Abstract
Viruses that spread transsynaptically provide a powerful means for studying interconnected circuits in the brain. Here we describe the use of adeno-associated virus, serotype 1 (AAV1) as a tool for achieving robust, anterograde transsynaptic spread in a variety of unidirectional pathways. A protocol for performing intracranial AAV1 injections in mice is presented, along with additional guidance in planning experiments, sourcing materials, and optimizing the approach to achieve the most successful outcomes. Following the methods presented here, researchers will be able to reveal postsynaptically connected neurons downstream of a given AAV1 injection site, and access these input-defined cells for subsequent mapping, recording, and manipulation to characterize their anatomical and functional properties.
Keywords: Viral tracing, AAV, anterograde transsynaptic virus
INTRODUCTION
A fundamental challenge in neuroscience is to define and experimentally access different cell-types within a circuit. Viral tools that spread transsynaptically greatly facilitate this process by naturally spreading to upstream neuronal populations defined by their postsynaptic targets (e.g. Rabies virus, Wickersham et al., 2007; PRV, DeFalco et al., 2001; Ekstrand et al., 2008) or downstream populations defined by their presynaptic inputs (e.g. Herpesvirus H129, Lo & Anderson, 2011; Zeng et al., 2017; VSV, Beier et al., 2011), and then driving the expression of viral transgenes that enable the study of these cells. However, while viruses that permit retrograde access to presynaptic neurons have been widely adopted, those that undergo anterograde transsynaptic spread remain under development due to their toxicity and bidirectional spread.
Recently, we demonstrated the capacity for AAV1 to spread transsynaptically to cells downstream of an injection site (Figure 1; Zingg et al., 2017; Zingg et al., 2020). This viral spread was restricted to first-order neurons in expected postsynaptic targets, did not appear to leak from axons-of-passage, exhibited preferential transmission through synaptic mechanisms, and demonstrated equal efficiency when tested in a variety of different cell-types and pathways. Thus, given these observations, along with its well established lack of toxicity, AAV1 shows great promise as an alternative to existing tools for manipulating and mapping input-defined cell populations. One important caveat, however, is that it must only be applied in unidirectional pathways due to its capacity to spread in a retrograde direction.
Figure 1. Anterograde transsynaptic spread of AAV1.
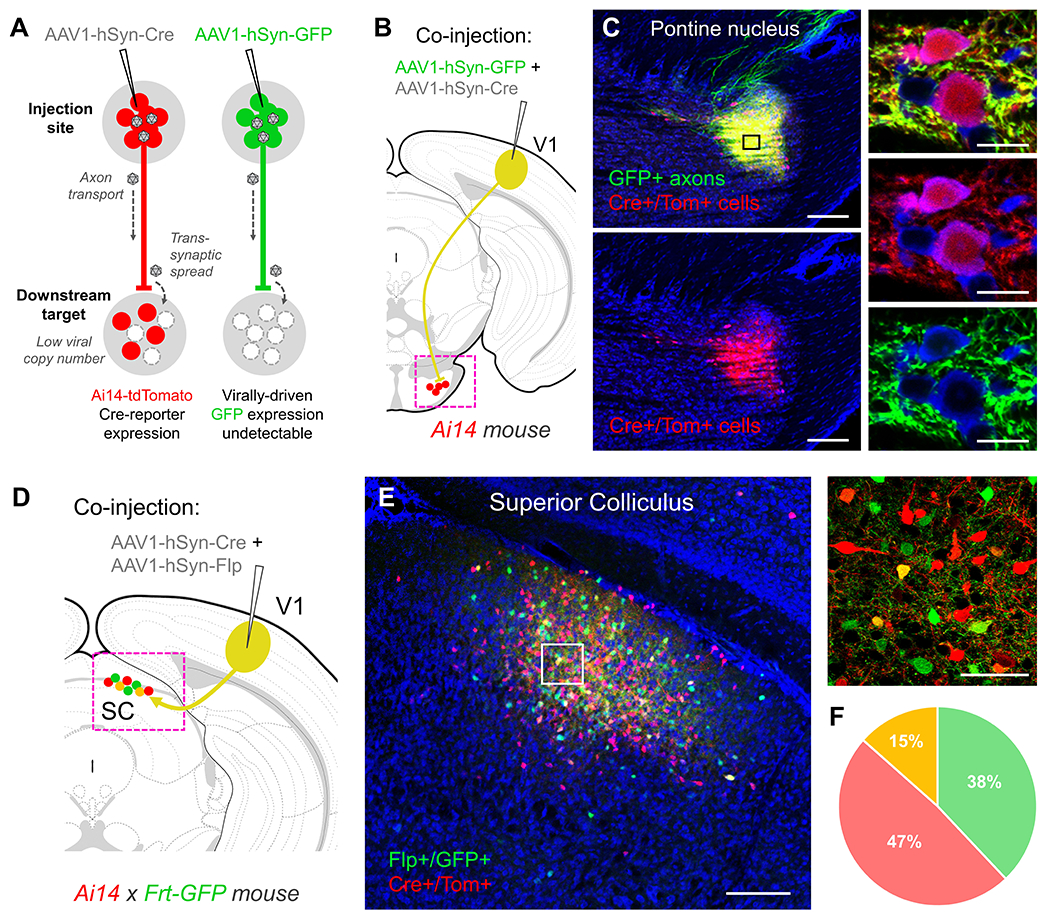
(A) Following high-titer injections of either AAV1-Cre (left) or AAV1-GFP (right) into the mouse brain (>1013 GC/mL, 50-100 nL total volume), a large number of viral particles are taken up locally at the soma and may undergo transport to the nucleus, leading to robust transduction of neurons at the injection site (red or green colored circles). In addition, a small fraction of viral particles may be trafficked down the axon and released postsynaptically, enabling transduction of downstream cell populations. This transsynaptic spread is revealed following injections of AAV1-Cre in fluorescent Cre-reporter mice (Ai14-tdTomato, left), but is rarely observed when using fluorescent protein-expressing AAV1 constructs alone (e.g. AAV1-GFP, right). This discrepancy may be due to the presumably small number of viral particles that are synaptically released, which may be insufficient to drive detectable levels of virally-driven fluorescence in postsynaptic cells. However, only trace amounts of virally-driven Cre may be required to successfully unlock tdTomato transgene expression in postsynaptic neurons in Ai14 mice. (B) Co-injection of a 1:1 mixture of AAV1-GFP and AAV1-Cre (80 nL total volume, 1.0 x 1013 GC/mL each) into V1 of an Ai14-tdTomato mouse results in postsynaptic labeling of Cre+/Tom+ cell bodies (red) that closely match the distribution of GFP+ axon terminal fields (green) (C). Higher magnification (40X, right panels) reveals brightly labeled GFP+ axon terminals surrounding Cre+/Tom+ cells, however native AAV1-GFP expression is undetectable in downstream cell bodies, despite sharing the same capacity for transsynaptic spread as the co-injected AAV1-Cre. Blue, fluorescent Nissl stain. Scale bar: 250 μm (left panels) and 50 μm (right panels). (D) Co-injection of a 1:1 mixture of AAV1-Cre and AAV1-Flp (80 nL total volume, 1.0 x 1013 GC/mL each) into V1 in a Cre- and Flp-reporter mouse (Ai14-tdTomato x Frt-GFP) results in (E) postsynaptic labeling of Cre+/Tom+ (red) and Flp+/GFP+ (green) cells in downstream targets. Despite equivalent chances for transsynaptic spread from the same starter cell population, the majority of downstream labeled neurons were either Cre+/Tom+ or Flp+/GFP+ (right panel), while only 15% of the total population were co-labeled. This further suggests a small fraction of viral particles are responsible for the observed transsynaptic spread, and the required threshold for unlocking reporter gene expression is just barely surpassed in many of these downstream cells. Blue, fluorescent Nissl stain. Scale bar: 250 μm (left panel) and 50 μm (right panel). (F) Quantification of co-labeled neurons in the superior colliculus. Flp+/GFP+ 219 cells (green), Cre+/Tom+ 271 cells (red), co-labeled 66 cells (yellow); n = 2 animals.
Here, we present a simple approach for accessing postsynaptic neurons using AAV1 constructs that express a particular recombinase protein, such as Cre or FlpO. These may be applied in combination with a wide variety of transgenic and viral resources to flexibly study the anatomical connections, functional response properties, or behavioral contributions of any given input-defined cell population of interest. Moreover, complementing existing transgenic and retrograde viral strategies for accessing cell-types, the anterograde transsynaptic transport properties of AAV1 uniquely enable the examination of topographically precise input-output relationships for a given structure, regardless of their small size or targeting difficulty (e.g. pallidal nuclei, Lee et al., 2020).
In this protocol, we focus primarily on the strategic planning and experimental design aspects of this method, highlighting the various combinatorial ways in which transsynaptic delivery of AAV1 may be used to study the connectivity and function of input-defined cell populations. We then present a basic protocol describing the procedures for intracranial injection of AAV1 in mice, and include additional steps for tissue processing and imaging. As these stereotaxic viral injection procedures have previously been reported in detail (Harris et al., 2012; Gore et al., 2013), we focus on any special considerations relevant to achieving optimal transsynaptic spread of AAV1 and subsequent detection of downstream labeling.
STRATEGIC PLANNING
This section offers guidance in choosing the best viruses and transgenic mice for AAV1 transsynaptic applications, identifies potential commercial sources for acquiring these materials, and addresses some of the important caveats that must be considered when designing experiments. In addition, we highlight several ways in which this technique may be used in combination with other viral tools and transgenic mice to determine the identity, connectivity, and functional properties of neurons labeled via transsynaptic delivery of AAV1.
Selection of AAV1 construct
There are a wide variety of commercially available AAV1 vectors that can be used for transsynaptic gene delivery (see Table 1). When choosing a particular viral construct, three features are important to consider for achieving optimal results: type of promoter, type of recombinase, and inclusion or exclusion of the woodchuck hepatitis virus post-transcriptional regulatory element (WPRE). Each are discussed separately below and recommendations are provided.
Table 1.
List of commercially available viruses and plasmids for AAV1 anterograde transsynaptic applications.
| Recommended commercially available AAV1 constructs for anterograde transsynaptic applications | |||||
|---|---|---|---|---|---|
| Virus name | Recombinase | Toxicity at injection site? | Originating Lab | Vendor name | Catalog number |
| AAV1-hSyn-Cre-WPRE | Cre | Yes, contains WPRE | James M. Wilson, UPENN | Addgene | 105553 |
| AAV1-Syn-iCre | iCre | No, lacks WPRE | - | SignaGen | SL101441 |
| AAV1-Syn-Cre | Cre | No, lacks WPRE | - | Vigene | CV17053-AV1 |
| scAAV1-hSyn-Cre | Cre | No, lacks WPRE | - | WZ Biosci. | AV204046-scAV1 |
| AAV1-Syn-FlpO | FlpO | No, lacks WPRE | - | SignaGen | SL101447 |
| AAV1-EF1a-DIO-FlpO-WPRE | Cre-dependent FlpO | Yes, but only in Cre+ cells at injection site | Li I. Zhang, USC | Addgene | 87306 |
| Recommended plasmids for custom AAV1 packaging | |||||
| Plasmid name | Recombinase | Notes | Originating Lab | Vendor name | Plasmid # |
| pAAV-hSyn-Cre | Cre | Lacks WPRE | James M. Wilson, UPENN | Addgene | 105555 |
| pAAV-hSyn-Cre-WPRE | Cre | Contains WPRE | James M. Wilson, UPENN | Addgene | 105553 |
| pAAV-hSyn-FlpO | FlpO | Lacks WPRE | Hongkui Zeng, Allen Institute | Addgene | 51669 |
| pAAV-hSyn-FlpO-WPRE | FlpO | Contains WPRE | Massimo Scanziani, UCSF | Addgene | 60663 |
| pAAV-hSyn-DreO | DreO | Lacks WPRE | Hongkui Zeng, Allen Institute | Addgene | 50363 |
| pAAV-hSyn-oNigri-WPRE | oNigri | Contains WPRE | Tanya Daigle, Allen Institute | Addgene | 163489 |
| pAAV-EF1a-fDIO-Cre-WPRE | FlpO-dependent Cre | Contains WPRE | Esteban Engel, Princeton Alexander Nectow, Columbia | Addgene | 121675 |
Choice of promoter:
The promoter sequence initiates transcription of the viral transgene and determines both the expression level and the type of tissue in which it takes place. Several promoters are commonly used, such as CAG, EF1a, and CMV, based on their ability to drive robust gene expression in the brain. Each of these, however, are capable of expressing in both neuronal and glial cell-types and may therefore result in astrocyte labeling when used in AAV1 transsynaptic experiments. To avoid this, we recommend using the human synapsin-1 promoter (commonly abbreviated as hSyn, hSyn1, Syn, or Syn1), which drives strong gene expression specifically in neurons, but not glia.
Choice of recombinase:
A growing number of recombinase options have become available to researchers, including the Cre/lox, FlpO/frt, DreO/rox, vCre/vlox, and oNigri/nox systems. A few considerations may be important when deciding which to use. These include the relative efficiency of recombination for a given system, the availability of “improved” or “codon-optimized” variants, and the potential for cross-reactivity when used in combination with other recombinase systems.
Recombinase proteins recognize and bind to specific sequences of DNA (e.g. loxP sites for Cre, frt sites for FlpO) and catalyze the excision or inversion of the DNA fragment between each recognition site (see Tian & Zhou, 2021 for a review). This capability has been exploited to achieve cell-type specific gene expression, whereby a recombinase is used to unlock an endogenous or virally-delivered transgene by rearranging or removing parts of the construct that normally prevent its expression. The efficiency with which different recombinase systems achieve this may vary, however, and this can factor into the overall efficiency of the transsynaptic labeling observed with any given AAV1 injection. In particular, FlpO has been reported to be somewhat less efficient than Cre (Ringrose et al., 1998), and in our experience injections of AAV1-hSyn-FlpO tend to yield slightly fewer postsynaptically labeled cells when compared with equivalent injections of AAV1-hSyn-Cre (Zingg et al., 2020). Given this variability, it may be important to test new recombinase systems relative to the performance of Cre before implementing them in AAV1 transsynaptic applications.
In addition, the genetic sequences of some recombinase proteins have been modified from their original form in order to improve expression in the mammalian system (Raymond & Soriano, 2007; Shimshek et al., 2002; Gustafsson et al., 2004). The modified versions are often denoted with an “O” for “codon-optimized” (e.g. FlpO, DreO, oNigri) or an “i” for “improved” (e.g. iCre). Opting for the use of these improved variants in the AAV1 construct may confer an increase in recombinase expression levels and result in greater numbers of transsynaptically labeled cells.
Lastly, cross-reactivity may occur between different recombinase systems when used together. This has been reported for Cre/lox and DreO/rox systems (Fenno et al., 2014; Madisen et al., 2015; Daigle et al., 2018), and thus should be avoided when designing intersectional experiments. On the other hand, Cre/lox and FlpO/frt have a well characterized lack of cross-reactivity and offer a safe choice for combinatorial studies (Madisen et al., 2015; Fenno et al., 2020). In light of this, controls to confirm the specificity of recombinase-dependent gene expression should be performed for any new pair of recombinases used.
Use of the WPRE enhancer:
WPRE is commonly included in AAV constructs to stabilize mRNA transcripts, which in turn boosts overall levels of protein expression from the viral transgene. Previous reports have cited as much as a sevenfold increase in gene products expressed from AAV constructs that include WPRE (Loeb et al., 2002). In our experience, the inclusion of WPRE in Cre- or FlpO-expressing AAV1 vectors greatly increases the amount of transsynaptic labeling observed, however this comes with the caveat of increased toxicity in cells at the injection site. This effect is likely due to the high level of recombinase expression achieved with WPRE, which in turn may increase the rate of off-target binding, recombination, and subsequent damage to DNA within the host cell (Loonstra et al., 2001; Thyagarajan et al., 2000; Janbandhu et al., 2014). On the other hand, at equivalent titers and volumes (1.0 x 1013 GC/mL, 80 nL total), injections of AAV1 constructs that exclude WPRE have all demonstrated a lack of toxicity at the injection site in our hands. This, however, comes at the cost of reduced numbers of transsynaptically labeled cells. Given these observations, in situations where transsynaptic delivery of AAV1 will be used to investigate the functional properties of a cell population, it may be advisable to avoid WPRE containing constructs that might damage upstream neurons and potentially introduce confounding physiological changes in the circuit.
AAV1 packaging and titer
Aside from optimizing recombinase expression through the use of strong promoters and enhancers, one of the most important factors in determining the efficiency of transsynaptic spread is viral titer. Previous work has shown that reducing the titer of AAV1 results in a proportional decrease in the amount of postsynaptic labeling, with spread eventually becoming undetectable at concentrations near or below 5.0 x 1011 GC/mL (see Zingg et al., 2017 for details). Given this dependence on titer, we recommend using AAV1 at the highest possible concentration in order to maximize the efficiency of viral spread. In general, AAV1 preparations that are at least 1.0 x 1013 GC/mL or higher are expected to perform well and yield useful numbers of cells for most tested pathways. In addition, the viral injection volume (typically 50 – 150 nL) may be flexibly adjusted as needed to increase spread to a particular target or compensate for lower viral titer. Most commercially available AAV1 stocks are provided at concentrations slightly above 1.0 x 1013 GC/mL and may be purchased from vendors such as Addgene (https://www.addgene.org/), SignaGen (https://signagen.com/), Vigene (https://www.vigenebio.com/), or WZ Biosciences (https://www.wzbio.com/). Table 1 lists several recommended options for Cre- or FlpO-based transsynaptic use.
Alternatively, researchers may prepare or request higher titer viral stock (e.g. up to 1.0 x 1014 GC/mL) using any available plasmids suitable for AAV packaging and expression. Several recommended plasmids for this purpose are also found in Table 1. These may be purchased from Addgene and sent to vendors such as SignaGen, Vigene, or WZ Biosciences for custom high titer packaging. In our experience, higher titer preparations of constructs that lack WPRE provide an ideal tool for achieving robust transsynaptic spread without inducing observable toxicity at the injection site.
Lastly, self-complementary packaging may provide a modest boost in labeling efficiency (McCarty et al., 2001; McCarty, 2008; Zingg et al., 2020) and may be requested with any custom packaging order from the aforementioned vendors. Table 1 indicates a potential source for pre-made self-complementary (sc)AAV1-hSyn-Cre stock.
Detection, mapping, and functional examination of downstream neurons
There are a variety of ways in which transsynaptic spread of AAV1 can be detected and used in a combinatorial fashion to investigate input-defined neurons. Some of these are highlighted in Figure 2 and discussed below.
Figure 2. Strategies for investigating input-defined cell populations using anterograde transsynaptic delivery of AAV1.
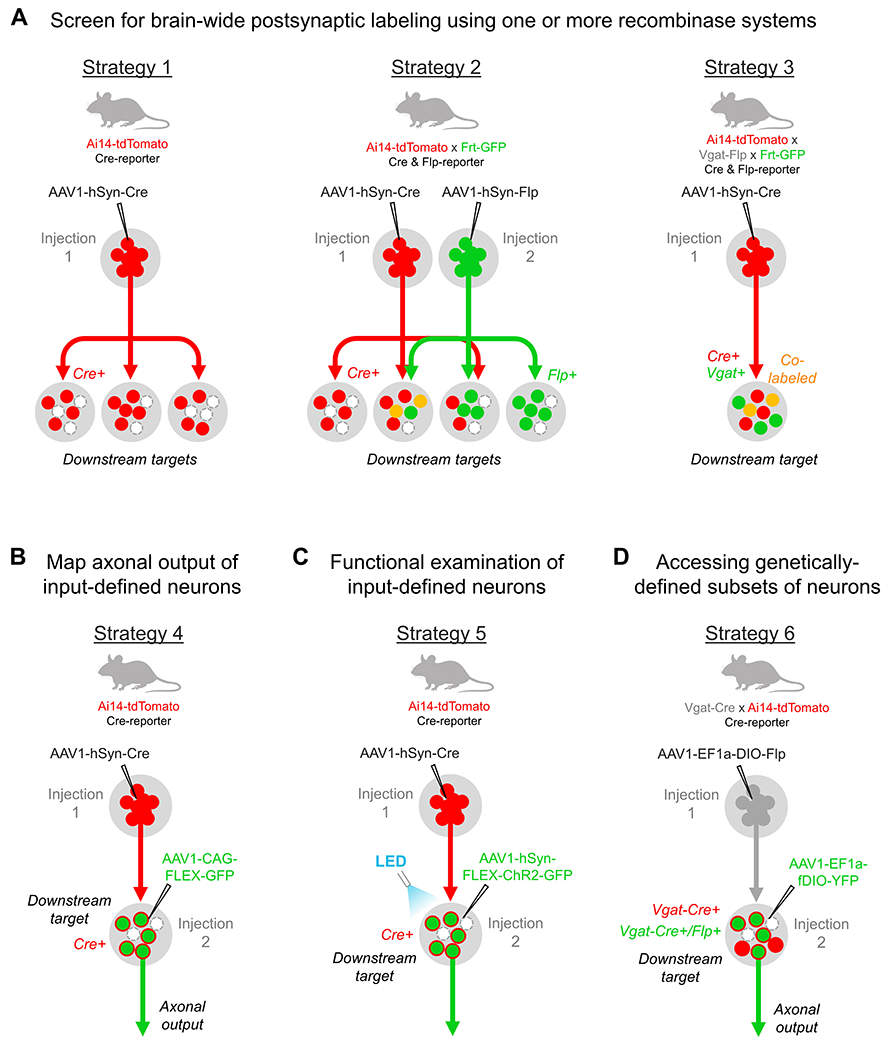
(A) Examples of one-step AAV1 injections for characterizing postsynaptic cell populations. A single injection of AAV1-Cre in a transgenic Cre-reporter mouse may be used to examine the morphology and distribution of cells downstream of a given brain region (Strategy 1). This approach may be expanded to include two or more different recombinase-expressing AAV injections (Strategy 2; e.g. AAV1-Cre and AAV1-Flp) in a transgenic mouse that reports the presence of each recombinase with a distinct fluorescent protein (e.g. Ai14-tdTomato x Frt-GFP mice). This allows for the direct demonstration of divergent (red or green circles) or convergent (yellow circles) inputs onto individual postsynaptic neurons from two or more upstream brain regions that share similar downstream targets. Lastly, the genetic identity of postsynaptic neurons may be screened (Strategy 3) by performing AAV1 injections (e.g. AAV1-Cre) in transgenic mice that fluorescently label a particular cell-type and report the presence of recombinase protein (e.g. Vgat-Flp x Frt-GFP x Ai14-tdTomato mice express GFP in all inhibitory neurons and tdTomato in all Cre+ neurons). Alternatively, AAV1-Flp may be used with comparable results in Cre-expressing transgenics crossbred with Ai14 x Frt-GFP mice. (B-D) Examples of two-step virus injections for the anatomical and functional examination of input-defined neurons. (B) The axonal output of a given input-defined population may be examined by injecting AAV1-Cre in an upstream region and AAV1-FLEX-GFP in a given downstream target (Strategy 4). Similar results may be achieved with other recombinase pairings (e.g. AAV1-Flp and AAV1-fDIO-tdTomato), and these may all be applied in a single animal to simultaneously compare multiple input-defined projection pathways in the same brain. (C) To functionally interrogate a given input-defined population (Strategy 5), the same procedure may be followed as in (B), however an AAV that conditionally expresses any desired functional tool for activating, silencing, or recording from the target population (e.g. AAV1-hSyn-FLEX-ChR2-GFP) is injected into the downstream region. (D) Lastly, the axonal output or function of a given input- and genetically-defined cell population may be examined (Strategy 6) by injecting a Cre-dependent Flp-expressing AAV1 (e.g. AAV1-EF1a-DIO-Flp) in an upstream region of a given Cre-expressing transgenic mouse (e.g. Vgat-Cre), followed by a second injection of Flp-dependent AAV (e.g. AAV1-fDIO-YFP) in a chosen downstream target. In this example, selective labeling of Vgat+ inhibitory neurons is achieved for the input-defined population. Similar injections may be applied in different transgenic mice to explore additional input-defined cell-types within the same target region (e.g. excitatory neurons in Vglut2-Cre mice).
Identifying and characterizing post-synaptic neurons:
Several methods are available for visualizing the transsynaptic spread of AAV1, however transgenic reporter mice offer the simplest and most effective means for detecting the distribution of postsynaptic cells across the entire brain (Fig. 2A, Strategy 1). These mice enable universal, recombinase-dependent expression of fluorescent protein in any cell that also co-expresses the required recombinase (Madisen et al., 2010; Madisen et al., 2015; Daigle et al., 2018). Moreover, in our experience, even low levels of recombinase may be sufficient to unlock reporter gene expression. This is important considering the efficiency of AAV1 transsynaptic spread is low and only a few viral particles may be responsible for the transduction of any given downstream neuron. As such, transgenic reporter mice provide a very sensitive means for revealing transsynaptic spread and may reliably report even weakly transduced postsynaptic neurons. Experiments shown in Figure 2A incorporate the use of two recommended reporter lines: Ai14, which provides Cre-dependent tdTomato expression (Madisen et al., 2010), and Frt-GFP (also known as RCE::FRT, Sousa et al., 2009) which drives Flp-dependent GFP expression. Their use has previously been described for the detection of transsynaptic AAV1-Cre or AAV1-FlpO expression, respectively (Zingg et al., 2020), and they may be bred together to create dual-reporter mice for the simultaneous detection of both (see below). Table 2 provides a list of additional reporter lines that may be useful for AAV1 detection, and all are available through The Jackson Laboratory (https://mice.jax.org/).
Table 2.
List of commercially available transgenic reporter mice for AAV1 anterograde transsynaptic applications.
| Transgenic name | Recombinase dependence | Fluorescent Reporter | Locus | Originating Lab | Vendor name | Stock # |
|---|---|---|---|---|---|---|
| Ai14 | Cre | tdTomato | ROSA26 | Hongkui Zeng, Allen Institute | JAX | 7914 |
| Ai140 | Cre | EGFP | TIGRE | Hongkui Zeng, Allen Institute | JAX | 34100 |
| RCE:loxP | Cre | EGFP | ROSA26 | Gord Fishell, Harvard | JAX | 32037 |
| RCE:FRT | FlpO | EGFP | ROSA26 | Gord Fishell, Harvard | JAX | 32038 |
| Ai65F | FlpO | tdTomato | ROSA26 | Hongkui Zeng, Allen Institute | JAX | 32864 |
| Ai193 | Cre or FlpO | Cre-dependent EGFP, FlpO-dependent tdTomato | TIGRE | Hongkui Zeng, Allen Institute | JAX | 34111 |
| RC::RLTG | DreO or DreO + Cre | DreO-dependent tdTomato, DreO + Cre-dependent EGFP | ROSA26 | Patricia Jensen, NIEHS | JAX | 26931 |
| Ai213 | Cre, FlpO, or oNigri | Cre-dependent EGFP, FlpO-dependent mOrange2, oNigri-dependent mKate2 | TIGRE | Hongkui Zeng, Allen Institute | JAX | 34113 |
As mentioned above, different transgenic reporter mice may be crossbred (e.g. Ai14 x Frt-GFP) or designed to report the presence of multiple recombinase systems using distinct fluorescent proteins (e.g. Ai193 dual-reporter or Ai213 triple-reporter mice, Table 2). As highlighted in Figure 2A. Strategy 2, injections of two different recombinase-expressing AAVs can be targeted to different brain regions in a dual-reporter mouse to directly compare the distribution of neurons downstream of each pathway. In structures that are co-targeted by each upstream injection, this strategy further provides an opportunity to assess the degree of convergence, or lack thereof, onto different classes of neurons in the target region. This approach may be extended to incorporate additional recombinase systems as more viral and transgenic resources become available.
Lastly, transgenic reporter mice can be bred with other cell-type specific Cre- or Flp-expressing transgenic lines for the purpose of screening the genetic identity of various transsynaptically labeled cell populations (Fig. 2A. Strategy 3). For example, Vgat-Flp mice (Jackson Laboratories, Stock 029591) may be crossed with both Ai14 and Frt-GFP lines to express GFP in all Vgat+ inhibitory neurons and tdTomato in Cre+ neurons. Then, following an injection of AAV1-Cre, transsynaptically labeled cells in a given target region may be screened for the presence of Vgat-GFP expression to determine their inhibitory identity. This provides a relatively simple approach for further classification of input-defined neurons based on their co-expression of functionally relevant marker genes.
If transgenic reporter mice are not available, the experimenter may rely on other methods for detecting transsynaptic spread in wild-type mice. These include immunostaining for the recombinase protein, performing systemic injections of a conditional, fluorophore-expressing AAV-PHP.eB vector (e.g. AAV-PHP.eB-hSyn-FLEX-GFP delivered via the retro-orbital sinus; Chan et al., 2017), or directly targeting of one or more downstream regions with injections of an AAV that conditionally expresses a fluorescent reporter protein. Regarding immunodetection, one potential caveat is that very low levels of Cre or FlpO expression may go undetected in the postsynaptic population and result in only partial reporting of the strongest labeled cells (Umaba et al., 2021). If this appears to be the case, researchers are encouraged to try retro-orbital injections of AAV-PHP.eB (typically a 50 uL injection volume containing ~1.0 x 1011 viral particles total; Chan et al., 2017; Graybuck et al., 2021) as an alternative approach for the global detection of labeling throughout the brain (see Yardeni et al., 2011 for a detailed protocol), or to use secondary injections of Cre- or Flp-dependent AAV as a more sensitive method for detecting spread in a particular target region, as described below.
Mapping axonal output of downstream neurons:
Transsynaptic delivery of AAV1-Cre or -Flp may be combined with secondary viral injections to selectively label input-defined cells in a chosen downstream target. As shown in Figure 2B, this may be achieved with an injection of AAV1-Cre in an upstream region, and AAV1-FLEX-GFP in a chosen postsynaptic target. This results in robust GFP expression specifically within the input-defined subset of cells and enables mapping of their corresponding axonal output to downstream targets. Given that many brain regions exhibit topographically specific patterns of input and output (e.g. cortico-ponto-cerebellar pathways; Henschke & Pakan, 2020; Zingg et al., 2020), this approach provides a valuable means for precisely demonstrating these relationships. As in other examples, AAV1-Cre and -Flp may be used simultaneously in two different pathways to directly demonstrate fine-scale topographical relationships in a single brain (e.g. see Lee et al., 2020 for a demonstration in striato-nigral and striato-pallidal circuits). Lastly, this approach may be further extended by using a dual-recombinase dependent AAV (e.g. INTRSECT, Fenno et al., 2014; Fenno et al., 2020) to map the axonal output of downstream neurons that receive input from two specified upstream sources.
Functional examination of downstream neurons:
The same two-step viral injection procedure described above may be used to functionally examine a given input-defined cell population. This simply involves substituting a fluorophore-expressing AAV with one that expresses a desired functional tool. For example, in Figure 2C, AAV1-FLEX-ChR2-GFP is injected into a specific region downstream of AAV1-Cre to conditionally express channelrhodopsin2 and GFP in post-synaptically labeled Cre+ cells. Following insertion of an optical fiber above the target region, the animal is recovered and several weeks later the input-defined cell population may be driven with pulses of blue LED light to examine any behavioral effects associated with activating this population (for additional examples, see Zingg et al., 2017; Beltramo & Scanziani, 2019; Cregg et al., 2020; and Li et al., 2021). A wide variety of Cre- and Flp-dependent AAV vectors are available for optogenetic (Kim et al., 2017) or chemogenetic (Roth, 2016) manipulation of a given input-defined cell population, and additional tools, such as Caspase3 (Gray et al., 2010; Yang et al., 2013) or diphtheria toxin receptor (Buch et al., 2005; Azim et al., 2014) may be expressed to selectively ablate these cells. Moreover, input-defined populations may instead be targeted for optical recording methods using virally expressed calcium or voltage sensors (Lin & Schnitzer, 2016; Chen et al., 2013; Dana et al., 2019; Piatkevich et al., 2018; Abdelfattah et al., 2019). Given the lack of toxicity associated with AAV infection in the downstream neurons, this approach is ideally suited for long term behavioral and functional studies. As previously mentioned, however, excessive recombinase expression in the upstream injection region should be mitigated, such as by avoiding vectors with WPRE, to avoid potential damage to these cells. Premade AAV stocks for nearly all of the functional tools highlighted here are readily available through Addgene (https://www.addgene.org/).
Accessing genetically-defined cell-types within a downstream neuronal population:
Lastly, each of the above two-step viral injection procedures may be performed in various transgenic Cre-driver mice to selectively access neurons based on both their input and their expression of a particular gene of interest. This may be achieved via transsynaptic delivery of an AAV1 vector that Cre-dependently expresses FlpO, followed by a second AAV injection that provides Flp-dependent expression of a fluorophore or functional tool in a given Cre-transgenic mouse (Zingg et al., 2017; Zingg et al., 2020). For example, in Figure 2D, AAV1-EF1a-DIO-FlpO (Addgene #87306, Table 1) is injected into an upstream region and AAV1-EF1a-fDIO-YFP is injected into a chosen downstream target in a Vgat-Cre mouse (Jackson Laboratories, Stock 029591) crossed with the Ai14-tdTomato line. Following anterograde transsynaptic spread of AAV1-EF1a-DIO-FlpO to the downstream target, FlpO will be expressed only in Vgat-Cre+ inhibitory cells that also receive presynaptic input from neurons in the upstream injection site. These cells will then unlock FlpO-dependent expression of AAV1-EF1a-fDIO-YFP to robustly label the dendrites and axonal projections of this input- and genetically-defined population. The same set of injections may then be repeated in different cell-type specific Cre-driver mice to explore the morphology, connectivity, and/or function of other genetically-defined subsets of neurons in the target region.
Selection of pathways for applying anterograde transsynaptic AAV1 spread
When choosing a particular target for the transsynaptic delivery of AAV1, several factors should be considered based on the known transport properties of AAV1, and its relative efficiency of spread through different cell-types and pathways in the brain.
AAV1 transport properties:
Previous studies have shown that AAV1 is capable of both retrograde and anterograde transsynaptic transport from a given injection site (Fig. 3A; Rothermel et al., 2013; Zingg et al., 2017; Zingg et al., 2020). In addition, its spread in the anterograde direction appears to be monosynaptic and always results in the labeling of cells in first order, but not second order, downstream targets (Fig. 3B; Zingg et al., 2017). Among these two properties, the former represents a major caveat that must be addressed when choosing a given pathway for transsynaptic experiments. In particular, given its capacity for bidirectional spread, AAV1 cannot be used in reciprocally connected circuits, since labeled cells resulting from the anterograde transsynaptic release of AAV1 may be indistinguishable from those labeled via retrograde transport. For example, most cortico-cortical pathways exhibit reciprocal connectivity at the regional level, which may result in both the labeling of pyramidal neurons that project to, and/or receive input from, different cell-types within the injection site. This complicates the interpretation of labeling results, though some insight may be gained through the use of combinatorial approaches to isolate specific cell populations in a target area. In particular, certain classes of pyramidal neurons, such as layer 6 (L6) corticothalamic, L5b corticofugal, or L5/6 near-projecting cells, as well as most inhibitory neurons, are not expected to form long-range intracortical axon projections (Harris & Shepherd, 2015; Tasic et al., 2018), and therefore are unlikely to be retrogradely labeled following a given AAV1 cortical injection. Anterograde transsynaptic inputs to these specific cell-types may then be identified by examining the co-expression of GFP and tdTomato in target cortical regions following injection of AAV1-hSyn-FlpO in a variety of different Cre-driver lines that label each cell class (e.g. Ntsr1-Cre, Sim1-Cre, Sst-Cre, etc.) and are crossed with Ai14 x Frt-GFP reporter mice (similar to Figure 2A, Strategy 3), or AAV1-EF1a-DIO-FlpO may be used instead to further target these input- and genetically-defined cells for Flp-dependent expression of tools for mapping and functional work (similar to Figure 2D, Strategy 6). Despite this potential workaround for accessing some postsynaptic cell populations within a reciprocal circuit, it remains a challenge to resolve the pre- or postsynaptic nature of connections among the many remaining cell-types in a given target that issue collaterals to the injection site. Application in cortico-cortical networks is therefore generally quite limited, and similar concerns also apply to cortico-thalamic circuits, given the high degree of reciprocal connectivity between the two structures.
Figure 3. Summary of AAV1 transport properties and efficiency of spread in different cell-types.
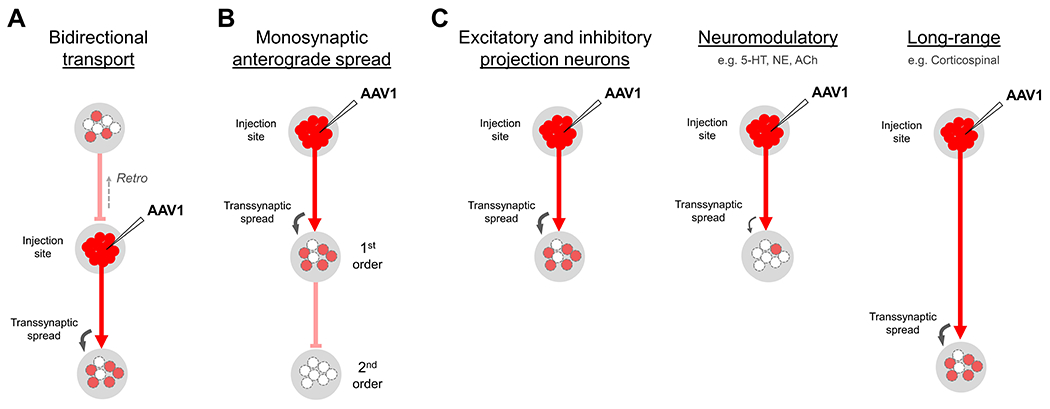
(A) Bidirectional spread. AAV1 exhibits both retrograde and anterograde transsynaptic transport from the injection site and should therefore be applied only in unidirectional pathways to avoid ambiguous labeling results. (B) Monosynaptic anterograde spread. Anterograde transsynaptic labeling is limited to first-order downstream cell populations and does not continue to spread beyond these initial cell targets, even after long survival times (e.g. 6 months). (C) Efficiency of anterograde transsynaptic spread in different cell-types. In all tested pathways, AAV1 appears to spread with equal efficiency in both excitatory and inhibitory cell populations (left), as well as in neurons that form long-range axonal projections, such as corticospinal neurons (right). However, transsynaptic spread appears to be inefficient in all neuromodulatory cell-types tested (middle), which includes serotonergic neurons in the dorsal raphe, noradrenergic neurons in the locus coeruleus, and cholinergic neurons in the basal forebrain. Dopaminergic neurons were not examined due to the bidirectional nature of their connections with the striatum and prefrontal cortex.
In light of this potential for retrograde transport, when designing a given experiment, it is important to avoid any potential ambiguity by first confirming the unidirectional nature of connections between a chosen AAV1 injection site and any given downstream target of interest. If the anatomy of the circuit is unclear, unidirectional connections should be verified by consulting available literature, reviewing relevant tracing examples found in online mapping databases (e.g. Allen Mouse Brain Connectivity Atlas, https://connectivity.brain-map.org/), or by directly performing retrograde tracer injections in the intended upstream AAV1 target (e.g. with fluorescently-conjugated cholera toxin subunit b, CTB-488, Thermo Fisher, Cat# C34775), followed by confirmation of the absence of retrograde cell body labeling in any of the given downstream targets of interest. This absence of retrograde labeling would imply a lack of reciprocal connectivity. In general, it is recommended to acquire as much anatomical information about the circuit as possible, as this can be useful in the experimental planning process to predict postsynaptic labeling outcomes for any chosen AAV1 injection (based on known axonal output for the region), and to provide confidence in which downstream targets will be unidirectional, and which may be reciprocally connected and must therefore be excluded from analysis.
AAV1 efficiency through different pathways:
Previous work has characterized the efficiency of AAV1 transsynaptic spread through a variety of cell-types and pathways in the brain (Zingg et al., 2017; Zingg et al., 2020). In general, AAV1 was observed to spread with equal efficiency through both excitatory and inhibitory projection neuron classes, and it was capable of robustly transducing cells downstream of long-range corticospinal projections, suggesting axon length may not diminish viral spread (Fig. 3C). These observations have since been supported by additional studies demonstrating its use in a variety of other unidirectional excitatory and inhibitory projection systems, including corticofugal (Yao et al., 2018; Henschke & Pakan, 2020; Li et al., 2021; Centanni et al., 2019; Bennet et al., 2019), pallidal (Lee et al., 2020; Foster et al., 2021), hippocampal (Trouche et al., 2019; Suarez et al., 2020; Umaba et al., 2021), thalamic (Huang et al., 2019; Watson et al., 2021), cerebellar (Fujita et al., 2020; Kelly et al., 2020), and brainstem pathways (Beltramo & Scanziani, 2019; Cregg et al., 2020; El-Boustani et al., 2020). Notably, however, transsynaptic spread of AAV1 through neuromodulatory cell populations, including cholinergic, serotonergic, and noradrenergic cell-types, was observed to be very inefficient, and should therefore be avoided (Fig. 3C; Zingg et al., 2020). This inefficient spread may be due to differences in viral uptake, intracellular trafficking, or the potential volume release of viral particles through axonal varicosities, rather than through direct synaptic contacts onto downstream neurons (Agnati et al., 1995). Overall, however, AAV1 is expected to perform well for excitatory or inhibitory pathways that densely innervate a given target, and satisfy the requirement of being unidirectional.
STEREOTAXIC INJECTION OF AAV1 FOR ANTEROGRADE TRANSSYNAPTIC SPREAD
The following protocol describes the process for stereotactically injecting AAV1 vectors in mice to achieve robust, anterograde transsynaptic transduction of downstream neurons. In addition, methods are presented for visualizing and characterizing the input-defined cell populations made accessible by this approach. Given the combinatorial possibilities associated with this method, we intend this protocol to serve as a general guide that may be adapted to address different scientific questions, depending on the investigator’s choice of pathways and use of viral and transgenic tools. Moreover, as the actual viral injection process is essentially the same as other previously published protocols, we present a basic overview here and refer readers to Harris et al., 2012 and Gore et al., 2013 for further details. Lastly, while this protocol describes procedures for mice, previous studies suggest AAV1 may also be used for anterograde transsynaptic experiments in rats (Umaba et al., 2021; Bennett et al., 2019; Suarez et al., 2020). Readers working in a rat model may refer to the Strategic Planning section of this protocol for background, and Testen et al., 2020 for a detailed stereotaxic injection protocol in rats.
Materials
AAV1 vector for anterograde transsynaptic use (should contain a neuron-specific promoter and express a chosen recombinase; titer should be ≥1.0 x 1013 GC/mL; aliquoted and stored at −80°C; see Table 1 for recommended viruses and commercial availability)
AAV vector for optional secondary injection in a downstream target (e.g. AAV-hSyn-FLEX-GFP, Addgene, Cat# 50457; should conditionally express a desired gene in neurons transsynaptically transduced by AAV1; titer should be ~1.0 x 1013 GC/mL and AAV serotypes 1 or 9 are recommended for strongest expression)
AAV vector for optional retro-orbital injection to detect transsynaptic labeling (e.g. AAV-PHP.eB-hSyn-FLEX-GFP, SignaGen, Cat# SL116009; or AAV-PHP.eB-CAG-FLEX-tdTomato, Addgene, Cat# 28306; prepared as a 50 μL solution containing 1.0 x 1011 viral particles total in PBS)
Wildtype or transgenic mice of a desired strain (see Table 2 for a list of recommended transgenic reporter mice for the detection of transsynaptic labeling)
Analgesic (e.g. Buprenorphine SR, 0.01 mg/mL in sterile saline)
Anesthetic (e.g. inhalable gas isoflurane, see Harris et al., 2012 for details)
Sterile 70% isopropyl alcohol prep pads (Vitality Medical, Cat# 58-204)
Betadine (Vitality Medical, Cat# MDS093944)
Mineral oil (VWR, Cat# J217-500ML)
Phosphate Buffered Solution (PBS) (Thermo Fisher, Cat# AM9624; dilute to 1X concentration before using)
4% paraformaldehyde (PFA) in PBS (Sigma-Aldrich, Cat# 441244)
Mounting medium (e.g. 60% glycerol in PBS, Sigma-Aldrich, Cat# G5516)
Glass pipets (Drummond Scientific, Cat# 3-000-203-G/X)
Micropipet puller (e.g. Sutter Instruments, Cat# P-30, or equivalent)
Vannas scissors (for trimming glass pipets, World Precision Instruments, Cat# 500086)
Stereotaxic apparatus (e.g. Kopf Model 1900, or equivalent)
Sterile insulin syringe, 0.5 mL, 29 G (for injecting analgesic, Fisher Scientific, Cat# 14-841-32)
Standard surgical instruments, sterilized (see Harris et al., 2012 for a complete list)
Microinjector (Nanoject III, Drummond Scientific, Cat# 3-000-207)
Sutures (VWR, Cat# 89219-134)
Heating pad (Kent Scientific, Cat# RT-JR-15)
Perfusion pump
Vibratome (e.g. Leica VT1000S, or equivalent)
Paintbrush (for collecting and mounting brain tissue, e.g. Blick Art Supply, Cat# 05857-1004)
Microscope slides (VWR, Cat# 16004-368)
Cover glass (VWR, Cat# 16004-312)
Epifluorescent or confocal microscope
NOTE: All protocols using live animals must first be reviewed and approved by an Institutional Animal Care and Use Committee (IACUC) and must follow officially approved procedures for the care and use of laboratory animals.
NOTE: Personal protective equipment should be used for this protocol, and all surgery should be done using aseptic and sterile techniques and equipment.
Choose an injection strategy and determine stereotaxic coordinates for each target
Refer to the Strategic Planning section and Figures 2 and 3 for guidance in designing a given experiment.
- Select one or more brain regions for anterograde transsynaptic injection of AAV1. If targeting multiple “starter” regions in a single brain, assign a different recombinase-expressing AAV1 vector (e.g. AAV1-hSyn-Cre or AAV1-hSyn-FlpO) to each injection site. Verify that downstream targets of interest are not bidirectionally connected with the AAV1 injection site.Due to the potential for retrograde transport of AAV1, it is critical to confirm the lack of reciprocal connectivity between the injection site region and any given downstream target. Consult literature, online brain mapping databases, or perform anterograde or retrograde tracer injections to verify this relationship before proceeding (see Strategic Planning for details).
- Choose a method for detecting viral spread in downstream neurons (e.g. by using transgenic reporter mice, systemic injections of AAV-PHP.eB, or secondary injections of recombinase-dependent AAV in a specific target).Immunostaining for recombinase protein may be used to confirm the location of AAV1 injection sites in wildtype mice. However, immunodetection of post-synaptic cells may be unreliable due to the low viral copy number and correspondingly low levels of recombinase expression found in many of these neurons.
- Determine stereotaxic coordinates for each injection target using a standard mouse brain reference atlas, such as the Allen Reference Atlas (Dong, 2008; https://atlas.brain-map.org/) or Allen Common Coordinate Framework (CCF) (Wang et al., 2020).When targeting a new structure, it may be useful to first test the accuracy of the chosen coordinates by performing an injection with an inexpensive fluorescent tracer, such as a conjugated dextran (e.g. Fluoro-Ruby, Cat# D1817, Thermo Fisher Scientific). Following injection, the animal may immediately be euthanized while still under anesthesia, transcardially perfused with 4% paraformaldehyde, and the brain processed and imaged, as described in more detail below. If the resulting injection site is off-target, apply a scale bar to the image, use this to calculate the distance to the desired target, and adjust the new coordinates accordingly.
- Acquire commercially available viruses for each planned injection site, or prepare your own. See Table 1 for a list of recommended AAV1 vectors for transsynaptic use, as well as several plasmids currently available for custom AAV1 production. Many additional AAV vectors for functional or anatomical characterization of post-synaptic cells are available through vector cores at Stanford University, University of North Carolina, or The Salk Institute for Biological Studies.For an introduction to producing AAV vectors in your own lab, see Grieger et al., 2006, Lock et al., 2010, and previously published protocols from Gray et al., 2011 and Negrini et al., 2020.For long-term storage of AAV stocks, aliquot into separate tubes containing 2-3 μL each and place in a −80°C freezer. It is important to avoid repeated freeze-thaw cycles that might damage the structural integrity of the virus and reduce the infectious titer of the stock over time. If stored properly, AAV stocks will remain viable for several years.
- Acquire the necessary mice based on the chosen experimental design. Consult Table 2 for a list of recommended transgenic recombinase-reporter mice.If needed, wildtype mice may be purchased directly from Jackson Laboratories (https://mice.jax.org/), and many cell-type specific Cre- or Flp-driver mice are also available through Jackson Laboratories or the Mutant Mouse Resource and Research Center (MMRRC; https://www.mmrrc.org/).
Prepare glass micropipettes for virus injection
-
7.
Using desired settings for length and taper, pull glass pipets with a micropipet puller.
-
8.Trim the pipets using a small pair of scissors (e.g. Vannas Scissors) to achieve a 20-25 μm inner diameter opening at the tip.For added control, it may be helpful to trim pipets under a dissection scope at 4X magnification. Tip diameter can be confirmed at 10X or 20X using a simple light microscope equipped with an eyepiece reticle.
-
9.Store pipets in a covered container for later use.To save time on the day of surgery, we recommend preparing all pipets in advance.
Inject viruses using pressure injection method
-
10.
Prepare the animal for stereotaxic surgery following previously published protocols (see Harris et al., 2012 and Gore et al., 2013 for detailed, step-by-step procedures). Briefly, the animal is anesthetized, positioned into a stereotaxic frame equipped with a heating pad, and administered an analgesic (e.g. Buprenorphine SR, 0.05 mg/kg) to minimize discomfort upon recovery. The scalp is then shaved, sterilized with 70% isopropyl alcohol and betadine, and an incision is made along the midline to expose the skull.
-
11.
Using predetermined stereotaxic coordinates, mark the location on the skull directly above the target area for each planned injection site.
-
12.
Drill a small hole (~0.5 mm diameter) in the skull for each target region.
-
13.Backfill a glass pipet with mineral oil, place into a programmable microinjector apparatus (e.g. Nanoject III), and withdraw into the pipet about 500 nL of a chosen AAV1 vector for use as an anterograde transsynaptic tracer (e.g AAV1-hSyn-Cre).Ensure that no air bubbles are present in the pipet. Air bubbles may compress during the injection and result in irregular or inefficient expulsion of viral solution from the pipet tip.
-
14.Lower the pipet to the desired targeting depth in the brain and slowly inject (~30 nL/min) a total of 50 – 150 nL of viral solution, depending on the desired size of the injection site and the desired strength of transsynaptic labeling.Iontophoresis may be used instead of the pressure injection approach for anterograde transsynaptic injections of AAV1, however it typically yields reduced numbers of postsynaptically labeled cells. Iontophoresis is therefore only recommended for experiments that require very discrete AAV1 injection sites with minimal spread to neighboring structures and minimal contamination along the pipet path. Preparation and targeting are the same as described above, and readers may refer to Harris et al., 2012 for details related to the iontophoretic injection process.
-
15.
After the injection is complete, allow the pipet to remain in place for ~1-2 mins before slowly withdrawing from the brain to minimize leakage of viral solution along the pipet path.
-
16.If another transsynaptic AAV1 injection is planned for a different recombinase-expressing vector (e.g. AAV1-hSyn-FlpO, Fig. 2A Strategy 2), complete this injection as described in steps 13 to 15.
-
17.If any secondary injections of recombinase-dependent AAV are planned for a given downstream target (e.g. AAV1-CAG-FLEX-GFP, Fig. 2B–D), these may be performed in the same surgical procedure as well. Fill pipets and target the planned injection sites as described in steps 13 and 14. Typically, a 50-100 nL injection volume is sufficient for good coverage and robust viral transgene expression in the target region.
-
18.
Complete all other remaining injections planned for the experiment following steps 13 to 15.
-
19.
If optogenetic manipulation is planned for a target region (Fig. 2C), the optical fiber may be implanted in the same procedure immediately following the last virus injection. Secure the fiber with a dental cement compound, and allow enough time for the cement to cure (typically ~15-20 mins, depending on the product used) before allowing the animal to recover from anesthesia.
-
20.
If no optogenetic work is planned, close the scalp incision with sutures.
-
21.If a retro-orbital injection of AAV-PHP.eB will be used to detect transsynaptically labeled neurons, perform this injection now before recovering the animal from anesthesia.This procedure typically requires less than 5 mins to complete. See the Strategic Planning section, Chan et al., 2017; and Yardeni et al., 2011 for a protocol describing the retro-orbital injection procedure in mice.
-
22.
Remove the mouse from anesthesia and allow time to recover in a clean cage placed on a heating pad.
-
23.
After all surgeries have been completed, return mice to their assigned holding room. Monitor their condition daily for 3 to 4 days after the procedure and provide post-operative care as needed to ensure healthy recovery.
Figure 4. Expected outcomes following one-step or two-step viral injection strategies.
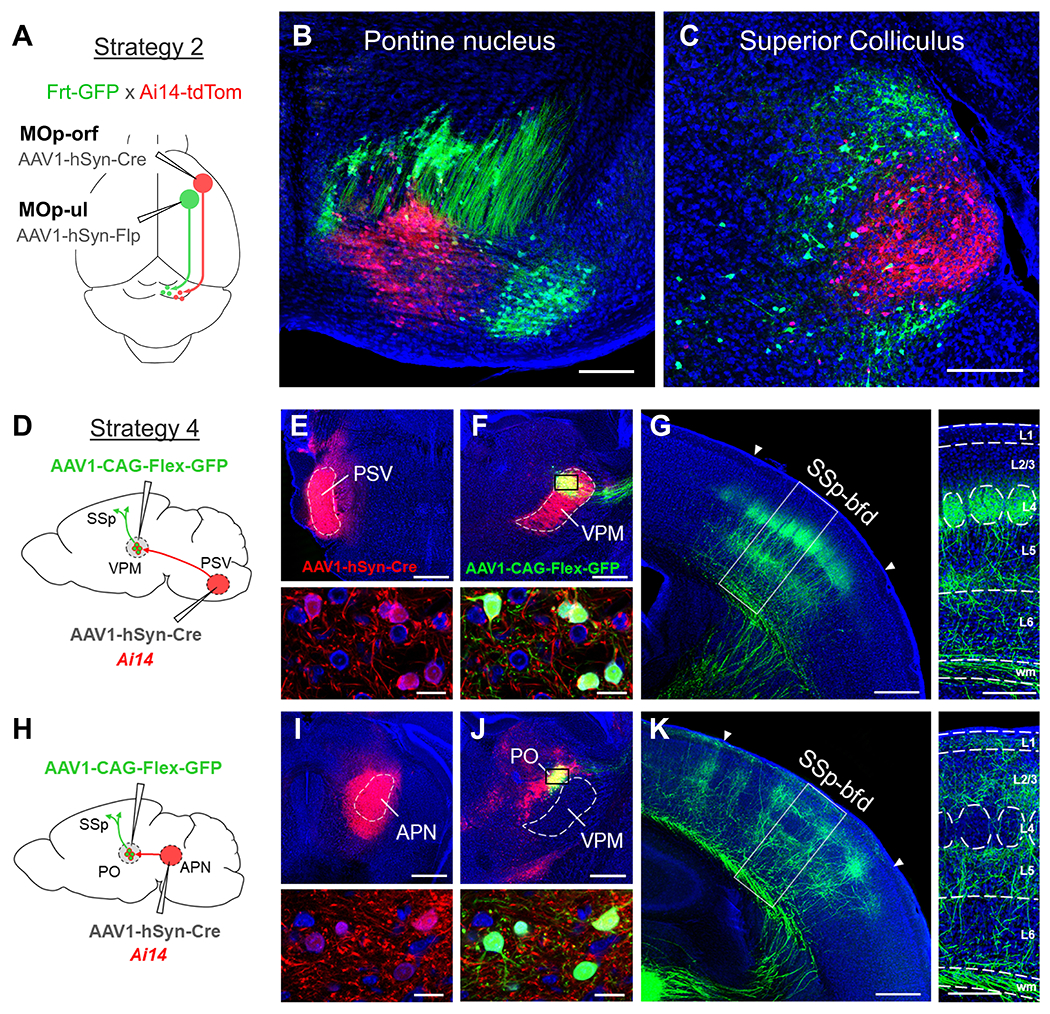
(A-C) An example of a one-step viral injection experiment in which single injections of AAV1-hSyn-Cre and AAV1-hSyn-Flp are applied in a dual reporter mouse (Frt-GFP x Ai14-tdTomato) to directly reveal the postsynaptic targets for each pathway (schematically shown in A, see Strategy 2). Following injection of AAV1-hSyn-Cre into MOp-orf and AAV1-hSyn-Flp into MOp-ul, topographically segregated populations of Cre+/tdTomato+ (red) and Flp+/GFP+ (green) cells were observed in downstream structures co-targeted by each pathway, such as the pontine nucleus (B) and the superior colliculus (C). Blue, fluorescent Nissl stain. Scale bar: 250 μm (B,C). (D-K) Examples of two-step viral injection experiments that compare the cortical targeting of two different input-defined thalamic populations: VPM (D-G) and PO (H-K). (D) Schematic of viral injections used to examine the axonal output of VPM thalamic neurons (see Strategy 4). AAV1-hSyn-Cre was injected into the contralateral PSV in an Ai14-tdTomato mouse (E) to postsynaptically label VPM, but not neighboring PO, neurons in the ipsilateral thalamus following a second injection of AAV1-CAG-FLEX-GFP into dorsal VPM (F). GFP (green) is expressed only in Cre+/tdTomato+ (red) VPM neurons (bottom panels, small box in F). Blue, fluorescent Nissl stain. Scale bar: 500 μm (E,F) and 25 μm (bottom panels). (G) Corresponding axonal projection to SSp-bfd from VPM neurons (green, left panel). Axons preferentially terminate in cortical layer 4 (L4) and at the border of L5/L6 (right panel). Scale bar: 500 μm (left panel) and 250 μm (right panel). (H) Schematic of viral injections used to examine the axonal output of PO thalamic neurons. AAV1-hSyn-Cre was injected into the APN (I) to postsynaptically label PO, but not adjacent VPM, neurons in the ipsilateral thalamus following secondary injection of AAV1-CAG-FLEX-GFP in PO (J). Bottom panels show close up of GFP expression in Cre+/tdTomato+ neurons in PO (small box in J). Scale bar: 500 μm (I,J) and 25 μm (bottom panels). (K) Corresponding axonal projection to SSp-bfd from PO neurons (left panel) and distinct laminar termination profile (right panel, L1, L2/3, L5), as compared with VPM neurons. Scale bar: 500 μm (left panel) and 250 μm (right panel). Abbreviations: APN, anterior pretectal nucleus; MOp-orf, primary motor cortex, orofacial part; MOp-ul, primary motor cortex, upper-limb part; PO, posterior nucleus of the thalamus; PSV, principal sensory trigeminal nucleus; SSp-bfd, somatosensory cortex, barrel field; VPM, ventral posteromedial nucleus of the thalamus
Post-injection survival and tissue processing
Following the injection procedure, we recommend allowing at least 2-3 weeks to achieve robust viral transgene expression. At this stage, functional and behavioral experiments can begin (e.g. using optogenetic, chemogenetic, or optical recording procedures to examine a postsynaptic cell population) and may proceed for any required length of time. Or, for anatomical experiments, mice may undergo transcardial perfusion at this timepoint, and their brains prepared for histological and imaging steps described below.
-
24.
Determine the experimental endpoint for each mouse and schedule transcardial perfusion times accordingly.
-
25.
Perfuse each mouse with PBS and 4% PFA according to standard protocols.
-
26.
Carefully dissect and remove the brain from the skull. Place the brain in 4% PFA and store at 4°C for 24 to 48 hours.
-
27.Section the brain at a desired thickness and orientation, and collect the tissue in wells containing PBS.We typically use a vibratome to cut 100 μm thick coronal sections across the entire brain.
-
28.Perform any desired immunostaining procedures according to standard protocols.As previously mentioned, immunostaining for Cre (or FlpO) may be used to visualize the location of AAV1 injection sites in wildtype mice. However, immunodetection of postsynaptic cell populations may be unreliable, given that many of these cells may only weakly express recombinase protein following transduction by just a few viral particles.To visualize cytoarchitectural information, we recommend treating sections with a fluorescent Nissl stain, such as NeuroTrace 640 (Thermo Fisher, Cat# N21483).
-
29.Mount sections onto glass slides and coverslip.For routine imaging of experiments, we use a simple mounting medium consisting of 60% glycerol in PBS.
-
30.
Store slides at 4°C and then image using a confocal or epifluorescent microscope.
COMMENTARY
Background Information
Given their natural ability to infect neurons and express desired genetic tools, a wide variety of neurotropic viruses have been adopted for use in the study of neural circuitry (Nassi et al., 2015; Bedbrook et al., 2018; Nectow & Nestler, 2020; Xu et al., 2020). These include viruses that locally transduce neurons (e.g. AAV, Chamberlin et al., 1998; Taymans et al., 2007; Ad5, Glover et al., 2002; and Lentivirus, Osten et al., 2006), distally transduce neurons via retrograde axonal uptake (e.g. CAV-2, Soudais et al., 2004; and AAVretro, Tervo et al., 2016), or spread transsynaptically and thus provide an important means for establishing connectivity between different populations of neurons across one or more synaptic steps (e.g. Rabies virus, Ugolini, 1995; Wickersham et al., 2007; PRV, DeFalco et al., 2001; Ekstrand et al., 2008; Herpes simplex virus H129, Lo & Anderson, 2011; Zeng et al., 2017; and VSV, Beier et al., 2011). Among these, rabies virus has proven to be especially useful for revealing presynaptically connected neurons upstream of a given starter cell population (Wickersham et al., 2007; Wall et al., 2010; Reardon et al., 2016), however viral tools that permit the forward mapping of postsynaptically connected cells (e.g. H129 and VSV) remain under development and may require further optimization to resolve issues related to their cytotoxicity and potential for bidirectional spread (Beier, 2019; Xu et al., 2020).
Recently, we demonstrated the capacity for AAV1 (and to some extent AAV9) to spread transsynaptically to cells downstream of an injection site (Zingg et al., 2017; Zingg et al., 2020). Given its broad neuronal tropism, efficient uptake at the soma, and lack of toxicity in host cells, AAV1 has become one of the most widely used vectors for driving stable, long-term gene expression in neurons locally transduced at the injection site. Despite this widespread use for local gene delivery, evidence indicating its capacity for transsynaptic spread has not been commonly reported (however, see Castle et al., 2014a; Hutson et al., 2016), and examples of robust viral gene expression in downstream neurons are rarely observed. This may be due to the possibility that only a small number of viral particles are transsynaptically released, and this results in undetectable levels of virally driven fluorescence in downstream neurons (see Figure 1 for details). However, when a recombinase-expressing AAV1 is injected into a transgenic reporter mouse, low levels of virally driven recombinase protein appear sufficient to unlock reporter gene expression in the postsynaptic population, thus revealing large numbers of fluorescently labeled cells in downstream targets and exposing this hidden capacity for AAV1 to spread transsynaptically. Importantly, the low levels of recombinase found in postsynaptic cells are also able to unlock strong expression of virally encoded fluorophores, opsins, or other tools delivered via secondary injections of AAV in a chosen downstream target. This greatly expands the application potential of AAV1 as a means for not only detecting, but also experimentally accessing postsynaptic cell populations to further characterize their functional and anatomical properties.
While the exact mechanism underlying transsynaptic spread of AAV1 remains unclear, previous studies have demonstrated that, following uptake at the cell body, AAV particles distribute broadly throughout the endosomal system, and a small fraction may undergo rapid, kinesin-2 dependent trafficking down the axon (Castle et al., 2014a; Castle et al., 2014b). Following delivery to synaptic terminals, exocytic vesicles containing AAV may then fuse with the presynaptic membrane and release their contents at, or near, the synaptic cleft, leading to viral uptake and transduction of downstream neurons. Using in vitro preparations, Castle et al. showed that viral trafficking down the axon occurs only after a high concentration of AAV has accumulated within the host cell. In addition, several different AAV serotypes (AAV1, 8, and 9) demonstrated a similar ability to traffic down the axon through common transport mechanisms, however, when delivered at equal concentrations, AAV1 was trafficked at a higher frequency due to its greater capacity for binding and entering host cells relative to AAV8 and 9 (Castle et al., 2014b). Taken together, these results corroborate our in vivo observations that AAV transsynaptic spread is specific to serotype 1, requires high viral titer to achieve, and scales in efficiency according to the total number of viral particles injected (Zingg et al., 2017). Moreover, while further studies are needed, these data provide a simple explanation for the observed serotype-specificity of transsynaptic spread. Rather than reflecting a serotype-dependent difference in intracellular trafficking, the unique ability of AAV1 to undergo anterograde transsynaptic transport may result from its more efficient import by host cells at equivalent concentrations, thus leading to more AAV particles available for trafficking down the axon, and a correspondingly greater numbers of particles released onto postsynaptic targets.
The synaptic specificity of AAV1 spread has been examined using a variety of different approaches. Initial observations revealed that AAV1 spreads only to expected first order targets downstream of an injection site, and the distribution of postsynaptically labeled cells closely matched the location and shape of presynaptic axon terminal fields (e.g. see Figure 1C). This, along with its demonstrated lack of spread from passing axons, suggested that AAV1 was most likely released at, or near, presynaptic terminals (Zingg et al., 2017). To further examine this, we (1) explored the specificity of spread to expected cell populations in anatomically well-defined circuits, (2) characterized the frequency with which postsynaptic cells received functional presynaptic input, and (3) revealed a dependence on synaptic vesicle machinery for viral release following tetanus toxin-mediated inhibition of synaptic vesicle fusion (see Zingg et al., 2020 for a detailed discussion). Collectively, these studies showed that AAV1 preferentially spreads to postsynaptically connected neurons in downstream targets. This synaptic spread was not entirely perfect, though, as an estimated 1-4% of cells were found to be labeled via extrasynaptic release of viral particles locally within the terminal field. Overall, however, these studies point to a high degree of selectivity in the spread of AAV1 to synaptically connected downstream cell populations.
Relative to other currently available tools for investigating anterograde transsynaptically defined circuits (e.g. H129 or VSV), AAV1 offers several advantages, including a well-established safety profile, widespread commercial availability, and a lack of toxicity that facilitates use in functional studies requiring the long-term expression of viral transgenes. Moreover, while transgenic Cre-driver mice and retrograde viral tracers enable the study of specific genetic- or target-defined cell-types within a circuit, viral tools that spread in an anterograde transsynaptic fashion provide a valuable alternative to these approaches, and offer unique access to input-defined cell populations that may otherwise be impossible to target using conventional approaches. Lastly, in terms of disadvantages, unlike H129 and VSV, anterograde transsynaptic spread of AAV1 cannot be initiated from a genetically specified starter cell population. Rather, AAV1 spread to downstream targets is expected to proceed through all projection neuron subtypes found within the injection site, and “starter cells” are therefore regionally specified with this technique and depend on the exact size and location of the viral injection. In addition, due to its potential for retrograde transport, transsynaptic use of AAV1 must be limited to use in unidirectional pathways (see Strategic Planning for additional discussion). Finally, given its inability to replicate and potential for variable uptake efficiency among different cell-types in the brain, AAV1 transsynaptic spread may not yield robust results for certain pathways, such as neuromodulatory cell-types, or projection neurons that form highly diffuse connections with large numbers of downstream targets. Future studies may seek to overcome these limitations by engineering novel capsid and host-cell receptor systems that enable selective uptake at the soma, but not local axon terminals, and facilitate robust transsynaptic spread from any genetically-defined starter cell population.
Critical Parameters
As previously discussed, the efficiency of anterograde transsynaptic spread of AAV1 depends on both the titer and strength of viral gene expression. If initial experiments yield insufficient transsynaptic spread for a given pathway, labeling results may be boosted simply by increasing the injection volume of the originally used virus. This will deliver a greater number of viral particles to the starter population, and may therefore increase the number of particles trafficked down the axons and released at postsynaptic sites. If the required injection volume is too large, however, specificity of the starter population may be lost, and tissue damage may occur at the injection site (e.g. with injections larger than 150 nL). In this case, acquiring a higher titer viral preparation (e.g. 1.0 x 1014 GC/mL) may be advantageous, as correspondingly large numbers of viral particles can now be delivered via a smaller injection volume. Lastly, if not previously implemented, use of a viral construct that incorporates the WPRE enhancer, along with a strong, neuron-specific promoter (e.g. hSyn), may further maximize transsynaptic transduction efficiency, especially if used at 1014 GC/mL. While this represents the highest performing viral preparation available, toxicity at the injection site will almost certainly occur due the excessively high levels of recombinase expression in the host cells. This may be problematic when used in functional and behavioral studies. Therefore, to mitigate this toxicity, researchers may opt to use a high titer preparation of a construct that lacks the WPRE enhancer. This will significantly reduce the recombinase expression levels in host cells at the injection site, and the final titer and injection volume can be adjusted to achieve the most optimal transsynaptic spread, while still preserving the health of the starter population.
Though not tested, another possible solution to eliminating recombinase-induced toxicity while still enabling use of maximal titer and gene expression strength, would be to inject an AAV1 construct that expresses one half of a split-Cre protein in a chosen upstream site, followed by injection of an AAV that expresses the other half of the split-Cre protein in a desired downstream target (Wang et al., 2012; Stanek et al., 2016). Following transsynaptic spread of AAV1 from the upstream injection, input-defined cells in the downstream target will express both halves of the protein, which then link to form a fully functional recombinase. As recombinase-induced toxicity likely results from off-target recombination and damage to host cell DNA (Loonstra et al., 2001; Thyagarajan et al., 2000; Janbandhu et al., 2014), expression of high levels of a non-functional half of this protein may not lead to toxic effects. In this case, the highest possible titer and strongest expressing construct could be used to maximize transsynaptic spread without causing damage to the starter cell population.
Finally, aside from optimizing spread and mitigating toxicity, one last consideration worth emphasizing again is that transsynaptic AAV1 injections must be used only in unidirectional pathways due to the potential for retrograde transduction of upstream neurons.
Troubleshooting
See Table 3 for a troubleshooting guide.
Table 3.
Troubleshooting guide for AAV1 anterograde transsynaptic injections.
| Problem | Possible Cause | Solution |
|---|---|---|
| Viral toxicity and cell death at the injection site | Excessively high recombinase protein expression | Use an AAV1 vector that lacks the WPRE enhancer element. Alternatively, reduce the viral titer and/or injection volume. Try an intersectional approach that avoids expression of functional recombinase at the injection site (e.g. split-Cre). |
| Astrocyte labeling | AAV1 promoter drives gene expression in both neurons and glial cells (e.g. EF1a, CAG, CMV) | Choose an AAV1 vector with a neuron-specific promoter (e.g. hSyn) |
| Insufficient spread of AAV1 to postsynaptic cell populations | Incomplete or insufficient viral injection volume | Try a larger injection volume (e.g. up to 150 nL) to increase transsynaptic spread. During the injection, monitor the movement of viral solution in the glass pipet to ensure the tip is not clogged. |
| Low viral titer | Ensure viral titer is ≥1.0 x 1013 GC/mL. Store aliquots at −80°C and avoid repeated freeze-thaw cycles to maintain viability. If possible, prepare or request higher titer stock (e.g. 1.0 x 1014 GC/mL), which can significantly increase transsynaptic labeling. | |
| Weak recombinase expression | If toxicity at the injection site is permitted, use an AAV1 vector that contains the WPRE enhancer. Avoid using vectors with weak promoters, such as CMV. Prepare or request self-complementary AAV1 packaging to further increase transduction efficiency. | |
| Inefficient recombinase | Use Cre instead of FlpO for a modest increase in transsynaptic labeling under equal conditions. Use codon optimized variants, when available. Test the performance of new recombinase options (e.g. DreO, oNigri) and choose the most efficient based on available information. |
Understanding Results
By following the procedures described in this protocol, researchers will be able to reveal and gain experimental access to input-defined neurons via transsynaptic injections of AAV1. Figure 4 provides examples of expected results following one-step or two-step viral injection procedures. In the first example, AAV1-hSyn-Cre and AAV1-hSyn-FlpO are injected into the orofacial- or forelimb-related regions of primary motor cortex in a dual Flp- and Cre-reporter mouse. Following transsynaptic spread from each injection site, the topographical distribution of postsynaptically labeled cells may be compared for each pathway. Both regions share common projections to several downstream targets, including the pontine nucleus (Fig. 4B) and superior colliculus (Fig. 4C), however the specific subsets of neurons innervated by each pathway appear separate from each other, and in turn each may have unique differences in connectivity and function that may be probed with follow up experiments.
The second example demonstrates results obtained from two-step injections used to map the axonal output of two different input-defined cell populations in the thalamus, VPM (Fig. 4D–G) and PO (Fig. 4H–K). Both regions are located close to each other and would be difficult to selectively target using conventional viral injection strategies. Following anterograde transsynaptic delivery of AAV1-hSyn-Cre from the PSV (Fig. 4E) or APN (Fig. 4I), however, postsynaptic Cre+ cells found specifically in VPM (Fig. 4F) or PO (Fig. 4J), respectively, can selectively be targeted with AAV1-CAG-FLEX-GFP to reveal their respective axonal projections to the primary somatosensory cortex (Fig. 4G,K). Each thalamic population has a unique targeting profile that innervates distinct layers of primary somatosensory cortex. Given this anatomical difference, follow up experiments can explore the function of these cells by substituting a Cre-dependent AAV that expresses an opsin, Ca2+ sensor, or other functional tool in each transsynaptically labeled thalamic population, as demonstrated in a similar study for this pathway (El-Boustani et al., 2020).
Time Considerations
The anticipated time required to complete each aspect of this protocol is listed below:
Perform one stereotaxic surgery: 1-2 hr, depending on number of targets
Perform a retro-orbital injection for detection of postsynaptic cell populations: ~5 min
Setup for the day: 30 min
Prepare all glass micropipets for a day of surgeries: ~45 min to prepare 12 pipets, assuming 3-6 surgeries, and 2-4 pipets used per animal.
Plan out a given surgery: ~30 min, this includes determining stereotaxic coordinates for each injection target and documenting basic information into a surgical log.
Post-injection survival time: 2-3 weeks for anatomical or ablation studies; optogenetic, chemogenetic, or optical recording experiments may begin at 2 weeks post-injection if local expression at the soma is sufficiently strong in the targeted population.
Transcardial perfusion and extraction of brain: 20 min per mouse
Post-fixation of brain tissue: 24-48 hr
Sectioning an entire brain: ~1 hr, cutting at 100 μm thickness
Immunostaining for recombinase protein or other cell markers: 3 days
Fluorescent Nissl staining: 2 hr at room temperature
Acquiring mice: Typically allow 1-2 weeks for orders of live mice to arrive, 10-14 weeks for cryopreserved mouse lines. If breeding, allow at least 3 months from the time of pairing to obtain adult offspring (≥2 months old) for use in experiments.
Acquiring viruses: Allow ~1 week for orders of premade AAV stock, and ~1-2 months for orders involving custom packaging of AAV expression plasmids.
Acknowledgements
This work was supported by NIH grants (R01DC008983) to L.I.Z., (R01EY019049) to H.W.T., and (U01MH116990) to L.I.Z., H.W.T., and H.W.D.
Footnotes
Conflict of interest statement:
The authors declare no competing interests.
Data Availability Statement:
The data, tools, and material (or their source) that support the protocol are available from the corresponding author upon reasonable request.
Literature Cited
- Abdelfattah AS, Kawashima T, Singh A, Novak O, Liu H, Shuai Y, Huang YC, Campagnola L, Seeman SC, Yu J, Zheng J, Grimm JB, Patel R, Friedrich J, Mensh BD, Paninski L, Macklin JJ, Murphy GJ, Podgorski K, … Schreiter ER (2019). Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science, 365(6454), 699–704. 10.1126/science.aav6416 [DOI] [PubMed] [Google Scholar]
- Agnati LF, Zoli M, Strömberg I, & Fuxe K (1995). Intercellular communication in the brain: Wiring versus volume transmission. Neuroscience, 69(3), 711–726. 10.1016/0306-4522(95)00308-6 [DOI] [PubMed] [Google Scholar]
- Azim E, Jiang J, Alstermark B, & Jessell TM (2014). Skilled reaching relies on a V2a propriospinal internal copy circuit. Nature, 508(7496), 357–363. 10.1038/nature13021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedbrook CN, Deverman BE, & Gradinaru V (2018). Viral Strategies for Targeting the Central and Peripheral Nervous Systems. Annual Review of Neuroscience, 41(1), annurev-neuro-080317–062048. 10.1146/annurev-neuro-080317-062048 [DOI] [PubMed] [Google Scholar]
- Beier KT (2019). Hitchhiking on the neuronal highway: Mechanisms of transsynaptic specificity. Journal of Chemical Neuroanatomy, 99(January), 9–17. 10.1016/j.jchemneu.2019.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier KT, Saunders A, Oldenburg IA, Miyamichi K, Akhtar N, Luo L, Whelan SPJ, Sabatini B, & Cepko CL (2011). Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. PNAS, 108(37), 15414–15419. 10.1073/pnas.1110854108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltramo R, & Scanziani M (2019). A collicular visual cortex: Neocortical space for an ancient midbrain visual structure. Science, 363(6422), 64–69. 10.1126/SCIENCE.AAU7052 [DOI] [PubMed] [Google Scholar]
- Bennett PJG, Maier E, & Brecht M (2019). Involvement of rat posterior prelimbic and cingulate area 2 in vocalization control. European Journal of Neuroscience, 50(7), 3164–3180. 10.1111/ejn.14477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJAJ, Kremer M, Wunderlich FT, Jung S, & Waisman A (2005). A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nature Methods, 2(6), 419–426. 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- Castle MJ, Perlson E, Holzbaur EL, & Wolfe JH (2014a). Long-distance Axonal Transport of AAV9 Is Driven by Dynein and Kinesin-2 and Is Trafficked in a Highly Motile Rab7-positive Compartment. Molecular Therapy: The Journal of the American Society of Gene Therapy, 22(3), 1–13. 10.1038/mt.2013.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle MJ, Gershenson ZT, Giles AR, Holzbaur ELF, & Wolfe JH (2014b). Adeno-associated virus serotypes 1, 8, and 9 share conserved mechanisms for anterograde and retrograde axonal transport. Human Gene Therapy, 25(8), 705–720. 10.1089/hum.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centanni SW, Morris BD, Luchsinger JR, Bedse G, Fetterly TL, Patel S, & Winder DG (2019). Endocannabinoid control of the insular-bed nucleus of the stria terminalis circuit regulates negative affective behavior associated with alcohol abstinence. Neuropsychopharmacology, 44(3), 526–537. 10.1038/s41386-018-0257-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin NL, Du B, De Lacalle S, & Saper CB (1998). Recombinant adeno-associated virus vector: Use for transgene expression and anterograde tract tracing in the CNS. Brain Research, 793(1–2), 169–175. 10.1016/S0006-8993(98)00169-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KY, Jang MJ, Yoo BB, Greenbaum A, Ravi N, Wu WL, Sánchez-Guardado L, Lois C, Mazmanian SK, Deverman BE, & Gradinaru V (2017). Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nature Neuroscience, 20(8), 1172–1179. 10.1038/nn.4593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr R. a, Orger MB, Jayaraman V, Looger LL, Svoboda K, & Kim DS (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature, 499, 295–300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Leiras R, Montalant A, Wanken P, Wickersham IR, & Kiehn O (2020). Brainstem neurons that command mammalian locomotor asymmetries. Nature Neuroscience. 10.1038/s41593-020-0633-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Li L, Tasic B, Walker M, Graybuck LT, Yao Z, Fong O, Nguyen TN, Garren E, Lenz GH, Mcgraw MJ, Ollerenshaw DR, Smith KA, Baker CA, & Ting JT (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Resource A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell, 465–480. 10.1016/j.cell.2018.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, Macklin JJ, Chen Y, Konnerth A, Jayaraman V, Looger LL, Schreiter ER, Svoboda K, & Kim DS (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature Methods, 16(7), 649–657. 10.1038/s41592-019-0435-6 [DOI] [PubMed] [Google Scholar]
- DeFalco J, Tomishima M, Liu H, Zhao C, Cai X, Marth JD, Enquist L, & Friedman JM. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001. Mar 30;291(5513):2608–13. doi: 10.1126/science.1056602 [DOI] [PubMed] [Google Scholar]
- Dong HW (2008). The Allen reference atlas: A digital color brain atlas of the C57Bl/6J male mouse. John Wiley & Sons Inc. [Google Scholar]
- Ekstrand MI, Enquist LW, & Pomeranz LE (2008). The alpha-herpesviruses: molecular pathfinders in nervous system circuits. Trends in Molecular Medicine, 14(3), 134–140. 10.1016/j.molmed.2007.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Boustani S, Sermet BS, Foustoukos G, Oram TB, Yizhar O, & Petersen CCH (2020). Anatomically and functionally distinct thalamocortical inputs to primary and secondary mouse whisker somatosensory cortices. Nature Communications, 11(1), 1–12. 10.1038/s41467-020-17087-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Mattis J, Ramakrishnan C, Hyun M, Lee SY, He M, Tucciarone J, Selimbeyoglu A, Berndt A, Grosenick L, Zalocusky K. a, Bernstein H, Swanson H, Perry C, Diester I, Boyce FM, Bass CE, Neve R, Huang ZJ, & Deisseroth K (2014). Targeting cells with single vectors using multiple-feature Boolean logic. Nature Methods, june. 10.1038/nmeth.2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno LE, Ramakrishnan C, Kim YS, Pichamoorthy N, Hong ASO, Deisseroth K, Fenno LE, Ramakrishnan C, Kim YS, Evans KE, Lo M, & Vesuna S (2020). Comprehensive Dual- and Triple-Feature Intersectional Single-Vector Delivery of Diverse Functional Payloads to Cells of Behaving Mammals. Neuron, 107(5), 836–853.e11. 10.1016/j.neuron.2020.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster NN, Barry J, Korobkova L, Garcia L, Gao L, Becerra M, Sherafat Y, Peng B, Li X, Choi J-H, Gou L, Zingg B, Azam S, Lo D, Khanjani N, Zhang B, Stanis J, Bowman I, Cotter K, … Dong H-W (2021). The mouse cortico–basal ganglia–thalamic network. Nature, 598(7879), 188–194. 10.1038/s41586-021-03993-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita H, Kodama T, & du Lac S (2020). Modular output circuits of the fastigial nucleus mediate diverse motor and nonmotor functions of the cerebellar vermis. ELife, 1–36. 10.1101/2020.04.23.047100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover CPJ, Bienemann AS, Heywood DJ, Cosgrave AS, & Uney JB (2002). Adenoviral-mediated, high-level, cell-specific transgene expression: A SYN1-WPRE cassette mediates increased transgene expression with no loss of neuron specificity. Molecular Therapy, 5(5), 509–516. 10.1006/mthe.2002.0588 [DOI] [PubMed] [Google Scholar]
- Gore BB, Soden ME, & Zweifel LS (2013). Manipulating gene expression in projection-specific neuronal populations using combinatorial viral approaches. Current Protocols in Neuroscience, 1(SUPPL.65), 1–20. 10.1002/0471142301.ns0435s65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray DC, Mahrus S, & Wells JA (2010). Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell, 142(4), 637–646. 10.1016/j.cell.2010.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray SJ, Choi VW, Asokan A, Haberman RA, McCown TJ, & Samulski RJ (2011). Production of recombinant adeno-associated viral vectors and use in in vitro and in vivo administration. Current Protocols in Neuroscience, SUPPL.57, 1–30. 10.1002/0471142301.ns0417s57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybuck LT, Daigle TL, Sedeño-Cortés AE, Walker M, Kalmbach B, Lenz GH, Morin E, Nguyen TN, Garren E, Bendrick JL, Kim TK, Zhou T, Mortrud M, Yao S, Siverts LA, Larsen R, Gore BB, Szelenyi ER, Trader C, … Tasic B (2021). Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron, 109(9), 1449–1464.e13. 10.1016/j.neuron.2021.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieger JC, Choi VW, & Samulski RJ (2006). Production and characterization of adeno-associated viral vectors. Nature Protocols, 1(3), 1412–1428. 10.1038/nprot.2006.207 [DOI] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, & Minshull J (2004). Codon bias and heterologous protein expression. Trends in Biotechnology, 22(7), 346–353. 10.1016/j.tibtech.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Harris JA, Wook Oh S, & Zeng H (2012). Adeno-associated viral vectors for anterograde axonal tracing with fluorescent proteins in nontransgenic and Cre driver mice. Current Protocols in Neuroscience, 1(April), 1–18. 10.1002/0471142301.ns0120s59 [DOI] [PubMed] [Google Scholar]
- Harris KD, & Shepherd GMG (2015). The neocortical circuit: themes and variations. Nature Neuroscience, 18(2), 170–181. 10.1038/nn.3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschke JU, & Pakan JM (2020). Disynaptic cerebrocerebellar pathways originating from multiple functionally distinct cortical areas. ELife, 9, 1–27. 10.7554/eLife.59148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Xi Y, Peng Y, Yang Y, Huang X, Fu Y, Tao Q, Xiao J, Yuan T, An K, Zhao H, Pu M, Xu F, Xue T, Luo M, So K-F, & Ren C (2019). A Visual Circuit Related to Habenula Underlies the Antidepressive Effects of Light Therapy. Neuron, 1–15. 10.1016/j.neuron.2019.01.037 [DOI] [PubMed] [Google Scholar]
- Hutson TH, Kathe C, & Moon LDF (2016). Trans-neuronal transduction of spinal neurons following cortical injection and anterograde axonal transport of a bicistronic AAV1 vector. Gene Therapy, 23(2), 231–236. 10.1038/gt.2015.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janbandhu VC, Moik D, & Fässler R (2014). Cre recombinase induces DNA damage and tetraploidy in the absence of LoxP sites. Cell Cycle, 13(3), 462–470. 10.4161/cc.27271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly E, Meng F, Fujita H, Morgado F, Kazemi Y, Rice LC, Ren C, Escamilla CO, Gibson JM, Sajadi S, Pendry RJ, Tan T, Ellegood J, Albert Basson M, Blakely RD, Dindot SV, Golzio C, Hahn MK, Katsanis N, … Tsai PT (2020). Regulation of autism-relevant behaviors by cerebellar–prefrontal cortical circuits. Nature Neuroscience, 23(9), 1102–1110. 10.1038/s41593-020-0665-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CK, Adhikari A, & Deisseroth K (2017). Integration of optogenetics with complementary methodologies in systems neuroscience. Nature Reviews Neuroscience, 18(4), 222–235. 10.1038/nrn.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Wang W, & Sabatini BL (2020). Anatomically segregated basal ganglia pathways allow parallel behavioral modulation. Nature Neuroscience, 23(11), 1388–1398. 10.1038/s41593-020-00712-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wei JX, Zhang GW, Huang JJ, Zingg B, Wang X, Tao HW, & Zhang LI (2021). Corticostriatal control of defense behavior in mice induced by auditory looming cues. Nature Communications, 12(1), 1–13. 10.1038/s41467-021-21248-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, & Schnitzer MJ (2016). Genetically encoded indicators of neuronal activity. Nature Neuroscience, 19(9), 1142–1153. 10.1038/nn.4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, & Anderson DJ (2011). A cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron, 72(6), 938–950. 10.1016/j.neuron.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock M, Alvira M, Vandenberghe LH, Samanta A, Toelen J, Debyser Z, & Wilson JM (2010). Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Human Gene Therapy, 21(10), 1259–1271. 10.1089/hum.2010.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JE, Cordier WS, Harris ME, Weitzman MD, & Hope TJ (2002). Enhanced Expression of Transgenes from Adeno-Associated Virus Vectors with the Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element: Implications for Gene Therapy. Human Gene Therapy, 10(14), 2295–2305. 10.1089/10430349950016942 [DOI] [PubMed] [Google Scholar]
- Loonstra A, Vooijs M, Beverloo HB, Allak B. Al, Van Drunen E, Kanaar R, Berns A, & Jonkers J (2001). Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America, 98(16), 9209–9214. 10.1073/pnas.161269798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Garner AR, Shimaoka D, Chuong AS, Klapoetke NC, Li L, van der Bourg A, Niino Y, Egolf L, Monetti C, Gu H, Mills M, Cheng A, Tasic B, Nguyen TN, Sunkin SM, Benucci A, Nagy A, Miyawaki A, … Zeng H (2015). Transgenic Mice for Intersectional Targeting of Neural Sensors and Effectors with High Specificity and Performance. Neuron, 85(5), 942–958. 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, and Zeng H (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature Neuroscience, 13(1), 133–140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DM (2008). Self-complementary AAV vectors; advances and applications. Molecular Therapy, 16(10), 1648–1656. 10.1038/mt.2008.171 [DOI] [PubMed] [Google Scholar]
- McCarty DM, Monahan PE, & Samulski RJ (2001). Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Therapy, 8(16), 1248–1254. 10.1038/sj.gt.3301514 [DOI] [PubMed] [Google Scholar]
- Nassi JJ, Cepko CL, Born RT, & Beier KT (2015). Neuroanatomy goes viral! Frontiers in Neuroanatomy, 9(July), 80. 10.3389/fnana.2015.00080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nectow AR, & Nestler EJ (2020). Viral tools for neuroscience. Nature Reviews Neuroscience, 21(12), 669–681. 10.1038/s41583-020-00382-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrini M, Wang G, Heuer A, Björklund T, & Davidsson M (2020). AAV Production Everywhere: A Simple, Fast, and Reliable Protocol for In-house AAV Vector Production Based on Chloroform Extraction. Current Protocols in Neuroscience, 93(1), 1–10. 10.1002/cpns.103 [DOI] [PubMed] [Google Scholar]
- Osten P, Dittgen T, & Licznerski P (2006). Lentivirus-based genetic manipulations in neurons in vivo. Front. Neurosci Chapter 13, 249–259. doi: 10.1201/9780203486283.ch13 [DOI] [PubMed] [Google Scholar]
- Piatkevich KD, Jung EE, Straub C, Linghu C, Park D, Suk HJ, Hochbaum DR, Goodwin D, Pnevmatikakis E, Pak N, Kawashima T, Yang CT, Rhoades JL, Shemesh O, Asano S, Yoon YG, Freifeld L, Saulnier JL, Riegler C, … Boyden ES (2018). A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nature Chemical Biology, 14(4), 352–360. 10.1038/s41589-018-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS and Soriano P (2007). High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PLoS ONE 2, e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, & Losonczy A (2016). Rabies Virus CVS-N2cΔG Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron, 89(4), 1–14. 10.1016/j.neuron.2016.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Lounnas V, Ehrlich L, Buchholz F, Wade R, & Stewart AF (1998). Comparative kinetic analysis of FLP and Cre recombinases: Mathematical models for DNA binding and recombination. Journal of Molecular Biology, 284(2), 363–384. 10.1006/jmbi.1998.2149 [DOI] [PubMed] [Google Scholar]
- Roth BL (2016). DREADDs for Neuroscientists. Neuron, 89(4), 683–694. 10.1016/j.neuron.2016.01.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimshek DR, Kim J, Hübner MR, Spergel DJ, Buchholz F, Casanova E, Stewart AF, Seeburg PH, & Sprengel R (2002). Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis, 32(1), 19–26. 10.1002/gene.10023 [DOI] [PubMed] [Google Scholar]
- Soudais C, Skander N, & Kremer EJ (2004). Long-term in vivo transduction of neurons throughout the rat CNS using novel helper-dependent CAV-2 vectors. The FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology, 18, 391–393. 10.1096/fj.03-0438fje [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, & Fishell G (2009). Characterization of Nkx6-2-derived neocortical interneuron lineages. Cerebral Cortex, 19(SUPPL. 1), 1–10. 10.1093/cercor/bhp038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanek E, Rodriguez E, Zhao S, Han B-X, & Wang F (2016). Supratrigeminal Bilaterally Projecting Neurons Maintain Basal Tone and Enable Bilateral Phasic Activation of Jaw-Closing Muscles. Journal of Neuroscience, 36(29), 7663–7675. 10.1523/JNEUROSCI.0839-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez AN, Liu CM, Cortella AM, Noble EE, & Kanoski SE (2020). Ghrelin and Orexin Interact to Increase Meal Size Through a Descending Hippocampus to Hindbrain Signaling Pathway. Biological Psychiatry, 87(11), 1001–1011. 10.1016/j.biopsych.2019.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasic B, Yao Z, Graybuck LT, Smith KA, Nguyen TN, Bertagnolli D, Goldy J, Garren E, Economo MN, Viswanathan S, Penn O, Bakken T, Menon V, Miller J, Fong O, Hirokawa KE, Lathia K, Rimorin C, Tieu M, … Zeng H (2018). Shared and distinct transcriptomic cell types across neocortical areas. Nature, 563(7729), 72–78. 10.1038/s41586-018-0654-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans JM, Vandenberghe LH, Van Den Haute C, Thiry I, Deroose CM, Mortelmans L, Wilson JM, Debyser Z, & Baekelandt V (2007). Comparative analysis of adeno-associated viral vector serotypes 1, 2, 5, 7, and 8 in mouse brain. Human Gene Therapy, 18(3), 195–206. 10.1089/hum.2006.178 [DOI] [PubMed] [Google Scholar]
- Tervo DGR, Hwang BY, Viswanathan S, Gaj T, Lavzin M, Ritola KD, Lindo S, Michael S, Kuleshova E, Ojala D, Huang CC, Gerfen CR, Schiller J, Dudman JT, Hantman AW, Looger LL, Schaffer DV, & Karpova AY (2016). A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron, 92(2), 372–382. 10.1016/j.neuron.2016.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testen A, Kim R, & Reissner KJ (2020). High-Resolution Three-Dimensional Imaging of Individual Astrocytes Using Confocal Microscopy. Current Protocols in Neuroscience, 91(1), 1–22. 10.1002/cpns.92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B, Guimarães MJ, Groth AC, & Calos MP (2000). Mammalian genomes contain active recombinase recognition sites. Gene, 244(1–2), 47–54. 10.1016/S0378-1119(00)00008-1 [DOI] [PubMed] [Google Scholar]
- Tian X, & Zhou B (2021). Strategies for site-specific recombination with high efficiency and precise spatiotemporal resolution. Journal of Biological Chemistry. 10.1016/j.jbc.2021.100509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouche S, Koren V, Doig NM, Ellender TJ, El-Gaby M, Lopes-Dos-Santos V, Reeve HM, Perestenko PV, Garas FN, Magill PJ, Sharott A, Dupret D (2019) A hippocampus-accumbens tripartite neuronal motif guides appetitive memory in space. Cell 176:1393–1406.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini G (1995). Specificity of rabies virus as a transneuronal tracer of motor networks: Transfer from hypoglossal motoneurons to connected second-order and higher order central nervous system cell groups. Journal of Comparative Neurology, 356(3), 457–480. 10.1002/cne.903560312 [DOI] [PubMed] [Google Scholar]
- Umaba R, Kitanishi T, & Mizuseki K (2021). Monosynaptic connection from the subiculum to medial mammillary nucleus neurons projecting to the anterior thalamus and Gudden’s ventral tegmental nucleus. Neuroscience Research, 171, 1–8. 10.1016/j.neures.2021.01.006 [DOI] [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, & Callaway EM (2010). Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proceedings of the National Academy of Sciences of the United States of America, 107(50), 21848–21853. 10.1073/pnas.1011756107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Ding SL, Li Y, Royall J, Feng D, Lesnar P, Graddis N, Naeemi M, Facer B, Ho A, Dolbeare T, Blanchard B, Dee N, Wakeman W, Hirokawa KE, Szafer A, Sunkin SM, Oh SW, Bernard A, Phillips JW, Hawrylycz M, Koch C, Zeng H, Harris JA, & Ng L. The Allen Mouse Brain Common Coordinate Framework: A 3D Reference Atlas. Cell. 2020. May 14;181(4):936–953.e20. doi: 10.1016/j.cell.2020.04.007, Epub 2020 May 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Chen T, Sakurai K, Han BX, He Z, Feng G, & Wang F (2012). Intersectional cre driver lines generated using split-intein mediated split-cre reconstitution. Scientific Reports, 2, 1–7. 10.1038/srep00497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson GDR, Hughes RN, Petter EA, Fallon IP, Kim N, Paolo F, Severino U, & Yin HH (2021). Thalamic projections to the subthalamic nucleus contribute to movement initiation and rescue of parkinsonian symptoms. Science Advances, February 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann KK, Young J. a T., & Callaway EM (2007). Monosynaptic Restriction of Transsynaptic Tracing from Single, Genetically Targeted Neurons. Neuron, 53, 639–647. 10.1016/j.neuron.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Holmes TC, Luo MH, Beier KT, Horwitz GD, Zhao F, Zeng W, Hui M, Semler BL, & Sandri-Goldin RM (2020). Viral Vectors for Neural Circuit Mapping and Recent Advances in Trans-synaptic Anterograde Tracers. Neuron, 107(6), 1029–1047. 10.1016/j.neuron.2020.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Chiang MC, Gray DC, Prabhakaran M, Alvarado M, Juntti SA, Unger EK, Wells JA, & Shah NM (2013). Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell, 153(4), 896–909. 10.1016/j.cell.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Zhang Q, Liao X, Li Q, Liang S, Li X, Zhang Y, Li X, Wang H, Qin H, Wang M, Li J, Zhang J, He W, Zhang W, Li T, Xu F, Gong H, Jia H, … Chen X (2018). A corticopontine circuit for initiation of urination. Nature Neuroscience, 21(11), 1541–1550. 10.1038/s41593-018-0256-4 [DOI] [PubMed] [Google Scholar]
- Yardeni T, Eckhaus M, & Morris HD (2011). Retro-orbital injections in mice. Lab Animal, 40(5), 155–160. 10.1038/laban0511-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WB, Jiang HF, Gang YD, Song YG, Shen ZZ, Yang H, Dong X, Tian YL, Ni RJ, Liu Y, Tang N, Li X, Jiang X, Gao D, Androulakis M, He X. Bin, Xia HM, Ming YZ, Lu Y, … Luo MH (2017). Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Molecular Neurodegeneration, 12(1), 1–17. 10.1186/s13024-017-0179-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng WB, Jiang HF, Gang YD, Song YG, Shen ZZ, Yang H, Dong X, Tian YL, Ni RJ, Liu Y, Tang N, Li X, Jiang X, Gao D, Androulakis M, He X. Bin, Xia HM, Ming YZ, Lu Y, … Luo MH (2017). Anterograde monosynaptic transneuronal tracers derived from herpes simplex virus 1 strain H129. Molecular Neurodegeneration, 12(1), 1–17. 10.1186/s13024-017-0179-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Chou X, Zhang Z, Mesik L, Liang F, Tao HW, & Zhang LI (2017). AAV-Mediated Anterograde Transsynaptic Tagging: Mapping Corticocollicular Input-Defined Neural Pathways for Defense Behaviors. Neuron, 93(1), 33–47. 10.1016/j.neuron.2016.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg B, Peng B, Huang J, Tao HW, & Zhang LI (2020). Synaptic Specificity and Application of Anterograde Trans-Synaptic AAV for Probing Neural Circuitry. The Journal of Neuroscience, 40(16), JN-RM-2158-19. 10.1523/jneurosci.2158-19.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data, tools, and material (or their source) that support the protocol are available from the corresponding author upon reasonable request.


