Abstract
Three-dimensional (3D) bioprinting systems serve as advanced manufacturing platform for the precise deposition of cells and biomaterials at pre-defined positions. Among the various bioprinting techniques, the drop-on-demand jetting approach facilitates deposition of pico/nanoliter droplets of cells and materials for study of cell-cell and cell-matrix interactions. Despite advances in the bioprinting systems, there is a poor understanding of how the viability of primary human cells within sub-nanoliter droplets is affected during the printing process. In this work, a thermal inkjet system is utilized to dispense sub-nanoliter cell-laden droplets, and two key factors – droplet impact velocity and droplet volume – are identified to have significant effect on the viability and proliferation of printed cells. An increase in the cell concentration results in slower impact velocity, which leads to higher viability of the printed cells and improves the printing outcome by mitigating droplet splashing. Furthermore, a minimum droplet volume of 20 nL per spot helps to mitigate evaporation-induced cell damage and maintain high viability of the printed cells within a printing duration of 2 min. Hence, controlling the droplet impact velocity and droplet volume in sub-nanoliter bioprinting is critical for viability and proliferation of printed human primary cells.
Keywords: 3D Bioprinting, 3D Printing, Biofabrication, Drop-on-demand printing, Sub-nanoliter cell printing
1. Introduction
The advances in three-dimensional (3D) bioprinting techniques enable the fabrication of highly-complex 3D patient-specific tissue-engineered constructs; the highly-automated manufacturing platform facilitates the precise patterning of living cells and biomaterials in a layer-by-layer approach to control the spatial arrangement of these functional components within the complex 3D tissue-engineered constructs[1-4]. The extracellular matrix (ECM) provides a suitable microenvironment for the living cells and plays important role in regulating cell-cell and cell-biomaterial interactions[5-8], and the 3D bioprinting techniques facilitates the fabrication of complex micro-architecture that closely resembles the ECM components within the 3D bioprinted constructs[9-14]. The 3D bioprinting techniques can be categorized into 3 distinct processes: material jetting[15-20], material extrusion[21-26], and vat polymerization[27-29]. Although the extrusion-based bioprinting approach is a commonly used technique for fabrication of 3D complex tissue constructs due to its wide range of printable materials and rapid fabrication speed, the drop-on-demand (DOD) material jetting approach is attractive for contactless deposition and patterning of different types of living cells and biomaterials within each layer to achieve improved cell-cell and cell-matrix interactions[30-33].
Despite the advances in DOD material jetting-based bioprinting techniques, there is limited understanding of how the viability of the printed cells are affected. Droplet impact of cell-laden bio-inks during DOD bioprinting process affects the quality of the printing outcome (through droplet splashing) and the cell viability. Although the droplet splashing phenomenon has been studied extensively for more than 140 years[34], in-depth and high-resolution studies were only conducted in the last 2 decades due to the advancement in the high-speed video technology[35]. The outcome of the droplet impact can be categorized into 6 different scenarios – deposition, prompt splash, corona splash, receding breakup, rebound, and partial rebound. To date, there are only limited studies that investigated the influence of droplet impact on the cell viability of the printed cells[36-39]. Several models have been proposed to simulate the cell viability post droplet impact, such as the Newtonian model where the cells and the droplets are assumed as Newtonian[37], or a compound droplet model with both the cells and the droplets modelled as viscoelastic fluids with different properties, and the ambient fluid modelled as Newtonian fluid[38]. Although these models provide some insights into how the cell viability might be affected by the fluid properties of the bio-ink, experimental results have not been collected to verify the accuracy of the models. The influence of droplet impact velocity on cell viability is a highly-complex phenomenon; an in-depth understanding of the droplet impact velocity on cell viability would be useful for DOD cell printing applications.
Another important consideration during DOD bioprinting of cell-laden bio-inks is the influence of droplet evaporation on the viability of the printed cells within the encapsulated droplets. This is a critical aspect of DOD bioprinting that has been overlooked, and there is a poor understanding of how the droplet evaporation influences the printed cell viability over time. The droplet evaporation mechanism is a highly-complex process, which is dependent on various parameters such as the evaporation mode of the deposited fluids, the physical parameters such as temperature and pressure, the property of the solvent, and the interactions between solvent, particles, and substrate[40].
In this study, we demonstrated using a DOD thermal inkjet bioprinting system that an increase in the cell concentration resulted in slower droplet impact velocity during jetting of sub-nanoliter cell-laden droplets. The decrease in the droplet impact velocity leads to higher viability of the printed cells and improves the printing outcome by mitigating droplet splashing. Furthermore, it is important to limit the printing duration for each printed layer within 2 min to prevent excessive droplet evaporation to maintain high cell viability. The cells were printed using the recommended printing parameters – cell concentration of 4 million cells/mL within printing duration of 2 min; the cells became elongated on day 1 and proliferated well over a period of 7 days to reach almost 90% cell confluency on day 7. The study has highlighted that controlling the droplet impact velocity and droplet evaporation is critical for achieving improved short-term cell viability and long-term cell proliferation of printed cells. The ability to maintain high cell viability and proliferation rate of the printed cells is useful for various bioprinting applications, such as fundamental studies of cell-cell or cell-matrix interactions, and fabrication of in-vitro tissue models.
2. Experimental section
2.1. Cell culture
Primary human dermal fibroblasts (HDF) were purchased from CellnTec Advanced Cell Systems and used in this study. The fibroblasts were cultured in CnT-Prime Fibroblast Proliferation Medium (CnT-PR-F, 1% serum medium supplemented with fully defined growth factors and co-factors) at a temperature of 37°C. The culture medium was changed once every 3 days. The cells were routinely passaged in tissue culture flasks (passages 3 – 5), and the adherent cells were harvested using CnT Accutase cell detachment solution (CnT-Accutase 100) at 90% confluency. Different concentrations of cell-laden bio-inks were prepared; the detached fibroblast cells were suspended in 1× phosphate-buffered saline (PBS) solution – HyClone™, 0.0067 M without calcium (Ca2+) and magnesium (Mg2+) to get the desired cell concentration (0 – 5 million cells/mL). The PBS solution was selected in this experiment for the following reasons: (i) it is a biocompatible medium to deposit cell-laden droplets for fundamental studies of cell-cell and cell-matrix interactions (whereby cell encapsulation in hydrogel matrix may not be desirable); and (ii) various hydrogels may be printed at low concentrations using the inkjet bioprinting approach; hence, the PBS solution was used to serve as a baseline to understand the influence of different printing conditions on cell viability.
2.2. Characterization of bio-inks
The printability of the bio-inks can be evaluated by determining the dimensionless Z value – the inverse of the Ohnesorge number (Oh), which can be defined as the ratio between the Reynolds number and the square root of the Weber number and is independent of the bio-ink velocity. To investigate the influence of cell concentration on the properties (Z values) of the cell-laden bio-inks, cell-laden bio-inks of varying cell concentrations (0 – 5 million cells/mL) were prepared. Measurements were performed on the different cell-laden bio-inks to investigate the influence of cell concentration on viscosity, surface tension, and density of the bio-inks and their respective Z values. The rheological properties of the cell-laden bio-inks were evaluated using the Discovery hybrid rheometer (TA instruments, New Castle, DE, USA). The values of the strain amplitude were first verified to ensure that all measurements were performed within the linear viscoelastic region. Next, the viscosities of different cell-laden bio-inks were evaluated for shear rates ranging from 102 to 104 s−1 at a constant temperature of 25°C. The surface tension of the bio-inks was measured using Optical contact angle system (OCA 15 EC, Data Physics Instrument), and a weighing balance was used to measure the density of the bio-inks (weight per mL of bio-ink). A sample size of 5 was used for all the measurements.
2.3. Evaluation of bio-inks
A thermal inkjet printer (HP D300e Digital Dispenser) was utilized for cell printing; cell-printing cassettes (specially-designed C-8 cassettes with 8 embedded thermal inkjet print-heads with nozzle orifice of 80 μm diameter were used in this study) with a high printing frequency of 1 kHz were used in this study. The thermal inkjet print-head dispensed a constant droplet volume of ~0.345 nL, and multiple droplets were printed at the same spot to achieve the desired droplet volume. The nozzle to substrate distance is approximately 15 mm. Different cell-laden bio-inks (1 – 5 million cells/mL) were printed directly onto dry tissue-treated 12 well plates at varying total dispensed volume of 20 nL, 40 nL, and 60 nL to evaluate its printability and printed cell output. Furthermore, the cell-laden bio-inks (1 – 5 million cells/mL) were also printed directly into tissue-treated 12-well plates that were filled close to the brim with 1× PBS solution to analyze the influence of thermal inkjet printing process on the viability of printed cells at varying cell concentrations.
2.4. High-speed imaging of droplet dispensing
A high-speed camera (Photron Nova S12 – up to 200,000 frames per second [fps]) was used to capture high-speed images of cell-laden droplets travelling from the nozzle orifice until it hits the substrate surface (~ 15 mm apart). As the number of recorded fps increases, it would lead to a smaller area of interest being captured. Hence, the number of recorded fps is selected based on the highest possible fps for the area of interest. To evaluate the droplet velocity profile, the high-speed images were captured at 100,000 fps, 1× zoom and 1/950,000 shutter speed to obtain the full profile of the droplets travelling along the nozzle-substrate distance of ~ 15 mm. The average droplet velocity profile can be obtained by calculating the distance travelled by the droplets (n = 15) between subsequent frames (10 μs apart). Furthermore, the camera is also focused on the substrate surface to capture high-speed images of the droplet impact on substrate surface at varying cell concentration (0 – 4 million cells per mL) using 144,000 fps, 5× zoom and 1/950,000 shutter speed.
2.5. Influence of droplet impact on printed cell viability
The thermal inkjet printer (HP D300e Digital Dispenser) was utilized to dispense cell-laden droplets (1 – 4 million cells/mL) directly onto dry tissue-treated culture plate across a nozzle-substrate distance of ~ 15 mm to investigate the influence of droplet impact on printed cell viability at varying cell concentrations. The cell-laden droplets were printed at 1 kHz frequency into tissue-treated 12-well plates to obtain 8 × 8 array of droplets (20 nL per spot) in each of the 12-well plate (both dry well-plates – original well-plates and the filled well-plates – original well-plates filled close to the brim with PBS solution). The total printing time for each cell-laden bio-inks (1 – 4 million cells/mL) is <2 min per well plate. The printed arrays of cell-laden droplets were immediately evaluated for its cell viability by adding Live/Dead Viability/Cytotoxicity kits (Invitrogen™ L3224, Thermo Fisher Scientific) directly and incubating for 10 min before fluorescence imaging. The stained green cells represent viable printed cells, whereas the stained red cells represent dead printed cells. The average printed cell viability (%) is obtained by calculating the ratio of viable green cells to dead red cells inside each printed droplet. The droplet impact velocity of each cell-laden bio-inks (1 – 4 million cells/mL) is then obtained from the high-speed images in earlier study to analyze the influence of droplet impact velocity on the viability of printed cells.
2.6. Influence of droplet evaporation on printed cell viability
The thermal inkjet printer (HP D300e Digital Dispenser) was utilized to dispense varying volume of cell-laden droplets (4 million cells/ml at 20, 30, and 40 nL per droplet position – the volume of each dispensed droplet is ~0.345 nL) directly onto dry tissue-treated culture plate to investigate the influence of droplet evaporation on printed cell viability. One of the key advantages of the inkjet bioprinting system is its high printing resolution through deposition of nanoliter droplets. However, there is limited studies that investigate the influence of droplet evaporation on printed cell viability in nanoliter droplets. The understanding of this phenomenon would help to implement a suitable printing duration for deposition of cell-laden droplets at nanoliter droplets with high cell viability. The cell-laden droplets were printed at 1 kHz frequency into tissue-treated 12-well plates to obtain 8 × 8 array of droplets (varying total dispensed volume of 20, 30, and 40 nL per droplet position). The total printing time for each study was limited to <1 min, and the time taken for this evaporation study was measured at the start of printing. The viability of printed arrays of cell-laden droplets (20 nL, 30 nL and 40 nL per spot) was evaluated by adding the Live/Dead Viability/Cytotoxicity kits (Invitrogen™ L3224, Thermo Fisher Scientific) at different time intervals (2, 4, 6, 8 and 10 min) post-printing and incubating the staining solution for 10 min before fluorescence imaging. The stained green cells represent viable printed cells, whereas the stained red cells represent dead printed cells. The average printed cell viability (%) is obtained by calculating the ratio of viable green cells to dead red cells inside each printed droplet for varying droplet volumes (20, 30, and 40 nL) at different time intervals (2, 4, 6, 8, and 10 min) post-printing.
2.7. Long-term printed cell proliferation study
The next important step was to demonstrate the long-term proliferation profile of the printed cells. The primary HDF cells were printed using optimal printing cell concentration and printing duration and cultured over a period of 7 days inside an incubator. For the long-term proliferation study, the PrestoBlue® assay was used to measure the proliferation profile of printed cells based on the normalized relative fluorescence units (RFUs) over a period of 7 days (day 1, 3 and 7) post-printing. Fresh culture medium was added to cells before the addition of the PrestoBlue® assay (10% of the total volume) in the ratio of 9:1, followed by incubation at 37°C for 2 h. A micro-plate reader was then used to excite the PrestoBlue® assay at 560 nm wavelength and measure its fluorescence emission at 590 nm wavelength. The measured fluorescence units for the different fluorescence measurements were then normalized to the control group on day 1 to obtain the normalized RFUs for easy comparison.
2.8. Statistical analysis
All experimental results were presented as mean ± standard deviation. Statistical comparisons were performed using Student’s t-test. Values are significantly different when P < 0.01. Significance levels are as follows: P < 0.001 (***) as the most significant and P < 0.01 (*) as the least significant.
3. Results and discussion
3.1. Characterization of bio-inks
The cell-laden bio-inks were prepared by mixing the primary HDFs directly with 1× PBS solution to obtain cell-laden bio-inks of varying cell concentrations (0 – 5 million cells/mL). In general, the cell-laden bio-inks can be categorized into 3 distinct classifications based on the cell volume fraction ɸ[41]: dilute bio-inks (ɸ ≤ 2%), semi-dilute bio-inks (ɸ ≤ 25%), and concentrated bio-inks (ɸ > 25%). The cell diameter of the HDFs is ~18.2 ± 1.6 μm, and the cell volume fractions of the cell-laden bio-inks (1 – 5 million cells/mL) used in this study were 0.337%, 0.674%, 1.011%, 1.348%, and 1.685%, respectively. Hence, the cell-laden bio-inks used in this study are considered dilute bio-inks. The rheological measurement demonstrates an increasing shear rate that results in lower average viscosity, and the decrease in bio-ink viscosities for all the cell-laden bio-inks (0 – 5 million cells/mL) becomes less significant at higher shear rates (> 103 s−1) (Figure 1). Hence, the measured average viscosity values at shear rate of 104 s−1 would be representative of the shear rate experienced by the cells during the printing process which is estimated to be > 104 s−1. An increase in cell concentration of the bio-inks resulted in higher average viscosity values from 0.687 mPa.s (0 million cells/mL) to 0.868 mPa.s (5 million cells/mL). The presence of suspended cells led to higher energy dissipation due to increased frictional force and distorted flow field experienced at the ink-cell interface during the printing process. Hence, an increase in cell concentration results in higher bio-ink viscosities and this observation is corroborated by earlier studies[42,43]. The measured surface tension of cell-laden bio-inks generally decreases with increasing cell concentration; the surface tension decreases from 72.12 ± 0.47 for 0 million cells/mL to 62.86 ± 1.00 for 5 million cells/mL. The increase in cell concentration leads to lower total free energy due to higher cell adsorption at the liquid-gas interface. Hence, the overall surface tension decreases with increasing cell concentration and this phenomenon is consistent with other prior works on particle-laden suspension[42-44]. The density of the 1× PBS solution is 1006.6 ± 2.2 kg/m3 and the overall density of the bio-inks generally increases with increasing cell concentration. The density of the cell-laden bio-inks increases from 1007.3 ± 2.6 kg/m3 at 1 million cells/mL to 1012. ± 2.2 kg/m3 at 5 million cells/mL.
Figure 1.
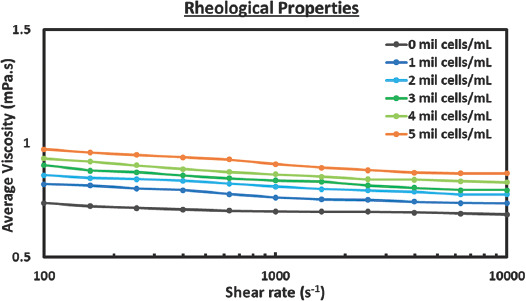
Influence of cell concentration on bio-ink properties. Characterization includes measurement of viscosity, surface tension, and density with a sample size of 5. Rheological characterization of cell-laden bio-inks ranging from 0 to 5 million cells/mL from shear rate of 100 to 10,000 s−1.
The physical properties (viscosity, surface tension and density) of the different cell-laden bio-inks were used to calculate the dimensionless Z value which helps to predict the printability of a bio-ink (Table 1). The dimensionless Z value is an inverse of the Ohnesorge number (Oh), which can be defined as the ratio between the Reynolds number and the square root of the Weber number, and is independent of the bio-ink velocity[15]. The viscous dissipation prevents droplet formation at low Z values (Z < 2), while undesirable satellite droplets form at high Z values (Z > 14)[45]. An increasing cell concentration generally leads to a lower Z value and the measured Z values of the cell-laden bio-inks in this study were within the range of 58.11 (5 million cells/mL) ≤ Z ≤ 72.92 (1 million cells/mL). The high Z values implied that all the cell-laden bio-inks (1 – 5 million cells/mL) were printable with formation of satellite droplets. Nevertheless, it is important to consider potential clogging of the cell-laden bio-inks (average cell diameter of ~18.2 ± 1.6 μm) in the 80 μm nozzle diameter used in this inkjet printing system.
Table 1.
Properties of cell-laden bio-inks ranging from 0 to 5 million cells/mL.
| Cell concentration (mil cells/mL) | Viscosity (mPa.s) | Surface tension (mN/m) | Density (kg/m3) | Nozzle radius (µm) | Z value |
|---|---|---|---|---|---|
| 0 | 0.687 | 72.12±0.47 | 1006.6±2.2 | 40 | 78.41 |
| 1.0 | 0.736 | 71.51±0.66 | 1007.3±2.6 | 40 | 72.92 |
| 2.0 | 0.776 | 66.52±0.90 | 1008.4±2.2 | 40 | 66.74 |
| 3.0 | 0.794 | 65.33±0.21 | 1009.2±2.5 | 40 | 64.67 |
| 4.0 | 0.828 | 63.48±0.82 | 1010.1±2.8 | 40 | 61.16 |
| 5.0 | 0.868 | 62.86±1.00 | 1012.0±2.2 | 40 | 58.11 |
Average viscosity at shear rate of 10,000 s−1, surface tension and density of the cell-laden bio-inks.
3.2. Evaluation of bio-inks
The different cell-laden bio-inks (1 – 5 million cells/mL) were evaluated for the jettability - the ability to eject a primary droplet out from the nozzle orifice. The cell volume fractions of the cell-laden bio-inks (1 – 5 million cells/mL) used in this study were 0.337%, 0.674%, 1.011%, 1.348%, and 1.685%, respectively. The clogging mechanism during the flow through narrow channels is an extremely complex phenomenon; clogging can occur even if the particles are an order of magnitude smaller than the nozzle diameter[46]. The maximum particle size that can be printed is limited by the nozzle diameter because of the potential agglomeration of particles inside the ink, which may lead to clogging of the nozzle. It has been reported that the printer nozzle diameter should be at least 100 times greater than the particle size to prevent potential clogging[47]. Other important considerations include the complexity of print-head design such as inner chamber height and the various types of coating within the firing chamber. The average cell diameter was ~18.2 ± 1.6 μm, and the ratio of nozzle diameter to cell diameter was ~4.396. It was observed that the cell-laden bio-ink containing 5 million cells/ml could not be ejected from the nozzle orifice possibly due to clogging issue. Hence, the subsequent experiments were conducted using printable cell-laden bio-inks (1 – 4 million cells/mL).
Next, the printed cell output per droplet volume was evaluated for all the cell-laden bio-inks (1 – 4 million cells/mL) at varying droplet volumes (20 nL, 40 nL and 60 nL). In general, the measured cell output per droplet volume is less than theoretical number of cells based on the cell concentration for all the cell-laden bio-inks (1 – 4 million cells/mL) at all droplet volumes (20 nL, 40 nL and 60 nL). It is likely that the cells adhere to the inner surface of the microchannel wall and accumulate over time, leading to lower-than-expected cell output. The printed cell output of all the cell-laden bio-inks at varying droplet volume was summarized in Figure 2A and Table 2. Furthermore, the cell-laden bio-inks (1 – 5 million cells/mL) were also printed directly into filled tissue-treated 12-well plates and compared against the non-printed cells to analyze the influence of thermal inkjet printing process on the viability of printed cells at varying cell concentrations. The viability of non-printed cells was determined to be at 97.4 ± 1.89%, and the cell suspension was adjusted to obtain various cell-laden bio-inks (1 – 5 million cells/mL) for printing experiments. Direct printing of cell-laden bio-inks into filled well plate helps to mitigate the damage from droplet impact to the encapsulated cells[39]. Although the printed cell viability decreases slightly with increasing cell concentration from 95.3 ± 3.80 % (1 million cells/mL) to 92.8 ± 2.82 % (4 million cells/mL), the influence of cell concentration on printed cell viability is not significant (Figure 2B).
Figure 2.
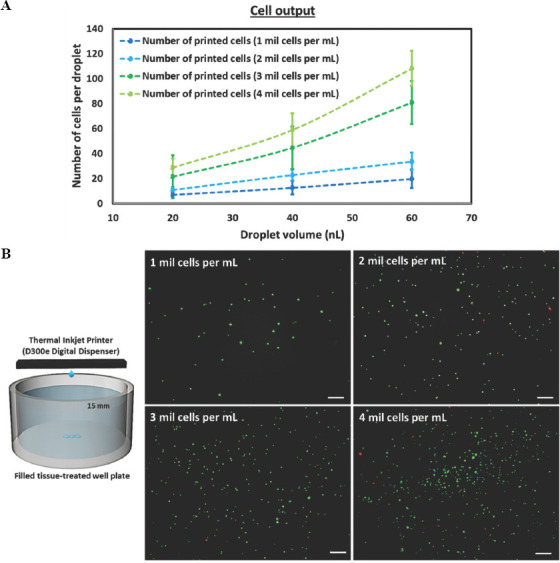
Evaluation of cell-laden bioinks in terms of printability, cell output and cell viability. (A) Printed cell output of cell-laden bio-inks ranging from 1 – 5 million cells/mL at varying droplet volume – 20, 40, and 60 nL on dry tissue-treated well plate. (B) Representative fluorescence images of printed cells stained using Live/Dead cell viability assay (green – viable cells, red – dead cells) to investigate the influence of shear stress in nozzle orifice on cell viability; the cell-laden bio-inks ranging from 1 – 5 million cells/mL are printed directly into a pool of PBS solution filled to the brim; scale bar = 200 μm.
Table 2.
Printed cell output, cell viability and printability of cell-laden bio-inks.
| Cell concentration (mil cells/mL) | No. of cells/droplet (20 nL) | No. of cells/droplet (40 nL) | No. of cells/droplet (60 nL) | Control:Non-printed cell viability (%) | Printing process on cell viability (%) | Printability |
|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | - | - | Yes |
| 1.0 | 6.83±2.75 | 12.6±5.42 | 19.76±7.50 | 97.4±1.89 | 95.3 ± 2.78 | Yes |
| 2.0 | 10.71±2.49 | 22.76±4.42 | 33.71±7.23 | 97.4±1.89 | 94.3±2.02 | Yes |
| 3.0 | 21.56±3.24 | 44.6±6.43 | 80.7±14.54 | 97.4±1.89 | 93.1±2.63 | Yes |
| 4.0 | 28.68±7.36 | 58.89±13.51 | 108.2±13.95 | 97.4±1.89 | 92.7 ± 2.38 | Yes |
| 5.0 | - | - | - | 97.4±1.89 | - | No |
3.3. High-speed imaging of droplet dispensing
A high-speed camera, Photron Nova S12 – up to 200,000 fps, was used to capture high-speed images of cell-laden droplets travelling between the nozzle orifice and substrate surface (~ 15 mm apart) to mimic printing of cell-laden droplets into standard tissue culture well plates. The high-speed images were captured at 100,000 fps to investigate the influence of cell concentration on droplet velocity before droplet impact. The average droplet velocity profile can be obtained by calculating the distance travelled by the droplets (n = 15) between subsequent frames (10 μs apart).
Representative images of the dispensed droplets travelling between the nozzle orifice and substrate surface at varying cell concentration (0 – 5 million cells/mL) are shown in Figure 3A. In general, an increase in cell concentration leads to slower initial droplet velocity from 23.43 m/s (0 million cells/mL), 16.95 m/s (1 million cells/mL), 14.76 m/s (2 million cells/mL), 13.25 m/s (3 million cells/mL) to 7.84 m/s (4 million cells/mL) and similar observations have been reported for printing of particle-laden suspensions[45,48]. An increase in cell concentration leads to higher viscous dissipation during droplet formation process; hence, it results in a lower overall droplet velocity at higher cell concentration. Furthermore, it was observed that the dispensed droplets for all cell concentrations decelerated at relatively similar rates over the nozzle-substrate distance of 15 mm (Figure 3B). The final droplet velocities just before droplet impact were 20.20 m/s (0 million cells/mL), 14.07 m/s (1 million cells/mL), 11.81 m/s (2 million cells/mL), 10.52 m/s (3 million cells/mL) to 5.77 m/s (4 million cells/mL).
Figure 3.
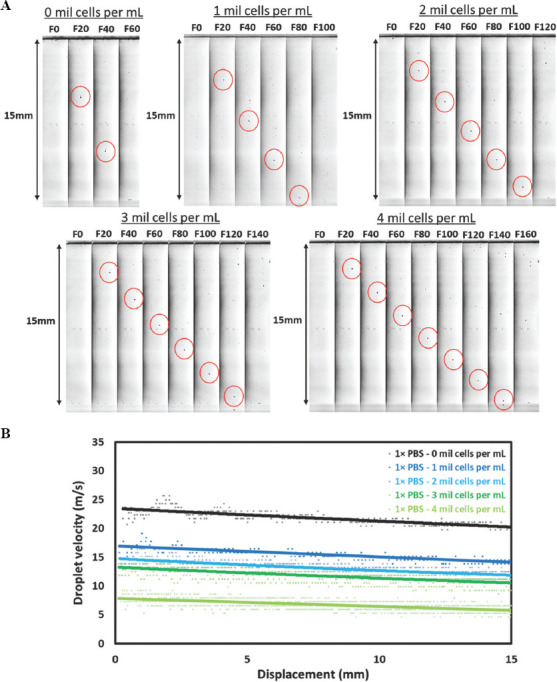
(A) Representative high-speed images of cell-laden droplets (highlighted by red cirle) under different cell concentrations ranging from 0 – 4 million cells/mL travelling across a nozzle-substrate vertical distance of ~ 15 mm at 100,000 fps and 1× zoom; the droplet images shown above are 20 frames apart (200 μs apart), and the droplet positions are highlighted by red circles. (B) Analysis of the droplet velocity profile containing different cell concentrations ranging from 0 – 4 million cells/mL.
The thermal inkjet print-head was used to dispense multiple droplets (constant droplet volume of ~0.345 nL) at the same spot to achieve the desired droplet volume across a nozzle-substrate distance of ~15 mm. The repeated droplet impacts quickly wet the substrate surface to form a layer of liquid film and transform the dynamics to droplet impacts on thin liquid films. The outcome of the droplet impact can be classified as (i) deposition, (ii) prompt splash, and (iii) corona splash. Droplet deposition is characterized by the absence of splashing (without any break-up) on droplet impact[49,50]. In contrast, prompt splash releases small droplets during the advancement of lamella immediately after impact[50,51] while the intact lamella forms a corona shape (bowl-like structure) ejecting multiple small droplets during corona splash[50,51].
The droplet impact on a dry substrate surface (0 – 4 million cells/mL) will always entrap a small air bubble under its center under atmospheric conditions; our observation was corroborated by a previous study that highlighted the lubrication pressure in the thin air layer becomes strong enough to facilitate deposition of sub-nanoliter droplets on non-wetted surface[51]. Subsequently, it was observed that the droplet impacts of all cell-laden bio-inks (1 – 4 million cells/mL) resulted in droplet deposition for droplet impact on wetted substrate surface, whereas droplet splashing was observed for 0 million cells/mL.
Repeated ejection of droplets on the pre-defined spot quickly wets the substrate surface to form a thin and continuous liquid film and this transforms the dynamics to droplet impacts on thin liquid films. Hence, high-speed images of the droplet impact on pre-wetted substrate surface were captured. Droplet splashing will occur when the ink properties and the process parameters exceed a threshold value, which is known as the splashing parameter, K[53]. The splashing parameter is related with several dimensionless numbers, such as the Reynolds number, Re=ρVi D0/m, the Weber number, Re=ρVi2 D0/gLV, and the Ohnesorge number, Oh = We0.5Re, where D0 is the initial droplet diameter before impact, Vi is the droplet’s impact velocity, and ρ, μ, and γLV are the density, viscosity, and surface tension of the droplet’s liquid. There are many variants to this splashing parameter for different boundary conditions[56], and the splashing parameter for this study can be expressed in the form of

where A, a, and b is a constant which is dependent on the boundary condition, and Kc is the splashing parameter, which the droplet will splash when Kc is exceeded. In these variants, the condition for droplet impact on thin liquid film is similar to our printing condition, the a and b constants were obtained from the best fit-line using experimental data to obtain equation (3)[55]:

It is to be noted that the above equation does not provide insight on the magnitude of splashing, but rather just qualitative information on whether splashing would occur. The calculated Oh Re1.17 value for different bio-inks in this study were 97.3 (0 million cells/mL), 63.2 (1 million cells/ml), 52.3 (2 million cells/mL), 46.4 (3 million cells/mL), and 23.1 (4 million cells/mL), respectively (Figure 4A). The high-speed images of droplet impact just above the substrate surface (0 – 4 million cells/mL) concurred with the above equation; droplet splashing was observed for only 0 – 1 million cells/mL (Oh Re1.17 > 63) and droplet deposition was observed for 2 – 4 million cells/mL (Oh Re1.17 < 63) (Figure 4B). The increase in cell concentration resulted in significantly slower droplet impact velocity (from 20.20 m/s for 0 million cells/mL to 5.77 m/s for 4 million cells/mL) which helped to mitigate droplet splashing and improve the printing accuracy.
Figure 4.
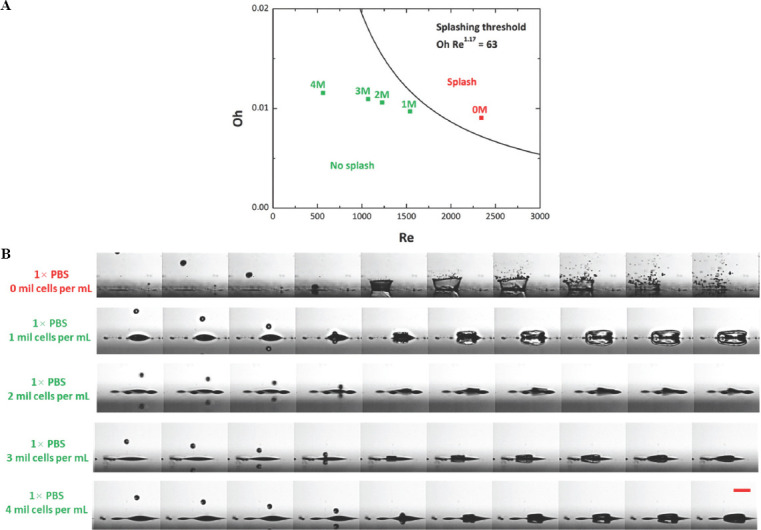
(A) Phase diagram of droplet splashing phenomenon computed using the splashing boundary conditions. (B) Representative high-speed images of ejected droplet hitting the substrate surface at varying cell concentration (0 – 4 million cells/mL) at 5× zoom and the images are taken at 144,000 fps. Increasing the cell concentration resulted in slower droplet velocity which helps to mitigate droplet splashing when hitting the pre-wetted surface; scale bar = 250 μm.
3.4. Influence of droplet impact on printed cell viability
Besides being subjected to the shear stress within the printing nozzle, the droplet impact during the printing process led to cell deformation, and this droplet deformation process has a significant effect on the viability of printed cells. The Live/Dead cell viability assay (green – viable cells, red – dead cells) was used to obtain fluorescence images of printed cells on dry well plates and analyze the influence of droplet impact velocity on cell viability during DOD cell printing applications.
We estimate the shear stress on the cells during droplet impact via the droplet spreading model (Table 3)[57]. This scaling model is based on the balance between the initial kinetic energy of the droplet,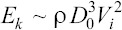 capillary energy,
capillary energy, 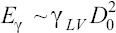 and viscous dissipation,
and viscous dissipation, 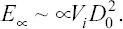 . Here, D0 is the initial droplet diameter prior to impact, Vi is the droplet’s impact velocity, and ρ, μ, and γLV are the density, viscosity, and surface tension of the droplet’s liquid. This model has been experimentally verified to correctly predict droplet spreading diameters as a function of impact velocity and fluid properties for impact Reynolds numbers (Re=ρVi
D0/μ) between 40 and 6300 and Weber numbers (We=ρVi2
D0/γLV) between 1.1 and 414 for impacts on both hard and soft, as well as smooth and rough surfaces[57]. This model predicts that the maximum spreading ratio βmax = Dmax/D0 as transcendental equation:
. Here, D0 is the initial droplet diameter prior to impact, Vi is the droplet’s impact velocity, and ρ, μ, and γLV are the density, viscosity, and surface tension of the droplet’s liquid. This model has been experimentally verified to correctly predict droplet spreading diameters as a function of impact velocity and fluid properties for impact Reynolds numbers (Re=ρVi
D0/μ) between 40 and 6300 and Weber numbers (We=ρVi2
D0/γLV) between 1.1 and 414 for impacts on both hard and soft, as well as smooth and rough surfaces[57]. This model predicts that the maximum spreading ratio βmax = Dmax/D0 as transcendental equation:
Table 3.
Summary of relevant splashing threshold.
| Splashing threshold | Discussion | Reference |
|---|---|---|
| Kc = Oh−0.37Re | This is the first existed splashing threshold model. However, this model is only appropriate for droplets on dry surface. Therefore, it is not suitable for our experiment. | [52] |
| Kc = Oh Re1.25, Kc is 57.7 |
This splashing threshold model is a variant for droplet on wetted rough surface (average roughness of 78 μm) and smooth surface (average roughness of 2.8 μm). However, this variant does not correlate well with our experiment as our glass slide substrate with average roughness in terms of nm. | [53] |
| Kc = Oh−0.4We, Kc is 10396 |
This splashing threshold is valid for droplets on wetted surface when the film thickness is thick enough such that the substrate roughness is negligible. However, the surface roughness of their substrate is 1 μm. | [54] |
| Kc = Oh Re1.17, Kc is 63 |
This variant of splashing threshold is used for droplets on thin film. The substrate of their experiment has an average roughness of 10 nm. Besides, the author had experimented with different liquids with a wide range of viscosity and surface tension, as opposed to the previous splashing threshold variants which is experimented on a single type of liquid. Therefore, this variant of splashing threshold model is more suitable for our application. | [55] |
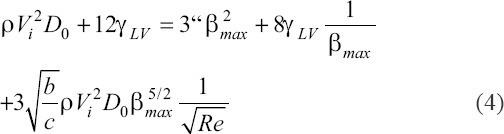
where c = 2 is a geometric parameter, Γ= γLV (1-cos θ) and θ is the dynamic contact angle at maximum spreading. To estimate characteristic shear stress, we approximate the shear stress τ based on the ratio of the characteristic spreading velocity scale and characteristic droplet height, h. We estimate the spreading velocity to scale as Dmax/tmax, where tmax is the time from impact to maximum spread, expressed as tmax=bDmax/Vi[57]. Here, b is the ratio of surface tension of drop liquid to that of water[57]. We estimate h from simple volume conservation, where we consider the spread droplet as a cylinder with height h and diameter Dmax. We obtain 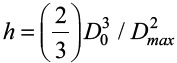 . Thus, we obtain a scaling for the characteristic shear stress as
. Thus, we obtain a scaling for the characteristic shear stress as
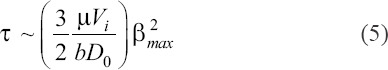
Substituting this into equation (4), we obtain a transcendental equation for shear stress,
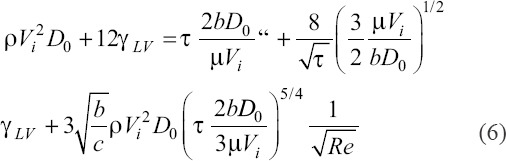
We plot shear stress as a function of impact velocity, initial droplet diameter, and droplet viscosity (Figure 5). This model predicts that shear stress increases strongly with droplet velocity, predicting lower cell viability at higher droplet velocities due to increased droplet spreading shear stress.
Figure 5.
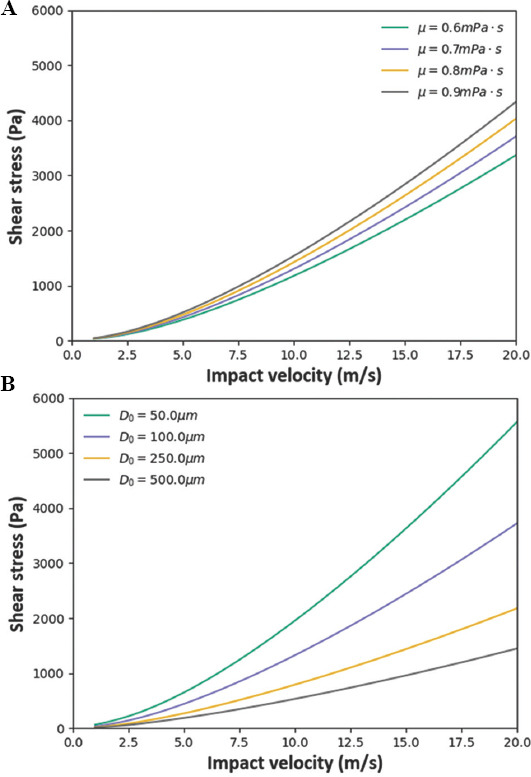
Theoretical results for droplet spreading model using the properties of 4 million cells/mL cell-laden bio-ink. (A) Droplet spreading shear stress as a function of impact velocity and droplet viscosity using 1× PBS solution with 4 million cells/mL with an initial droplet diameter of 80 μm. (B) Droplet spreading shear stress as a function of impact velocity and initial droplet diameter for droplet viscosity of 0.868 mPa.s. (B) For this plot ρ = 1010 kg/m3, γLv = 63.48 mN/m, b = 0.868, c = 2, θ = 93°.
This agrees with our observations that decreasing droplet impact velocity resulted in improved cell viability (Figure 6A). It was also observed that the decrease in average cell viability (%) increases exponentially with increasing droplet impact velocity (Figure 6B). The measured average cell viability (%) from the Live/Dead cell viability assay showed a reduction of 0.44% average cell viability for droplet impact velocity of 5.77 m/s to a reduction of 27.9 % average cell viability for droplet impact velocity of 14.07 m/s (Table 4). The experimental data from this study showed that the droplet impact velocity has a significant effect on the average cell viability of the printed sub-nanoliter cell-laden droplets when the bio-ink viscosity is low (in the order of 1 mPa.s). Hence, controlling the droplet impact velocity (< 5.77 m/s) of low-viscosity cell-laden droplets is critical in achieving high cell viability of more than 90 %.
Figure 6.

Increasing cell concentration leads to slower average droplet impact velocity. Cell-laden bio-inks of varying cell concentration (1 – 4 million cells per mL) were used to investigate the influence of droplet impact velocity on printed cell viability. (A) Representative fluorescence images of printed cells (20 nL per spot) on dry well-plates stained using Live/Dead cell viability assay (green – viable cells, red – dead cells); scale bar = 200 μm. (B) The influence of droplet impact velocity on printed cell viability before and after hitting the substrate surface on dry tissue-treated well plate.
Table 4.
Average droplet impact and printed cell viability of different cell-laden bio-inks (1 – 4 million cells per mL) before and after hitting the substrate surface.
| Cell concentration (mil cells/mL) | Average droplet impact velocity (m/s) | Influence of printing process on cell viability (%) | Influence of droplet impact on cell viability (%) | Change in cell viability (%) |
|---|---|---|---|---|
| 1.0 | 14.07 | 95.3±2.78 | 67.4±3.86 | −27.9% |
| 2.0 | 11.81 | 94.3±2.02 | 82.5±1.74 | −11.8 |
| 3.0 | 10.52 | 93.1±2.63 | 87.3±1.97 | −5.92 |
| 4.0 | 5.77 | 92.7±2.38 | 92.2±1.99 | −0.44 |
Our model also predicts that shear stress decreases with increasing droplet diameter, suggesting that it is prudent to dispense cells using a small number of larger drops rather than a large number of small drops to improve cell viability. Finally, shear stress increases only moderately with droplet viscosity in the relevant viscosity range, predicting that higher cell concentration bio-inks might result in moderately lower cell viability, due to increase in viscosity causing an increase in droplet spreading shear stress. However, we believe the effect of viscosity in decreasing droplet impact velocity dominates in practice; and hence, we observed higher cell viability when a cell-laden bio-inks of higher cell concentration are used.
3.5. Influence of droplet evaporation on printed cell viability
In general, the different variants of droplet evaporation mode are constant contact radius (CCR) evaporation mode, stick-slide (SS) evaporation mode, or a mixed mode of both CCR and SS evaporation modes[58]. The pinning (CCR evaporation mode) and depinning (SS evaporation mode) of the droplet’s contact line depends on the Young’s unbalance force. A low Young’s unbalanced force leads to CCR evaporation mode, whereas a high Young’s unbalanced force leads to SS evaporation mode. In the SS evaporation mode, the contact line remains pinned for a period and then slides to form a smaller radius repeatedly. The sliding of contact line is triggered when the unbalanced Young’s force is too high, and a new equilibrium is reached with a smaller drop radius due to the less deviation from the equilibrium contact angle[59]. The contact angle of the liquid drop reduces as the evaporation progress, resulting in the increase in the unbalanced Young’s force. CCR evaporation mode experiences a constant evaporation flux over time, whereas SS evaporation mode experiences decreasing evaporation flux over time[60]. Hence, droplets of the same volume would evaporate faster under CCR evaporation mode as compared to SS evaporation mode.
Although different hydrogels may be printed together with the cells in the thermal inkjet print-head, the printable polymer concentration is typically low at 1% w/v or lower. Hence, the PBS solution used in this study serves as a baseline to understand the influence of droplet evaporation on cell viability. It was observed that the cell-laden droplets experienced CCR evaporation mode as the contact line remained pinned during the evaporation of the cell-laden droplets. As a result, the evaporation flux remained constant throughout the evaporation mode as there is no change in the droplet diameter. The evaporation flux for CCR evaporation mode in this study can be expressed as[61]:
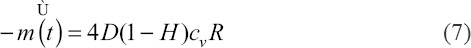
where D is the diffusivity of the vapor in the air, H is the relative humidity of the ambient air, cv is the vapor concentration and R is the contact-line radius. The 10 nL droplets evaporate rapidly within 2 min, hence only larger droplets are used for the droplet evaporation study. Although a larger droplet volume would experience a larger evaporation flux, an increase in the droplet volume from 20 nL to 40 nL (2×) only resulted in a ~1.3× change in diameter from 594.2 ± 4.6 μm to 772.4 ± 5.3 μm. Hence, the 40 nL droplets would take ~ 50% more time to reach complete dryness as compared to the 20 nL droplets. All the printed cell-laden droplets maintained a cell viability of > 92% during the first 2 min of droplet evaporation. After which, there is a significant influence of droplet volume on the viability of printed cells and the viability of printed cells decreased significantly from 88.3 ± 2.45% for 40 nL droplets to 48.2 ± 3.54% for 20 nL droplets at 4-min interval (Figure 7A). Low cell viability of <50% was observed for all cell-laden droplets (20 – 40 nL) at 6-min interval and beyond (Figure 7B). The constant droplet evaporation led to a more hypertonic, i.e., ”high salt” environment and thus resulted in higher cell apoptosis over time[62]. Hence, it is important to strike a balance between achieving high printing resolution and maintaining high cell viability. A higher printing resolution (smaller droplet volume) would lead to lower cell viability due to the droplet evaporation process that leads to an unfavorable hypertonic environment for the encapsulated cells. Hence, it is recommended to deposit a minimum droplet volume of 20 nL and limit the printing time of cell-laden droplets for each printed layer within 2 min for 20 – 30 nL droplets and within 4 min for 40 nL droplets to achieve a high cell viability of > 85%.
Figure 7.
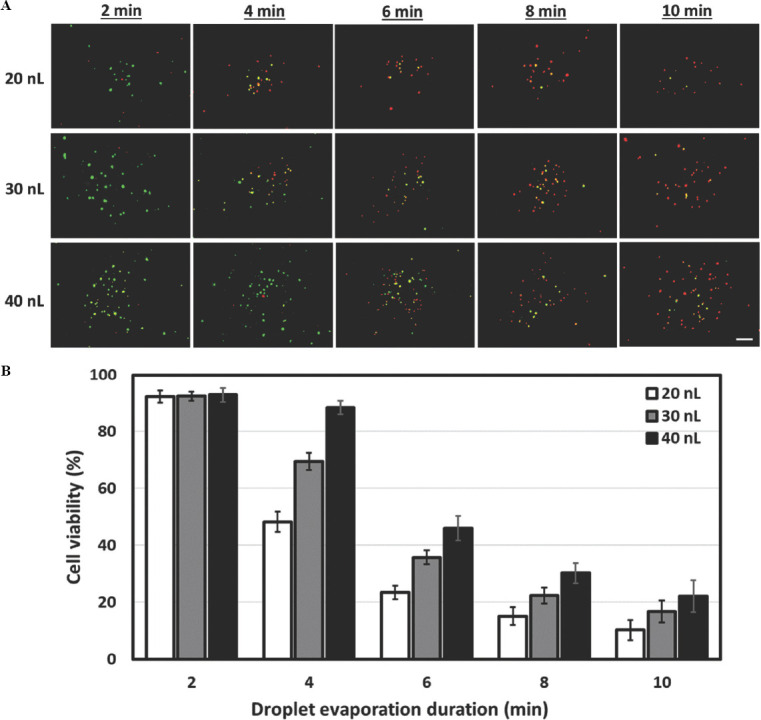
Influence of time on evaporation and viability of nanoliter cell-laden droplets (4 million cells/mL at varying total dispensed volumes of 20, 30, and 40 nL) over a period of 10 min. (A) Representative fluorescence images of printed cells on dry well-plates stained using Live/Dead cell viability assay (green – viable cells, red – dead cells) at different time points (2, 4, 6, 8 and 10-min interval); scale bar = 200 μm. (B) A graph showing the change in cell viability (%) for different total dispensed volumes at different droplet evaporation duration.
3.6. Long-term printed cell proliferation study
Two critical steps (droplet impact velocity and droplet volume) within the DOD bioprinting process have been identified in this study that play important role in influencing the viability of the printed cells. A cell-laden bio-ink with higher cell concentration (4 million cells/mL) leads to an overall slower droplet impact velocity (5.77 m/s); this helps to mitigate the degree of droplet impact-induced damage to the encapsulated cells. Next, the droplet evaporation study in this work has shown that droplet evaporation over time leads to an unfavorable hypertonic environment which causes potential cell death that is apoptosis process (Figure 8A). Hence, the following parameters were selected to evaluate the long-term cell proliferation profile of the printed cells: a cell-laden bio-ink with a concentration of 4 million cells/mL (to achieve the lowest droplet impact velocity) and a printing duration of <2 min (to mitigate the dehydration of printed cells). The cell-laden droplets were printed as 8 × 8 array of droplets (30 nL droplet volume per spot) and cultured over a period of 7 days to evaluate its proliferation profile. There is no negative control (non-printed cells) for this study as the manual hand-held micropipettes dispense cell-laden droplets in the range of microliter (mL) volume and hence, no fair comparison can be obtained between the printed and non-printed cells. The representative fluorescence images showed that most of the printed HDFs showed normal cell morphology and became elongated on day 1 and they proliferated well to reach almost ~90% confluency on day 7 (Figure 8B). Quantitative analysis was performed using the PrestoBlue® assay; the measured RFUs are directly proportional to the number of living cells and all the measured values are normalized to day 1 for easy comparison. The measured normalized RFUs over a period of 7 days have confirmed that the printed HDFs are continually proliferating (Figure 8C). The study has confirmed that controlling the droplet impact velocity and droplet volume leads to high short-term cell viability and long-term cell proliferation of printed HDFs.
Figure 8.
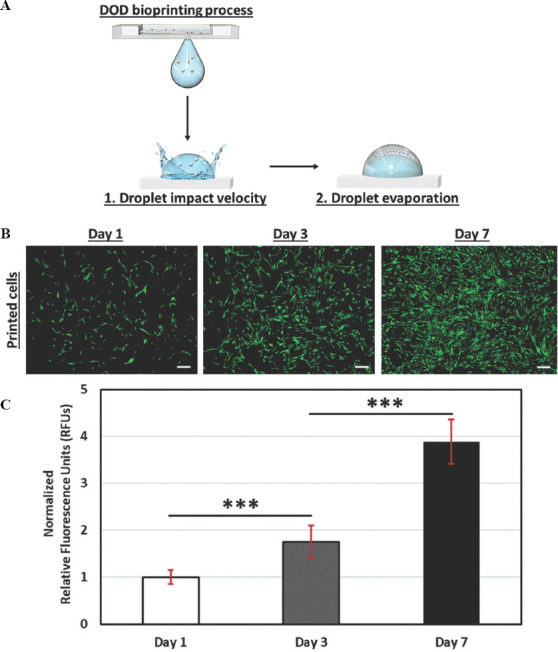
Cell proliferation profile over time. (A) Schematic drawing of the various steps that influence the cell viability during the DOD bioprinting process. (B) Representative fluorescence images of the printed primary HDFs (30 nL droplet volume per spot) printed using the optimal parameters of 4 million cells/mL cell-laden bio-inks printed within 2-min printing duration over a period of 7 days; scale bar = 200 μm. (C) Analysis of the cell proliferation profile using normalized RFUs from the PrestoBlue® assay at different time intervals (day 1, 3 and 7).
4. Conclusions
This work pioneers the investigation of droplet impact velocity and droplet evaporation on viability of printed primary human cells during the DOD thermal inkjet bioprinting process. It provides a better understanding on the different factors that affect the viability of printed cells in sub-nanoliter droplets. A systematic approach was used to first determine the influence of cell concentration on the bio-ink’s physical properties (viscosity, surface tension, and density) and printability. The printability range of cell concentration using a thermal inkjet printer (HP D300e Digital Dispenser) and its cell-printing cassettes (specially-designed C-8 cassettes with 8 embedded thermal inkjet print-heads) with nozzle diameter of 80 mm is between 1 – 4 million cells/mL, and a change in the cell concentration (1 – 4 million cells/mL) has no significant effect on the viability of printed cells during the printing process. Next, the evaluation of droplet velocity profile using the high-speed camera revealed that an increase in the cell concentration leads to significantly slower droplet impact velocity. A slower droplet impact velocity helps to mitigate droplet splashing, improve the printing accuracy and significantly enhance the viability of printed cells within the sub-nanoliter droplets. Furthermore, the PBS solution serves as a baseline to understand the influence of droplet evaporation on cell viability and a minimum droplet volume of 20 nL per spot and total printing duration of 2 min for each printed layer are recommended for maintaining a high cell viability of > 90%. The generated results from this work using cell-laden PBS droplets serve as a baseline for other droplet-based bioprinting techniques that involve contactless jetting of nano-liter cell-laden droplets across a nozzle-substrate distance. The ability to maintain high cell viability and proliferation rate of the printed cells by controlling the droplet impact velocity and droplet evaporation is useful for various bioprinting applications, such as fundamental studies of cell-cell or cell-matrix interactions and fabrication of in-vitro tissue models through precise patterning of cell-laden droplets at the pre-defined positions within the 3D tissue models.
Acknowledgments
This study is supported under the RIE2020 Industry Alignment Fund – Industry Collaboration Projects (IAF-ICP) Funding Initiative, as well as cash and in-kind contribution from the industry partner, HP Inc., through the HP-NTU Digital Manufacturing Corporate Lab. We would also like to acknowledge and thank the D300e HP team for supplying the C8 cell-dispensing cassettes for the experiments and Professor Zhou Kun’s group for the use of their rheometer.
Appendix
Publisher’s note
Whioce Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Conflict of interest
The authors declare no potential conflicts of interest.
Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
References
- 1.Ng WL, Chua CK, Shen YF. Print Me An Organ!Why We Are Not There Yet. Prog Polym Sci. 2019;97:101145. https://doi.org/10.1016/j.progpolymsci.2019.101145. [Google Scholar]
- 2.Murphy SV, Atala A. 3D Bioprinting of Tissues and Organs. Nat Biotechnol. 2014;32:773–85. doi: 10.1038/nbt.2958. https://doi.org/10.1038/nbt.2958. [DOI] [PubMed] [Google Scholar]
- 3.Lee JM, Ng WL, Yeong WY. Resolution and Shape in Bioprinting:Strategizing Towards Complex Tissue and Organ Printing. Appl Phys Rev. 2019;6:011307. https://doi.org/10.1063/1.5053909. [Google Scholar]
- 4.Ng WL, Chan A, Ong YS, et al. Deep Learning for Fabrication and Maturation of 3D Bioprinted Tissues and Organs. Virtual Phys Prototyp. 2020;15:340–58. https://doi.org/10.1080/17452759.2020.1771741. [Google Scholar]
- 5.Wan AC. Recapitulating Cell-Cell Interactions for Organoid Construction-are Biomaterials Dispensable? Trends Biotechnol. 2016;34:711–21. doi: 10.1016/j.tibtech.2016.02.015. https://doi.org/10.1016/j.tibtech.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Gruene M, Pflaum M, Hess C, et al. Laser Printing of Three-dimensional Multicellular Arrays for Studies of Cell-Cell and Cell-Environment Interactions. Tissue Eng Part C Methods. 2011;17:973–82. doi: 10.1089/ten.tec.2011.0185. https://doi.org/10.1089/ten.tec.2011.0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ng WL, Lee JM, Zhou M, et al. Hydrogels for 3-D Bioprinting-based Tissue Engineering. In: Narayan R, editor. Rapid Prototyping of Biomaterials. Chapel Hill, NC; Elsevier: 2020. pp. 183–204. https://doi.org/10.1016/b978-0-08-102663-2.00008-3. [Google Scholar]
- 8.Kathawala MH, Ng WL, Liu D, et al. Healing of Chronic Wounds an Update of Recent Developments and Future Possibilities. Tissue Eng Part B Rev. 2019;25:429–44. doi: 10.1089/ten.TEB.2019.0019. https://doi.org/10.1089/ten.teb.2019.0019. [DOI] [PubMed] [Google Scholar]
- 9.Rose JC, De Laporte L. Hierarchical Design of Tissue Regenerative Constructs. Adv Healthc Mater. 2018;7:1701067. doi: 10.1002/adhm.201701067. https://doi.org/10.1002/adhm.201701067. [DOI] [PubMed] [Google Scholar]
- 10.Ng WL, Goh MH, Yeong WY, et al. Applying Macromolecular Crowding to 3D Bioprinting:Fabrication of 3D Hierarchical Porous Collagen-based Hydrogel Constructs. Biomater Sci. 2018;6:562–74. doi: 10.1039/c7bm01015j. https://doi.org/10.1039/c7bm01015j. [DOI] [PubMed] [Google Scholar]
- 11.Lee JM, Suen SK, Ng WL, et al. Bioprinting of Collagen:Considerations, Potentials, and Applications. Macromol Biosci. 2020;21:2000280. doi: 10.1002/mabi.202000280. https://doi.org/10.1002/mabi.202000280. [DOI] [PubMed] [Google Scholar]
- 12.Osidak EO, Kozhukhov VI, Osidak MS, et al. Collagen as Bioink for Bioprinting:A Comprehensive Review. Int J Bioprint. 2020;6:270. doi: 10.18063/ijb.v6i3.270. https://doi.org/10.18063/ijb.v6i3.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng WL, Ayi TC, Liu YC, et al. Fabrication and Characterization of 3D Bioprinted Triple-layered Human Alveolar Lung Models. Int J Bioprint. 2021;7:332. doi: 10.18063/ijb.v7i2.332. https://doi.org/10.18063/ijb.v7i2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JM, Sing SL, Tan EY, et al. Bioprinting in Cardiovascular Tissue Engineering:A Review. Int J Bioprint. 2016;2:136–45. https://doi.org/10.18063/ijb.2016.02.006. [Google Scholar]
- 15.Saunders RE, Derby B. Inkjet Printing Biomaterials for Tissue Engineering:Bioprinting. Int Mater Rev. 2014;59:430–48. https://doi.org/10.1179/1743280414y.0000000040. [Google Scholar]
- 16.Ng WL, Lee JM, Yeong WY, et al. Microvalve-based Bioprinting Process, Bio-inks and Applications. Biomater Sci. 2017;5:632–47. doi: 10.1039/c6bm00861e. https://doi.org/10.1039/c6bm00861e. [DOI] [PubMed] [Google Scholar]
- 17.Ng WL, Yeong WY, Naing MW. Microvalve Bioprinting of Cellular Droplets with High Resolution and Consistency. Proceedings of the International Conference on Progress in Additive Manufacturing. 2016:397–402. [Google Scholar]
- 18.Koch L, Brandt O, Deiwick A, et al. Laser Assisted Bioprinting at Different Wavelengths and Pulse Durations with a Metal Dynamic Release Layer:Aa Parametric Study. Int J Bioprint. 2017;3:42–53. doi: 10.18063/IJB.2017.01.001. https://doi.org/10.18063/ijb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo F, Li P, French JB, et al. Controlling Cell-Cell Interactions Using Surface Acoustic Waves. Proc Natl Acad Sci U S A. 2015;112:43–8. doi: 10.1073/pnas.1422068112. https://doi.org/10.1073/pnas.1422068112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choe YE, Kim GH. A PCL/Cellulose Coil-shaped Scaffold Via a Modified Electrohydrodynamic Jetting Process. Virtual Phys Prototyp. 2020;15:403–16. https://doi.org/10.1080/17452759.2020.1808269. [Google Scholar]
- 21.Ozbolat IT, Hospodiuk M. Current Advances and Future Perspectives in Extrusion-Based Bioprinting. Biomaterials. 2016;76:321–43. doi: 10.1016/j.biomaterials.2015.10.076. https://doi.org/10.1016/j.biomaterials.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 22.Ng WL, Yeong WY, Naing MW. Development of Polyelectrolyte Chitosan-gelatin Hydrogels for Skin Bioprinting. Proc CIRP. 2016;49:105–12. https://doi.org/10.1016/j.procir.2015.09.002. [Google Scholar]
- 23.Ng WL, Yeong WY, Naing MW. Polyelectrolyte Gelatin-chitosan Hydrogel Optimized for 3D Bioprinting in Skin Tissue Engineering. Int J Bioprint. 2016;2:53–62. https://doi.org/10.18063/ijb.2016.01.009. [Google Scholar]
- 24.Zhuang P, Ng WL, An J, et al. Layer-by-layer Ultraviolet Assisted Extrusion-Based (UAE) Bioprinting of Hydrogel Constructs with High Aspect Ratio for Soft Tissue Engineering Applications. PLoS One. 2019;14:e0216776. doi: 10.1371/journal.pone.0216776. https://doi.org/10.1371/journal.pone.0216776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng WL, Yeong WY, Naing MW. Potential of Bioprinted Films for Skin Tissue Engineering. Proceedings of the 1st International Conference on Progress in Additive Manufacturing. 2014:441–6. https://doi.org/10.3850/978-981-09-0446-3_065. [Google Scholar]
- 26.Meng Z, He J, Li J, et al. Melt-based, Solvent-free Additive Manufacturing of Biodegradable Polymeric Scaffolds with Designer Microstructures for Tailored Mechanical/Biological Properties and Clinical Applications. Virtual Phys Prototyp. 2020;15:417–44. https://doi.org/10.1080/17452759.2020.1808937. [Google Scholar]
- 27.Ng WL, Lee JM, Zhou M, et al. Vat Polymerization-based Bioprinting-process, Materials, Applications and Regulatory Challenges. Biofabrication. 2020;12:022001. doi: 10.1088/1758-5090/ab6034. https://doi.org/10.1088/1758-5090/ab6034. [DOI] [PubMed] [Google Scholar]
- 28.Li W, Mille LS, Robledo JA, et al. Recent Advances in Formulating and Processing Biomaterial Inks for Vat Polymerization-Based 3D Printing. Adv Healthc Mater. 2020;9:2000156. doi: 10.1002/adhm.202000156. https://doi.org/10.1002/adhm.202000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieto D, Corrales JA, de Mora AJ, et al. Fundamentals of Light-Cell-Polymer Interactions in Photo-cross-linking Based Bioprinting. APL Bioeng. 2020;4:041502. doi: 10.1063/5.0022693. https://doi.org/10.1063/5.0022693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gudupati H, Dey M, Ozbolat I. A Comprehensive Review on Droplet-based Bioprinting:Past, Present and Future. Biomaterials. 2016;102:20–42. doi: 10.1016/j.biomaterials.2016.06.012. https://doi.org/10.1016/j.biomaterials.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Ng WL, Tan ZQ, Yeong WY, et al. Proof-of-concept:3D Bioprinting of Pigmented Human Skin Constructs. Biofabrication. 2018;10:025005. doi: 10.1088/1758-5090/aa9e1e. https://doi.org/10.1088/1758-5090/aa9e1e. [DOI] [PubMed] [Google Scholar]
- 32.Ng WL, Wang S, Yeong WY, et al. Skin Bioprinting:Impending Reality or Fantasy? Trends Biotechnol. 2016;34:689–99. doi: 10.1016/j.tibtech.2016.04.006. https://doi.org/10.1016/j.tibtech.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 33.Ng WL, Yeong WY. The Future of Skin Toxicology Testing 3D Bioprinting Meets Microfluidics. Int J Bioprint. 2019;5:237. doi: 10.18063/ijb.v5i2.1.237. https://doi.org/10.18063/ijb.v5i2.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Worthington AM. XXVIII. On the Forms Assumed by Drops of Liquids Falling Vertically on a Horizontal Plate. Proc R Soc London. 1877;25:261–72. https://doi.org/10.1098/rspl.1876.0048. [Google Scholar]
- 35.Thoroddsen ST, Etoh TG, Takehara K. High-Speed Imaging of Drops and Bubbles. Annu Rev Fluid Mech. 2008;40:257–85. https://doi.org/10.1146/annurev.fluid.40.111406.102215. [Google Scholar]
- 36.Lepowsky E, Muradoglu M, Tasoglu S. Towards Preserving Post-printing Cell Viability and Improving the Resolution:Past, Present, and Future of 3D Bioprinting Theory. Bioprinting. 2018;11:e00034. https://doi.org/10.1016/j.bprint.2018.e00034. [Google Scholar]
- 37.Tasoglu S, Kaynak G, Szeri AJ, et al. Impact of a Compound Droplet on a Flat Surface:A Model for Single Cell Epitaxy. Phys Fluids. 2010;22:082103. doi: 10.1063/1.3475527. https://doi.org/10.1063/1.3475527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nooranidoost M, Izbassarov D, Tasoglu S, et al. A Computational Study of Droplet-based Bioprinting:Effects of Viscoelasticity. Phys Fluids. 2019;31:081901. https://doi.org/10.1063/1.5108824. [Google Scholar]
- 39.Hendriks J, Visser CW, Henke S, et al. Optimizing Cell Viability in Droplet-based Cell Deposition. Sci Rep. 2015;5:11304. doi: 10.1038/srep11304. https://doi.org/10.1038/srep11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gorr HM, Zueger JM, McAdams DR, et al. Salt-induced Pattern Formation in Evaporating Droplets of Lysozyme Solutions. Colloids Surf B Biointerfaces. 2013;103:59–66. doi: 10.1016/j.colsurfb.2012.09.043. https://doi.org/10.1016/j.colsurfb.2012.09.043. [DOI] [PubMed] [Google Scholar]
- 41.Rutgers IR. Relative Viscosity of Suspensions of Rigid Spheres in Newtonian liquids. Rheol Acta. 1962;2:202–10. [Google Scholar]
- 42.Ng WL, Yeong WY, Naing MW. Polyvinylpyrrolidone-Based Bio-Ink Improves Cell Viability and Homogeneity during Drop-On-Demand Printing. Materials. 2017;10:190. doi: 10.3390/ma10020190. https://doi.org/10.3390/ma10020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu C, Zhang M, Huang Y, et al. Study of Droplet Formation Process during Drop-on-Demand Inkjetting of Living Cell-Laden Bioink. Langmuir. 2014;30:9130–8. doi: 10.1021/la501430x. https://doi.org/10.1021/la501430x. [DOI] [PubMed] [Google Scholar]
- 44.Dong L, Johnson D. Surface Tension of Charge-stabilized Colloidal Suspensions at the Water Air Interface. Langmuir. 2003;19:10205–9. https://doi.org/10.1021/la035128j. [Google Scholar]
- 45.Jang D, Kim D, Moon J. Influence of Fluid Physical Properties on Ink-jet Printability. Langmuir. 2009;25:2629–35. doi: 10.1021/la900059m. https://doi.org/10.1021/la900059m. [DOI] [PubMed] [Google Scholar]
- 46.Lee A, Sudau K, Ahn KH, et al. Optimization of Experimental Parameters to Suppress Nozzle Clogging in Inkjet Printing. Ind Eng Chem Res. 2012;51:13195–204. https://doi.org/10.1021/ie301403g. [Google Scholar]
- 47.Srichan C, Saikrajang T, Lomas T, et al. Inkjet Printing PEDOT:PSS Using Desktop Inkjet Printer. 2009 6th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology, IEEE. 2009:465–8. https://doi.org/10.1109/ecticon.2009.5137049. [Google Scholar]
- 48.Derby B, Reis N. Inkjet Printing of Highly Loaded Particulate Suspensions. MRS Bull. 2003;28:815–18. https://doi.org/10.1557/mrs2003.230. [Google Scholar]
- 49.Yarin AL. Drop Impact Dynamics:Splashing, Spreading, Receding, Bouncing. Annu Rev Fluid Mech. 2006;38:159–92. https://doi.org/10.1146/annurev.fluid.38.050304.092144. [Google Scholar]
- 50.Rioboo R, Tropea C, Marengo M. Outcomes from a Drop Impact on Solid Surfaces. Atomization Sprays. 2001;11:155–66. https://doi.org/10.1615/atomizspr.v11.i2.40. [Google Scholar]
- 51.Josserand C, Thoroddsen ST. Drop Impact on a Solid Surface. Annu Rev Fluid Mech. 2016;48:365–91. https://doi.org/10.1146/annurev-fluid-122414-034401. [Google Scholar]
- 52.Stow CD, Hadfield MG. An Experimental Investigation of Fluid Flow Resulting from the Impact of a Water Drop with an Unyielding Dry Surface. Proc R Soc London A Math Phys Sci. 1981;373:419–41. https://doi.org/10.1098/rspa.1981.0002. [Google Scholar]
- 53.Mundo C, Sommerfeld M, Tropea C. Droplet-wall Collisions:Experimental Studies of the Deformation and Breakup Process. Int J Multiphase Flow. 1995;21:151–73. https://doi.org/10.1016/0301-9322(94)00069-v. [Google Scholar]
- 54.Yarin AL, Weiss DA. Impact of Drops on Solid Surfaces:Self-similar Capillary Waves, and Splashing as a New Type of Kinematic Discontinuity. J Fluid Mech. 1995;283:141–73. https://doi.org/10.1017/s0022112095002266. [Google Scholar]
- 55.Wal RL, Berger GM, Mozes SD. The Splash/Non-splash Boundary Upon a Dry Surface and thin Fluid Film. Exp Fluids. 2006;40:53–9. https://doi.org/10.1007/s00348-005-0045-1. [Google Scholar]
- 56.Moreira AL, Moita AS, Panão MR. Advances and challenges in explaining fuel spray impingement:How much of single droplet impact research is useful? Prog Energy Combust Sci. 2010;36:554–80. https://doi.org/10.1016/j.pecs.2010.01.002. [Google Scholar]
- 57.Lee JB, Derome D, Guyer R, et al. Modeling the Maximum Spreading of Liquid Droplets Impacting Wetting and Nonwetting Surfaces. Langmuir. 2016;32:1299–308. doi: 10.1021/acs.langmuir.5b04557. https://doi.org/10.1021/acs.langmuir.5b04557. [DOI] [PubMed] [Google Scholar]
- 58.Shaikeea AJ, Basu S, Tyagi A, et al. Universal Representations of Evaporation Modes in Sessile Droplets. PLoS One. 2017;12:e0184997. doi: 10.1371/journal.pone.0184997. https://doi.org/10.1371/journal.pone.0184997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goh GL, Saengchairat N, Agarwala S, et al. Sessile Droplets Containing Carbon Nanotubes:A Study of Evaporation Dynamics and CNT Alignment for Printed Electronics. Nanoscale. 2019;11:10603–14. doi: 10.1039/c9nr03261d. https://doi.org/10.1039/c9nr03261d. [DOI] [PubMed] [Google Scholar]
- 60.Birdi K, Vu D. Wettability and the Evaporation Rates of Fluids from Solid Surfaces. J Adhes Sci Technol. 1993;7:485–93. https://doi.org/10.1163/156856193x00808. [Google Scholar]
- 61.Hu H, Larson RG. Evaporation of a Sessile Droplet on a Substrate. J Phys Chem B. 2002;106:1334–44. [Google Scholar]
- 62.Maeno E, Takahashi N, Okada Y. Dysfunction of Regulatory Volume Increase is a Key Component of Apoptosis. FEBS Lett. 580:6513–7. doi: 10.1016/j.febslet.2006.10.074. https://doi.org/10.1016/j.febslet.2006.10.074. [DOI] [PubMed] [Google Scholar]


