ABSTRACT
Stony coral tissue loss disease (SCTLD) is decimating Caribbean corals. Here, through the metatranscriptomic assembly and annotation of two alphaflexivirus-like strains, we provide genomic evidence of filamentous viruses in SCTLD-affected, -exposed, and -unexposed coral colonies. These data will assist in clarifying the roles of viruses in SCTLD.
ANNOUNCEMENT
Viral infections of endosymbiotic dinoflagellates (family Symbiodiniaceae) within coral tissues are hypothesized to play a role in stony coral tissue loss disease (SCTLD) (1), a widespread disease that affects Caribbean stony corals (2–4). Here, we present high-quality draft genome sequences for two viruses in the family Alphaflexiviridae, coral holobiont-associated alphaflexvirus (CHFV) 1 and 2 (Fig. 1A), that were assembled from metatranscriptomes from SCTLD-affected, SCTLD-exposed, and control (unexposed) coral holobionts sampled during a SCTLD transmission experiment (5). The field collections were authorized by the Department of Planning and Natural Resources Coastal Zone Management under permit number DFW19057U.
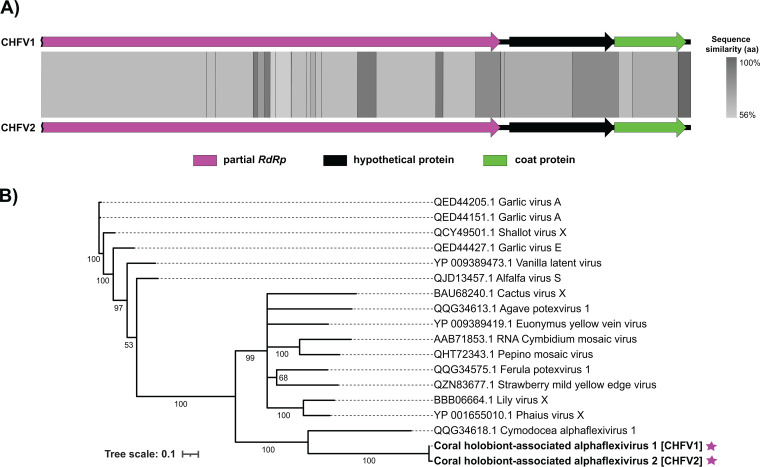
FIG 1.
(A) Visualized tBLASTx pairwise alignment of the two coral holobiont-associated alphaflexivirus (CHFV) genomes reported in this study. The arrows represent the predicted genes; the arrow color corresponds to the annotation type. The gray-scale shading between the two genomes represents the percent amino acid (aa) sequence similarity. (B) Maximum likelihood phylogeny generated from translated alphaflexivirus RdRp amino acid sequences from the CHFVs (purple stars) reported in this study, as well as previously described plant-associated alphaflexiviruses. Translated alphaflexivirus RdRp amino acid sequences were aligned using MUSCLE v5 (36) and trimmed using trimAl (37). The phylogeny was constructed using IQTREE v2 (38) with the LG+I+G4 substitution model (determined by ModelFinder [39]), and support was assessed using 1,000 nonparametric bootstrap replicates. The tree was visualized using the Interactive Tree of Life v5 (40); branches with bootstrap support values of <50 were collapsed. The tree scale indicates the number of amino acid substitutions per site.
Tissue samples were harvested from 12 frozen fragments of three coral species (Montastraea cavernosa, Porites astreoides, and Pseudodiploria strigosa) collected from St. Thomas, U.S. Virgin Islands (Table 1). Total RNA was extracted using the RNAqueous-4PCR total RNA isolation kit (Invitrogen, Life Technologies AM1914). Tissues were lysed using a refrigerated Qiagen TissueLyser II microcentrifuge at 30 oscillations per second for 30 s. The elution stage consisted of two consecutive 30-μL elutions. Contaminating DNA and chromatin were removed from the total RNA using the Ambion DNase I (RNase-free) kit (Invitrogen, Life Technologies AM2222). Samples were preprocessed by Novogene Co., Ltd. (Davis, CA, USA) for mRNA enrichment using polyA tail capture; the mRNA libraries underwent 150-bp, paired-end sequencing on an Illumina NovaSeq 6000 instrument using the NEBNext Ultra II RNA library prep kit.
TABLE 1.
Sample information and RNA sequencing results for libraries with reads that contributed to the generation of coral holobiont-associated alphaflexivirus genome assemblies (CHFV1 and CHFV2)
| Coral speciesa | Sample ID | SRA accession no. | No. of raw reads (millions) | No. of cleaned reads (millions) | No. of noncoral/non-Symbiodiniaceae reads (millions)b | Colony health statusc | No. of reads mapped to CHFV1 | No. of reads mapped to CHFV2 |
|---|---|---|---|---|---|---|---|---|
| Montastraea cavernosa | Mcav_c3 | SRR17230326 | 72.450238 | 72.148886 | 19.297152 | Control | 2,872 | 142 |
| Mcav_c6 | SRR17230325 | 43.625450 | 43.554690 | 12.021936 | Control | 780 | 46 | |
| Mcav_c7 | SRR17230323 | 61.253870 | 60.945362 | 17.604312 | Control | 300 | 3,524 | |
| Mcav_d2 | SRR17230322 | 58.179614 | 57.916530 | 15.643962 | Disease exposed | 26 | 152 | |
| Mcav_d3 | SRR17230321 | 55.231168 | 54.982506 | 15.468590 | Disease affected | 834 | 92 | |
| Mcav_d4 | SRR17230320 | 55.800824 | 55.586294 | 15.306024 | Disease exposed | 16 | 12 | |
| Mcav_d6 | SRR17230319 | 51.396670 | 50.957730 | 15.184224 | Disease affected | 3,282 | 142 | |
| Mcav_d8 | SRR17230318 | 62.688392 | 62.423220 | 18.170028 | Disease exposed | 34 | 56 | |
| Porites astreoides | Past_c6 | SRR17230317 | 56.484214 | 56.032382 | 12.568462 | Control | 16 | 16 |
| Past_d4 | SRR17230316 | 62.052534 | 61.792644 | 13.390588 | Disease exposed | 316 | 72 | |
| Past_d6 | SRR17239955 | 38.818898 | 38.603064 | 8.295652 | Disease affected | 16 | 0 | |
| Pseudodiploria strigosa | Pstrig_d5 | SRR17230324 | 55.559408 | 55.277124 | 20.791874 | Disease exposed | 4,736 | 1,422 |
On shallow reefs in the U.S. Virgin Islands, M. cavernosa typically harbors Symbiodiniaceae in the genus Cladocopium, P. astreoides typically harbors Symbiodiniaceae in the genus Symbiodinium, and P. strigosa is typically dominated by Symbiodiniaceae in the genus Breviolum (6, 7) but can also be dominated by Cladocopium symbionts (8).
Reads not mapping to coral or Symbiodiniaceae transcriptomes and retained for further analysis using BBSplit (within BBMap v38.90) (9). A genome-guided M. cavernosa transcriptome was generated using the draft genome from reference 10, and de novo Porites astreoides and Pseudodiploria strigosa transcriptomes were assembled using Trinity v2.11.0 (11). These reference transcriptomes were generated by the Mydlarz lab (University of Texas at Arlington, Arlington, TX, USA) for internal use but will be made available upon request. Symbiodiniaceae transcriptomes representing the genera Symbiodinium, Breviolum, Cladocopium, and Durusdinium were sourced from reference 12, “Kb8 Sequences” (http://medinalab.org/zoox/), reference 13, “S. minutum” (http://zoox.reefgenomics.org/download/), reference 14, “Clade C1 Symbiodinium” (http://ssid.reefgenomics.org/download/), and reference 15, “Dtrenchii_rnaseq_assembly_v1.0” (https://datadryad.org/stash/dataset/doi:10.5061/dryad.12j173m), respectively.
“Disease-affected” colony health status indicates corals that showed active lesions at the time of sampling; “disease-exposed” indicates coral fragments that were exposed to SCTLD but showed no signs of disease by the end of the experiment; “control” indicates that fragments were never exposed to SCTLD and never developed lesions during the course of the experiment.
All bioinformatic tools were run using default parameters unless otherwise specified. BBSplit (BBMap v38.90) was used to map quality-filtered (fastp v0.20.1 [16]) reads to coral or Symbiodiniaceae transcriptomes (9) and generate three read files: (i) coral, (ii) Symbiodiniaceae, and (iii) noncoral/non-Symbiodiniaceae. Noncoral/non-Symbiodiniaceae reads were combined and normalized using BBnorm.sh within BBMap (Table 1). Normalized reads were assembled using the program TransPi (17). Multiple assemblies were generated using rnaSPADES v3.14.0 (kmer: 75,85,91,107 nucleotides) (18), Trans-ABySS v2.0.1 (kmer: 25,35,55,75,85 nucleotides) (19), SOAPdenovo-Trans v1.03 (kmer: 25,35,55,75,85 nucleotides) (20), Trinity v2.9.1 (kmer: 35 nucleotides) (11), and Velvet v1.2.12/Oases v0.2.09 (kmer: 65,71,81,87,91,97,101 nucleotides) (21, 22). The multiple assemblies were concatenated into a single file, and the EvidentialGene tr2aacds pipeline v2019.05.14 (23, 24) was used to collapse duplicates and remove misassembled contigs from the assembly file. VirSorter2 (25) was used to detect RNA viruses from the nonredundant metatranscriptome-assembly file (minimum length, 300 nucleotides). Viral genomes similar to known members of the Alphaflexiviridae were identified by aligning translated open reading frames (ORFs) to the proteic version of the Reference Virus Database (26, 27) with DIAMOND BLASTx v2.0.11.149 in “ultra-sensitive” mode (28, 29). Cenote-Taker 2 (30) was used to annotate identified viral genomes with similarity to the Alphaflexiviridae and calculate the genome coverage using the normalized reads. The alphaflexivirus read count per sample library was estimated by mapping nonnormalized reads to the nonredundant assembly using bowtie2 (31) with the align_and_estimate_abundance.pl script (11; Table 1).
The CHFV1 and CHFV2 genomes are linear, share 85.9% genome-wide nucleotide identity, and are 6,228 and 6,227 nucleotides long with 42.4% and 42.0% G+C content, respectively. Coverages for the CHFV1 and CHFV2 assemblies are estimated at 334.9× and 123.4×, respectively. CheckV (32) was used to identify the genomes as high quality with 90% completeness (average amino acid identity-based [medium-confidence]). Visualization of a tBLASTx (33) pairwise alignment between the CHFV genomes was conducted using Easyfig (34) and depicted the genomes’ three shared ORFs (Fig. 1A). The closest relative of the CHFV genomes, as determined using Cenote-Taker 2, is strawberry mild yellow edge virus (NCBI protein accession number NP_620642.1) (35), sharing ∼33.5% amino acid similarity for the RNA-dependent RNA polymerase (RdRp) (ORF1).
A phylogenetic tree was generated from translated RdRp sequences from the two CHFVs and 16 plant-associated alphaflexiviruses (Fig. 1B). The CHFV replicase sequences formed a clade with the RdRp sequence of an unclassified alphaflexivirus that infects Cymodocea nodosa seagrass (Fig. 1B).
The CHFV genomes reported here constitute genomic-based evidence of filamentous viruses from coral colonies. Quantitative PCR primer sets can be developed from these genome assemblies to support the critical next step of characterizing the presence/absence and abundance of coral holobiont-associated alphaflexiviruses across coral colonies, to further clarify the potential role of viruses in SCTLD.
Data availability.
Coral holobiont-associated alphaflexivirus 1 and 2 have been deposited at NCBI’s GenBank (accession numbers OM030231 and OM030232). The raw reads from the transcriptome sequencing (RNA-Seq) libraries were deposited at NCBI’s Sequence Read Archive (SRA) under BioProject accession number PRJNA788911 (Table 1).
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation (Biological Oceanography) award numbers 1928753 to M.E.B. and T.B.S., 1928609 to A.M.S.C., 1928817 to E.M.M., 19228771 to L.D.M., 1927277 to D.M.H., and 1928761 to A.A., as well as by VI EPSCoR (NSF numbers 0814417 and 1946412).
This work could not have been completed without the hours of lab and fieldwork dedicated by Adam Glahn, Danielle Lasseigne, Amanda Long, Bradley Arrington, Daniel Mele, Kathryn Cobleigh, Bradford Dimos, Nicholas Macknight, Naomi Huntley, and Alexandra Gutting. We also thank Samantha Coy for her input regarding CHFV transcripts, as well as Nikoloas Schizas and the Marine Genomic Biodiversity Lab of the Department of Marine Sciences at the University of Puerto Rico—Mayaguez for providing us with access to computational resources used to conduct this study.
This is contribution number 229 from the Center for Marine and Environmental Studies at the University of the Virgin Islands.
Contributor Information
A. M. S. Correa, Email: ac53@rice.edu.
Jelle Matthijnssens, KU Leuven.
REFERENCES
- 1.Work TM, Weatherby TM, Landsberg JH, Kiryu Y, Cook SM, Peters EC. 2021. Viral-like particles are associated with endosymbiont pathology in Florida corals affected by stony coral tissue loss disease. Front Mar Sci 8:750658. doi: 10.3389/fmars.2021.750658. [DOI] [Google Scholar]
- 2.NOAA. 2018. Stony coral tissue loss disease case definition. https://floridadep.gov/sites/default/files/Copy%20of%20StonyCoralTissueLossDisease_CaseDefinition%20final%2010022018.pdf.
- 3.Precht WF, Gintert BE, Robbart ML, Fura R, van Woesik R. 2016. Unprecedented disease-related coral mortality in southeastern Florida. Sci Rep 6:31374. doi: 10.1038/srep31374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt ME, Ennis RS, Meiling SS, Townsend J, Cobleigh K, Glahn A, Quetel J, Brandtneris V, Henderson LM, Smith TB. 2021. The emergence and initial impact of stony coral tissue loss disease (SCTLD) in the United States Virgin Islands. Front Mar Sci 8:715329. doi: 10.3389/fmars.2021.715329. [DOI] [Google Scholar]
- 5.Meiling SS, Muller EM, Lasseigne D, Rossin A, Veglia AJ, MacKnight N, Dimos B, Huntley N, Correa AMS, Smith TB, Holstein DM, Mydlarz LD, Apprill A, Brandt ME. 2021. Variable species responses to experimental stony coral tissue loss disease (SCTLD) exposure. Front Mar Sci 8:670829. doi: 10.3389/fmars.2021.670829. [DOI] [Google Scholar]
- 6.Correa AMS, Brandt ME, Smith TB, Thornhill DJ, Baker AC. 2009. Symbiodinium associations with diseased and healthy scleractinian corals. Coral Reefs 28:437–448. doi: 10.1007/s00338-008-0464-6. [DOI] [Google Scholar]
- 7.Cunning R, Gates RD, Edmunds PJ. 2017. Using high-throughput sequencing of ITS2 to describe Symbiodinium metacommunities in St. John, US Virgin Islands. PeerJ 5:e3472. doi: 10.7717/peerj.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LaJeunesse TC. 2002. Diversity and community structure of symbiotic dinoflagellates from Caribbean coral reefs. Mar Biol 141:387–400. doi: 10.1007/s00227-002-0829-2. [DOI] [Google Scholar]
- 9.Bushnell B. 2014. BBTools software package. http://sourceforge.net/projects/bbmap.
- 10.Rippe JP, Dixon G, Fuller ZL, Liao Y, Matz MV. 2021. Environmental specialization and cryptic genetic divergence in two massive coral species from the Florida Keys Reef Tract. Mol Ecol 30:3468–3484. doi: 10.1111/mec.15931. [DOI] [PubMed] [Google Scholar]
- 11.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bayer T, Aranda M, Sunagawa S, Yum LK, DeSalvo M, Lindquist E, Schwarz J, Coffroth MA, Voolstra CR, Medina M. 2012. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 7:e35269. doi: 10.1371/journal.pone.0035269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson JE, Baumgarten S, Michell CT, Baums IB, LaJeunesse TC, Voolstra CR. 2016. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol Evol 8:665–680. doi: 10.1093/gbe/evw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies SW, Ries JB, Marchetti A, Castillo KD. 2018. Symbiodinium functional diversity in the coral Siderastrea siderea is influenced by thermal stress and reef environment, but not ocean acidification. Front Mar Sci 5:150. doi: 10.3389/fmars.2018.00150. [DOI] [Google Scholar]
- 15.Bellantuono AJ, Dougan KE, Granados-Cifuentes C, Rodriguez-Lanetty M. 2019. Free-living and symbiotic lifestyles of a thermotolerant coral endosymbiont display profoundly distinct transcriptomes under both stable and heat stress conditions. Mol Ecol 28:5265–5281. doi: 10.1111/mec.15300. [DOI] [PubMed] [Google Scholar]
- 16.Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rivera-Vicéns RE, Garcia-Escudero CA, Conci N, Eitel M, Wörheide G. 2021. TransPi—a comprehensive TRanscriptome ANalysiS PIpeline for de novo transcriptome assembly. bioRxiv https://www.biorxiv.org/content/10.1101/2021.02.18.431773v3. [DOI] [PubMed]
- 18.Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. GigaScience 8:giz100. doi: 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman SD, Mungall K, Lee S, Okada HM, Qian JQ, Griffith M, Raymond A, Thiessen N, Cezard T, Butterfield YS, Newsome R, Chan SK, She R, Varhol R, Kamoh B, Prabhu AL, Tam A, Zhao Y, Moore RA, Hirst M, Marra MA, Jones SJ, Hoodless PA, Birol I. 2010. De novo assembly and analysis of RNA-seq data. Nat Methods 7:909–912. doi: 10.1038/nmeth.1517. [DOI] [PubMed] [Google Scholar]
- 20.Xie Y, Wu G, Tang J, Luo R, Patterson J, Liu S, Huang W, He G, Gu S, Li S, Zhou X, Lam T-W, Li Y, Xu X, Wong GK-S, Wang J. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics 30:1660–1666. doi: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- 21.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz MH, Zerbino DR, Vingron M, Birney E. 2012. Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28:1086–1092. doi: 10.1093/bioinformatics/bts094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert DG. 2013. Gene-omes built from mRNA seq not genome DNA. 7th Annu Arthropod Genom Symp. University of Notre Dame, Notre Dame, IN. http://arthropods.eugenes.org/EvidentialGene/about/EvigeneRNA2013poster.pdf. [Google Scholar]
- 24.Gilbert DG. 2019. Longest protein, longest transcript or most expression, for accurate gene reconstruction of transcriptomes? bioRxiv doi: 10.1101/829184. [DOI]
- 25.Guo J, Bolduc B, Zayed AA, Varsani A, Dominguez-Huerta G, Delmont TO, Pratama AA, Gazitúa MC, Vik D, Sullivan MB, Roux S. 2021. VirSorter2: a multi-classifier, expert-guided approach to detect diverse DNA and RNA viruses. Microbiome 9:37. doi: 10.1186/s40168-020-00990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodacre N, Aljanahi A, Nandakumar S, Mikailov M, Khan AS. 2018. A reference viral database (RVDB) to enhance bioinformatics analysis of high-throughput sequencing for novel virus detection. mSphere 3:e00069-18. doi: 10.1128/mSphereDirect.00069-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bigot T, Temmam S, Pérot P, Eloit M. 2019. RVDB-prot, a reference viral protein database and its HMM profiles. F1000Res 8:530. doi: 10.12688/f1000research.18776.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buchfink B, Reuter K, Drost H-G. 2021. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat Methods 18:366–368. doi: 10.1038/s41592-021-01101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 30.Tisza MJ, Belford AK, Domínguez-Huerta G, Bolduc B, Buck CB. 2021. Cenote-Taker 2 democratizes virus discovery and sequence annotation. Virus Evol 7:veaa100. doi: 10.1093/ve/veaa100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nayfach S, Camargo AP, Schulz F, Eloe-Fadrosh E, Roux S, Kyrpides NC. 2021. CheckV assesses the quality and completeness of metagenome-assembled viral genomes. Nat Biotechnol 39:578–585. doi: 10.1038/s41587-020-00774-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelkmann W, Maiss E, Martin RR. 1992. The nucleotide sequence and genome organization of strawberry mild yellow edge-associated potexvirus. J Gen Virol 73:475–479. doi: 10.1099/0022-1317-73-2-475. [DOI] [PubMed] [Google Scholar]
- 36.Edgar RC. 2021. MUSCLE v5 enables improved estimates of phylogenetic tree confidence by ensemble bootstrapping. bioRxiv doi: 10.1101/2021.06.20.449169. [DOI]
- 37.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol 37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Coral holobiont-associated alphaflexivirus 1 and 2 have been deposited at NCBI’s GenBank (accession numbers OM030231 and OM030232). The raw reads from the transcriptome sequencing (RNA-Seq) libraries were deposited at NCBI’s Sequence Read Archive (SRA) under BioProject accession number PRJNA788911 (Table 1).