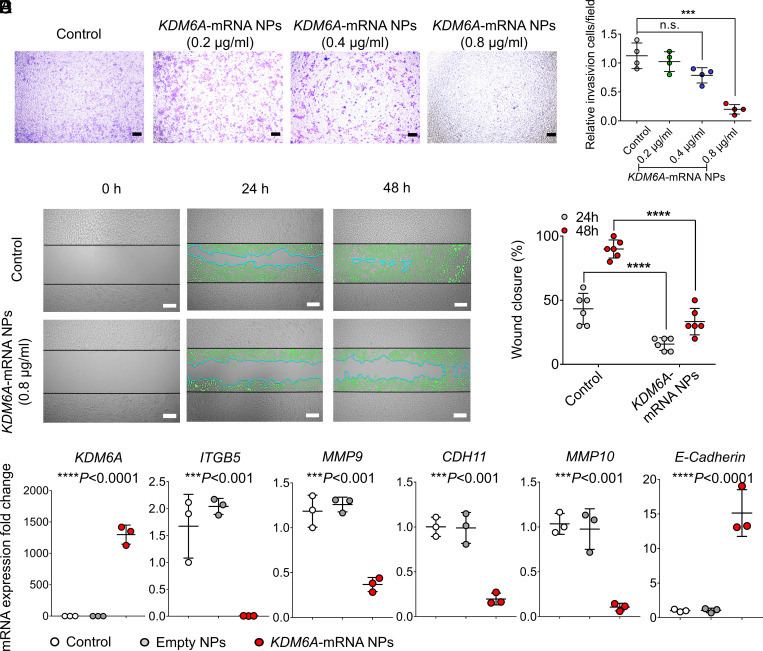
Next, we further examined whether introducing KDM6A in Kdm6a-null KU19-19 cells could lead to the inhibition of cell proliferation, invasion, and migration. Both the colony formation study and the proliferation assays demonstrated that the introduction of KDM6A via mRNA NPs had an antiproliferation effect on Kdm6a-null KU19-19 cells (SI Appendix, Fig. S6), and this inhibition effect can be enhanced by increasing the concentrations of KDM6A-mRNA NPs. A transwell assay was performed in vitro to simulate the invasion and migration of Kdm6a-null BCa cells with or without treatment of mRNA NPs. Our results demonstrated that the treatment of KDM6A-mRNA NPs effectively reduced the invasion and migration of Kdm6a-null KU19-19 cells in a dosage-dependent manner (Fig. 2 A and B). In vitro wound healing assays further confirmed that mRNA NP–mediated introduction of KDM6A significantly inhibited the migration of KU19-19 cells (Fig. 2 C and D). Hence, it is possible that loss of KDM6A promotes invasion and migration of BCa cells in vitro, and our data showed that reintroducing the functional KDM6A proteins in Kdm6a-null BCa cells could effectively alleviate the tendency toward invasion and migration of these BCa cells.
Fig. 2.
mRNA NPs restores KDM6A in Kdm6a-null BCa cells, inhibits metastasis-related pathways, and prevents the invasion and migration of BCa cells in vitro. (A) Representative images of Kdm6a-null KU19-19 BCa cells after treatment with KDM6A-mRNA NPs at different concentrations in the 24-transwell system from transwell invasion assays. KU19-19 cells without NP treatment were assigned to the control group (Scale bars, 160 µm). (B) Quantitative analysis of the transwell invasion assays. Invasive cells were counted in five representative magnified views per transwell. Experiments were carried out four times. Statistical significance was defined by ***P < 0.001. (C) Representative images of Kdm6a-null KU19-19 cells with or without treatment with KDM6A-mRNA NPs from in vitro wound healing assays (incubation time: 24 and 48 h). Green cells represent invasive and migrating cells. Blue lines represent the borderlines (Scale bars, 200 µm). (D) Quantitative analysis of the in vitro wound healing assays. Statistical significance was defined by ****P < 0.0001. (E) qRT-PCR was carried out to measure the expression of KDM6A, ITGB5, MMP9, CDH11, MMP10, and E-Cadherin. Statistical significance was defined as ***P < 0.001 and ****P < 0.0001 between control group and KDM6A-mRNA NPs Group.
In addition, the PCR assay also showed that, compared with control group and empty NPs group, the introduction of KDM6A via mRNA NPs in KU19-19 cells significantly down-regulated the expression of ITGB5, MMP9, CDH11, and MMP10 (Fig. 2E), which are actively involved in the tumor progression and invasion pathways. Moreover, the expression of E-Cadherin was significantly up-regulated after the treatment of KDM6A-mRNA NPs. E-Cadherin is a critical determinant of tumor progression through suppressing tumor invasion and metastasis (31, 32). Up-regulation of E-Cadherin level via KDM6A-mRNA NP treatment in KU19-19 cells indicates that the metastasis-related pathways in epithelial mesenchymal transition (EMT) might be inhibited, which may further take a role in preventing their invasion and migration in vitro.
Transcriptomic Analysis on the Relationship between KDM6A Restoration via mRNA NPs in Kdm6a-null BCa Cells and Their Metastasis.
In this section of the study, to examine the effect of KDM6A on BCa metastasis, we analyzed the gene expressions at a transcriptomic level. Similarly, it was interesting to identify a large number of genes that were differentially expressed due to the loss of Kdm6a in KU19-19 cells (versus Kdm6a-wild-type RT-4 cells) (SI Appendix, Fig. S7A). Among them, metastasis-related genes were significantly up-regulated in Kdm6a-null KU19-19 cells (versus Kdm6a-wild-type RT-4 cells) (SI Appendix, Fig. S7B). A subsequent gene ontology analysis of the gene expression differences between Kdm6a-null KU19-19 cells and Kdm6a-wild-type RT-4 cells revealed that loss-of-Kdm6a strengthened the focal adhesion and cell adhesion molecules (CAMs) pathways (SI Appendix, Fig. S7C). Because both pathways are closely related to tumor metastasis, these results indicate a potential promoting role of loss-of-Kdm6a in the metastasis of BCa. Meanwhile, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis also found that the genes linked to adhesion pathways were among the most significantly expressed in Kdm6a-null KU19-19 cells (SI Appendix, Fig. S7D). Moreover, the results confirmed by the gene set enrichment analysis (GSEA) also demonstrated that both the focal adhesion signaling pathway (***P < 0.001) and the CAMs signaling pathway (*P < 0.05) were significantly up-regulated in Kdm6a-null KU19-19 cells compared to those in Kdm6a-wild-type RT-4 cells (SI Appendix, Fig. S7 E and F).
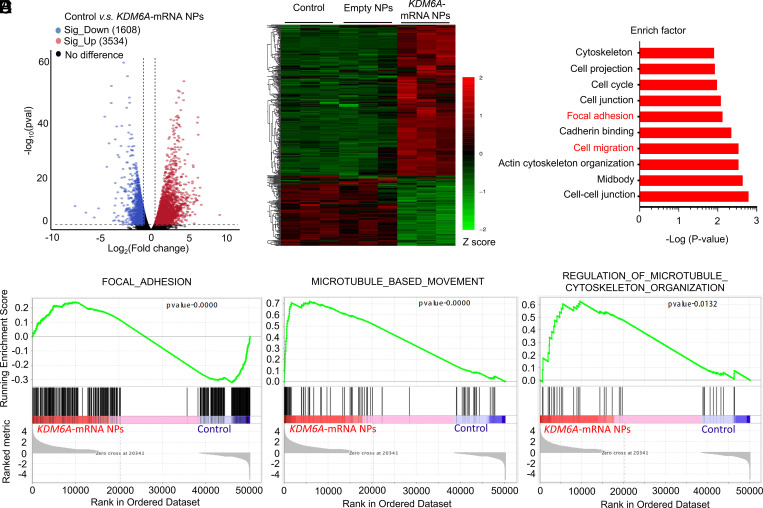
To further investigate the KDM6A’s role in BCa metastasis, we also performed a transcriptomic analysis between Kdm6a-null KU19-19 cells (control group) and KDM6A-mRNA NP–treated Kdm6a-null KU19-19 cells (KDM6A-mRNA NPs group). After the restoration of KDM6A via mRNA NPs in Kdm6a-null KU19-19 cells, 1,608 genes were significantly down-regulated and 3,534 genes were significantly up-regulated (versus control group) (Fig. 3A). Among them, metastasis-associated genes (such as ITGB5, MMP9, CDH11, MMP10, etc.) were significantly down-regulated in Kdm6a-null KU19-19 cells after the treatment of KDM6A-mRNA NPs (Fig. 3B), compared with both the control group and the empty NPs group. In agreement with the previous results, the KEGG pathway analysis demonstrated that the genes linked to focal adhesion and cell migration pathways were among the most significantly expressed in Kdm6a-null KU19-19 cells, compared with the Kdm6a-null KU19-19 cells after the treatment of KDM6A-mRNA NPs (Fig. 3C). In addition, the GSEA results also confirmed that the focal adhesion signaling pathway (****P < 0.0001), microtubule-based movement signaling involved in the cell migration pathway (****P < 0.0001), and the regulation of microtubule cytoskeleton organization signaling involved in the cell migration pathway (*P < 0.05) were significantly down-regulated in Kdm6a-null KU19-19 cells after the treatment of KDM6A-mRNA NPs, compared with the control group (Fig. 3 D–F). Overall, these transcriptomic-level findings support that loss of Kdm6a might promote the metastasis of BCa, while restoration of KDM6A via mRNA NPs could inhibit the invasiveness tendency.
Fig. 3.
Transcriptomic analysis between Kdm6a-null KU19-19 cells (control group) and KDM6A-mRNA NP–treated Kdm6a-null KU19-19 cells (KDM6A-mRNA NPs Group). (A) Volcano plot of gene expression (Control versus KDM6A-mRNA NPs; fold changes ≥ 2; P < 0.05). (B) Hierarchical clustering and heatmap of representative gene expression resulting from control group, Empty NPs Group (i.e., empty NP-treated Kdm6a-null KU19-19 cells), and KDM6A-mRNA NPs Group through transcriptomic sequencing. Row-scaled z-scores of quantile-normalized gene expression (from >5,000 genes; P < 0.0001; Green: down-regulated genes; Red: up-regulated genes). (C) KEGG pathway analysis on the significant gene expression. (D) GSEA of the genes associated with the focal adhesion signaling (P < 0.0001) in control group and KDM6A-mRNA NPs Group. (E) GSEA of the genes associated with the microtubule-based movement signaling (P < 0.0001) in control group and KDM6A-mRNA NPs Group. (F) GSEA of the genes associated with the regulation of microtubule cytoskeleton organization signaling (P = 0.0132) in control group and KDM6A-mRNA NPs Group.
Engineering of Surface Mucoadhesive mRNA NPs for Effective Intravesical Delivery of mRNA.
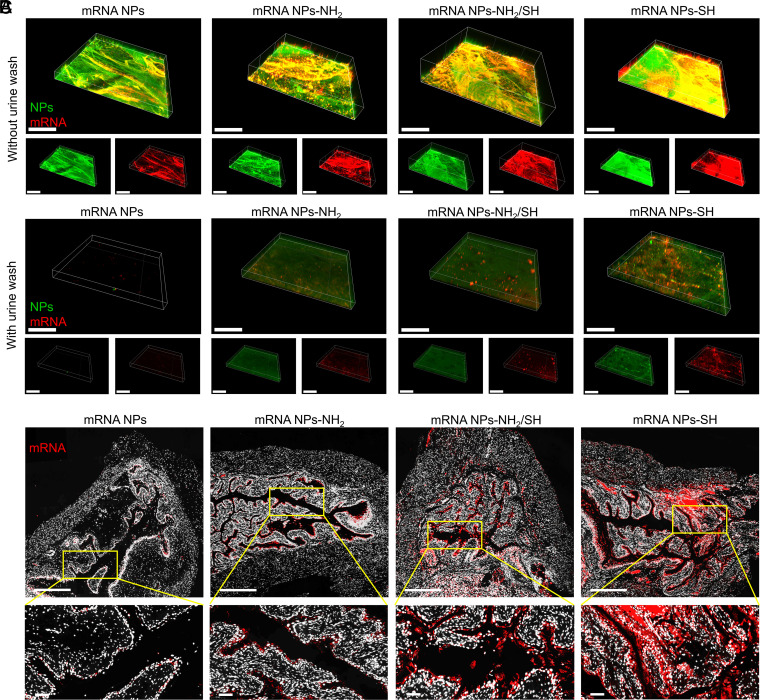
To prolong the dwell time of mRNA NPs in the bladder and improve their uptake and penetration, we further functionalized the surface of our NPs to be mucoadhesive, enabling them to adhere to the bladder for sustained and efficient delivery of mRNA in the bladder. Therefore, instead of coating the NP with nonfunctionalized DSPE-PEG, we used functionalized DSPE-PEG-NH2 (100%), DSPE-PEG-NH2/DSPE-PEG-SH hybrids (50 versus 50%), or DSPE-PEG-SH (100%) to confer mucoadhesive properties. As shown in the TEM images (SI Appendix, Fig. S8A), compared with the nonmucoadhesive mRNA NPs, the obtained mucoadhesive mRNA NPs tend to cluster together with the increased ratios of thiol-terminated (-SH) surface modification. To further assess the mucoadhesivity of mRNA NPs with different surface functionalizations, we performed ex vivo dual-staining experiments on mouse bladder tissues after different mRNA NP treatments. Fluorescein (FITC)-labeled PLGA and Cy5-labeled mRNA were used to form these different NPs, allowing us to simultaneously track both NPs via green signals and mRNA via red signals. PBS containing dual-labeled NPs (i.e., nonmucoadhesive mRNA NPs, mucoadhesive mRNA NPs-NH2, mucoadhesive mRNA NPs-NH2/SH, or mucoadhesive mRNA NPs-SH) was used to individually incubate the mouse bladder at 37 °C for 2 h. After simple PBS washing, CLSM was performed to observe the signals of different mRNA NPs on the bladder wall (Fig. 4A). All three kinds of mucoadhesive mRNA NPs showed higher signals on the bladder walls than nonmucoadhesive mRNA NPs. Meanwhile, the bladder wall receiving the mRNA NPs-SH treatment exhibited the highest signals among the three kinds of mucoadhesive mRNA NPs.
Fig. 4.
Engineering of surface mucoadhesive mRNA NPs for effective intravesical delivery of mRNA. (A) CLSM volume view images of mouse bladder walls after incubation (2 h) with nonmucoadhesive mRNA NPs, mucoadhesive mRNA NPs-NH2, mucoadhesive mRNA NPs-NH2/SH, or mucoadhesive mRNA NPs-SH. mRNA was labeled with Cy5 (red fluorescence), and NPs were labeled with Fluorescein (FITC) (green fluorescence) (Scale bar, 400 μm). (B) CLSM volume view images of mouse bladder walls after 2 h of incubation with different mRNA NPs, followed by another 3 h of incubation in urine (i.e., urine wash). mRNA was labeled with Cy5 (red fluorescence), and NPs were labeled with FITC (green fluorescence) (Scale bar, 400 μm). (C) Sections of the bladder tissues from mice treating with nonmucoadhesive mRNA NPs, mucoadhesive mRNA NPs-NH2, mucoadhesive mRNA NPs-NH2/SH, or mucoadhesive mRNA NPs-SH via intravesical administration (Scale bars in raw images, 500 µm; Scale bars in enlarged images, 50 µm). mRNA was labeled with Cy5 (red fluorescence).
To better mimic the activity in the clinical bladder treatment, after the same treatment procedure with different NPs, we individually incubated the bladders with urine for an additional 3 h at 37 °C followed by simple PBS washing (Fig. 4B). Nonmucoadhesive mRNA NPs could easily be washed away by urine, as shown almost all signals on the bladder wall were disappeared. However, all three kinds of mucoadhesive mRNA NPs created detectable signals on the bladder walls. Moreover, our data indicated that mRNA NPs-SH showed the highest adhesive capacity and uptake among the three kinds of mucoadhesive mRNA NPs. Similar results were further confirmed through the sections from bladder tissues of mice after intravesical administration of different Cy5-mRNA NPs (Fig. 4C) through an in vivo bladder tissue adhesion and mucoadhesive assay. These results could be explained by the fact that thiolated-NPs were able to form -S-S- bonds with cysteine-rich domains of the mucus glycoproteins (24, 33), and such covalent bonds had much stronger interactions than the noncovalent interactions (e.g., van der Waals forces, hydrogen bonds, and ionic interactions with the anionic substructures of the mucus layer) (33, 34). Because the mRNA NPs-SH showed the best mucoadhesive properties for effective intravesical delivery of mRNA, we used them in our subsequent studies.
Characterization of the Mucoadhesive mRNA NPs with Thiol-Surface Functionalization.
The NP solution via bladder perfusion would usually be excreted by urine within several hours. Therefore, they should be relatively stable in the solution at least for several hours (i.e., together with urine at 37 °C) and then attached to the bladder tissues to realize the designed functions. We then tested the stability of our mucoadhesive mRNA NPs-SH in the urine at 37 °C. The results showed that the size of mRNA NPs-SH did not have obvious changes within 24 h (SI Appendix, Fig. S8B), and the stable time is enough for intravesical delivery. In terms of storage in the real clinic application, the prepared mRNA NP solution is usually frozen and thawed before use. Therefore, they should be stable in the solution after being thawed before use. We thus stored the prepared mRNA NPs-SH in both PBS solution and PBS solution containing 10% fetal bovine serum (FBS) and immediately froze these fresh mRNA NPs-SH in a −20 °C condition. After different hours of storage/frozen, the mRNA NPs-SH were thawed, and their size changes were subsequently monitored. As can be observed from our results, the thawed mucoadhesive mRNA NPs-SH were stable enough in both PBS solution (SI Appendix, Fig. S8C) and PBS solution containing 10% FBS (SI Appendix, Fig. S8D) at the tested time points.
We also checked if the thiolated-surface modification would affect the endosome escape ability of mRNA NPs-SH. Similar to the results from nonmucoadhesive mRNA NPs (Fig. 1C), the mRNA (red) carried by the mucoadhesive NPs-SH also effectively escaped the endosomes (green) with an increase in incubation time (SI Appendix, Fig. S9A), indicating the mucoadhesive surface modification does not affect the endosome escape ability of these NPs. We further studied the cellular uptake and intracellular transport pathways of mRNA NPs-SH via commonly used inhibitors (10, 26) and their effects on the transfection efficiency of mRNA NPs-SH in Kdm6a-null KU19-19 cells. The results from KU19-19 cells pretreated with different inhibitors (SI Appendix, Fig. S9B) showed that the macropinocytosis inhibitor pretreatment significantly decreased the transfection, but the pretreatment with the caveolae-mediated inhibitor or the clathrin-mediated inhibitor did not affect the transfection, indicating the macropinocytosis mainly mediated the cellular internalization of mRNA NPs-SH. Moreover, the pretreatment with the proton-pump inhibitor also markedly decreased the transfection, indicating that the NPs-SH could release the mRNA cytosolically via inducing a proton-sponge effect after entering the cells via macropinocytosis.
Mucoadhesive mRNA NPs Restore KDM6A in Mice Bearing Orthotopic Kdm6a-null BCa Tumors and Inhibit Metastasis to Adjacent Tissues and the Lymphatic System.
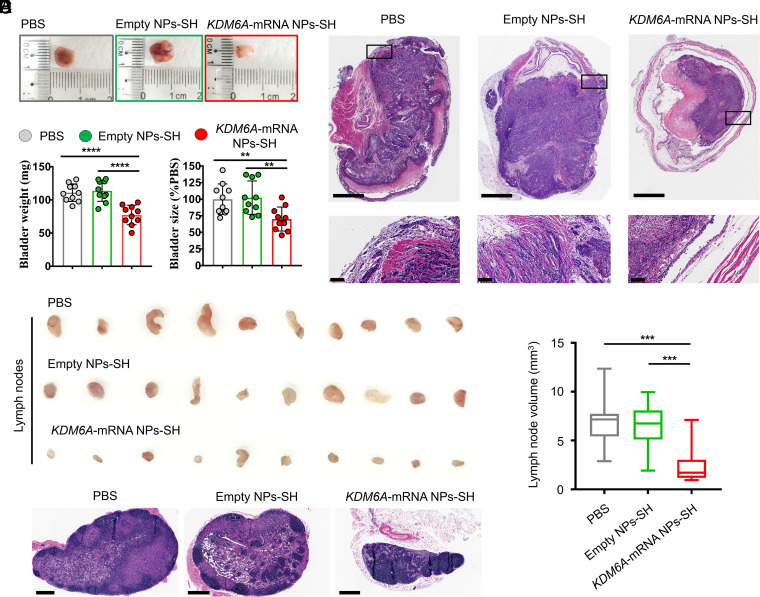
To verify the functional consequence of KDM6A restoration via mucoadhesive mRNA NPs in BCa, we developed an orthotopic bladder tumor model in athymic nude mice with Kdm6a-null KU19-19 cells. The orthotopic bladder tumor models were established through transurethral instillation of Kdm6a-null KU19-19 BCa cells into the bladders of immunodeficient nude mice. After 1 wk of implantation, the mice bearing orthotopic bladder tumors (with similar sizes) received the treatments of PBS, empty NPs-SH, or KDM6A-mRNA NPs-SH, respectively, via intravesical administration every 3 d, followed by ultrasonic monitoring on days 0, 4, 8, 12, and 16. The high-frequency ultrasound images showed that KDM6A-mRNA NPs-SH treatment effectively inhibited the growth of orthotopic primary bladder tumors compared with those receiving PBS or empty NPs-SH treatment (SI Appendix, Fig. S10). The immunohistochemistry (IHC) staining assays showed that KDM6A proteins were widely expressed in orthotopic KU19-19 tumor sections after treatment with KDM6A-mRNA NPs-SH, while no expression of KDM6A was found in the tumor sections from animals receiving treatment with either PBS or empty NPs-SH (SI Appendix, Fig. S11). Because the introduction of KDM6A tumor suppressor significantly inhibited the proliferation of KU19-19 cells in vitro, it is not surprising that the orthotopic tumor growth in the KDM6A-mRNA NPs-SH group was inhibited after the high expression of KDM6A in KU19-19 tumors. Moreover, intravesical delivery of KDM6A-mRNA via mucoadhesive NPs also significantly reduced the number of mice with metastasis (SI Appendix, Fig. S12A) as well as the number of metastatic lymph nodes (SI Appendix, Fig. S12B), indicating the therapeutic potential of KDM6A introduced by mucoadhesive mRNA NPs in inhibiting the metastasis of BCa in vivo. At 2 d after the last treatment, the bladders bearing orthotopic primary BCa tumors were entirely collected from the mice after euthanasia for further evaluation. Compared with the KDM6A-mRNA NPs-SH group, the orthotopic primary BCa tumors in the PBS group and the empty NPs-SH group expanded the bladders much larger, and the normal structures of bladders were severely damaged by the orthotopic primary BCa tumors in these two groups (Fig. 5 A and B). The hematoxylin and eosin (H&E) staining assay showed more direct evidence that KDM6A-mRNA NPs-SH treatment via intravesical bladder instillation was able to effectively inhibit the growth of orthotopic primary BCa tumors and prevented the invasiveness of primary BCa cells into the mucosal layer and the muscle layer (Fig. 5C), avoiding the BCa metastasis and protecting the bladder structures. It was also found that KDM6A-mRNA NPs-SH treatment could effectively inhibit the blood vascular (BV) invasion and the lymphatic vessel (LV) invasion (SI Appendix, Fig. S13). In addition, the in vivo expression of KDM6A via KDM6A-mRNA NPs-SH could also have significantly inhibited the BCa metastasis to lymph nodes (Fig. 5 D–F), indicating the therapeutic potential of KDM6A in treating the metastasis of BCa.
Fig. 5.
Introduction of KDM6A via mucoadhesive mRNA NPs in mice bearing orthotopic BCa tumors could inhibit metastasis potential of BCa. (A) Representative images of the entire bladders bearing orthotopic primary BCa tumors in different groups. (B) The weight (mg) and the size changes (% compared with the PBS group) of the harvested bladders from different groups. Data shown as means ± SEM (n = 10) with a two-tailed Student’s t test (**P < 0.01, ****P < 0.0001). (C) Representative H&E staining sections from the entire bladders bearing orthotopic primary BCa tumors in different groups (Scale bars in raw images, 1.25 mm; Scale bars in enlarged images, 100 µm). (D) Representative images of the maximum-volume lymph nodes collected from each group (n = 10). (E) Histogram analysis of the volume calculated from all the lymph nodes in different groups. Data shown as means ± SEM (n = 10) with a two-tailed Student’s t test (***P < 0.001). (F) Representative H&E staining images of the lymph nodes from different groups (Scale bars, 400 μm).
To further verify that the introduction of KDM6A via mRNA NPs in BCa tumors can inhibit metastasis in vivo, we established another luciferase-expressing orthotopic bladder tumor model via transurethral instillation of Kdm6a-null luciferase–expressing KU19-19 (KU19-19-Luc) cells into the bladders of athymic nude mice. Tumor growth and adjacent tissue metastasis were monitored via evaluating the average radiance within tumor sites by bioluminescence imaging. After 1 wk, mice with similar bioluminescence signals were randomly divided into three groups and treated with PBS, empty NPs-SH, or KDM6A-mRNA NPs-SH via intravesical administration every 3 d. Bioluminescence imaging studies were performed on days 0, 4, 8, 12, and 16. Compared with the control group (PBS) and the control NP group (empty NPs-SH), intravesical treatment with KDM6A-mRNA NPs-SH effectively reduced the orthotopic tumor burden and adjacent tissue metastasis (SI Appendix, Figs. S14 and S15). The BCa metastasis majorly occurred in the abdominal regions of the mice at our tested experiment levels as reflected by the bioluminescence signals (SI Appendix, Fig. S14A). Taken all together, we demonstrated that introduction of KDM6A via mucoadhesive mRNA NPs in mice bearing orthotopic Kdm6a-null BCa tumors can inhibit adjacent tissue metastasis and lymphatic metastasis, further indicating the key therapeutic role of KDM6A in BCa metastasis.
Mucoadhesive mRNA NP–Mediated Strategy on Mechanistic Understanding of KDM6A’s Role in Inhibiting the BCa Metastasis In Vivo.
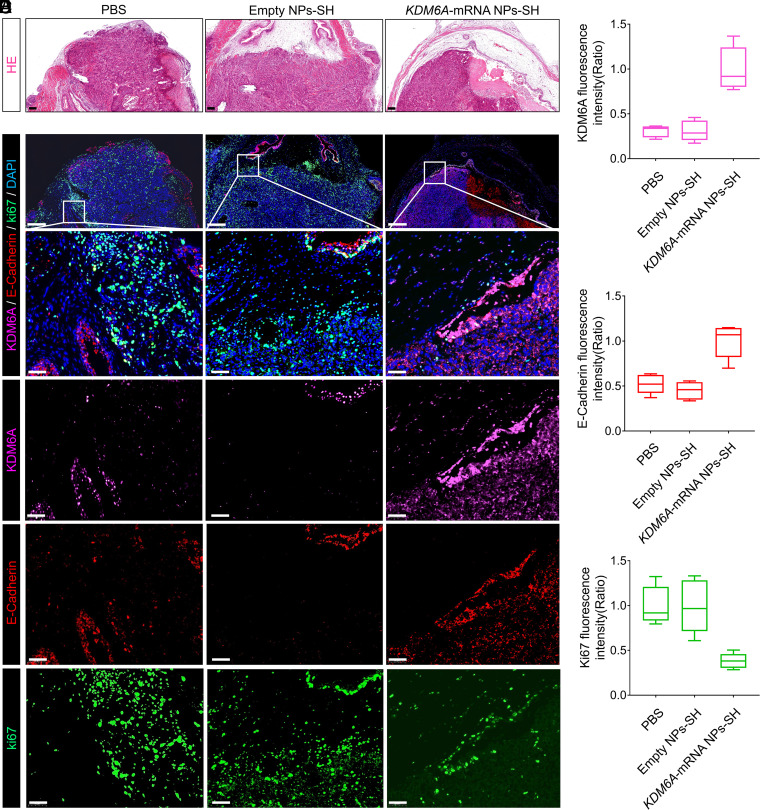
To clarify the potential mechanism of KDM6A’s role in inhibiting the BCa metastasis in vivo, the orthotopic BCa tumors from the mice after different treatments were collected (Fig. 6A), and IF staining analysis was further performed with these tumors. Consistent with the previous in vitro data (Fig. 2E), we observed that the KDM6A (pink color) introduced in tumor sections by KDM6A-mRNA NPs-SH effectively up-regulated the expression of E-Cadherin (epithelial marker; red color) (Fig. 6 B–D). To further confirm the role of KDM6A in inhibiting BCa metastasis, we also performed Western blot (WB) analysis to detect the expression of several typical metastatic phenotype markers in the orthotopic BCa tumors after different treatments. Along with the re-expression of KDM6A in Kdm6a-null orthotopic BCa tumors via KDM6A-mRNA NPs-SH, the increased expression of E-Cadherin was also solidly confirmed via WB analysis (SI Appendix, Fig. S16), indicating that the metastasis potential of BCa might be inhibited by KDM6A-mediated E-Cadherin up-regulation. In addition, restoration of KDM6A in vivo inhibited the mesenchymal markers Vimentin, N-Cadherin, SNAIL, and SLUG expressions at varying degrees. As N-Cadherin usually takes an active role in promoting tumor migration via EMT (35), the introduction of KDM6A via KDM6A-mRNA NPs-SH may also successfully stop the tumor migration process via down-regulating N-Cadherin expression. Taken together, these data suggest that KDM6A restoration via mucoadhesive mRNA NP–enabled intravesical delivery of KDM6A-mRNA may suppress BCa metastasis via signaling pathways in EMT (summarized in SI Appendix, Fig. S17).
Fig. 6.
Local restoration of KDM6A in Kdm6a-null BCa tumors via a mucoadhesive mRNA NP–mediated strategy for the mechanistic understanding of KDM6A’s role in inhibiting BCa metastasis in vivo. (A) H&E staining sections of the bladders bearing orthotopic primary BCa tumors that used for the following IF analysis (Scale bars, 200 µm). (B) IF analysis of the KDM6A (pink), E-Cadherin (red), and Ki67 (green) expression in orthotopic Kdm6a-null KU19-19 tumors after different treatments (Scale bars in raw images, 200 µm; Scale bars in enlarged images, 50 µm). The nucleus was stained by DAPI (blue). Quantitative analysis of (C) KDM6A (pink), (D) E-Cadherin (red), and (E) Ki67 (green) expression in the bladders bearing orthotopic primary BCa tumors from different groups.
As KDM6A is a well-recognized tumor suppressor, it is not surprising to find that its introduction to Kdm6a-null orthotopic BCa tumors via KDM6A-mRNA NPs-SH can down-regulate the proliferation markers Ki-67 (green color) (Fig. 6 B and E). To verify the in vivo safety of this strategy, healthy mice receiving treatment with mucoadhesive KDM6A-mRNA NPs-SH (via intravesical perfusion; local delivery) were euthanized and major organs (heart, liver, spleen, lung, and kidneys) were harvested for H&E staining. We did not find any obvious histological differences between the organ sections from mice receiving PBS treatments and those receiving KDM6A-mRNA NPs-SH treatments (SI Appendix, Fig. S18), suggesting that the local introduction of KDM6A via our mucoadhesive mRNA NPs can be a safe strategy for in vivo use.
Clinical Significance of KDM6A on Metastasis of BCa and its Potential Combination with Elemene.
As can be summarized from the Cancer Genome Atlas database, Kdm6a shows the highest rate of mutation (29.37%) in BCa among the different cancer types (SI Appendix, Fig. S19A), indicating its potentially critical role in BCa (36, 37). To examine the clinical relevance of KDM6A as a potential target in treating human BCa invasiveness and metastasis, we collected a cohort of 110 BCa patients and initially assessed the expression of KDM6A proteins by IHC. As could be directly observed from the IHC images (SI Appendix, Fig. S20A), tumor sections (red arrow) from the bladder tissues in the non-MI (NMI) BCa patients (i.e., stages Ta to T1; nonmetastatic BCa) showed extensive expression of KDM6A, while those from MI BCa patients exhibited low to undetectable levels of KDM6A. Clear boundaries of mucosa (area I), submucosa (area II), and muscle (area III) could be observed in the bladder tissues from NMI BCa patients. However, due to the invasiveness and metastasis, no such boundaries could be distinguished clearly in the bladder tissues from MI BCa patients. Moreover, both the numbers (n) and the percentages (%) of BV invasive and LV invasive are significantly higher in KDM6A low-expression BCa patients than those of KDM6A high-expression BCa patients (SI Appendix, Fig. S20B), further suggesting that clinical significance of KDM6A in the invasiveness and metastasis of BCa in patients.
The correlations between clinicopathological signatures of the patients with BCa and KDM6A expression were further analyzed (SI Appendix, Figs. S19B and S20C). These results clearly showed that the invasive and metastatic BCa have much lower KDM6A expression. For example, 97.06% of patients with MI BCa (stages T2 to T4) exhibited low expression of KDM6A (versus 81.58% of those in stages T1 to T2), as did a full 100% of the BCa patients in TNM stage II to IV (versus 81.93% of those in TNM stage I). Similarly, 90.32% of the BCa patients with lymphatic metastasis exhibited low KDM6A expression (versus 84.81% of those without lymphatic metastasis). Moreover, KDM6A expression was remarkably correlated with 3-y overall survival (OS) for patients with BCa (SI Appendix, Fig. S20D). Compared with BCa patients exhibiting high KDM6A expression, low expression of KDM6A was associated with a significantly lower percentage of 3-y OS in all patients. Interestingly, within our sample, all female BCa patients had low expression of KDM6A and a poor OS rate (<40%). It is worth to mentioning that we also observed that intravesical treatments with mucoadhesive KDM6A-mRNA NPs-SH effectively improved the OS of BCa mice and these mucoadhesive mRNA NPs might be applied in combination therapy with other therapeutics (e.g., elemene, a clinical used chemotherapeutic drug that can be used for intravesical infusion therapy of BCa) for enhanced therapeutic effects (SI Appendix, Fig. S21). Together, these results indicate that low expression of KDM6A is likely associated with an increased risk of invasiveness, metastasis, and mortality (i.e., poor prognosis) in BCa patients. Therefore, restoration of KDM6A via our mucoadhesive mRNA NP strategy might possess significant clinical impacts on treating BCa patients and preventing invasiveness and metastasis.
Discussion
As a leading cause of tumor-associated deaths, BCa is more prevalent in developed than in developing countries (38–40) and is 3 to 4 times more common in males than in females. NMI BCa (i.e., a more common type associated with a favorable prognosis) and MI BCa (i.e., a less prevalent type, typically associated with a poor prognosis) are two broad subgroups of this disease (41, 42). Notably, BCa is not only among the most expensive malignancies to treat (40), but little progress has been made in metastatic BCa treatment over the past two decades. Recent studies suggest that Kdm6a inactivation may be a powerful driver in bladder oncogenesis, and a majority of Kdm6a mutations are predicted to result in a total a loss-of-function or loss-of-expression in the JmjC domain of KDM6A (over 70% of the mutations lead to a total loss-of-function in this tumor type) (37). Therefore, the Kdm6a gene and the KDM6A proteins that it encodes are of great significance in BCa. As a well-recognized and key tumor suppressor in BCa (1–3), KDM6A has been revealed to play a vital role in both mechanistic and translational studies of BCa, which may offer potential solutions to the current problems in the treatment of BCa and provide deeper insights into the understanding of its metastasis. However, until now, KDM6A’s function in the metastasis of BCa remained largely unexplored.
The most straightforward strategy to understand the role of KDM6A is to verify the functional consequences after its 1) inactivation/deletion and 2) reactivation in the biological system. While the former can be achieved simply using gene knockout mice/cell lines, the latter remains difficult, especially for in vivo introduction of KDM6A in the local site(s). At present, DNA transfection is the only reported strategy for introducing the exogenous KDM6A (1, 5–7), and the most commonly used tools are either nonviral vector lipofectamine reagents or virus vectors. However, both these useful tools have formidable drawbacks: 1) because of poor stability, nonviral lipofectamine reagents can be used only for cellular experiments, preventing their wider application in animals and translational studies and 2) concerns such as low packaging capacity, high production cost, high immunogenicity, and potential transmission into the environment may still exist for virus vector–based technologies. Moreover, the risks of genomic integration and mutagenesis may still present a major hurdle in the transition of DNA transfection strategy. Therefore, alternative approaches that can effectively introduce KDM6A while avoiding these problems will likely hold great promise, from mechanistic understandings to translational cancer research.
Recently, emerging mRNA technologies have attracted considerable attention in various areas including vaccines against infectious diseases including COVID-19 and beyond (12, 43, 44), drugs for regenerative angiogenesis in diabetes patients (45), tumor-suppressor restoration for cancer therapy (9, 10), targeted treatment for inherited metabolic liver disorder arginase deficiency (46), and others. mRNA is essentially a single-stranded nucleic acid molecule. Unlike DNA transfection, the retention time of mRNA is temporary, and it will be degraded once the specific functions are realized, which leads to “on-demand” up-regulation of the specific proteins to manipulate the targeted pathways only when needed. Because nuclear entry is not required for mRNA to fulfill its transfection activity, integration into the host genome and potential detrimental genotoxicity are no longer concerns. mRNA-based strategies also work rapidly and cost effectively. Once the specific target is identified, the design and production of mRNA drugs are procedural and straightforward. In principle, one mRNA drug can express multiple proteins, which offers convenience for multistage vaccines or multiprotein combination treatments. At this writing, there are 60 different pharmaceutical mRNAs in different stages of clinical trials. The first mRNA therapeutic has also been urgently approved in humans in order to tackle the pandemic caused by COVID-19 (for example, mRNA Prophylactic Vaccine mRNA-1273 by Moderna, Inc.) (11, 47). Due to the distinctive properties of mRNA-based strategies, chemically modified mRNA can be a biocompatible, straightforward, and highly promising modality for KDM6A introduction in target cells or sites. Nevertheless, an effective method of delivery of mRNA to in situ bladder tumors for target protein expression remains a major challenge.
Emerging nanotechnology has provided opportunities to improve the cytosolic delivery of different RNA therapeutics into cancer cells, and some NPs have been used for systemic mRNA delivery (9, 10, 48). However, for patients with BCa, intravesical perfusion has the potential to selectively deliver high concentrations of mRNA to in situ bladder tumors, minimize systemic exposure (23, 24), and offer greater benefit than systemic mRNA delivery. To the best of our knowledge, the use of NPs for intravesical delivery of mRNA to specific in situ bladder tumors had remained untested. For effective protein introduction at the local site(s), NPs need to overcome different barriers associated with intravesical mRNA delivery, such as nuclease degradation, rapid clearance through the voiding of urine, lack of dwell time for sustained delivery, and insufficient mRNA escape from endosomes to the cytoplasm (9, 10, 22, 49). Herein, we designed a lipid–polymer hybrid NP platform in which 1) positively charged G0-C14 was used to absorb the mRNA and promote endosomal escape (9, 50, 51) and 2) the FDA-approved PLGA polymer was applied to load the mRNA/C0-C14 complexes and avoid serum nuclease degradation of mRNA (10, 52). DSPE-PEG-SH, DSPE-PEG-NH2, or their hybrid was used to construct mucoadhesive mRNA NPs with prolonged dwell time in the bladder and improved uptake into BCa tissue for the intravesical delivery of mRNA (23, 24). It is also worth mentioning that such a straightforward and simple surface modification strategy, changing FDA-approved DSPE-PEG to DSPE-PEG-NH2 (or -SH) to generate the mucoadhesive mRNA NPs, will provide maximum benefit in terms of reproducibility and clinical translation. Our results demonstrate that KDM6A-mRNA can be efficiently loaded into mucoadhesive NPs, which can adhere to the bladder for delivery of mRNA into Kdm6a-null in situ bladder tumors and express KDM6A. In the context of our previous studies using nonmucoadhesive NPs for the systemic delivery of mRNA (9, 10), this study extends the application of mRNA NPs to specific delivery for in situ bladders and expands our mechanistic understanding of a critical target in the metastasis of BCa. Moreover, our results also indicate the potential application of these mucoadhesive mRNA NPs in combination therapy, which might expand their potential application in clinic in the future. Nevertheless, as the current design is limited to the surface engineering of mRNA NPs for enhanced mucoadhesive properties, surface modification with additional targeting ligands to achieve targeted or more specific delivery of mRNA to tumor cells could be needed to further advance the clinical application of this mRNA delivery platform in treating BCa. For example, neurotensin (NT) peptide, which can efficiently target BCa cells via its specific binding to NT receptor type 1 overexpressed on the BCa membrane (53, 54), could be conjugated on the mRNA NPs-SH surface at an optimized ratio to simultaneously balance the mucoadhesive properties and targeted delivery. In addition, to further enhance the transfection efficacy and reduce the dosage in future clinical applications, more robust formulation strategies may be optimized to improve the stability of these mucoadhesive mRNA NPs in urine at 37 °C for a longer time.
In summary, we not only provide evidence of KDM6A’s role in inhibiting the metastasis of BCa via an mRNA strategy but also develop a specific mucoadhesive NP platform for intravesical mRNA delivery to in situ bladder tissues. Considering the potential role of KDM6A in promoting the antitumor immune response in treating cancers including BCa (55, 56), it might be worthwhile to expect the possible application of these KDM6A-mRNA NPs in cancer immunotherapy. Moreover, given 1) the critical role of KDM6A in controlling chromatin and cell fate, development, various cancers, and other diseases (57–61) and 2) the frequent inactivation of KDM6A in these diseases (37, 62–67), this study might also provide an effective and straightforward strategy to express KDM6A via NPs for both mechanistic explorations and therapeutic applications in these diseases. We expect that this mucoadhesive mRNA nanotechnology could be used to specifically up-regulate other target proteins within mucosal organs in situ for various applications.
Materials and Methods
Detailed materials and methods are provided in SI Appendix, Materials and Methods, including the cell culture and major reagents; in vitro transcription of chemically modified KDM6A-mRNA; xCELLigence RTCA invasion and migration assays; RNA library construction and sequencing; mass spectrometry sample processing and analysis; transwell and in vitro wound healing assays; WB assay; qRT-PCR; colony formation assays; synthesis of nonmucoadhesive mRNA NPs and mucoadhesive mRNA NPs; physicochemical characterization of mRNA NPs; endosomal escape of the mRNA delivered by NPs in Kdm6a-null BCa cells; expressing exogenous and functional proteins via mRNA NPs in Kdm6a-null BCa cells; ex vivo bladder tissue adhesion and mucoadhesive assay; generation of KU19-19 cells with luciferase expression (KU19-19-Luc) for orthotopic BCa model; intravesical delivery of mRNA via mucoadhesive NPs in the in vivo orthotopic BCa mouse model; transabdominal high-frequency micro-ultrasound imaging; IHC and IF staining; OS study and combination treatment strategy with elemene; and statistical analysis. Animal protocol was approved by the Institutional Animal Care and Use Committee at Harvard Medical School/Brigham and Women’s Hospital and Hangzhou Normal University.
Supplementary Material
Acknowledgments
This work is supported by grants from the National Natural Science Foundation of China (Grant Nos. 81730108 and 81973635 to T.X.), Zhejiang Provincial Natural Science Foundation of China for Distinguished Young Scholars (grant No. LR18H160001, to X.S.), US METAvivor Early Career Investigator Award (Grant No. 2018A020560 to W.T.), and Harvard Medical School/Brigham and Women’s Hospital Department of Anesthesiology-Basic Scientist Grant (Grant No. 2420 BPA075 to W.T.). G.W. is a Fellow of The Leukemia & Lymphoma Society. W.T. is a recipient of the Khoury Innovation Award (Award No. 2020A003219) and American Heart Association Collaborative Sciences Award (Award No. 2018A004190). W.T. also received a start-up package (for 3 y) from the Department of Anesthesiology, Perioperative and Pain Medicine to establish his independent research laboratory at Harvard Medical School and Brigham and Women’s Hospital. We thank our department for this generous support. We thank Qi Zhang from Zhejiang Provincial People’s Hospital for providing the human BCa tissue microarray. The IRB/EC approval number is No. 2021QT414.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. L.Z. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2112696119/-/DCSupplemental.
Data Availability
All study data are available within this manuscript and the associated SI Appendix.
References
- 1.Roy D. M., Walsh L. A., Chan T. A., Driver mutations of cancer epigenomes. Protein Cell 5, 265–296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunford A., et al. , Tumor-suppressor genes that escape from X-inactivation contribute to cancer sex bias. Nat. Genet. 49, 10–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ler L. D., et al. , Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci. Transl. Med. 9, eaai8312 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Kaneko S., Li X., X chromosome protects against bladder cancer in females via a KDM6A-dependent epigenetic mechanism. Sci. Adv. 4, eaar5598 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Haaften G., et al. , Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41, 521–523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lang A., et al. , Contingencies of UTX/KDM6A action in urothelial carcinoma. Cancers (Basel) 11, E481 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho A. S., et al. , The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 45, 791–798 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer G., et al. , Multicenter prospective phase II trial of neoadjuvant dose-dense gemcitabine plus cisplatin in patients with muscle-invasive bladder cancer. J. Clin. Oncol. 36, 1949–1956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong N., et al. , Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci. Transl. Med. 11, eaaw1565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam M. A., et al. , Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2, 850–864 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Z., et al. , Insights from nanotechnology in COVID-19 treatment. Nano Today 36, 101019 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang Z., et al. , A materials-science perspective on tackling COVID-19. Nat. Rev. Mater. 5, 1–14 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin H., et al. , Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541–555 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Quabius E. S., Krupp G., Synthetic mRNAs for manipulating cellular phenotypes: An overview. N. Biotechnol. 32, 229–235 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Dammes N., Peer D., Paving the road for RNA therapeutics. Trends Pharmacol. Sci. 41, 755–775 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee J., Boczkowski D., Nair S., Programming human dendritic cells with mRNA. Methods Mol. Biol. 969, 111–125 (2013). [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto A., Kormann M., Rosenecker J., Rudolph C., Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 71, 484–489 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Leonhardt C., et al. , Single-cell mRNA transfection studies: Delivery, kinetics and statistics by numbers. Nanomedicine (Lond.) 10, 679–688 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Ligon T. S., Leonhardt C., Rädler J. O., Multi-level kinetic model of mRNA delivery via transfection of lipoplexes. PLoS One 9, e107148 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Veiga N., et al. , Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 9, 4493 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granot-Matok Y., Kon E., Dammes N., Mechtinger G., Peer D., Therapeutic mRNA delivery to leukocytes. J. Control. Release 305, 165–175 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Tyagi P., Tyagi S., Kaufman J., Huang L., de Miguel F., Local drug delivery to bladder using technology innovations. Urol. Clin. North Am. 33, 519–530, x (2006). [DOI] [PubMed] [Google Scholar]
- 23.Mugabe C., et al. , In vivo evaluation of mucoadhesive nanoparticulate docetaxel for intravesical treatment of non-muscle-invasive bladder cancer. Clin. Cancer Res. 17, 2788–2798 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Zhang Q., et al. , Functionalized mesoporous silica nanoparticles with mucoadhesive and sustained drug release properties for potential bladder cancer therapy. Langmuir 30, 6151–6161 (2014). [DOI] [PubMed] [Google Scholar]
- 25.GuhaSarkar S., Banerjee R., Intravesical drug delivery: Challenges, current status, opportunities and novel strategies. J. Control. Release 148, 147–159 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Zhu X., et al. , Long-circulating siRNA nanoparticles for validating Prohibitin1-targeted non-small cell lung cancer treatment. Proc. Natl. Acad. Sci. U.S.A. 112, 7779–7784 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X., et al. , Surface De-PEGylation controls nanoparticle-mediated siRNA delivery in vitro and in vivo. Theranostics 7, 1990–2002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suk J. S., Xu Q., Kim N., Hanes J., Ensign L. M., PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 99 (Pt A), 28–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurmeister S., et al. , MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol. Cell. Biol. 32, 633–651 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazzone M., et al. , Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell 136, 839–851 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeanes A., Gottardi C. J., Yap A. S., Cadherins and cancer: How does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onder T. T., et al. , Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 68, 3645–3654 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Bravo-Osuna I., Vauthier C., Farabollini A., Palmieri G. F., Ponchel G., Mucoadhesion mechanism of chitosan and thiolated chitosan-poly(isobutyl cyanoacrylate) core-shell nanoparticles. Biomaterials 28, 2233–2243 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Yin L., et al. , Drug permeability and mucoadhesion properties of thiolated trimethyl chitosan nanoparticles in oral insulin delivery. Biomaterials 30, 5691–5700 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Cao Z. Q., Wang Z., Leng P., Aberrant N-cadherin expression in cancer. Biomed. Pharmacother. 118, 109320 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas Research Network, Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Shilatifard A., UTX mutations in human cancer. Cancer Cell 35, 168–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegel R. L., Miller K. D., Jemal A., Cancer statistics, 2018. CA Cancer J. Clin. 68, 7–30 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Bray F., et al. , Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi T., Owczarek T. B., McKiernan J. M., Abate-Shen C., Modelling bladder cancer in mice: Opportunities and challenges. Nat. Rev. Cancer 15, 42–54 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaufman D. S., Shipley W. U., Feldman A. S., Bladder cancer. Lancet 374, 239–249 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Dinney C. P., et al. , Focus on bladder cancer. Cancer Cell 6, 111–116 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Zhang N. N., et al. , A thermostable mRNA vaccine against COVID-19. Cell 182, 1271–1283.e16 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richner J. M., et al. , Modified mRNA vaccines protect against Zika virus infection. Cell 169, 176 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Gan L. M., et al. , Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 10, 871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Truong B., et al. , Lipid nanoparticle-targeted mRNA therapy as a treatment for the inherited metabolic liver disorder arginase deficiency. Proc. Natl. Acad. Sci. U.S.A. 116, 21150–21159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang Z., et al. , A materials-science perspective on tackling COVID-19. Nat. Rev. Mat. 5, 847–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenblum D., Joshi N., Tao W., Karp J. M., Peer D., Progress and challenges towards targeted delivery of cancer therapeutics. Nat. Commun. 9, 1410 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang M. R., et al. , Intravesical delivery of small activating RNA formulated into lipid nanoparticles inhibits orthotopic bladder tumor growth. Cancer Res. 72, 5069–5079 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Xu X., et al. , Tumor microenvironment-responsive multistaged nanoplatform for systemic RNAi and cancer therapy. Nano Lett. 17, 4427–4435 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X., et al. , Multifunctional envelope-type siRNA delivery nanoparticle platform for prostate cancer therapy. ACS Nano 11, 2618–2627 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tao W., et al. , siRNA nanoparticles targeting CaMKIIγ in lesional macrophages improve atherosclerotic plaque stability in mice. Sci. Transl. Med. 12, eaay1063 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minervini A., Siena G., Falciani C., Carini M., Bracci L., Branched peptides as novel tumor-targeting agents for bladder cancer. Expert Rev. Anticancer Ther. 12, 699–701 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Bao Q., Hu P., Ren W., Guo Y., Shi J., Tumor cell dissociation removes malignant bladder tumors. Chem 6, 2283–2299 (2020). [Google Scholar]
- 55.Chen X., et al. , Significance of KDM6A mutation in bladder cancer immune escape. BMC Cancer 21, 635 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yi J., Shi X., Xuan Z., Wu J., Histone demethylase UTX/KDM6A enhances tumor immune cell recruitment, promotes differentiation and suppresses medulloblastoma. Cancer Lett. 499, 188–200 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chakraborty A. A., et al. , Histone demethylase KDM6A directly senses oxygen to control chromatin and cell fate. Science 363, 1217–1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Laarhoven P. M., et al. , Kabuki syndrome genes KMT2D and KDM6A: Functional analyses demonstrate critical roles in craniofacial, heart and brain development. Hum. Mol. Genet. 24, 4443–4453 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee S., Lee J. W., Lee S. K., UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev. Cell 22, 25–37 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lederer D., et al. , Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. Am. J. Hum. Genet. 90, 119–124 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dupere-Richer D., et al. , Loss of KDM6A/UTX accelerate the development of multiple myeloma. Blood 132 (suppl. 1), 1004 (2018). [Google Scholar]
- 62.Gozdecka M., et al. , UTX-mediated enhancer and chromatin remodeling suppresses myeloid leukemogenesis through noncatalytic inverse regulation of ETS and GATA programs. Nat. Genet. 50, 883–894 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andricovich J., et al. , Loss of KDM6A activates super-enhancers to induce gender-specific squamous-like pancreatic cancer and confers sensitivity to BET inhibitors. Cancer Cell 33, 512–526.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hurst C. D., et al. , Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 32, 701–715.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gao Y. B., et al. , Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 46, 1097–1102 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Varela I., et al. , Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li X., et al. , UTX is an escape from X-inactivation tumor-suppressor in B cell lymphoma. Nat. Commun. 9, 2720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are available within this manuscript and the associated SI Appendix.