Abstract
The reported influence of donor Killer-cell Immunoglobulin-like Receptor (KIR) genes on the outcomes of haematopoietic cell transplantation (HCT) are contradictory, in part due to diversity of disease, donor sources, era and conditioning regimens within and between different studies. Here, we describe the results of a retrospective clinical analysis establishing the effect of donor KIR motifs on the outcomes of 119 HLA-matched, unrelated donor HCT for adult acute myeloid leukaemia (AML) using myeloablative conditioning (MAC) in a predominantly T cell deplete (TCD) cohort. We observed that HCT involving donors with at least one KIR B haplotype were more likely to result in non-relapse mortality (NRM) than HCT involving donors with two KIR A haplotypes (p=0.019). Upon separation of KIR haplotypes into their centromeric (Cen) and telomeric (Tel) motif structures, we demonstrated that the Cen-B motif was largely responsible for this effect (p=0.001). When the cause of NRM was investigated further, infection was the dominant cause of death (p=0.006). No evidence correlating donor KIR B haplotype with relapse risk was observed. The results from this analysis confirm previous findings in the unrelated, TCD, MAC transplant setting and imply a protective role for donor-encoded Cen-A motifs against infection in allogeneic HCT recipients.
Introduction
Despite developments in the treatment of patients with haematological malignancies to specifically target diseased cells, achieving long term remission in adult acute myeloid leukaemia (AML) remains challenging and haematopoietic cell transplantation (HCT) continues as the mainstay of treatment for high risk patients1. Selection of volunteer unrelated donors (VUD) for allogeneic HCT is primarily based on HLA allele matching at the HLA-A, -B, -C, -DRB1 and -DQB1 loci, although many centres have also recently adopted a permissible matching model including the HLA-DPB1 locus2–5. However, even in recipients of well-matched grafts, five year overall survival (OS) remains <50%, with both relapse and death from transplant-related complications remaining significant problems1, 6. As such, investigation into secondary donor characteristics have been performed and confirmed the importance of non-HLA factors, particularly donor age and CMV matching, in reducing non-relapse mortality (NRM)4, 7, 8.
In addition to these secondary donor characteristics, selection of donors for non-HLA genetic factors has also been explored as a method to improve HCT outcomes. The Killer-cell Immunoglobulin-like Receptors (KIR), predominantly expressed on the surface of natural killer (NK) cells, are amongst the most promising non-HLA candidate gene families. KIR form a family of activating and inhibitory receptors which, upon binding their cognate HLA ligand, may elicit, or inhibit, an immune response. The genes encoding these proteins can be grouped into two main haplotypes: KIR A haplotypes are conserved in gene content and encode only one activating KIR gene (KIR2DS4) in combination with multiple inhibitory genes (KIR2DL1, KIR2DL3, KIR2DL4, KIR3DL1, KIR3DL2 and KIR3DL3). By contrast, KIR B haplotypes have a more variable gene content and encode at least one of the alternative KIR genes9. In addition, KIR haplotypes may be further defined according to their centromeric (Cen) or telomeric (Tel) gene motifs10.
The relevance of KIR-mediated immunity in HCT to treat AML was first discovered by investigating disparity between donor and recipient inhibitory KIR ligands, subsets of HLA class I molecules encoding the HLA-C1, -C2 and -Bw4 motifs, in haploidentical T cell-depleted (TCD) transplantations11. Ruggeri et al. (2002)12, demonstrated protection from disease relapse without concurrent increase in frequency of graft versus host disease (GVHD) in AML recipients whose grafts were derived from donors possessing KIR ligands that were not present in the recipient, often referred to as “missing self’. As such, they proposed that graft versus leukaemia (GVL) alloreactivity could be mediated by donor NK cells when KIR ligand disparity was present. Importantly, this effect appeared to be limited to AML recipients as the same effect was not observed in acute lymphoblastic leukaemia (ALL) patients. Following this, several studies have confirmed this model in haploidentical and other HLA-mismatched allogeneic transplant settings13, 14.
In addition to relapse and GVHD, infection remains a major contributor to the high mortality rates associated with HCT. In addition to de novo infections acquired during the extended periods of immunosuppression, viral reactivation is also a common cause of morbidity and mortality. In the UK, frequent use of TCD as GVHD prophylaxis, often utilising alemtuzumab, may exacerbate this issue15. NK cells are the first lymphocyte subset to reconstitute following HCT and are known to target virally-infected cells. However, NK cell reactivity resulting from KIR-ligand mismatching has, in contrast to its findings in relapse, been proposed to increase patients’ susceptibility to infection-related mortality16, 17.
Although mismatches between donor and recipient KIR ligands are not possible in HLA-matched transplants, KIR-mediated alloreactivity may still exist, as donor NK cells may express inhibitory KIR specific for ligands that are not encoded by either the patient or donor. This represents a “missing ligand” condition that has been shown to increase the risk of acute GVHD (aGVHD) but decrease the risk of relapse, ultimately increasing OS and disease-free survival (DFS)18–23. In addition, there are KIR molecules whose ligands are yet to be defined which may also permit KIR-mediated alloreactivity.
The most recent KIR-mediated alloreactivity model has been proposed based on findings from a large cohort of T cell replete, myeloablative conditioning (MAC) transplants. Using this model, a scale of alloreactivity is established based on the activating KIR content of the graft, reflected by the donor’s KIR haplotypes. This has shown that OS can be increased by selecting donors who encode at least one copy of the KIR B haplotype (KIR Bx)24. Upon further investigation, it was discovered that Cen-B motifs were predominantly associated with this outcome, and their presence correlated with a significant reduction in relapse and improved DFS, particularly in HLA-C mismatched transplants where the recipient encodes the HLA-C1 ligand10, 25. However, when a similar comparison investigating Cen motifs was performed in a large cohort of transplants utilising reduced intensity conditioning (RIC) regimens, no significant difference was observed18, 20.
The effect of KIR genotype polymorphism on HCT outcomes is therefore controversial and appears highly dependent on a variety of transplant characteristics. To reduce heterogeneity within the cohort, this study focusses only on the outcomes of a specific group of HCT recipients: TCD, HLA-matched, adult, myeloablative transplants to treat AML. Thereafter, we have investigated the influence of donor KIR genotypes on the outcomes of HCT within this UK cohort.
Materials and Methods
Study cohort
One hundred and nineteen HCT recipients and their respective VUDs were included in this study. All transplants took place between December 1996 and June 2011. Transplant inclusion criteria were as follows: i) UK-based adult transplanted to treat AML, ii) MAC regimen, iii) stem cells provided from an Anthony Nolan VUD and iv) complete allele-level HLA matching for HLA-A, -B, -C, -DRB1 and –DQB1, as described previously26. Clinical outcomes data were obtained in collaboration with the British Society of Blood and Marrow Transplantation. Ethical approval was obtained from the National Research Ethics Service (www.nres.nhs.uk, application number: MREC 01/8/31). The project was approved by Anthony Nolan medical and scientific committees. Informed consent was obtained from all participants prior to donation of blood or buccal cell samples for genetic analysis.
DNA extraction
Genomic DNA was extracted from whole blood or buccal swab samples. When extracted from blood, DNA was obtained either from salting-out27 or paramagnetic bead-based DNA purification (Promega, Madison, WI, USA). When extracted from buccal swabs, DNA was obtained using Gentra Puregene Buccal Cell Kit (QIAGEN, Hilden, Germany).
KIR genotyping
Briefly, presence or absence of 16 individual KIR genes was analysed using a polymerase chain reaction sequence-specific priming (PCR-SSP) approach described previously28. No distinction was made between the presence of KIR2DL5A or KIR2DL5B. The presence of at least one KIR B haplotype-specific locus indicated that the genotype contained at least one B haplotype. Such samples were depicted as KIR Bx. All samples that lacked the presence of all KIR B loci were assigned the AA genotype designation (KIR AA). Centromeric (Cen) and telomeric (Tel) gene motifs were assigned as described previously10. HLA-C1, -C2 and -Bw4 epitope ligands for KIR molecules were inferred from previous HLA typing.
Statistical analysis
Survival and DFS probability curves were calculated by the method of Kaplan-Meier29. Groups were compared using the log-rank test, whilst multivariate analysis was performed by Cox regression30. Several analyses incurred competing risks. The competing risk in relapse analysis was non-relapse mortality (NRM), whilst relapse was the competing risk in NRM analysis. When comparing the risk of infectious mortality between different groups, relapse or death due to any other cause were the competing risks. For these competing risk analyses, univariate probabilities were calculated using the cumulative incidence function31. Multivariate competing risk analysis was performed using the method by Fine and Gray32. A forward stepwise selection of covariates for multivariate analysis was performed using p≤0.05 inclusion criteria. Statistical significance was denoted at p≤0.05, whilst statistical trend was signified by p≤0.1. All univariate and multivariate analyses were performed using ‘R’ software (version 3.4.2).
Results
Patient and donor characteristics
Donor and recipient demographics and HCT conditions are given in Table 1. Of the 84 donors encoding at least one KIR B haplotype, 65 encoded at least one Cen-B motif (Cen-Bx, Figure 1). The remaining 54 donors (45%) encoded only Cen-A haplotype motifs (Cen-AA). When comparing the Cen-AA and Cen-Bx donor groups, the only statistically significant difference was between donor-recipient gender matching, by which gender-matched transplants were more likely to utilise Cen-Bx donors. As donor KIR genotyping was not performed prior to donor selection, this criterion was not knowingly selected. No other significant differences in clinical or prognostic factors were observed between those transplants using donors encoding Cen-AA or Cen-Bx.
Table 1 –
Recipient and donor demographics
| Variable | Donor KIR Cen-AA | % | Donor KIR Cen-BX | % | P-value |
|---|---|---|---|---|---|
|
| |||||
| Donor age, years | |||||
|
| |||||
| Median (Range) | 34 (20-49) | 35 (19-60) | 0.88 | ||
|
| |||||
| ≤30 | 17 | 31.5 | 22 | 33.8 | 0.94 |
| >30 | 37 | 68.5 | 43 | 66.2 | |
|
| |||||
| Recipient age, years | |||||
|
| |||||
| Median (Range) | 34 (18-64) | 37 (18-67) | 0.17 | ||
|
| |||||
| ≤40 | 40 | 74.1 | 45 | 69.2 | 0.71 |
| >40 | 14 | 25.9 | 20 | 30.8 | |
|
| |||||
| Donor sex | |||||
|
| |||||
| Female | 10 | 18.5 | 7 | 10.8 | 0 35 |
| Male | 44 | 81.5 | 58 | 89.2 | |
|
| |||||
| Recipient sex | |||||
|
| |||||
| Female | 22 | 40.7 | 24 | 36.9 | 0.81 |
| Male | 32 | 59.3 | 41 | 63.1 | |
|
| |||||
| Recipient-donor sex matching | |||||
|
| |||||
| Matched | 26 | 48.1 | 44 | 67.7 | 0.049 |
| Mismatched | 28 | 51.9 | 21 | 32.3 | |
|
| |||||
| Recipient-donor CMV | |||||
|
| |||||
| Matched | 43 | 79.6 | 48 | 73.8 | 0.57 |
| Mismatched | 10 | 18.5 | 16 | 24.6 | |
| Missing | 1 | 1.9 | 1 | 1.5 | |
|
| |||||
| Donor positive, Recipient positive | 9 | 16.7 | 6 | 9.2 | 0.32 |
| Donor positive, Recipient negative | 0 | 0.0 | 4 | 6.2 | |
| Donor negative, Recipient positive | 10 | 18.5 | 12 | 18.5 | |
| Donor negative, Recipient negative | 34 | 63.0 | 42 | 64.6 | |
| Missing | 1 | 1.9 | 1 | 1.5 | |
|
| |||||
| Transplant era | |||||
|
| |||||
| 1996-1999 | 9 | 16.7 | 6 | 9.2 | 0.69 |
| 2000-2003 | 19 | 35.2 | 25 | 38.5 | |
| 2004-2007 | 17 | 31.5 | 22 | 33.8 | |
| 2008-2011 | 9 | 16.7 | 12 | 18.5 | |
|
| |||||
| T cell deplete | |||||
|
| |||||
| Yes | 43 | 79.6 | 54 | 83.1 | 0.41 |
| No | 4 | 7.4 | 2 | 3.1 | |
| Missing | 7 | 13.0 | 9 | 13.8 | |
|
| |||||
| Disease risk – EBMT score | |||||
|
| |||||
| Good | 19 | 35.2 | 32 | 49.2 | 0.20 |
| Intermediate/Poor | 34 | 63.0 | 33 | 50.8 | |
| Missing | 1 | 1.9 | 0 | 0.0 | |
|
| |||||
| Stem cell source | |||||
|
| |||||
| BM | 26 | 48.1 | 28 | 43.1 | 0.71 |
| PBSC | 28 | 51.9 | 37 | 56.9 | |
|
| |||||
| Previous autografts | |||||
|
| |||||
| 0 | 50 | 92.6 | 62 | 95.4 | 0.70 |
| ≥1 | 4 | 7.4 | 3 | 4.6 | |
CMV = Cytomegalovirus, BM = bone marrow, PBSC = peripheral blood stem cells.
Categorical variables were compared by Chi-squared test (or Fisher’s Exact test when n≤5 for any subgroup). Continuous variables were compared by Mann-Whitney test. Statistically significant p-values are denoted in italics.
Figure 1:
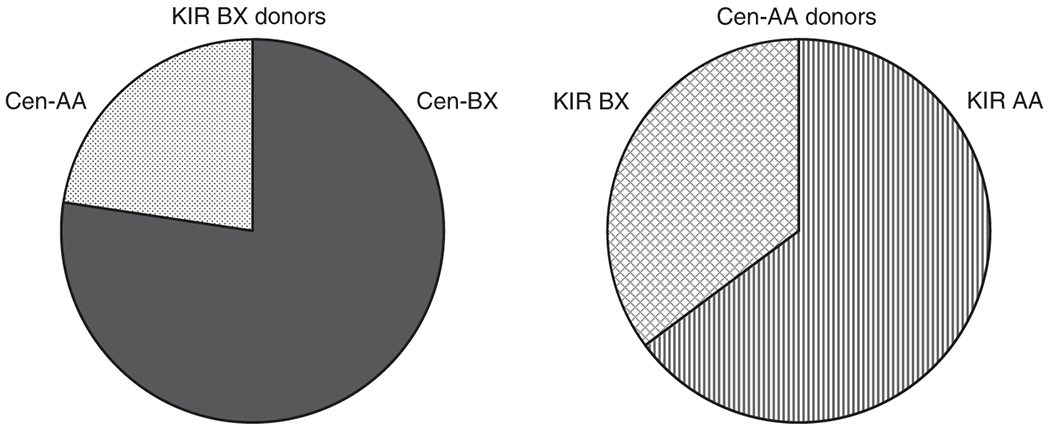
Charts demonstrating the proportions of different centromeric motif structures within donor subgroups. Over 75% of KIR BX donors encode at least one Cen-B motif (Cen-BX, solid grey). The remainder all encode two copies of the Cen-A motif (Cen-AA, dotted). Of the Cen-AA donors, approximately one third encode the KIR BX genotype (hashed), whilst the remainder encode KIR AA (striped).
For the whole cohort, the probabilities of survival and relapse at five years post-transplant were 38.6% and 34.5% respectively, whilst the probability of NRM at one year post-transplant was 23.0%. All such univariate analyses were performed using methods of Kaplan-Meier and cumulative incidence as described in the Materials and Methods. When assessing the impact of the clinical variables on these outcomes of HCT, several factors demonstrated trends and borderline significance with detrimental outcomes. Older recipients (>40 years) had decreased OS at five years post-transplant (p=0.049), as did recipients with a history of previous autografts (p=0.028).
Presence of donor KIR B haplotypes increase incidence of non-relapse mortality
Univariate analysis of the effect of donor KIR haplotypes on the outcomes of HCT associated the presence of donor-encoded KIR B haplotype with an increase in the incidence of NRM after one year post-transplant (KIR AA: 9%, 95% confidence interval [CI]=2.9-26.1 vs KIR Bx: 29%, 0=20.6-40.6; p=0.019; Figure 2A, Table 2). This increase in NRM was associated with statistical trends towards decreased OS (KIR AA: 49%, CI=34.5-69.4 vs KIR Bx: 34%, CI=25.4-46.6; p=0.06) and DFS (KIR AA: 46%, CI=32.2-66.9 vs KIR Bx: 31%, CI=22.5-43.4; p=0.087) at five years post-transplant. Interestingly, despite most previous analyses implicating KIR-mediated differences in relapse risk, no statistically significant differences were observed in this dataset (Table 2).
Figure 2:
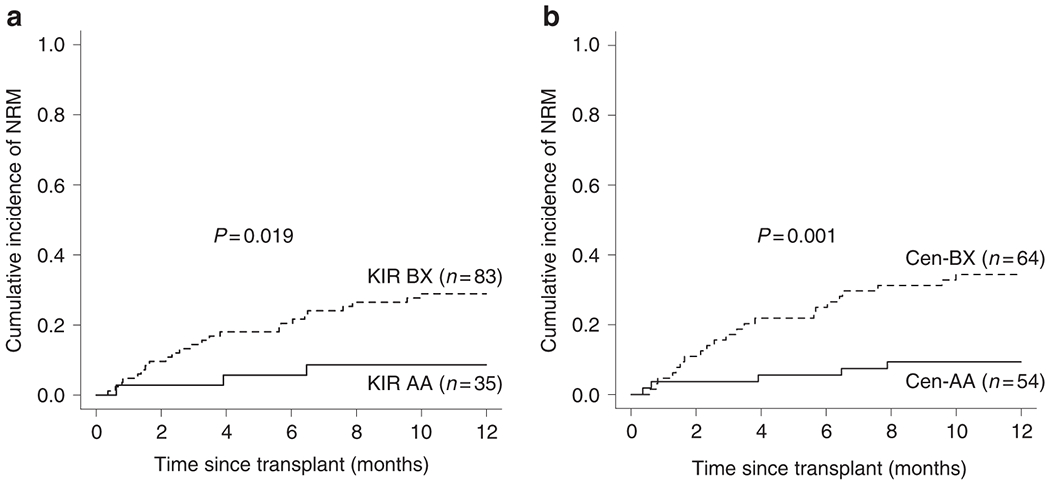
Donor KIR B genotype increases NRM. A) Univariate probability of NRM at one year post-transplant for groups based on the presence of at least one donor-encoded KIR B haplotype. This demonstrates that a significant increase in NRM is associated with donors encoding the KIR BX haplotype structure. B) When the haplotype structure is refined according to centromeric motif structure, donor-encoded Cen-B appears culpable for the increase in NRM. As described in the footer of Table 2, the total number of transplants included in this NRM analysis is one less than listed in Table 2 as a result of one transplant missing relapse data.
Table 2 –
Univariate analyses of recipient and donor factors on OS, relapse, DFS and NRM
| Variable | Valid cases (n) | 5 year OS | 5 year relapse§ | 5 year DFS§ | 1 year NRM§ | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| % | P-value | % | P-value | % | P-value | % | P-value | ||
| Donor age, years | |||||||||
|
| |||||||||
| <30 | 39 | 42.2 | 0.67 | 24.2 | 0.12 | 42.9 | 0.37 | 28.6 | 0.36 |
| >30 | 80 | 37.2 | 39.2 | 32.6 | 20.2 | ||||
|
| |||||||||
| Recipient age, years | |||||||||
|
| |||||||||
| <40 | 85 | 42.6 | 0.049 | 34.3 | 0.79 | 38.4 | 0.083 | 19.2 | 0.097 |
| >40 | 34 | 28.5 | 35.3 | 29.1 | 32.4 | ||||
|
| |||||||||
| Donor sex | |||||||||
|
| |||||||||
| Female | 17 | 35.9 | 0.99 | 43.7 | 0.66 | 26.9 | 0.53 | 29.4 | 0.49 |
| Male | 102 | 38.8 | 33.1 | 37.3 | 21.9 | ||||
|
| |||||||||
| Recipient sex | |||||||||
|
| |||||||||
| Female | 46 | 39.0 | 0.97 | 37.9 | 0.47 | 32.5 | 0.59 | 19.8 | 0.51 |
| Male | 73 | 38.3 | 32.3 | 37.9 | 25.0 | ||||
|
| |||||||||
| Recipient-donor sex matching | |||||||||
|
| |||||||||
| Matched | 70 | 41.4 | 0.41 | 35.4 | 0.86 | 38.0 | 0.54 | 21.7 | 0.69 |
| Mismatched | 49 | 34.6 | 33.3 | 32.6 | 24.7 | ||||
|
| |||||||||
| Recipient-donor CMV matching | |||||||||
|
| |||||||||
| Matched | 91 | 40.8 | 0.17 | 32.8 | 0.33 | 38.2 | 0.14 | 21.1 | 0.52 |
| Mismatched | 26 | 29.4 | 43.5 | 25.4 | 26.9 | ||||
|
| |||||||||
| Transplant era | |||||||||
|
| |||||||||
| 1996-1999 | 15 | 60.0 | 0.45 | 28.6 | 0.049 | 50.0 | 0.60 | 21.4 | 0.11 |
| 2000-2003 | 44 | 34.1 | 50.0 | 31.8 | 13.6 | ||||
| 2004-2007 | 39 | 35.6 | 20.5 | 33.1 | 35.9 | ||||
| 2008-2011† | 21 | 38.6 | 31.2 | 40.7 | 19.9 | ||||
|
| |||||||||
| T cell deplete | |||||||||
|
| |||||||||
| Yes | 97 | 37.5 | 0.28 | 34.0 | 0.46 | 34.9 | 0.22 | 24.1 | 0.63 |
| No | 6 | 66.7 | 16.7 | 66.7 | 16.7 | ||||
|
| |||||||||
| Disease risk – EBMT score | |||||||||
|
| |||||||||
| Good | 51 | 36.7 | 0.89 | 26.7 | 0.12 | 31.2 | 0.72 | 28.0 | 0.30 |
| Intermediate/Poor | 67 | 39.3 | 40.8 | 38.1 | 19.6 | ||||
|
| |||||||||
| Stem cell source | |||||||||
|
| |||||||||
| BM | 54 | 46.0 | 0.13 | 37.7 | 0.59 | 39.5 | 0.49 | 18.9 | 0.41 |
| PBSC | 65 | 31.88 | 31.6 | 32.1 | 26.4 | ||||
|
| |||||||||
| Previous autografts | |||||||||
|
| |||||||||
| 0 | 112 | 40.1 | 0.028 | 34.0 | 0.62 | 37.2 | 0.063 | 21.7 | 0.18 |
| ≥1 | 7 | 14.3 | 42.9 | 14.3 | 42.9 | ||||
|
| |||||||||
| Donor KIR genotype | |||||||||
|
| |||||||||
| KIR AA | 35 | 48.9 | 0.060 | 38.7 | 0.60 | 46.5 | 0.087 | 8.7 | 0.019 |
| KIR BX | 84 | 34.4 | 32.8 | 31.3 | 28.9 | ||||
|
| |||||||||
| Donor Tel motif pattern | |||||||||
|
| |||||||||
| Tel-AA | 74 | 36.2 | 0.42 | 33.6 | 0.77 | 34.2 | 0.47 | 27.6 | 0.13 |
| Tel-BX | 45 | 42.3 | 36.1 | 38.2 | 15.6 | ||||
|
| |||||||||
| Donor Cen motif pattern | |||||||||
|
| |||||||||
| Cen-AA | 54 | 47.7 | 0.024 | 38.0 | 0.45 | 44.6 | 0.045 | 9.3 | 0.001 |
| Cen-BX | 65 | 31.2 | 31.5 | 28.6 | 34.4 | ||||
| Cen-AA | 54 | 47.7 | 0.010 | 38.0 | 0.75 | 44.6 | 0.031 | 9.3 | 0.005 |
| Cen-AB | 53 | 36.8 | 31.2 | 33.7 | 32.7 | ||||
| Cen-BB | 12 | 8.3 | 33.3 | 8.3 | 41.7 | ||||
NRM/DFS/Relapse data missing for one transplant.
Estimated incidence of OS, relapse and DFS at latest clinical follow-up (4 years) reported.
Statistically significant results (≤0.05) are italicized. OS = Overall survival, NRM = Non-relapse mortality, CMV = Cytomegalovirus, BM = bone marrow, PBSC = peripheral blood stem cells
Following the observation that the presence of donor KIR B haplotypes was associated with increased NRM probability, donor genotypes were stratified by their Cen and Tel motif patterns. Outcomes in patients receiving HCT from donors encoding the Tel-Bx motif were not associated with any difference when compared to Tel-AA donor transplants (Table 2). Presence of the Cen-B motif within donors, however, was associated with a significant increase in the probability of NRM at one year post-transplant (Cen-AA: 9%, CI=4.0-21.7 vs Cen-Bx: 34%, CI=24.4-48.4; p=0.001, Figure 2B). This observation correlated with significantly improved five year OS (Cen-AA: 48%, CI=35.7-63.7 vs Cen-Bx: 31%, CI=21.6-45.1; p=0.024) and DFS (Cen-AA: 45%, CI=32.9-60.5 vs Cen-Bx: 29%, CI=19.3-42.6; p=0.045, Table 2). In a multivariate regression analysis, the significant difference between outcomes of Cen-AA and Cen-Bx donor transplants was preserved (OS: Cen-Bx hazard ratio [HR]=1.9, CI=1.2-3.1, p=0.01; NRM: Cen-Bx HR=4.2, CI=1.6-11.0, p=0.004, Table 3).
Table 3 –
Multivariate analysis of OS, NRM and death by infection
| Variable | 5 year OS | 1 year NRM† | 1 year death by infection†‡ | |||
|---|---|---|---|---|---|---|
|
| ||||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Recipient age, years | ||||||
|
| ||||||
| <40 | 1.00 | - | 1.00 | - | 1.00 | - |
| >40 | 1.91 (1.15-3.16) | 0.012 | 1.81 (0.82-4.01) | 0.15 | 2.28 (0.91-5.69) | 0.078 |
|
| ||||||
| Transplant era | ||||||
|
| ||||||
| 1996-1999 | 1.00 | - | ||||
| 2000-2003 | 1.15 (0.15-8.99) | 0.89 | ||||
| 2004-2007 | 5.27 (0.84-32.9) | 0.075 | ||||
| 2008-2011 | 0.74 (0.05-9.93) | 0.82 | ||||
|
| ||||||
| Previous autografts | ||||||
|
| ||||||
| 0 | 1.00 | - | 1.00 | - | ||
| ≥1 | 3.05 (1.30-7.15) | 0.010 | 2.45 (0.55-10.92) | 0.24 | ||
|
| ||||||
| Donor Cen motif pattern | ||||||
|
| ||||||
| Cen-AA | 1.00 | - | 1.00 | - | 1.00 | - |
| Cen-BX | 1.90 (1.17-3.10) | 0.010 | 4.16 (1.58-11.00) | 0.004 | 5.50 (1.49-20.32) | 0.011 |
Statistically significant results (≤0.05) are italicized. OS = Overall survival, NRM = Non-relapse mortality
NRM data missing for one transplant.
Cause-of-death data missing for three transplants.
When compared to the Cen-AA motif structure, the impact of each additional Cen-B motif was also assessed. This revealed a dose effect, whereby the more copies of donor-encoded Cen-B motif, the higher the risk of NRM at one year post-transplant (Cen-AA: 9%, CI=4.0-21.7 vs Cen-AB: 33%, CI=22.0-48.5 vs Cen-BB: 42%, CI=20.5-84.8; p=0.005, Figure 3A). This corresponded with significant differences in OS (Cen-AA: 48%, CI=35.7-63.7 vs Cen-AB: 37%, CI=25.7-52.7 vs Cen-BB: 8%, CI=1.3-54.4; p=0.01, Figure 3B) and DFS (Cen-AA: 45%, CI=32.9-60.5 vs Cen-AB: 34%, CI=22.9-49.8 vs Cen-BB: 8%, CI=1.3-54.4; p=0.031, Table 2) at five years post-transplant.
Figure 3:
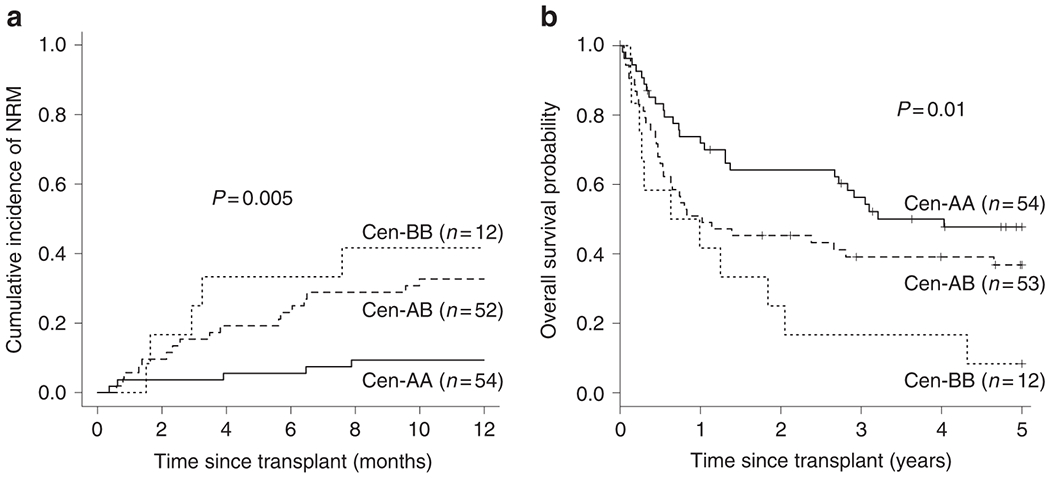
Effect of donor Cen-B is dose-dependent. A) Univariate probability of NRM at one year post-transplant for groups based on donor-encoded Cen-B motif copy number. With each additional Cen-B motif, risk of NRM increases. B) When OS is assessed with the same grouping strategy, the detrimental effect of donor Cen-B is also evident. As described in the footer of Table 2, the total number of transplants included in this NRM analysis is one less than listed in Table 2 as a result of one transplant missing relapse data.
Cause-of-death analysis implicates donor Cen-B with impaired viral protection
To further investigate how donor-encoded centromeric motif structure affects NRM risk, the 27 transplants resulting in NRM were stratified by cause-of-death. Infection was recorded as a cause-of-death in 19 recipients, whilst GVHD was implicated in only five (cause-of-death in one recipient included both GVHD and infection). One transplant resulted in NRM without infection or GVHD, and data was missing for three further transplants. Accordingly, a competing risk analysis assessing the risk of death by infection at one year between transplants utilising Cen-AA and Cen-Bx donors was performed and revealed a strong protective effect of donor-encoded Cen-AA (Cen-AA: 6%, CI=1.8-17.0 vs Cen-Bx: 25%, CI=15.8-38.4; p=0.006). This withstood multivariate analysis as the only remaining statistically significant factor (Cen-Bx: HR=5.5, CI=1.5-20.3, p=0.011, Table 3). Of the 15 instances where data on the type of infection was available, 13 cases (87%) involved viral infection.
Discussion
The relevance of matching between donor and recipient HLA types has been well-documented and is a key determinant of HCT success3, 4. However, the KIR genotype of the donor, encoding receptors for these hyperpolymorphic HLA, is not routinely considered in VUD selection. Previous studies in T cell replete MAC cohorts have implicated donor-encoded Cen-B haplotype motif presence with a beneficial reduction in relapse risk, leading to improved OS and DFS10, 25. By contrast, the results obtained in this predominantly TCD cohort fail to indicate any beneficial reduction in AML relapse associated with donor-encoded Cen-B motifs, and instead implicate these motifs with increased NRM risk, leading to decreased OS and DFS.
Although our findings contradict these apparently similar studies, the different T cell content between the grafts may be responsible for the conflicting outcomes. These data may support an orchestrated role for NK cell interaction with T cells33, interpreted as innate NK cells playing a coordinating role for early T cell reconstitution after transplant. This NK cell-T cell interaction is likely to be common to all HCT, but the effects may be more apparent after TCD where T cell function is impaired or delayed. In addition, our findings concur with the study by Kröger et al. (2006)17, whereby a higher number of different activating KIRs encoded by the donor corresponded with increased NRM in a MAC, TCD cohort. Furthermore, another study investigating the effect of TCD on KIR-mediated immunity following HCT also observed elevated NRM as a result of increased infection-related mortality, theorising the observation as a result of increased targeting of antigen-presenting dendritic cells by activated NK cells16, 34.
When the cause of death was investigated in the study presented here, infection, particularly viral infection, was strongly associated with increased mortality in Cen-Bx donor transplants, whereas a greater level of protection against infection-related mortality was offered by Cen-AA donors. This, again, contrasts with studies in T cell replete transplants where increasing numbers of activating KIR, and particularly KIR2DS2 (restricted to the Cen-B motif), were demonstrated to aid control of human cytomegalovirus (CMV) reactivation35. Viruses, such as CMV, display a range of functions aimed to modulate NK cell reactivity, including the upregulation of expression of the inhibitory ligand, HLA-E36, as well as sequestration of activating ligands such as major histocompatibility complex class I polypeptide-related sequence B (MICB)37. However, viral downregulation of HLA class I antigen expression, as a means of evading T cell-mediated immunity, can also stimulate NK cell activation via the recognition of “missing-self”38, 39. Licensed NK cells, which are more functional owing to expression of at least one inhibitory receptor for a host-encoded HLA class I molecule, recognize the lack of inhibition and mount an immune response.
The strong avidity offered by alleles of KIR2DL2/3 commonly located on the Cen-B haplotype motif has been shown to correspond with functionally stronger licensing than KIR2DL2/3 alleles which tend to reside on the Cen-A motif40, 41. This increased level of licensing, when tested in cells lines that fail to express any HLA class I on the cell surface, is capable of stimulating an increased response. However, complete absence of HLA class I expression is unlikely to be environmentally plausible during viral infection. As such, presence of high avidity Cen-B KIR2DL2/3 alleles in combination with downregulated HLA-C may actually offer a greater level of inhibition than the equivalent interaction between Cen-A KIR2DL2/3 alleles and downregulated HLA-C. The increased inhibition would require a greater activating signal to supersede it, resulting in decreased NK cell reactivity. In addition, the delayed reconstitution of KIR2DL1 following HCT may place additional burden on KIR2DL2/3 licensed NK cell immunity42. Differential NK cell inhibition via KIR2DL2/3 has also been proposed as a theory to explain the observation that increasing copies of KIR2DL3-HLA-C1 (typically weak avidity interactions) results in improved resolution of hepatitis C virus infection43, 44. Additionally, evidence that NK cell education via activating KIRs (such as those which define the Cen-B motif) renders NK cells hyporesponsive may also indicate improved NK cell reactivity associated with the Cen-A haplotype motif45.
Several limitations to the study mean that the results must be approached with some caution. Although care was taken to maximise cohort homogeneity, the retrospective, multicentre aspect of this study introduces the caveat of variable transplant protocols and presented difficulties in collecting complete clinical follow-up data, including those relating to co-morbidities and the types of viral infections that occurred post-transplant. In addition, the era of transplants ranged considerably, from 1996 to 2011. Amongst other factors, significant evolution of antiviral and antifungal agents has occurred over this time period. Furthermore, the relatively small sample size and event incidence may be underpowered to resolve some compound variables. The KIR locus itself introduces a range of complexities not accounted for in this study. For example, the highly polymorphic nature of each KIR gene introduces variety in the expression and functionality of each locus. The implementation of high resolution, allelic-level KIR typing is warranted to resolve these issues in the future46. Finally, the scope of this analysis has been limited to only investigate the KIR-mediated aspect of immunity, ignoring other NK cell receptor-ligand signalling pathways and alloreactivity mediated by T and B cells. Future, well-defined prospective studies using uniform transplant conditions may help to clarify the effects of the combinations of donor KIR and recipient ligands on HCT outcomes.
In summary, we have demonstrated that donor-encoded KIR genes can affect the NRM risk following VUD HCT. Specifically, the presence of donor-encoded Cen-B haplotype motifs conveys a significant risk of infectious mortality, which in turn equates to a significant reduction in OS. Multivariate analysis adjusting for other transplant characteristics suggested that donor KIR centromeric genotype was the only significant determinant for NRM risk. However, these findings may only be applicable to cases of HLA-matched, unrelated donor, MAC, TCD transplants to treat adult AML, as differing HCT scenarios have repeatedly generated contradictory findings, including observations in our own TCD, RIC cohort (unpublished data). This highlights the important differences between transplant scenarios and suggests that, when selecting donors based on KIR genotype information, it is unlikely that a ‘one-size-fits-all’ donor KIR genotype exists. Instead, these findings support the selection of VUDs based on KIR genotype, but only when considered in parallel with other transplant factors.
Acknowledgements
This project was supported in part by funding provided by the National Institutes of Health (NIH, P01-CA-111412) whilst further funding and support was provided by Anthony Nolan.
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
None of the authors declare any conflicts of interest.
References
- 1.Mohty M Acute Myeloid Leukaemia. In: Apperley JF, Carreras E, Gluckman E, Masszi T (eds). The EBMT Handbook on Haematopoietic Stem Cell Transplantation, 6th edn. Forum Service Editore: Genoa, 2012, pp 316–329. [Google Scholar]
- 2.Crivello P, Zito L, Sizzano F, Zino E, Maiers M, Mulder A et al. The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 2015; 21(2): 233–241. doi: 10.1016/j.bbmt.2014.10.017 [DOI] [PubMed] [Google Scholar]
- 3.Lee SJ, Klein J, Haagenson M, Baxter-Lowe LA, Confer DL, Eapen M et al. High-resolution donor-recipient HLA matching contributes to the success of unrelated donor marrow transplantation. Blood 2007; 110(13): 4576–4583. doi: 10.1182/blood-2007-06-097386 [DOI] [PubMed] [Google Scholar]
- 4.Shaw BE, Mayor NP, Szydlo RM, Bultitude WP, Anthias C, Kirkland K et al. Recipient/donor HLA and CMV matching in recipients of T-cell-depleted unrelated donor haematopoietic cell transplants. Bone Marrow Transplant 2017; 52(5): 717–725. doi: 10.1038/bmt.2016.352 [DOI] [PubMed] [Google Scholar]
- 5.Fleischhauer K, Shaw BE, Gooley T, Malkki M, Bardy P, Bignon JD et al. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol 2012; 13(4): 366–374. doi: 10.1016/S1470-2045(12)70004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Souza A, Fretham C. Current Uses and Outcomes of Hematopoietic Cell Transplantation (HCT): CIBMTR Summary Slides. In, 2017.
- 7.Shaw BE, Logan BR, Spellman SR, Marsh SGE, Robinson J, Pidala J et al. Development of an Unrelated Donor Selection Score Predictive of Survival after HCT: Donor Age Matters Most. Biol Blood Marrow Transplant 2018; 24(5): 1049–1056. doi: 10.1016/j.bbmt.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kollman C, Spellman SR, Zhang MJ, Hassebroek A, Anasetti C, Antin JH et al. The effect of donor characteristics on survival after unrelated donor transplantation for hematologic malignancy. Blood 2016; 127(2): 260–267. doi: 10.1182/blood-2015-08-663823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vierra-Green C, Roe D, Jayaraman J, Trowsdale J, Traherne J, Kuang R et al. Estimating KIR Haplotype Frequencies on a Cohort of 10,000 Individuals: A Comprehensive Study on Population Variations, Typing Resolutions, and Reference Haplotypes. PLoS One 2016; 11(10): e0163973. doi: 10.1371/journal.pone.0163973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Le CT et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010; 116(14): 2411–2419. doi: 10.1182/blood-2010-05-283051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K et al. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood 1999; 94(1): 333–339. [PubMed] [Google Scholar]
- 12.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 2002; 295(5562): 2097–2100. doi: 10.1126/science.1068440 [DOI] [PubMed] [Google Scholar]
- 13.Giebel S, Locatelli F, Lamparelli T, Velardi A, Davies S, Frumento G et al. Survival advantage with KIR ligand incompatibility in hematopoietic stem cell transplantation from unrelated donors. Blood 2003; 102(3): 814–819. doi: 10.1182/blood-2003-01-0091 [DOI] [PubMed] [Google Scholar]
- 14.Mancusi A, Ruggeri L, Urbani E, Pierini A, Massei MS, Carotti A et al. Haploidentical hematopoietic transplantation from KIR ligand-mismatched donors with activating KIRs reduces nonrelapse mortality. Blood 2015; 125(20): 3173–3182. doi: 10.1182/blood-2014-09-599993 [DOI] [PubMed] [Google Scholar]
- 15.Kroger N, Shaw B, Iacobelli S, Zabelina T, Peggs K, Shimoni A et al. Comparison between antithymocyte globulin and alemtuzumab and the possible impact of KIR-ligand mismatch after dose-reduced conditioning and unrelated stem cell transplantation in patients with multiple myeloma. Br J Haematol 2005; 129(5): 631–643. doi: 10.1111/j.1365-2141.2005.05513.x [DOI] [PubMed] [Google Scholar]
- 16.Schaffer M, Malmberg KJ, Ringden O, Ljunggren HG, Remberger M. Increased infection-related mortality in KIR-ligand-mismatched unrelated allogeneic hematopoietic stem-cell transplantation. Transplantation 2004; 78(7): 1081–1085. [DOI] [PubMed] [Google Scholar]
- 17.Kroger N, Binder T, Zabelina T, Wolschke C, Schieder H, Renges H et al. Low number of donor activating killer immunoglobulin-like receptors (KIR) genes but not KIR-ligand mismatch prevents relapse and improves disease-free survival in leukemia patients after in vivo T-cell depleted unrelated stem cell transplantation. Transplantation 2006; 82(8): 1024–1030. doi: 10.1097/01.tp.0000235859.24513.43 [DOI] [PubMed] [Google Scholar]
- 18.Sobecks RM, Wang T, Askar M, Gallagher MM, Haagenson M, Spellman S et al. Impact of KIR and HLA Genotypes on Outcomes after Reduced-Intensity Conditioning Hematopoietic Cell Transplantation. Biol Blood Marrow Transplant 2015; 21(9): 1589–1596. doi: 10.1016/j.bbmt.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanga U, Mourya M, Seth T, George J, Sood P, Sharma R et al. Role of killer immunoglobulin-like receptor-ligand interactions in human leukocyte antigen-matched sibling hematopoietic stem cell transplantation. Transplant Proc 2012; 44(4): 919–921. doi: 10.1016/j.transproceed.2012.03.036 [DOI] [PubMed] [Google Scholar]
- 20.Venstrom JM, Pittari G, Gooley TA, Chewning JH, Spellman S, Haagenson M et al. HLA-C-dependent prevention of leukemia relapse by donor activating KIR2DS1. N Engl J Med 2012; 367(9): 805–816. doi: 10.1056/NEJMoa1200503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S, Kim K, Jang JH, Kim SJ, Kim WS, Kang ES et al. KIR alloreactivity based on the receptor-ligand model is associated with improved clinical outcomes of allogeneic hematopoietic stem cell transplantation: Result of single center prospective study. Hum Immunol 2015; 76(9): 636–643. doi: 10.1016/j.humimm.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 22.Leung W, Iyengar R, Turner V, Lang P, Bader P, Conn P et al. Determinants of antileukemia effects of allogeneic NK cells. J Immunol 2004; 172(1): 644–650. [DOI] [PubMed] [Google Scholar]
- 23.Neuchel C, Furst D, Niederwieser D, Bunjes D, Tsamadou C, Wulf G et al. Impact of Donor Activating KIR Genes on HSCT Outcome in C1-Ligand Negative Myeloid Disease Patients Transplanted with Unrelated Donors-A Retrospective Study. PLoS One 2017; 12(1): e0169512. doi: 10.1371/journal.pone.0169512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J, Le CT et al. Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 2009; 113(3): 726–732. doi: 10.1182/blood-2008-07-171926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T, Marsh SGE et al. Donor killer cell Ig-like receptor B haplotypes, recipient HLA-C1, and HLA-C mismatch enhance the clinical benefit of unrelated transplantation for acute myelogenous leukemia. J Immunol 2014; 192(10): 4592–4600. doi: 10.4049/jimmunol.1302517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayor NP, Hayhurst JD, Turner TR, Szydlo RM, Shaw BE, Bultitude WP et al. Recipients Receiving Better HLA-Matched Hematopoietic Cell Transplantation Grafts, Uncovered by a Novel HLA Typing Method, Have Superior Survival: A Retrospective Study. Biol Blood Marrow Transplant 2019; 25(3): 443–450. doi: 10.1016/j.bbmt.2018.12.768 [DOI] [PubMed] [Google Scholar]
- 27.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16(3): 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens 2007; 70(5): 415–422. doi: 10.1111/j.1399-0039.2007.00923.x [DOI] [PubMed] [Google Scholar]
- 29.Kaplan EL, Meier P. Nonparametric Estimation from Incomplete Observations. Journal of the American Statistical Association 1958; 53(282): 457–481. doi: 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- 30.Cox DR. Regression Models and Life-Tables. Journal of the Royal Statistical Society. Series B (Methodological) 1972; 34(2): 187–220. [Google Scholar]
- 31.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann. Statist 1988; 16(3): 1141–1154. [Google Scholar]
- 32.Fine JP, Gray RJ. A Proportional Hazards Model for the Subdistribution of a Competing Risk. Journal of the American Statistical Association 1999; 94(446): 496–509. doi: 10.1080/01621459.1999.10474144 [DOI] [Google Scholar]
- 33.Cooley S, McCullar V, Wangen R, Bergemann TL, Spellman S, Weisdorf DJ et al. KIR reconstitution is altered by T cells in the graft and correlates with clinical outcomes after unrelated donor transplantation. Blood 2005; 106(13): 4370–4376. doi: 10.1182/blood-2005-04-1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith LE, Olszewski MA, Georgoudaki AM, Wagner AK, Hagglof T, Karlsson MC et al. Sensitivity of dendritic cells to NK-mediated lysis depends on the inflammatory environment and is modulated by CD54/CD226-driven interactions. J Leukoc Biol 2016; 100(4): 781–789. doi: 10.1189/jlb.3A0615-271RR [DOI] [PubMed] [Google Scholar]
- 35.Zaia JA, Sun JY, Gallez-Hawkins GM, Thao L, Oki A, Lacey SF et al. The effect of single and combined activating killer immunoglobulin-like receptor genotypes on cytomegalovirus infection and immunity after hematopoietic cell transplantation. Biol Blood Marrow Transplant 2009; 15(3): 315–325. doi: 10.1016/j.bbmt.2008.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomasec P, Braud VM, Rickards C, Powell MB, McSharry BP, Gadola S et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 2000; 287(5455): 1031. [DOI] [PubMed] [Google Scholar]
- 37.Welte SA, Sinzger C, Lutz SZ, Singh-Jasuja H, Sampaio KL, Eknigk U et al. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur J Immunol 2003; 33(1): 194–203. doi: 10.1002/immu.200390022 [DOI] [PubMed] [Google Scholar]
- 38.Halenius A, Hauka S, Dolken L, Stindt J, Reinhard H, Wiek C et al. Human cytomegalovirus disrupts the major histocompatibility complex class I peptide-loading complex and inhibits tapasin gene transcription. J Virol 2011; 85(7): 3473–3485. doi: 10.1128/JVI.01923-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ljunggren HG, Karre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today 1990; 11(7): 237–244. [DOI] [PubMed] [Google Scholar]
- 40.Frazier WR, Steiner N, Hou L, Dakshanamurthy S, Hurley CK. Allelic variation in KIR2DL3 generates a KIR2DL2-like receptor with increased binding to its HLA-C ligand. J Immunol 2013; 190(12): 6198–6208. doi: 10.4049/jimmunol.1300464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bari R, Thapa R, Bao J, Li Y, Zheng J, Leung W. KIR2DL2/2DL3-E(35) alleles are functionally stronger than -Q(35) alleles. Sci Rep 2016; 6: 23689. doi: 10.1038/srep23689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer JC, Ottinger H, Ferencik S, Sribar M, Punzel M, Beelen DW et al. Relevance of C1 and C2 epitopes for hemopoietic stem cell transplantation: role for sequential acquisition of HLA-C-specific inhibitory killer Ig-like receptor. J Immunol 2007; 178(6): 3918–3923. [DOI] [PubMed] [Google Scholar]
- 43.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 2004; 305(5685): 872–874. doi: 10.1126/science.1097670 [DOI] [PubMed] [Google Scholar]
- 44.Vidal-Castineira JR, Lopez-Vazquez A, Diaz-Pena R, Alonso-Arias R, Martinez-Borra J, Perez R et al. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol 2010; 84(1): 475–481. doi: 10.1128/JVI.01285-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fauriat C, Ivarsson MA, Ljunggren HG, Malmberg KJ, Michaelsson J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood 2010; 115(6): 1166–1174. doi: 10.1182/blood-2009-09-245746 [DOI] [PubMed] [Google Scholar]
- 46.Bultitude WP, Gymer AW, Robinson J, Anthias C, Potter MN, Russell NH et al. The effect of donor KIR2DL1 allelic diversity on the outcomes of HSCT is influenced by conditioning regimen. HLA 2019; 94(2): 122–123. [Google Scholar]


