Abstract
During a survey of Group G and C streptococcal infections of humans two epidemiologically unrelated Group G streptococcal isolates were identified, one from a case of bacteremia and one from a wound infection. These isolates were atypical among this sample in that the emm gene could not be amplified from them by PCR. Biochemical characterization identified the isolates as Streptococcus canis, an organism normally associated with animal hosts. The biochemical identification was confirmed by sequencing of the 16S rRNA gene from both isolates and comparison with sequences of the S. canis type strain and other related streptococci of animals and humans. Comparative sequencing of fragments of two other housekeeping genes, sodA and mutS, confirmed that the isolates are most closely related to S. canis. The identification of two isolates of S. canis from a relatively small sample set suggests that the practice of identifying streptococci only by the Lancefield serological group may result in underestimation of the presence of S. canis in the human population.
Streptococcus canis is a species originally proposed in 1986 (3) for streptococci isolated from dogs and cows possessing the Lancefield Group G antigen. The species has since been isolated from a variety of other animals including cats, rats, mink, mice, rabbits, and foxes. Studies of human and animal Group G streptococci (GGS) based on whole-cell protein profiles (13) and multilocus enzyme electrophoresis (14) have confirmed that S. canis forms a clear taxonomic group within the pyogenic streptococci. The species appears distinct from the most closely related species that include other pyogenic streptococci associated with humans and animals. These include Streptococcus dysgalactiae subsp. equisimilis, normally associated with humans and which can belong to GGS or Group C streptococci (GCS); the animal-associated GCS Streptococcus dysgalactiae subsp. dysgalactiae; and the major pathogen of humans, the Group A streptococcus (GAS) Streptococcus pyogenes. Although S. canis isolates may often represent commensal flora of the canine skin and mucosa, they have been implicated in a variety of canine diseases associated with urinary tract infections, abortion, vaginitis, metritis, mastitis, and skin infections. In addition, severe invasive S. canis infections in dogs, analogous to human streptococcal toxic shock syndrome and necrotizing fasciitis associated predominantly with GAS, have been reported (4, 9).
S. canis infection of humans is thought to be rare. The single reliably confirmed report concerns a case of septicemia in a 77-year-old man where the portal of entry was thought to be leg ulcers (2). Two other possible reports, concerning a case of meningitis (7) and a case of peritonitis (8), have been considered unconfirmed due to incomplete or contradictory data (2). However, it remains possible that human infections associated with this organism may be underestimated as routine identification of β-hemolytic streptococci is generally based only on Lancefield serological grouping, which is not species specific in the case of GGS. In addition it remains possible that colonization with S. canis may be essentially unrecognized if there are rarely clinical implications for the host.
As part of a study examining over 200 isolates of GGS and GCS associated with human infection, two epidemiologically unrelated isolates, CG5 and CG40, were obtained. They were unusual in that the emm gene, encoding the M protein, could not be amplified from them by PCR using primers 1 and 2 described previously (16) and purified chromosomal DNA isolated as described previously (15). These primers are routinely used in the emm typing of GAS (1) and in this survey of some 200 isolates of human GGS and GCS were found to successfully amplify emm from all confirmed isolates of S. dysgalactiae subsp. equisimilis examined (data not shown). Isolate CG5 was obtained in pure culture from two separate blood specimens taken from a 76-year-old male leukemia patient in Burton-upon-Trent, United Kingdom. The isolate was identified as a Group G β-hemolytic streptococcus and was sensitive to penicillin, ampicillin, and erythromycin. Isolate CG40 was obtained from a wound swab on the ear of 50-year-old female in Newcastle-upon-Tyne, United Kingdom, from which a scanty growth of hemolytic GGS was isolated along with heavy Staphylococcus aureus growth. The isolate was sensitive to penicillin, flucloxacillin, erythromycin, trimethoprim, chloramphenicol, and amoxicillin but resistant to gentamicin. The extent and nature of any domestic animal contact of these two patients are unknown.
As a result of the inability to amplify emm, isolates CG5 and CG40 were subjected to identification on the basis of their biochemical characteristics using API Rapid ID32 Strep System strips. Both of the isolates were identified as S. canis (identification, 99.9%). A number of biochemical characteristics are known to be useful in differentiating human GGS (S. dysgalactiae subsp. equisimilis) and animal GGS (S. canis), as illustrated in Table 1 (2, 3, 5, 6). The most informative discriminatory tests are recognized to be the production of α-galactosidase and β-galactosidase and the production of acid from trehalose. All these biochemical tests were consistent with identification of the two isolates as S. canis.
TABLE 1.
Biochemical properties used to differentiate human and animal group G streptococci
| Activity or production | Result for:
|
|||
|---|---|---|---|---|
| Human-infecting GGSa | Animal-infecting GGSb | |||
| CG5 | CG40 | |||
| α-Galactosidase | − | + | + | + |
| β-Galactosidase | − | + | + | + |
| β-Glucuronidase | + | vc | − | − |
| Acid from trehalose | + | − | − | − |
S. dysgalactiae subsp. equisimilis.
S. canis.
v, variable.
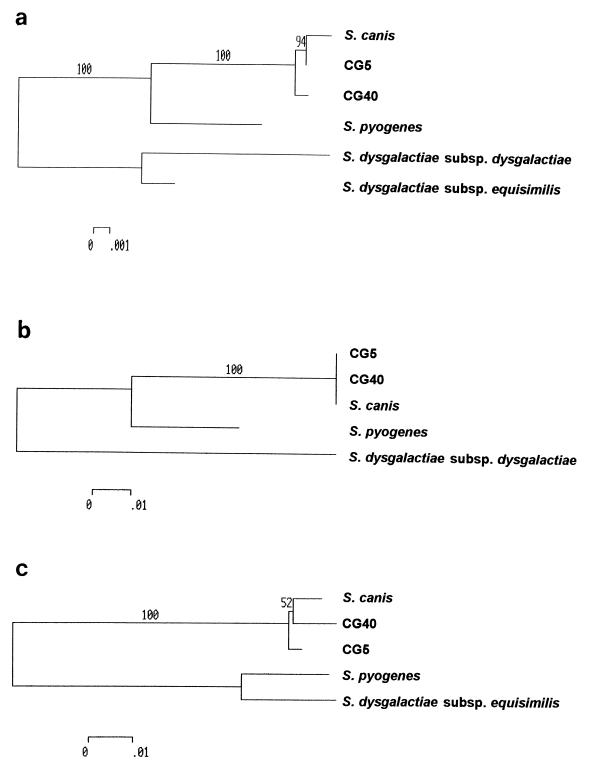
In order to confirm the identities of these isolates on the basis of genetic data the 16S rRNA gene was amplified from CG5 and CG40 using primers 16Sup (5′ AGAGTTTGATCCTGGCTC 3′) and 16Sdown (5′ CACCTTAGGCGGCTGGCT 3′). The sequence corresponding to the vast majority of the 16S rRNA gene (1,322 bp) was obtained from CG5, CG40, and the S. canis type strain NCTC12191 using a series of internal primers and the Beckman CEQ200 sequencing system. These sequences were compared with the corresponding extant sequences in GenBank from the S. pyogenes, S. dysgalactiae subsp. dysgalactiae, and S. dysgalactiae subsp. equisimilis type strains. As illustrated in Fig. 1a, showing an alignment of variable sites relative to the S. canis sequence and the corresponding phylogenetic tree (Fig. 2a) constructed using the MEGA package, and in agreement with the phenotypic identification, both isolates are closely related to S. canis. Sequences of species representing other human and animal pyogenic GCS and GGS and those of GAS are clearly distinct.
FIG. 1.
Analysis of polymorphic sites in housekeeping genes of CG5 and CG40 and other closely related streptococci relative to the S. canis type strain sequences. Sites identical to those seen in S. canis are indicated by dots, while alignment gaps are indicated by dashes. Sequences of fragments of the 16S rRNA gene (A), sodA (B), and mutS (C) are shown. Superscript numbers indicate accession numbers as follows: 1, AB002521; 2, AB002485; 3, AB008926; 4, Z99175; 5, Z95915; 6, Z95900.
FIG. 2.
Phylogenetic trees corresponding to the sequence data shown in Fig. 1: 16S rRNA gene (A); sodA (B); mutS (C). Analysis was performed using the MEGA package, and trees were constructed using the neighbor-joining method with the Jukes-Cantor correction. The percent bootstrap confidence levels of internal branches were calculated from 500 resamplings of the original sequence data.
Two further housekeeping genes were examined in order to confirm the genetic backgrounds of the isolates. First, sodA, encoding superoxide dismutase, was chosen because sequences of this gene have been published from some of the type strains used in the 16S rRNA gene analysis described above (10). The sodA gene was amplified using primers d1 and d2 described previously (10), and 384 bp of sequence, corresponding to bases 37 to 420 of the S. canis database sequence (Z99175), was obtained. A comparison of the variable sites, as shown in Fig. 1b, demonstrates that CG5 and CG40 share identical sequence with the S. canis type strain but are divergent from the S. pyogenes and S. dysgalactiae subsp. dysgalactiae sequences (no sodA sequence from S. dysgalactiae subsp. equisimilis is available). Analysis of a second housekeeping gene, mutS, encoding DNA mismatch repair protein, carried out by amplifying this gene using primers mutSextup (5′ CGTTATGGAACAGCAGAATT 3′) and mutSextdn (5′ GCCATACCATCATAAGTTGC 3′) confirmed that the CG5 and CG40 sequences, though not identical to the S. canis sequence, are far more closely related to S. canis than to the other species examined (Fig. 1c and 2c). Thus, analysis of two genes distinct from the 16S rRNA gene confirms that the genetic background of CG5 and CG40 most closely resembles that of S. canis.
Both phenotypic and genotypic analyses described above confirm the presence of S. canis in human infections. The isolates described in this report were initially identified on the basis of the inability to amplify an emm gene product by PCR. The question of the presence of the emm gene-encoded M protein in S. canis has been the subject of some controversy. In agreement with our findings, a number of studies have suggested that isolates of S. canis do not possess M protein on the basis of lack of hybridization with emm probes (9, 11, 12). However, other studies have shown hybridization with M protein probes (4), although the presence of motifs conserved among many gram-positive surface proteins means that such experiments need to be interpreted with caution. If some isolates of S. canis do possess the M protein-encoding gene, they would not have been identified in this study and in this case there could have been other isolates of S. canis among the 200 examined. Based on our findings, it seems likely that S. canis colonization of humans may be more common than is currently recognized and that the practice of simply applying Lancefield grouping procedures to identify streptococci may be masking the occurrence of this organism in the population. Clearly the two organisms, isolated at least 150 miles apart, are not epidemiologically related, and sequence diversity within both the 16S rRNA and mutS sequences confirms that they do not represent members of a clone. Determining whether there are particular isolates of S. canis which preferentially infect humans, rather than animals, or whether these isolates represent occasional reverse zoonoses requires a much more detailed molecular epidemiological analysis. The pathogenic potential of S. canis in humans remains unclear. One of our isolates was obtained from a wound swab in which bacterial numbers suggested that S. aureus was the primary pathogenic species, with S. canis most likely representing a secondary invading species. However, the second isolate, obtained in pure culture from blood, represents a second confirmed report of invasive S. canis infection of humans (2). Clearly, clinical microbiology laboratories should be aware of the possibility of S. canis infection, particularly when dealing with elderly or immunocompromised patients in contact with domestic animals.
Nucleotide sequence accession numbers.
All novel sequences described in this study have been deposited in the EMBL database and assigned accession numbers as follows: S. canis NCTC12191 16S rRNA gene, AJ413203; CG5 16S rRNA gene, AJ413204; CG40 16S rRNA gene, AJ413205; CG5 sodA, AJ413206; CG40 sodA, AJ413207; S. canis NCTC12191 mutS, AJ413208; CG5 mutS, AJ413209; CG40 mutS, AJ413210; S. pyogenes SF370 mutS, AJ413211; S. dysgalactiae subsp. equisimilis NCFB1356 mutS, AJ413212.
Acknowledgments
A.M.W is supported by a Wellcome Trust Research Fellowship in Biodiversity (grant no. 053589). This work was funded in part by an award from the University of Warwick Research and Teaching Development Fund.
We thank Malcolm West and Jan Wheeler for collection of the strains described in this report.
REFERENCES
- 1.Beall B, Facklam R, Thompson T. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J Clin Microbiol. 1996;34:953–958. doi: 10.1128/jcm.34.4.953-958.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bert F, Lambert-Zechovsky N. Septicemia caused by Streptococcus canis in a human. J Clin Microbiol. 1997;35:777–779. doi: 10.1128/jcm.35.3.777-779.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devriese L A, Hommez J, Kilpper-Bälz R, Schleifer K H. Streptococcus canis sp. nov.: a species of group G streptococci from animals. Int J Syst Bacteriol. 1986;36:422–425. [Google Scholar]
- 4.DeWinter L M, Low D E, Prescott J F. Virulence of Streptococcus canis from canine streptococcal toxic shock syndrome and necrotising fasciitis. Vet Microbiol. 1999;70:95–110. doi: 10.1016/s0378-1135(99)00128-5. [DOI] [PubMed] [Google Scholar]
- 5.Efstratiou A. Pyogenic streptococci of Lancefield groups C and G as pathogens in man. J Appl Microbiol. 1997;83:72S–79S. doi: 10.1046/j.1365-2672.83.s1.8.x. [DOI] [PubMed] [Google Scholar]
- 6.Efstratiou A, Coleman G, Hahn G, Timoney J F, Boeufgras J M, Monget D. Biochemical differences among human and animal streptococci of Lancefield group C or group G. J Med Microbiol. 1994;41:145–148. doi: 10.1099/00222615-41-2-145. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs J A, de Kron M C T, Kellens J T C, Stobberingh E E. Meningitis and sepsis due to the Group G streptococcus. Eur J Clin Microbiol Infect Dis. 1993;12:224–225. doi: 10.1007/BF01967119. [DOI] [PubMed] [Google Scholar]
- 8.Khan J A, Evans H E, Macabuhay M R, Lee Y-E, Werner R. Primary peritonitis due to Group G streptococcus: a case report. Pediatrics. 1975;56:1078–1079. [PubMed] [Google Scholar]
- 9.Miller C W, Prescot J F, Mathews K A, Betschel S D, Yager J A, Guru V, DeWinter L, Low D E. Streptococcal toxic shock syndrome in dogs. J Am Vet Med Assoc. 1996;209:1421–1426. [PubMed] [Google Scholar]
- 10.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnitzler N, Podbielski A, Baumgarten G, Mignon M, Kaufhold A. M or M-like protein gene polymorphisms in human group G streptococci. J Clin Microbiol. 1995;33:356–363. doi: 10.1128/jcm.33.2.356-363.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson W J, Robbins J C, Cleary P P. Evidence for group A-related M protein genes in human but not animal-associated group G streptococcal pathogens. Microb Pathog. 1987;3:339–350. doi: 10.1016/0882-4010(87)90004-0. [DOI] [PubMed] [Google Scholar]
- 13.Vandamme P, Pot B, Falsen E, Kersters K, Devriese L A. Taxonomic study of Lancefield streptococcal groups C, G and L (Streptococcus dysgalactiae) and proposal of S. dysgalactiae subsp. equisimilis subsp. nov. Int J Syst Bacteriol. 1996;46:774–781. doi: 10.1099/00207713-46-3-774. [DOI] [PubMed] [Google Scholar]
- 14.Vieira V V, Teixeira L M, Zahner V, Momen H, Facklam R R, Steigerwalt A G, Brenner D J, Castro A C D. Genetic relationships among the different phenotypes of Streptococcus dysgalactiae strains. Int J Syst Bacteriol. 1998;48:1231–1243. doi: 10.1099/00207713-48-4-1231. [DOI] [PubMed] [Google Scholar]
- 15.Whatmore A M. Streptococcus pyogenes sclB encodes a putative hypervariable surface protein with a collagen-like repetitive structure. Microbiology. 2001;147:419–429. doi: 10.1099/00221287-147-2-419. [DOI] [PubMed] [Google Scholar]
- 16.Whatmore A M, Kehoe M A. Horizontal gene transfer in the evolution of group A streptococcal emm-like genes: gene mosaics and variation in the Vir regulon. Mol Microbiol. 1994;11:363–374. doi: 10.1111/j.1365-2958.1994.tb00316.x. [DOI] [PubMed] [Google Scholar]