Significance
The PD-1/PD-L1 immunoinhibitory axis plays a key role in immune evasion of cancer cells, and therapies targeting PD-1/PD-L1 show high efficacy in certain cancer types. Understanding how cancer drivers regulate immune surveillance will enable development of novel therapeutic strategies targeting cancer-mediated immune evasion, and identification of new biomarkers of response. Here, we utilized genetic screening with a curated library of 500 tumor suppressor genes to identify cohesin subunits and CTCF among the most significant suppressors of PD-L1. We report upregulation of additional key immune-regulatory molecules PD-L2 and MHC-I, and describe transcriptional consequences of loss of the STAG2 cohesin subunit, including an induction of IFN and NF-κB responses, which have implications for biology and treatment of cohesin-deficient tumors.
Keywords: cancer, cohesin, PD-L1, CRISPR
Abstract
Immune evasion is a significant contributor to tumor evolution, and the immunoinhibitory axis PD-1/PD-L1 is a frequent mechanism employed to escape tumor immune surveillance. To identify cancer drivers involved in immune evasion, we performed a CRISPR-Cas9 screen of tumor suppressor genes regulating the basal and interferon (IFN)-inducible cell surface levels of PD-L1. Multiple regulators of PD-L1 were identified, including IRF2, ARID2, KMT2D, and AAMP. We also identified CTCF and the cohesin complex proteins, known regulators of chromatin architecture and transcription, among the most potent negative regulators of PD-L1 cell surface expression. Additionally, loss of the cohesin subunit RAD21 was shown to up-regulate PD-L2 and MHC-I surface expression. PD-L1 and MHC-I suppression by cohesin were shown to be conserved in mammary epithelial and myeloid cells. Comprehensive examination of the transcriptional effect of STAG2 deficiency in epithelial and myeloid cells revealed an activation of strong IFN and NF-κB expression signatures. Inhibition of JAK-STAT or NF-κB pathways did not result in rescue of PD-L1 up-regulation in RAD21-deficient cells, suggesting more complex or combinatorial mechanisms at play. Discovery of the PD-L1 and IFN up-regulation in cohesin-mutant cells expands our understanding of the biology of cohesin-deficient cells as well as molecular regulation of the PD-L1 molecule.
Cancer cells acquire genetic and epigenetic changes that promote escape from normal tissue homeostasis and enable uncontrolled growth. In addition to impaired differentiation, increased proliferation, and prosurvival properties, cancer cells can acquire the ability to evade the immune system (1). Mechanisms of immune evasion commonly acquired by cancer cells include but are not limited to suppression of T-cell function and infiltration, down-regulation of antigen presentation, recruitment of immunosuppressive cell types, and release of immunosuppressive cytokines (2). Detection of virally infected and cancerous cells by CD8+ T cells occurs through the interaction between the T-cell receptor and antigens presented on MHC molecules on the surface of target cells. Antigen recognition leading to activation of CD8+ T cells is accompanied by up-regulation of inhibitory receptors such as PD-1 (programmed death-1), which, when engaged by cognate ligands such as PD-L1 (programmed death-ligand 1) on target cells, prevents immune overreaction by limiting the activity of CD8+ T cells. In this way, inhibitory receptors, also known as checkpoint molecules, prevent tissue damage and help maintain self-tolerance (3, 4).
CD8+ T cells secrete type II interferon, interferon gamma (IFN-γ), and regulate antigen presentation as well as expression of immunoinhibitory ligands. These opposing responses are thought to have differential kinetics that allow an appropriate modulation of response during infection. IFN-γ is the most potent PD-L1 inducer in the cancer microenvironment (5, 6). Activation of JAK-STAT signaling downstream of the IFN receptor IFNGR2 results in up-regulation of IRF1, which binds directly to the PD-L1 promoter inducing its expression (6). IRF2 is a basally expressed competitive repressor of IRF1, which negatively regulates PD-L1 (7, 8). PD-L1 expression can also be induced by type I interferons, IFN-α and IFN-β, albeit to a lesser degree (6). Cancer cells exploit this off switch by up-regulating expression of immunoinhibitory molecules such as PD-L1 and PD-L2. Expression of PD-L1 on cancer cells and immune infiltrates drives immune escape in a context-dependent manner (9–11). PD-L1/PD-1 checkpoint blockade has been shown to be a highly effective treatment strategy in a specific subset of tumor types, and PD-L1 levels and presence of immune infiltrates act as predictors of response to PD-L1/PD-1 checkpoint blockade therapies in certain contexts (12, 13).
Understanding how cancers activate immune evasion could provide additional insights into new therapeutic strategies to circumvent cancer-mediated immune evasion along with biomarkers that could aid application of those therapeutic strategies. Some of the mechanisms by which tumor cells may activate immune evasion are likely driven by cancer driver mutations themselves. Here, we perform an unbiased CRISPR-Cas9 screen of basal and inducible PD-L1 expression levels using human mammary epithelial cells (HMECs) with a curated single-guide (sgRNA) library targeting 500 tumor suppressor genes. We identify the cohesin complex–related components and CTCF, known regulators of three-dimensional chromatin organization and gene expression, as negative regulators of PD-L1 cell surface expression. Our comprehensive analysis of gene expression changes induced by deficiency of the cohesin subunit STAG2 reveals an induction of strong IFN and NF-κB responses. Our findings thus have significant implications for understanding genetic regulators of PD-L1 expression as well as biology of cohesin-mutant cells.
Results
Genetic Screen Identifies Subunits of the Cohesin Complex and CTCF as Strong Negative Regulators of PD-L1.
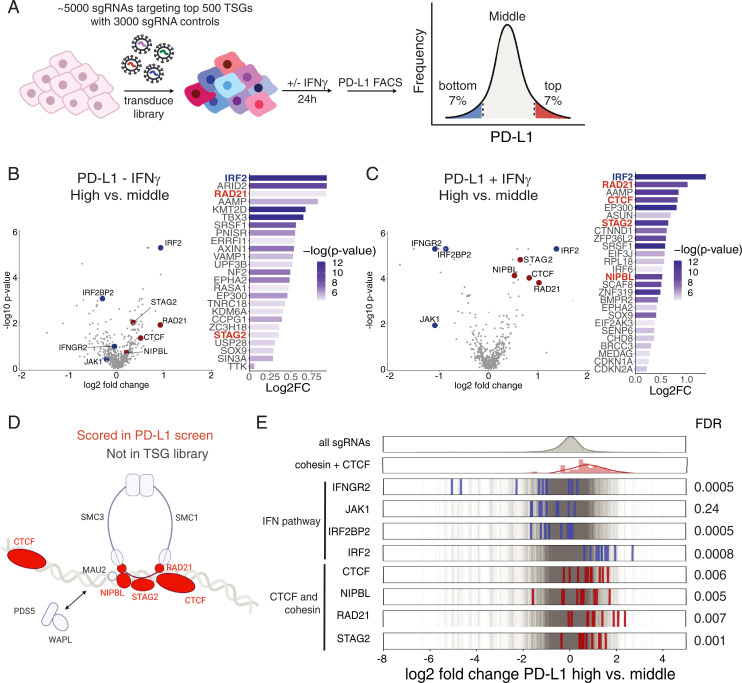
Given the role of the immune system in shaping cancer evolution, we hypothesized that mutations in a subset of frequently mutated cancer drivers would promote immune evasion. Therefore, to identify regulators of basal and IFN-γ–inducible PD-L1 expression in cancer, we performed a CRISPR-Cas9 genetic screen utilizing a tumor suppressor gene (TSG) sgRNA library. This library contains 10 sgRNAs targeting each of the top 500 genes whose mutational patterns in tumors are suggestive of a possible role as a TSG by the Tumor Suppressor and Oncogene Explorer (TUSON) algorithm (14). We used HMECs expressing hTERT, given their high basal level of PD-L1 expression inducible by IFN-γ treatment (SI Appendix, Fig. S1A) in our screen. HMECs infected with the TSG CRISPR library were stained with an anti–PD-L1 antibody and sorted into three bins based on PD-L1 expression (PD-L1 low [bottom 7%], PD-L1 middle, and PD-L1 high [top 7%]) (Fig. 1 A and B). To identify regulators of inducible IFN-γ expression, we also performed the screen in the presence of IFN-γ, where we treated HMECs infected with the TSG library with IFN-γ for 24 h (Fig. 1C). We hypothesized that sgRNAs targeting negative regulators of PD-L1 expression, such as IRF2, would be enriched in the PD-L1 high-expressing cells while sgRNAs targeting positive regulators of PD-L1 expression, such as components of the JAK-STAT pathway, would be enriched in the PD-L1 low-expressing cells.
Fig. 1.
CRISPR-Cas9 screen of basal and inducible regulators of PD-L1 cell surface expression. (A) Graphical representation of the FACS-based PD-L1 screen with the TSG CRISPR-Cas9 library. (B and C) Log2 fold-changes (log2FC) and −log10(P values) for the top 25 genes with the lowest P value enriched in the high (top 7%) versus middle PD-L1 sort (middle ∼80%) in the presence and absence of IFN-γ. Volcano plots of the CRISPR-Cas9 screen of PD-L1 negative regulators in the absence and presence of IFN-γ. Log2FC and P values calculated with MAGeCK of high- versus middle-bin HMECs infected with TSG CRISPR-Cas9 library. Cohesin and CTCF genes are depicted in red, and IFN pathway genes are depicted in blue. (D) Diagram of the components of the cohesin complex and CTCF DNA-binding proteins, which were present in the TSG library (red) and scored in the FACS-based CRISPR-Cas9 screen. Created with BioRender. (E) Frequency histograms of enrichment or depletion (log2 fold-change PD-L1 sort high versus middle) of all sgRNAs in gray, IFN-γ pathway sgRNAs in blue, and cohesin and CTCF pathway sgRNAs in red.
We identified known positive and negative regulators of PD-L1 basal and inducible cell surface expression. Components of the JAK-STAT signaling pathway present in the TSG library, including the IFN-γ receptor IFNGR2 and JAK1 kinase, were depleted in the PD-L1 high bin compared to the PD-L1 middle bin only in the presence of exogenous IFN-γ (Fig. 1 B and C). We also identified that IRF2 is the most potent suppressor of PD-L1 in the TSG library in both the presence and absence of IFN-γ (Fig. 1 B and C). Moreover, IRF2BP2, a negative regulator of IRF2, was found to positively regulate PD-L1 expression (Fig. 1 B and C). The screen thus revealed components of the IFN-γ signaling pathway and known regulators of PD-L1 downstream of IFN-γ signaling as the most significant regulators of basal and inducible PD-L1 cell surface expression, as previously reported. Several previously unknown negative regulators of PD-L1 expression were identified, including the extracellular protein AAMP, subunit of the PBAF chromatin remodeling complex ARID2, and histone methyltransferase KMT2D (Fig. 1 B and C).
Among the previously unappreciated genes scoring in this screen were all three cohesin complex–related proteins present in the TSG library, including the cohesin ring subunits STAG2 and RAD21 and the cohesin-loading protein NIPBL, as well as CTCF, which marks the boundaries of cohesin-mediated loop extrusion. The cohesin ring is a multimeric protein complex formed by SMC3, SMC1, RAD21, and the partially redundant STAG2 and STAG1 proteins that play several essential cellular functions, including sister chromatid cohesion, chromatin organization, transcriptional regulation, and DNA damage repair (15, 16). NIPBL and MAU2 are components of the cohesin-loading complex necessary for cohesin loading onto DNA (15). Mutations in cohesin ring subunits and modulators have been found to be recurrent genetic drivers across multiple cancer types, including myeloid malignancies and breast cancer (17).
Cohesin and CTCF genes all behaved as potent negative regulators of PD-L1 surface levels in the presence and absence of IFN-γ (Fig. 1 B–E and SI Appendix, Fig. S1B). RAD21, NIPBL, STAG2, and CTCF all scored among top 15 hits in the presence of IFN-γ (Fig. 1 B and C). As RAD21, NIPBL, and CTCF are essential genes, hypomorphic alleles of these genes were likely generated by CRISPR. This is consistent with the fact that cohesin subunits in tumors are generally heterozygous predicted loss-of-function mutations exhibiting haploinsufficiency, except for STAG2, which is an X-linked gene and has a partially redundant paralog STAG1 (17, 18). Altogether, our CRISPR-Cas9 screen of basal and inducible regulators of PD-L1 cell surface expression utilizing a TSG library has identified multiple TSGs which regulate PD-L1 cell surface expression. Interestingly, we identify an unexpected role of CTCF and the cohesin complex proteins as some of the most significant suppressors of PD-L1 cell surface expression.
Loss of the Cohesin Subunits and CTCF Results in Up-regulation of PD-L1 Cell Surface Expression.
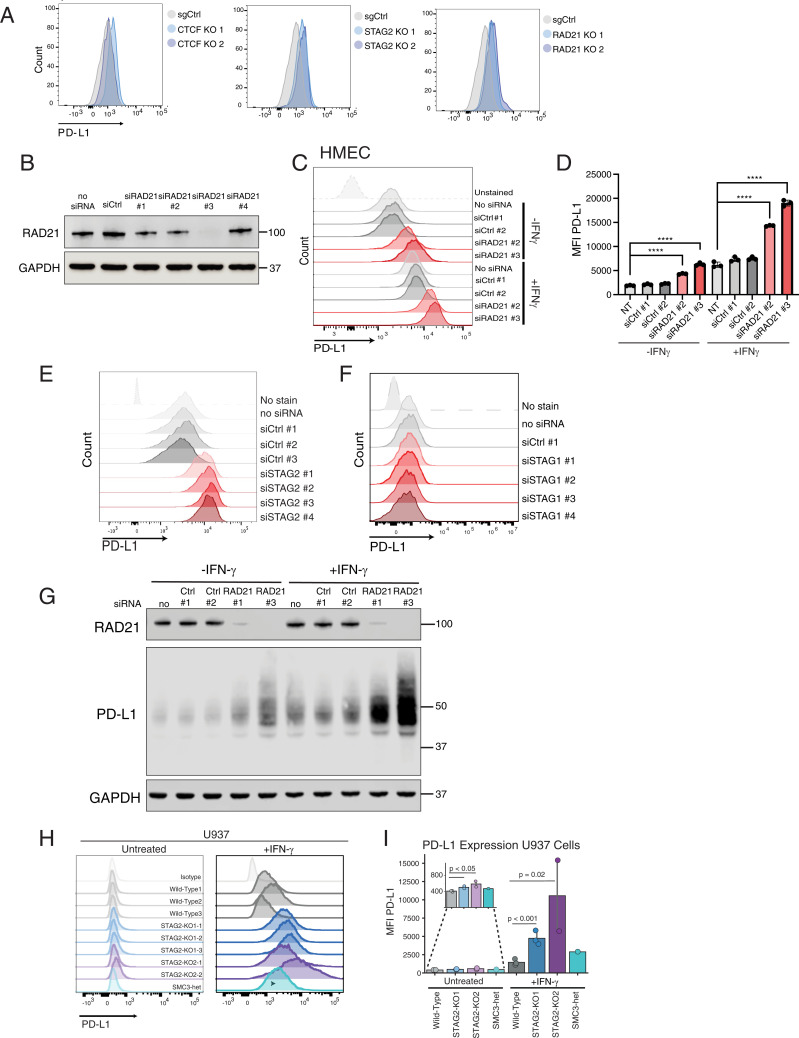
To validate negative regulation of PD-L1 by the components of the cohesin ring and CTCF, we generated loss-of-function mutations of individual cohesin ring subunits STAG2 and RAD21, as well as CTCF, using CRISPR-Cas9 in HMEC cells. Efficacy of CRISPR-Cas9–mediated reduction of CTCF, RAD21, and STAG2 protein levels was confirmed via Western blot analysis (SI Appendix, Fig. S2A). Mutation of STAG2, RAD21, and CTCF resulted in up-regulation of PD-L1 surface expression relative to the negative control sgRNA targeting the AAVS1 locus (Fig. 2A). Since CTCF and RAD21 are essential genes, we also utilized short interfering RNA (siRNA)-mediated depletion to transiently deplete CTCF, STAG2, and RAD21 messenger RNA (mRNA). siRNA-mediated depletion of RAD21, STAG2, and CTCF recapitulated the findings of CRISPR-mediated knockout (KO) in HMECs (Fig. 2 B–E and SI Appendix, Fig. S2 B and C). The increase in PD-L1 cell surface expression in cohesin-depleted cells was accompanied by PD-L1 protein up-regulation (Fig. 2G). CTCF depletion had a weaker PD-L1 up-regulation phenotype than STAG2 or RAD21 depletion in both CRISPR-Cas9–mediated mutation and siRNA-mediated depletion experiments, suggesting that the cohesin ring may play a more important role in PD-L1 regulation than CTCF.
Fig. 2.
CTCF and cohesin suppress PD-L1 expression levels. (A) FACS analysis of PD-L1 cell surface expression after CRISPR-Cas9 mutation of CTCF, STAG2, and RAD21 in HMECs compared to cells treated with targeting sgAAVS1 sgRNA control (sgCtrl). (B) Western blot demonstrating successful generation of RAD21-depleted HMECs using four different siRNAs compared to cells infected with negative control scrambled siRNA and no siRNA control. (C) FACS analysis of PD-L1 expression in control and RAD21 siRNA–depleted HMECs cultured in the presence and absence of 20 ng/mL IFN-γ for 24 h prior to FACS analysis. (D) MFI of PD-L1 staining in siRAD21-treated HMECs from three biological replicates. One-way ANOVA with Dunnett’s correction was used to calculate differences between means (****P value < 0.0001) of each group and no siRNA control for each condition (untreated, +IFN-γ). Data are represented as the means ± SD. (E) FACS analysis of cell surface PD-L1 expression in control and STAG2 siRNA–depleted HMECs. (F) FACS analysis of cell surface PD-L1 expression in control and STAG1 siRNA–depleted HMECs. (G) Western blot analysis of RAD21, PD-L1, and GAPDH protein expression in control and RAD21 siRNA–depleted HMECs in the presence and absence of 20 ng/mL IFN-γ. (H) FACS analysis of PD-L1 cell surface expression in WT, STAG2-mutant (STAG2 KO1-1,2,3, STAG2 KO2-1,2) and SMC3 heterozygous (SMC3-het) U937 cells cultured in the presence or absence of 2,000 IU/mL IFN-γ for 72 h. (I) MFI of PD-L1 staining in STAG2-mutant, SMC3-het, and WT control cells quantified from replicates shown in H. Kruskal–Wallis H test (P = 0.013) with post hoc analysis was used to determine significance between groups.
The STAG2 subunit of the cohesin complex is the most frequently mutated subunit of cohesin, while mutations in its paralog, STAG1, are not frequently found in cancer (17, 18). STAG1- and STAG2-bound cohesin have many overlapping functions including sister chromatid cohesion and a few distinct roles (16, 19). We were thus interested in whether STAG2 and STAG1 share the function of suppressing PD-L1 expression. Interestingly, STAG1 deficiency did not result in up-regulation of PD-L1 (Fig. 2F). PD-L1 up-regulation by STAG2 deficiency, but not STAG1 deficiency, suggests that PD-L1 suppression is a selective function of STAG2-bound cohesin in these cells.
To determine whether this regulatory circuit is conserved in other tissue types, we next examined different models of myeloid malignancies in which cohesin mutations are common genetic drivers of transformation (20). Mutations in the components of the cohesin complex and its modulators occur in ∼13% of patients with de novo acute myeloid leukemia (AML) and 20% of patients with high-risk myelodysplastic syndromes (MDS) and secondary AML (17–22). We examined the levels of PD-L1 expression in two different AML cell lines that were engineered with CRISPR-Cas9 technology with heterozygous mutations in SMC3 and RAD21 and homozygous mutations in STAG2 (SI Appendix, Fig. S2 D and E) (19). Unlike HMECs, which have a relatively high basal level of PD-L1 expression, U937 and K562 cell lines express PD-L1 at very low basal levels. However, treatment with IFN-γ resulted in significant up-regulation of PD-L1 expression of both cell lines (Fig. 2 H and I and SI Appendix, Fig. S2 F and G). Similar to what we observed in HMECs, loss-of-function mutations in STAG2 and SMC3 led to up-regulation of PD-L1 cell surface expression in the absence and presence of IFN-γ treatment in AML cell lines (Fig. 2 H and I and SI Appendix, Fig. S2 F and G). Next, we explored the effects of cohesin mutations in additional cell line models. Analysis of a published whole-genome CRISPR screen that identified regulators of PD-L1 cell surface expression in human near-haploid HAP1 cells (23) revealed that cohesin components STAG2 and RAD21 and cohesin-loading complex components NIPBL and MAU2 scored among the most potent negative regulators of PD-L1 expression (SI Appendix, Fig. S2H). Mutation of STAG1 did not influence PD-L1 expression in this screen, which is consistent with our results for STAG1 depletion in HMECs (Fig. 2F). Altogether, these data suggest that the STAG2–cohesin complex acts as a negative regulator of basal and inducible PD-L1 expression in different tissue types.
Loss-of-Function Cohesin Mutations Result in Up-regulation of IFN Response Gene Signatures in Breast and Myeloid Cells.
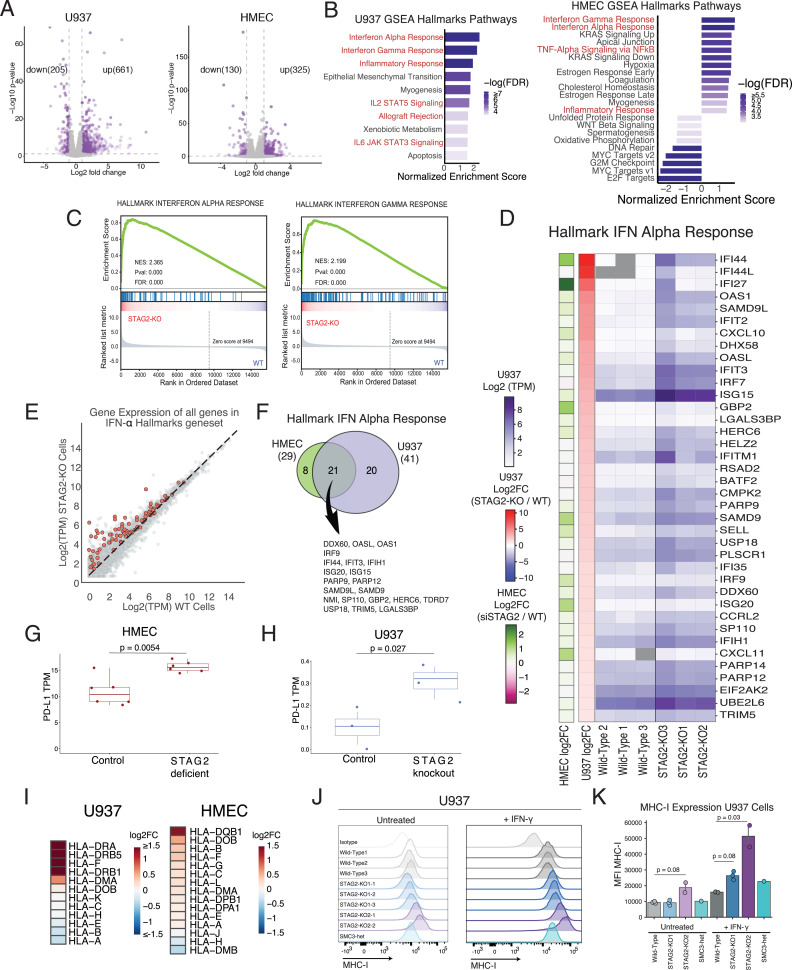
To comprehensively examine the transcriptional response to loss of cohesin proteins, we performed RNA-sequencing (RNA-seq) analysis on wild-type (WT) and STAG2-mutant U937 and STAG2 siRNA-depleted HMECs. Hundreds of differentially expressed genes were identified (Fig. 3A). Gene set enrichment analysis (GSEA) of the Hallmark gene sets revealed multiple up-regulated pathways in both cell lines (Fig. 3B). Several Hallmark gene sets involved in immune-related functions were identified (Fig. 3B). IFN-α and IFN-γ signatures were the top two most significantly enriched gene sets in both cell types (Fig. 3 B–F and SI Appendix, Fig. S3 A and B). Several components of the IFN-signaling pathway, IFN response factors (IRFs), and IFN-stimulated genes (ISGs) were found to be up-regulated in STAG2-deficient HMECs and STAG2-mutant U937 cells (Fig. 3D). Several ISGs were found to be shared between STAG2-deficient epithelial and myeloid cells, but there were also tissue-specific differences in IRF utilization and ISG induction (Fig. 3F).
Fig. 3.
Cohesin deficiency results in up-regulation of IFN transcriptional signature. (A) Volcano plots of the RNA-seq analysis in STAG2 siRNA-depleted and control HMECs, and STAG2-mutant and WT U937 cells. Log2FC and P values calculated with DESeq2. Down-regulated and up-regulated genes (absolute value of log2FC > 1 and P value < 0.05) depicted in purple. (B) GSEA hallmark gene sets enriched or depleted in the RNA-seq analysis of STAG2-mutant and WT control U937 cells and control and siSTAG2-treated HMECs (FDR < 0.05). Immune-related pathways are highlighted in red. (C) Top two enriched GSEA hallmark gene sets in the RNA-seq analysis of STAG2-mutant and WT control U937 cells showing up-regulation of IFN-α and IFN-γ pathway in STAG2-mutant cells. (D) Heatmap of gene expression (TPM values in purple and log2FC values in red-blue) for genes in the IFN-α expression signature with absolute value of log2FC >1 between the STAG2-mutant and WT U937 control cells. Log2FC changes in siSTAG2 HMECs compared to controls depicted in green-pink. (E) Gene expression [Average Log2(TPM)] of STAG2-mutant and WT control cells with genes present in IFN-α gene set highlighted in red and all other genes colored in gray. (F) Overlap between the genes in the IFN-α hallmark gene set enriched in the RNA-seq analysis of STAG2-mutant U937 and STAG2-deficient HMECs (log2FC > 0.2 and P value < 0.05). (G) TPM values of PD-L1 in RNA-seq analysis of STAG2-deficient and control HMECs. Student’s t test was used to calculate differences between means. (H) TPM values of PD-L1 in RNA-seq analysis of STAG2-mutant and control U937 cells. Student’s t test was used to calculate differences between means. (I) Heatmap showing the log2FC of mRNA expression levels of HLA genes in STAG2-mutant U937 and STAG2 HMECs determined by RNA-seq analysis. (J) Flow cytometry analysis of MHC-I expression in WT, STAG2-mutant (STAG2 KO1-1,2,3, STAG2 KO2-1,2), and SMC3-heterozygous (SMC3-het) U937 cells in the presence and absence of IFN-γ treatment (2,000 IU/mL for 72 h). Staining with the isotype antibody shown as a control. (K) Quantification of MFI of MHC-I expression in J. Kruskal–Wallis H test (P = 0.016) with post hoc analysis was used to determine significance between groups.
PD-L1 was found to be transcriptionally up-regulated in the RNA-seq analysis of STAG2-deficient HMECs (Fig. 3G), suggesting that PD-L1 up-regulation in cohesin-deficient cells is at least partially transcriptional. Although the levels of PD-L1 expression in IFN-γ–untreated U937 cells are very low, loss of STAG2 resulted in transcriptional PD-L1 up-regulation (Fig. 3H). Similarly, PD-L1 mRNA levels are up-regulated in RAD21-depleted HMECs (SI Appendix, Fig. S3C). These results suggest that PD-L1 is regulated by cohesin deficiency in at least a partially transcriptional manner. The degree of transcriptional up-regulation of PD-L1 likely does not account for the total protein-level and cell surface–level up-regulation, and we suspect that there are other nontranscriptional mechanisms of PD-L1 up-regulation in cohesin-deficient cells.
Up-regulation of ISGs has many protective roles against pathogens and tumorigenesis. One of the key roles of IFN stimulation is up-regulation of class I histocompatibility gene (MHC-I) expression to enhance antigen presentation and T-cell killing of target cells. We were therefore interested to see if cohesin-mutant cells up-regulate MHC-I. HLA genes were found to be transcriptionally up-regulated in RNA-seq of STAG2-deficient HMEC and STAG2-mutant U937 cells (Fig. 3I). We examined cell surface levels of MHC-I in U937 and K562 AML cells in the presence and absence of IFN-γ in WT, SCM3, or STAG2-mutant cells. While we observed mild effects of cohesin mutations on basal MHC-I expression, there was a significant increase of MHC-I expression in cohesin-mutant cells in the presence of IFN-γ (Fig. 3 J and K and SI Appendix, Fig. S3 D and E). We similarly observed MHC-I up-regulation in RAD21-depleted HMECs (SI Appendix, Fig. S3F). Furthermore, loss of RAD21 and STAG2 in the presence of IFN-γ led to increased expression of PD-L2, a second immunoinhibitory ligand of PD-1 (SI Appendix, Fig. S3G). We thus identified up-regulation of the IFN-α signaling pathway in cohesin-mutant cell lines as well as up-regulation of several key targets of IFN signaling, including PD-L1, MHC-I, and PD-L2.
STAG2-Deficient Cells Activate JAK-STAT and NF-κB Pathways, Which Do Not Contribute to PD-L1 Up-regulation.
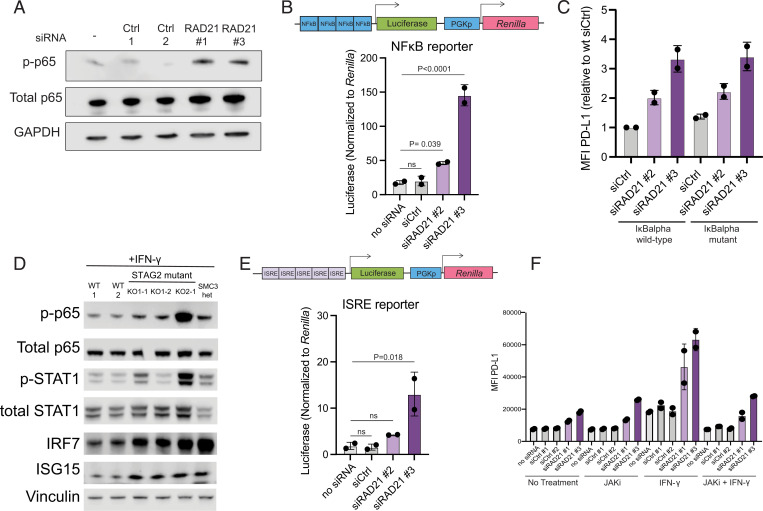
We next explored possible regulators of transcriptional activation of immune genes in cohesin-deficient cells. RNA-seq analysis in STAG2-deficient cells revealed activation of the hallmark NF-κB transcriptional targets (Fig. 3B). This was corroborated by activation of the NF-κB pathway as assessed by p65 phosphorylation and activation of the NF-κB luciferase reporter (consisting of a minimal promoter containing five NF-κB binding motifs upstream from firefly luciferase) in RAD21-siRNA–depleted HMECs (Fig. 4 A and B) as well as STAG2- and SMC3-mutant U937 cells (Fig. 4D). NF-κB pathway activation has been reported to result in transcriptional and posttranslational PD-L1 up-regulation in several cell types (24, 25). To determine if the NF-κB pathway is required for PD-L1 up-regulation in response to cohesin deficiency, we inhibited the pathway by expressing the mutant “superrepressor” IκBα allele in HMECs. The IκBα superrepressor harbors two amino acid substitutions (S32A/S36A), which prevent its phosphorylation and degradation and thus inhibit nuclear translocation of NF-κB. We confirmed that expression of the mutant IκBα superrepressor prevented nuclear translocation of NF-κB upon TNF-α treatment (SI Appendix, Fig. S4A). However, mutant IκBα did not suppress PD-L1 up-regulation in RAD21-deficient HMECs (Fig. 4C), suggesting that activation of the NF-κB pathway in RAD21-deficient HMECs does not contribute to PD-L1 up-regulation.
Fig. 4.
Cohesin mutations result in activation of JAK-STAT signaling and NF-κB. (A) Western blot analysis of p-p65, p65, and GAPDH loading control in siRAD21 or control HMECs. (B) Firefly luciferase quantification of NF-κB luciferase reporter normalized to Renilla luciferase in control and siRAD21-treated HMECs. One-way ANOVA with Dunnett’s correction analysis was used to calculate differences between means of each group and no siRNA control. (C) FACS analysis of PD-L1 cell surface levels in IκBα superrepressor mutant-expressing cells or IκBα WT-expressing HMECs treated with siRAD21 or control siRNA. (D) Western blot analysis of components of the NF-κB (p-p65 and total p65) and IFN response (p-STAT1, STAT1, IRF7, and ISG15) pathways in STAG2-mutant and SMC3-heterozygous U937 cells. (E) Firefly luciferase quantification of ISRE luciferase reporter normalized to Renilla luciferase in control and siRAD21-treated HMECs. One-way ANOVA with Dunnett’s correction analysis was used to calculate differences between means of each group and no siRNA control. (F) FACS analysis of PD-L1 cell surface levels in control or siRAD21-treated HMECs treated with JAKi (0.5 μM for 48 h), IFN-γ (20 ng/mL for 24 h), or combination of JAKi and IFN-γ treatment. MFI of PD-L1 surface staining is plotted on the y-axis. Data are represented as the means ± SD in all panels.
We next examined the role of the JAK-STAT signaling pathway in activating IFN-stimulated gene expression. Cohesin-mutant AML cells significantly up-regulate p-STAT1 levels, consistent with activation of the JAK-STAT pathway (Fig. 4D). We confirmed protein up-regulation of several IFN-inducible factors including IRF7 and ISG15 in STAG2- and SMC3-mutant U937 cells (Fig. 4D). RNA-seq analysis revealed increased transcriptional levels of IFN-β transcripts in STAG2-mutant U937 cells, a potential source for the activation of JAK-STAT signaling (SI Appendix, Fig. S4B). Additionally, we utilized an interferon-stimulated response element (ISRE) luciferase reporter, consisting of a minimal promoter with five ISRE motifs upstream of the luciferase gene, to measure activation of interferon response factors in HMECs (Fig. 4E). ISRE motifs are recognized by interferon response factors whose transcription is induced by activation of JAK-STAT signaling downstream of the IFN receptor. We detected increased reporter activity in RAD21-deficient HMECs stably expressing the ISRE luciferase reporter, confirming activation of IRFs in cohesin-deficient HMECs (Fig. 4E).
Type I and type II IFNs and downstream activation of JAK-STAT signaling are known positive regulators of PD-L1 expression. To test whether activation of JAK-STAT signaling is responsible for PD-L1 up-regulation in STAG2-deficient cells, we treated RAD21-depleted cells with the JAK inhibitor ruxolitinib. Treatment with ruxolitinib suppressed induction of PD-L1 expression in the presence of IFN-γ, but it did not prevent PD-L1 up-regulation in RAD21-deficient HMECs (Fig. 4F). These data suggest that activation of JAK-STAT signaling does not drive up-regulation of PD-L1 expression in RAD21-deficient HMECs. Consistent with this observation, we did not observe up-regulation of the key downstream PD-L1 regulator IRF1 (6) or IRF8 in cohesin-deficient HMECs or cohesin-mutant AML cells (SI Appendix, Fig. S4C). We suspect that the failure to induce IRF1 might explain why strong IFN response does not seem to contribute to PD-L1 up-regulation across our cohesin-deficient AML and HMEC models.
Altogether, our data demonstrate strong activation of JAK-STAT and NF-κB signaling in STAG2-deficient and RAD21-deficient breast epithelial and myeloid cell lines. Intriguingly, inhibition of the JAK/STAT or NF-κB pathways does not rescue PD-L1 up-regulation in RAD21-deficient cells, suggesting that PD-L1 induction might be independent of IFN and NF-κB activation in the context of RAD21 deficiency.
Discussion
In this study, we performed a CRISPR-Cas9 screen with a curated sgRNA library targeting tumor suppressor genes to identify negative regulators of PD-L1 expression. Unexpectedly, we identified CTCF and multiple cohesin genes, STAG2, RAD21, and NIPBL, as the top negative regulators of IFN-γ–inducible PD-L1 up-regulation within the library of the 500 most frequently mutated tumor suppressor genes. STAG2 deficiency was previously reported to up-regulate PD-L1 levels in U2OS cells (26), although whether this was due to its role in cohesion or a distinct function of STAG2 had not been clear. Here, we identify additional subunits of the cohesin complex and CTCF as negative regulators of PD-L1 through unbiased screening and strongly implicate the cohesin complex in regulating PD-L1 expression levels.
PD-L1 regulation by the cohesin complex is at least in part transcriptional. The ability of the cohesin ring to extrude loops of DNA has been found to play an important role in shaping the three-dimensional organization of the genome (27, 28). The cohesin complex and CTCF play an essential role in the generation and maintenance of topologically associated domains that regulate gene expression by facilitating pairing between promoters and their cognate enhancers (15, 29). CTCF has no known role in sister chromatid cohesion but phenocopies regulation of PD-L1 expression by cohesin, which suggests that their shared roles in chromatin organization and regulation of gene expression likely affect PD-L1 regulation. It remains to be elucidated whether cohesin and CTCF loss directly impacts the transcriptional regulation of PD-L1 locus or whether it has an indirect effect on transcription of a PD-L1 regulator.
Deficiency of individual cohesin ring subunits has previously been reported to up-regulate or down-regulate IFN signaling in different cellular models (30, 31).We comprehensively analyzed the transcriptional impact of STAG2 deficiency in primary breast and myeloid cancer cell lines and detected up-regulation of type I IFN response genes. However, several canonical IFN response genes were instead found to be down-regulated by cohesin deficiency despite a strong activation of IFN signature. These down-regulated factors include IRF8 and IRF1, key positive regulators of IFN-inducible transcriptional programs. Down-regulation of a number of IFN response genes could perhaps be explained by the previously reported necessity of the cohesin complex for transcriptional activation of certain IFN-inducible genes. Recently, loss of Rad21 in primary mouse macrophages was shown to lead to rapid transcriptional down-regulation of IFN response genes, suggesting a necessity for the cohesin complex for the transcriptional activation of certain subsets of ISGs (31). Based on these observations, we hypothesize that cohesin deficiency results in IFN response induction in epithelial and myeloid human cell lines, but the necessity of the cohesin complex for transcriptional up-regulation of certain IFN response genes, such as IRF1 and IRF8, results in their down-regulation. We speculate that this might result in a “noncanonical” IFN response phenotype with unique consequences on the tumor microenvironment of cohesin-deficient cells.
Recurrent somatic mutations in genes encoding cohesin subunits and modulators have been identified across a wide spectrum of human malignancies, and STAG2 is one of only a dozen human genes found to be significantly mutated in four or more distinct cancer types (32). Our previous analysis of mutations in PanCancer The Cancer Genome Atlas (TCGA) datasets revealed that there is a strong selective pressure for loss-of-function mutations in CTCF and STAG2 (14). CTCF ranked as the 20th, and STAG2 as the 29th, most potent TSG based on distortion of the mutational signature toward loss-of-function mutations. In support of our observations that CTCF and cohesin play roles in regulating immune-related phenotypes, we have recently identified CTCF as one of the most potent regulators of immune evasion in both triple-negative breast cancer and colon cancer cell lines using in vivo CRISPR screening in WT and immunodeficient mice (33). Future studies will be needed to confirm the role of loss of CTCF and cohesin components in cancer immune evasion in vivo, but we speculate that up-regulation of JAK-STAT signaling, NF-κB pathway, and PD-L1 expression will play a key role. While these mechanisms remain to be elucidated, the discovery of the PD-L1 and IFN up-regulation in cohesin-mutant cells expands our understanding of transcriptional regulation of PD-L1 as well as the biology of cohesin-deficient cells, both of which may have important implications for our understanding of biology and treatment of cohesin-deficient tumors.
Materials and Methods
Cell Lines.
HMECs (Lonza catalog no. CC-2551) were immortalized with hTERT as previously reported and were maintained in HuMEC media (Thermo Fisher Scientific). The 293T cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS) (HyClone), 100 units/mL penicillin, and 0.1 mg/mL streptomycin (Invitrogen, Thermo Fisher Scientific).
U937 cells were obtained from ATCC. STAG2- and SMC3-mutant U937 single cell–derived clones were generated as previously reported (19). Genotypes of STAG2- and SMC3-mutant U937 single cell–derived clones are available in SI Appendix, Table S1. The cells were grown in Roswell Park Memorial Institute (RPMI) Medium (Invitrogen, Thermo Fisher Scientific) supplemented with 10% fetal calf serum (MilliporeSigma), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, Thermo Fisher Scientific).
CRISPR-Cas9 Screen of Regulators of PD-L1 Cell Surface Expression.
A library of ∼5,000 sgRNAs targeting the top 500 TSGs with 3,000 sgRNA controls was designed as previously described (35). Cells were transduced with lentivirus containing the CRISPR TSG library in duplicate at multiplicity of infection (MOI) of 0.2 and representation of 1,000 cellular integrations per sgRNA. After selection in 3 μg/mL puromycin for 3 d and expansion for 2 d, cells were treated with 10 ng/mL IFN-γ (Invitrogen no. PHC4031) or phosphate buffered saline (PBS) for 24 h, followed by an additional treatment with 10 ng/mL IFN-γ or PBS 1 h prior to harvesting cells. Cells were then stained with an allophycocyanin (APC)-conjugated CD274 monoclonal antibody (eBioscience, clone MIH1) at final concentration of 1.5 μg/million cells, fixed with BD Cytofix Fixation Buffer (Fisher catalog no. 554655), subjected to fluorescence activated cell sorting (FACS) at FACSAria Flow Cytometer (BD Biosciences), and pelleted for analysis. Gates were set so that the PD-L1high and PD-L1low populations received ∼7% of stained cells each. For the PD-L1 + IFN-γ screen, four populations were collected from sorting: PD-L1high, PD-L1med, PD-L1low, and PD-L1verylow. Gates were again set so that the PD-L1high and PD-L1low populations received ∼7% of stained cells each. The PD-L1verylow contained ∼3% of cells and was excluded from further analysis.
Analysis of Screen Results.
Genomic DNA was isolated from cell pellets by four rounds of phenol:chloroform extraction using Phaselock tubes (5 PRIME) and three rounds of chloroform extraction. A total of 25 μg/mL Rnase A (Qiagen catalog no. 19101) was added and incubated for 4 h at 37 °C. DNA was ethanol precipitated, recovered by centrifugation, washed three times with 70% ethanol, and resuspended in 10 mM Tris-Cl pH 8.5.
sgRNA barcode sequences were PCR amplified from resuspended genomic DNA and adapted for Illumina NextSeq500 sequencing. Reads were aligned to the reference library using Bowtie (36). CRISPR read counts were analyzed by Model-based Analysis of Genome-wide CRISPR-Cas9 Knockout (MAGeCK) to calculate gene rank lists, P values, and false discovery rates (FDRs) for each gene (37). Results of MAGeCK analysis of the high versus middle bin in the PD-L1–IFN-γ screen are available in Dataset S1. Results of MAGeCK analysis of the high versus middle bin in the presence of IFN-γ screen are available in Dataset S2.
Plasmid Construction.
sgRNAs targeting human CTCF, STAG2, and RAD21 were cloned into lentiCRISPR version 2 puro vector (Addgene catalog no. 52961). The sequences of sgRNAs utilized are available as SI Appendix, Table S2.
To construct luciferase reporter plasmids, the complementary DNA (cDNA) for Renilla luciferase followed by a T2A was synthesized as a gene block (Integrated DNA Technologies). The Renilla-T2A sequence was cloned into pHAGE DEST PGK Blast (38) via Gibson assembly to yield pHAGE DEST PGK Renilla-T2A-Blast. Next, the NF-κB-TA-firefly luciferase sequence, a gift from DarrellKotton, Boston University, Boston, Massachusetts (Addgene plasmid catalog no. 49343) (39), was cloned into pHAGE DEST PGK Renilla-T2A-Blast via Gibson assembly to generate the NF-κB reporter pHAGE NF-κB-TA-firefly luciferase PGK Renilla-T2A-Blast. The IFN-stimulated response element (ISRE) reporter was made by annealing complementary oligos containing five ISRE motifs flanked by NheI and NcoI sites followed by ligation into the pHAGE NF-κB reporter backbone digested with NheI and NcoI. Plasmids were verified by Sanger sequencing.
WT (Addgene plasmid catalog no. 15290) and superrepressor IκBα (Addgene plasmid catalog no. 15291) plasmids were a gift from William Hahn, Harvard Medical School, Boston, Massachusetts (40). IκBα WT and superrepressor genes were then cloned into pHAGE-EF1α-DEST-mAmetrine vector.
Lentiviral Transduction and CRISPR-Cas9–Mediated Knockout in HMECs.
Virus was prepared by transfecting 293T cells with the sgRNA lentiCRISPR version 2 plasmid and psPax2 and pMD2.G lentiviral packaging vectors. Viral supernatants were harvested at 48 h posttransfection. The amount of virus that resulted in ∼30% cell viability (MOI ∼0.3) following puromycin selection was used for infection to ensure low MOI. Since knockouts of CTCF and RAD21 are lethal, cell pellets were harvested 48 h after selection. Efficacy of CRISPR-Cas9–mediated mutation for reducing protein levels was confirmed via Western blotting.
The same methodology was used for lentiviral production and transduction of ISRE and NF-κB luciferase reporters and WT and superrepressor IκBα constructs. After transduction with IκBα constructs, mAmetrine positive cells were FACS sorted.
siRNA-Mediated Depletion.
RAD21, STAG2, CTCF, and STAG1 depletion was performed using siRNA technology. Four siRNAs were tested per gene, and the siRNA with the highest levels of knockdown as determined by Western blotting was utilized for downstream analysis. Approximately 600,000 HMECs were transfected with 5 μL per 6-well plate of Lipofectamine RNAiMAX Transfection Reagent (Thermo catalog no. 13778075) and 40 nM siRNA duplex. Four siRNAs were tested per gene utilizing siRNA from Dharmacon (CTCF targeting: LQ-020165-00-0002, RAD21 targeting: catalog no. LQ-021351-00-0002, STAG2 targeting: LQ-021351-00-0002, STAG1 targeting: LQ-010638-01-0002). Catalog numbers for individual siRNAs are available in SI Appendix, Table S3. The degree of siRNA-mediated depletion was confirmed via Western blot analysis. The cells were split after 24 h and treated with 20 ng/mL IFN-γ (Peprotech catalog no. 300-02) the same day (for RAD21) or the second day (for STAG2, STAG1, and CTCF). The cells were harvested 2 d after transfection (for RAD21) or 3 d after transfection (for STAG2, STAG1, and CTCF) and utilized for downstream analysis.
Western Blotting.
All cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer with protease and phosphatase inhibitors (Thermo catalog no. 78441). Protein was quantified by Bradford assay (Bio-Rad). Protein lysates were separated on 4 to 12% Bis-Tris gels (Invitrogen catalog no. NP0335BOX) and transferred to nitrocellulose membranes (Bio-Rad). Membranes were blocked with 5% milk in Tris-buffered saline with Tween 20 (TBS-T) (Cell Signaling catalog no. 997S) and incubated with the following antibodies overnight at 4 °C at 1:1,000 dilution unless indicated otherwise: Vinculin (1:10,000, catalog no. V9131) from Sigma, STAG1 (catalog no. 14015-1-AP) from Proteintech and RAD21 (catalog no. 4321), CTCF (catalog no. 2899S), STAG2 (catalog no. 4239), PD-L1 (catalog no. 29122), p-p65 (catalog no. 3033T), and p65 (catalog no. 8242), all from Cell Signaling. Antibodies used in U937 and K562 lysates were incubated at 4 °C overnight at 1:1,000 dilution unless indicated otherwise: STAG1 (catalog no. 4455) from Abcam, and STAG2 (catalog no. 5882), SMC3 (catalog no. 5696), actin (1:10,000, catalog no. 8H10D10), p-p65 (catalog no. 3033T), p65 (catalog no. 8242), p-STAT1 (catalog no. 8183), STAT1 (catalog no. 9172), IRF7 (catalog no. 4290), and ISG15 (catalog no. 2743) all from Cell Signaling.
RNA-seq Library Preparation and Sequencing.
For U937 cells, total RNA was isolated from 1 million cells using the RNeasy Plus Isolation Kit (Qiagen) following the manufacturer instructions. The library was prepared using the TruSeq mRNA kit (Illumina) according to the standard protocol. Sequencing was performed on HiSeq 4000 using 100-bp paired-end reads, and 50 million reads were obtained per sample. For HMECs, total RNA was isolated from ∼1 million cells using the RNeasy Plus Isolation Kit (Qiagen) following the manufacturer instructions. RNA-seq was performed on two replicates of cells treated with no siRNA, two replicates treated with control siRNA #1, two replicates with control siRNA #2 and three replicates each of cells treated with siRNA STAG2 #2 and siRNA STAG2 #3. The library was prepared using the NEBnext Ultra II Directional RNA Library kit according to the standard protocol from 1 μg isolated RNA. Barcoded libraries were pooled at molar ratios, and 86-bp single-end sequencing was performed using Ilumina NextSeq 500.
RNA-seq Data Processing.
For U937, RNA-seq reads were mapped using STAR version 2.4.2a. The STAR genome index was generated from human reference sequence hg19 (FASTA downloaded from the University of California, Santa Cruz genome browser) and coding transcripts from GENCODE Release 19 (GTF downloaded from https://www.gencodegenes.org/). Reads mapping to genes were quantified with featureCounts from the R package Subread (version 2.0.1). Transcripts per million (TPM) were quantified from the count data. The sequencing reads and processed TPM are available on Gene Expression Omnibus (GEO) (accession no. GSE189818). For HMECs, RNA-seq reads were mapped using STAR version 2.7.0a to the hg19 genome (41). Reads mapping to genes were quantified with featureCounts. Raw read counts were analyzed using DESeq2 (42)
GSEA.
A matrix of TPM values was used as the input for GSEA with three replicates per genotype for U937 cells and six replicates for HMECs. Pathway enrichment in the STAG2-mutant U937 and STAG2-deficient HMECs was determined for the Hallmark (C1) gene sets. A summary table of the normalized enrichment score and FDR for each of the hallmark gene sets is included in Datasets S3 and S4.
HMEC IFN-γ Treatment and Flow Cytometry.
Approximately 1 million RAD21 siRNA–, STAG2 siRNA–, and siRNA-treated control cells were harvested. Cells were washed with Cell Staining Buffer (BioLegend catalog no. 420201) and stained for 20 min with the relevant antibody: APC anti-human CD274 (BioLegend catalog on. 329708), PE anti-human CD273 (BioLegend catalog no. 329605), and Brilliant Violet 421 anti-human HLA-A2 Antibody (BioLegend catalog no. 343326). The cells were then washed twice with Cell Staining Buffer and analyzed on a BD LSR II fluorescent cell analyzer. Analysis of cell populations and mean fluorescence intensity (MFI) was performed in FlowJo.
U937 and K562 Cell IFN-γ Treatment and Flow Cytometry.
U937 and K562 cells were treated with 2,000 IU/mL of human IFN-γ for 72 h (PeproTech catalog no. 300-02). A total of 500,000 cells were collected per condition and washed with FCS buffer (PBS + 2% FBS). Staining was performed with 1:100 anti-PDL1 (PE) (BioLegend catalog no. 329706) and 1:100 anti-MHC1 (FITC) (BioLegend catalog no. 311404). Isotype controls used were mouse IgG2b, k-PE (BioLegend catalog no. 400313), and mouse IgG2a, k-FITC (BioLegend catalog no. 400208). Cells were washed twice after staining and analyzed immediately on a BD LSR Fortessa. Analysis of cell populations and MFI calculation were performed in FlowJo.
RT-PCR.
RNA from RAD21 and control siRNA-treated cells was isolated using the RNeasy Plus Mini Kit (Qiagen catalog no. 74134). The cDNA was synthesized from 1 ug total RNA using SuperScript IV Reverse Transcriptase (Invitrogen catalog no. 18090050). A total of 1 μL RT reaction product was used for RT-PCR analysis using SYBR Green PCR Master Mix (Thermo Fisher catalog no. 4344463) with the following primers: β-actin (Forward: 5′-CACCATTGGCAATGAGCGGTTC-3′, Reverse: 5′-AGGTCTTTGCGGATGTCCACGT-3′) and PD-L1 (Forward: 5′-GGTGCCGACTACAAGCGAAT-3′ Reverse: 5′-AGCCCTCAGCCTGACATGTC-3′). Each sample was run with three technical replicates.
Immunofluorescence and Microscopy.
Approximately 20,000 HMECs expressing IκBα WT or IκBα superrepressor were grown in tissue culture chamber slides. Cells were treated with 20 ng/mL TNF-α (PreproTech catalog no. 300-01A) for 20 min prior to fixation. Cells were then washed twice with PBS and fixed for 15 min at room temperature in 4% paraformaldehyde (PFA) (Electron Microscopy Science catalog no. 15713). Cells were permeabilized with 0.5% Triton-X for 10 min at room temperature. Cells were blocked in 3% bovine serum albumin (BSA) (Amresco catalog no. 0332). Primary antibody against p65 (Cell Signaling catalog no. 8242) was diluted 1:200 in 3% BSA and incubated for 1 h at room temperature. Secondary antibody was diluted 1:500 in 3% BSA and incubated for 1 h at room temperature. Slides were mounted with DAPI Fluoromount-G (Southern Biotech catalog no. 0100-20). Slides were imaged on a wide-field Nikon Ti inverted microscope.
Luciferase Reporter Assays.
Approximately 1 million HMEC cells expressing ISRE and NF-κB reporters were passively lysed, and Firefly luciferase and Renilla luciferase activity was analyzed using the Dual-Luciferase Reporter Assay System (Promega catalog no. E1960) following the manufacturer’s instructions. Each sample was run with three technical replicates. Firefly luciferase activity was normalized to Renilla luciferase activity, which is driven by the PGK constitutive promoter.
Statistical Analysis.
The significance was analyzed using one-way ANOVA with post hoc test or Student’s t test. Differences were considered significant when P <0.05.
Supplementary Material
Acknowledgments
We thank P. Bruno for critical reagents, E. Watson for help with RNA-seq protocol, C. O’Leary for reagents and insights about IFN signaling, and the other members of the S.J.E. and Z.T. laboratories for critical comments on the manuscript and advice. We thank microscopy core facility staff and the North Quad (MicRoN) core at Harvard Medical School for microscopy resources. The work of Z.T. was supported by NIH Grant K08HL140138-01, The Burroughs Wellcome Fund, The Doris Duke Charitable Foundation. E.C.W. is supported by NIH Grant F32 HL159905-01.
Footnotes
Reviewers: R.L., Memorial Sloan-Kettering; and L.Z., Harvard University, Massachusetts General Hospital.
Competing interest statement: S.J.E. is a cofounder of TSCAN Therapeutics, MAZE Therapeutics, and ImmuneID; serves on the Scientific Advisory Board of the above entities and Homology Medicines, Inc.; and is an advisor for MPM Capital. Z.T. receives research funding from Novartis. None of these interests in any way are connected to this study.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2120540119/-/DCSupplemental.
Data Availability
RNA-seq data are available on the Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession: GSE189818) (43). All other study data are included in the article and/or supporting information.
References
- 1.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Garner H., de Visser K. E., Immune crosstalk in cancer progression and metastatic spread: A complex conversation. Nat. Rev. Immunol. 20, 483–497 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Sun C., Mezzadra R., Schumacher T. N., Regulation and function of the PD-L1 checkpoint. Immunity 48, 434–452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wherry E. J., Kurachi M., Molecular and cellular insights into T cell exhaustion. Nat. Rev. Immunol. 15, 486–499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee S. J., et al. , Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-γ-induced upregulation of B7-H1 (CD274). FEBS Lett. 580, 755–762 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Diaz A., et al. , Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep. 19, 1189–1201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorand R. D., et al. , Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science 353, 399–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harada H., et al. , Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell 58, 729–739 (1989). [DOI] [PubMed] [Google Scholar]
- 9.Noguchi T., et al. , Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol. Res. 5, 106–117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau J., et al. , Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nat. Commun. 8, 14572 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juneja V. R., et al. , PD-L1 on tumor cells is sufficient for immune evasion in immunogenic tumors and inhibits CD8 T cell cytotoxicity. J. Exp. Med. 214, 895–904 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber J., et al. ; CheckMate 238 Collaborators, Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med. 377, 1824–1835 (2017). [DOI] [PubMed] [Google Scholar]
- 13.Robert C., et al. ; KEYNOTE-006 investigators, Pembrolizumab versus ipilimumab in advanced melanoma. N. Engl. J. Med. 372, 2521–2532 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Davoli T., et al. , Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155, 948–962 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yatskevich S., Rhodes J., Nasmyth K., Organization of chromosomal DNA by SMC complexes. Annu. Rev. Genet. 53, 445–482 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Losada A., Cohesin in cancer: Chromosome segregation and beyond. Nat. Rev. Cancer 14, 389–393 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Jann J. C., Tothova Z., Cohesin mutations in myeloid malignancies. Blood 138, 649–661 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waldman T., Emerging themes in cohesin cancer biology. Nat. Rev. Cancer 20, 504–515 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Tothova Z., et al. , Cohesin mutations alter DNA damage repair and chromatin structure and create therapeutic vulnerabilities in MDS/AML. JCI Insight 6, e142149 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kon A., et al. , Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat. Genet. 45, 1232–1237 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Papaemmanuil E., et al. , Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley T. J., et al. , Cancer Genome Atlas Research Network, Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mezzadra R., et al. , Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549, 106–110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim S. O., et al. , Deubiquitination and stabilization of PD-L1 by CSN5. Cancer Cell 30, 925–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gowrishankar K., et al. , Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-κB. PLoS One 10, e0123410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie Z., et al. , STAG2 loss-of-function mutation induces PD-L1 expression in U2OS cells. Ann. Transl. Med. 7, 127 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieberman-Aiden E., et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagano T., et al. , Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 59–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ren G., et al. , CTCF-mediated enhancer-promoter interaction is a critical regulator of cell-to-cell variation of gene expression. Mol. Cell 67, 1049–1058.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding S., et al. , STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat. Commun. 9, 1485 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cuartero S., et al. , Control of inducible gene expression links cohesin to hematopoietic progenitor self-renewal and differentiation. Nat. Immunol. 19, 932–941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lawrence M. S., et al. , Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin T. D., et al. , The adaptive immune system is a major driver of selection for tumor suppressor gene inactivation. Science 373, 1327–1335 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Solimini N. L., et al. , Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science 337, 104–109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liao S., et al. , A genetic interaction analysis identifies cancer drivers that modify EGFR dependency. Genes Dev. 31, 184–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B., Trapnell C., Pop M., Salzberg S. L., Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li W., et al. , MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koren I., et al. , The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell 173, 1622–1635.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson A. A., et al. , Lentiviral delivery of RNAi for in vivo lineage-specific modulation of gene expression in mouse lung macrophages. Mol. Ther. 21, 825–833 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boehm J. S., et al. , Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129, 1065–1079 (2007). [DOI] [PubMed] [Google Scholar]
- 41.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.E. Oreskovic et al., Genetic analysis of cancer drivers reveals cohesin and CTCF as suppressors of PD-L1. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE189818. Deposited 29 November 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data are available on the Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/ (accession: GSE189818) (43). All other study data are included in the article and/or supporting information.