Abstract
A bacterium was isolated from the blood and empyema of a cirrhotic patient. The cells were facultatively anaerobic, nonsporulating, gram-negative, seagull shaped or spiral rods. The bacterium grows on sheep blood agar as nonhemolytic, gray colonies 1 mm in diameter after 24 h of incubation at 37°C in ambient air. Growth also occurs on MacConkey agar and at 25 and 42°C but not at 4, 44, and 50°C. The bacterium can grow in 1 or 2% but not 3, 4, or 5% NaCl. No enhancement of growth is observed with 5% CO2. The organism is aflagellated and nonmotile at both 25 and 37°C. It is oxidase, catalase, urease, and arginine dihydrolase positive, and it reduces nitrate. It does not ferment, oxidize, or assimilate any sugar tested. 16S rRNA gene sequencing showed that there are 91 base differences (6.2%), 112 base differences (7.7%), and 116 base differences (8.2%) between the bacterium and Microvirgula aerodenitrificans, Vogesella indigofera, and Chromobacterium species, respectively. The G+C content (mean and standard deviation) is 68.0% ± 2.43%, and the genomic size is about 3 Mb. Based on phylogenetic affiliation, the bacterium belongs to the Neisseriaceae family of the β-subclass of Proteobacteria. For these reasons, a new genus and species, Laribacter hongkongensis gen. nov., sp. nov., is proposed, for which HKU1 is the type strain. Further studies should be performed to ascertain the potential of this bacterium to become an emerging pathogen.
Since the discovery of PCR and DNA sequencing, comparison of the gene sequences of bacterial species has shown that the 16S rRNA gene is highly conserved within a species and among species of the same genus and hence can be used as the new gold standard for the identification of bacteria to the species level. Using this new standard, phylogenetic trees based on base differences between species can be constructed, and bacteria can be classified and reclassified into new genera (7, 8). Furthermore, noncultivable organisms and organisms with ambiguous biochemical profiles can be classified and identified (10, 11). Recently, this technique was used for the identification of a strain of Mycobacterium neoaurum with ambiguous biochemical and whole-cell fatty acid profiles isolated from a patient with acute lymphoblastic leukemia (18), a strain of Escherichia coli with an ambiguous biochemical profile isolated from a bone marrow transplant recipient (16), a strain of Enterobacter cloacae with an ambiguous biochemical profile isolated from a renal transplant recipient (14), a strain of tube coagulase-negative Staphylococcus aureus isolated from a patient with refractory anemia with excessive blasts in transformation (19), a strain of Arcobacter cryaerophilus isolated from a traffic accident victim (13), and a noncultivable strain of Pseudomonas veronii from a patient with a pseudotumor (3).
In this study, we report the isolation of a bacterial strain from the blood and empyema of a cirrhotic patient. The strain, named HKU1, exhibited phenotypic characteristics that do not fit into the patterns of any known genus and species. 16S rRNA gene sequencing showed that there was only 93.8% base identity between the 16S rRNA gene of HKU1 and that of the most closely related species. On the basis of these studies, we propose a new genus and species, Laribacter hongkongensis gen. nov., sp. nov., to describe this bacterium.
MATERIALS AND METHODS
Patient and microbiological methods.
All clinical data were collected prospectively as described in a previous publication (5). The BACTEC 9240 blood culture system (Becton Dickinson, Cockeysville, Md.) was used. All isolates were identified by standard conventional biochemical methods (6). All tests were performed in triplicate with freshly prepared media on separate occasions. In addition, the Vitek system (bioMerieux Vitek, Hazelwood, Mo.) and the API system (bioMerieux Vitek) were used for the identification of the bacterial isolates in this study. Antimicrobial susceptibility was tested by the Kirby-Bauer disk diffusion method, and results were interpreted according to NCCLS criteria for E. coli (2).
SEM.
The isolates were grown in brain heart infusion broth at 37°C. Bacterial cells were washed twice using Milli-Q water. A suspension of the bacterium was allowed to settle on a polycarbonate membrane (Nuclepore) with a pore size of 0.8 μm for 5 min. The membrane was fixed in 2.5% (wt/vol) glutaraldehyde for 30 min and washed once in 0.1 M sodium cacodylate buffer. Fixed material was dehydrated through a graded ethanol series from 20% to 80% in 20% steps, followed by two changes of absolute ethanol. Each of the stepwise changes was for 15 min. Dehydrated material in absolute ethanol was critical point dried in a BAL-TEC CPD O30 critical point drier using carbon dioxide as the drying agent. Critical-point-dried material was mounted on an aluminum stub and coated with palladium by use of a BAL-TEC SCD 005 scanning electron microscopy (SEM) coating system. Coated material was examined in a Leica Cambridge Stereoscan 440 scanning electron microscope operating at 12 kV and with the specimen stage tilted at 0°.
Extraction of bacterial DNA for 16S rRNA gene sequencing.
Bacterial DNA extraction was modified from a previously published protocol (17). Briefly, 80 μl of NaOH (0.05 M) was added to 20 μl of bacterial cells suspended in distilled water, and the mixture was incubated at 60°C for 45 min. Then, 6 μl of Tris-HCl (pH 7.0) was added, achieving a final pH of 8.0. The resultant mixture was diluted 10-fold, and 5 μl of the diluted extract was used for PCR.
PCR, gel electrophoresis, and 16S rRNA gene sequencing.
PCR amplification and sequencing of the 16S rRNA gene were performed as described in previous publications (3, 13, 14, 15, 16, 18, 19). Briefly, DNase I-treated distilled water and PCR master mix (which contains deoxynucleoside triphosphates, PCR buffer, and Taq polymerase [see below]) were used in all PCRs by adding 1 U of DNase I (Pharmacia, Uppsala, Sweden) to 40 μl of distilled water or PCR master mix and incubating the mixture at 25°C for 15 min and subsequently at 95°C for 10 min to inactivate the DNase I. The bacterial DNA extract and control were amplified with 0.5 μM primers (LPW264, 5′-GAGTTTGATCMTGGCTCAG-3′, and LPW265, 5′-GNTACCTTGTTACGACTT-3′) (Gibco BRL, Rockville, Md.). The PCR mixture (50 μl) contained bacterial DNA, PCR buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 2 mM MgCl2, 0.01% gelatin), 200 μM each deoxynucleoside triphosphate, and 1.0 U of Taq polymerase (Boehringer Mannheim, Mannheim, Germany). The mixture was amplified for 40 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min and a final extension at 72°C for 10 min in an automated thermal cycler (Perkin-Elmer Cetus, Gouda, The Netherlands). DNase I-treated distilled water was used as the negative control. Ten microliters of each amplified product was electrophoresed in a 1.0% (wt/vol) agarose gel with a molecular size marker (lambda DNA AvaII digest; Boehringer) in parallel. Electrophoresis in Tris-borate-EDTA buffer was performed at 100 V for 1.5 h. The gel was stained with ethidium bromide (0.5 μg/ml) for 15 min, rinsed, and photographed under UV light illumination.
The PCR product was gel purified using a QIAquick PCR purification kit (QIAgen, Hilden, Germany). Both strands of the PCR product were sequenced twice using an ABI 377 automated sequencer according to the manufacturer's instructions (Perkin-Elmer, Foster City, Calif.) with PCR primers LPW264 and LPW265 and additional sequencing primers (LPW266, 5′-TCCCAGTGTGGCAGATCAT-3′, and LPW267, 5′-GAAAGGGAGCGGTAACGCA-3′). The sequence of the PCR product was compared with known 16S rRNA gene sequences in the GenBank database by multiple sequence alignment using the CLUSTAL W program (12).
Determination of G+C content.
Genomic DNA was prepared according to a previously published protocol (1), and the G+C content was determined by thermal denaturation (4). Briefly, the temperature of the genomic DNA in SSC (0.15 M NaCl plus 0.015 M sodium citrate) buffer (25 μg/ml) was increased slowly (0.5°C/min) from 25°C, and the absorbance of the solution at 260 nm was monitored continuously against that of a blank containing SSC buffer only. The Tm of the DNA was defined as the temperature at 50% hyperchromicity. The G+C content of the genomic DNA was calculated with the following formula: percent G+C = 2.44Tm − 169.
Determination of genomic size.
The genomic size of HKU1 was determined by pulsed-field gel electrophoresis using a CHEF Mapper XA system (Bio-Rad Laboratories, Hercules, Calif.). The gel was subjected to electrophoresis for 72 h at 14°C, at 3 V/cm, with an included angle of 120°, and with pulse times of 3 to 15 min in 0.5× TBE (0.045 M Tris-borate, 0.001 M EDTA [pH 8.0]) buffer. After electrophoresis, the gel was stained with ethidium bromide, and the genomic DNA was visualized with a UV transilluminator. The size of the genomic DNA was determined by comparing the distance of migration with those of the CHEF DNA size markers, Hansenula wingel chromosomes and Saccharomyces cerevisiae chromosomes (Bio-Rad).
Phylogenetic characterization.
The phylogenetic relationships of strain HKU1 to other members of the β-subclass of Proteobacteria was determined using the CLUSTAL method with MegAlign 4.00 (DNAstar Inc., Madison, Wis.). A total of 1,398 nucleotide positions were included in the analysis.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of HKU1 has been deposited in the GenBank sequence database under accession no. AF389085.
RESULTS
Patient.
A 54-year-old Chinese man was hospitalized because of fever and shortness of breath for 4 days. He had alcoholic cirrhosis complicated by recurrent ascites. Examination showed an oral temperature of 40.0°C, hepatosplenomegaly, ascites, and right pleural effusion. A chest radiograph and a contrast CT scan of the thorax showed right empyema and collapse-consolidation of the lower lobe of the right lung. The hemoglobin level was 10.8 g/dl; the total white cell count was 13.0 × 109/liter, with neutrophils at 11.7 × 109/liter, lymphocytes at 0.3 × 109/liter, and monocytes at 0.8 × 109/liter; and the platelet count was 75 × 109/liter. The serum bilirubin level was 50 μmol/liter, the albumin level was 20 g/liter, the alkaline phosphatase level was 95 U/liter, the alanine aminotransferase level was 13 U/liter, the aspartate aminotransferase level was 37 U/liter, and the γ-glutaryltransferase level was 521 U/liter. The serum urea and creatinine levels were within normal limits. The random serum glucose level was 15.9 mmol/level, the prothrombin time was 15.1 s, and the activated partial thromboplastin time was 38.4 s. Blood culturing, thoracocentesis, and abdominal paracentesis were performed, and empirical intravenous cefuroxime and netilmicin were administered. Pleural fluid examination revealed a white cell count of 18,540 × 106/liter, with 93% neutrophils and 7% lymphocytes; a protein level of 50 g/liter; a glucose level of 4.9 mmol/liter; a lactate dehydrogenase level of 5,510 U/liter; and pH 7.0. Peritoneal fluid examination revealed a white cell count of 750 × 106/liter, with 35% neutrophils, 3% lymphocytes, and 62% monocytes; a protein level of 7.0 g/liter; and a glucose level of 16.1 mmol/liter.
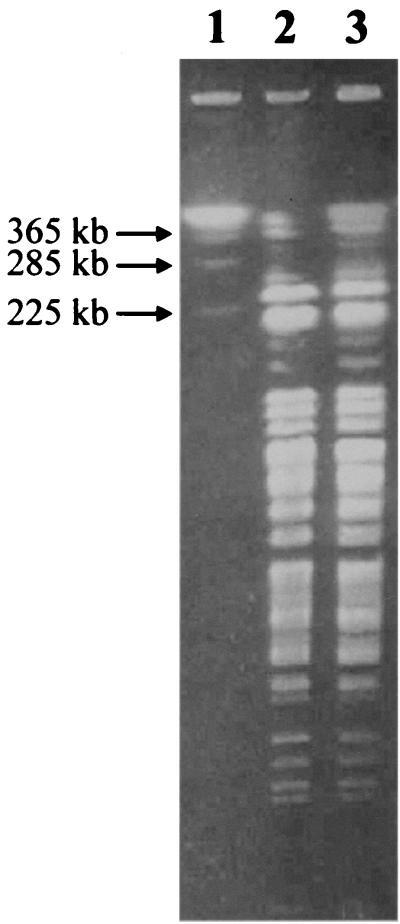
On day 3 postincubation, the blood culture turned positive with a gram-negative, seagull-shaped organism (strain HKU1). The same organism was also recovered from a pleural fluid culture (as demonstrated by the same biochemical profile, 16S rRNA gene sequence, and pulsed-field gel electrophoresis pattern after XbaI digestion) (Fig. 1) but not in peritoneal fluid. The patient responded to cefuroxime and netilmicin and adequate drainage of the empyema and was discharged after 38 days.
FIG. 1.
Pulsed-field gel electrophoresis of genomic DNA of HKU1 recovered from blood (lane 2) and empyema (lane 3) after XbaI digestion and S. cerevisiae chromosomal DNA size marker (lane 1).
Phenotypic characteristics.
Strain HKU1 is a seagull-shaped, gram-negative, non-spore-forming bacterium. It grows on sheep blood agar as nonhemolytic, gray colonies 1 mm in diameter after 24 h of incubation at 37°C in ambient air. No enhancement of growth is observed with 5% CO2. It also grows on MacConkey agar, in a microaerophilic or anaerobic environment, and at 25 and 42°C but not at 4, 44, and 50°C. It can grow in 1 or 2% NaCl but not in 3, 4, or 5% NaCl. It is nonmotile at both 25 and 37°C. The biochemical profile of strain HKU1 is shown in Table 1. It produces catalase, cytochrome oxidase, urease, and arginine dihydrolase and reduces nitrate. It does not ferment, oxidize, or assimilate any sugar tested. It is sensitive to ampicillin, cephalothin, cefuroxime, ceftazidime, ceftriaxone, imipenem, azetreonam, erythromycin, clarithromycin, gentamicin, amikacin, ciprofloxacin, levofloxacin, chloramphenicol, tetracycline, co-trimoxazole, and polymyxin B but resistant to vancomycin, clindamycin, metronidazole, and 0/129.
TABLE 1.
Biochemical profile determined for strain HKU1 by conventional biochemical tests and Vitek GNI+, API 20E, and API 20NE systems
| Biochemical reaction or enzyme | Resulta in the following test or system:
|
|||
|---|---|---|---|---|
| Conventional | Vitek GNI+ | API 20E | API 20NE | |
| Catalase | + | |||
| Cytochrome oxidase | + | |||
| Nitrate reduction | + | + | + | |
| β-Galactosidase | − | − | − | − |
| Arginine dihydrolase | + | + | + | + |
| Lysine decarboxylase | − | − | − | |
| Ornithine decarboxylase | − | − | − | |
| Citrate utilization | − | − | − | − |
| Malonate utilization | − | − | ||
| Mineral base medium acetate utilization | − | |||
| Acetamide utilization | − | |||
| H2S | − | − | − | |
| Urease | + | + | + | + |
| Tryptophan deaminase | − | − | − | |
| Indole | − | − | − | |
| Acetoin | − | − | ||
| Gelatinase | − | − | − | |
| Fermentation, oxidation, or assimilation of: | ||||
| Glucose | − | − | − | − |
| Mannitol | − | − | − | − |
| Inositol | − | − | − | |
| Sorbitol | − | − | − | |
| Rhamnose | − | − | − | |
| Sucrose | − | − | − | |
| Melibiose | − | − | ||
| Amygdalin | − | − | ||
| Arabinose | − | − | − | − |
| Lactose | − | − | ||
| Maltose | − | − | − | |
| Xylose | − | − | ||
| Raffinose | − | − | ||
| Mannose | − | − | ||
| Adonitol | − | − | ||
| Dulcitol | − | |||
| Salicin | − | |||
| N-Acetylglucosamine | − | |||
| Gluconate | − | |||
| Caprate | + | |||
| Adipate | + | |||
| Malate | + | |||
| Phenylacetate | − | |||
| Indoxyl-β-d-glucoside metabolism | − | |||
| Glucose fermentation in the presence of p-coumaric acid | − | |||
| Esculin hydrolysis | − | − | − | |
| Polymyxin B resistance | − | − | ||
| 2,4,4′-Trichloro-2′-hydroxydiphenylether resistance | − | |||
| Identification (%b) | Nonfermenting gram-negative Bacillus sp. (asaccharolytic) (75); Comamonas acidovorans (11) | Photobacterium damselae (81.7); Bordetella sp., Alcaligenes sp., or Moraxella sp. (14.3) | Comamonas testosteroni or Pseudomonas alcaligenes (99.3); Bordetella bronchiseptica (0.6) | |
+, positive; −, negative.
Percent confidence of identification.
SEM.
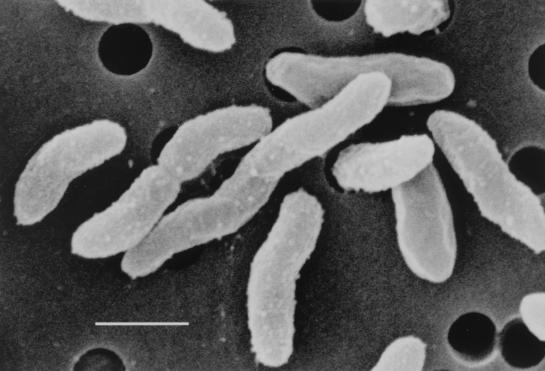
A scanning electron micrograph of L. hongkongensis is shown in Fig. 2. Bacterial cells were aflagellated, spiral, slender rods which multiplied by longitudinal division.
FIG. 2.
Scanning electron micrograph of L. hongkongensis. Cells vary in length from 0.79 to 2.5 μm. The bacterium has a spiral curvature and is aflagellated. Bar, 1 μm.
Molecular characterization by 16S rRNA gene sequencing, determination of G+C content and genomic size, and phylogenetic characterization.
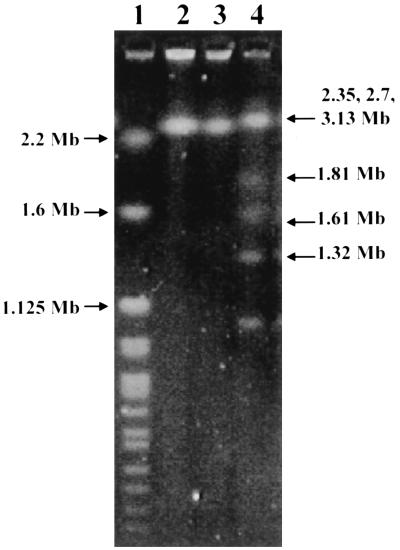
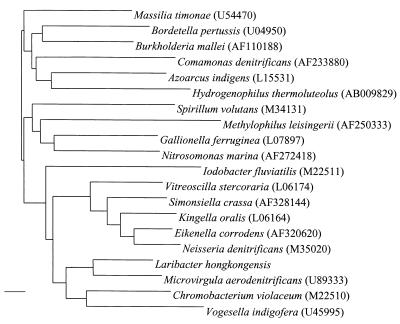
PCR of the 16S rRNA gene of strain HKU1 showed a band at 1,495 bp. There were 91 base differences (6.2%) between strain HKU1 and Microvirgula aerodenitrificans (GenBank accession no. U89333.1), 112 base differences (7.7%) between strain HKU1 and Vogesella indigofera (GenBank accession no. AB021385.1), 119 base differences (8.2%) between strain HKU1 and the β-subclass of Proteobacteria (GenBank accession no. AB017489.1), and 116 base differences (8.2%) between strain HKU1 and Chromobacterium species (GenBank accession no. AB017487.1). The G+C content of strain HKU1 (mean and standard deviation) was 68.0% ± 2.43%. The genomic size of strain HKU1 was about 3 Mb (Fig. 3). Based on phylogenetic affiliation, HKU1 belongs to the Neisseriaceae family of the β-subclass of Proteobacteria (Fig. 4).
FIG. 3.
Pulsed-field gel electrophoresis of genomic DNA of HKU1 recovered from blood (lane 2) and empyema (lane 3), H. wingel chromosomal DNA size marker (lane 1), and S. cerevisiae chromosomal DNA size marker (lane 4), showing that the genome size of HKU1 is about 3 Mb.
FIG. 4.
Phylogenetic tree showing the relationship of L. hongkongensis gen. nov., sp. nov., to the other members of the β-subclass of Proteobacteria. A total of 1,398 nucleotide positions in each 16S rRNA gene were included in the analysis. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes-Cantor correction; the length of the bar represents 1 substitution. Names and accession numbers are given as cited in the GenBank database.
DISCUSSION
We report the isolation of HKU1 from the blood and empyema of a Chinese cirrhotic patient. The clinical significance of the bacterium was evident by its isolation from sterile body fluids and the patient's local and systemic responses (high fever, leukocytosis, and neutrophilia) to the bacterium. 16S rRNA gene sequence analysis unambiguously placed HKU1 in the β-subclass of the Proteobacteria. However, the 16S rRNA gene of HKU1 exhibited less than 94% nucleotide identity with the 16S rRNA gene of all previously described members of this subclass. The most closely related species is M. aerodenitrificans, a new genus and species described in 1998; this organism was recovered from an upflow denitrifying filter inoculated with activated sludge and has never been implicated as a cause of human infections (9).
HKU1 exhibited phenotypic and genotypic characteristics that are very different from those of the other members of the β-subclass of the Proteobacteria, as well as members of other morphologically related pathogenic genera (Table 2). The G+C content of HKU1 is 68%, similar to those of M. aerodenitrificans and Chromobacterium violaceum, two other members of the β-subclass of the Proteobacteria, but very different from those of representative species of other morphologically related pathogenic genera. The major characteristics for distinguishing HKU1 from M. aerodenitrificans are that HKU1 is nonmotile, assacharolytic, and urease and arginine dihydrolase positive. Phenotypically, HKU1 is most closely related to Photobacterium damselae (identified by API 20E with 81.7% confidence) and Comamonas testosteroni and Pseudomonas alcaligenes (identified by API 20NE with 99.3% confidence). The major characteristics for distinguishing HKU1 from P. damselae are that HKU1 is assacharolytic and lysine decarboxylase negative but P. damselae is glucose fermenting and lysine decarboxylase positive; those for distinguishing HKU1 from C. testosteroni or P. alcaligenes are that HKU1 is arginine dihydrolase and urease positive but C. testosteroni or P. alcaligenes is arginine dihydrolase and urease negative. Since HKU1 is assacharolytic but urease and arginine dihydrolase positive, it probably utilizes proteins instead of carbohydrates as its energy sources. It is interesting that HKU1 has a seagull or spiral shape but is nonmotile, as all the other famous members of the Proteobacteria with spiral or curved shapes (e.g., M. aerodenitrificans of the β-subclass, Vibrio spp. of the γ-subclass, and Campylobacter spp., Helicobacter spp., and Arcobacter spp. of the ɛ-subclass) are all highly motile, and it is believed that the curved or spiral shape actually assists in the movement of the organisms. Because of its interesting phenotypic characteristics, unique G+C content, and genome size and because it could not be assigned to any currently recognized genus, we propose a new genus and a new species for HKU1.
TABLE 2.
Comparison of phenotypic and genotypic characteristics of L. hongkongensis and representative species of morphologically and/or phylogenetically related pathogenic generaa
| Organism | G+C content (mol %) | Genome size (Mb) | Morphology | Oxygen requirement | Motility | Sugar utilization | Urease | 2,4-Diamino- 6,7-diisopropylpteridine (0/129) |
|---|---|---|---|---|---|---|---|---|
| Campylobacter jejuni | 30.6 | 1.64 | Helical | Microaerophilic | Motile | Asaccharolytic | − | NA |
| Arcobacter cryaerophilus | 27–30 | NA | Curved | Microaerophilic and aerobic | Motile | Asaccharolytic | − | NA |
| Vibrio cholerae | 47.3 | 4.03 | Curved | Facultatively anaerobic | Motile | Saccharolytic | − | Sensitive |
| Chromobacterium violaceum | 65–68 | NA | Straight | Facultatively anaerobic | Motile | Saccharolytic | Variable | NA |
| Helicobacter pylori | 39 | 1.64 | Helical | Microaerophilic | Motile | Asaccharolytic | + | NA |
| Microvirgula aerodenitrificans | 65 | NA | Curved | Aerobic | Motile | Asaccharolytic | − | NA |
| Laribacter hongleongensis | 68 | 3 | Seagull shaped | Facultatively anaerobic | Nonmotile | Asaccharolytic | + | Resistant |
NA, not available. −, negative; +, positive.
The reservoir of HKU1 and its route of transmission in our patient remain unknown. We speculate that the patient may have aspirated the bacteria into his lungs, thereby developing empyema and subsequent bacteremia. Endogenously, the most likely potential reservoir would be the oral cavity or the gastrointestinal tract. However, screening of throat swab and stool samples from 30 volunteers did not show the presence of this bacterium (data not shown). The environment is another possible source of the bacterium, and its tolerance to 2% NaCl makes aquatic environments possible reservoirs for HKU1. However, surveillance of water samples with different NaCl contents also failed to reveal the presence of this bacterium (data not shown).
Description of Laribacter hongkongensis gen. nov., sp. nov.
Laribacter means seagull-shaped rod; hongkongensis, in honor of Hong Kong, means the place where the bacterium was discovered.
Cells are facultatively anaerobic, nonsporulating, gram-negative, seagull-shaped or spiral rods. The bacterium grows on sheep blood agar as nonhemolytic, gray colonies 1 mm in diameter after 24 h of incubation at 37°C in ambient air. Growth also occurs on MacConkey agar and at 25 and 42°C but not at 4, 44, and 50. It can grow in 1 or 2% NaCl but not in 3, 4, or 5% NaCl. No enhancement of growth is observed with 5% CO2. The organism is aflagellated and is nonmotile at both 25 and 37°C. It is oxidase, catalase, urease, and arginine dihydrolase positive, and it reduces nitrate. It does not ferment, oxidize, or assimilate any sugar tested (Table 1). The moles percent G+C content of the DNA of the strain is 68.0% ± 2.43%. The genomic size of the strain is about 3 Mb. The organism was isolated from the blood and empyema of a cirrhotic patient. The type strain of L. hongkongensis is strain HKU1. Its 16S rRNA gene sequence has been deposited within the GenBank sequence database under accession no. AF389085.
ACKNOWLEDGMENTS
This work was partly supported by the University Research Grant Council and the Committee for Research and Conference Grants, The University of Hong Kong.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1998. pp. 2.4.1–2.4.2. [Google Scholar]
- 2.Bauer A W, Kirby W M M, Sherris J C, Turck M. Antibiotic susceptibility testing by a single disc method. Am J Clin Pathol. 1966;45:493. [PubMed] [Google Scholar]
- 3.Cheuk W, Woo P C Y, Yuen K Y, Yu P H, Chan J K C. Intestinal inflammatory pseudotumor with regional lymph node involvment: identification of a new bacterium as the etiologic agent. J Pathol. 2001;192:289–292. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH767>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Goodfellow M. Chemical methods in bacterial systematics. London, England: Academic Press Ltd.; 1985. pp. 67–93. [Google Scholar]
- 5.Luk W K, Wong S S, Yuen K Y, Ho P L, Woo P C Y, Lee R A, Chau P Y. Inpatient emergencies encountered by an infectious disease consultative service. Clin Infect Dis. 1998;26:695–701. doi: 10.1086/514591. [DOI] [PubMed] [Google Scholar]
- 6.Murray P R, Baro E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. [Google Scholar]
- 7.Olsen G J, Overbeek R, Larsen N, Marsh T L, McCaughey M J, Maciukenas M A, Kuan W M, Macke T J, Xing Y, Woese C R. The ribosomal database project. Nucleic Acids Res Suppl. 1992;20:2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olsen G J, Woese C R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 9.Patureau D, Godon J J, Dabert P, Bouchez T, Bernet N, Delgenes J P, Moletta R. Microvirgula aerodenitrificans gen. nov., sp. nov., a new gram-negative bacterium exhibiting co-respiration of oxygen and nitrogen oxides up to oxygen-saturated conditions. Int J Syst Bacteriol. 1998;48:775–782. doi: 10.1099/00207713-48-3-775. [DOI] [PubMed] [Google Scholar]
- 10.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis. An approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 11.Relman D A, Schmidt T M, MacDermott R P, Falkow S. Identification of the uncultured bacillus of Whipple's disease. N Engl J Med. 1992;327:293–301. doi: 10.1056/NEJM199207303270501. [DOI] [PubMed] [Google Scholar]
- 12.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo P C Y, Chong K T K, Leung K W, Que T L, Yuen K Y. Identification of Arcobacter cryaerophilus isolated from a traffic accident victim with bacteraemia by 16S ribosomal RNA gene sequencing. Diagn Microbiol Infect Dis. 2001;40:125–127. doi: 10.1016/s0732-8893(01)00261-9. [DOI] [PubMed] [Google Scholar]
- 14.Woo P C Y, Cheung E Y L, Leung K W, Yuen K Y. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species with ambiguous biochemical profile from a renal transplant recipient. Diagn Microbiol Infect Dis. 2001;39:85–93. doi: 10.1016/s0732-8893(01)00206-1. [DOI] [PubMed] [Google Scholar]
- 15.Woo P C Y, Fung A M, Wong S S Y, Tsoi H W, Yuen K Y. Isolation and characterization of a Salmonella enterica serotype Typhi variant and its clinical and public health implications. J Clin Microbiol. 2001;39:1190–1191. doi: 10.1128/JCM.39.3.1190-1194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo P C Y, Leung P K L, Leung K W, Yuen K Y. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol Pathol. 2000;53:211–215. doi: 10.1136/mp.53.4.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo P C Y, Lo C Y, Lo S K, Siau H, Peiris J S M, Wong S S Y, Luk W K, Chan T M, Lim W W, Yuen K Y. Distinct genotypic distributions of cytomegalovirus (CMV) envelope glycoprotein in bone marrow and renal transplant recipients with CMV disease. Clin Diagn Lab Immunol. 1997;4:515–518. doi: 10.1128/cdli.4.5.515-518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo P C Y, Tsoi H W, Leung K W, Lum P N L, Leung A S P, Ma C H, Kam K M, Yuen K Y. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J Clin Microbiol. 2000;38:3515–3517. doi: 10.1128/jcm.38.9.3515-3517.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woo P C Y, Leung A S P, Leung K W, Yuen K Y. Identification of slide-coagulase positive, tube-coagulase negative Staphylococcus aureus by 16S ribosomal RNA gene sequencing. Mol Pathol. 2001;54:244–247. doi: 10.1136/mp.54.4.244. [DOI] [PMC free article] [PubMed] [Google Scholar]