Abstract
Cryo-electron microscopy (cryo-EM) has become a leading approach for protein structure determination, but it remains challenging to accurately model atomic structures with cryo-EM density maps. We propose a hybrid method, CR-I-TASSER, which integrates deep neural-network learning with I-TASSER assembly simulations for automated cryo-EM structure determination. The method is benchmarked on 778 proteins with simulated and experimental density maps, where CR-I-TASSER constructs models with a correct fold (TM-score>0.5) for 643 targets that is 64% higher than the best of other de novo and refinement-based approaches on high-resolution data samples. Detailed data analyses showed that the major advantage of CR-I-TASSER lies in the deep-learning based position prediction, which significantly improves the threading template quality and therefore boosts the accuracy of final models through optimized fragment assembly simulations. These results demonstrate a new avenue to determine cryo-EM protein structures with high accuracy and robustness covering various target types and density-map resolutions.
INTRODUCTION
Knowledge of 3D structures of proteins is crucial for understanding their biological functions. Over the past decades, nuclear magnetic resonance (NMR) spectroscopy1, X-ray crystallography2 and electron microscopy (EM)3 have been widely employed to obtain protein structures. However, NMR can only be used for relatively small proteins, whereas X-ray crystallography is often constrained by the difficulty of protein crystallization4. Although EM can overcome some of these limitations, it suffers from sample damage due to high-energy radiation, or low signal-to-noise ratio when very low electron doses are used5. The idea of cryogenic electron microscopy (cryo-EM) was first proposed in the 1980s to reduce sample damage through frozen specimens6. Over the last decade, various technological innovations, such as single particle analysis and direct electron detection cameras5, 7, 8, have made cryo-EM a practical means for probing protein structures without crystallization (X-ray) or size limitations (NMR). However, the success rate of cryo-EM is low with low-resolution density map data and more than half of cryo-EM samples in the EMDataResource have no atomic structure determined9.
To help cryo-EM structure determination, a variety of computational structure modeling methods have been proposed, which can be generally categorized into two groups. The first group of approaches, such as Rosetta-Ref10, Flex-EM11, iMODFIT12, MDFF13, Situs14 and EM-Refiner15, are built on structure refinement guided by correlations between the atomic model and cryo-EM maps. Despite the relative simplicity, most of the refinement programs require predefined model and map superposition, and the success rate critically depends on the quality of initial models and the superposition. The second group is referred to as ‘de novo’ modeling which constructs models from sequence and density map alone. One such example is Rosetta de novo (Rosetta-dn)16, 17 which creates the initial model from a density map followed by RosettaES17 beam growing and Rosetta folding refinement. Another example is MAINMAST18 which constructs initial backbone models from local dense points and then refines the models with the MDFF program13. Although these de novo approaches are capable of creating models from density maps alone, their success is highly sensitive to the resolution level of density maps. Additionally, methods such as MAINMAST require manual tuning and combination of multiple parameter-sets, rendering the programs less convenient to be automatedly implemented.
We present a hybrid pipeline, CR-I-TASSER (CRyo-EM Iterative Threading ASSEmbly Refinement), for fully automated protein structure determination. While it is a de novo type approach in terms of creating models from sequence and density maps alone, CR-I-TASSER does utilize multithreading algorithms to identify homologous and analogous templates from the PDB to facilitate structural assembly. Technically, most existing de novo and refinement-based approaches rely on model-to-map correlations to guide the structural modeling simulations, but such correlation information is not precise and specific when the map resolution is low. In CR-I-TASSER, we extend deep residual convolutional neural networks (CNN)19 to create high-accuracy atom trace models from experimental density maps, providing a specific set of target atom positions that can be used to significantly improve threading template quality. In addition, the deep-learning boosted threading models are further assembled with cutting-edge I-TASSER folding simulations, under the guidance of specific CNN models and the highly optimized I-TASSER knowledge-based force field20. Our large-scale benchmark tests show a significant advantage of CR-I-TASSER over the traditional de novo and refinement-based approaches in assembling atomic cryo-EM protein structures. The online server and standalone package of CR-I-TASSER have been made publicly available at https://zhanggroup.org/CR-I-TASSER/.
RESULTS
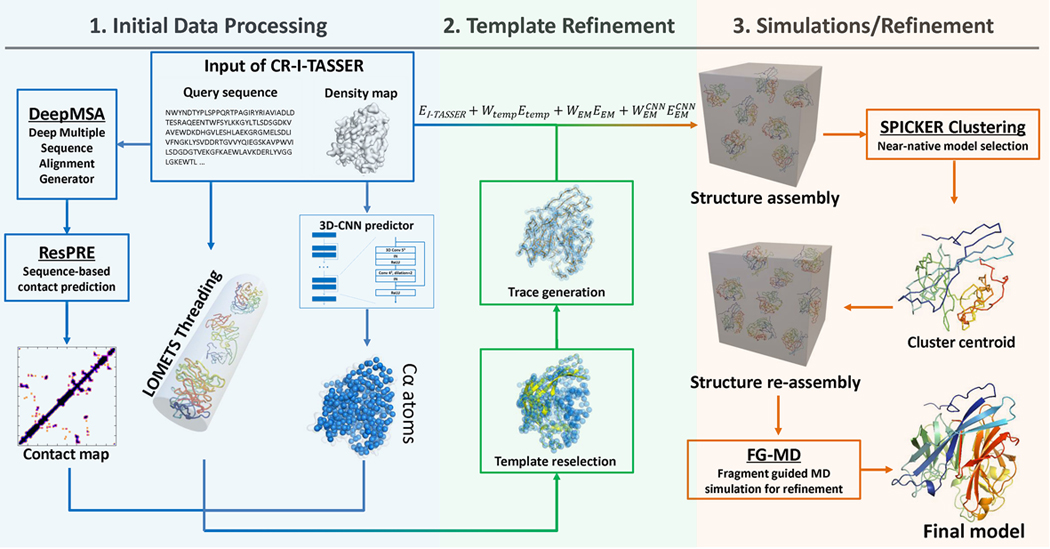
CR-I-TASSER is a hybrid method for determining atomic-level protein structures from cryo-EM density maps. As outlined in Fig. 1, CR-I-TASSER starts with the creation of a sequence-order independent conformation by deep convolutional neural network (3D-CNN) training from density maps. The conformation is then used to improve the threading templates created by LOMETS21, using multiple heuristic iteration algorithms designed to match the query and template sequences with the conformation for template reselection and trace regeneration. Finally, the iterative threading assembly refinement method (I-TASSER20) is extended to assemble atomic structure models under the guidance of both cryo-EM density map correlation and deep-learning boosted template restraints. Here, although CR-I-TASSER is built on I-TASSER and LOMETS21, the development of a deep-learning approach for cryo-EM based atom prediction and the integration of sequence-order independent models with advanced structure assembly methods represent the major novelty of the pipeline. Although there were prior efforts in applying deep-learning techniques to extract structural information from cryo-EM density maps22, 23, CR-I-TASSER marks the first pipeline utilizing sequence-order independent positions to improve threading alignments and regenerate order-dependent trace models, so that the deep-learning derived cryo-EM models can be directly used for guiding atomic-level structural assembly simulations. See Supplementary Text 1 for details of CR-I-TASSER datasets.
Figure 1.
CR-I-TASSER pipeline. Starting with a query sequence and cryo-EM density map, CR-I-TASSER constructs atomic models through 3 consecutive steps: 1. Initial data processing to generate 3D-CNN conformation, LOMETS threading and ResPRE contact-map prediction; 2. Density-map based template reselection and trace generation; 3. Density-map guided fragment reassembly simulations and model refinements.
Density-map based significantly improve template quality
A key component of CR-I-TASSER is the deep neural-network based atom prediction from cryo-EM density maps, which is used to guide both template regeneration and structure folding simulations. Since the predicted atoms from 3D-CNN do not have indexes, we define CRscore to estimate the similarity between the predicted atoms and the native structure by
| (1) |
where L is the target length. is the distance between ith atom in the 3D-CNN model and jth atom in the native structure, where the i-j correspondence is established by a greedy method selecting the non-redundant i-j pairs of the shortest distance (see Supplementary Text 2). is a distance scale taken from TM-score to rule out length dependence24. Here, the index information (and index connectivity) of both structures is completely ignored when computing CRscore since we establish the i-j correspondence by using their coordinate information only (see Supplementary Text 2).
In Supplementary Fig. 1a, we list the average CRscore of 3D-CNN models on the 530 test proteins in different resolution ranges. The average CRscore is >0.95 when the resolution is high (<5 Å), but slightly decreases when the resolution becomes lower (>10 Å). This is consistent with the trend of RMSD shown in Supplementary Fig. 1b, which is around 2–3 Å for high-resolution density maps but rises to 3–5 Å for low-resolution maps. As a comparison, we employ an established algorithm, MAINMAST, which can generate locations from the density map. In addition, we create atom models by a naïve greedy procedure which picks atom positions of the highest density values not in an excluded volume (see Supplementary Text 3). As shown in Supplementary Fig. 1, the average CRscore and RMSD from our 3D-CNN models are considerably better than MAINMAST and the naïve greedy procedure when resolution is high to medium (1–8 Å), and the scores become much better as the resolution drops, demonstrating the efficiency of deep-learning training process for position prediction.
Using the 3D-CNN models, CR-I-TASSER creates two types of templates by either density-map based template reselection or trace regeneration, followed by score re-ranking. In Supplementary Table 2, we compare TM-scores of the templates from LOMETS with those after 3D-CNN based refinement, where TM-score is a metric defined to assess structural similarity of two structures, which has values ranged in (0,1] with a higher value indicating closer similarity24 (see Supplementary Text 4 for a more detailed description of TM-score). In general, 3D-CNN makes the largest improvement for Hard targets in which traces deduced from 3D-CNN models have a significantly higher TM-score (0.690 and 0.527 with high- and low-resolution density maps respectively) than that of the original LOMETS (0.283). Combining both Easy and Hard targets, the TM-score of the first models by 3D-CNN (0.707) is 45% higher than that by the original LOMETS (0.487), which corresponds to a p-value=1.3×10−174 using Student’s t-test, showing that the template quality improvement brought by 3D-CNN is statistically highly significant.
CR-I-TASSER on high-resolution simulated density maps
To examine the efficiency of the CR-I-TASSER pipeline, we first apply it to the 301 Hard targets from our benchmark set that lack homologous templates in the PDB. Overall, CR-I-TASSER creates models with average TM-score=0.772 and RMSD=4.4 Å. If we count the targets with TM-score >0.5, which corresponds to a model with correct fold25, CR-I-TASSER creates correct folds for 251 targets, which is 9.3 times of that obtained by I-TASSER (=27, see Table 1), showing a significant impact of cryo-EM density maps on I-TASSER based structure modeling.
Table 1.
Modeling results by CR-I-TASSER and other methods on 778 benchmark test proteins involving different density map types and resolutions. P-values are calculated using two-tailed Student’s t-tests between the TM-scores produced by CR-I-TASSER and the other methods. Bold fonts highlight the performer which obtained the best average result in each category.
| Methods | N (TM>TM0)4 | (Å) | P-value | |
|---|---|---|---|---|
| 301 Hard targets with high-resolution density map (resolution in 2–5 Å) (TM0=0.5) | ||||
|
| ||||
| I-TASSER1 | 0.345 | 27 | 12.0 | 8.0×10−91 |
| Flex-EM2 | 0.318 | 22 | 12.4 | 3.8×10−96 |
| iMODFIT2 | 0.340 | 25 | 11.9 | 6.6×10−91 |
| MDFF2 | 0.331 | 26 | 12.1 | 3.4×10−91 |
| EM-Refiner2 | 0.315 | 18 | 12.2 | 6.9×10−96 |
| Rosetta-Ref2 | 0.297 | 30 | 14.0 | 1.2×10−99 |
| MAINMAST3 | 0.438 | 121 | 10.2 | 9.8×10−47 |
| Rosetta-dn3 | 0.419 | 94 | 12.2 | 8.7×10−52 |
| Phenix3 | 0.466 | 134 | 8.6 | 8.7×10−42 |
| CR-I-TASSER3 | 0.772 | 251 | 4.4 | -- |
|
| ||||
| 229 Easy targets with high-resolution density map (resolution in 2–5 Å) (TM0=0.9) | ||||
|
| ||||
| I-TASSER1 | 0.762 | 16 | 5.1 | 8.4×10−75 |
| Flex-EM2 | 0.824 | 66 | 4.4 | 4.6×10−35 |
| iMODFIT2 | 0.799 | 43 | 4.7 | 5.6×10−48 |
| MDFF2 | 0.857 | 104 | 4.1 | 4.8×10−21 |
| EM-Refiner2 | 0.846 | 76 | 4.0 | 3.5×10−37 |
| Rosetta-Ref2 | 0.851 | 103 | 4.0 | 6.9×10−21 |
| MAINMAST3 | 0.439 | 9 | 11.8 | 5.7×10−78 |
| Rosetta-dn3 | 0.474 | 17 | 12.0 | 8.0×10−77 |
| Phenix3 | 0.493 | 8 | 8.4 | 1.4×10−76 |
| CR-I-TASSER3 | 0.950 | 198 | 1.4 | -- |
|
| ||||
| 301 Hard targets with low-resolution density map (resolution in 5–15 Å) (TM0=0.5) | ||||
|
| ||||
| I-TASSER1 | 0.345 | 27 | 12.0 | 2.0×10−48 |
| Flex-EM2 | 0.303 | 13 | 12.3 | 1.2×10−61 |
| iMODFIT2 | 0.316 | 23 | 12.0 | 2.0×10−56 |
| MDFF2 | 0.319 | 29 | 11.8 | 6.8×10−55 |
| EM-Refiner2 | 0.305 | 19 | 12.1 | 2.3×10−60 |
| Rosetta-Ref2 | 0.268 | 18 | 13.9 | 1.6×10−70 |
| MAINMAST3 | 0.204 | 3 | 14.3 | 2.1×10−86 |
| Rosetta-dn3 | 0.201 | 7 | 14.6 | 6.7×10−91 |
| Phenix3 | 0.180 | 0 | 12.5 | 5.5×10−95 |
| CR-I-TASSER3 | 0.597 | 191 | 6.3 | -- |
|
| ||||
| 229 Easy targets with low-resolution density map (resolution in 5–15 Å) (TM0=0.9) | ||||
|
| ||||
| I-TASSER1 | 0.762 | 16 | 5.1 | 8.4×10−75 |
| Flex-EM2 | 0.666 | 0 | 5.3 | 3.5×10−90 |
| iMODFIT2 | 0.767 | 34 | 4.4 | 4.0×10−29 |
| MDFF2 | 0.788 | 46 | 4.3 | 5.5×10−23 |
| EM-Refiner2 | 0.739 | 21 | 4.7 | 5.3×10−42 |
| Rosetta-Ref2 | 0.714 | 14 | 4.9 | 7.5×10−49 |
| MAINMAST3 | 0.202 | 0 | 15.6 | 5.7×10−311 |
| Rosetta-dn3 | 0.225 | 1 | 9.2 | 1.5×10−238 |
| Phenix3 | 0.174 | 0 | 13.8 | 3.2×10−309 |
| CR-I-TASSER3 | 0.898 | 137 | 2.1 | -- |
|
| ||||
| 178 targets with experimental density map (resolution in 2–5 Å) (TM0=0.9) | ||||
|
| ||||
| I-TASSER1 | 0.647 | 6 | 8.3 | 4.0×10−15 |
| Flex-EM2 | 0.681 | 24 | 8.5 | 3.6×10−9 |
| iMODFIT2 | 0.695 | 19 | 7.8 | 6.8×10−8 |
| MDFF2 | 0.709 | 37 | 7.3 | 4.9×10−6 |
| EM-Refiner2 | 0.690 | 32 | 8.3 | 2.5×10−7 |
| Rosetta-Ref2 | 0.688 | 40 | 8.5 | 7.1×10−7 |
| MAINMAST3 | 0.323 | 2 | 15.2 | 7.4×10−72 |
| Rosetta-dn3 | 0.353 | 5 | 15.7 | 1.4×10−60 |
| Phenix3 | 0.349 | 1 | 13.3 | 2.7×10−63 |
| CR-I-TASSER3 | 0.810 | 75 | 4.9 | -- |
|
| ||||
| 70 targets with experimental density map (resolution in 5–10 Å) (TM0=0.5) | ||||
|
| ||||
| I-TASSER1 | 0.612 | 49 | 9.2 | 2.7×10−3 |
| Flex-EM2 | 0.546 | 45 | 9.3 | 4.3×10−7 |
| iMODFIT2 | 0.603 | 48 | 8.9 | 1.7×10−3 |
| MDFF2 | 0.573 | 46 | 8.7 | 5.9×10−5 |
| EM-Refiner2 | 0.576 | 45 | 8.8 | 9.7×10−5 |
| Rosetta-Ref2 | 0.554 | 43 | 9.3 | 9.7×10−6 |
| MAINMAST3,5 | 0.221 | 0 | 16.1 | 2.0×10−31 |
| Rosetta-dn3 | 0.176 | 1 | 15.6 | 5.4×10−41 |
| Phenix3 | 0.118 | 0 | 18.3 | 1.5×10−43 |
| CR-I-TASSER3 | 0.714 | 63 | 6.2 | -- |
Protein structure prediction methods
Cryo-EM based structure refinement methods
Cryo-EM based de novo structure modeling methods
TM0=0.5 for simulated Hard targets and low-resolution experimental targets, =0.9 for simulated Easy targets or high-resolution experimental targets
Only 61 targets are solved with MAINMAST, probably due to the low resolution and experimental noise
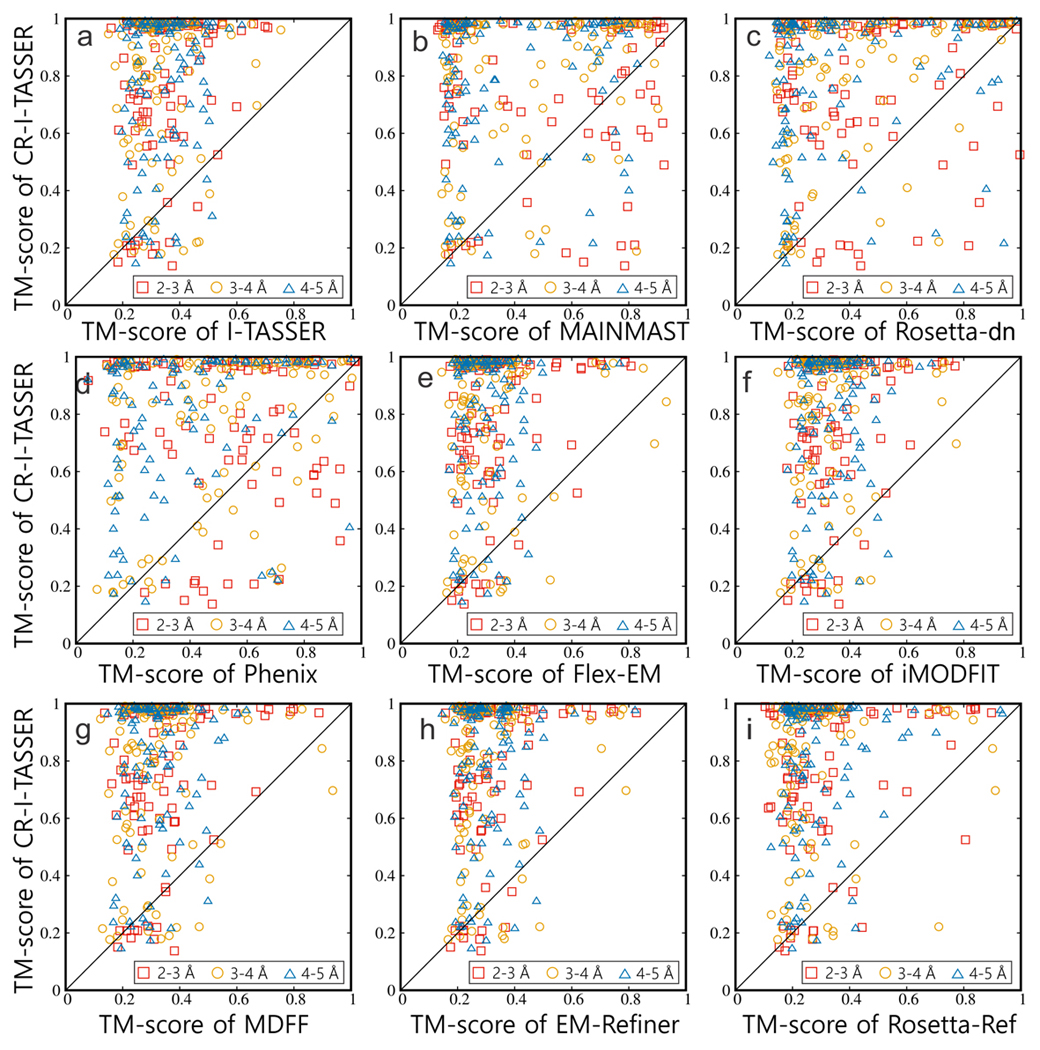
As a comparison, we list in Table 1 (Rows 9–11) the results from three de novo programs, MAINMAST18, Rosetta-dn16, 17 and Phenix26, which create models from the same set of density map data (see Supplementary Texts 5-7 for setting). It shows that CR-I-TASSER outperforms these programs significantly with the average TM-score 76% higher than MAINMAST (0.438), 84% higher than Rosetta-dn (0.419), and 66% higher than Phenix (0.466). In Figs. 2b-d, we present a head-to-head TM-score comparison of CR-I-TASSER with the three control programs, where CR-I-TASSER has a higher TM-score in 259/270/252 cases than MAINMAST/Rosetta-dn/Phenix and the latter does so only in 42/31/49 cases. In Figs. 2e-i, we also list the modeling results by five state-of-the-art cryo-EM refinement programs from Flex-EM11, iMODFIT12, MDFF13, EM-Refiner15 and Rosetta-Ref10, which start with the I-TASSER models after superposition of the density maps using Situs14 (see Supplementary Texts 8-12). Overall, the refinement programs do not work well for the Hard targets, where their TM-scores are even lower than that of the initial I-TASSER models, probably due to the poor quality of the initial I-TASSER models for the Hard proteins that have an average TM-score of 0.345. This result is consistent with a previous observation15, which showed that the correlation between model quality and model-to-map correlation coefficient (CC) vanishes when the TM-score of the initial models <0.5, and therefore there is no sufficient CC gradient to guide the programs for refining structures. We also benchmarked CR-I-TASSER on 229 Easy targets, where it outperforms other control groups with a significantly higher TM-score (0.949; p<10−20 in all cases, Student’s t-test). Details can be found in Supplementary Text 13.
Figure 2.
TM-score comparisons of CR-I-TASSER with I-TASSER and eight other control methods on 301 Hard targets with 2–5 Å resolution simulated density maps. CR-I-TASSER versus (a) I-TASSER; (b) MAINMAST; (c) Rosetta-dn; (d) Phenix; (e) Flex-EM; (f) iMODFIT; (g) MDFF; (h) EM-Refiner; (i) Rosetta-Ref. The symbols with different colors and shapes denote different ranges of resolution: red square: 2–3 Å; yellow circle: 3–4 Å; blue triangle: 4–5 Å.
In addition to the global structure quality listed in Table 1, we also calculate the local structure scores, including clashes and Molprobity27, in Supplementary Table 3. CR-I-TASSER achieves the second-best clash and Molprobity scores following Rosetta-Ref, indicating that the CR-I-TASSER models have a reasonable local structure quality. Moreover, we demonstrate that improvement of template quality plays a critically important role in CR-I-TASSER structure assembly (Supplementary Text 14), and benchmarked CR-I-TASSER under Gaussian noises added by Xmipp28 (see Supplementary Texts 15 and 16 for details). Furthermore, in Supplementary Fig. 3, we present an illustrative example from polyomavirus VP1 pentamer protein (PDB ID: 1vps-A), which demonstrates that the template regeneration process can create high-quality templates from the 3D-CNN traces and result in much improved full-length structure models, even though the initial threading templates are completely incorrect (see Supplementary Text 17 for details).
CR-I-TASSER on low-resolution simulated density maps
While cryo-EM experiments are now achieving increasingly good resolutions, it is still of importance to model structures from medium- and low-resolution density maps, especially for molecules with high flexibility or conformational/compositional heterogeneity5. In Table 1 (Rows 25–34), we examine the performance of CR-I-TASSER on the 301 Hard proteins with resolution ranging from 5 to 15 Å. Compared to the models with high-resolution density maps (2–5 Å), the overall performance of CR-I-TASSER is reduced in the low-resolution set with an average TM-score=0.597; this is mainly due to the reduction of the 3D-CNN model quality with lower map resolution, as shown in Supplementary Fig. 1. Nevertheless, the TM-score of CR-I-TASSER is significantly higher than the de novo programs by MAINMAST (0.204), Rosetta-dn (0.201) and Phenix (0.180), as well as the refinement programs by Flex-EM (0.303), iMODFIT (0.316), MDFF (0.319), EM-Refiner (0.305) and Rosetta-Ref (0.268). A similar trend can be found on the 229 Easy targets as summarized in Table 1 (Rows 36–47); see Supplementary Text 18 for details.
In Supplementary Figs. 4a-b, we list a head-to-head TM-score comparison of CR-I-TASSER with the best de novo and refinement programs, where CR-I-TASSER outperforms MAINMAIST/MDFF in 296/265 cases, while the latter does so only in 5/36 cases. If we count the number of cases with TM-score >0.5, CR-I-TASSER constructs the correct fold for 191 out of the 301 targets, which is 63 times of that by MAINMAST (3) and 7.3 times of that by MDFF (26). As an illustration, we present in Supplementary Figs. 4c-h the modeling results on Q6MIM9 from Bdellovibrio bacteriovorus, which highlights that the hybrid effects of both template reselection and regeneration processes, as well as the optimized structure assembly simulations, make a major contribution to the modeling of a Hard target with very low-resolution density maps (see Supplementary Text 19).
Overall, although the average TM-score of CR-I-TASSER drops for low-resolution maps in 530 Hard/Easy targets, the magnitude of the TM-score reduction for CR-I-TASSER (by 17% from 0.849 to 0.727) is much smaller than that of the other de novo methods, including MAINMAST (54%), Rosetta-dn (53%) and Phenix (73%). Even with the low-resolution maps, the average TM-score of CR-I-TASSER is 87% higher than that of the second-best method (MDFF) for Hard targets, and 14% (299%) higher than other refinement-based (de novo) methods for Easy targets. This advantage on low-resolution data modeling is mainly attributed to the integration of multi-threading alignments and the deep Cα trace learning with the BFGS and MC assembly simulations, which makes CR-I-TASSER a robust pipeline for a wide range of map densities.
Structure modeling on experimental density maps
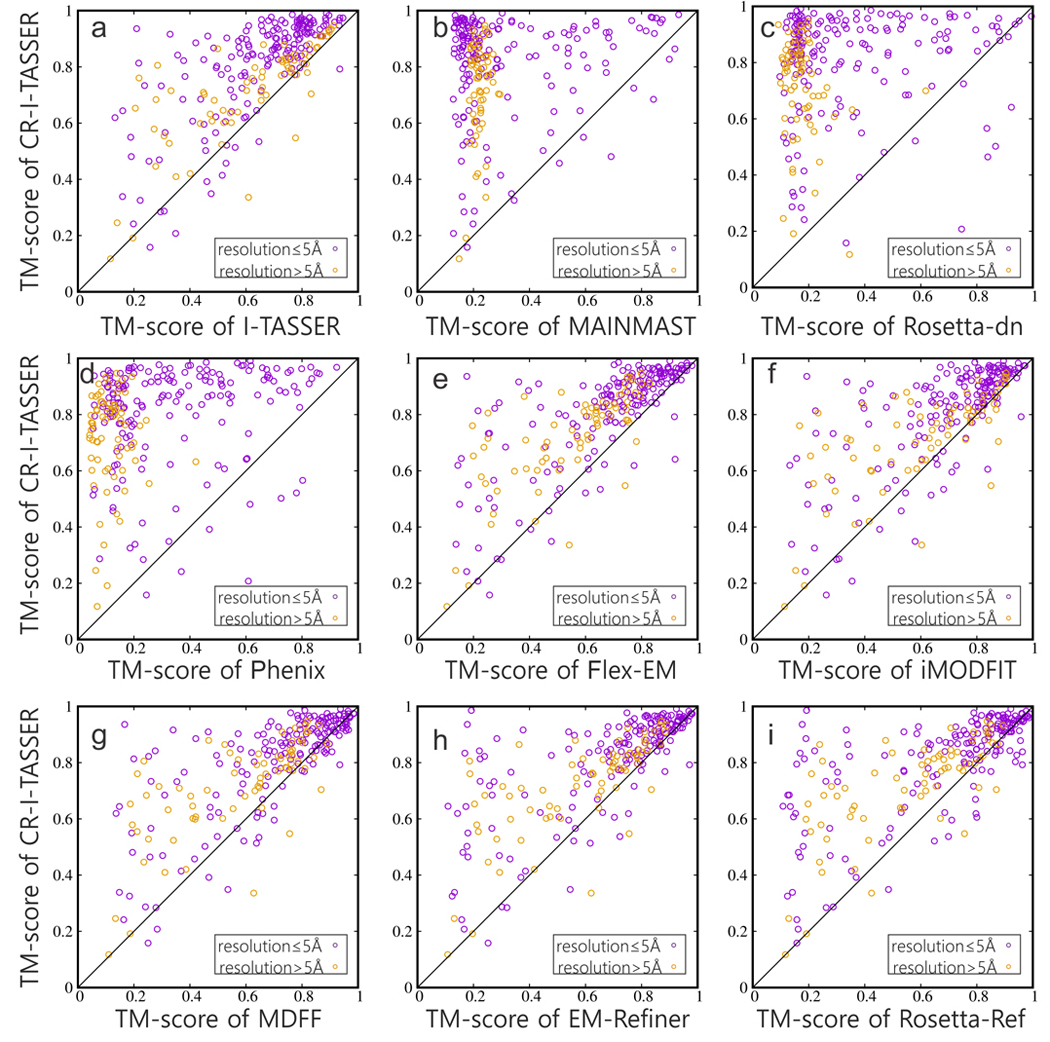
To examine our pipeline in a realistic setting, we further tested CR-I-TASSER on 248 non-redundant proteins with experimental density maps; see Supplementary Text 1 for details of dataset. On average, CR-I-TASSER achieves an average TM-score=0.783 for the 248 EMDataResource targets, which is 158% higher than the best de novo program Rosetta-dn (0.303) and 17% higher than the best refinement program MDFF (0.671). In Fig. 3, we present a head-to-head comparison of CR-I-TASSER with I-TASSER and other control programs, where CR-I-TASSER outperforms the control methods (including I-TASSER) in most of the cases. Especially, CR-I-TASSER outperforms the sequence-based I-TASSER method in 228 out of 248 cases (92%). The average TM-score of CR-I-TASSER (0.783) is 23% higher than that of I-TASSER (0.637), which corresponds to a p-value=3.8 in Student’s t-test, showing significant impact of the introduction of cryo-EM data in the cutting-edge structure assembly simulations. If we count the number of cases with TM-score >0.5/0.9 for low-/high-resolution targets, CR-I-TASSER achieves good predictions in 138 cases, which is 23 and 1.7 times of that by the best de novo program (Rosetta-dn, 6) and the best refinement program (MDFF, 83), respectively. In the bottom of Table 1 (rows 46–67), we split the data samples into high- and low-resolution, where a similar trend of the superiority of CR-I-TASSER over other methods is seen. The gap between CR-I-TASSER and the comparison methods, as assessed by TM=TM-scoreCR-I-TASSER-TM-scoreother, is slightly larger for the low-resolution (0.543/0.141 for Rosetta-dn/MDFF) than the high-resolution samples (0.457/0.101), despite that all methods perform better for high- than low-resolution samples. This is probably due to the fact that TM-scores of the control methods for low-resolution samples are lower and therefore have more room for improvement. Furthermore, we specifically checked whether any particular secondary structure components would affect the performance of CR-I-TASSER. As shown in Supplementary Fig. 5, although CR-I-TASSER performs better in high-resolution than in low-resolution maps, there is no obvious correlation between the average TM-score and the ratio of secondary components for both high- and low-resolution cases. More benchmark results (e.g., template homology cutoff, different network trainings, full maps etc.) can be found in Supplementary Text 20.
Figure 3.
Modeling results on 248 targets with experimental density maps by different methods. CR-I-TASSER versus (a) I-TASSER; (b) MAINMAST; (c) Rosetta-dn; (d) Phenix; (e) Flex-EM; (f) iMODFIT; (g) MDFF; (h) EM-Refiner; (i) Rosetta-Ref. The symbols with different colors denote different ranges of resolution: purple: 2–5 Å; yellow: 5–10 Å.
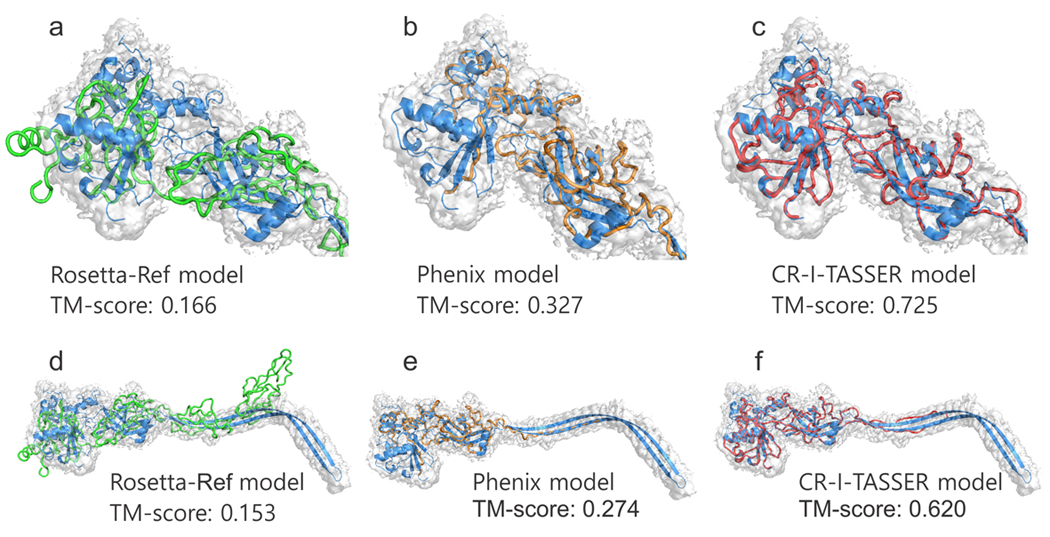
As a further case study focusing on difficult targets, we examine in detail a hard example from the anthrax toxin antigen pore protein (PDB ID: 3j9c-A) in Fig. 4. This target consists of 423 residues and the cryo-EM density map has a resolution of 2.6 Å. In this case, LOMETS failed to locate good templates (the best template has a TM-score=0.257), which resulted in an incorrect fold of the final I-TASSER model with a TM-score=0.132. Therefore, the superposition from Situs is nearly random. Consequently, all refinement-based methods failed to model the target and have the final model with TM-score=0.144, 0.132, 0.136, 0.143 and 0.153 for Flex-EM, iMODFIT, MDFF, EM-Refiner and Rosetta-Ref, respectively. As illustrated in Figs. 4a and 4d, the Rosetta-Ref model does not match the native structure both globally and locally. On the other hand, Phenix built a model from density map alone which fits the global conformation with the density map. However, there are multiple misconnections and disordered local structures in the model, resulting in an incorrect topology and sequence mapping with a TM-score=0.274 (Figs. 4b and 4e). Similar results were obtained by MAINMAST and Rosetta-dn with TM-score=0.165 and 0.245, respectively.
Figure 4.
Structure modeling results on a protective antigen pore protein (PDB ID: 3j9c-A) with high-resolution (2.9 Å) density map. (a-c) Predicted models by Rosetta-Ref (green), Phenix (orange) and CR-I-TASSER (red) are shown along with the native structure on the head globular domain (Residues 1–98; 185–423, blue). (d-f) The corresponding full-length models including the stem region. The predicted conformations and connection pattern can be found in Supplementary Fig. 6.
Given the high resolution of the density map, 3D-CNN generated a well-predicted conformation with CRscore=0.947. Benefitting from this high-quality prediction, the template regeneration algorithm created a reasonable trace model with TM-score=0.534. Following the CR-I-TASSER reassembly, the final model achieves a TM-score=0.725) for the head globular domain (Fig. 4c) and TM-score=0.620 for the overall chain (Fig. 4f), which are both significantly higher than that by all template and cryo-EM based modeling programs.
It is notable that the TM-score of the sequence-ordered trace model in CR-I-TASSER is considerably lower than the CRscore calculated from the order-independent conformation in the anthrax toxin antigen pore protein case. This is mainly due to the extreme complexity of target structure consisting of a 3-domain globular head flanked with a long beta-hairpin stem that form an antigen pore with other homo-chains; such structural complexity not only introduces noise to position predictions due to the high flexibility of the long stem, but also results in a huge conformational space of fragment connection patterns, which makes the true backbone difficult to trace. As shown in Supplementary Fig. 8, there are many mis-predicted atoms around the long stem. Additionally, the connection conformational space is huge because the two long beta strands are close to each other, making it hard for the fragment-tracing program to interpret the correct connection patterns. Given the specific local structures, however, this issue could be amended by using the density-map-based secondary structure prediction models because the backbone conformational space could be significantly reduced by excluding the zigzag connection patterns in the predicted beta zone. A separate computational pipeline implementing real-space secondary structure prediction powered with deep-learning is currently under development, which may in the future highly benefit modeling for targets with extremely low-resolution maps as well.
End-to-end studies on protein complexes EMD-10564/EMD-30703
As end-to-end case studies from raw density map to final structure, we first present an illustrative example in Figs. 5a-f and Supplementary Figs. 9a-c for a large-size homo-tetramer complex Beta-galactosidase (PDB ID: 6tsk), with each chain consisting of 1040 residues. The corresponding density map EMD-10564 has a resolution of 2.3 Å and is segmented by Phenix segment_and_split_map that has been integrated in the CR-I-TASSER pipeline (see Supplementary Text 22), resulting in a reasonable segmentation model as shown in Supplementary Fig. 9a. Here, we construct 4 models from the 4 segmented density maps separately and look specifically into chain A. As shown in Supplementary Fig. 9b, 3D-CNN creates a high-quality model with CRscore=0.946, which is subsequently used for template reranking and selection from the LOMETS alignment pool (outlined in Supplementary Fig. 12) and for trace generation with the trace connection algorithm (outlined in Supplementary Fig. 14). In this case, the best template with a TM-score=0.666 was identified by both LOMETS and the predicted trace conformation, as shown in Supplementary Fig. 9c. However, the rest of the threading templates are not as good as the best one, resulting in an average TM-score=0.446 for the top-40 LOMETS templates. By combining the template reranking and trace generation processes, CR-I-TASSER improved the TM-score from 0.446 to 0.513 for the top-40 templates.
Figure 5.
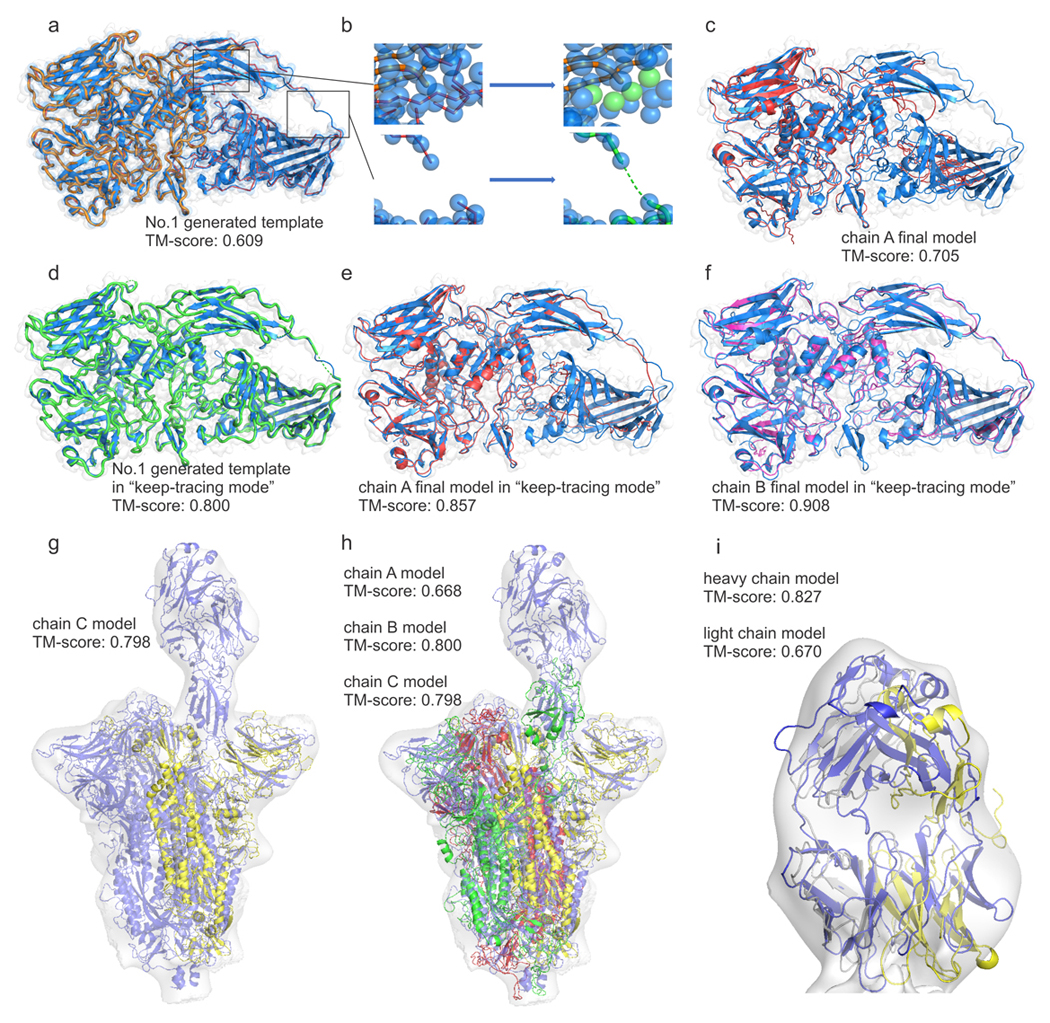
Illustrative examples of end-to-end structural modeling by CR-I-TASSER from unsegment maps. Through all pictures, native structures are shown in blue overlaid on density map in gray. (a-f) Beta-galactosidase in complex with L-ribose (PDB ID: 6tsk) from density map (EMD-10564, resolution 2.3 Å). (a) Best trace model (orange) superposed with the native. (b) Zoom-in pictures of breaking connections can be remedied by the “keep-tracing mode” (see Supplementary Fig. 15 for details). (c) Full-length model by CR-I-TASSER with default setting (red). (d) trace model generated with “keep-tracing mode” (green). (e) Full-length model by CR-I-TASSER with “keep-tracing mode” (red); (f) Full-length model with the highest eTM-score among 4 chains (magenta). (g-i) the SARS-CoV-2 spike protein with receptor-binding domains (RBD) bound with a 2H2 Fab (PDB ID: 7dk5) from density map (EMD-30703, resolution 13.5 Å). (g) First CR-I-TASSER model (yellow) built on the map as in the chain C location; (h) Models of chains A (green), B (red) and C (yellow) built on the map; (i) Final CR-I-TASSER models of heavy/light chains of 2H2 Fab (gold/silver) using complex-based superposition process described in Supplementary Text 24.
These templates are submitted to the structural assembly simulations which are guided by the restraint-enhanced I-TASSER force field and the density-map correlations. Eventually, CR-I-TASSER constructed the final model with TM-score=0.705 (Fig. 5c), which is 41% higher than that of the original I-TASSER prediction (0.500). Due to the size and complexity of the model, Situs does not correctly superpose the I-TASSER model into the density map, resulting in the general low quality from the refinement-based programs with TM-score=0.476, 0.474, 0.343, 0.359 and 0.353 for Flex-EM, iMODFIT, MDFF, EM-Refiner and Rosetta-Ref, respectively. Meanwhile, the de novo programs that we tested are also unsuccessful in creating correct folds because of the complexity of tracing/building such a large protein, resulting in final TM-scores of 0.194, 0.105 and 0.251, for MAINMAST, Rosetta-dn and Phenix, respectively.
Although CR-I-TASSER successfully built a model with the highest TM-score among the state-of-the-art programs, there is still room for improvement. In fact, the final model in Fig. 5c shows that the structure of the three domains in the left side of the picture is very close to the native, but that for the remaining two domains in the right side is poor. This is partly because the correct LOMETS alignments are mostly located in the left domains. However, the connection patterns of the trace model shown in Fig. 5a overlaps well with the target structure, indicating the connections are mostly correct. A closer view shows that there are several small flaws of misconnections in beta sheets of the right part, where these misconnections can terminate the growth of the long traces as the target atoms may be out of the probing radius of the last atom, as shown in the zoom-in figure of Fig. 5b. The probing radius request is employed as the default in CR-I-TASSER to ensure the reasonability of the tracing models for general sequences. Nevertheless, if we use the option of “keep-tracing mode” provided in the CR-I-TASSER pipeline, which allows for the end point of current trace to break the connection patterns (see Supplementary Text 23), the created trace models are greatly improved with the average TM-score increased from 0.446 to 0.708 for this case, where the TM-score of the first template is improved from 0.666 to 0.749. These high-quality trace templates lead to a much-improved full-length model with TM-score=0.857 (Fig. 5e). Despite the improved performance for this case, the “keep-tracing mode” is not used as default setting in CR-I-TASSER as the drop off of the probing radius could increase the connection uncertainty and reduce the average performance for regular proteins. Additionally, since we have separately modeled 4 segmented chains, we could choose a possibly better model by examining the estimated TM-scores (see Eq. 8 in Methods), which are 0.777, 0.912, 0.834 and 0.856 for chain A, B, C and D, respectively. By selecting the model for chain B, we obtained the final full-length model with a TM-score of 0.908 as shown in Fig. 5f.
Overall, this example demonstrates the practicality of CR-I-TASSER for generating high-quality models from unsegmented raw density map data, but also exposes the potential weaknesses of the default CR-I-TASSER pipeline which is sometime too conservative when generating traces for targets involving long loops/tails and disorder regions, where the “keep-tracing mode” may help provide an alternative solution for better tracing and final model constructions for these cases when the first try fails.
In Figs. 5g-i, we present another example of models built from raw low-resolution density map (13.5 Å), which is for the complex of the SARS-CoV-2 spike protein with a 2H2 Fab (PDB ID: 7dk5). In this complex, three large homo-chains (each with 1261 residues) are bound with the two heavy/light chains of a 2H2 Fab with 214/218 residues. Due to the low resolution, it is not feasible to automatically segment with only density map information. Thus, we attempted to build models on the whole map. Given that CR-I-TASSER performs better for the cases with higher protein-map size ratio as shown in Supplementary Fig. 7b, we first tried to build a long spike protein chain in the map. In this case, LOMETS recognize the top-1 template with TM-score=0.562, where the CR-I-TASSER re-ranked the alignments and chose a better first-rank template with TM-score=0.671. As shown in Fig. 5g and Supplementary Fig. 9d, CR-I-TASSER superposed the first-rank template into the low-resolution density map correctly and built a final model with TM-score=0.798 to the deposited structure in the chain C position, where the model built by I-TASSER has only a TM-score=0.682. After that, the density map was masked by deleting the part which overlaps with the model just built. The remaining density map was then used by CR-I-TASSER to build the second and third spike chains subsequently by repeating this process. As shown in Fig. 5h and Supplementary Figs. 9e, CR-I-TASSER eventually built three spike protein models on the low-resolution map with TM-scores of 0.668, 0.800 and 0.798 for the chain A (with up receptor-binding domain, RBD) and chain B/C with down RBDs, respectively (compared to 0.599, 0.677 and 0.682 by I-TASSER). Although the resolution is low, CR-I-TASSER still assembles spikes with up/down RBD conformations in the correct position.
Following the long-chain structure modeling for the spike proteins, we further attempted to build models of the heavy/light chains of 2H2 Fab. Since these two chains are of similar length but not identical, it is hard to tell which one should be built first. By randomly selecting the heavy chain to start, CR-I-TASSER created models with TM-scores of 0.702 and 0.518 for the heavy and light chains respectively, which are marginally better than I-TASSER (TM-score=0.524 and 0.571), where the positions of the two chains on the map are apparently incorrect (see Supplementary Figs. 9f-g). The failure for improvement is partly because the native structures of these two chains share similar folds (TM-score=0.730 by TM-align32), and hence they have very similar density maps, which make it harder to locate the correct position in such a low-resolution map. Instead of one-by-one modeling, a better strategy may be to introduce complex modeling. Here, we slightly extended the current pipeline to simultaneously superpose the templates from two chains and choose the best combination poses (see details in Supplementary Text 24). With this, good templates for both chains were correctly ranked and superposed in the density map as shown in Supplementary Fig. 9h. These templates were then submitted to CR-I-TASSER simulations separately, which resulted in the final models with higher TM-scores (0.827/0.670 for heavy/light chains, see Fig. 5i and Supplementary Fig. 9i). Despite the simplicity of the strategy, this result demonstrates the feasibility to extend CR-I-TASSER for complex-based structural modeling on full density maps.
CONCLUSION
We present a hybrid pipeline, CR-I-TASSER, for automated protein structure modeling from cryo-EM density map. The core component of the pipeline is the density-map based trace predictions from deep convolutional neural networks, which are used for threading template selection and initial model generations through fragment tracing. The advanced I-TASSER folding simulation platform is then extended to reassemble the template and trace models, under the guidance of an optimized force field combining 3D-CNN density-map and template restraints with the advanced knowledge-based energy potentials.
CR-I-TASSER was benchmarked on a large-scale data set containing 778 proteins with both computer-simulated and experimental density maps, compared to three state-of-the-art de novo (Rosetta-dn16, 17, MAINMAST18 and Phenix26) and five refinement-based (Flex-EM11, iMODFIT12, MDFF13, EM-Refiner15 and Rosetta-Ref10) methods. Overall, CR-I-TASSER generates models with an average TM-score=0.839 when high-resolution (2–5 Å) density maps are used, which is 88% higher than the best de novo modeling program (Phenix) and 41% higher than the best refinement program (MDFF), with a p-value <10−66 in Student’s t-test for both comparisons. When the medium-to-low resolution (5–15 Å) maps are used, although the average TM-score of CR-I-TASSER is slightly reduced (=0.726), it still generates correct fold with a TM-score >0.5 for 482 cases, which is 66% higher than the best of other methods (289 by MDFF program). Detailed data analyses showed that the density-map based deep-learning trace models from 3D-CNN play a critical role in the structure quality improvement. Since deep-learning can derive specific and precise information on atoms from density map, the 3D-CNN trace models can therefore be used to more efficiently constrain both initial template regeneration and CR-I-TASSER model assembly simulations, compared to traditional de novo and refinement-based approaches that are guided solely by model-to-map correlations. Thus, CR-I-TASSER provides currently best-in-class performance for automated structure prediction from cryo-EM density maps.
Despite the encouraging results, it is important to note that the current CR-I-TASSER pipeline relies on the success of 3D-CNN on trace prediction, and we observe that the accuracy can decrease on low-resolution data. There are also issues in converting positions into ordered tracing models when the target structure involves long loops/tails or disordered regions. Given the exciting progress witnessed in hybrid deep-learning and evolution-based protein structure prediction29–31, the combination of 3D-CNN with deep multiple sequence alignments collected from metagenome databases should help further improve the 3D-CNN trace and CR-I-TASSER model accuracy. Additionally, a new module of CR-I-TASSER aimed to further enhance its performance on low-resolution data is in development, in which we employ density-map based real-space secondary structure modeling powered by deep neural-network learning to assist cryo-EM model construction. The preliminary result is encouraging and shows that since secondary structure is “coarser” than positions, the models are easier to learn and can provide more relevant information to improve the modeling accuracy for the targets with poorer resolution maps. Meanwhile, CR-I-TASSER mainly focuses on monomer proteins, for which the density maps need to be segmented manually in the first place. We expect that it will be possible to combine CR-I-TASSER in a modular fashion with improved upstream or downstream tools for other modeling tasks (e.g., segmentation or refinement) to further enhance future performance. Given that a major advantage of cryo-EM is on large-size protein complex structure determination, however, an important next step is to extend the deep-learning based structure assembly simulations for protein-protein/protein-nucleic acid complex structure modeling and determination. While one of the current state-of-the-art segmentation programs has been integrated into CR-I-TASSER, new algorithms built on I-TASSER homology modeling and heuristic structure-map alignment iterations32 can be a meaningful solution; investigations along these lines are under progress.
ONLINE METHODS
CR-I-TASSER is a hierarchical method integrating I-TASSER with cryo-EM density maps for high-accuracy protein structure determination. As outlined in Fig. 1, the pipeline consists of three consecutive steps: (1) initial data processing; (2) deep learning-based template refinement and regeneration; (3) density map guided structural reassembly simulations.
Initial data processing
Starting from query sequence and cryo-EM density map, CR-I-TASSER extracts three parts of information.
Predicting Cα locations using deep neural-network learning.
Deep convolutional neural network (3D-CNN) with a residual network architecture19 (see Supplementary Text 25 for details) is employed to predict atom locations in a grid system, where the input of the 3D-CNN is the cryo-EM density map, and the output is the grid values ranging from 0 to 1 representing the possibility of atoms at the grids. The overall 3D-CNN architecture is shown in Supplementary Fig. 10a, where the density map in 3D grid space is taken as input signal to send through a 3D convolutional layer followed by instance normalization and ReLU and extended to 32 channels. Next, 10 basic blocks with residual network architecture are used to enhance the network capability of learning essential information of density maps. Eventually, the signal goes through the last layer which contains a 3D convolutional layer with 2 output channels and a SoftMax layer. The final outputs of 3D-CNN contain two complementary probability maps with the same size of the input density map, in which one map represents the probability of class 1 (“having atom”) while the other one stands for class 0 (“not having atom”). Since only a few grids are with atoms around them, these two classes are highly imbalanced. Specifically, if we look at the central part (instead of marginal part) of density maps where proteins are located, the ratio of the numbers of class 0/1 in experimental training set is 440,462,749/9,537,251, which is approximately 50/1 (see Supplementary Table 1). Therefore, to make the training process more balanced, we set the weights as 1.0 and 50.0 for class 0 and 1 respectively when computing the loss function, for which the Cross Entropy Loss is employed. Although the weights are important in imbalance training and can affect the training process, the slightly different weights (e.g. 1/25 or 1/75 for class 0/1) will have negligible effect on the final training result and hence we only used the weights that are most naturally derived from statistics result. During the training, Adam optimizer is employed to enhance the learning efficiency with a learning rate of 0.0005. To reduce overfitting, random dropout is also used with a drop_rate=0.2, and the batch sizes are set to 1.
The network was trained on two datasets to obtain two network models separately: To obtain the first training dataset, we download the file “cullpdb_pc20_res1.6_R0.25_d190404_chains3470.gz” from http://dunbrack.fccc.edu/Guoli/pisces_download.php, which contains 3,470 non-redundant proteins and was then randomly split into a training (3,088 proteins) and a validation (382 proteins) set with a ratio ~9:1 to prevent overfitting. The density map for the first dataset is simulated by
| (2) |
where with R being the resolution parameter randomly taken from [1, 15Å], is the coordinate vector of the density map, and indicate the coordinate vector and atomic number of ith atom of the protein, respectively. The second training dataset contains 3,600 targets with experimental density maps whose resolutions range from 2.1 Å to 10.0 Å. These experimental maps were generated from 36 large complexes with well-superposed experimental structures by randomly segmenting them into small maps with a size of . To make the training process focus more on atoms, we set a filter of these small maps by containing at least 250 atoms. This can avoid the issue of containing too few atoms in a map, which could happen in the marginal parts of experimental maps. Through the 3D-CNN networks, the first model was trained on the simulated training set with more than 720 epochs. We calculated the average CRscore loss from the validation set every 30 epochs and stopped the training if: (1) training epochs > 500 and max average CRscore > 0.8 and the latest average CRscore is 0.02 less than the max average CRscore, or (2) training epochs > 2000. After stopping training, we selected the model with the max CRscore (708 epochs, see Supplementary Fig. 11a). The second model started from the first model and was trained on the experimental training set for 217 more epochs, where the average loss against training epochs is shown in Supplementary Fig. 11b. The loss in the first model starts to saturate around 600–700 epochs, while that in the second model does so after 800 epochs, probably because of the relatively higher complexity associated with the experimental maps.
Following the 3D-CNN model, a quick procedure is designed to convert the possibility map into atom coordinates (Supplementary Fig. 10b). The procedure first locates the grid with the highest possibility and labels it as the first atom. It then iteratively searches for the next atom with the highest possibility at the grids with distance no less than 3.3Å from all the labeled atoms. The procedure repeats to ensure at least L (=query length) atoms are located. It will continue until 1.2*L atoms are located if the next highest possibility is >0.9.
Initial template identification by LOMETS.
We employed LOMETS21, a meta-threading method containing 11 leading fold-recognition programs, to identify homologous and analogous templates from the PDB. For each query sequence, top 300 templates are collected based on the normalized Z-score (), which measures the significance of query-template alignments by each program. Accordingly, a target will be defined as ‘Easy’ if there is on average one or more good templates with >1 for each program, while others are labeled ‘Hard’ due to the lack of good templates.
Inter-residue contact map prediction.
ResPRE33 is used to predict the residue-residue contact maps. From a query sequence, ResPRE first uses DeepMSA34 to collect multiple sequence alignments (MSAs) from the whole-genome and metagenome sequence databases, where the inter-residue contact maps are then predicted from the inverse covariance matrix derived from the MSAs, based on deep residual convolutional network training19.
Deep learning-based template selection and regeneration
We design two procedures utilizing the deep-learning based conformations to improve initial template quality of CR-I-TASSER through template reselection and model regeneration, respectively.
Template reselection by and density map matching.
LOMETS creates multiple threading templates, but the best templates do not always rank at the top by the Z-score. We re-rank the top 300 template structures based on their match with the conformations predicted by the 3D-CNN from cryo-EM density map, using a procedure outlined in Supplementary Fig. 12. Because the 3D-CNN conformation has no sequence index assigned, the matching procedure starts with the calculation of the “fingerprint” for each atom in a given LOMETS template and conformation, where a fingerprint vector of ith atom is defined as a set of 20 ascending-ranking intra-distances between ith atom and 20 nearest atoms in the template (or conformation). A pairing score of ith atom at template with jth atom at conformation is then calculated by
| (3) |
The lower is, the more similar environment two atoms (i, j) are in, indicating a higher probability for () to be correctly paired. Therefore, we initially select the atom pairs with the minimum and pair them in the ascending order, where each atom can only be paired once. Generally, if ith and iith atoms from the template are correctly paired to jth and jjth atoms from the conformation, the intra-distance between ith and iith atoms, , should be close to that between jth and jjth atoms, . Based on this assumption, we further refine the initial pairing using a weighted matching score defined by
| (4) |
Here, , where is the weight for ith atom from the template which is initially set as 1 and updated iteratively by an algorithm outlined in Supplementary Fig. 13. After the convergence, only the pairs with a matching score are selected, where the threshold is defined by the 2-mean clustering of the matching scores. Based on the selected pairing, the Kabsch RMSD superposition of template and conformation is performed35, where the inter-chain distance <10 Å will be used as a new condition to select pairing in addition to Eqs. (3–4). This new pairing will be used as the input of pairing refinement and Kabsch superposition to generate a newer pairing. The procedure will repeat until the final pairing and structure superposition converge (Supplementary Fig. 12). Overall, the idea of the superposition process described above is to identify the correct pairs of atoms between conformation (index-free) and template alignments (indexed) by comparing their intra environments.
Finally, the CRscore is calculated for each template with the 3D-CNN conformation based on the selected pairing, where the 300 LOMETS templates selected by Z-score are re-ranked based on the calculated CRscores. A template will be defined as a ‘good’ template if the CRscore >0.5. Up to 30 good templates () are selected from this template reselection procedure.
Initial model generation from 3D-CNN conformations.
Since CR-I-TASSER uses 40 replicas in the replica-exchange Monte Carlo (REMC) simulations and each replica starts with different templates, we generate new templates directly from the 3D-CNN conformations; this contains two steps of -trace connection and sequence-trace mapping (Supplementary Fig. 14).
For -trace connection, we first connect all neighboring atoms which have a distance below a bond-length . All connections to a atom that has the number of connections () are considered as ‘true’ connections (e.g., connections to Atoms-1, 3, 5, 7 and 8 in Supplementary Fig. 14a), while all other connections that contradict with the true connections and make for other atoms are removed (e.g., connection 2–4 in Supplementary Fig. 14a). After this scan, if a still contains >2 connections, this atom will be removed from the trace (e.g., Atom 6 in Supplementary Fig. 14a). As shown in Supplementary Fig. 14b, the remaining trace pattern will depend on the selection of . In CR-I-TASSER, we implement the procedure under eighteen different cutoffs of 3.8, 3.9, …, 5.5 Å separately, and keep only the connections with a frequency of occurrence >40% in the final connection.
This connection procedure creates multiple fragments, where up to 1,000,000 traces are generated by randomly connecting the fragments, until no atom is available for the next connection. The latter could happen at the true end of the protein, or if there is no available atom in the probing radius (5.5 Å), or if there are other atoms but are already fully connected in an unused fragment. Although the constraints involved in the connection process can help improve the accuracy of the template generation on average, it cannot always result in trace model with full length because the growth could stop anywhere under the constraints. To address this issue, CR-I-TASSER provides an alternative “keep-tracing mode” to improve fragment tracing success rate for some special cases by partially releasing some of the restraints or additional iterations (see Supplementary Text 23 for details).
Assuming that each fragment is continuous, we map the query sequence to each trace by gapless threading and calculate the contact map using a distance cutoff Å. Top 300 traces are selected based on the Pearson correlation coefficient (PCC) of the contact map with the predicted contact map from ResPRE, as well as the PCC of the template structure with the target density map (see Supplementary Text 26). Finally, templates are selected from the 300 traces based on the PCC of the template structure with the target density map. This PCC is also employed to re-rank all top-40 templates including those from template reselection and regeneration.
It is noted that two 3D-CNN models have been trained on the simulated and experimental density-map datasets separately, which generates two sets of conformations for each target. If the two conformations are close, i.e., with the CRscore between them >0.85, which usually indicates good quality of the conformations, we will take the average for each atom pair to generate the final conformation and use it for the template reselection and regeneration as described above. In case the conformations are different (CRscore<0.85), which while rare, happens in some cases with low-resolution experimental cases and usually indicates that the predicted conformation is not reliable, we skip the conformation-based template reselection and regeneration. Instead, we match each of the LOMETS templates directly with the density maps using BFGS algorithm (Supplementary Text 27) followed by a short Metropolis Monte Carlo simulation under the guidance of template-density correlation as defined in Supplementary Text 26, with movements including 2,000 rigid-body translations/rotations. The top 40 templates are then selected based on the correlation coefficients from high to low.
Density-map guided structural assembly simulations
CR-I-TASSER performs REMC simulations to assemble full-length structure models, under a composite energy force field of
| (5) |
where is the inherent knowledge-based potential extended from I-TASSER20 and described in Supplementary Eqs. S2-33 in Supplementary Text 28, contains four aspects of distance and contact restraints collected from the top templates determined by LOEMTS and 3D-CNN models (Supplementary Eqs. S34-43 in Supplementary Text 29). counts for the global correlation between structure conformation and experimental density map ρ0 by
| (6) |
where is calculated by Eq. (2). The counts for the correlation between structure conformation and the 3D-CNN predicted conformation:
| (7) |
where is the density maps calculated by Eq. (2) for the 3D-CNN conformation. This term is performed only when CRscore between the two 3D-CNN conformations is >0.85, which is designed to enhance the convergence of simulations to the consensus conformations. It is noted that the negative cross correlation in Eqs. (6–7) instead of PCC defined in Supplementary Text 26 is implemented because the former is computed faster than the latter. Additionally, benefit from the linear combination form of Eqs. (6–7), energy terms need to be computed only for the local segment involved in each movement, which is significantly faster than the calculations on the entire chain after each movement. The resolution for and calculations is automatically detected and set by a short-trained 3D-CNN predictor for resolution prediction. Our benchmark results showed that the final model quality is not sensitive to the value of setting resolution. The weight parameters in Eq. (5), as well as those in the inherent knowledge-based I-TASSER force field, are determined in a separate training protein dataset, which is non-homologous to the test proteins of this work, by maximizing the average TM-score of the final models.
Final model selection and model quality estimation
The structure conformations generated by CR-I-TASSER (referred as ‘decoys’) in eight low-temperature replicas are clustered by SPICKER to select the states corresponding to the lowest free energy states36. Specifically, an all-to-all RMSD matrix is calculated among all decoys where a pair of decoys are considered as neighbors if their RMSD is within a cutoff. The decoy with the largest number of neighbors is selected as the center of the first cluster and the representative centroid model for the cluster is obtained by averaging all decoys included. The second cluster is obtained in a similar way on the remaining decoys after excluding all decoys from the first cluster, and the procedure repeats till five clusters are obtained. Thus, a decoy cluster captures the inherent statistics of the Monte Carlo process, i.e., the larger the size of the decoy cluster is, the higher the convergence is, and accordingly the less uncertainty the model sampling is. As the cluster centroid models from SPICKER often contain steric clashes, the centroids of the five biggest clusters are reassembled by a second round of REMC simulation to improve the hydrogen-bonding network and local structural geometry. The lowest energy conformations are selected from the second-round simulations and further refined at atomic level by the fragment-guided molecular dynamics (FG-MD)37 to create final models.
To evaluate the quality of predicted structures, we calculate the estimated TM-score (eTM-score) of the mth CR-I-TASSER model relative to the target structure by
| (8) |
where TM-scoremn is the TM-score between mth and nth predicted models. The confidence score Cm is defined as
| (9) |
where is the total number of decoy conformations submitted to SPICKER, is the number of decoys at mth cluster, is the average RMSD of the decoys to the cluster centroid, and the CRscorem is the matching score of the model with the 3D-CNN predicted conformation by Eq. (1).
Supplementary Fig. 16 displays the data of eTM-score versus the actual TM-scores on the first predicted models of all 530 test proteins with high-/low-resolution density maps, where most of the data points are located near the diagonal line, showing a strong linear correlation. The PCC and cosine similarity between eTM-score and TM-score are 0.858 and 0.989, respectively. If we use eTM-score=0.5 as cutoff to split “Positive”/“Negative” cases, the numbers of cases for True Positive (TP), False Negative (FN), True Negative (TN) and False Positive (FP) are 856, 44, 119 and 41, respectively, which correspond the TP, FN, TN and FP rates of 95.1%, 4.9%, 74.4% and 25.6%, and the overall Matthews correlation coefficient (MCC) = 0.710. The strong correlation indicates that eTM-score can be used to reliably estimate the quality of predicted models.
In addition to the eTM-score for overall quality estimation, we introduce two metrics, local PCC and local confidence, to estimate the local agreement to the density for the final models. First, the local PCC for ith-residue modeling quality from the mth predicted model is defined as
| (10) |
where is the density on grid y calculated by Eq. (2) but only from the ith residue of the mth predicted model. Eq. (10) is very similar to the normal PCC (see Supplementary Text 26) except that we use a modified density instead of the experimental density :
| (11) |
The reason we use the modified density to compute local PCC for ith residue is because the experimental density on grid y contains contributions from all residues, where Eq. (11) is designed to decouple the experimental density for ith residue specifically. Toy model results shown in Supplementary Fig. 17 demonstrate that the is more reasonable than when computing the local PCC.
Second, the local confidence for ith-residue from the mth predicted model is defined by integrating eTM-score and local PCC:
| (12) |
where is defined as
| (13) |
Here, is the distance of ith residue between mth and nth models, and is a scaling parameter from TM-score (see Supplementary Text 4). is the number of final models predicted by CR-I-TASSER which is no more than five.
As an illustration, Supplementary Fig. 18 displays the local PCC and local confidence scores on two end-to-end study proteins (6tsk-B and 7dk5), where Supplementary Table 5 lists the average correlation coefficients between the local quality scores and the local error of predicted models from the experimental structure for all 248 test proteins with experimental density maps. The data show that both scores can be used for local model quality estimation. Although the local confidence shows a slightly higher correlation with the local modeling errors, CR-I-TASSER output both scores for alternative local quality estimations. In addition, CR-I-TASSER produces up to five models, which allow user to estimate the global/local quality using other methods such as ensemble structure comparison.
Data Availability
All training and testing data are available at https://zhanggroup.org/CR-I-TASSER/ and https://zenodo.org/record/5774563#.YbWYaNPJeXA (DOI: 10.5281/zenodo.5774563).
Code Availability
The standalone package of the CR-I-TASSER programs, including library and manual documents, are available to download at https://zhanggroup.org/CR-I-TASSER/download.html and https://zenodo.org/record/5774535#.YbWYudPJeXA (DOI: 10.5281/zenodo.5774535).
Supplementary Material
Acknowledgements
We thank Drs. Yang Li, Chengxin Zhang and Xiaogen Zhou for stimulating discussions. This work is supported in part by the National Institute of General Medical Sciences (GM136422, S10OD026825 to Y.Z.), the National Institute of Allergy and Infectious Diseases (AI134678 to Y. Z.), and the National Science Foundation (IIS1901191, DBI2030790, MTM2025426 to Y. Z.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. This work used computing resources from the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by the National Science Foundation (ACI1548562).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Wuthrich K. Protein structure determination in solution by nuclear magnetic resonance spectroscopy. Science 243, 45–50 (1989). [DOI] [PubMed] [Google Scholar]
- 2.Drenth J. Principles of protein X-ray crystallography. (Springer Science & Business Media, 2007). [Google Scholar]
- 3.Henderson R. & Unwin PNT Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32 (1975). [DOI] [PubMed] [Google Scholar]
- 4.Chayen NE & Saridakis E. Protein crystallization: from purified protein to diffraction-quality crystal. Nature methods 5, 147–153 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Nogales E. The development of cryo-EM into a mainstream structural biology technique. Nat Methods 13, 24–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knapek E. & Dubochet J. Beam damage to organic material is considerably reduced in cryo-electron microscopy. Journal of Molecular Biology 141, 147–161 (1980). [DOI] [PubMed] [Google Scholar]
- 7.Cheng Y. Single-particle cryo-EM at crystallographic resolution. Cell 161, 450–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaeser RM How good can cryo-EM become? Nature methods 13, 28 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Lawson CL et al. EMDataBank unified data resource for 3DEM. Nucleic acids research 44, D396–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang RY-R et al. Automated structure refinement of macromolecular assemblies from cryo-EM maps using Rosetta. Elife 5, e17219 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joseph AP et al. Refinement of atomic models in high resolution EM reconstructions using Flex-EM and local assessment. Methods 100, 42–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez-Blanco JR & Chacon P. iMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J Struct Biol 184, 261–270 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Singharoy A. et al. Molecular dynamics-based refinement and validation for sub-5 Å cryo-electron microscopy maps. Elife 5, e16105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kovacs JA, Galkin VE & Wriggers W. Accurate flexible refinement of atomic models against medium-resolution cryo-EM maps using damped dynamics. BMC Struct Biol 18, 12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B, Zhang X, Pearce R, Shen H-B & Zhang Y. A New Protocol for Atomic-Level Protein Structure Modeling and Refinement Using Low-to-Medium Resolution Cryo-EM Density Maps. Journal of Molecular Biology 432, 5365–5377 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang RY-R et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature methods 12, 335–338 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frenz B, Walls AC, Egelman EH, Veesler D. & DiMaio F. RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat Methods 14, 797–800 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terashi G. & Kihara D. De novo main-chain modeling for EM maps using MAINMAST. Nature communications 9, 1–11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He K, Zhang X, Ren S. & Sun J. in Proceedings of the IEEE conference on computer vision and pattern recognition 770–778 (2016).
- 20.Yang J. et al. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12, 7–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng W. et al. LOMETS2: improved meta-threading server for fold-recognition and structure-based function annotation for distant-homology proteins. Nucleic acids research 47, W429–W436 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Si D. et al. Deep Learning to Predict Protein Backbone Structure from High-Resolution Cryo-EM Density Maps. Sci Rep 10, 4282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maddhuri Venkata Subramaniya SR, Terashi G. & Kihara D. Protein secondary structure detection in intermediate-resolution cryo-EM maps using deep learning. Nat Methods 16, 911–917 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Y. & Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins 57, 702–710 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Xu J. & Zhang Y. How significant is a protein structure similarity with TM-score = 0.5? Bioinformatics 26, 889–895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afonine PV et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D Struct Biol 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica. Section D, Biological crystallography 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De la Rosa-Trevín J. et al. Xmipp 3.0: an improved software suite for image processing in electron microscopy. Journal of structural biology 184, 321–328 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Yang J. et al. Improved protein structure prediction using predicted interresidue orientations. Proceedings of the National Academy of Sciences 117, 1496–1503 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Senior A. et al. & Penedones H(2020). Improved protein structure prediction using potentials from deep learning. Nature 577. [DOI] [PubMed] [Google Scholar]
- 31.Zheng W. et al. Deep‐learning contact‐map guided protein structure prediction in CASP13. Proteins: Structure, Function, and Bioinformatics 87, 1149–1164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y. & Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic acids research 33, 2302–2309 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Hu J, Zhang C, Yu D-J & Zhang Y. ResPRE: high-accuracy protein contact prediction by coupling precision matrix with deep residual neural networks. Bioinformatics 35, 4647–4655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang C, Zheng W, Mortuza S, Li Y. & Zhang Y. DeepMSA: constructing deep multiple sequence alignment to improve contact prediction and fold-recognition for distant-homology proteins. Bioinformatics 36, 2105–2112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kabsch W. A solution for the best rotation to relate two sets of vectors. Acta Crystallographica Section A 32, 922–923 (1976). [Google Scholar]
- 36.Zhang Y. & Skolnick J. SPICKER: a clustering approach to identify near‐native protein folds. Journal of computational chemistry 25, 865–871 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Liang Y. & Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure 19, 1784–1795 (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All training and testing data are available at https://zhanggroup.org/CR-I-TASSER/ and https://zenodo.org/record/5774563#.YbWYaNPJeXA (DOI: 10.5281/zenodo.5774563).