SUMMARY
The immune checkpoint receptor PD-1 on T follicular helper (Tfh) cells promotes Tfh:B cell interactions and appropriate positioning within tissues. Here we examined the impact of regulation of PD-1 expression by the genomic organizer SATB1 on Tfh cell differentiation. Vaccination of CD4CreSatb1f/f mice enriched for antigen-specific Tfh cells, and TGF-β-mediated repression of SATB1 enhanced Tfh differentiation of human T cells. Mechanistically, high Icos expression in Satb1−/− CD4+ T cells promoted Tfh cell differentiation by preventing T follicular regulatory cell skewing, and resulted in increased isotype-switched B cell responses in vivo. Ovarian tumors in CD4CreSatb1f/f mice accumulated tumor antigen-specific, LIGHT+CXCL13+IL-21+ Tfh cells and tertiary lymphoid structures (TLS). TLS formation decreased tumor growth in a CD4+ T cell and CXCL13-dependent manner. Transfer of Tfh cells, but not naïve CD4+ T cells, induced TLS at tumor beds and decreased tumor growth. Thus, TGF-β-mediated silencing of Satb1 licenses Tfh cell differentiation, providing insight into the genesis of TLS within tumors.
Keywords: Tertiary Lymphoid Structure, Tumor Immunology, B cell cancer, T Follicular Helper Cell, SATB1, Immuno-Oncology
Graphical Abstract

eTOC
Tertiary lymphoid structures (TLS) are associated with improved outcomes for cancer patients. Chaurio et al. demonstrate that TGF-β-mediated regulation of the genomic organizer Satb1 governs the differentiation of T follicular helper (Tfh) cells and T follicular regulatory cells. Loss of Satb1promotes Icos expression and thereby Tfh cell differentiation, resulting in the assembly of intra-tumoral TLS.
INTRODUCTION
Immuno-oncology today is sharply focused on T cells. However, B cells can present antigens and provide co-stimulation to T cells and coordinated T and B cell activation needs to occur to maximize immune protection. In the tumor microenvironment, T and B cells frequently interact (Biswas et al., 2021), but cooperation between humoral and cellular responses remains incompletely understood (Conejo-Garcia et al., 2020). T follicular helper (Tfh) cell-driven B cell responses play a role in the effectiveness of checkpoint inhibitors (Hollern et al., 2019). However, the drivers of Tfh conversion at tumor beds remain unknown. Some T:B cell intra-tumoral interactions take place in highly organized conglomerates that recapitulate the architecture of secondary lymph nodes, termed tertiary lymphoid structures (TLS) (Sautes-Fridman et al., 2019). Similar to TLS in autoimmune conditions, some intra-tumoral TLS contain germinal centers with interdigitating follicular dendritic cells, plus an adjacent T cell zone composed by CD4+ and CD8+ lymphocytes and high endothelial venules (Sautes-Fridman et al., 2019). The presence of TLS is associated with positive outcomes in multiple human malignancies, including ovarian (Iglesia et al., 2014; Kroeger et al., 2016; Montfort et al., 2017), pancreatic (Hiraoka et al., 2015), colorectal (Di Caro et al., 2014), bladder (Pfannstiel et al., 2019) and non-small cell lung cancer (Silina et al., 2018).
Because TLS are rarely found in mouse models, the sequence of events leading to the assembly of these structures remains incompletely understood. Nonetheless, a conducive chemokine niche, able to attract diverse hematopoietic components is generally accepted as a seminal event in TLS formation. Accordingly, delivery of the cytokine LIGHT promotes TLS assembly in vivo by inducing the production of CCL21 by tumor endothelial cells (Johansson-Percival et al., 2017). Additional studies also underscore the importance of other chemokines driving the recruitment of the immune cells that partake in the build-up of TLS during dendritic cell: T cell interactions. Mulé and colleagues, for instance, identified a 12-chemokine signature across multiple TLS+ human tumors (Messina et al., 2012), while seminal studies by the Engelhard and Storkus laboratories support the importance of lymphotoxin produced by effector lymphocytes, on the one hand, and IL-36 produced by dendritic cells, on the other, as drivers of the recruitment of the specialized cellular types that interact to generate TLS (Peske et al., 2015) (Weinstein and Storkus, 2015). Fibroblast activation is also important for TLS genesis (Buckley et al., 2015).
While the orchestration of the right cytokine and chemokine milieu seems to be crucial for TLS assembly, Tfh cells must be also generated for the formation and maintenance of germinal centers (Crotty, 2014, 2019). BCL6+ Tfh cells represent a different lineage of CD4+ T cells that secrete CXCL13, a marker of germinal center activity that attracts CXCR5+ B cells (Gu-Trantien et al., 2013; Havenar-Daughton et al., 2016), as well as IL-21, which elicits proliferation and maturation of B cells (Walters and Vinuesa, 2016). In turn, B cells act as antigen-presenting cells for CD4+ T cells initially primed for Tfh cell differentiation (Crotty, 2019). Proper priming of CD4+ T cells to skew their differentiation into Tfh cells could therefore be at the genesis of TLS assembly.
High expression of the immune checkpoint receptor PD-1 in Tfh cells is required for their positioning within tissues and fostering their interaction with B cells, which takes place through PD-L1:PD-1 interactions and CXCR3 repression (Shi et al., 2018). We have shown that the genomic organizer SATB1 governs epigenetic repression of PD-1 in T cells, and that PD-1 expression is induced downstream of TGF-β signaling by SMAD3-mediated repression of Satb1 expression (Stephen et al., 2017). SATB1 creates loops in heterochromatin and serves as a hub for the recruitment of epigenetic modifiers in multiple immune cell types, thus driving their phenotype and, subsequently, inflammatory and immune responses (Cai et al., 2006; Kakugawa et al., 2017; Kitagawa et al., 2017; Naik and Galande, 2019; Stephen et al., 2017; Tesone et al., 2016).
Interestingly, Tfh cells express lower amounts of SATB1 (Georgiev et al., 2018). Given the importance of PD-1 in Tfh:B cell interactions and the role of SATB1 in PD-1 repression, we investigated how SATB1 expression regulates Tfh cell differentiation and, subsequently, TLS formation at tumor beds. Deletion of SATB1 licensed Tfh cell differentiation and the induction of protective isotype-switched antibody responses. Enhanced Tfh differentiation and abrogated regulatory T cell (Treg) conversion of Satb1−/− CD4+ T cells resulted in spontaneous formation of intra-tumoral TLS and decreased tumor growth. Transfer of Tfh cells to tumor-bearing mice was sufficient to elicit TLS formation at tumor beds. Our findings provide insight into TLS assembly in cancer, and suggest that the TGF-b-SATB1axis may be manipulated to enhance anti-tumor immunity.
RESULTS
Repression of Satb1 licenses activated CD4+ T cells for effective Tfh cell differentiation
To define the role of the genomic organizer Satb1 in Tfh cell differentiation, we first vaccinated >4 week-old CD4CreSatb1f/f mice with ovalbumin (OVA). In this model, OVA-reactive T cells progress through thymic selection and only a slight reduction in the proportions of T cell splenocytes is found in adult mice, with no difference in proliferative responses (Stephen et al., 2017) and recovery of regulatory T cell (Treg) function in adult mice (Kitagawa et al., 2017). Strikingly, >3-fold increases in the proportions of CD3+CD4+PD-1hiCxcr5hiBcl6+ Tfh cell subsets were consistently found in mice with Satb1−/− T cells in both spleens (Figure 1A) and inguinal lymph nodes (Figure 1B) in independent experiments.
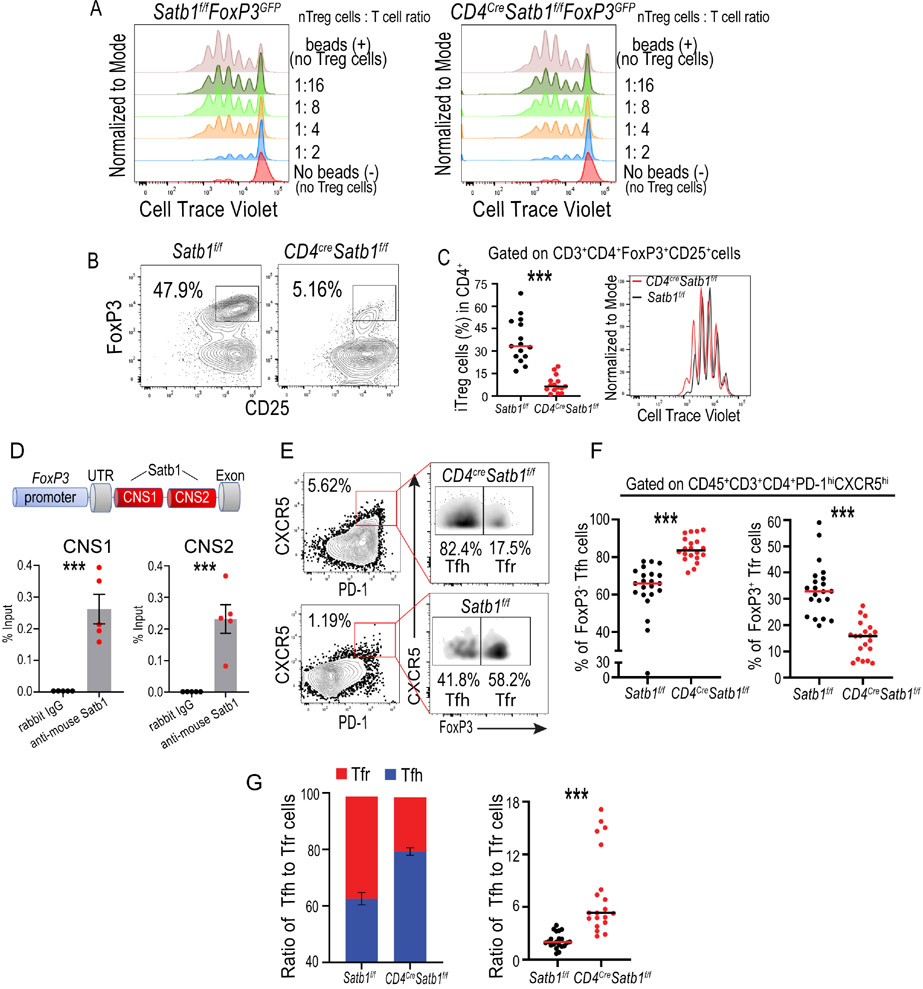
Figure 1. Repression of Satb1 licenses activated CD4+ T cells for effective Tfh cell differentiation.
(A, B) Quantification of the proportions of Tfh cells in the spleens (A) and inguinal lymph node (B) of OVA-vaccinated CD4CreSatb1f/f mice and Satb1f/f control littermates (n≥20), after 14 days. (C) Corresponding Satb1-dependent increases in OVA antigen-specific Tfh cells obtained from Ly5.1+ mice after transfer of purified naïve OT-II Satb1f/f (n=14) or OT-II CD4CreSatb1f/f (n=15) T cells, followed by immunization with NP-OVAL and analysis 5 days later. (D, E) Production of IL-21 (n=9) (D) and CXCL13 (n=13) (E) quantified through intracellular flow cytometry staining after stimulation with PMA (100 ng/mL), ionomycin (0.5 μg/mL) and brefeldin A (10 μg/mL) for 6 hours in CD3+CD4+Foxp3−CD62L−CD44+PD1hiCXCR5+ Tfh and naïve CD3+CD4+Foxp3−CD62L+CD44− cells from the spleens of vaccinated mice as in A&B. (F) Serum levels of CXCL13 in OVA-vaccinated mice as in A&B. (G) Production of LIGHT/TNFSF14 quantified as in D&E (n=11). (H) Differential OVA-specific titers of total IgG responses in the serum of Satb1+/Satb1− (n=12) vaccinated mice as in A&B. Serum from naïve mice were used as control (n=6). (I) Quantification by flow cytometry of surface IgG1 and the GL7 and FAS markers of germinal center activity in splenic CD19+B220+ B cells in CD4CreSatb1f/f mice and littermates from A. Data are presented as median. A-E, G and H are pooled from ≥2 independent experiments. F&I are representative of at least three independent experiments. *p<0.05, **p≤0.01, ***p≤0.001.
These responses were antigen-specific because corresponding Satb1-dependent increases in Tfh cell differentiation were identified in (OVA-specific) OT-II+ T cells injected at the time of immunization (Figure 1C). In contrast, although we observed a trend towards basal increases in Tfh cells in CD4creSatb1f/f mice, Satb1−/− antigen-specific (OT-II) cells did not proliferate in the absence of the OVA-antigen (Suppl. Figure 1A&B).
Differentiated lymphocytes were functional and not merely expressing Tfh cell surface markers, as they produced more IL-21 and CXCL13, compared to naïve and Tfh cells in identically vaccinated Satb1-competent mice; both at the protein (Figure 1D&E) and mRNA abundance (Suppl. Figure 1C). Accordingly, higher levels of serum CXCL13, a marker of germinal center activity (Gu-Trantien et al., 2013; Havenar-Daughton et al., 2016), were also found in the serum of OVA-vaccinated mice carrying Satb1−/− T cells (Figure 1F). Interestingly, we also found that (Satb1-competent) Satb1f/f and Satb1−/− CD4+ Tfh cells produced similar levels of the Light/TNFSF14 cytokine (Figure 1G), which is sufficient to trigger TLS formation in cancer through the activation of tumor endothelium (Johansson-Percival et al., 2017). Furthermore, increases in the titers of serum OVA-specific IgG were identified in CD4CreSatb1f/f mice, compared to Satb1+ littermates (Figure 1H). Hence, upon OVA immunization, CD19+CD20+ B cells from CD4CreSatb1f/f mice showed higher proportions of the cell surface IgG1 isotype compared to control littermates, along with positive expression of FAS and GL7, markers of germinal center activity (Williams et al., 2015), (Figure 1I).
Together, these results indicate that Satb1 silencing in CD4+ T cells intrinsically promotes their differentiation into functional antigen-specific Tfh cells, resulting in a chemokine milieu conducive of B-cell recruitment and activation and, accordingly, antigen-specific, isotype-switched humoral responses.
Decreased expression of Satb1 promotes Tfh differentiation through Icos de-repression and decreased T follicular regulatory cell (Tfr) formation
To define possible drivers of SATB1 downregulation during Tfh cell differentiation, we focused on TGF-β, known to decrease Satb1 expression in T cells (Stephen et al., 2017) while also promoting Tfh cell polarization (Schmitt et al., 2014). As shown in Figure 2A&B, under skewing conditions that included known drivers of Tfh vs. Th17 cell differentiation (Schmitt et al., 2014), human CD4+ T cells differentiated more effectively into CXCR5+PD-1hiICOS+BCL6+ Tfh cells in the presence of TGF-β (Crotty, 2014; Marshall et al., 2015; Schmitt et al., 2014), with a consistent reduction of SATB1 (Figure 2C&D). Challenging the notion that TGF-β is constantly immunosuppressive in cancer (Park et al., 2016; Stephen et al., 2017; Stephen et al., 2014), ovarian tumors from the TCGA database expressing higher levels of TGFB exhibited concurrent increases not only in markers of protective humoral responses (Biswas et al., 2021), (Suppl. Figure 2A); but also in markers of denser T cell infiltration (CD3ε, CD8α) or enhanced cytolytic activity (IFNG, PRF1); (Suppl. Figure 2B).
Figure 2. TGF-β-mediated repression of SATB1 expression promotes Tfh cell phenotypes.
Representative dot-plots (left) and histograms (right) of the effect of TGF-β on human Tfh cell differentiation and SATB1 repression, respectively. Increased in vitro expression of human CXCR5+PD-1hi Tfh cells markers from peripheral naïve CD4+ T cells in the presence of TGF-β, under different Tfh cell skewing conditions: (A) (lower panel) hIL-23 plus hIL-10; (upper panel) hIL-23; (B) (lower panel) hIL-12 plus hIL-10; (upper panel) hIL-12. (C) TGF-β-dependent Tfh cell differentiation was associated with lower levels of SATB1 (D), as determined by intracellular staining. A-D data (n=3) are representative of two independent experiments with similar results and represent mean ± SEM. *p<0.05, **p≤0.01, ***p≤0.001.
To understand how Satb1 down-regulation licenses Tfh cell differentiation, we first observed that Satb1−/− CD4+ T cells from OVA-vaccinated mice exhibited higher levels of ICOS, a crucial signal for Tfh cell formation (Crotty, 2014) (Figure 3A-C). Supporting a direct role for Satb1 in repressing Icos, chromatin immuno-precipitation (ChIP) experiments identified occupancy of Satb1 around 60 bp upstream of the Icos Transcription Start Sites (Figure 3D), consistent with the silencing activity of Satb1 on other genes (Stephen et al., 2017).
Figure 3. Decreased expression of Satb1 promotes Tfh cell differentiation by derepressing Icos.
(A) Higher expression of Icos in Satb1−/− CD4+ T cells (n=11). (B) Higher proportions of Icos+CD4+ T cells among lymphocytes in the absence of Satb1 (n≥19). (C) Higher proportions of Icos+ Tfh cells after OVA vaccination in the presence of Satb1−/− CD4+ T cells (n=22). (D) (Left) Schematic depiction of Satb1 occupancy at the Icos promoter. Location of primers used for ChIP-PCR is indicated. (Right) Chromatin was immunoprecipitated with anti-Satb1 or control IgG from negatively immunopurified Satb1-competent mouse CD4+ T cells. Percent of input before immunoprecipitation (2.5% input values) was quantified by Real-Time Q-PCR (n=7). (E) anti-Icos / IgG (200 μg/mouse) was administered intraperitoneally in congenic Ly5.1+ mice reconstituted with OT-II-Satb1f/f vs. OT-II-CD4CreSatb1f/f splenocytes followed by immunization with NP-OVAL in alum, and Tfh cell differentiation was quantified in spleens 5 days later (n≥12). Data are presented as median and pooled from ≥2 independent experiments. **p≤0.01, ***p≤0.001.
As expected from the role of ICOS signaling in Tfh cell differentiation, ICOS blockade in OVA-vaccinated mice reduced, but did not fully eliminate, Satb1-dependent differences in the proportions of OVA-specific Tfh cells (Figure 3E).
To identify additional mechanisms by which Satb1 silencing could promote Tfh cell differentiation, we also investigated the role of Satb1 in the formation of PD-1hiCXCR5+FoxP3+ Tfr cells, which inhibit the function of Tfh cells (Sage et al., 2013; Sage et al., 2016) and, through neuritin, B cells (Gonzalez-Figueroa et al., 2021). In agreement with previous reports (Kitagawa et al., 2017), natural Treg function was preserved in adult CD4CreSatb1f/f mice (Figure 4A). However, the capacity of CD3+CD4+Foxp3−CD62L+CD44− naïve T cells sorted from CD4CreSatb1f/fFoxF3GFP mice to differentiate into FoxP3+ Treg cells under skewing conditions was dramatically reduced, while their Satb1-competent counterparts effectively acquired FoxP3 (Figure 4B). Lack of Treg conversion cannot be attributed to a proliferation defect (Stephen et al., 2017), as similar cell counts and Cell-Trace Violet dilution were found under differentiation conditions for both populations (Figure 4C). Mechanistically, ChIP-PCR demonstrated that Satb1 bound to both the CNS1 and CNS2 intronic enhancers in the FoxP3 gene (Figure 4D), originally identified by the Rudensky laboratory (Zheng et al., 2010). Accordingly, the formation of PD-1hiCXCR5hiBCL6+FoxP3+ Tfr cells was reduced in OVA-vaccinated mice carrying Satb1−/− T cells, with higher ratios of Tfh to Tfr cells, compared to Satb1-competent littermates (Figure 4E-G).
Figure 4. Satb1 is required for the generation of induced regulatory T cells.
(A) Natural CD3+CD4+FoxP3+ Treg from the spleens of adult (week 10) Satb1−/− mice effectively suppressed CD3/CD28-induced T cell splenocyte activation in a dose-dependent manner. Representative of two independent experiments. (B) CD3+CD4+Foxp3−CD62L+CD44− naïve T cells sorted from CD4CreSatb1f/fFoxP3GFP mice or control Satb1f/fFoxP3GFP littermates were in vitro differentiated to Treg cells for 3 days. Conversion into FoxP3+ Treg cells was compromised in CD4CreSatb1f/fFoxP3GFP cells. (C) (Left) Satb1-dependent differences in Treg conversion (n=15); (Right) Similar proliferation rates were observed under differentiation conditions for Satb1+/Satb1− populations. (D) ChIP-PCR for the CNS1 (left panel) and CNS2 (right panel) enhancers in Foxp3. Chromatin was immunoprecipitated with anti-Satb1 or control IgG from FoxP3+ Treg cells, in vitro differentiated from naïve Satb1-competent CD4+ T cells. Percent of input before immunoprecipitation (2.5% input values) was quantified by Real-Time Q-PCR (n=5). Mean and SEM are shown. (E) Representative dot plots showing the gating strategy for Tfh and Tfr cells in OVA-vaccinated mice carrying Satb1−/− T cells, compared to Satb1-competent littermates. (F) Reduced generation of PD-1hiCXCR5+Foxp3+ Tfr cells with corresponding increases in FoxP3− Tfh cells (n≥20). (G) Higher ratios Tfh to Tfr cells in Satb1−/− T cells compared to control littermates (n≥20). Data are presented as median and pooled from ≥2 independent experiments. ***p≤0.001.
Taken together, these results demonstrate that Satb1 governs CD4+ T cell differentiation into Treg/Tfr and Tfh cells in opposite ways: On the one hand, although Satb1 is dispensable for natural Treg-dependent tolerance in adult mice (Kitagawa et al., 2017), probably due to constitutive de-methylation of CNS2 in thymic Treg, it is required for efficient Treg conversion. On the other hand, Satb1 represses the Icos promoter. Accordingly, Satb1 silencing licensed Tfh cell formation while preventing the generation of Tfr cells.
Satb1-dependent Tfh differentiation drives enhanced B cell responses and TLS formation in orthotopic ovarian tumors
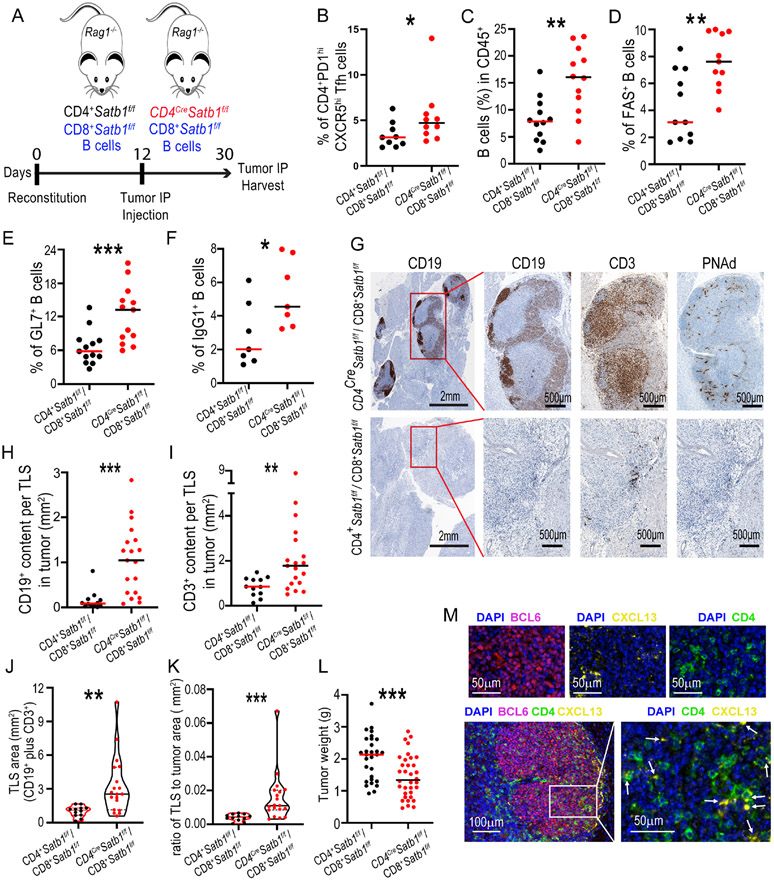
To understand the consequences of enhanced Tfh cell formation in Satb1−/− T cells in cancer, we challenged CD4CreSatb1f/f mice and Satb1-competent littermates with intraperitoneal UPK10 tumors, a mildly immunogenic model of p53/KRas-driven ovarian carcinosarcoma that progresses as solid tumor masses in peritoneal lining (Cubillos-Ruiz et al., 2015; Rutkowski et al., 2015; Scarlett et al., 2012; Tesone et al., 2016). As expected, splenic (Figure 5A) and intratumoral (Figure 5B) CD3+CD4+PD-1hiCXCR5hiBCL6+ Tfh cells accumulated in greater proportions when Satb1 was specifically absent in T cells, which resulted in higher Tfh to Tfr cell ratios in the absence of Satb1 expression in T cells (Suppl. Figure 2C&D). As we observed in the OVA-vaccinated mice, Satb1−/− Tfh cells expressed higher levels of ICOS in both spleen (Suppl. Figure 2E&F) and at tumor beds (Figure 5C&D). Interestingly, intraperitoneal tumors from CD4CreSatb1f/f mice were strongly infiltrated by CD45+CD19+B220+ B cells (Suppl. Figure 2G). Moreover, the accumulation of isotype-switched B cells both in the spleen and tumor beds was enhanced in CD4CreSatb1f/f mice (Figure 5E&F), indicative of enhanced humoral responses. Accordingly, a higher anti-tumor humoral (IgG1) response was observed in serum from CD4CreSatb1f/f tumor-bearing mice, compared to the Satb1-sufficient littermates (Suppl. Figure 3A&B). These responses appear to be maintained at tumor beds, because immunization of naïve mice with irradiated tumor cells did not increase the titer of circulating antibodies (Suppl. Figure 3C).
Figure 5. Satb1-dependent Tfh cell differentiation promotes B cell responses and TLS formation in ovarian cancer.
(A, B) CD4CreSatb1f/f mice accumulated more Tfh cells in the spleen (A) and at UPK10 intraperitoneal tumor beds (B), compared to control Satb1f/f mice; (n≥16). (C) Higher ICOS expression in intra-tumoral Satb1−/− Tfh cells (n=17) and controls (n=18). (D) Corresponding increases in the accumulation of ICOS+CD4+ T cells at tumor beds (n=15). (E, F) Increased accumulation of isotype-switched B cells in the spleen (E) and at tumor beds (F) in CD4CreSatb1f/f mice, compared to Satb1-competent controls; (n=12). (G) B cell depletion using anti-B220 antibodies IP (200 μg/mouse; days −2, 0, 2, 4, 6 after tumor challenge) accelerates UPK10 tumor growth in Satb1f/f mice compared to isotype control (n≥9). (H) Spontaneous large PNAd+ TLS formation in intraperitoneal tumors in CD4CreSatb1f/f mice (top), compared to Satb1f/f mice (bottom). Representative of ≥3 independent experiments. (I) Quantification of TLS (defined as adjacent conglomerates of T cells and B-cells at tumor beds), based on the area covered by CD19+ cells plus the area covered by CD3+ cells). (J) Ratio of the area occupied by TLS to the entire tumor section. For I&J: Each point represents a single TLS in the CD4CreSatb1f/f (n=9 mice) and Satb1f/f n=11 mice) groups. (K) TLS assembly was abrogated upon CXCL13 blockade using anti-CXCL13 antibodies IP (150 μg/mouse; days 3, 6, 9 after tumor challenge). (L) Quantification of TLS after CXCL13 blocking as performed in I (2 independent experiments). (M) (Upper panel) Schematic depiction of the experiment. A chunk of IP tumor from a donor mouse was isolated and s.c. transplanted to a naïve recipient. (Lower panel) TLS undergo major changes in the distribution of T and B cells over time. Data are presented as median and pooled from ≥2 independent experiments. *p<0.05, **p≤0.01, ***p≤0.001.
As in ovarian cancer patients (Biswas et al., 2021; Montfort et al., 2017), B cell responses in our model were associated with immune protection, because B cell depletion using anti-B220 antibodies accelerated UPK10 tumor growth (Figure 5G). Interestingly, depletion of CD8+ T cells in UPK10 tumor-bearing mice also accelerated malignant progression (Suppl. Figure 3D), which supports that both arms of the adaptive immune system work in coordination to achieve superior immune pressure against the progression of human ovarian cancer (Biswas et al., 2021).
We next assessed tumors histologically to gain deeper insight in the formation of TLS. Strikingly, we found enhanced spontaneous formation of TLS in intraperitoneal tumors in CD4CreSatb1f/f mice, in multiple independent experiments. We defined those structures as >0.1 mm2 conglomerates of B cells adjacent to accumulations of T cells, clearly identified inside tumor masses and co-stained for peripheral node addressin (PNAd), with a central core of T cells (Figure 5H). These structures contained CD21+ follicular dendritic cells, BCL6+ B cells (Suppl. Figure 4A), and CD23, a marker representative of fully mature secondary-follicle-like TLS (Suppl. Figure 4B). The presence of TLS correlates with higher densities of B cells and a trend towards increased T cell infiltration outside TLS (Suppl. Figure 3E).
TLS assembly was not restricted to UPK10 tumors, as it was also obvious in intraperitoneal Brpkp110 tumors (Allegrezza et al., 2016) (Suppl. Figure 4C). Supporting previous reports on spontaneous TLS assembly in peritoneal tumors (Engelhard et al., 2018; Rodriguez et al., 2018), we also found scattered conglomerates of T and B cells in tumors growing in Satb1-competent mice, although they were less frequent, poorly organized and smaller (Figure 5H-J). Importantly, TLS assembly was dependent on CXCL13, because CXCL13 blockade abrogated TLS formation in CD4CreSatb1f/f intraperitoneal tumor-bearing mice (Figure 5K&L), with higher tumor growth (Suppl. Figure 3F).
Enhanced tumor immunogenicity also promoted TLS formation, because intraperitoneal UPK10 tumors expressing low levels of OVA (UPK10-OVAlo, quasi-universally orchestrated these structures in WT mice (Figure 5M).
To define possible changes in the distribution of T and B cells in TLS over time, we transplanted fragments of intraperitoneal OVAlo UPK10 tumors into the flank of naïve WT mice. As shown in Figure 5M, the central distribution of T cells in early-stage TLS changed to the periphery, as B cells occupied the TLS center over time, which is the structure more frequently observed in advanced human tumors (Cillo et al., 2020). Together, these results indicate that increased differentiation of Tfh cells in cancer drives enhanced humoral responses in ovarian cancer, which promotes spontaneous orchestration of TLS where T and B cells dynamically interact at tumor beds.
TLS assembly is driven by antigen-specific Tfh cells and delays ovarian cancer progression
A limitation of the CD4CreSatb1f/f model is that deregulation of PD-1 expression abrogates the effector activity of tumor-reactive CD8+ T cells (Stephen et al., 2017). To segregate the role of Satb1 specifically in CD4+ T cells, and to avoid contamination of secondary lymphoid organs, we reconstituted Rag1−/− mice with a mix of syngeneic Satb1+ wild-type CD8+ T cells and B cells, plus either Satb1+ or Satb1−/− CD4+ T cells. Both cohorts of mice were subsequently challenged with UPK10 intraperitoneal tumors and followed for malignant progression (Figure 6A), Tfh cell differentiation and potential anti-tumor B cell responses. As expected, increases in CD3+CD4+CXCR5+PD-1hiBCL6+ Tfh cells were found at both spleens (Suppl. Figure 5A) and tumor beds (Figure 6B) in mice reconstituted with Satb1−CD4+ T cells. Likewise, increased Tfh cell differentiation translated into increased B cell accumulation at both spleens (Suppl. Figure 5B) and in the tumor (Figure 6C), including increases in CD19+ cells expressing markers of germinal center activity, such as FAS (Figure 6D & Suppl. Figure 5C) and GL7 (Figure 6E & Suppl. Figure 5D). These responses were associated with corresponding increases in isotype-switched B cells (Figure 6F & Suppl. Figure 5E). Notably, enhanced Tfh cell conversion in the absence of Satb1 was sufficient to induce the orchestration of large TLS, which again at this early stage exhibited a central core of T cells surrounded by B cells (Figure 6G), while small conglomerates of T- and B-cells were found in mice reconstituted with Satb1-competent CD4+ T cells (Figure 6G-K).
Figure 6. Tfh cell-driven, TLS-associated humoral responses are associated with delayed malignant progression in ovarian cancer.
(A) Schematic depiction of the experiment. Rag1−/− mice were reconstituted with a mix of syngeneic wild-type CD8+ T cells, wild-type B-cells and either Satb1-competent or Satb1−/− CD4+ T cells. Mice were subsequently challenged with UPK10 intraperitoneal tumors and followed for malignant progression and humoral responses. (B-F) Flow cytometry quantification of (B) Tfh cells, (C) CD19+ B cells (n=12); (D, E) Germinal center markers FAS (n=11) and GL7 (n=13), and (F) B cell isotype switching at tumor beds in reconstituted Rag1−/− mice (n=7). (G) Spontaneous orchestration of large PNAd+ TLS in intraperitoneal tumors in Rag1−/− mice reconstituted with CD4CreSatb1f/f CD4+ T cells (Upper panel), but not upon reconstitution with Satb1f/f control CD4+ T cells (Lower panel). (H, I) Intra-tumoral TLS, defined as adjacent conglomerates (>0.1mm2) of CD3+ T cells and CD19+ B-cells at tumor beds, were quantified based on the (H) B cell content (CD19+ area) and (I) T cells (CD3+ area) inside the structure. (J) At least 60% of the Satb1−/− CD4+ T cells reconstituted mice, developed large TLS (>2 mm2) quantified as CD19+ plus CD3+ area. (K) Ratio of the area occupied by TLS to the entire tumor section. (L) Differences in IP tumor burden for the mice depicted in A. Each point represents the sum of total tumor content in a single mouse (n≥20). (M) Multiplex immunofluorescence for CD3+CD4+BCL6+CXCL13+ Tfh cells inside intra-tumoral TLS. G&M are representative of ≥3 independent experiments. From graphics H to K (n=10 mice) for both CD4CreSatb1f/f and Satb1f/f experimental groups. For J&K each point represents a single TLS. *p<0.05, **p≤0.01, ***p≤0.001.
TLS in mice reconstituted with Satb1-defective CD4+ T cells also contained CD21+ dendritic cells, BCL6+ B-cells (Suppl. Figure 6A&B) and CD23+ cells (Suppl. Figure 6C) that characterize bona fide lymphoid organs, and were infiltrated by CD3+CD4+BCL6+ Tfh cells producing CXCL13 (Figure 6M), which chemoattracts CXCR5+ B cells.
CD4+ T cell-driven B cell responses were again associated with a reduction in tumor growth, further supporting the protective role of humoral activity in ovarian cancer (Figure 6L & Suppl. Figure 5F). Because the only difference in this experiment is the expression of Satb1 in CD4+ T cells, these results indicate that Tfh cells are crucial for TLS formation and pressure against malignant progression.
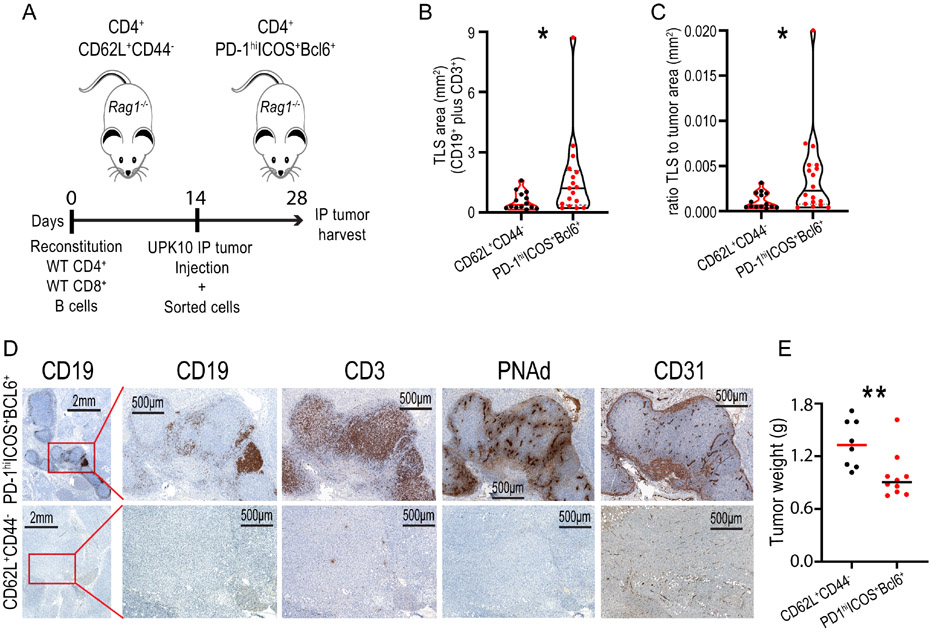
To confirm that Tfh cell conversion is sufficient to drive TLS assembly in cancer, we reconstituted Rag1−/− mice with WT T- and B-cells, and challenged them with UPK10 tumors, admixed with either CD3+CD4+PD-1hiICOS+Bcl6+ Tfh cells (Suppl. Figure 6D) or naïve CD3+CD4+CD62L+CD44− T lymphocytes from the UPK10 tumor-draining inguinal lymph nodes from a different cohort of mice (Figure 7A). The addition of Tfh cells resulted not only in more TLS formation (Figure 7B&C), but also promoted the orchestration of large TLS (>2 mm2) in 50% of mice, while no TLS of this size was found in tumors developed in the presence of naïve control CD4+ T cells from the same donors (Figure 7D). Tfh cell-induced TLS exhibited CD31+PNAd+ HEVs, primarily located in the B-cell area of the germinal center (Figure 7D). Finally, tumors in mice with TLS induced by Tfh cell transfer grew less, compared to those developed in mice receiving control CD4+ T cells (Figure 7E). Together, these results further support that Satb1-dependent conversion of Tfh cells is the genesis of TLS formation in cancer, and open new avenues for eliciting TLS formation in unresectable tumors.
Figure 7. Satb1-dependent Tfh cells trigger the formation of large TLS at tumor beds.
(A) Scheme of the experiment: Rag1−/− mice were reconstituted with a mix of syngeneic wild-type CD4+ and CD8+ T cells, and wild-type B-cells. Fourteen days later, mice were subsequently challenged with UPK10 intraperitoneal tumors, admixed with either CD3+CD4+PD-1hiICOS+(BCL-6+) Tfh cells or CD4+CD3+CD62L+CD44− naïve T cells, sorted from the tumor-draining inguinal lymph nodes of syngeneic tumor-bearing donor mice. TLS assembly was assessed after 14 days. BCL-6 expression was verified in sorted populations independently. (B) Quantification of total TLS area (TLS defined as adjacent conglomerates of CD3+ T cells and CD19+ B-cells) at tumor beds. (C) Ratio of the area occupied by TLS to the entire tumor section. (D) (Upper panel) Representative IHC of adjacent tumor sections showing large TLSs in Rag1−/− mice reconstituted with Tfh cells, consisting of defined areas of CD19+ and CD3+ aggregations with corresponding PNAd and CD31 positive signal, and its controls (Lower panel). (E) Differences in IP tumor growth for the mice depicted in A. Each point represents the sum of total tumor content in a single mouse (n≥8). Pooled from two independent experiments. For B&C each point represents a single TLS for both Tfh cell (n=4 mice) and naïve T cell (n=4 mice) groups. Data are presented as median and representative of two independent experiments with similar results. *p<0.05, **p≤0.01.
DISCUSSSION
We found that decreased expression of Satb1 in CD4+ T cells licensed antigen-specific Tfh cell differentiation through Icos de-repression and by impairing Tfr cell formation, driving isotype-switched antibody responses. Enhanced generation of Tfh cells created a chemokine niche that promoted spontaneous assembly of TLS at tumor beds, while Tfh cells were sufficient to elicit TLS formation in the tumor microenvironment.
SATB1 is a genomic organizer with multiple activities in different immune cell types (Cai et al., 2006; Kakugawa et al., 2017; Kitagawa et al., 2017; Naik and Galande, 2019; Stephen et al., 2017; Tesone et al., 2016). By creating loops in heterochromatin and recruiting epigenetic modifiers to enhancers and promoters, SATB1 governs complex phenotypes that go beyond the narrower range of genes typically regulated by individual transcription factors. Our results unveil Icos as a direct target of Satb1-dependent repression. In addition, Treg cell conversion was abrogated when CD4+ T cells lack Satb1. While Sakaguchi and colleagues identified an initial defect in thymic Treg in the absence of Satb1 specifically in T cells, differences in immunosuppressive activity are corrected in adulthood (Kitagawa et al., 2017). In contrast, our results show that adult CD4+ T cells remained unable to differentiate into FoxP3+ cells without Satb1 under TGF-β-dependent skewing conditions, suggesting a different role in thymic versus peripheral Treg cells. It is likely that the binding of Satb1 to the intronic enhancers of FoxP3 (Kitagawa et al., 2017) becomes irrelevant once DNA de-methylation has been established in natural Treg cells, but remains necessary for FoxP3 expression in converted Treg lymphocytes. Nevertheless, our study unveils that the combined activities of SATB1 include a negative role in Tfh cell differentiation, along with other previously identified genetic repressors (Wang et al., 2014), and show that SATB1 is a necessary player in Treg conversion.
Another key finding of our study is that Tfh cells produced the cytokine milieu that is at the genesis of intratumoral TLS. Tfh cells were consistently located at the central core of TLS, which we propose represented the initial stage of TLS formation, before germinal centers changed this architecture. Other studies point to fibroblasts as pivotal drivers of TLS formation at the earliest phases of TLS establishment, preceding lymphocyte infiltration in non-tumor tissues (Buckley et al., 2015; Nayar et al., 2019). The importance of fibroblasts in cancer TLS is supported by studies from the Mulé group, where a lymph node-derived stromal cell line induced the formation of TLS in immunogenic tumor models (Zhu et al., 2018). Fibroblast reticular cells in secondary lymphoid organs produce CCL21, which mediates homing of T cells, but not B lymphocytes. Consistently, CCL21 has been included in a 12-chemokine signature that reliably identifies TLS in multiple human tumors (Messina et al., 2012). However, CCL21 production by different cell types (i.e., tumor endothelial cells) can be induced through signaling by the TNF-α family member cytokine LIGHT (Johansson-Percival et al., 2017). We found that Tfh cells were significant producers of LIGHT, so that Tfh cells at tumor beds could indeed provide the primordial signal for TLS assembly in cancer, besides their necessary role in the formation of germinal centers.
Another element found to be critical for TLS formation at tumor beds by other authors is lymphotoxin secreted by CD8+ T cells and NK cells, which acts on tumor blood vessels to transform them into “lymph node-like” vasculature (Peske et al., 2015). This recapitulates the importance of lymphotoxin derived from lymphoid tissue inducer cells (LTi) during the development of secondary lymphoid organs (Germain et al., 2015). However, we found that Tfh cells were substantial producers of the lymphotoxin homologous TNFSF14/LIGTH cytokine, while activated CD4+ T cells can substitute LTi cells in TLS generated in autoimmune conditions (Marinkovic et al., 2006), further supporting that Tfh cells are a crucial player in the assembly of primordial intratumoral TLS.
Overall, our results point to a model whereby activated CD4+ T cells that decrease expression of SATB1 in the tumor microenvironment (i.e., in response to TGF-β signaling) secrete LIGHT, which activates CCL21 production by endothelial cells and likely fibroblasts; CXCL13, which drives the recruitment of CXCR5+ B cells (Gu-Trantien et al., 2017); and IL-21, which activates B cells in situ. A crosstalk between T and B cells is established through ICOS:ICOS-ligand and CD40:CD40L interactions, while B cells sustain T cell activation through antigen presentation, eventually leading to the recruitment, differentiation and distribution of dendritic cells, high endothelial venules, fibroblasts, plasma cells, neutrophils and macrophages that compose canonical TLS in cancer. Intra-tumoral administration of autologous antigen-specific Tfh cells in metastatic cancers or tumors adhered to unresectable locations could therefore promote the generation of TLS where anti-tumor T cells could find a protective niche to exert immune pressure against the progression of advanced malignancies, including supporting existing immunotherapies.
Limitations of the study
Our study unveiled Tfh cell differentiation as a seminal event during the assembly of TLS in cancer. We cannot rule out that there are multiple ways of orchestrating TLS, but intratumoral Tfh administration was sufficient to elicit TLS formation. Since Tfh cells came from the lymph nodes of tumor-bearing mice, future studies should determine whether these lymphocytes need to be tumor antigen-specific or not. It will be also important to confirm these findings in other tumor context, as well as in human cancer patients.
We also show that CXCL13 blockade abrogated intratumoral TLS assembly. A caveat is that CXCL13 is also produced by stromal cells such as follicular dendritic cells and marginal reticular cells, so these effects cannot be exclusively attributed to Tfh cells.
Finally, we unveiled changes in the distribution of T and B cells during the progression of TLS in vivo. A limitation is that TLS evolved at a different anatomical location (the flank vs. the peritoneal cavity), which could drive specific architectural patterns.
STAR METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for supporting data and resources should be directed to and will be fulfilled upon reasonable request by the Lead Contact, Jose R. Conejo-Garcia, MD, PhD (jose.conejo-garcia@moffitt.org).
Materials availability
This study did not generate new unique reagents. The authors declare that all the results supporting the findings of this study are available within the paper and its figures.
Data and code availability
This paper does not report original code. Any additional information required to reanalyze the data reported, as well as raw image for TLS IHC studies, are available from the lead contact upon request.
EXPERIMENTAL MODELS
Mice and human cells
Healthy human buffy coats were obtained from LifeSouth (Gainesville, FL, USA). Studies were exempted from Institutional Review Board (IRB) approval.
Satb1−/− mice were generated in the Wistar Institute’s transgenic facility on a C57BL/6 background (Tesone et al., 2016). Satb1f/f mice were crossed with either CD4Cre mice (Taconic, 4196M) or FoxP3GFP (Jackson Labs.) to generate CD4CreSatb1f/f or Satb1f/fFoxP3GFP respectively. The CD4CreSatb1f/f mice were crossed with Satb1f/fFoxP3GFP to generate CD4CreSatb1f/fFoxP3GFP and control littermates. The OT-II CD4CreSatb1f/f mice were generated by crossing Satb1f/f mice with the OT-II (C57BL/6-Tg (TcraTcrb)425Cbn/Crl, strain code 643 (Charles River) to generate OT-II Satb1f/f in a C57BL/6 background. After three generations, OT-II Satb1f/f were crossed with CD4CreSatb1f/f to generate triple-transgenic OT-II CD4CreSatb1f/f and control littermates. 6–8-week-old female Rag1-deficient (Rag1−/−) mice of the same age group were procured from Jackson Laboratory. Mice were maintained by the animal facility of the Moffitt Cancer Center, with a 12 light/12 dark cycle, at 18–23 °C with 40–60% humidity and maintained in pathogen-free barrier facilities. All animal studies were reviewed and approved by the Institutional Animal Care and Use Committee at the University of South Florida Research Integrity and Compliance department.
METHOD DETAILS
In vivo tumor challenge
In vitro culture of UPK10 cells, a p53/KRas-driven ovarian carcinosarcoma cell line, was done in complete R10 medium defined as RPMI-1640 (Sigma), supplemented with L-glutamine (2 mM, Gen Clone), penicillin (100 U/mL, Lonza), streptomycin (100 μg/mL Lonza) and 10% fetal bovine serum (Sigma) at 37°C, 5% CO2. Unless otherwise indicated, UPK10-induced-intraperitoneal (IP) tumors were initiated by injecting 6 × 106 cells/mouse in PBS into the peritoneal cavity. Animals were monitored for health concerns and euthanized after 14 days. IP tumor contents were extracted for either tumor dissociation or prepared for immunohistochemistry analysis. For flow cytometry analysis, tumor tissues were dissected mechanically into single-cell suspensions in complete medium. UPK10 flank tumors were generated by subcutaneous injections of 107 cells in the posterior inferior flank of mouse. Animals were monitored and euthanized after 14 days. When necessary, inguinal lymph nodes were extracted for either flow cytometry analysis or cell sorting. Gross tumor mass was determined by weighing excised tissues in an analytical balance (Mettler Toledo) with necessary taring.
Mice reconstitution and immunizations
Rag1−/− mice were retro-orbitally reconstituted with a mix of syngeneic wild-type CD8+ T cells (1 × 106), wild-type B-cells (6 × 106) and either Satb1+ or Satb1−/− CD4+ T cells (2 × 106). Immunopurified mouse cells were isolated by using the CD8+ (19853), B (19854) and CD4+ (19852) negative selection kits (StemCell Technologies), respectively. Unless mentioned otherwise, immunization experiments were performed by a subcutaneous (s.c.) injection of Ovalbumin (albumin from chicken egg white; Sigma, A5503), at 50 μg into each flank resuspended in Complete Freund’s Adjuvant (Sigma), resulting in 100 μg per mouse.
In studies investigating antigen cellular-specific responses, 1 × 106 of either naïve OT-II CD4CreSatb1f/f or OT-II Satb1f/f cells, were isolated with the Mouse Naïve CD4+ T Cell negative selection Kit (StemCell Technologies: 9765), and retro-orbitally injected in the recipient B6 CD45.1 congenic mice (The Jackson Labs.). Immediately after, mice were IP immunized with 4-Hydroxy-3-nitrophenylacetyl Ovalbumin (NP-OVAL, Biosearch Technologies) (50 μg/mouse) in Imject™ Alum Adjuvant (ThemoFisher Scientific). Five days after immunization, mice were euthanized for spleen isolation and flow cytometry analysis. In the case of immunizations with ovarian tumor cells, (20 × 106) UPK10 cells were X-ray irradiated in PBS with a total dose of 40 Gy in a XRAD 160. Immediately after, cells were centrifuged, and IP injected in 0.3 ml PBS. Serums were isolated from whole blood after 14 days.
In vivo mouse antibodies for cells and cytokine depletion
B cells were depleted in vivo in Satb1f/f mice by IP injections (200 μg/mouse) of anti-mouse B220 (RA3.3A1/6.1 (TIB-146), BioXCell) vs. rat IgG isotype control (200 μg/mouse) 2 days before tumor challenge and every other day, at days 0, 2, 4 and 6. CD8+ T cells were in vivo depleted in Satb1f/f mice by IP injections (150 μg/mouse) of anti-mouse CD8α (YTS 169.4, BioXCell) vs. rat IgG2b (LTF-2) isotype control (150 μg/mouse) 2 days before tumor challenge and every two days, at days 0, 2, 4, 6 and 8. CXCL13 was blocked by IP injections (150 μg/mouse) of anti-mouse CXCL13 antibody (143614, R&D) vs. control rat IgG2A, every 3 days at days 3, 6 and 9 from the tumor challenge. All injections were done in 300 μl phosphate buffered saline (PBS).
Subcutaneous tumor implantation
Animals were prepared for surgery and anesthetized in according to the IACUC approved protocols. A 0.5 cm skin incision was made on the flank, 0.2-0.5 cm to the proximal femur of the animal. Freshly IP isolated tumor chunks (0.5x0.5 cm2) from a donor tumor bearing mouse were kept in PBS. Afterwards, a tumor piece per mouse flank was implanted subcutaneously, and the incision closed with stainless steel wound clips. A piece of tumor from the donor mouse was preserved as a control, for IHC studies, just before the s.c flank tumor implantation. Mice were regularly monitored and euthanized after 14 days of tumor implantation or when tumor size reached 2 cm2.
Flow cytometry assays
Samples were prepared as single cell suspensions and resuspended in 1% Fetal Bovine Serum (FBS) in PBS prior to staining. Cells were stained with fluorescent conjugated antibodies in 1% FBS. The conjugated antibodies and probes used for flow cytometry are listed in the Key Resources Table. Studies involving intracellular staining were performed with the Foxp3 Transcription Factor Staining Buffer Set (eBioscience, ThermoFisher Scientific) following manufacturer’s instructions. In case of the mouse cytokines IL-21, CXCL13 and Light/Tnfsf14, total splenocytes from s.c OVA-vaccinated mice were stimulated with PMA (100 ng/mL), ionomycin (0.5 μg/mL) and brefeldin A (10 μg/mL) for 6 hours in complete medium prior to the staining.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Mouse Antibodies | ||
| anti-mouse B220 (RA3.3A1/6.1 (TIB-146)) | BioXCell | RRID: AB_1107651 Cat#BE0067 |
| anti-mouse CD8α (YTS 169.4) | BioXCell | RRID: AB_10950145 Cat#BP0117 |
| BUV395 Hamster anti-mouse CD3e (145-2C11) | BD Biosciences | RRID: AB_2738278 Cat#563565 / 565992 |
| BV711 rat anti-mouse CD45 (30-F11) | BD Biosciences | RRID: AB_2687455 Cat# 563709 |
| Alexa Fluor® 700 rat anti-mouse CD44, (IM7) | BD Biosciences | RRID: AB_1727480 Cat# 560567 |
| BUV737 anti-mouse CD4 (RM4-5) | BD Biosciences | RRID: AB_2870165 Cat# 612843 / 612844 |
| PE Mouse anti-Bcl-6 (K112-91) | BD Biosciences | RRID: AB_2869985 Cat# 566979 |
| APC rat anti-mouse IgG1 (A85-1) | BD Biosciences | RRID: AB_1645625 Cat# 560089 |
| BV711 mouse anti-mouse CD45.1 (A20) | BD Biosciences | RRID: AB_2872214 Cat# 747742 |
| PerCP/Cyanine5.5 anti-mouse/human GL7 Antigen (GL7) | Biolegend | RRID: AB_2562978 Cat# 144609 / 144610 |
| Brilliant Violet 605 anti-mouse CD45 Antibody (30-F11) | Biolegend | RRID: AB_2562341 Cat# 103139 / 103155 / 103140 |
| Brilliant Violet 510 anti-mouse CD62L (MEL-14) | Biolegend | RRID: AB_2561537 Cat# 104441 |
| PE/Dazzle 594 anti-mouse CD279 (PD-1) (29F.1A12) | Biolegend | RRID: AB_2566005 Cat# 135127 / 135228 |
| Brilliant Violet 605 anti-mouse CD45.2 Antibody (104) | Biolegend | RRID: AB_2563485 Cat# 109841 |
| Brilliant Violet 421 anti-mouse CD185 (CXCR5) (L138D7) | Biolegend | RRID: AB_2562127 Cat# 145511 / 145512 |
| Brilliant Violet 785 anti-human/mouse/rat CD278 (ICOS) (C398.4A) | Biolegend | RRID: AB_2629729 Cat# 313533 / 313534 |
| Alexa Fluor 488 anti-mouse/rat/human FOXP3 Antibody (150D) | Biolegend | RRID: AB_439747 Cat# 320011 / 320012 |
| Brilliant Violet 510 anti-mouse/human CD45R/B220 Antibody (RA3-6B2) | Biolegend | RRID: AB_2561394 Cat# 103247 / 103248 |
| Brilliant Violet 605 anti-mouse CD95 (Fas) Antibody (SA367H8) | Biolegend | RRID: AB_2728202 Cat# 152612 |
| APC anti-mouse CD25 Antibody (PC61) | Biolegend | RRID: AB_312860 Cat# 102011 / 102012 |
| Brilliant Violet 785 anti-mouse I-A/I-E Antibody (M5/114.15.2) | Biolegend | RRID: AB_2565977 Cat# 107645 |
| Brilliant Violet 605 anti-mouse CD11c Antibody (N418) | Biolegend | RRID: AB_11204262 Cat# 117333 / 117334 |
| PE-Cyanine7 anti-mouse CD19 (1D3) | Tonbo Biosciences | RRID: AB_2621840 Cat# 60-0193 |
| Anti-mouse IFN-gamma (XMG1.2) | Tonbo Biosciences | RRID: AB_2621810 Cat# 50-7311 |
| Anti-mouse IL-4 (11B11) | Tonbo Biosciences | RRID: AB_2621465 Cat# 40-7041 |
| Anti-Mouse CD3e (145-2C11) | Tonbo Biosciences | RRID: AB_2621659 Cat# 35-0031 |
| Anti-mouse CD28 (37.51) | Tonbo Biosciences | RRID: AB_2621492 Cat# 70-0281 |
| Rat anti-mouse CXCL13/BLC/BCA-1 MAb antibody (143614) | R&D Systems | RRID: AB_2086062 Cat#MAB470 |
| rat IgG2A (Isotype) Alexa Fluor 647 (54447) | R&D Systems | Cat# IC006R |
| rat IgG2B (Isotype) Alexa Fluor 647 (141945) | R&D Systems | Cat# IC013R |
| Alexa Fluor 647 anti-mouse LIGHT/TNFSF14 (885310) | R&D Systems | Cat# FAB17942R |
| Alexa Fluor 647 Rat anti mouse CXCL13 (RM0205-19N1) | Novus Biologicals | Cat#NBP2-12223AF647 |
| IL-21 Monoclonal Antibody (Clone FFA21), PE, eBioscience. | ThermoFisher Scientific | RRID: AB_1834466 Cat# 12-7211-82 |
| anti-mouse CD23 (Invitrogen, SP23) | ThermoFisher Scientific | RRID: AB_10986348 Cat# MA5-14572 |
| rabbit anti-mouse CD3 (SP7) | Abcam | RRID: AB_443425 Cat# ab16669 |
| anti-mouse CD31 | Abcam | RRID: AB_726362 Cat# ab28364 |
| anti-mouse PNAd (MECA-79) | Millipore | Cat# MABF2050 |
| rabbit anti-mouse CD19 (D4V4B) | Cell Signaling | RRID: AB_2800152 Cat# 90176 |
| PE anti-mouse H-2Kb bound to SIINFEKL | Biolegend | RRID: AB_10895905 Cat#141604 |
| Human Antibodies | ||
| BV421 mouse anti-Bcl-6 (K112-91) | BD Biosciences | RRID: AB_2738159 Cat# 563363 |
| BV786 mouse anti-human CD45 (HI30) | BD Biosciences | RRID: AB_2716864 Cat# 563716 |
| BB515 rat anti-human CXCR5 (CD185)(RF8B2) | BD Biosciences | RRID: AB_2738871 Cat# 564624 / 564625 |
| BV421 Mouse IgG1 k Isotype Control (X40) | BD Biosciences | RRID: AB_11207319 Cat# 562438 |
| BV650 Mouse anti-human CD279 (PD-1), (EH12.1) | BD Biosciences | RRID: AB_2738595 Cat# 564104 |
| Alexa Fluor 647 mouse anti-SATB1 (14/SATB1) | BD Biosciences | RRID: AB_11153310 Cat# 562378 |
| BUV395 mouse anti-human CD3 (SK7) | BD Biosciences | RRID: AB_2744382 Cat# 564000 / 564001 |
| Brilliant Violet 785 anti-human CD4 (RPA-T4) | Biolegend | RRID: AB_2564381 Cat# 300553 / 300554 |
| PE-Dazzle 594 anti-human/mouse/rat CD278 (ICOS), (C398.4A) | Biolegend | RRID: AB_2566129 Cat# 300553 / 300554 |
| Brilliant Violet 711 anti-human CD25 (BC96) | Biolegend | RRID: AB_11219793 Cat# 300553 / 300554 |
| PE Anti-Human CD25 (BC96) | Tonbo Biosciences | RRID: AB_2621758 Cat# 50-0259 |
| Other Antibodies | ||
| Normal Rabbit IgG control isotype | Cell Signaling | RRID: AB_1031062 Cat# 2729 |
| InVivoMAb polyclonal rat IgG | BioXCell | RRID: AB_1107795 Cat# BE0094 |
| InVivoPlus rat IgG2b isotype control (LTF-2) | BioXCell | RRID: AB_2891360 Cat# BP0090 |
| Rat IgG2A Isotype Control (54447) | R&D Systems | RRID: AB_357349 Cat# MAB006 |
| ε-CD3 monoclonal antibody (SP7) | ThermoFisher Scientific | RRID: AB_10982026 Cat# MA5-14524 |
| Recombinant anti-CD20 antibody [SP32] | Abcam | RRID: AB_1139386 Cat# ab64088 |
| Recombinant anti-CD19 antibody [SP291] - C-terminal | Abcam | Cat# ab245235 |
| Anti-Bcl6 antibody | Abcam | Cat# ab272859 |
| Recombinant anti-CD21 antibody [EP3093] | Abcam | RRID: AB_1523292 Cat# ab75985 |
| Recombinant anti-SATB1 antibody [EPR3951] | Abcam | RRID: AB_10862207 Cat# ab109122 |
| Anti-Satb1 - C-terminal | Abcam | Cat# ab228772 |
| Recombinant DNA | ||
| Plasmid: pLV-EF1a-IRES-Blast | Addgene | Cat# 85133 |
| pcDNA3-deltaOVA | Addgene | Cat# 64595 |
| VSV.G | Addgene | Cat# 14888 |
| pCMV delta R8.2 | Addgene | Cat# 12263 |
| Bacterial and virus strains | ||
| One Shot TOP10 Chemically Competent E. coli | ThyermoFisher SCIENTIFIC | Cat# C404003 |
| Chemicals, peptides, and recombinant proteins | ||
| Rainbow Fluorescent Particles, 1 peak (3.0-3.4 μm) Mid Range Intensity | BioLegend | Cat#422905 |
| Zombie NIR™ Fixable Viability Kit | BioLegend | Cat#423106 |
| 4-Hydroxy-3-nitrophenylacetyl Ovalbumin (NP-OVAL) | Biosearch Technologies | N-5051-100 |
| Dako antibody diluent (Agilent) | Carpenteria, CA | Cat#SO809 |
| Ionomycin (calcium salt) | Cayman Chemical Co. | Cat# 11932 |
| Phorbol 12-myristate 13-acetate (PMA) | Cayman Chemical Co. | Cat# 10008014 |
| Brefeldin A | Cell Signaling | Cat# 9972S |
| Ficoll-Paque gradient | GE Healthcare Systems | Cat# 17-1449-02 |
| EDTA pH 9.0 buffer | Invitrogen | Cat# 15575-038 |
| Recombinant Human IL-2 | PeproTech | Cat# 200-02 |
| rhIL-10 | PeproTech | Cat# 200-10 |
| rhIL-12 p70 | PeproTech | Cat# 200-12 |
| rhIL-23 | PeproTech | Cat# 200-23 |
| Recombinant Human TGF-β1 | R&D Systems | 7754-BH-100/CF |
| Ovalbumin (albumin from chicken egg white) | Sigma | Cat# A5503 |
| anti-CD3/CD28 mouse T-activator beads | ThermoFisher Scientific | Cat# 11456D |
| Cell Trace Violet | ThermoFisher Scientific | Cat# C34571 |
| Dynabeads™ Human T-Activator CD3/CD28 | ThermoFisher Scientific | Cat# 11161D |
| Foxp3 Transcription Factor Staining Buffer Set | ThermoFisher Scientific | Cat# 00-5523-00 |
| Imject™ Alum Adjuvant | ThermoFisher Scientific | Cat# 77161 |
| SYBR Green PCR Master Mix | ThermoFisher Scientific | Cat# 4344463 |
| DEPC-treated Water | Thermo Scientific | Cat# R0601 |
| Gel RED Nucleic Acid Stain | Biotium | Cat# 41002 |
| Agarose LE, Quick Dissolve | Genesee Scientific | Cat# 20-102QD |
| Pharmco Products Ethyl Alcohol (200 Proof) | Fisher Scientific | Cat# 111000200 |
| β-Mercaptoethanol, BME | Sigma | Cat#M6250-10ML |
| Bacto AGAR | BD | Cat# 240230 |
| Difco LB Broth, Lennox | BD | Cat# 240110 |
| S.O.C. Medium | ThermoFisher (Invitrogen™) | Cat#15544034 |
| Cell Conditioning 1 (CC1) | Ventana products | Cat #950-124 |
| Discovery EZ Prep wash solution | Ventana products | Cat #950-510 |
| Buffered Formalde-Fresh 10% formalin | Fisher Chemical | Cat# SF93-20 |
| Hematoxylin | Sigma Aldrich | Cat#H9627 |
| Blasticidine S hydrochloride | Sigma Aldrich | Cat#15205 |
| EcoRI | New England Biolabs | Cat# R0101S |
| HpaI | New England Biolabs | Cat# R0105S |
| JetPRIME | Life science research. PolyPlus. | Ref#101000027 |
| Critical commercial assays | ||
| OPAL TM 7-Color Automation IHC kit | AKOYA Biosciences | Cat# NEL821001KT |
| Mouse anti-Ovalbumin Quantitative ELISA Kit | Alpha Diagnostic International | Cat# 600-105-OGG |
| BLC/CXCL13 Mouse ELISA Kit | ThermoFisher Invitrogen | Cat# EMCXCL13 |
| RNeasy Plus Micro Kit | QIAGEN | Cat#74034 |
| EasySep™ Mouse CD8+ T Cell Isolation Kit | StemCell Technologies | Cat# 19853 |
| EasySep™ Mouse B Cell Isolation Kit | StemCell Technologies | at# 19854 |
| EasySep™ Mouse CD4+ T Cell Isolation Kit | StemCell Technologies | Cat# 19852 |
| Mouse Naïve CD4+ T Cell negative selection Kit | StemCell Technologies | Cat# 9765 |
| Mouse T Cell negative selection kit | StemCell Technologies | Cat# 19851 |
| Human Naïve CD4+ T Cell isolation kit | StemCell Technologies | Cat# 17555 |
| High-Capacity Reverse Transcription kit, Applied Biosystems | ThermoFisher Scientific | Cat# 4368814 |
| Ventana ChromoMap kit | Ventana products | Cat# 760-159 |
| Experimental models: Cell lines | ||
| UPK10 ovarian carcinosarcoma | PMID: 22351930 PMID: 25533336 | N/A |
| UPK10-OVAlo ovarian carcinosarcoma | This paper | N/A |
| Brpkp110 breast-p53-KRas-p110alpha | PMID: 27803104 | N/A |
| HEK293T | ATCC | Cat# CRL-3216 |
| Biological Samples | ||
| Blood buffy coat units | LifeSouth (Gainesville, FL, USA) | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6 | The Jackson Laboratory/Charles River | Strain: 000664/027 |
| Mouse: B6 Cd45.1 (B6.SJL-Ptprca) | The Jackson Laboratory | Strain: 002014 |
| Mouse: B6 Rag1 KO (B6.129S7-Rag1tm1Mom/J) | The Jackson Laboratory | Strain: 002216 |
| Mouse: C57BL/6 Tg(TcraTcrb)425Cbn/Crl OT-II | Charles River | Strain: 643 |
| Mouse: B6 CD4CreSatb1f/f | PMID: 26876172 PMID: 28099864 |
N/A |
| Mouse: B6 OT-II+- CD4CreSatb1f/f | This paper | N/A |
| Mouse: B6 Satb1f/fFoxp3GFP | This paper | N/A |
| Mouse: B6 CD4CreSatb1f/fFoxp3GFP | This paper | N/A |
| Oligonucleotides | ||
| Primer sequence (5′→3′) mouse Il-21 F: AATCAAGCCCCCAAGGGCCA, R: AGCTGGGCCACGAGGTCAAT | Integrated DNA Technologies, IDT | N/A |
| Primer sequence (5′→3′) mouse Cxcl13 F: AAGTTACGCCCCCTGGGAATGGCT, R: ACTCACTGGAGCTTGGGGAGTTG | Integrated DNA Technologies, IDT | N/A |
| Primer sequence (5′→3′) mouse Light/Tnfsf14 F: ATCTCACCAGGCCAACCCAGCA, R: ACGCCGCTCAGCTGCACTTT | Integrated DNA Technologies, IDT | N/A |
| Primer sequence (5′→3′) Gapdh (mRNA normalization) F: CCTGCACCACCAACTGCTTA, R: AGTGATGGCATG GACTGTGGT | Integrated DNA Technologies, IDT | N/A |
| Primer sequence (5′→3′) Icos promoter F: TAC ATC ACC GGG TAC TTG CCA T; R: GCT CAA AAG TGT CAG TGA AGT CGT | Integrated DNA Technologies, IDT | N/A |
| Primer sequence (5′→3′) CSN1 region F: ACA AGG CCA AAC AAC AAT GAT AGA CA, R: TCA ACA TGA GGA ACA GTG CAG GA. | Integrated DNA Technologies, IDT | N/A |
| Primer sequence (5′→3′) CSN2 region F:GAC AAC AGG GCC CAG ATG TAG AC; R: GGC GTT CCT GTT TGA CTG TTT CT. | Integrated DNA Technologies, IDT | N/A |
| Software and algorithms | ||
| Prism, v8 | GraphPad | N/A |
| FlowJo, v10.8 | BD Biosciences | N/A |
| Aperio Image Scope, v12.4 | Leica Biosystems | N/A |
| Definiens Tissue Studio, v4.4.2 | Definiens Inc. | N/A |
Cell proliferation assays were performed by resuspending cells in 1 μM Cell Trace Violet in PBS for 15 min at 37 °C (ThermoFisher) and washed twice in complete medium. Cell viability was assessed through staining with the Zombie NIR dye (Biolegend) for 15 min in PBS. The reaction was stopped by washing with R10.
Samples were run either on a BD LSRII for flow analysis or sorted using a BD FACS Aria. All sorted populations were confirmed to be >98% pure through post-sort analysis. For assays evaluating protein expression levels using MFI, we used the Rainbow Fluorescent Particles, (Biolegend) in order to normalize voltage variance between measurements. Data were analyzed using FlowJo software v10.7.
Immunosuppression assay
Splenic CD3+ T cells were immunopurified from Satb1f/f mice (Mouse T Cell negative selection kit, 19851; StemCell Technologies) and labeled with the proliferation tracker Cell Trace Violet as indicated. T cell proliferation was stimulated by adding mouse specific anti-CD3/CD28 dynabeads (ThermoFisher) at a 1:1 T cell to bead ratio according to the manufacturer’s protocol. CD3+CD4+FoxP3GFP natural Treg cells (nTreg cells) were sorted from both CD4CreSatb1f/fFoxP3GFP and Satb1f/fFoxP3GFP control littermates. Immunopurified CD3+ T cells (2×105) were subsequently co-cultured at 1:2, 1:4, 1:8, and 1:16 T cell to nTreg cells ratios and incubated for 3 days prior to flow cytometry analysis.
Mouse inducible Treg cell differentiation
CD3+CD4+Foxp3−CD62L+CD44− naïve sorted splenocytes from CD4CreSatb1f/fFoxP3GFP mice and Satb1f/fFoxP3GFP control littermates were plated with anti-CD3/CD28 mouse T-activator beads (ThermoFisher) for 3 days in R10 under the following skewing conditions: IL-2, 100 U/mL (PeproTech), recombinant mouse TGF- β1, 10 ng/mL (R&D Systems), anti-mouse IFN-gamma (XMG1.2), and anti-mouse IL-4 (11B11), both (5 μg/mL) from Tonbo Biosciences.
Cytokine stimulation of naïve human T cells
Naïve human cells were isolated by using Human Naïve CD4+ T Cell isolation kit (Stemcell Technologies, 17555) from peripheral blood mononuclear cells (PBMCs) after Ficoll-Paque gradient (GE Healthcare Systems). Naïve human CD4+ T cells were incubated with Dynabeads™ Human T-Activator CD3/CD28 (ThermoFisher, 11161D) for 3 days in R10 under the following skewing conditions: a) rhIL-23 (25 ng/ml, PeproTech); b) rhIL-23 (25 ng/ml, PeproTech) plus rhIL-10 (10 ng/ml PeproTech); c) rhIL-12 p70 (1 ng/ml, PeproTech) and d) rhIL-12 p70 (1 ng/ml, PeproTech) plus rhIL-10 (10 ng/ml PeproTech). The former conditions were cultured in the presence/absence of hTGF-β (5 ng/ml) (R&D Systems).
Multiplex immunofluorescence
Formalin-fixed and paraffin-embedded (FFPE) tissue samples were immunostained using the AKOYA Biosciences OPAL TM 7-Color Automation IHC kit (Waltham, MA) on the BOND RX autostainer (Leica Biosystems, Vista, CA). The OPAL 7-color kit uses tyramide signal amplification (TSA)-conjugated to individual fluorophores to detect various targets within the multiplex assay. Sections were baked at 65°C for one hour then transferred to the BOND RX (Leica Biosystems). All subsequent steps (ex., deparaffinization, antigen retrieval) were performed using an automated OPAL IHC procedure (AKOYA). OPAL staining of each antigen occurred as follows: heat induced epitope retrieval (HIER) was achieved with EDTA pH 9.0 buffer for 20 min at 95°C before the slides were blocked with AKOYA blocking buffer for 10 min. Then slides were incubated with primary antibody, CD20 (AbCAM, SP291, 1:50, dye 520) at RT for 60 min followed by OPAL RbHRP polymer and one of the OPAL fluorophores during the final TSA step. Individual antibody complexes are stripped after each round of antigen detection. This was repeated four more times using the following antibodies, ε-CD3 (ThermoFisher, SP7, HIER- EDTA pH 9.0, 1:100, dye540), CD19 (AbCAM, SP291, HIER- EDTA pH 9.0, 1:200, dye620), BCL6 (AbCAM, Lot# ab272859, HIER- EDTA pH 9.0, 1:600, dye550), CD21 (AbCAM, EP3093, HIER- EDTA pH 9.0, 1:300, dye690); CD4 (Cell Signaling Technology, D7D2Z, 1:100), BCA1 (AbCAM , ab199043, EPR19259-147, 1:50) and CD34 (AbCAM , ab81289 EP373Y, 1:300).
After the final stripping step, DAPI counterstain was applied to the multiplexed slide and later removed from BOND RX for coverslipping with ProLong Diamond Antifade Mountant (ThermoFisher Scientific). Autofluorescence slides (negative control) were included, which use primary and secondary antibodies omitting the OPAL fluors and DAPI. All slides were scanned with the Vectra®3 Automated Quantitative Pathology Imaging System and the data from the multispectral camera were accessed by the imaging InForm software. Respective positive-control tissue adjacent sections were stained with isotype control antibodies to rule out false-positive staining. Positivity thresholds were determined per marker based on published nuclear or cytoplasmic staining patterns.
Analysis of TCGA data
Molecular and clinical data from TCGA for ovarian serous cystadenocarcinoma (designated OV) were downloaded from the cBio Cancer Genomics Portal (http://www.cbioportal.org/), Broad Firehose website (https://gdac.broadinstitute.org/) and Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). A total of 428 patients with matched clinical information and tumor RNA-seq data was used in this study. Raw RNA-seq reads were aligned to the GRCh37 human transcriptome using STAR24 (v.2.5.3a). Uniquely aligned reads were counted against Gencode v.19 using htseq-count31 (v.0.6.1) and then normalized using DESeq2 taking into account batches and RNA composition bias. All statistical analysis and visualization were performed on normalized count.
Immunohistochemistry, TLS definition and quantification.
In all cases, the whole mouse IP tumor contents were processed for TLS analysis. Tumor tissues were fixed in 10% formalin, embedded in paraffin, and serially sectioned for H&E and immunohistochemistry staining. All tumor sections slides were systematically stained using a Ventana Discovery XT automated system (Ventana Medical Systems, Tucson, AZ) as indicated by the manufacturer. Briefly, slides were deparaffinized with EZ Prep solution (Ventana). Heat-induced antigen retrieval method was used. Either primary rabbit anti-mouse CD3 (ab16669, Abcam, Cambridge, MA) was used at 1:200 dilution in Dako antibody diluent (Carpenteria, CA) or rabbit anti-mouse CD19 (90176, Cell Signaling, Danvers, MA) at 1:1000 dilution, and incubated for 30 min. For the case of the anti-mouse PNAd (MABF2050, Millipore, Billerica, MA) we used at 1:50 dilution in Dako antibody diluent and was incubated for 60 min. For the anti-mouse CD23 (Invitrogen, SP23) was used at 1:25 dilution. Retrieval was Cell Conditioning 1 (Ventana product) and incubation for 3 hours. The anti-mouse CD31 (Abcam ab28364) was used at 1:100. Retrieval was cell conditioning 1 and incubation was 32 minutes. The Ventana OmniMap anti-rabbit secondary antibody was used for 15 min. The detection system used was the Ventana ChromoMap kit and slides were counterstained with Hematoxylin. Finally, stained slides were dehydrated and coverslipped.
Slides were scanned using the Aperio™ ScanScope AT2 (Leica Biosystems, Vista, CA) with a 20x/0.75NA objective lens and an optical doubler to bring the final magnification to 40X. These images and meta-data were then imported into the Definiens Tissue Studio (Munich, Germany) software suite for segmentation (Payne et al., 2020). In Tissue Studio a machine learning algorithm was used to segment each tissue section image into positive CD19, CD3 areas and negative areas. The results of the segmentation were further refined using proximity classifiers, meaning positive clusters within 20 μm were merged. This refinement step was necessary to help to get the total positive CD19, CD3 area per TLS. We defined TLS as >0.1 mm2 conglomerates of CD19+B cells adjacent to accumulations of CD3+ T cells, clearly identified inside tumor masses and co-stained for peripheral node addressin (PNAd), with a central core of T cells. Likewise, large TLS were defined as regions >2 mm2. TLS areas were calculated by summing up the CD3 and CD19 positively stained zones in proximity (CD19+ plus CD3+ contents).
In addition to the TLS quantification, an overall estimate of the tumors’ density infiltration by both CD3+ T cells and CD19+ B cells outside the TLS, in tumors developed in mice with Satb1-competent vs. Satb1−/− T cells, was performed using an Aperio Positive Pixel Count® v9.0 algorithm with the following thresholds: [Hue Value =.1; Hue Width =.5; Color Saturation Threshold =0.04; IWP(High) = 220; Iwp(Low)=Ip(High) = 175; Ip(low) =Isp(High) =100 Isp(Low) =0] to segment positive staining of various intensities. The algorithm was applied to the entire digital image to determine the percentage of positive biomarker staining by applicable area. The TLS area was then subtracted from total biomarker area in order to determine the amount of positive area outside of the TLS regions.
Quantitative real time PCR
RNA was isolated (RNeasy Plus Micro Kit, QIAGEN) from CD3+CD4+CD62L−CD44+PD-1hiICOS+CXCR5+ Tfh and CD3+CD4+CD62L+CD44− naive cells immediately after sorting from total splenocytes and inguinal lymph nodes from s.c OVA-vaccinated CD4CreSatb1f/f mice and Satb1f/f control littermates at day 14. Reverse transcription was carried out using High-Capacity Reverse Transcription kit (Applied Biosystems). SYBR Green PCR Master Mix (Applied Biosystems) was used in a 7900 HT Fast Real-Time PCR System Software analysis (2.4), (Applied Biosystems). The following primer sequences were used (5′→3′): mouse Il-21 F: AATCAAGCCCCCAAGGGCCA, R: AGCTGGGCCACGAGGTCAAT; mouse Cxcl13 F: AAGTTACGCCCCCTGGGAATGGCT, R: ACTCACTGGAGCTTGGGGAGTTG; mouse Light/Tnfsf14 F: ATCTCACCAGGCCAACCCAGCA, R: ACGCCGCTCAGCTGCACTTT; and Gapdh (mRNA normalization) F: CCTGCACCACCAACTGCTTA, R: AGTGATGGCATGGACTGTGGT. The average of 3 independent replicas was calculated using the ΔΔ threshold cycle (Ct) method and was normalized to the endogenous reference control gene GAPDH.
Chromatin immunoprecipitation and Chip-PCR
Immunopurified CD4+ T cells from spleens (19852, EasySep) of naïve Satb1f/f mice were stimulated in vitro with immobilized anti-mouse CD3e (5 μg/mL) (145-2C11) (Tonbo Biosciences) and soluble anti-mouse CD28 (1 μg/mL) (37.51) (Tonbo Biosciences) for 30 hours. ChIP-PCR assays were performed as we previously reported (Stephen et al., 2017; Tesone et al., 2016), following standard methods with a chromatin shearing protocol of three cycles of 10 minutes each one, at high intensity (320W), consisting each minute as follows: (ON 30 seconds, OFF 30 seconds), followed by 5 minutes OFF between cycles, set up in a Diagenode Bioruptor 300. Shearing chromatin fragments of 100 – 300 bp were immunoprecipitated overnight with the anti-Satb1 - C-terminal (Abcam, ab228772) and the rabbit IgG as a control isotype (Cell Signaling; 2729). The percent input method, in this case 2.5 % of starting chromatin, was used. The following primers were used for the quantification of the Icos promoter: (5′→3′): F: TAC ATC ACC GGG TAC TTG CCA T; R: GCT CAA AAG TGT CAG TGA AGT CGT. For the case of the CNS1 and CNS2 regions, we used in vivo differentiated iTreg cells (kept for 6 days under skewing conditions) from immunopurified naïve CD4+ sorted cells (StemCell Technologies: 9765). Likewise, samples were immunoprecipitated overnight with the anti-Satb1 - C-terminal (Abcam, ab109122). The following primers were used for the quantification of the CSN1 region: (5′→3′): F: ACA AGG CCA AAC AAC AAT GAT AGA CA, R: TCA ACA TGA GGA ACA GTG CAG GA. Likewise, the following primers were used for the quantification of the CSN2 region: (5′→3′): F:GAC AAC AGG GCC CAG ATG TAG AC; R: GGC GTT CCT GTT TGA CTG TTT CT. Input and immunoprecipitated DNA were analyzed with a SYBR Green PCR Master Mix (Applied Biosystems) in a 7900 HT Fast Real-Time PCR System Software analysis (2.4), (Applied Biosystems).
Generation of the UPK10-OVAlow cell line
The lentiviral constitutive expression vector pLV-EF1a- OVAdeltaN-2A-IRES-blast was created by cloning chicken ovalbumin lacking the sequence coding for amino acids 1-49 from the vector pcDNA3-deltaOVA (Addgene 64595) into the pLV-EF1a IRES-blast vector backbone (Addgene #85133) using 5′ EcoRI and 3′ HpaI restriction sites by using standard techniques. UPK10 cells were transduced to express OVA using the packing cell line HEK293T and the packaging plasmid, VSVG (Addgene #14888) and Delta 8.2 (Addgene # 12263), (following the JetPRIME reagent protocol). Two days after transduction, UPK10 cells expressing OVA were selected under Blasticidin (6ug/mL). UPK10-OVAlow expressing cells were sorted after staining with the PE anti-mouse H-2Kb bound to SIINFEKL. Sorted fractions were further expanded in R10 supplemented with Blasticidin (6ug/mL).
Enzyme-linked immunosorbent assays
Quantification of total IgG and CXCL13 from serum of subcutaneous OVA-vaccinated CD4CreSatb1f/f mice and Satb1f/f control littermates, harvested at day 14, was done according to manufacturer protocol of the following kits: a) mouse anti-Ovalbumin Quantitative ELISA Kit (Alpha Diagnostic International, 600-105-OGG); b) BLC/CXCL13 Mouse ELISA Kit (ThermoFisher Scientist EMCXCL13).
Statistics analysis
Unless mentioned otherwise, all data are presented as median. Unpaired two-tailed t-test was used for calculating differences between means between two experimental groups. One-way ANOVA with Tukey’s post hoc test or two-way ANOVA (mixed model) followed by Sidak’s test, were used for calculating differences between means using multiple comparisons between groups. Analyses were carried out in Graph Pad Prism (v.8.0). The number of replicates per experiment is indicated in the legends. A significance threshold (0.05) for p values was used. Unless noted otherwise, all experiments were repeated at least twice and with similar results.
Supplementary Material
HIGHLIGHTS.
Satb1−/− CD4+ T cells show heightened antigen-specific Tfh cell differentiation.
SATB1-mediated suppression of Icos expression is required for Tfr cell differentiation.
CD4CreSatb1f/f mice spontaneously develop intra-tumoral tertiary lymphoid structures.
Tfh cell transfer elicits intra-tumoral TLS assembly and reduces tumor growth.
ACKNOWLEDGEMENTS
Support for Shared Resources was provided by Cancer Center Support Grant (CCSG) CA076292 to H. Lee Moffitt Cancer Center. This study was supported by R01CA157664, R01CA124515, R01CA178687, R01CA211913 and U01CA232758 to JRCG; R01CA184185 to PCR. KKP was supported by T32CA009140 and The American Cancer Society Postdoctoral Fellowship. We are especially grateful to Advanced Analytical and Digital Pathology (J. Nguyen), Flow Cytometry and Tissue Core Shared Resources at Moffitt Cancer Center, for exceptional support. We would like to thank the staff members of Comparative Medicine in the Stabile Research Building vivarium for their husbandry and technical assistance. We also thanks to Johana Melendez, Jonathan Semidey-Hurtado, Mary Jane Perkins, Noel D Clark, Neelkamal Chaudhary and Haley E. Bruns for technical support.
Footnotes
COMPETING INTERESTS
JRCG has stock options with Compass Therapeutics, Anixa Biosciences and Alloy Therapeutics, receives honorarium from Anixa Biosciences, Alloy Therapeutics and Leidos, and has sponsored research with Anixa Biosciences. RL: Clinical trial protocol committee – CG Oncology; Scientific advisor/consultant – BMS, Ferring, Fergene, Arquer Diagnostics. B.A.P. has completed Advisory Board with AstraZeneca and have Research Support from BMS. JR is currently an employee of STEMCELL Technologies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Allegrezza MJ, Rutkowski MR, Stephen TL, Svoronos N, Perales-Puchalt A, Nguyen JM, Payne KK, Singhal S, Eruslanov EB, Tchou J, and Conejo-Garcia JR (2016). Trametinib Drives T cell-Dependent Control of KRAS-Mutated Tumors by Inhibiting Pathological Myelopoiesis. Cancer Res 76, 6253–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Mandal G, Payne KK, Anadon CM, Gatenbee CD, Chaurio RA, Costich TL, Moran C, Harro CM, Rigolizzo KE, et al. (2021). IgA transcytosis and antigen recognition govern ovarian cancer immunity. Nature 591, 464–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CD, Barone F, Nayar S, Benezech C, and Caamano J (2015). Stromal cells in chronic inflammation and tertiary lymphoid organ formation. Annu Rev Immunol 33, 715–745. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, and Kohwi-Shigematsu T (2006). SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet 38, 1278–1288. [DOI] [PubMed] [Google Scholar]
- Cillo AR, Kurten CHL, Tabib T, Qi Z, Onkar S, Wang T, Liu A, Duvvuri U, Kim S, Soose RJ, et al. (2020). Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 52, 183–199 e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conejo-Garcia JR, Biswas S, and Chaurio R (2020). Humoral immune responses: Unsung heroes of the war on cancer. Semin Immunol 49, 101419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2014). T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S (2019). T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity 50, 1132–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos-Ruiz JR, Silberman PC, Rutkowski MR, Chopra S, Perales-Puchalt A, Song M, Zhang S, Bettigole SE, Gupta D, Holcomb K, et al. (2015). ER Stress Sensor XBP1 Controls Anti-tumor Immunity by Disrupting Dendritic Cell Homeostasis. Cell 161, 1527–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Caro G, Bergomas F, Grizzi F, Doni A, Bianchi P, Malesci A, Laghi L, Allavena P, Mantovani A, and Marchesi F (2014). Occurrence of tertiary lymphoid tissue is associated with T cell infiltration and predicts better prognosis in early-stage colorectal cancers. Clin Cancer Res 20, 2147–2158. [DOI] [PubMed] [Google Scholar]
- Engelhard VH, Rodriguez AB, Mauldin IS, Woods AN, Peske JD, and Slingluff CL Jr. (2018). Immune Cell Infiltration and Tertiary Lymphoid Structures as Determinants of Antitumor Immunity. J Immunol 200, 432–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev H, Ravens I, Papadogianni G, Halle S, Malissen B, Loots GG, Forster R, and Bernhardt G (2018). Shared and Unique Features Distinguishing Follicular T Helper and Regulatory Cells of Peripheral Lymph Node and Peyer's Patches. Front Immunol 9, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain C, Gnjatic S, and Dieu-Nosjean MC (2015). Tertiary Lymphoid Structure-Associated B Cells are Key Players in Anti-Tumor Immunity. Front Immunol 6, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Figueroa P, Roco JA, Papa I, Nunez Villacis L, Stanley M, Linterman MA, Dent A, Canete PF, and Vinuesa CG (2021). Follicular regulatory T cells produce neuritin to regulate B cells. Cell. [DOI] [PubMed] [Google Scholar]
- Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, Ravoet M, Le Buanec H, Sibille C, Manfouo-Foutsop G, et al. (2013). CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest 123, 2873–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu-Trantien C, Migliori E, Buisseret L, de Wind A, Brohee S, Garaud S, Noel G, Dang Chi VL, Lodewyckx JN, Naveaux C, et al. (2017). CXCL13-producing Tfh cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havenar-Daughton C, Lindqvist M, Heit A, Wu JE, Reiss SM, Kendric K, Belanger S, Kasturi SP, Landais E, Akondy RS, et al. (2016). CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci U S A 113, 2702–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka N, Ino Y, Yamazaki-Itoh R, Kanai Y, Kosuge T, and Shimada K (2015). Intratumoral tertiary lymphoid organ is a favourable prognosticator in patients with pancreatic cancer. Br J Cancer 112, 1782–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollern DP, Xu N, Thennavan A, Glodowski C, Garcia-Recio S, Mott KR, He X, Garay JP, Carey-Ewend K, Marron D, et al. (2019). B Cells and T Follicular Helper Cells Mediate Response to Checkpoint Inhibitors in High Mutation Burden Mouse Models of Breast Cancer. Cell 179, 1191–1206 e1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesia MD, Vincent BG, Parker JS, Hoadley KA, Carey LA, Perou CM, and Serody JS (2014). Prognostic B-cell signatures using mRNA-seq in patients with subtype-specific breast and ovarian cancer. Clin Cancer Res 20, 3818–3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson-Percival A, He B, Li ZJ, Kjellen A, Russell K, Li J, Larma I, and Ganss R (2017). De novo induction of intratumoral lymphoid structures and vessel normalization enhances immunotherapy in resistant tumors. Nat Immunol 18, 1207–1217. [DOI] [PubMed] [Google Scholar]
- Kakugawa K, Kojo S, Tanaka H, Seo W, Endo TA, Kitagawa Y, Muroi S, Tenno M, Yasmin N, Kohwi Y, et al. (2017). Essential Roles of SATB1 in Specifying T Lymphocyte Subsets. Cell Rep 19, 1176–1188. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, Yasuda K, Motooka D, Nakamura S, Kondo M, et al. (2017). Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol 18, 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroeger DR, Milne K, and Nelson BH (2016). Tumor-Infiltrating Plasma Cells Are Associated with Tertiary Lymphoid Structures, Cytolytic T-Cell Responses, and Superior Prognosis in Ovarian Cancer. Clin Cancer Res 22, 3005–3015. [DOI] [PubMed] [Google Scholar]
- Marinkovic T, Garin A, Yokota Y, Fu YX, Ruddle NH, Furtado GC, and Lira SA (2006). Interaction of mature CD3+CD4+ T cells with dendritic cells triggers the development of tertiary lymphoid structures in the thyroid. J Clin Invest 116, 2622–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall HD, Ray JP, Laidlaw BJ, Zhang N, Gawande D, Staron MM, Craft J, and Kaech SM (2015). The transforming growth factor beta signaling pathway is critical for the formation of CD4 T follicular helper cells and isotype-switched antibody responses in the lung mucosa. Elife 4, e04851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messina JL, Fenstermacher DA, Eschrich S, Qu X, Berglund AE, Lloyd MC, Schell MJ, Sondak VK, Weber JS, and Mule JJ (2012). 12-Chemokine gene signature identifies lymph node-like structures in melanoma: potential for patient selection for immunotherapy? Sci Rep 2, 765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montfort A, Pearce O, Maniati E, Vincent BG, Bixby L, Bohm S, Dowe T, Wilkes EH, Chakravarty P, Thompson R, et al. (2017). A Strong B-cell Response Is Part of the Immune Landscape in Human High-Grade Serous Ovarian Metastases. Clin Cancer Res 23, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik R, and Galande S (2019). SATB family chromatin organizers as master regulators of tumor progression. Oncogene 38, 1989–2004. [DOI] [PubMed] [Google Scholar]
- Nayar S, Campos J, Smith CG, Iannizzotto V, Gardner DH, Mourcin F, Roulois D, Turner J, Sylvestre M, Asam S, et al. (2019). Immunofibroblasts are pivotal drivers of tertiary lymphoid structure formation and local pathology. Proc Natl Acad Sci U S A 116, 13490–13497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BV, Freeman ZT, Ghasemzadeh A, Chattergoon MA, Rutebemberwa A, Steigner J, Winter ME, Huynh TV, Sebald SM, Lee SJ, et al. (2016). TGFbeta1-Mediated SMAD3 Enhances PD-1 Expression on Antigen-Specific T Cells in Cancer. Cancer Discov 6, 1366–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne KK, Mine JA, Biswas S, Chaurio RA, Perales-Puchalt A, Anadon CM, Costich TL, Harro CM, Walrath J, Ming Q, et al. (2020). BTN3A1 governs antitumor responses by coordinating alphabeta and gammadelta T cells. Science 369, 942–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peske JD, Thompson ED, Gemta L, Baylis RA, Fu YX, and Engelhard VH (2015). Effector lymphocyte-induced lymph node-like vasculature enables naive T cell entry into tumours and enhanced anti-tumour immunity. Nat Commun 6, 7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannstiel C, Strissel PL, Chiappinelli KB, Sikic D, Wach S, Wirtz RM, Wullweber A, Taubert H, Breyer J, Otto W, et al. (2019). The Tumor Immune Microenvironment Drives a Prognostic Relevance That Correlates with Bladder Cancer Subtypes. Cancer Immunol Res 7, 923–938. [DOI] [PubMed] [Google Scholar]
- Rodriguez AB, Peske JD, and Engelhard VH (2018). Identification and Characterization of Tertiary Lymphoid Structures in Murine Melanoma. Methods Mol Biol 1845, 241–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski MR, Stephen TL, Svoronos N, Allegrezza MJ, Tesone AJ, Perales-Puchalt A, Brencicova E, Escovar-Fadul X, Nguyen JM, Cadungog MG, et al. (2015). Microbially Driven TLR5-Dependent Signaling Governs Distal Malignant Progression through Tumor-Promoting Inflammation. Cancer Cell 27, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage PT, Francisco LM, Carman CV, and Sharpe AH (2013). The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol 14, 152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage PT, Ron-Harel N, Juneja VR, Sen DR, Maleri S, Sungnak W, Kuchroo VK, Haining WN, Chevrier N, Haigis M, and Sharpe AH (2016). Suppression by Tfr cells leads to durable and selective inhibition of B cell effector function. Nat Immunol 17, 1436–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sautes-Fridman C, Petitprez F, Calderaro J, and Fridman WH (2019). Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer 19, 307–325. [DOI] [PubMed] [Google Scholar]
- Scarlett UK, Rutkowski MR, Rauwerdink AM, Fields J, Escovar-Fadul X, Baird J, Cubillos-Ruiz JR, Jacobs AC, Gonzalez JL, Weaver J, et al. (2012). Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J Exp Med 209, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K, Banchereau J, and Ueno H (2014). The cytokine TGF-beta co-opts signaling via STAT3-STAT4 to promote the differentiation of human Tfh cells. Nat Immunol 15, 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Hou S, Fang Q, Liu X, Liu X, and Qi H (2018). PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity 49, 264–274 e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silina K, Soltermann A, Attar FM, Casanova R, Uckeley ZM, Thut H, Wandres M, Isajevs S, Cheng P, Curioni-Fontecedro A, et al. (2018). Germinal Centers Determine the Prognostic Relevance of Tertiary Lymphoid Structures and Are Impaired by Corticosteroids in Lung Squamous Cell Carcinoma. Cancer Res 78, 1308–1320. [DOI] [PubMed] [Google Scholar]
- Stephen TL, Payne KK, Chaurio RA, Allegrezza MJ, Zhu H, Perez-Sanz J, Perales-Puchalt A, Nguyen JM, Vara-Ailor AE, Eruslanov EB, et al. (2017). SATB1 Expression Governs Epigenetic Repression of PD-1 in Tumor-Reactive T Cells. Immunity 46, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen TL, Rutkowski MR, Allegrezza MJ, Perales-Puchalt A, Tesone AJ, Svoronos N, Nguyen JM, Sarmin F, Borowsky ME, Tchou J, and Conejo-Garcia JR (2014). Transforming Growth Factor beta-Mediated Suppression of Antitumor T Cells Requires FoxP1 Transcription Factor Expression. Immunity 41, 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesone AJ, Rutkowski MR, Brencicova E, Svoronos N, Perales-Puchalt A, Stephen TL, Allegrezza MJ, Payne KK, Nguyen JM, Wickramasinghe J, et al. (2016). Satb1 Overexpression Drives Tumor-Promoting Activities in Cancer-Associated Dendritic Cells. Cell Rep 14, 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters GD, and Vinuesa CG (2016). T Follicular Helper Cells in Transplantation. Transplantation 100, 1650–1655. [DOI] [PubMed] [Google Scholar]
- Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, Tas J, Choi YS, Takata H, Day TJ, et al. (2014). The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol 15, 667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein AM, and Storkus WJ (2015). Therapeutic Lymphoid Organogenesis in the Tumor Microenvironment. Adv Cancer Res 128, 197–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JM, Bonami RH, Hulbert C, and Thomas JW (2015). Reversing Tolerance in Isotype Switch-Competent Anti-Insulin B Lymphocytes. J Immunol 195, 853–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, and Rudensky AY (2010). Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T cell fate. Nature 463, 808–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Nemoto S, Mailloux AW, Perez-Villarroel P, Nakagawa R, Falahat R, Berglund AE, and Mule JJ (2018). Induction of Tertiary Lymphoid Structures With Antitumor Function by a Lymph Node-Derived Stromal Cell Line. Front Immunol 9, 1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This paper does not report original code. Any additional information required to reanalyze the data reported, as well as raw image for TLS IHC studies, are available from the lead contact upon request.