Abstract
Rapidly changing and transient protein-protein interactions regulate dynamic cellular processes in the cardiovascular system. Traditional methods, including affinity purification and mass-spectrometry (MS), have revealed many macromolecular complexes in cardiomyocytes and the vasculature. Yet these methods often fail to identify in vivo or transient protein-protein interactions. To capture these interactions in living cells and animals with subsequent MS identification, enzyme-catalyzed proximity labeling techniques have been developed in the past decade. Although the application of this methodology to cardiovascular research is still in its infancy, the field is developing rapidly, and the promise is substantial. In this review, we outline important concepts and discuss how proximity proteomics has been applied to study physiological and pathophysiological processes relevant to the cardiovascular system.
INTRODUCTION
Resolving spatial organization and dynamic protein interaction networks within the cell has long been an aspirational goal. Traditional techniques, namely immuno-precipitation (IP) coupled with mass spectrometry (MS), have yielded important insights (Fig. 1A). These studies, however, are limited because they require high-quality antibodies and may miss transient or low-affinity interactions, in part because the purification is performed after detergent solubilization. Moreover, capturing local protein-interaction dynamics from homogenized cell lysates is often impossible. Crosslinking can stably fix some of these interactions 1, but complicates MS analysis due to chemical protein modification and background crosslinking. Subcellular fractionation followed by mass spectrometry has offered insights into the constituents of various cellular compartments (Fig 1B), but information on protein-protein interactions in specific sub-cellular compartments remained limited 2. The Contaminant Repository for Affinity Purification (CRAPome) 3 and Significance Analysis of INTeractome (SAINT) 4 have cataloged many putative false positives using these approaches.
Figure 1. Standard methods for identifying protein-protein interactions.
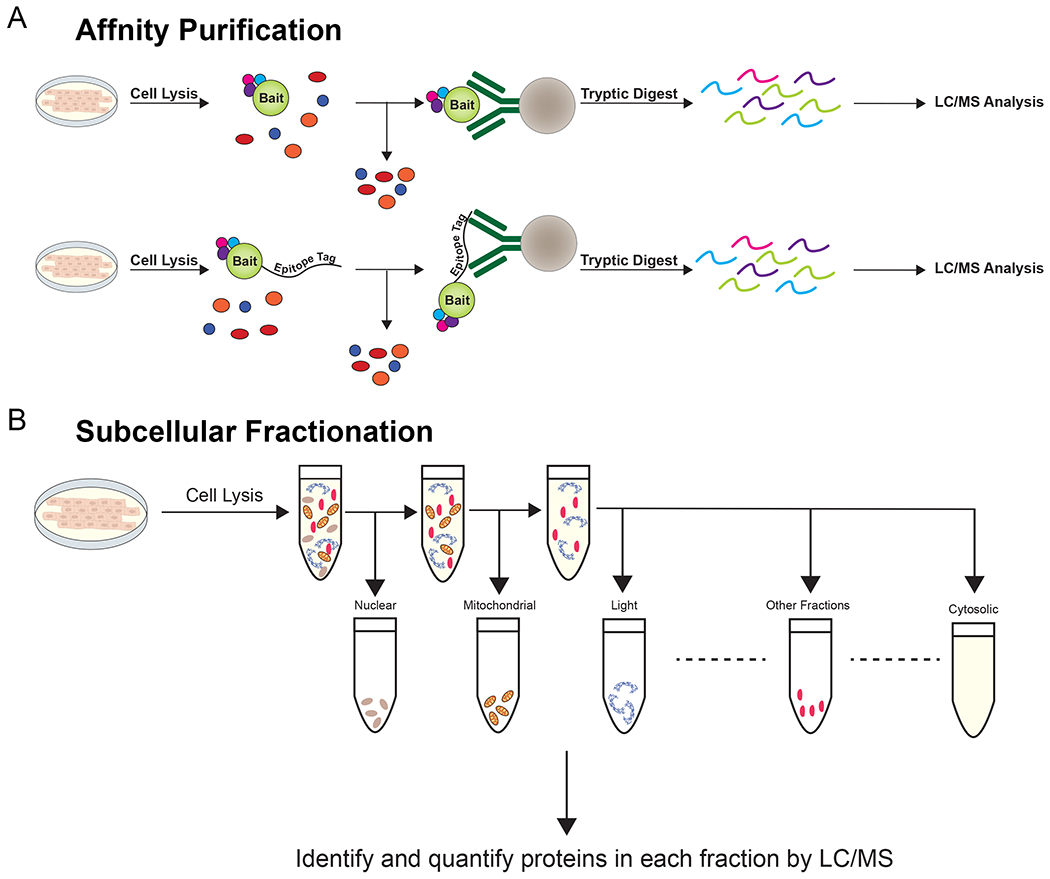
(A) In affinity purification, cell lysates are exposed to antibodies against the bait protein. These antibodies are precipitated and interacting proteins are digested and identified by LC/MS. To study interactors of bait proteins with universal epitope-tag directed antibodies, the fusion of a standard epitope tag can overcome the need for protein-specific antibodies. (B) In lysates subjected to sucrose gradient centrifugation, organelles and subcellular locations of interest are enriched based on their densities.
Proximity-dependent labeling has been developed to resolve the nano-environment (a.k.a. “neighborhood”) of target proteins in situ 5, 6. In most studies, the protein of interest is expressed as a fusion to the labeling enzyme in cultured cells or organisms 5, 7–11, although some approaches have been developed using an antibody against the protein of interest to target biotin conjugation to adjacent proteins in fixed cells, primary tissues and zebrafish 12, 13. For proximity-dependent approaches, proteins near the protein of interest are labeled with biotin, with the labeling radius primarily dependent on the reactivity of the biotinylating agent: biotin-phenoxy radicals for peroxidases and biotinoyl-AMP esters for biotinylation-enzyme derivatives. Labeling also depends in part on the steric accessibility of reactive amino acids. Peroxidase-catalyzed labeling can be rapidly stopped and quenched, allowing short 30-60 second labeling pulses. Biotinylated proteins can be efficiently affinity-purified with streptavidin and quantified by mass spectrometry. Compared to traditional antibody affinity purification, proximity labeling can capture weak and transient interactions, even of trans-membrane proteins that require membrane insertion to maintain an interaction, which are disrupted by detergent solubilization 14–17. In contrast to antibody- or epitope-based affinity purifications, enrichment of biotinylated proteins is compatible with harsh protein extraction, denaturing and stringent wash conditions since labeling is completed in situ prior to enrichment and biotin-streptavidin affinity is very high. In this review, we describe the rapid adoption of proximity-dependent labeling for quantification of the molecular protein environment, focusing on studies that have used the methodology to reveal composition and dynamics of macromolecular complexes and signaling pathways in the cardiovascular system.
FOUNDATIONAL STUDIES OF PROXIMITY LABELING
There are two classes of enzymes used for proximity labeling, derivatives of biotin-ligases, and peroxidases, both of which make use of biotin to covalently modify nearby proteins (Fig. 2A–B). Biotin, also known as vitamin B7, is a cofactor for carboxylase enzymes that transfer carbon dioxide to organic acids and is essential for all life forms from archaea to eukaryotes 18, 19. Three different classes of enzymes rely on biotin as cofactor: carboxylases, including CoA carboxylases, decarboxylases and transcarboxylases 19. In eukaryotes, these enzymes are in the cytoplasm and mitochondria and participate in carbohydrate, lipid, and amino acid metabolism and energy transduction. Native biotin ligases display high lysine specificity of only few biotinylation-target proteins 5, 18. Due to the limited number of endogenous biotinylated proteins and the exceptional strength of the non-covalent biotin-avidin interaction (Kd = 10−14), an early iteration of enzyme catalyzed proximity labeling used a mutant Escherichia coli biotin ligase BirA R118G, also known as BirA* and named BioID for “biotin identification” 5 (Table 1).
Figure 2. Proteomic mapping by proximity labeling.
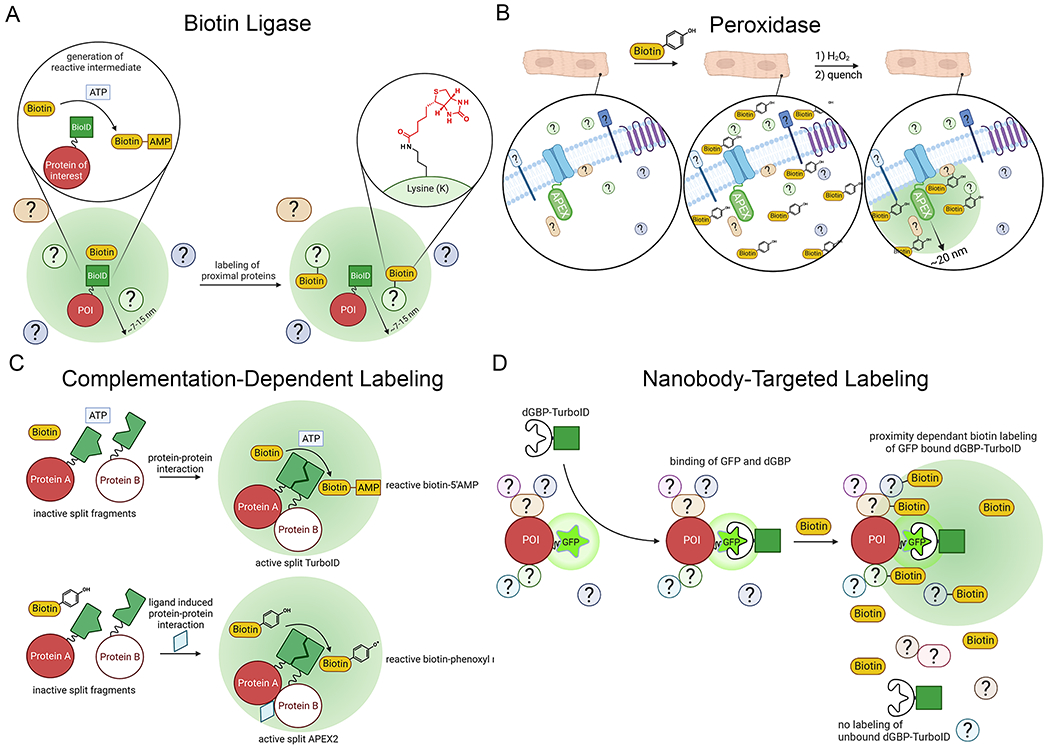
(A) In an ATP dependent process, biotin ligases covalently modify basic residues such as lysine with biotin. (B) In cells loaded with biotin-phenol and treated with H2O2, peroxidases generate biotin-phenoxy radicals which attack nearby electron-rich amino acids, preferentially tyrosine. The labeling radius is determined by the longevity of these radicals prior to their quenching. (C) Cleaved or Split-TurboID and Split-APEX2 systems have been developed such that complementation and subsequent biotin-phenol/biotin dependent-proximity labeling occurs when the cleaved enzyme fusions are close together 28, 29. This complementation increases labeling specificity for the cellular domain where the fused proteins interact. (D) In this modular labeling system, a nanobody to GFP that is destabilized when not bound to GFP was conjugated to TurboID (dGFP-TurboID), which is co-expressed with a GFP-fused protein of interest (POI) 12. The destabilization of the nanobody ensures high labeling specificity for the POI. Illustration Credit: Ben Smith.
Table 1:
| Enzymes | APEX28 | HRP6, 23 | BioID5 | BioID222 | TurboID9 |
|---|---|---|---|---|---|
| Type | Peroxidase | Peroxidase | Biotin ligase | Biotin ligase | Biotin ligase |
| Size (kDa) | 28 | 44 | 35 | 27 | 35 |
| Active region | intracellular | extracellular, secretory pathway, inactive in cytosol | intracellular | intracellular | intracellular |
| Model organisms | mammalian cell culture, bacteria, yeast, flies, mice | mammalian cell culture, flies, primary human tissue | mammalian cell, culture, yeast, flies, plants, mice | mammalian cell culture, mice | mammalian cell, culture, yeast, flies, plants, worms |
| Labeling time | 30 sec-1 min | 1 min | 18 h | 18 h | 10 min |
| Modification sites | preferentially Tyr, also Trp, Cys, His | Tyr, Trp, Cys, His | Lys | Lys | Lys |
| Substrates for protein labeling | biotin-phenol, H2O2 | biotin-phenol, H2O2 | biotin, ATP | biotin, ATP | biotin, ATP |
| Half-life of labeling | <1msec | <1msec | minutes | minutes | minutes |
| Advantages | High temporal resolution; versatility for both protein and RNA labeling | High temporal resolution; versatility for both protein and RNA labeling | Non-toxic for in vivo applications | Non-toxic for in vivo applications | Highest activity biotin ligase; non-toxic for in vivo applications |
| Limitations | Limited application in vivo because of the toxicity of H2O2 and low biotin-phenol permeability. H2O2 quickly decomposes. | Limited application in vivo because of the toxicity of H2O2; limited to secretory pathway and extracellular applications | Poor temporal resolution because of low catalytic activity | Poor temporal resolution because of low catalytic activity | Potentially less control of labeling window because of high biotin affinity, potential toxicity in long-term experiments |
| Notes | Can be used for electron microscopy | Can be used for electron microscopy; can be used when conjugated to antibodies | Reduced activity below 37 °C | Reduced activity below 37 °C | Can be used when conjugated to antibodies |
BirA requires both biotin and ATP to generate a reactive biotinoyl-5’AMP ester intermediate, which it uses to transfer biotin to specific lysine residues on bacterial carboxylase proteins. The R118G mutation of BirA loosens a loop normally preventing diffusion of biotinoyl-5’AMP, markedly reducing the enzyme’s affinity for this reactive intermediate, thus enabling the promiscuous, substrate-nonspecific labeling of lysine residues on proximal proteins (Fig. 2A) 20. BirA* was expressed as a fusion protein with lamin-A, an intermediate filament protein, that is a constituent of the nuclear lamina in the nuclear envelope 5. BirA*-lamin-A was expressed in HeLa cells and was used to perform in situ labeling. Upon the addition of 50 μM biotin to the tissue culture medium, BirA* was capable of substrate-nonspecific but proximity-dependent biotin ligation within a ~10 nm radius 5, 21. Numerous protein-protein interactors, including novel ones such as SLAP75, were identified by combining streptavidin-based affinity purification of biotinylated proteins with liquid chromatography/mass spectrometry 5. BirA* fusion, which adds 321 amino acids to the protein of interest, may influence trafficking or function of the target protein. A smaller biotin ligase from Aquifex aeolicus was modified for superior expression in human cells and mutated to create BioID2, which was shown to alter localization to a lesser extent and displayed robust biotinylation at lower biotin concentrations 22 (Table 1). BioID has been applied in diverse models, including the heart 11.
BioID is not optimal for the study of fast dynamic processes in the seconds to minute range because of its low catalytic efficiency and relatively long labeling time. A directed-evolution variant of BioID, called TurboID, was developed to overcome some of these limitations of BioID and specially to improve labeling kinetics 9 (Table 1). Removing the N-terminus of TurboID generated an even smaller version of TurboID, termed miniTurbo. Compared to BioID and miniTurbo, TurboID has faster kinetics, which confers increased temporal resolution for resolving evolving changes in the domain of interest. Potential drawbacks for TurboID are increased protein instability, an increase in the labeling radius, and persistent biotinylation in the absence of exogenously supplemented biotin with potentially harmful effects on organism viability 9, 25. This was noted in the improved viability of drosophila pupae given supplemental biotin, presumably complementing biotin depletion by TurboID conjugation 9.
Peroxidase-catalyzed proximity labeling is based on the expression of either horseradish peroxidase (HRP) or an engineered soybean ascorbate peroxidase (APEX) and its engineered derivatives such as APEX2 (Table 1) 6–8, 26. HRP is inactive when expressed in mammalian cytosol, likely due to the reduction of its disulfide bonds and Ca2+-dependence 6. After the addition of H2O2 for 1 minute in cells or tissues loaded with the substrate biotin-phenol (biotin-tyramide), the peroxidase generates biotin-phenoxy radicals that react with electron-rich side chains of amino acids, preferentially tyrosines, of nearby proteins (Fig. 2B). The phenoxy radical, which is believed to be membrane impermeant, has a short half-life of less than 1 ms, limiting the radius of labeling of approximately 20 nm for APEX. The fast-labeling kinetics of APEX and the limited labeling intervals can be exploited to probe very rapid and dynamic events during cell signaling 14, 17, 27.
Mitochondrial matrix-targeted APEX was initially used to identify proteins within the human mitochondrial matrix. Using stable isotope labeling of experimental and control samples and tandem mass spectrometry, 495 proteins were identified including 31 proteins not previously linked to mitochondria 26. Although the depth of coverage was high, approximately 15% of known mitochondrial proteins were not identified, likely because they were sterically inaccessible to phenoxy radicals 26. Thereafter, APEX was used to characterize the mitochondrial intermembrane space proteome, which was previously inaccessible with traditional purification approaches 7. To discriminate protein proximity-specific biotinylation from unwanted background, a stable isotope labeling by amino acids in cell culture (SILAC) based ratiometric tagging strategy was developed. The ratio of biotinylation by two different APEX fusion proteins, one within the region of interest, namely the mitochondrial intermembrane space, and one outside of the region of interest, cytosolic APEX, was utilized 7. With this approach a high-quality mitochondrial intermembrane space proteome of 127 proteins, with >94% specificity and 65% coverage, was identified.
To increase spatial specificity, reduce background labeling from endogenous peroxidase activity, and conditionally control proximity ligation activity, split enzymes for APEX2, BioID and TurboID approaches have been developed 10, 28–30. When N- and C-terminal paired fragments of the enzymes are complemented, enzymatic activity is restored (Fig. 2C). Split-APEX2 and split-TurboID have been used to characterize the proteome of the contact region between mitochondria and endoplasmic reticulum 10, 28, 29.
Distinguishing interacting proteins from bystanders
Several methods have been developed to distinguish interacting proteins from bystanders, which are compartment-specific proteins residing in the local neighborhood but not physically interacting with the bait. Two approaches were used to elucidate signaling through G-protein coupled receptors, which are critical regulators of physiological responses. In theory, agonist-induced changes in G-protein coupled receptor signaling would induce dynamic changes in the interactome but likely not in the abundance of bystanders. One study focused on quantification of dynamic changes of the proximity proteome by isobaric tandem mass tag (TMT) MS, without distinguishing constant interactors from constant bystanders 17. Proximity labeling with biotin-phenol, denaturing streptavidin purification and TMT labeling of multiple individual samples in parallel was followed by pooling of samples and analysis in a single mass spectrometry experiment to achieve accurate relative quantification of protein abundance, enabling the recording of time courses of GPCR signaling 17. Dynamic and rapid changes in the interactome could be resolved because multiplexed TMT-MS allows massive parallel and time-resolved quantitative analysis 31, 32, now in up to 18 channels 33. In HEK293 cells, type 1 angiotensin II receptor and β2-adrenergic receptor (β2-AR) were expressed as APEX2 fusion proteins. Using a time course of agonist ligand binding and proximity labeling, followed by TMT-MS, the tracking of activation and internalization of these receptors and the identification of novel and functionally relevant interaction partners, such as LMBRD2, after ligand binding was achieved 17. Thus, the combined use of APEX2 proximity labeling and multiplexed quantitative mass spectrometry is ideally suited to study changes in perturbed biological systems 16. These studies formed the basis of our utilization of APEX2-based proximity labeling to elucidate the molecular mechanisms underlying adrenergic regulation of CaV1.2 in the heart 14.
Similarly, APEX2 was used to capture snapshots of β2-AR and the δ-opioid receptor interactome 27. In an alternative approach to distinguish the specific protein-protein interaction network from bystanders, spatially specific APEX reference labeling probes were developed: Lyn11 (plasma membrane targeting), 2xFYVE (early endosome targeting), and GFP (cytoplasmic). It is critically important to select the correct spatial reference and ensure achievement of the anticipated localization pattern. For instance, the known receptor network components for β2-adrenergic receptor could not be distinguished from bystanders using GFP-APEX or 2xFYVE-APEX at 1 minute after agonist stimulation, when receptors should still be in the plasma membrane. Ten minutes after agonist stimulation, the endosomal 2xFYVE-APEX spatial reference differentiated the β2-adrenergic receptor network from bystanders. Thus, label-free mass spectrometry-based quantification with spatial references can be used to distinguish an interaction network from bystanders.
Off-the-shelf proximity-based biotinylation methods
The “standard” approaches for proximity-based labeling rely on the knock-in or expression via plasmids of the biotinylation enzyme fused to the bait protein in target cells of interest. Creating animals expressing proximity-based labeling enzymes is labor- and resource-intensive and carries the risk that conjugation of the APEX2 enzyme alters bait protein function. Moreover, proximity-based labeling using the expression of a fusion gene cannot be used on primary human tissue samples. Several methods have been developed to facilitate biotin-based proximity labeling without the need for genetic engineering or transfection of cultured cells. Prior to the development of TurboID or APEX, proximity labeling using local biotin conjugation was based on antibody-guided targeting of HRP 34–37. This approach was further developed to study fixed and permeabilized tissue samples, in which a primary antibody binds to the target protein and in the presence of biotin-phenol and H2O2, a secondary HRP-conjugated antibody creates biotin-phenoxy radicals that covalently conjugate to nearby proteins 13, which can be identified and quantified using mass spectrometry. Another approach is to use Protein A fused to the TurboID enzyme (ProtA-Turbo). In permeabilized fixed or non-fixed mammalian cells, including clinical samples, the ProtA-Turbo enzyme can be specifically targeted to bait proteins using antibodies 38. Upon the addition of exogenous biotin, proteins proximal to the bait are subsequently biotinylated and analyzed by MS 38.
The applicability of proximity labeling in animals has been limited by the necessity to tag each protein of interest with an engineered biotin ligase or peroxidase, and then generate transgenic organisms. A more adaptable system was developed by combining the ability to use existing transgenic zebrafish expressing GFP-tagged proteins and a biotin-ligase conjugated to a GFP-binding nanobody for proteomic comparison of different tissues and/or different proteins (Fig. 2D) 12, 39. Initial attempts to use BirA* and BioID2 in zebrafish were unsuccessful and did not demonstrate detectable in vivo biotinylation 12 (although see 40, in which BioID2 was successfully used in zebrafish, discussed below). In contrast, TurboID induced strong biotin labeling as detected by streptavidin blotting. In a novel approach to labeling, a TurboID-conjugated nanobody directed at GFP was created, in which the GFP-binding nanobody is degraded when not bound to GFP (‘destabilized GFP binding protein’ or dGBP) (Fig. 2D). Transgenic zebrafish were created with expression, under the control of a broadly expressing actin promoter, of a TurboID-dGBP fusion with a P2A- red-fluorescent protein element (termed “biotin labeling in tagged zebrafish” or BLITZ) 12. Double-positive TurboID-dGBP and GFP-transgenic fish were selected by fluorescence. In double-positive fish treated with supplemental biotin, in vivo biotinylation co-localized with the same subcellular distribution of the GFP-tagged proteins. Streptavidin affinity purification was able to precipitate known interactors of the GFP-tagged proteins. Streptavidin affinity purification of double-positive transgenic embryos expressing GFP-tagged Cavin4B, a protein associated with striated muscle and transverse (T)-tubules, enriched muscle specific proteins including SPEG, LDB3 and CMYA5, which have also been enriched in dyad protein proximity proteomes from transgenic BioID and APEX2 mouse model systems 14, 41, 42.
CARDIOVASCULAR APPLICATIONS OF PROXIMITY PROTEOMICS
Proximity proteomics to identify Kir2.1 macromolecular complexes in HEK cells
The strong inward rectifying potassium (K+) current imparted by Kir2.1 is an important regulator of the resting membrane potential. KCNJ2 gain-of-function mutations cause short QT syndrome, which increases the risk of sudden cardiac death 43. Kir2.1WT and Kir2.1Δ314-315, an Andersen-Tawil Syndrome-associated mutant that blocks Kir2.1 Golgi export, were conjugated to BirA* and stably expressed in HEK293T cells 44. HEK cells expressing a yeast transmembrane domain conjugated to BirA* (TM-CTRL) were used as a negative control. The proteomic analysis, which did not utilize mass tag quantification, resulted in the identification of 218 high-confidence Kir2.1 BioID hits. Of 24 previously identified Kir2.1 interactors, 10 (~42%) were identified by BioID as well. Of the 218 proteins identified, 75 interacted preferentially with Kir2.1WT, 66 interacted preferentially with Kir2.1Δ314-315 and the remaining 77 hits interacted with both Kir2.1WT and Kir2.1Δ314-315. Kir2.1WT channel interactors were enriched for protein families involved in cell adhesion, and the top Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways included arrhythmogenic right ventricular cardiomyopathy, dilated cardiomyopathy, and hypertrophic cardiomyopathy. The Kir2.1Δ314-315 that is retained in the Golgi preferably interacts with protein families involved in intracellular transport, ESCRT-0, and lysosomal processes, suggesting that the ATS1 mutation leads to increased targeting of Kir2.1 to lysosomal degradation. The functional role of one Kir2.1 interactor, Plakophilin 4 (PKP4), was validated as a neighbor and functional interactor, respectively, using co-immunofluorescence in isolated rat ventricular myocytes, and in a stable HEK293 cell line expressing Kir2.1, in which PKP4 expression increased current density, whereas depletion of PKP4 reduced current density. Thus, proximity labeling identified a new Kir2.1 interactor PKP4, which functions as a positive regulator of Kir2.1 density 44.
Use of proximity labeling in cultured neonatal cardiomyocytes
i. Identifying the adherens junction proteome using N-cadherin-BioID
The adherens junction (AJ), the primary anchor for myofibrils and linker of actin filaments from adjacent cells, senses mechanical forces, enables cells to maintain shape, and transduces signals related to the actin cytoskeleton 45. The transmembrane protein N-cadherin is the main component of the AJs, homodimerizing with N-cadherin from adjacent cells in the extracellular space. To identify novel components of cardiomyocyte AJs, a fusion protein of N-cadherin-BioID2 was expressed using adenovirus in cultured primary cardiomyocytes isolated from neonatal (P1-P3) mice 46. The cadherin-BioID2 fusion proteins trafficked to the AJ, like endogenous cadherin. Upon biotin supplementation in the culture media, biotinylated proteins were observed along cell-cell contacts and, to a lesser extent, at Z-disks. 917 biotinylated proteins were identified by mass spectrometry. Excluding proteins with a single unique peptide reduced the list to 487 proteins. The list was further refined by including only proteins enriched by ≥ 10-fold, compared to control, non-infected cardiomyocytes, further reducing it to 365 proteins. The most abundant proteins were known components of nearby macromolecular complexes: the AJ, including β-, Δ-, and α-catenin, and junction plakoglobin; desmosomal proteins, desmoglein2 and plakophilin2, reflecting the proximity of AJs and desmosomes; and actin-binding proteins, vinculin and afadin, reflecting the importance of these proteins linking the AJ to the myofibril network. Comparing this refined list of cadherin-BioID2 enriched proteins with the published epithelial cadherin (E-cadherin) interactome in epithelia, one finds many of the closest N-cadherin interactors are shared with E-cadherin, indicating that the core cardiomyocyte AJ protein interactome is similar to that of epithelia, whereas specialization seems to occur in the less-enriched, deeper, N-cadherin specific interaction network.
In the N-cadherin proteome network, ~140 curated intercalated disk proteins were not detected including connexin 43 46. The absence of these proteins could be due to the range of biotinylation, the absence of surface lysine residues, the occlusion of access to biotinylation by other macromolecular components, or the specifics of the method (for instance, the use of cultured neonatal versus freshly isolated cardiomyocytes). Of the 185 N-cadherin hits, only 13 are curated intercalated disk proteins. As in other proximity labeling studies, some hits may not be located at the intercalated disk but rather reflect that biotin-labeling can occur through all stages of the fusion-protein life cycle.
ii. Defining the caveolin 3 interactome
Caveolae, small invaginations in the lipid bilayer, are created by the oligomerized scaffolding protein caveolin 47, 48. They have important roles as scaffolding proteins acting on intracellular signal transduction, endocytosis, and mechano-modulation. To identify caveolin 3 interactions in cardiomyocytes, N-terminal tagged V5-APEX2-caveolin 3 was expressed using adenovirus in cultured neonatal rat ventricular myocytes 49. Expression of V5-APEX2 via adenovirus served as a control. Ratiometric proteomic analysis was performed using 3-state isotope labeling by amino acids in cell culture (SILAC) labeling for 13 days. Adenoviral expression of either V5-APEX2-caveolin 3 or V5-APEX2 occurred on day 11. Biotinylated proteins were enriched by affinity purification and MS identified 1131 bound proteins, of which 101 proteins were significantly enriched by V5-APEX2-caveolin 3. All essential and muscle-specific components of the core caveolar complex were identified including cavin1 and cavin4. Other proteins identified included caveolin 1, myosin light chain, actin, troponin, Na+-K+ ATPase α1 and β1 subunits, and Na+-Ca2+-exchanger. Proteins that are involved in transmembrane substrate transport were also detected, including monocarboxylate transporter (McT1) and transferrin receptor (TfR1), which were speculated to be new caveolin 3 proximity candidates. The monocarboxylate transporter is a major pathway for transmembrane lactate and pyruvate transport in the heart 50. To validate these findings, caveolin 3 knockout human iPSC-CMs were created. Surface expression of the monocarboxylate transporter was decreased in the knockout cardiomyocytes, causing reduced extracellular acidification through the lactate/proton shuttle. These results demonstrate the power of proximity labeling in identifying new caveolin partners 51.
iii. Defining the phospholamban proteomic subdomain
Calcium handling in the heart requires the coordinated activities of ion channels and pumps. The sarcoplasmic reticulum (SR) Ca2+-ATPase (SERCA) pumps Ca2+ from the cytosol into the SR during diastole. Its activity is regulated by the inhibitory protein, phospholamban (PLB). Phosphorylation of PLB relieves the inhibition of PLN, leading to accelerated relaxation and increased contraction 52, 53. PLN is phosphorylated at Ser16 by PKA and at Thr17 by Ca2+-calmodulin-dependent kinase (CaMKII) 54.
To explore the proteomic neighborhood of SERCA and PLB, proximity labeling and mass spectrometry with APEX2-conjugated PLB was performed in neonatal rat cardiomyocytes 55. The cardiomyocytes were cultured for 13 days in heavy, medium, and light SILAC (stable isotope labeling by amino acids in cell culture) medium and thereupon transduced with adenoviral vectors for expression of V5-APEX2-PLB, V5-APEX2 fused to green fluorescent protein (negative controls for background binding to avidin) or V5-APEX2 fused to PLB lacking its N-terminal domain (non-specific APEX2-mediated labeling). Although PLB, protein phosphatase 1 and Hsp20 were enriched, SERCA was not detected. Moreover, six of seven 14-3-3 proteins were labeled, consistent with precipitation studies showing a complex between 14-3-3 and phosphorylated PLB. The binding of 14-3-3 to phosphorylated PLB prevents the rapid dephosphorylation of PLB, stabilizing the increased SERCA activity. The authors propose that 14-3-3 binding to PLB creates a molecular memory of kinase activity.
Use of proximity labeling in cultured iPSC-CMs.
Actinin, an integral member of the actin cytoskeleton, interacts with multiprotein complexes at Z-disks and focal adhesions, which both function as hubs for mechanical force generators and sensors. Using CRISPR, BirA* (BioID) was fused with a hemagglutinin (HA) tag to the C-terminus of endogenous ACTN2 in a wildtype human iPSC line and a troponin T2 gene knock-out (TNNT2ko/ko) iPSC line (Fig. 3A). While both iPSC lines can be differentiated to cardiomyocytes, TNNT2ko/ko cardiomyocytes exhibit sarcomere structural and functional deficits such that Z-disk assembly is blocked at the Z-body stage, and twitch force does not occur. Using these two models, actinin nano-environments were studied 56. Biotin supplementation (50 μM) induced strong biotinylation, and actinin proximity partners were identified by TMT quantitative mass spectrometry. 324 neighboring proteins were enriched, principally involved in cell adhesion, anchoring junction, actin cytoskeleton, contractile fiber, and RNA binding. 24 actinin partners were exclusive to the Z-disk stage and one was exclusive to the Z-body stage. Using gene ontology (GO) analysis, the study also identified 99 proteins with general RNA-binding functions, such as ribosomal assembly, transcription/translation and RNA localization and metabolism. Many of these RNA-binding proteins 56, excluding IGF2BP2, were also identified in the CaV1.2-APEX list 14.
Figure 3. Use of BioID to probe the cardiac actinin neighborhood.
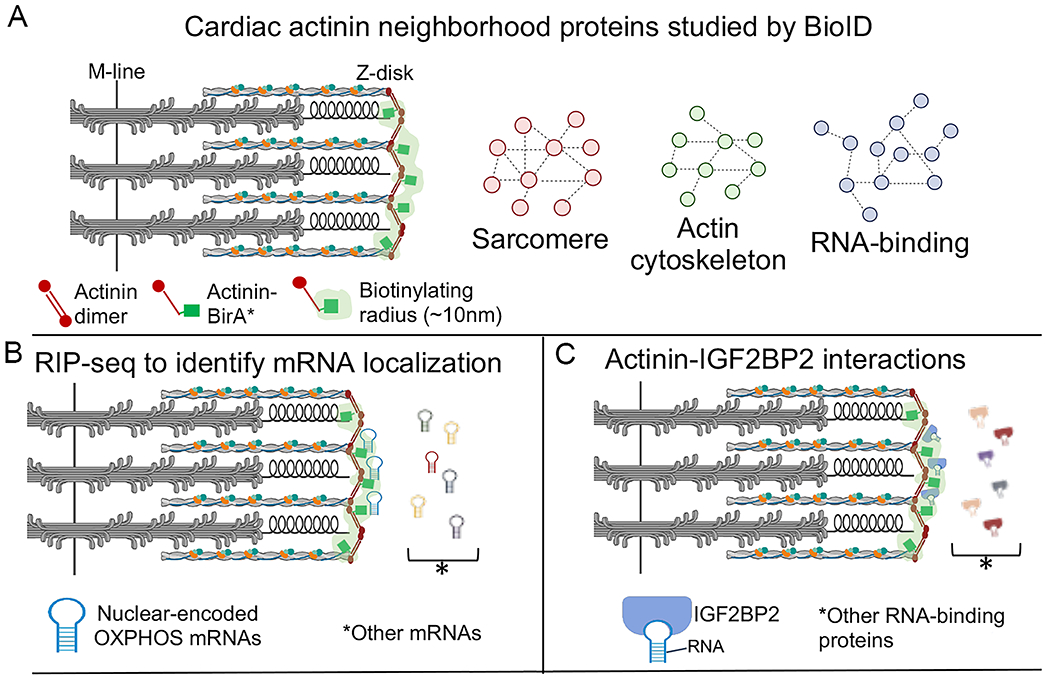
(A) Actinin-BirA* was expressed, via CRISPR knock-in, in human inducible pluripotent stem cell derived cardiomyocytes. Biotinylated proteins were purified and identified by mass spectrometry. Gene ontology analysis of the proteins revealed functions in actin cytoskeleton, sarcomere, and RNA binding. The light-green shading indicates an estimate of the zone of biotinylation. (B) RNA precipitation followed by sequencing (RIP-seq) using streptavidin affinity purification to pull-down RNA-binding proteins that were biotinylated within ~10 nm of actinin-BirA*. Gene ontology analysis indicated enrichment of oxidative phosphorylation (OXPHOS) transcripts. (C) Actinin interacts directly with insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), localizing IGF2BP2 to Z-disks. Adapted from 56.
Proximity labeling methods can also be applied to elucidate subcellular RNA localization either by direct RNA-labeling 57, or through proximity labeling of RNA-binding proteins followed by RNA precipitation and sequencing (RIP-seq) 58. To reveal gene transcripts bound to the RNA-binding proteins in the actinin sub-proteome, biotinylated RNA-binding proteins were affinity purified using streptavidin followed by RIP-seq (Fig. 3B). Differential expression analysis identified 945 transcripts bound to biotinylated RNA-binding proteins, and GO analysis revealed enrichment of electron transport chain (ETC) and ribosome transcripts. Additional biochemical and protein interaction studies revealed that Insulin Like Growth Factor 2 mRNA Binding Protein 2 (IGF2BP2), an RNA-binding protein with metabolic functions, directly interacted with actinin, and IGF2BP2 knock-down impaired ETC transcript localization to Z-disks but not overall transcript stability (Fig. 3C). To study the function of actinin-IGF2BP2 interactions in cardiomyocytes, the actinin missense mutation glutamic acid at residue 445 substituted for alanine (E445A) was studied because it disrupted actinin-IGF2BP2 interactions without disrupting actinin dimerization and sarcomere assembly. Cardiomyocytes expressing E445A actinin exhibited diminished oxygen consumption rates and cell survival following pathological sarcomere activation induced by a hypertrophic cardiomyopathy-causing mutation in TNNT2. Overall, the use of proximity labeling in human iPSC-CMs enabled the identification of new actinin interaction partners including through sarcomere assembly, and their essential role in the metabolic responses to pathological sarcomere activation 56.
Expression of proximity labeling enzyme-conjugated proteins in hearts of mice
i. Elucidating the molecular mechanism of adrenergic CaV1.2 modulation
Voltage-gated Ca2+ channels in heart cells mediate the initiation of cardiac excitation-contraction coupling, control action potential duration, and regulate gene expression. Cardiac CaV1.2 channels are prominently up-regulated by β-adrenergic agonists via activation of PKA 59, 60. This regulation is believed to contribute to the increased contractility of the heart during exercise, and as a component of the physiological ‘fight-or-flight’ response. The elucidation of mechanisms underlying β-adrenergic regulation of cardiac Ca2+ channels has resisted decades of investigation, marked by the inability, until recently, to recapitulate this regulation in heterologous expression systems 61. Since phosphorylation of the core α1C and β2 subunits were shown to be dispensable in the process 14, 62–65, we speculated that other members of the CaV1.2 macromolecular complex were the functional protein kinase A (PKA) target(s). A long-standing barrier to discover this basic mechanism regulating Ca2+ in the heart was the inability to reconstitute this regulation in the heart, precluding initial investigations using heterologous expression. To explore the nano-domain of the CaV1.2 macromolecular complex in heart, we created two transgenic mouse lines, one with inducible, cardiomyocyte-specific expression of dihydropyridine-resistant, APEX2 and V5-epitope-tagged α1C subunit and the other with inducible, cardiomyocyte-specific expression of APEX2 and V5-epitope-tagged β2B subunit 14. Importantly, fusing APEX2 to either α1C or β2B altered neither trafficking nor function of CaV1.2 channels, nor adrenergic stimulation of Ca2+ channels. We reasoned that the use of two APEX2-conjugated target proteins, one transmembrane and the other cytoplasmic, would report intersecting and distinct neighboring proteins. Instead, the proteomic compositions were remarkably similar 14. Using triple-stage mass spectrometry, we quantified hundreds of biotinylation-enriched proteins although many are possibly bystanders rather than physically interacting proteins. Many of these proteins could theoretically be the functional PKA target and identifying the correct one is challenging. Furthermore, a substantial number of these proteins may not be located within the macromolecular complex of a fully matured channel at the surface membrane. Proximity labeling may occur in several cellular compartments and likely reflects, to some extent, synthesis, maturation and trafficking with or adjacent to CaV1.2 channel subunits.
The speed of APEX2-mediated labeling has enabled quantitative analysis of G-protein coupled receptor (GPCR) signaling, allowing parallel time-resolved measurement of the dynamic changes in the proximity of hundreds to thousands of proteins 17, 27. Work on β2AR-APEX functioned as a proof of concept showing that it is possible to identify signal transduction components via target-APEX2 fusion even in the absence of prior knowledge in signaling effectors 17. We reasoned that β-adrenergic agonist induced PKA-dependent stimulation of Ca2+ current in the heart, which occurs rapidly, could alter the macromolecular complex of CaV1.2 such that PKA phosphorylation of an activator or inhibitor could be either recruited or depleted respectively. Following biotin-phenol incubation, we induced proximity labeling with H2O2 during the exposure of isoproterenol to either isolated cardiomyocytes or retrograde perfused heart (Fig. 4A–B). We confirmed that in the presence of biotin-phenol and H2O2, PKA signaling was preserved by assessing the phosphorylation status of phospholamban, and for the retrograde-perfused hearts, the effects of isoproterenol on the heart rate. The biotinylated proteins were affinity-purified under denaturing conditions using streptavidin and their relative abundance was quantified by tandem mass tag synchronous precursor selection triple stage MS (TMT SPS MS3). β-adrenergic stimulation induced changes in the biotinylation of several proteins, including an increased biotinylation of the PKA catalytic subunit and decreased biotinylation of Rad (Ras associated with diabetes) 14, a member of the RGK (Rad, Rem, Rem2, Gem/Kir) Ras-family of proteins (Fig. 4C). These findings indicate that more PKA catalytic subunit and less Rad are near CaV1.2 channels upon exposure to isoproterenol.
Figure 4. APEX2-based proximity labeling identifies mechanism underlying adrenergic regulation of CaV1.2 in heart.
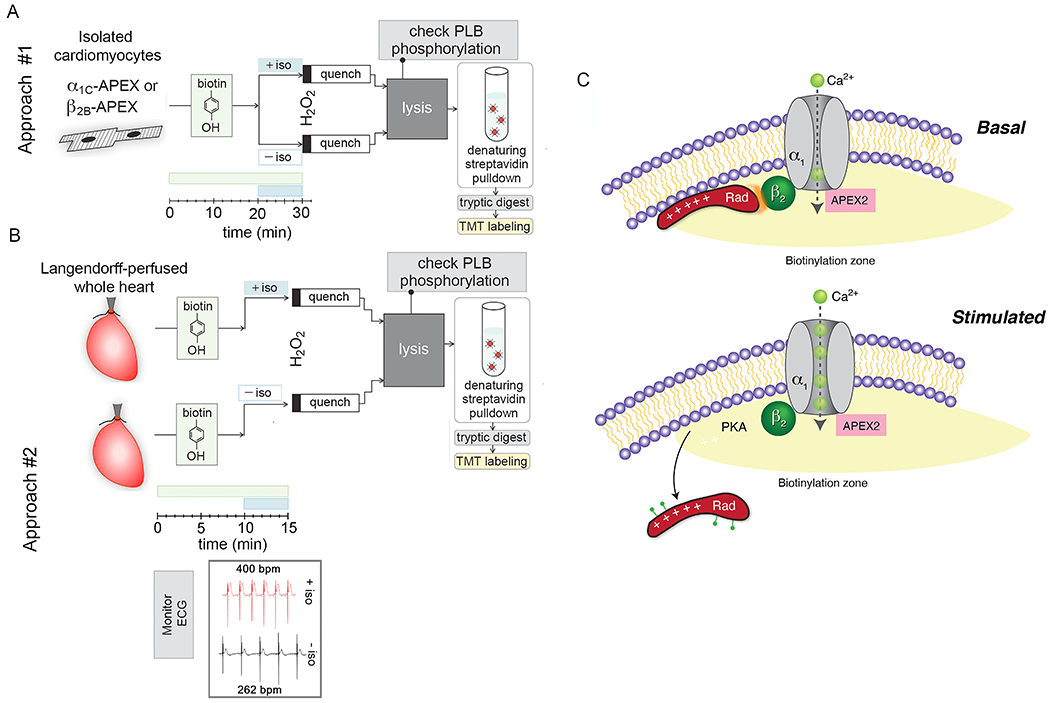
(A-B) Workflow for processing proximity-labeling in isolated cardiomyocytes and retrograde-perfused hearts. V5 epitope and APEX2 cDNA were conjugated to the N-terminus of dihydropyridine-resistant α1C and wild-type β2B subunits. Transgenic mice with non-targeted insertion of tetracycline-regulated cDNAs were bred with cardiac-specific codon-optimized reverse transcriptional transactivator (rtTA) mice 74. Transgene expression did not require doxycycline due to low basal binding of rtTA to the Tet operator sequences. In approach #1, ventricular myocytes were isolated by enzymatic digestion and proximity labeling was performed as previously described for isolated cells except for the addition of 1 μM isoproterenol for 10 minutes prior to labeling with H2O2. For each heart, samples were split into 2 groups, one with isoproterenol and one without isoproterenol. After lysis, the effect of PKA on phospholamban (PLB) was determined by western blotting to ensure viability of cardiomyocytes during labeling. In approach #2 (B), samples could not be paired, and hearts were labeled in the absence or presence of isoproterenol. Incubation with biotin-phenol was reduced to 15 minutes and isoproterenol for 5 minutes. Bpm= beats per minutes. (C) Schematic demonstrating zone of biotinylation around CaV1.2 channels. PKA is recruited to and Rad is depleted from the channel neighborhood after isoproterenol. Adapted from 14.
Further studies revealed the functional relevance of these dynamic changes in the macromolecular complex of CaV1.2 channels 14, 66. At baseline, the association of Rad with CaV1.2 inhibits a subpopulation of Ca2+ channels. Upon PKA phosphorylation of Rad, in response to β-adrenergic agonists, the interaction of Rad with the channel is diminished, releasing the inhibition of Ca2+ channels and thereby increasing Ca2+ influx. Co-expression of multiple classes of voltage-gated Ca2+ channels with Rad in HEK cells demonstrated increased Ca2+ current in response to stimulation with forskolin, an adenylate cyclase and PKA activator. Thus, through the use of proximity labeling and quantitative mass-spectrometry, the long-sought mechanism underlying adrenergic control of Ca2+ influx in the heart was discovered 14, 61, 66.
ii. Identifying the cardiac dyadic proteome
Junctophilin 2 (JPH2) spans the cleft from the transverse tubule (t-tubule) to the sarcoplasmic reticulum, tethering these two membrane structures to facilitate normal cardiac Ca2+ handling. Mutations in JPH2 are associated with cardiomyopathy in humans 67–69. Mice with BioID2 fused to the Jph2 coding sequence were created using a CRISPR-Cas9 knock-in strategy 42. The 3XFLAG-BioID2-Jph2 proteins were expressed at levels equivalent to Jph2 protein. Saturation of proximity labeling was achieved in homozygous knock-in mouse hearts after several days of intraperitoneal injection of biotin. Using label-free tandem MS, 550 biotinylated proteins were identified including RyR2 and the CaV1.2 subunits, α1C and β2. Gene ontology enrichment analysis showed that the top cellular components were sarcolemma, cation channel complexes and t-tubule. A comparison with an IP-MS analysis of Jph2 interactors in a transgenic mouse line overexpressing Jph2-HA fusion protein showed that of the 18 proteins suggested to be direct interactors of Jph2 70, only 2 were found in the enriched Jph2-BioID2 list (Fig. 5B and 5D), namely RyR2 and Striated Muscle Preferentially Expressed Protein Kinase (SPEG). The study demonstrates that a BioID2 knockin can be used to identify potentially novel protein-protein interactions in the heart.
Figure 5: Proximity proteomes of the cardiac Z-disk and cardiac dyad.
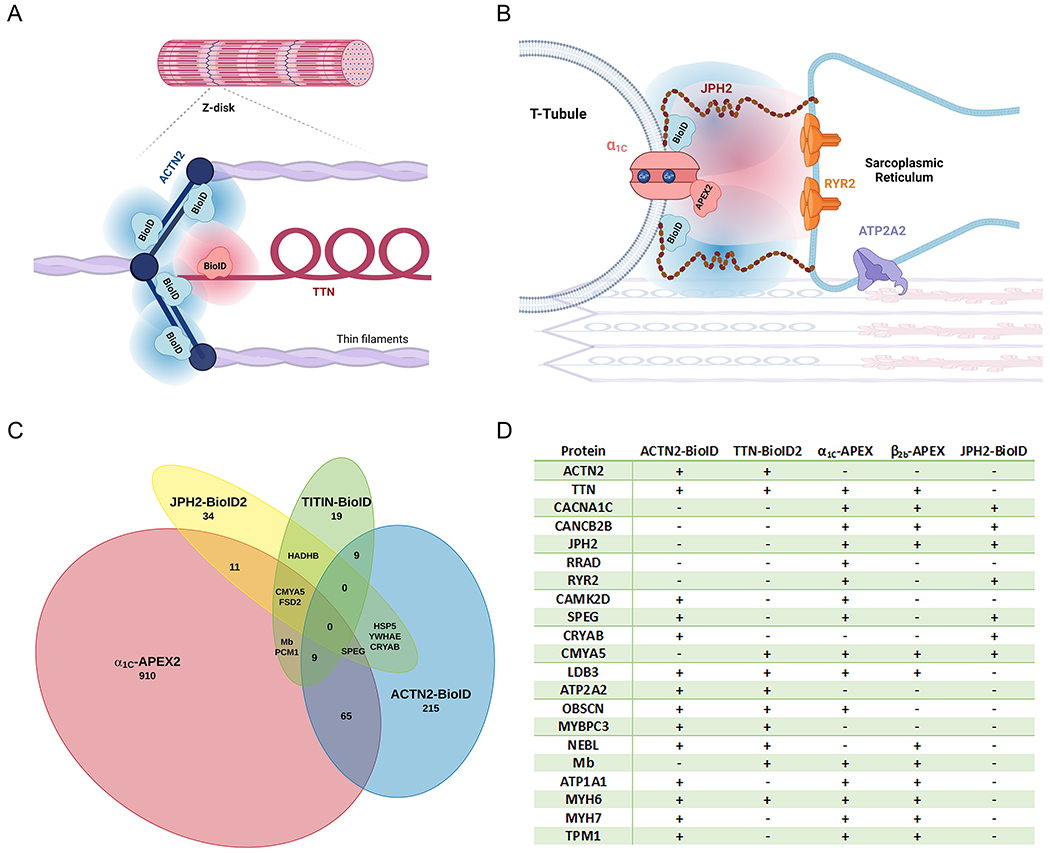
(A-B) Schematics illustrating the foci of published proteomic mapping studies located at the Z-disk (A) and the overlying sarcolemma (B). (C) Overlapping proteomic mapping of sarcolemmal and Z-disk protein neighborhoods. Proteins included were described by authors as showing significant enrichment in the neighborhood of their protein of interest. In vivo proximity labeling occurred in knock-in mice with BioID fused to Ttn, BioID2 fused to Jph2, human iPSC-derived cardiac myocytes with knock-in of BioID fused to ACTN2. Live in-situ ex-vivo proximity labeling was examined in mice with inducible transgenic overexpression of the CaV1.2 α1C subunit fused to APEX2. (D) Enrichment of proteins critical for cell structure, Ca2+ homeostasis, contraction and signaling were noted across multiple proximity proteomes, including those from isolated myocytes from mice overexpressing CaV1.2 β2B-APEX2. Created in part with LucidChart and Biorender.com 14, 41, 42, 56.
iii. Identifying the sarcomere proteome using titin-BioID
Using homologous recombination BioID was inserted into exon 28 of the titin (ttn) gene, corresponding to a location on the titin filament located at the transition from the Z-disc to I-band (Fig. 5A) 41. Mice tolerated this insertion and displayed normal growth and heart size. The most prominently biotinylated protein was titin with the expected hotspot of biotinylated sites near the BioID insertion site, yielding an estimated radius of biotinylation of 7-15 nm. In addition to biotinylated titin, which accounted for > 90% of total biotinylated peptides, other proteins were identified related to muscle filament assembly, myocyte function, α-actinin binding, and metabolism. In adults, 478 sarcomeric or sarcomere-associated proteins were identified in heart and skeletal muscle – more than twice the number of known sarcomeric proteins. BioID-mediated labeling can be applied in unique ways leading to new insights into positioning and function of titin, greater understanding of myofilament dynamics, and, by comparing the neonatal versus adult sarcomere proteome, capturing a snapshot of the shift to adult energy metabolism. For instance, the distribution of biotinylation sites suggested that titin forms a hairpin as the sarcomere contracts, which may contribute to the elastic properties of the sarcomere 41.
Interaction networks in zebrafish
Given the high energy requirements and low rate of proliferation of post-natal mammalian cardiomyocytes, the heart is an organ with limited capacity for regeneration after injury. Proximity labeling can identify protein networks during development and in animal models of tissue regeneration. Two transgenic zebrafish lines were created with cardiac-specific expression driven myosin light chain 7 promoter; one with BioID2 fused to GFP and the other with BioID-GFP fused to a membrane-localizing CAAX motif 40. Robust biotinylation was not detected by streptavidin blotting of hearts from fish in biotin-supplemented water. Rather adult fish 3-8 months old were given three sequential daily intraperitoneal injections of biotin (3.7μg biotin in total), demonstrating that effective biotinylation in zebrafish only occurs after injection. Homogenates of pooled BioID expressing fish hearts were affinity purified on neutravidin beads and exposed to stringent detergent and denaturing conditions prior to quantitative mass spectrometry. From this study 1113 proteins were quantified, 343 of which were enriched at least 2.5-fold in the transgenic fish with BioID2-CAAX fusion 40.
BioID2 fish were crossed with transgenic fish with tamoxifen-inducible expression of diphtheria toxin A in the heart, which has been shown to ablate 60% of myocytes 71. Fish were biotin supplemented 11-13 days after ablation and the BioID2 and BioID2-CAAX proteomes were examined during cardiac regeneration. Here 173 proteins were changed at least 1.5-fold in the injured compared to BioID-expressing uninjured hearts. Validating this approach, Seta, a protein expressed in injured heart, was enriched only in the BioID2-CAAX hearts after injury. Similarly, the abundance of Epb41l5 was reduced in recovering BioID2-CAAX heart but increased in BioID2 fish without CAAX. Confocal microscopy confirmed localization of this protein changes from membrane-restricted to global after injury. Gene ontology term analysis showed injured hearts expressed more proteins involved in wound healing, cardiac development, and muscle differentiation while there were diminished proteins involved in energy metabolism, particular oxidative phosphorylation, identified.
The proximity proteome of ErbB2, a co-receptor of the mitogen NRG1 shown to stimulate myocyte proliferation in the adult heart 72, was explored using a transgenic zebrafish line expressing the HA-tagged fusion protein ErbB2-BioID2. Crossed with diphtheria toxin A transgenic fish, quantitative mass spectrometry analysis of ErbB2-BioID2 hearts 14 days after myocyte ablation showed 1.5-fold enrichment of 108 proteins among the 667 identified. The most enriched protein was Rho A, a Rho GTPase which was HA-precipitated from injured hearts but not from healthy hearts. Follow-up pharmacologic and genetic inhibition studies showed reduced Rho A function inhibited repair in a resection-model of zebrafish heart injury. Taken together, these proximity proteomic experiments in zebrafish demonstrate that the effects of Nrg1 and other mitogens are mediated by RhoA in cardiomyocytes during regeneration 40.
Proximity labeling to identify secretomes
Proximity-based biotinylation has also been used as a platform to capture tissue-specific secretome. HA-tagged BioID2 with a C-terminal KDEL endoplasmic reticulum (ER) retention sequence (ER-BioIDHA) was expressed using lentiviral transduction in cultured endothelial cells 73. Biotin supplementation of lentivirally transduced cultured endothelial cells resulted in ER-localized anti-HA and streptavidin staining, indicating successful biotinylation of proteins in the ER. In contrast to media from cells transduced with ER-retained GFP (ER-GFP) and supplemented with biotin, streptavidin blotting of electrophoretically resolved media from ER-BioID transduced cells identified multiple proteins. Additionally, ER-BioID was detected in cell lysates but not in the media, suggesting that biotinylation of secreted protein occurred only intracellularly. This technique was used to characterize the changing secretome of an in vitro model of the endothelial mesenchymal transition.
To study the proteome of secreted proteins in animals, transgenic mice with ER-BioIDHA expression could be induced by excision of a loxP-flanked enhanced GFP 73. When these mice were crossed with mice expressing Cre-recombinase in endothelium or skeletal muscle, the enhanced GFP is excised and ER-BioIDHA is expressed. From serum of mice supplemented with biotin, biotinylated proteins were purified and analyzed by mass-spectrometry. In mice expressing ER-BioIDHA in the endothelium, several well-known proteins were identified from the serum including endothelial cell surface receptors and von-Willebrand factor. Proteins not commonly associated with the endothelium were also identified, such as fibrocystin. To obtain sufficient biotinylation of proteins, mice expressing ER-BioIDHA in muscle required biotin-supplementation via multiple routes of administration (intraperitoneal, subcutaneous, and oral feeding) for 5 days. Mice were separated into cages without and with a running wheel to determine how the muscle-derived secretome changed with voluntary exercise. Exercise altered the abundance of several proteins including myostatin which modestly decreased and sarcalumenin which increased 73. These findings demonstrate the feasibility and sensitivity of using proximity proteomics to detect, in a largely unbiased fashion, alterations in secreted proteins distant from the site of their generation, in changing pathophysiologic states.
CONCLUSIONS AND FUTURE DIRECTIONS
With the rapid advancement of proximity labeling techniques and mass spectrometry, elucidation of novel protein-protein interactions and their dynamics is now feasible. Given the individual strengths of these proximity labeling techniques, the optimal system of choice depends on the biologic question involved (see Table 1). Biotin ligases, with their smaller labeling radiuses and lower catalytic efficiency are well-suited to mapping neighborhoods with relatively high specificity, at different ages, in different genetic backgrounds and in health versus disease. Peroxidase-based labeling, in contrast, is particularly suited to investigate rapid signaling events. For signaling events that are suspected to involve recruitment or removal of an interactor, APEX2 labeling, combined with TMT multiplexing has exceptional specificity—potentially hundreds of neighboring proteins that remain stationary during the perturbation are unchanged during quantification.
To date, we are aware of four studies that have probed the t-tubule and underlying Z-disk with proximity proteomics, examining separately the interaction networks of ACTN2, TTN with BirA* in its Z-disk domain, JPH2, and CaV1.2, in human iPSC myocytes, knock-in and transgenic overexpression mice (Fig. 5A–B) 14, 41, 42, 56. BioID proteomes were compiled according to the authors’ criteria for significant enrichment, generally comparing biotin supplemented BioID animals or cells versus control. For ACTN2 303 such proteins were identified, 50 for TTN, and 52 for JPH2. For the α1C-APEX mouse cardiomyocyte proteome, we included the top 1000 proteins, which were all at least 4-fold enriched over non H2O2 treated controls. Both Z-disk proteins, ACTN2 and TTN identified the other in its nanodomain, as did the all three sarcolemmal proteomes, including that of β2B-APEX (examining its top 1000 β2B-APEX hits, also at least 4-fold enriched). Despite the close-proximity of the bait, there were no proteins in common among all 4 proteomes, though there was abundant overlap (Fig. 5C–D).
Although APEX2-labeled neighborhoods can be revealing, we found that proximity labeling using CaV1.2-APEX2 fusion proteins expressed in mice were particularly effective to identify dynamic neighborhoods, namely how β-adrenergic agonists alter the proteomic neighborhood around CaV1.2 14. Through our experiments we identified the Ca2+ channel inhibitor Rad as a key mediator in β-adrenergic stimulation of cardiac CaV1.2 channels. Rad was experimentally confirmed as the regulator of voltage-gated Ca2+-channel that permits their adrenergic regulation, after its identification in the α1C-APEX and β2B-APEX “neighborhood”. This, and its absence from other published proteomes, makes detection of Rad an instructive example of the relative of merits of proximity labeling methods. Though this protein’s primary site of regulation is the sarcolemma of myocytes, a space that measures a mere 12 nm across, recombinant dyad proteins fused to biotin ligases did not identify significant enrichment of Rad 42, 56. On the other hand, Rad was found in a list of proteins identified by mass-spectrometry after HA-antibody affinity purification of heart lysates from transgenic HA-tagged junctophilin 70. Although not compared head-to-head in the same experiments, the greater catalytic efficiency of peroxidase-based systems may lead to the detection of more rare labeling events, at the cost of identifying more bystanders as well.
More broadly, proximity-mediated labeling represents an exciting method for continued probing into the microenvironments of diverse proteins, advancing the discovery of key mediators of cardiac function in health and disease, such as during the development of heart failure and hypertrophy. Techniques and methodological approaches to distinguish interacting proteins from bystanders, and methods for high throughput screening for potential interactions will accelerate adoption and successful implementation of proximity-dependent biotinylation studies.
Sources of funding
The research was supported by K08 HL151969 and AHA SDG to JSK, NHLBI R01 HL142787 and U01 HL156349 to JT Hinson; R01 HL146149, R01 HL155377, and R01 HL121253 to SOM, NHLBI R01 HL160089 to GSP and SOM.
Non-standard Abbreviations and Acronyms:
- IP
immuno-precipitation
- MS
mass spectrometry
- HRP
horseradish peroxidase
- APEX
ascorbate peroxidase
- TMT
tandem mass tag
- Rad
Ras associated with diabetes
- RGK
Rad, Rem, Rem2, Gem/Kir
- PLB
phospholamban
- PKA
protein kinase A
- CaMKII
Ca2+-calmodulin-dependent kinase
- ER
endoplasmic reticulum
- β2-AR
β2-adrenergic receptor
- GFP
Green fluorescent protein
- dGBP
‘destabilized GFP binding protein
- t-tubule
transverse tubule
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- AJ
adherens junction
- E-cadherin
epithelial cadherin
- SERCA
sarcoplasmic reticulum (SR) Ca2+-ATPase
- HA
hemagglutinin
- GO
gene ontology
- RIP-seq
RNA precipitation and sequencing
- ETC
electron transport chain
- IGF2BP2
Insulin Like Growth Factor 2 mRNA Binding Protein 2
- GPCR
G-protein coupled receptor
- JPH2
Junctophilin 2
- CAAX
C is cysteine residue, AA are two aliphatic residues, and X represents any C-terminal amino acid
Footnotes
Competing Interests
The authors declare no competing interests
REFERENCES:
- 1.Agou F, Veron M. In vivo protein cross-linking. Methods Mol Biol. 2015;1278:391–405 [DOI] [PubMed] [Google Scholar]
- 2.Christopher JA, Stadler C, Martin CE, Morgenstern M, Pan Y, Betsinger CN, Rattray DG, Mahdessian D, Gingras AC, Warscheid B, et al. Subcellular proteomics. Nat Rev Methods Primers. 2021;1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, et al. The crapome: A contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi H, Liu G, Mellacheruvu D, Tyers M, Gingras AC, Nesvizhskii AI. Analyzing protein-protein interactions from affinity purification-mass spectrometry data with saint. Curr Protoc Bioinformatics. 2012;Chapter 8:Unit8 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol. 2012;30:1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hung V, Zou P, Rhee HW, Udeshi ND, Cracan V, Svinkina T, Carr SA, Mootha VK, Ting AY. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric apex tagging. Mol Cell. 2014;55:332–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. Directed evolution of apex2 for electron microscopy and proximity labeling. Nat Methods. 2015;12:51–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY. Efficient proximity labeling in living cells and organisms with turboid. Nat Biotechnol. 2018;36:880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho KF, Branon TC, Udeshi ND, Myers SA, Carr SA, Ting AY. Proximity labeling in mammalian cells with turboid and split-turboid. Nat Protoc. 2020;15:3971–3999 [DOI] [PubMed] [Google Scholar]
- 11.Samavarchi-Tehrani P, Samson R, Gingras AC. Proximity dependent biotinylation: Key enzymes and adaptation to proteomics approaches. Mol Cell Proteomics. 2020;19:757–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiong Z, Lo HP, McMahon KA, Martel N, Jones A, Hill MM, Parton RG, Hall TE. In vivo proteomic mapping through gfp-directed proximity-dependent biotin labelling in zebrafish. Elife. 2021;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar DZ, Atkatsh K, Tavarez U, Erdos MR, Gruenbaum Y, Collins FS. Biotinylation by antibody recognition-a method for proximity labeling. Nat Methods. 2018;15:127–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu G, Papa A, Katchman AN, Zakharov SI, Roybal D, Hennessey JA, Kushner J, Yang L, Chen BX, Kushnir A, et al. Mechanism of adrenergic cav1.2 stimulation revealed by proximity proteomics. Nature. 2020;577:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin W, Cho KF, Cavanagh PE, Ting AY. Deciphering molecular interactions by proximity labeling. Nat Methods. 2021;18:133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalocsay M Apex peroxidase-catalyzed proximity labeling and multiplexed quantitative proteomics. Methods Mol Biol. 2019;2008:41–55 [DOI] [PubMed] [Google Scholar]
- 17.Paek J, Kalocsay M, Staus DP, Wingler L, Pascolutti R, Paulo JA, Gygi SP, Kruse AC. Multidimensional tracking of gpcr signaling via peroxidase-catalyzed proximity labeling. Cell. 2017;169:338–349 e311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chapman-Smith A, Cronan JE Jr. The enzymatic biotinylation of proteins: A post-translational modification of exceptional specificity. Trends Biochem Sci. 1999;24:359–363 [DOI] [PubMed] [Google Scholar]
- 19.Tong L Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci. 2013;70:863–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by escherichia coli biotin protein ligase. Protein Sci. 2004;13:3043–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci U S A. 2014;111:E2453–2461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ. An improved smaller biotin ligase for bioid proximity labeling. Mol Biol Cell. 2016;27:1188–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martell JD, Yamagata M, Deerinck TJ, Phan S, Kwa CG, Ellisman MH, Sanes JR, Ting AY. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat Biotechnol. 2016;34:774–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bosch JA, Chen CL, Perrimon N. Proximity-dependent labeling methods for proteomic profiling in living cells: An update. Wiley Interdiscip Rev Dev Biol. 2021;10:e392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.May DG, Scott KL, Campos AR, Roux KJ. Comparative application of bioid and turboid for protein-proximity biotinylation. Cells. 2020;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobingier BT, Huttenhain R, Eichel K, Miller KB, Ting AY, von Zastrow M, Krogan NJ. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell. 2017;169:350–360 e312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han Y, Branon TC, Martell JD, Boassa D, Shechner D, Ellisman MH, Ting A. Directed evolution of split apex2 peroxidase. ACS Chem Biol. 2019;14:619–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho KF, Branon TC, Rajeev S, Svinkina T, Udeshi ND, Thoudam T, Kwak C, Rhee HW, Lee IK, Carr SA, et al. Split-turboid enables contact-dependent proximity labeling in cells. Proc Natl Acad Sci U S A. 2020;117:12143–12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schopp IM, Amaya Ramirez CC, Debeljak J, Kreibich E, Skribbe M, Wild K, Bethune J. Split-bioid a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat Commun. 2017;8:15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by ms/ms. Anal Chem. 2003;75:1895–1904 [DOI] [PubMed] [Google Scholar]
- 32.Pappireddi N, Martin L, Wuhr M. A review on quantitative multiplexed proteomics. Chembiochem. 2019;20:1210–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J, Cai Z, Bomgarden RD, Pike I, Kuhn K, Rogers JC, Roberts TM, Gygi SP, Paulo JA. Tmtpro-18plex: The expanded and complete set of tmtpro reagents for sample multiplexing. J Proteome Res. 2021;20:2964–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotani N, Gu J, Isaji T, Udaka K, Taniguchi N, Honke K. Biochemical visualization of cell surface molecular clustering in living cells. Proc Natl Acad Sci U S A. 2008;105:7405–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hashimoto N, Hamamura K, Kotani N, Furukawa K, Kaneko K, Honke K, Furukawa K. Proteomic analysis of ganglioside-associated membrane molecules: Substantial basis for molecular clustering. Proteomics. 2012;12:3154–3163 [DOI] [PubMed] [Google Scholar]
- 36.Li XW, Rees JS, Xue P, Zhang H, Hamaia SW, Sanderson B, Funk PE, Farndale RW, Lilley KS, Perrett S, et al. New insights into the dt40 b cell receptor cluster using a proteomic proximity labeling assay. J Biol Chem. 2014;289:14434–14447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rees JS, Li XW, Perrett S, Lilley KS, Jackson AP. Selective proteomic proximity labeling assay using tyramide (spplat): A quantitative method for the proteomic analysis of localized membrane-bound protein clusters. Curr Protoc Protein Sci. 2015;80:19 27, 11–18 [DOI] [PubMed] [Google Scholar]
- 38.Santos-Barriopedro I, van Mierlo G, Vermeulen M. Off-the-shelf proximity biotinylation for interaction proteomics. Nat Commun. 2021;12:5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ariotti N, Hall TE, Rae J, Ferguson C, McMahon KA, Martel N, Webb RE, Webb RI, Teasdale RD, Parton RG. Modular detection of gfp-labeled proteins for rapid screening by electron microscopy in cells and organisms. Dev Cell. 2015;35:513–525 [DOI] [PubMed] [Google Scholar]
- 40.Pronobis MI, Zheng S, Singh SP, Goldman JA, Poss KD. In vivo proximity labeling identifies cardiomyocyte protein networks during zebrafish heart regeneration. Elife. 2021;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudolph F, Fink C, Huttemeister J, Kirchner M, Radke MH, Lopez Carballo J, Wagner E, Kohl T, Lehnart SE, Mertins P, et al. Deconstructing sarcomeric structure-function relations in titin-bioid knock-in mice. Nat Commun. 2020;11:3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng W, Liu C, Spinozzi S, Wang L, Evans SM, Chen J. Identifying the cardiac dyad proteome in vivo by a bioid2 knock-in strategy. Circulation. 2020;141:940–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Priori SG, Pandit SV, Rivolta I, Berenfeld O, Ronchetti E, Dhamoon A, Napolitano C, Anumonwo J, di Barletta MR, Gudapakkam S, et al. A novel form of short qt syndrome (sqt3) is caused by a mutation in the kcnj2 gene. Circ Res. 2005;96:800–807 [DOI] [PubMed] [Google Scholar]
- 44.Park SS, Ponce-Balbuena D, Kuick R, Guerrero-Serna G, Yoon J, Mellacheruvu D, Conlon KP, Basrur V, Nesvizhskii AI, Jalife J, et al. Kir2.1 interactome mapping uncovers pkp4 as a modulator of the kir2.1-regulated inward rectifier potassium currents. Mol Cell Proteomics. 2020;19:1436–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vermij SH, Abriel H, van Veen TA. Refining the molecular organization of the cardiac intercalated disc. Cardiovasc Res. 2017;113:259–275 [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Merkel CD, Zeng X, Heier JA, Cantrell PS, Sun M, Stolz DB, Watkins SC, Yates NA, Kwiatkowski AV. The n-cadherin interactome in primary cardiomyocytes as defined using quantitative proximity proteomics. J Cell Sci. 2019;132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bastiani M, Parton RG. Caveolae at a glance. J Cell Sci. 2010;123:3831–3836 [DOI] [PubMed] [Google Scholar]
- 48.Drab M, Verkade P, Elger M, Kasper M, Lohn M, Lauterbach B, Menne J, Lindschau C, Mende F, Luft FC, et al. Loss of caveolae, vascular dysfunction, and pulmonary defects in caveolin-1 gene-disrupted mice. Science. 2001;293:2449–2452 [DOI] [PubMed] [Google Scholar]
- 49.Peper J, Kownatzki-Danger D, Weninger G, Seibertz F, Pronto JRD, Sutanto H, Pacheu-Grau D, Hindmarsh R, Brandenburg S, Kohl T, et al. Caveolin3 stabilizes mct1-mediated lactate/proton transport in cardiomyocytes. Circ Res. 2021;128:e102–e120 [DOI] [PubMed] [Google Scholar]
- 50.Garcia CK, Goldstein JL, Pathak RK, Anderson RG, Brown MS. Molecular characterization of a membrane transporter for lactate, pyruvate, and other monocarboxylates: Implications for the cori cycle. Cell. 1994;76:865–873 [DOI] [PubMed] [Google Scholar]
- 51.Van Eyk JE. The world of protein interactions: Defining the caveolin 3 cardiac interactome. Circ Res. 2021;128:720–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu G, Lester JW, Young KB, Luo W, Zhai J, Kranias EG. A single site (ser16) phosphorylation in phospholamban is sufficient in mediating its maximal cardiac responses to beta -agonists. J Biol Chem. 2000;275:38938–38943 [DOI] [PubMed] [Google Scholar]
- 53.Kranias EG, Solaro RJ. Phosphorylation of troponin i and phospholamban during catecholamine stimulation of rabbit heart. Nature. 1982;298:182–184 [DOI] [PubMed] [Google Scholar]
- 54.Fujii J, Maruyama K, Tada M, MacLennan DH. Expression and site-specific mutagenesis of phospholamban. Studies of residues involved in phosphorylation and pentamer formation. J Biol Chem. 1989;264:12950–12955 [PubMed] [Google Scholar]
- 55.Menzel J, Kownatzki-Danger D, Tokar S, Ballone A, Unthan-Fechner K, Kilisch M, Lenz C, Urlaub H, Mori M, Ottmann C, et al. 14–3-3 binding creates a memory of kinase action by stabilizing the modified state of phospholamban. Sci Signal. 2020;13 [DOI] [PubMed] [Google Scholar]
- 56.Ladha FA, Thakar K, Pettinato AM, Legere N, Ghahremani S, Cohn R, Romano R, Meredith E, Chen YS, Hinson JT. Actinin bioid reveals sarcomere crosstalk with oxidative metabolism through interactions with igf2bp2. Cell Rep. 2021;36:109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, Ting AY. Atlas of subcellular rna localization revealed by apex-seq. Cell. 2019;178:473–490 e426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaewsapsak P, Shechner DM, Mallard W, Rinn JL, Ting AY. Live-cell mapping of organelle-associated rnas via proximity biotinylation combined with protein-rna crosslinking. Elife. 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kamp TJ, Hell JW. Regulation of cardiac l-type calcium channels by protein kinase a and protein kinase c. Circ Res. 2000;87:1095–1102 [DOI] [PubMed] [Google Scholar]
- 60.Reuter H, Scholz H. The regulation of the calcium conductance of cardiac muscle by adrenaline. J Physiol. 1977;264:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Tsien RW. Suspect that modulates the heartbeat is ensnared. Nature. 2020;577:624–626 [DOI] [PubMed] [Google Scholar]
- 62.Yang L, Katchman A, Samad T, Morrow J, Weinberg R, Marx SO. Beta-adrenergic regulation of the l-type ca2+ channel does not require phosphorylation of alpha1c ser1700. Circ Res. 2013;113:871–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang L, Katchman A, Weinberg RL, Abrams J, Samad T, Wan E, Pitt GS, Marx SO. The pdz motif of the alpha1c subunit is not required for surface trafficking and adrenergic modulation of cav1.2 channel in the heart. J Biol Chem. 2015;290:2166–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katchman A, Yang L, Zakharov SI, Kushner J, Abrams J, Chen BX, Liu G, Pitt GS, Colecraft HM, Marx SO. Proteolytic cleavage and pka phosphorylation of alpha1c subunit are not required for adrenergic regulation of cav1.2 in the heart. Proc Natl Acad Sci U S A. 2017;114:9194–9199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang L, Katchman A, Kushner J, Kushnir A, Zakharov SI, Chen BX, Shuja Z, Subramanyam P, Liu G, Papa A, et al. Cardiac cav1.2 channels require beta subunits for beta-adrenergic-mediated modulation but not trafficking. J Clin Invest. 2019;129:647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papa A, Kushner J, Hennessey JA, Katchman AN, Zakharov SI, Chen BX, Yang L, Lu R, Leong S, Diaz J, et al. Adrenergic cav1.2 activation via rad phosphorylation converges at alpha1c i-ii loop. Circ Res. 2021;128:76–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsushita Y, Furukawa T, Kasanuki H, Nishibatake M, Kurihara Y, Ikeda A, Kamatani N, Takeshima H, Matsuoka R. Mutation of junctophilin type 2 associated with hypertrophic cardiomyopathy. J Hum Genet. 2007;52:543–548 [DOI] [PubMed] [Google Scholar]
- 68.Landstrom AP, Weisleder N, Batalden KB, Bos JM, Tester DJ, Ommen SR, Wehrens XH, Claycomb WC, Ko JK, Hwang M, et al. Mutations in jph2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J Mol Cell Cardiol. 2007;42:1026–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sabater-Molina M, Navarro M, Garcia-Molina Saez E, Garrido I, Pascual-Figal D, Gonzalez Carrillo J, Gimeno Blanes JR. Mutation in jph2 cause dilated cardiomyopathy. Clin Genet. 2016;90:468–469 [DOI] [PubMed] [Google Scholar]
- 70.Quick AP, Wang Q, Philippen LE, Barreto-Torres G, Chiang DY, Beavers D, Wang G, Khalid M, Reynolds JO, Campbell HM, et al. Speg (striated muscle preferentially expressed protein kinase) is essential for cardiac function by regulating junctional membrane complex activity. Circ Res. 2017;120:110–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J, Panakova D, Kikuchi K, Holdway JE, Gemberling M, Burris JS, Singh SP, Dickson AL, Lin YF, Sabeh MK, et al. The regenerative capacity of zebrafish reverses cardiac failure caused by genetic cardiomyocyte depletion. Development. 2011;138:3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gemberling M, Karra R, Dickson AL, Poss KD. Nrg1 is an injury-induced cardiomyocyte mitogen for the endogenous heart regeneration program in zebrafish. Elife. 2015;4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Jang JY, Pirooznia M, Liu S, Finkel T. The secretome mouse provides a genetic platform to delineate tissue-specific in vivo secretion. Proc Natl Acad Sci U S A. 2021;118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Valencik ML, McDonald JA. Codon optimization markedly improves doxycycline regulated gene expression in the mouse heart. Transgenic Res. 2001;10:269–275 [DOI] [PubMed] [Google Scholar]


