Abstract
A new epidemic of acute respiratory viral pneumonia was discovered in central China at the end of 2019. The disease was given the name coronavirus disease 2019 (COVID-19), and the virus that caused this disease was known as severe acute respiratory syndrome coronavirus (SARS-CoV-2). So far, diagnostic methods have been focused on (a) human antibody detection, (b) viral antigen detection and (c) viral gene detection, the latter using RT-PCR being the most accurate approach. In this paper, we present a summary of the COVID-19 pandemic, clinical features and epidemiology and pathogenesis. Also, we focus on the recent advances in bioanalytical diagnostic methods based on various techniques for SARS-CoV-2 sensing that have recently been published (2020–2021). Furthermore, we present the mechanisms, advantages and disadvantages of the most common biosensors for COVID-19 detection, which include optical, electrochemical and piezoelectric biosensors as well as wearable and smart nanobiosensors, immunosensors, aptasensors and genosensors.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00604-022-05167-y.
Keywords: Coronaviruses, Biosensors, Bioanalytical methods, Diagnosis, SARS-CoV-2, COVID-19 pandemic
Introduction
The World Health Organization declared an epidemic on January 30, 2020, following the outbreak of the SARS-CoV-2 virus in Wuhan, Hubei Province, China, and its rapid spread to 25 countries. This happened just 1 month after the announcement of the first case of the disease on December 31, 2019 [1–4]. Coronaviruses are positive single-stranded RNA viruses that belong to the coronavirus family and are genetically classified into four genera: α, β, γ and δ coronavirus [5–8]. The basis of the SARS-CoV-2 coronavirus is in the beta-coronavirus genus. These viruses often infect animals such as birds and mammals and usually cause mild respiratory infections in humans. Due to the SARS-CoV-2 RNA content and its high potential for emergence, respiratory infections caused by the virus have recently led to deadly endemics in humans, such as SARS and measles. The causative agent of these two types of coronavirus diseases is zoonotic and belongs to the genus β-coronavirus of the coronaviridae family [9–11]. The first case of Middle East respiratory syndrome (MERS) was observed in Saudi Arabia in 2011–2012, of which 2495 cases have been reported since then, of which 858 cases were associated with death and the death rate was estimated at 34.4%. While no new MERS-CoV cases have been reported since 2004, the SARS-CoV-2 outbreak occurred unexpectedly in 2020 [11, 12]. SARS-CoV-2 is the seventh member of the coronavirus family, which, like MERS-CoV, SARS-CoV causes respiratory disease in humans with a genome size of 27–35 kb and belongs to the beta-coronavirus species. Like other coronaviruses, it encodes several structural proteins and non-structural proteins. Spike glycoprotein (S) of and nucleocapsid protein (N), membrane protein (M) and coating protein (E) are among its structural proteins [13]. The virus coat is one of the hardest coatings in the coronavirus family, and it appears that SARS-CoV-2 is more resistant to body fluids and the environment than other viruses in its family, including MERS-CoV and SARS-CoV, and has a longer lead time. MERS-CoV and SARS-CoV remain outside the environment and require fewer viral particles for infection [14].
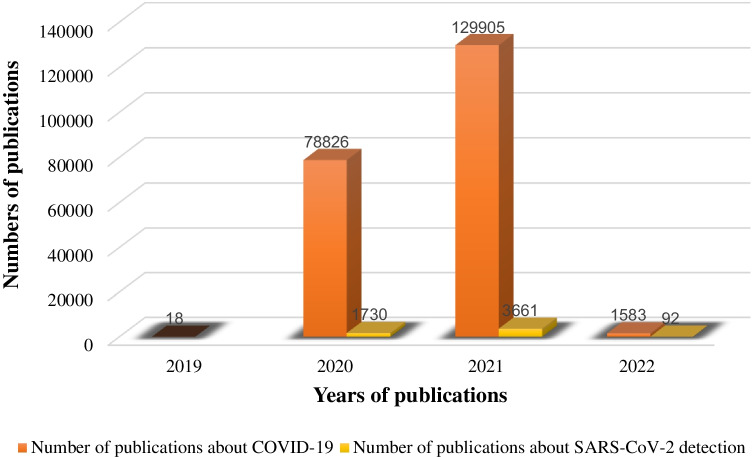
SARS-CoV-2: It is assumed that the virus is transmitted via droplets, close contact, aerosols and possibly faecal-oral transmission and patients pending the incubation period can transmit the virus to others. In addition, an infected person can spread more active viral particles [15, 16]. Research on the epidemiology, clinical, laboratory and radiological features of patients has shown that the virus causes a severe SARS-like respiratory illness that occurs throughout. Sequence analysis of the SARS-CoV-2 genome taken from patients during the epidemic shows a sequence almost identical to that of SARS-CoV [17]. Using its genome, which is closely related to SARS-CoV and MERS-CoV, and the clinical and experimental data collected on previous viruses, it is possible to predict how the host’s immune system will respond to this specific virus, and how the virus may escape the host’s immune responses [18]. Diagnosis of SARS-CoV-2 virus is crucial to stopping the spread and prompt treatment. Hence, a growing interest in developing bioanalytical methods for the detection of SARS-CoV-2 virus has been observed from 2019 until now (Scheme 1).
Scheme 1.
Statistics of the number of publications per year related to COVID-19
What are coronaviruses?
Coronaviruses are single-stranded, enveloped RNA viruses 80–120 nm in diameter and they are classified into four groups: α, β, γ and δ. Prior to the identification of COVID-19, only six types of coronavirus could infect humans, and COVID-19, a member of the β-coronavirus family, was the seventh. Of these viruses, four coronaviruses, HCoV-OC43, HCoV-229E, HCoV-NL63 and HCoVHKU1, they are less pathogenic and cause only mild respiratory diseases, but the two coronaviruses, SARS-CoV and MERS-CoV, are better than humans, respectively. They transmitted two fatal epidemics. Meanwhile, the homology and pathogenesis mechanism of SARS-CoV is very similar to COVID-19. Due to the adaptation of COVID-19 in bats, which have a higher temperature than the human body, this virus is more resistant to temperature than SARS-CoV [3, 19].
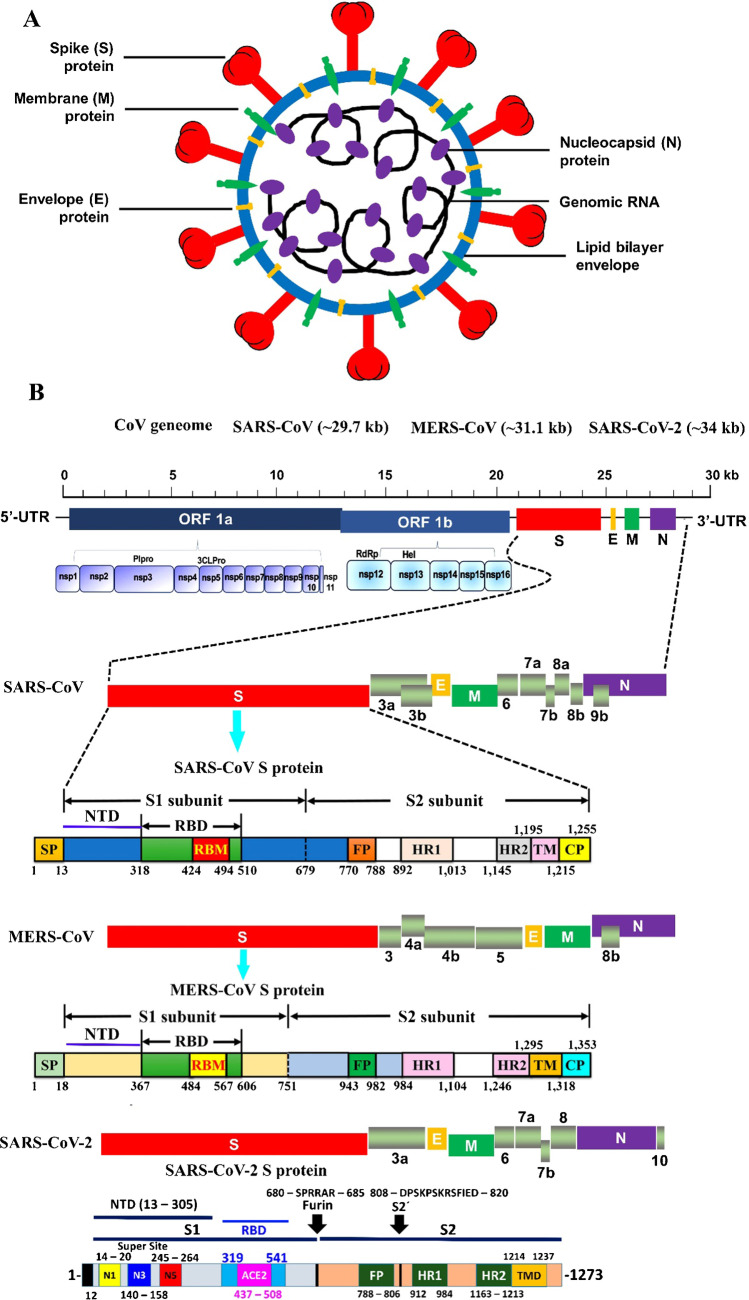
Coronavirus has four major structural proteins called S (Spike), E (Envelope), N (Nucleocapsid) and M (Membrane) as shown in Fig. 1. COVID-19 uses angiotensin-converting enzyme-type-2 (ACE2) as a receptor and infects cells with ACE2 via a receptor-binding domain (RBD) in the spike protein. The ACE2 receptor is found in alveolar cells, myocytes and vascular endothelial cells due to the high affinity of COVID-19 for ACE2. Genital pathology, including testes and ovaries, is also effective. COVID-19 probably affects sperm production and reduces its number, and also produces sex hormones and can reduce sexual desire [20, 21].
Fig. 1.
A Schematic structure of coronaviruses [127]. B Schematic representation of the genome organization and functional domains of S protein for SARS-CoV, MERS-CoV and SARS-CoV-2. Reproduced with permission from [127, 128]. The genomes of CoVs consist of 2 partially overlapping replicase open reading frames (OFR1a and 1b) and several downstream ORFs that encode viral functional structural proteins and other proteins with unknown function. 50-UTR and 30-UTR, untranslated regions at the N and C-terminal regions, respectively. Kb, kilobase; pair SP, signal peptide; NTD, the N-terminal domain; RBD, receptor-binding domain; RBM, receptor-binding motif; FP, fusion peptide; HR1 and HR2, heptad repeat 1 and 2; TM, transmembrane domain; CP, cytoplasmic domain
Spike is a homotrimer S glycoprotein that binds to the type-1 integral membrane in ACE2; then, pH-dependent endocytosis occurs. The acidic pH of lysosomes and endosomes activates the enzymes cathepsin B and L and breaks down S glycoprotein into two subunits, S1 and S2. Subunit S1 is used for binding and S2 for integration with the host cell membrane. The virus also needs an acidic environment with a pH of about 3 to enter the cell cytosol. In the cell cytoplasm, the virus begins to multiply with the help of its RNA polymerase, which infects infected cells after leaving the cell. Host cells also cause the membrane to attach to adjacent cells by forming spike proteins on their surface, creating a syncytium. In addition to disrupting the function of the organ involved, the formation of cynicium also provides the basis for further spread of the virus and escape from the immune system [8, 22, 23]. By creating bilayer vesicles, coronaviruses block the expression of pattern recognition receptors (PRRs) and, as a result, their natural immune system does not know them and continue to multiply within the vesicles. They disrupt the manufacture of type I interferons as one of the most important antiviral agents. Coronaviruses also interfere with the delivery of antigen by immune cells [23].
In a study using protected amplitude analysis, homologous modelling and molecular docking techniques, Liu and colleagues showed that ORF8 (open reading frame) domains and COVID-19 surface glycoproteins could bind to porphyrubin in porphyrubin. The ORF1ab, ORF10 and ORF3a domains of the virus can also remove iron from porphyrins. They claimed that chloroquine could block the release of iron by binding to the ORF1ab, ORF10 and ORF3a domains, thereby helping red blood cells to properly load oxygen. The ORF1ab domain is a coronavirus-specific protein that plays an important role in RNA replication and virus replication. Therefore, ORF1ab is a target for vaccine production and drug design. Haemoglobin is made up of two alpha globulin chains and two beta-globulin chains that bind together. Each haemoglobin molecule has one globin molecule and four heme molecules, so that each heme molecule is attached to one of the globin chains. Heme is composed of a porphyrin ring with a divalent iron atom at its centre [11, 24].
Types of coronavirus
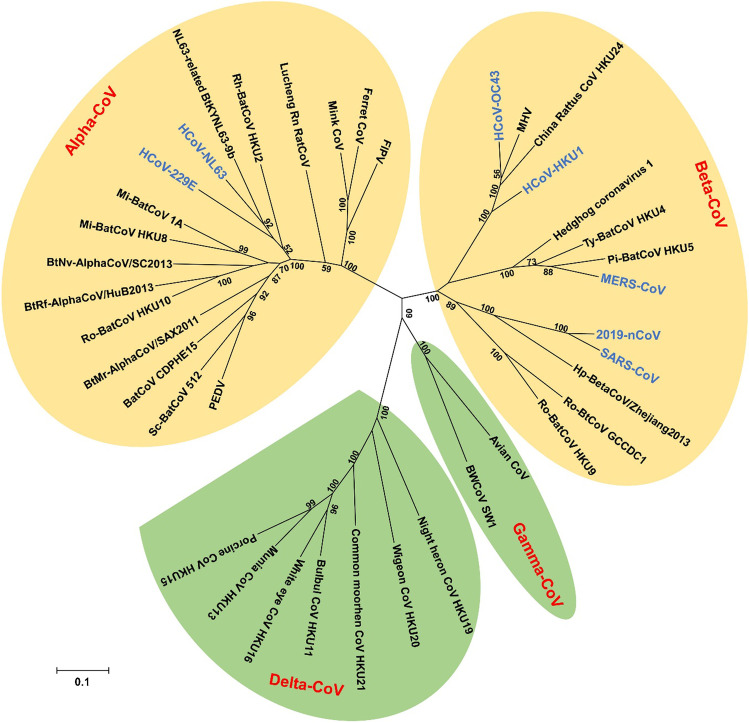
Coronaviruses are classified into four subtypes: alpha, beta, gamma and delta, which scientists use these classifications to classify different species. Seven coronaviruses have been linked to human disease. The upper and lower airways, nose, sinuses, mouth and lungs are affected by four of these, including human coronavirus 229E and NL63 (which belong to alpha-CoVs) [25], as well as human coronavirus OC43, and HKU1 (which belong to beta-CoVs) [26] as shown in Fig. 2. These viruses are widespread in the world, accounting for 15–30% of all common colds. They only spread to the lower respiratory tract in a small percentage of cases.
Fig. 2.
Phylogenetic tree of coronaviruses (CoVs) based on the nucleotide sequences of RNA-dependent RNA polymerase (RdRp). The tree, with 1000 bootstrap values, was constructed by the maximum likelihood method using MEGA 6. The four main phylogenetic clusters correspond to genera alpha-CoV, beta-CoV, gamma-CoV and delta-CoV. Each CoV genus contains different subgenera. The letters in blue indicate human CoVs. Reproduced with permission from [127]
Three more coronaviruses emerged from animal infections. These viruses developed over time and were ultimately passed on to humans. These coronaviruses are more harmful to people’s health. They are listed as below:
Origin and spread of COVID-19
Coronaviruses, which are single-stranded and positive-sense RNA viruses, have the longest genome of any known RNA virus, with a genomic content (GC) ranging from 32 to 43% [29]. They have a spherical shape with protruding branches and a crown, and this spatial shape has led to the naming of this viral family as coronavirus. The root of this name is derived from the Latin word corona, meaning crown [30]. Coronaviruses cause 15% of respiratory illnesses and usually do not cause an acute form of the disease, but they can cause mild upper respiratory infections. This viral family infects a wide range of animals (mammals) and humans [30, 31].
According to research on coronaviruses since 1965, these viruses have the potential to infect animals and humans, and some have the ability to transmit from animals to humans or vice versa [32]. Studies since the SARS epidemic have shown that bats carry various coronaviruses that have the potential to infect humans [30]. Researchers extracted the genetic material of the new virus from people who studied it and found that the new corona virus had a similar origin to the SARS virus and may have originated in bats or snakes, but that other animals could still be mediated hosts [13]. These viruses change in the bat’s body, and the bat itself does not show any signs of disease, making it easier to transmit the virus. Usually, in viruses that are transmitted from animals to humans, other human-to-human transmission does not occur or rarely occurs, but there are exceptions. Some animal viruses that can be transmitted from human to human, such as the human immunodeficiency virus (HIV), the Ebola virus (EBOLA) and some coronaviruses, can cause more deadly disease. When these viruses first infect human society, there is usually no cure or vaccine against them; thus, early generations of the outbreak can be fatal and cause significant casualties, as has been observed during the HIV and EBOLA epidemics [30].
Clinical features
The most common clinical signs of COVID-19 infection are fever (9.87%), cough (7.67%) and fatigue (1.38%); diarrhoea (7.3%) and vomiting (5%) are rare symptoms that are similar to other coronaviruses of animal origin. Acute respiratory distress syndrome (ARDS) occurs about 9 days after the onset of infection. Except for the lungs, the virus damages other tissues, including the heart, kidneys, liver, eyes and nervous system [33]. Dizziness, forgetfulness, weakening and disappearance of olfactory and taste powers, and nerve pain up to seizures and strokes are among the neurological symptoms of this virus, which are related to hypoxia and inflammation of the brain. Inflammation of the brain can be indirectly caused by a cytokine storm (autoimmune encephalitis) or directly caused by a broken blood–brain barrier by a virus (viral encephalitis) [34, 35]. COVID-19 also causes cardiovascular complications by causing thrombosis in the arteries and veins. The mechanism of thrombosis in this disease is inflammation, platelet activation, vascular dysfunction and vasoconstriction. On this basis, antithrombotic drugs are prescribed to patients [36]. Some people with COVID-19 also experience other viral infections, the most common of which are respiratory syncytial virus, rhinovirus, enterovirus and SARS-CoV-2 coronaviridae [37]. Lymphopenia, thrombocytopenia, elevated levels of D-dimer, c-reactive protein (CRP), erythrocyte sedimentation rate (ESR), lactate dehydrogenase (LDH), creatine kinase (CK), ferritin and prolonged prothrombin time (PT) are seen in a significant number of patients, while elevated levels of transaminases, cardiac enzymes and creatinine are lower. Thrombocytopenia and high D-dimer levels increase the need for a ventilator, ICU admission and mortality. High levels of interleukin 6 (IL-6) and IL-10, as well as low levels of CD4 + T and CD8 + T cells, are consistent with the severity of the disease. CD8 + T cells, which are the most important virus-killing cells in the immune system, increase the number of CD8 + markers on their surface to compensate for their decline to maximize their activity. CD8 + T cells work with human leukocyte antigen (HLA) class I and CD4 + T cells work with HLA class II. HLA is inherited from one’s parents and makes people more susceptible or resistant to certain pathogens. Decreasing the number of CD4 + T cells can interfere with the production of B memory cells [17, 38–40]. In this disease, the window period lasts about 7 days and that is when the antibody has not been made yet. IgM is synthesized as the first antibody on the seventh day and disappears around the 21st day of the disease. IgG begins to be synthesized on day 14 and production continues. The asymptomatic period is from the time of infection until the fifth day and the onset period of clinical symptoms is from the fifth to the eighth day. The disease begins to subside on day 14, which corresponds to the onset of a humoral immune response shift from IgM to IgG. The disease resolves by day 28. Not all antibodies produced against the corona virus are neutralizing or neutralizing the virus, and in some cases even increase the infection of antibody receptor (FcR) cells, including macrophages. Dysfunctional (non-neutralizing) antibodies bind to the virus via their own Fab and via their Fc to cells with FcR. In fact, the antibody complex with FcR mimics the role of the virus receptor for the virus to enter the cell [41, 42].
Epidemiology and pathogenesis
Coronavirus has increased interspecific transmission due to its extensive genetic diversity and frequent recombination [43]. The natural host of the virus is the bat, and penguins and snakes act as intermediate hosts. The first transfer took place through the use of natural hosts and interfaces. Direct and respiratory contact drops are the most common ways of transmitting the virus in society. It has an average incubation period of 3 days (ranging from 0 to 24 days), and the average time from the onset of the first symptom to death is 14 days [1, 44]. Fortunately, intrauterine transmission of the virus from an infected mother to the foetus has not been reported [45]. COVID-19 is coated and less stable than uncovered viruses in the gastrointestinal tract, and therefore less likely to contaminate surface and groundwater. The risk of transmitting the virus through the faeces of an infected person is also low. High temperatures, low or high pH and sunlight reduce the number of viruses. The survival rate of the virus varies at different levels between 2 h and 9 days. Factors affecting the survival of the virus are surface type, temperature and relative humidity. Common disinfectants such as ethanol kill 70% and hypochlorite kills 0.1% of the virus within 1 min [46]. Asymptomatic carriers play a major role in the transmission of the disease from person to person. Information about carriers of the disease who do not show symptoms is limited. People under the age of 15 make up a significant percentage of these carriers. Clinical signs and CT imaging do not assist much in distinguishing asymptomatic carriers, and the best way to distinguish these individuals is with real-time PCR, because most of them have no clinical signs and a normal CT image [41].
Bioanalytical diagnostic methods
Prior to testing, proper collection of the respiratory tract sample at the appropriate time and from the appropriate anatomical location is critical for rapid and accurate molecular diagnosis of the virus. Five to six days after the onset of the disease, the virus level in the upper and lower respiratory tract is high and nasopharyngeal and oropharyngeal swab specimens are appropriate. When sampling is delayed, the best sample is obtained from the lower respiratory tract through bronchoalveolar lavage. Exposure of the swab or sample for 30 min at 56 °C destroys the virus RNA and shows false-negative results in real-time reverse transcription-polymerase chain reaction (RT-PCR). After the test, the most important thing is to accurately interpret the results using molecular and serological methods together [23, 47].
Detection of virus nucleic acids by real-time RT-PCR is a standard method for determining coronavirus infection, although this method has a high specificity as well as low sensitivity, so it has a false negative and is time-consuming [48]. Negative results led to the conclusion that the combined use of CT imaging and laboratory tests could help in the early distinguish of COVID-19 pneumonia. Antibody-based immunoassay techniques are complemented by molecular techniques as complementary tools. These techniques are fast and inexpensive, but have low sensitivity and are the best tools in epidemiological studies of the disease and diagnosis of asymptomatic patients.
For molecular detection, the WHO recommends screening the sample with the Envelope corona gene and then confirming it with the RdRp gene (RNA-dependent RNA polymerase). In the USA, two proteins, N1 and Nucleocapsid N2 (virus), are being tested. In the immunoassay method, anti-N protein antibody, which is an immunodominant antigen, is used for early detection of the disease [47].
Biosensors
Several molecular diagnostic platforms are being used for diagnosing SARS-CoV-2, such as polymerase chain reaction (PCR) and quantitative real-time PCR (qRT-PCR), clustered regularly interspaced short palindromic repeats (CRISPR), computed tomography imaging (CT imaging) and reverse transcription loop-mediated isothermal amplification (RT-LAMP). RT-PCR testing has become employed as one of the most reliable ways for detecting novel viruses; nevertheless, this procedure is time-consuming, labour-intensive and needs the employment of qualified laboratory employees. Furthermore, false negatives have been documented despite its high sensitivity and specificity, particularly in non-nasopharyngeal swab samples with lower virus loads [49]. Therefore, providing individuals in underdeveloped nations with mass testing is difficult. On the other hand, many serology biosensors are now being used for SARS-CoV-2 diagnostics, such as electrochemical, optical, microfluidic and lateral flow immunoassay (LFIA). Biosensor science is interdisciplinary between biology, chemistry, electronics and engineering that is currently one of the most attractive and dynamic fields of research in analytical chemistry. There is usually no need to prepare a sample when using biosensors. Biosensors are generally evaluated based on sensitivity, limit of detection (LOD), linear range, reproducibility, selection and interference response [50, 51]. Biosensors are capable of real-time analysis, so they are important for the rapid measurement of body analytes. Biosensors promise cheap, fast and simple tools. They are a wide range of emerging technologies that are ideally suited for health analysis [52–54].
Electrochemical biosensors
Electrochemical biosensors are a subset of sensors that are more popular than other methods due to their lower-cost, portability and lower diagnostics. The electrochemical reaction is performed on the electrode surface. To session the growing demand for hypersensitivity, researchers have developed several electrochemical methods for producing nanocomposite-based electrochemical sensors. With remarkable success, nanoscience and nanotechnology have demonstrated that nanomaterial-based electrochemical signal reinforcement has considerable potential to improve both sensitivity and selectivity for electrochemical sensors and biosensors. Furthermore, it can be used to construct modern and modified sensing devices, in particular electrochemical sensors and biosensors [53, 55–60].
As compared to other biosensors, electrochemical transduction has many advantages, including low cost, ease of miniaturization for point of care (POC) use, high sensitivity and relatively easy instrumentation. Electrochemical biosensors (especially based on differential pulse voltammetric and amperometric detection techniques) normally use an electroactive label since antibody/antigen and DNA hybridization reactions do not produce a significant signal on their own. As a redox probe, ferro/ferricyanide is used in several studies recorded in the literature. The major drawbacks of this form of biosensor are current signals resulting from non-specific adsorption of proteins or other biomaterials, as well as biofouling of the electrode surface. In 2021, Yakoh et al. have shown an advantageous paper-based electrochemical platform for diagnosing COVID-19. The sensing scheme depends on the disruption of the redox conversion ([Fe (CN)6]3−/4−) caused by immunocomplex formation between the captured immunoglobulins generated in response to SARS-CoV-2 in humans with the immobilized spike protein of SARS-CoV-2. Because of the label-free electrochemical system, no antibodies are needed in this work, unlike other lateral flow immunoassay (LFIA) platforms that use multiple antibodies to control the reporter. The colorimetric LFA was found to be more sensitive (3 orders of magnitude) than the rapid (30 min) and sensitive detection of SARS-CoV-2 antibodies, with a detection limit of 1 ng/mL. Furthermore, this paper-based device has adequate sensitivity and specificity for detecting targeted antibodies in clinical sera from patients (100% and 90%, respectively). They improved the functionality of the paper-based electrochemical platform by allowing direct detection of the SARS-CoV-2 spike protein antigen [61].
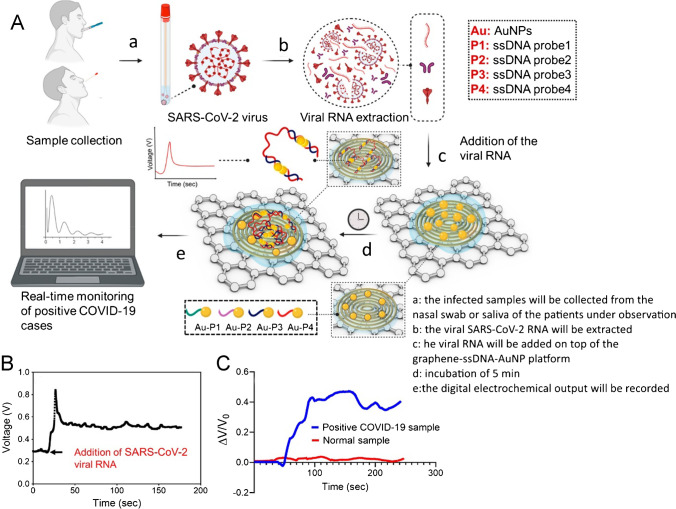
Alafeef et al. developed a low-cost, fast (less than 5 min) and simple-to-implement paper-based electrochemical sensor chip for the digital sensing of SARS-CoV-2 genetic content. The biosensor employs gold nanoparticles, captured with highly specific ssDNA (specific antisense oligonucleotides), aimed at phosphoprotein virus nucleocapsides (N-gene) as shown in Fig. 3A. The sensing samples are immobilized to produce a nuclear acid testing device, which can be read with a single, readable manual reader, on a paper-based electrochemical platform. Samples from Vero cells infected with the SARS-CoV-2 virus and biological fluids have been tested for the biological sensor chip. In the presence of a target (SARS-CoV-2 RNA), the sensor significantly improves the output signal after less than 5 min of incubation (Fig. 3B), resulting in an LOD of 6.9 copies/µL and a sensitivity value of 231 (copies µL−1) −1 without the need for further amplification. The performance of the sensor chip has been evaluated by a clinically validated FDA-approved RT-PCR COVID-19 diagnostic kit with 22 COVID-19-positive patients and 26 healthy asymptomatic individuals. With approximately 100% accuracy, sensitivity and specificity, the sensor reliably separates positive samples of COVID-19 from negative samples and shows a small increase in the output signal for samples with no SARS-CoV-2 viral target (e.g. MERS-CoV, SARS-CoV or negative COVID-19 samples collected from healthy people) as shown in Fig. 3C. The sensor is also assured by the design of the ssDNA-conjugated AuNPs, which target two distinct areas with the identical SARS-CoV-2 N gene, even during a genome mutation by the virus [62].
Fig. 3.
A Schematic illustrations of the principle of the COVID-19 electrochemical sensing platform. B Sensor output signal as a function of time with the addition of SARS-CoV-2 viral RNA load. C Real-time response of the sensor chip toward COVID-19-positive clinical samples and healthy samples. Reproduced with permission from [62]
Fabiani et al. reported an electrochemical immunoassay to identify SARS-CoV-2 coronavirus in saliva in a quick and accurate manner. The electrochemical approach is based on the use of secondary antibodies with alkaline phosphatase and magnetic beads as immunological labels and immunological chain support, respectively, to identify spike (S) or nucleocapsid (N) proteins. Carbon black nanomaterial-modified screen-printed electrodes were used to detect the enzymatic by-product 1-naphtol. The analytical properties of the electrochemical immune test were examined in the case of the S and N protein standard solution in buffer and non-treated saliva with a detection limit of 19 ng/mL and 8 ng/mL in untreated saliva, respectively in respect of S and N protein. Its efficacy has been evaluated in biosafety level 3 using cultured viruses with the comparison of data using real-time PCR nasopharyngeal swab. This analysis tool has a high potential for market entry as the first highly sensitive electrochemical immunoassay for the detection of SARS-CoV-2, thanks to its data agreement, low-detection limit, rapid analysis (30 min), miniaturization and portability combined with ease of use and non-invasive sampling [63].
Some of the bioanalytical methods used for the detection of SARS-CoV-2 are given in Table S1.
Optical biosensors
Optical biosensors are based on changes in optical properties, and are a powerful tool in identification and analysis in different aspects such as biomedicine, health, pharmaceuticals, environmental monitoring and security [64].
Optical biosensors are resistant to electromagnetic interference and can also be operated remotely due to this resistance. In fluorescence-based identification, both the target molecule and the biomarker are labelled with fluorescent labels such as dyes, and the intensity of the fluorescence indicates the presence of target molecules and the strong bond between the biosensor molecules and the target. Although fluorescence detection is sensitive, it is limited to the detection of a single molecule. The molecule labelling step impairs the function of the molecule, and the number of fluorophores on each molecule is not properly controlled, interfering with quantitative calculations. Unlike the fluorescence method, in free-label detection, the target molecules are not altered or labelled and are detected in their natural state. This method of identification is relatively easy and inexpensive and allows the quantitative and kinetic measurement of intermolecular interactions. This method of identification is the basis of the work of biosensors of surface plasmon resonance [65, 66]. Optical biosensors are very important for detecting pathogenic bacteria due to their sensitivity. The most common methods of optical detection are surface plasmon resonance and fluorescence. In addition to these methods, the use of fibre optics, lasers and prisms are also used to identify pathogens [67].
Optical biosensors are very important for detecting pathogenic bacteria and viruses due to their sensitivity. The most common methods of optical detection are surface plasmon resonance (SPR) and fluorescence. The localized surface plasmon resonance (LSPR) detection concept is based on the local refractive index changes surrounding the metal nanostructures owing to the biomolecule binding events (i.e. antigen–antibody interaction). This causes a red shift of the noble metal nanostructure LSPR peak, which proportionates directly to the antibody level of the target [68]. Another benefit of LSPR-based sensing is that the short electromagnetic field decay duration in localized surface plasmons substantially decreases the interfering influences on the bulk solution, which is desired in the analysis of complicated samples like fibrinogen-containing blood plasma or serum and globulins [69]. In addition to these methods, the use of fibre optics, lasers and prisms are also used to identify pathogens. The SARS-CoV-2 detection by the “naked eye” is highly desirable, and it can be validated without the use of advanced instrumental methods. In this regard, Moitra et al. reported the development of a colorimetric assay based on gold nanoparticles (AuNPs) that, when capped with suitably developed thiol-modified antisense oligonucleotides (ASOs) specific for the SARS-CoV-2 N-gene (nucleocapsid phosphoprotein), could be used to diagnose positive COVID-19 cases within 10 min from isolated RNA samples. In the presence of its target RNA sequence of SARS-CoV-2, the thiol-modified ASO-capped AuNPs agglomerate selectively and exhibit a change in surface plasmon resonance. Furthermore, the addition of RNaseH cleaves the RNA strand from the RNA–DNA hybrid, resulting in a visually visible precipitate from the solution, which is mediated by additional AuNP agglomeration. The assay’s selectivity was tested in the presence of MERS-CoV viral RNA, with a detection limit of 0.18 ng/L of RNA with SARS-CoV-2 viral load. Thus, without the use of sophisticated instrumental techniques, the current study reports a selective and visual “naked-eye” detection of COVID-19 causative virus, SARS-CoV-2 [70].
Funari et al. designed an opto-microfluidic sensing device for the detection of antibodies against SARS-CoV-2 spike protein based on the LSPR principles. This optofluidic technology comprises of an Au nanospike-coated glass substrate embedded in a microfluidic chip connected with a reflection probe for monitoring the presence of antibodies against the SARS-CoV-2 spike protein in diluted human plasma (1:1000) within 30 min. The LSPR wavelength peak shift of gold nanospikes induced by the local refractive index change owing to antigen–antibody interaction could be associated with the target antibody concentration. The LOD of this label-free microfluidic platform is 0.08 ng/mL (0.5 pM), which is below the clinically relevant concentration range. The proposed opto-microfluidic technology can be used as a point-of-care testing tool to supplement traditional serological assays and make quantitative SARS-CoV-2 diagnoses simpler, faster and cheaper [71].
Qiu et al. proposed an alternative and promising approach for clinical diagnoses COVID-19 via the combination of the plasmonic photothermal (PPT) effect with the LSPR sensing transduction. Through nucleic acid hybridization, two-dimensional gold nanoislands (AuNIs) functionalized with complementary DNA receptors may detect targeted sequences from the SARS-CoV-2. The thermoplasmonic heat is produced by its plasmonic resonance frequency on the same AuNI chip to improve sensing performance. The localized PPT heat allows the hybridisation of two similar gene sequences in situ to increase and improve precise discrimination. Our dual-function LSPR biosensor demonstrates an increased sensitivity for chosen SARS-CoV-2 sequences with a lower level of detection up to 0.22 pM and allows the specified target to be accurately detected in a multigene mixture. The study provides insight into the improvement of thermoplasmonics and its usefulness in the testing for nucleic acid and detection of viral diseases [72].
In another work, Qiu et al. demonstrated a thermoplasmonic-assisted dual-mode transducing (TP-DMT) concept to provide a sensible and self-validating plasmonic nanoplatform for quantifying SARS-CoV-2 trace amounts for a period of 30 min, using an amplification-free direct viral RNA detection and an amplification-based cyclic fluorescence probe cleavage detection (CFPC). Endonuclease IV identified the synthetic abasic site in the CFPC detection and cleavaged the fluorescent samples of the hybridized duplex. The shorter fluorescent probes were dehybridized by nanoscale thermoplasmonic heating, which aided the cyclical binding–cleavage–dissociation (BCD) process, which may give a very sensitive amplification-based response. This TP-DMT method was effectively verified by analyzing clinical COVID-19 patient samples, indicating that it may be used for rapid clinical infection screening and real-time environmental monitoring [73].
Peng et al. have developed a unique phase-modulation plasmonic biosensor that works in the near-infrared (NIR) range and can be used to sensitively detect SARS-CoV-2 and its spike (S) glycoprotein. The suggested plasmonic biosensor was developed by integrating the transparent indium tin oxide (ITO) film with the two-dimensional (2D) Van der Waals heterostructures such as tellurene and carboxylic functionalized molybdenum disulphide (MoS2). Under excitation of 1550 nm, the thickness of the ITO film and tellurene-MoS2 heterostructures may be optimized, resulting in outstanding biosensing performance. The optimal plasminic configuration of 121 nm ITO film/3-layer tellurene/10-layer MoS2-COOH can provide a maximum detecting sensitivity of 8.4 × 104 degrees/RIU for a sensing interface refractive index that changes to as low as 0.0012 RIU (RIU, refractive index unit). More significant, a MoS2, COOH layer, which is an excellent adsorption place to selectively bind SAR S-CoV-2 S glycoprotein, may capture angiotensin-converting enzyme II. The linear detection range of glycoprotein S and SARS-CoV-2 samples is 0.00–301.67 nM and 0.00–67.88 nM, respectively. As a result, this work provides an alternate method for swiftly diagnosing new coronaviruses in clinical settings [74].
One approach developed by Karakuş et al. is the colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with gold nanoparticle-based biosensors. Due to antibody–antigen interaction, gold nanoparticles aggregated swiftly and permanently in the presence of the SARS-CoV-2 spike antigen, changing colour from red to purple, as clearly visible with the naked eye or UV–Vis spectrometry through spectral red shifting with an LOD of 48 ng/mL. Furthermore, electrochemical detection was accomplished by simply dropping produced probe solution (gold nanoparticles (AuNPs)–SARS-CoV-2 spike monoclonal antibody (mAb) (AuNP–mAb)) onto a commercially available and disposable screen-printed gold electrode, which required no electrode preparation and modification, taking only a few minutes. The approach detected SARS-CoV-2 spike antigen at 1 pg/mL and demonstrated a linear response to the antigen at concentrations ranging from 1 pg/mL to 10 ng/mL. Even at high concentrations, both techniques were highly specific for identifying the SARS-CoV-2 spike antigen but not for other antigens such as Influenza A (H1N1), MERS-CoV or Streptococcus pneumoniae [75].
El-Said et al. synthesized reduced porous graphene oxide (rPGO) coated with gold nanoparticles (Au NPs) for the modification of ITO electrode. The extremely uniform Au NPs@rPGO-modified ITO electrode was then employed as a surface-enhanced Raman spectroscopy-active surface as well as a working electrode. The combination of Au nanoparticles and porous graphene improves Raman signals as well as electrochemical conductivity. The COVID-19 protein-based biosensor was designed by immobilising anti-COVID-19 antibodies onto a modified electrode and using it as a probe to capture the COVID-19 protein. The developed biosensor demonstrated the capacity to monitor the COVID-19 protein at concentrations ranging from 100 nmol/L to 1 pmol/L, with a limit of detection (LOD) of 75 fmol/L. Additionally, COVID-19 protein was identified using electrochemical methods at concentrations ranging from 100 nmol/L to 500 fmol/L, with an LOD of 39.5 fmol/L. Finally, three COVID-19 protein concentrations spiked in human serum were examined. As a result, the current sensor detected COVID-19 with great efficiency [76].
Piezoelectric biosensors
A piezoelectric (PZ) sensor is an effective device that measures variations in pressure, temperature, acceleration, force or strain and converts these changes into an electrical signal using the piezoelectric effect. An antibody-coated piezoelectric sensor increases the mass of the crystal surface by placing it in a solution containing antigens and binding between the antigen and the antibody, which changes the frequency. The most common piezoelectric materials include SiO2 and LiTaO3. In a piezoelectric sensor, the surface used must have chemical resistance and a high level of stabilized biologically active agents. In addition, the surface coating should be as thin as possible [77]. Compared to electrochemical and fibre optic sensors, these types of sensors are used to a lesser extent to identify pathogens [67]. The difficulty of coating and stabilizing biological agents on the crystal surface is an important factor in the limitations of these sensors [78].
Piezoelectric quartz crystal nanobiosensors have received a great level of interest for chemical and biological applications, in particular for the detection of viruses [79, 80], because of their simple model, immediate identification and real-time output. “Bulk wave” (BW) and surface acoustic wave (SAW) are the two kinds of piezoelectric sensors [81, 82]. These biosensors can detect biological entities and convert mechanical energy into electricity, resulting in a useable energy output for the user in response to a specified measurement input [81]. For example, a piezoelectric immunosensor has also been presented to detect SARS-CoV in the gas phase of sputum. PZ polyclonal antibodies against SARS-CoV were attached to the surface of a PZ crystal in an oriented state via protein A to create this biosensor. The antigen sample, on the other hand, was atomized into an aerosol using an ultrasonicator, allowing the antibodies on the PZ crystal’s surface to specifically adsorb the SARS-CoV antigen, resulting in a change in crystal mass and, potentially, a change in frequency. Changes in frequency at optimal conditions were linearly related to antigen concentrations ranging from 0.6 to 4 g/mL. The device has a short analyzing time (less than 2 min), is feasible, specific, simple, stable (the immunosensor has been stable for more than 2 months when stored at 4–6 °C over silica gel blue) and has high reproducibility [83].
Field-effect transistor biosensor
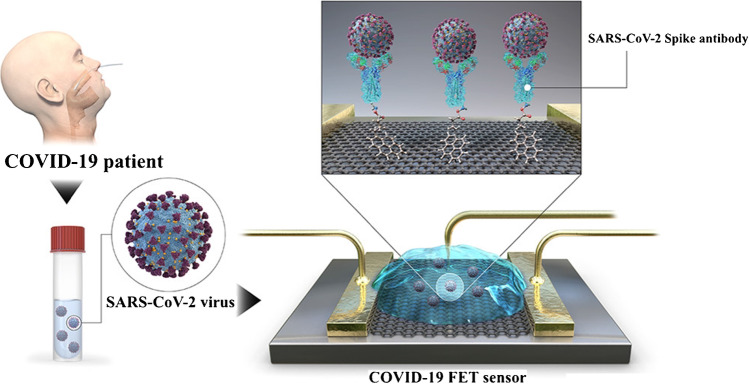
The field-effect transistor (FET) is a kind of transistor used for controlling current flows in a two-dimensional (2D) semiconductor (e.g. black phosphorous (BP), MoS2 and graphene) using an electric field. FETs are 3-terminal devices consisting of a source, a gate and a drain. Applying a voltage to the gateway that affects the conductivity between the drain and the source controls the current flow in FETs. The biosensor for the FET (Bio-FET) consists of a layer of isolation (for example, SiO2), which is selected as a transducer from the biological recognition element to the target molecule. Once charged molecules, such as biomolecules, attach to the FET gate, which is typically made of a dielectric compound, the charge distribution of the underlying semiconductor material changes, resulting in a change in channel conduction [84]. Ease of use, cheap cost and fast response are all advantages of FET sensors, which are monitored in real time with low-cost metres that can be calibrated for various applications. By anchoring particular probes on the conducting channel, FET biosensors may achieve high sensitivity and selectivity for specific biomolecules, which is a crucial factor for FET sensor performance [85]. A Bio-FET is made up of two primary compartments: one for organic recognition and the other for field-effect transistors. For the detection of SARS-CoV-2 in medical samples, a FET-based biosensing device was designed by Kim and co-workers (Fig. 4). The FET graphene sheets were coated with a specific antibody against SARS-CoV-2 spike protein to create the sensor. Antigen protein, cultured virus and nasopharyngeal swab specimens from COVID-19 patients were used to test the sensor’s functionality. At quantities of 1 fg/mL in phosphate-buffered saline and 100 fg/mL in clinical transport medium, the proposed FET device detected the SARS-CoV-2 spike protein. Furthermore, the FET sensor effectively identified SARS-CoV-2 in both culture media (LOD: 1.6 × 101 pfu/mL) and clinical samples (LOD: 2.42 × 102 copies/mL). This device is an immunologically highly sensitive method for diagnosing COVID-19 that does not need pretreatment or labelling [86].
Fig. 4.
Scheme of the COVID-19 field-effect transistor (FET)-based biosensor. Reproduced with permission from [86]
Wearable, microfluidic and smart (nano)biosensors
The development of nanotechnology has led to the production of nanoscale biosensors that are extremely sensitive and adaptable. The purpose of nanosensors is to find biochemical and biophysical signals related to a particular disease at the level of a molecule or cell. Detection of disease-related biomolecules, such as disease-particular metabolites, nucleic acids and proteins, is essential for biomedical research involving drug discovery. Advances in nanotechnology are rapidly improving our current diagnostic biological speed in terms of features, speed and cost by increasing sensitivity and reducing instrument size. The reduced sensor size provides high flexibility for use in multiple, portable, wearable and even implantable medical devices. Biomedical applications of nanosensors are widespread [87].
Customized point of care (POC), without the need of complex tools, offers quick access to health monitoring. Recently, miniaturized biosensors with portable characteristics have been increasingly employed as POC devices for their reliable, sensitive and quick detection [88]. Wearable biosensors deliver continuous and real-time physiological information through dynamic and non-invasive evaluations of biochemical markers in biological liquids such as sweat, tears, saliva and interstitial liquids. The changes, along with electrochemical and optical biosensors, have intensified with the development of non-invasive monitoring of biomarkers such as metabolites, bacteria and hormones. Wearable biosensors are promising, but better acceptance of the relationship between the concentration of analytes in the blood and non-invasive fluids is needed to increase confidence. A wide range of bio-correlation methods in the body and more measurement strategies are required to examine more biomarkers. Accurate and reliable real-time measurement of physiological information using wearable biosensor technologies will have a profound effect on daily life [89].
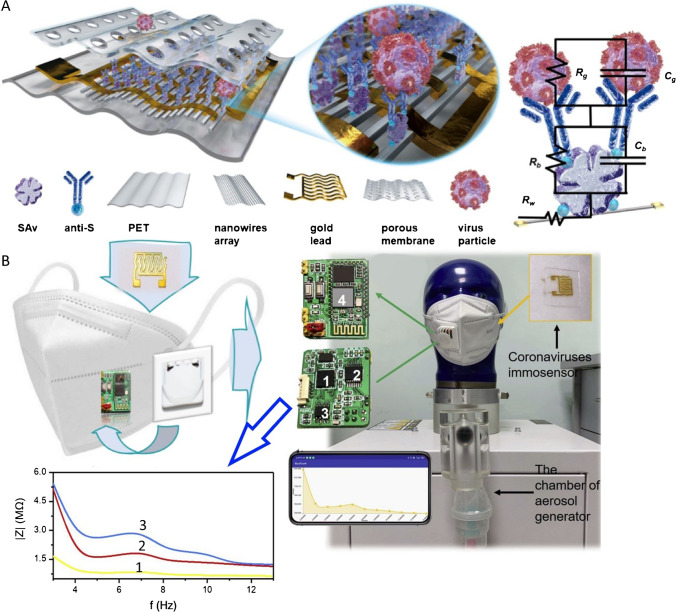
The use of a testing instrument that allows for on-site virus identification could cut down on sample-to-answer time, which is crucial in preventing the pandemic. Xue et al. suggested an intelligent face mask that included a flexible immunosensor based on a high-density conductive nanowire array, a downsized impedance circuit and wireless communication components (Fig. 5). The nanowire arrays’ sub-100 nm size and the distance between neighbouring nanowires make it easier to confine tiny virus particles and boost detection efficiency. In simulated human breath, a point-of-care (POC) device was used to identify coronavirus “spike” protein and complete virus aerosol. In approximately 5 min, viral concentrations as low as 7 pfu/mL were detected from an atomized sample of coronavirus aerosol mimic. The POC systems can be used on-site for basic screening of coronavirus infections and may aid in understanding the course of COVID-19 while a patient is on treatment [90].
Fig. 5.
Schematic representation of A the design nanoscale sensor includes PET, gold lead, porous membrane, nanowires array, anti-S, Sav and virus particle. B Virus sensing mechanism (Rc: the equivalent capacitance of surface binding antibody; Rg: the equivalent resistance of surface binding antigen; Rb: the equivalent resistance of surface binding antibody; Cg: the equivalent capacitance of surface binding antigen; Rw: the equivalent resistance of nanowires). C The images of the constituted intelligent face mask. D The setup of the breath simulation experiment. Reproduced with permission from [90]
A cotton-tipped electrochemical immunosensor for detecting the virus antigen of the SARS-CoV-2 was developed by Eissa et al. [90]. Unlike previously described methods, we combined sample collection and detection instruments on a single platform by coating screen-printed electrodes with absorbent cotton padding. The immunosensor was created by immobilising the viral nucleocapsid (N) protein on carbon nanofiber-modified screen-printed electrodes and functionalizing them with diazonium electro-grafting. Swabbing was used to identify the virus antigen, which was then followed by a competitive assay with a set amount of N protein antibody in the solution. A voltammetric square wave detection technique was utilized. The detection limit was 0.8 pg/mL for SARS-CoV-2, suggesting very excellent sensitivity to the sensor for the electrochemical biosensor. The biosensor displayed no significant over-reactivity, indicating high selectivity with other viral antigens such as influenza A and HCoV. In addition, the biosensor was successfully employed in spike samples, demonstrating good recovery percentages (91–95.5%) for the identification of the virus antigen. Our electrochemical immunosensor is therefore a promising diagnostic instrument to detect the COVID-19 virus directly and quickly and does not require any transfer of samples or pretreatment [91].
In 2021, Zhao et al. described a calixarene functionalized graphene oxide-based ultrasensitive electrochemical detection technology for SARS-CoV-2 RNA targeting. The technology was validated to detect the RNA of SARS-CoV-2 without nucleic acid amplification or reverse transcription using a portable electrochemical smartphone, based on a supersandwich-type recognition strategy. During in silico analysis and actual tests, the biosensor demonstrated high specificity and selectivity. The biosensor discovered a total of 88 RNA extracts from 25 SARS-CoV-2-confirmed patients and eight recovery patients. The detectable ratios were higher than those obtained using RT-qPCR (85.5% and 46.2%, respectively). The clinical specimen’s LOD was 200 copies/mL, the lowest LOD of any SARS-CoV-2 RNA measurement published to date. In addition, each assay only requested two copies (10 L) of SARS-CoV-2. As a result, they designed an ultrasensitive, precise and simple assay for SARS-CoV-2 detection, which could be used as a point-of-care test [92].
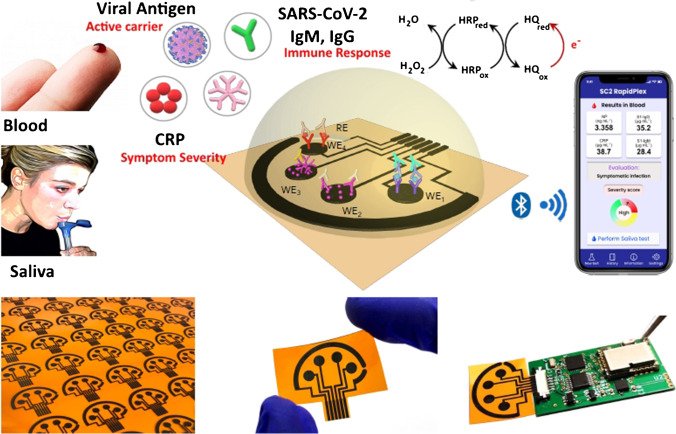
From the perspective of fast, sensitive and selective simultaneous interrogation of viral antigen nucleocapsid protein, S1-IgG and-IgM isotype antibodies and the inflammatory biomarker C-reactive protein (CRP) of serum and saliva biopsies from healthy, and RT-PCR confirmatory COVID-19 patients, Torrente-Rodrigez et al. have designed and implemented the first multiplexed electrochemical graphene platform SARS-CoV-2 RapidPlex (Fig. 6). The integration of advantageous properties in the graphene material and a sensitive and specific immunoassay strategy makes a promising diagnostic device for the SARS-CoV-2 RapidPlex platform for effective sensing of COVID-19 serum infection and non-invasive biofluids without the requirement for complex sample pretreatment. In a single and rapid experiment (target capture would be as low as 1 min), measuring the selected targets can provide detailed data not only on early COVID-19 infections through viral antigen detection and IgM isotype detection, but also on disease severeness, through the assessment of CRP and possible immunity acquired through IgG isotype quantification. The SARS-CoV-2 RapidPlex’s ability to quickly assess illness severity on-site provides the unprecedented benefit of instantaneous COVID-19 triaging. Due to the ease of use, compatibility of saliva samples and the quick time to results, the SARS-CoV-2 RapidPlex platform has a great promise for POC deployment for patient triages, as well as home telemedicine and remote monitoring applications. In addition, the wireless telemedicine diagnostic platform may give full information on a person’s health state during the COVID-19 pandemic when combined with developing wearable biosensors, to continually monitor vital signs and other chemicals. Based on basic well-established methods and sensing principles for surface functionalization processes, it is possible to translate easily to detect additional highly informational SARS-CoV-2 reporters by simply switching the capture reception. Therefore, the designed platform is a high-value test technique to combat current and future pandemics that will contribute to the end of one of the most serious global health, economical and international problems in recent times [93].
Fig. 6.
Schematic illustration of the SARS-CoV-2 RapidPlex multisensor telemedicine platform for detection of SARS-CoV-2 viral proteins. Reproduced with permission from [93]
Mavrikou et al. developed a new SARS-CoV-2 S1 spike protein biosensor for ultrafast detection. This approach is based on Vero mammalian cells which have been electroinserted with the human chimeric spike S1 antibodies. A generic cell-based test principle for the determination of analytes, including biomolecules, is based upon the special and selective interaction between target analytes and cellular element biorecognition, the surfaces of which are modified by electroinserting of target-specific analyses. This approach is known as Molecular Identification by Membrane Engineering. Binding the analytes to the membrane-bound antibodies led to a detectable change in the electrical characteristics, in particular hyperpolarization of the designed cell membrane, for the biorecognition components. Any appropriate bioelectric/bioelectrochemical sensor can be used to monitor these changes. The current work shows that binding the SARS-CoV-2 S1 protein to the corresponding antibody led to a significant and selective change in the membrane-engineered cell characteristics, which were evaluated using a cell-biosensor setup in accordance with the Bioelectric Recognition Assay (BERA) principles. The proposed biosensors had ultrafast results (3 min), an LOD of 1 fg/mL and a semi-linear response interval of 10 fg to 1 µg/mL. Moreover, no cross-reactivity with the SARS-CoV-2 nucleocapsid protein has been reported. Furthermore, the biosensor has been designed as a ready-to-use platform with a portable smartphone/tablet read-out device. Thus, it can be demonstrated that without prior sample processing, the new biosensor may possibly be used to screen masses of SARS-CoV-2 surface antigens such that a potential solution for prompt monitoring and ultimate management of the pandemic can be obtained [94]. In a similar manner, the same researchers used the new biosensor on patient-derived nasopharyngeal samples in a healthcare setting, with no sample preparation. More notably, membrane-engineered cells were pre-immobilized in a unique biomatrix, allowing for long-term storage prior to use and greatly enhancing their ease-of-handling as test consumables. When compared to RT-PCR, the plug-and-play new biosensor detected the virus in positive samples with a 92.8% success rate. There were no false negatives found. These results indicate that the biosensor has the potential to be used for early, routine mass screening of SARS-CoV-2 on a scale that has yet to be achieved [95].
For the precise detection of COVID-19 at POCs, a fast and ultrasensitive microfluidic-based biosensor was developed. COVID-19 was successfully detected using the rolling circle amplification (RCA) method using a nylon mesh media. DNA hydrogels are extensively produced on the surface of the nylon mesh linked to the glass tube by RCA and rapidly and readily restrict the flow channel in the mesh-attached tube. The major benefit of this technique is the small effective surface area between the source (nylon mesh) and the pathogen target, as well as the continual rotation of the magnet bar, which allows the most pathogen molecules to be attached. Furthermore, the amount of DNA hydrogel needed to block the glass tube or channel was lowered since the flow routes through the nylon mesh were substantially reduced due to the high density of the mesh’s micro-hole structures. As a result, the current microfluidic technology can detect SARS-CoV-2 with an exceptional LOD (3 aM in 15 min or 30 aM in 5 min), which is the lowest LOD to date. Furthermore, because the current approach makes use of RCA as a molecular diagnostic tool, it is extremely selective and accurate. The current microfluidic system’s practical implementation may be simply utilized for screening tests at airports and other areas where infectious pathogens are often transmitted. The utilization of a microfluidic platform for simultaneous multiplex detection of different infectious viruses, especially those responsible for influenza A, influenza B and COVID-19, is the subject of continuing study. The examined samples were not clinical samples acquired from diverse biofluids such as nasopharyngeal swabs, mucus and saliva, but synthetic nucleic acid templates, which is a limitation of the current work. For the full credit of the proposal, a clinical assessment must be done with IRB permission and a biosafety facility [96].
Immunoassay
Immunosensors are biosensors based on specific interactions between antibodies and antigens that have medical and bioanalytical applications [97, 98]. In 2021, Mojsoska et al. presented a label-free electrochemical immunoassay for the fast detection of SARS-CoV-2 virus via the spike surface protein as a proof-of-concept. The assay is made up of a graphene working electrode that has been functionalized with anti-spike antibodies. The immunosensor’s principle is to detect signal perturbation derived from ferri/ferrocyanide measurements after antigen binding during 45 min of incubation. The detection range of the spike protein was determined by measuring the absolute change in the [Fe(CN)6]3/4− current as antigen concentrations on the immunosensor surface increased. The sensor detected a special signal above 260 nM (20 g/mL) of recombinant spike protein subunit 1. Furthermore, it detected SARS-CoV-2 at a concentration of 5.5 × 105 PFU/mL, which is within the physiologically important concentration range. The novel immunosensor has a much faster analysis time than standard qPCR and is powered by a portable device, allowing for on-site infection diagnosis [99]. In another work, Rahmati et al. detected SARS-CoV-2-specific viral antibodies using recombinant SARS-CoV-2 spike protein antigen (spike protein) as a special receptor. In this regard, the screen-printed carbon electrode (SPCE) surface was effectively electrodeposited with a layer of nickel hydroxide nanoparticles (Ni(OH)2 NPs) to enable greater loading of spike protein on the SPCE surface. Through a specific and persistent binding response, differential pulse voltammetry (DPV) revealed signals that were inversely proportional to antibody concentrations (from 1 fg/mL to 1 µg/mL). The test took 20 min and had a detection limit of 0.3 fg/mL. Compared to non-specific antibodies, this biological device showed great sensitivity and specificity. In addition, the SARS‐CoV‐2 antibody, which does not require any labelling, might be regarded as a very sensitive immunological diagnostic tool. In order to detect antibodies in real samples of blood serum, the manufactured hand-held bio-device demonstrated an average good recovery rate of about 99–103% for usage with individual quality serological monitoring. Also, the biological device has been evaluated utilizing the enzyme-linked immunosorbent test (ELISA) technique and has demonstrated satisfactory results using genuine patients’ and healthy human samples [100].
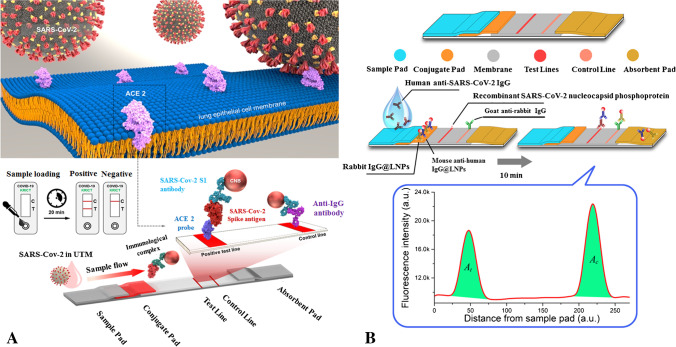
Based on the utilization of the SARS-CoV-2 receptor ACE2 which may form pairs with commercially accessible antibodies, a novel fast detection technique for SARS-CoV-2 spike 1 (S1) protein has been developed. In a lateral flow immunoassay (LFIA), ACE2 and S1-mAb were coupled for capture and detection and did not cross-react with MERS-CoV spike 1 or SARS-CoV spike 1 protein (Fig. 7A). The ACE2-based LFIA has been identified with the SARS-CoV-2 S1 (< 5 ng recombinant protein/reaction). In the clinical specimen COVID-19 patients without cross-responsiveness to nasal swabs from healthy participants, the limits of detection of the ACE2-LFIA were 1.86 × 105 copies/mL. This is the first study to use an LFIA in conjunction with a matched combination of ACE2 and antibody to identify SARS-CoV-2 S1 antigen [101].
Fig. 7.
A The designed ACE2-based LFIA for the detection of SARS-CoV-2 spike 1 antigens [101]. B Lanthanide-doped nanoparticle-based LFIA for sensing of anti-SARS-CoV-2 IgG [102]
In another work, Chen et al. developed a fast and sensitive LFIA that detects anti-SARV-CoV-2 IgG in human serum using lanthanide-doped polystyrene nanoparticles (LNPs). To capture specific IgG, a recombinant nucleocapsid phosphoprotein of SARS-CoV-2 was distributed over a nitrocellulose membrane (Fig. 7B). Self-assembled LNPs that acted as fluorescent reporters were used to label mouse anti-human IgG antibodies. For this test, a 100-L aliquot of blood samples (1:1000 dilution) was utilized, and the entire detection procedure took 10 min. The validation experiment’s results satisfied the criteria for clinical diagnostic reagents. By testing 51 normal samples, a cutoff value of 0.0666 was determined. The tests included 7 reverse transcription (RT-) PCR-positive samples, and 12 negative but clinically suspect anti-SARS-CoV-2 IgG samples. One negative sample was found to be positive for SARS-CoV-2 IgG, whereas the findings were compatible with RT-PCR for the other samples. This test can therefore quickly and sensitively monitor anti-SARS-CoV-2 IgG in human serum, allowing positive identification in suspected cases. Also, it can be used to monitor the advancement of the COVID-19 test and to evaluate the patients’ response to therapy [102].
In order to identify SARS-CoV-2 antibodies (IgG and IgM) rapidly and simultaneously in clinical blood samples within 15 min, the lateral flow combined IgG–IgM immunochromatographic assays are being developed. In samples of blood from COVID-19 hosts, the clinical sensitivity and specificity of the test strips are examined. The sensitivity of this test and its specificity are respectively 85.29% and 100.00%. The combined immunochromatographic IgG-IgM strip test offers greater sensitivity compared to a single IgG and IgM test. The results of this study indicate that the combination IgG–IgM immunochromatographene strip is suitable in confirmed COVID-19 patients, suspicious patients and asymptomatic SARS-CoV-2 carriers for fast screening of SARS-CoV-2 infection [103].
Rashed et al. described a non-faradic capacitive immunoassay test utilizing a commercially available impedance detection device that employs ACEA Biosciences’ customized well plates with integrated sensing electrodes (Fig. 8). The 16-well plate xCELLigence system (RTCA S16) from ACEA Biosciences was designed for non-invasive EIS detection of cell growth, morphological change and attachment quality. Each well comprises an array of interdigitated electrodes that have been carefully developed and fused to polyethylene terephthalate. The plate interface handles each well separately to obtain single frequency measurements (10 kHz) every several seconds. The observed mechanism correlates to binding kinetics between the SARS-CoV-2 spike protein receptor-binding domain (RBD) and the anti-SARS-CoV-2 antibody, allowing SARS-CoV-2 antibodies to be detected effectively. This method yields rapid and consistent results, is lightweight and portable and has the potential to be built up (simultaneous scanning of up to four 384 well plates is possible). As a result, this method is an excellent option for screening serum samples for SARS-CoV-2 antibodies quickly [104].
Fig. 8.
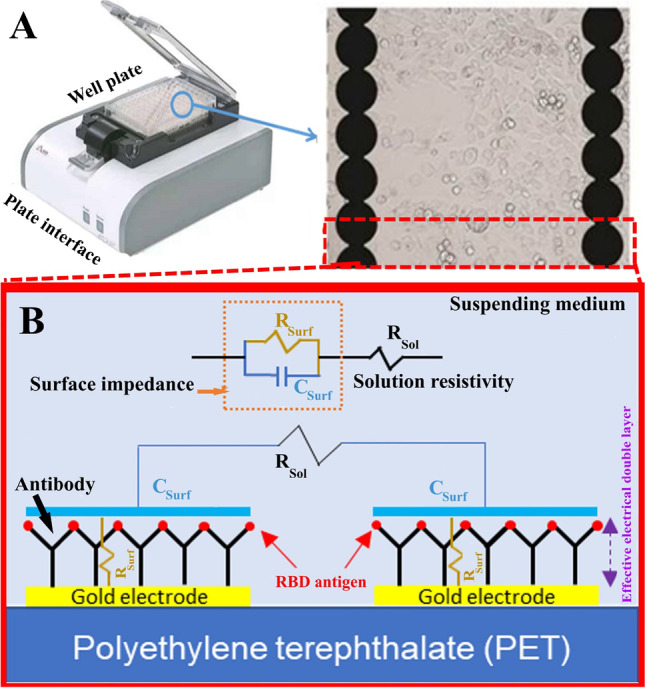
A Image of ACEA Bioscience’s 96-well platform. B Schematic of electrical impedance equivalent circuit model of the protein/antibody in solution. Reproduced with permission from [104]
The monitoring of nucleocapsid and spike antibodies against SARS-CoV-2 in dried blood spots (DBS) and human serum, plasma was reported by Djailleb et al. based on the development of surface plasmin resonance (SPR) sensors and corresponding ELISAs. When exposed to SARS-CoV-2 or SARS-CoV-2 vaccination, the immune system reacts with antibodies at levels to detect and monitor for identifying the portion of the population possibly resistant to SARS-CoV-2 and to assist strategic vaccine deployment efforts. Human anti-SARS-CoV-2 IgG antibodies were identified in clinical samples using an SPR sensor covered with a peptide monolayer and functionalized with several sources of SARS-CoV-2 recombinant proteins produced in different cell lines. The sensitivity of the tests did not alter much when nucleocapsid was produced in various cell lines, but using a CHO cell line to express spike ectodomain resulted in good results. This bioassay was carried out using a portable SPR device that could measure four biological samples in less than 30 min after sample/sensor contact and could be regenerated at least nine times. The in-house and commercial ELISAs were then used to undertake multi-site validation, which demonstrated good cross-correlations with Pearson’s coefficients surpassing 0.85 in all cases for detection plasma and DBS. In the context of viral infection and vaccination effectiveness monitoring, this method paves the door for point-of-care and fast antibody testing [105].
Aptasensing and nucleic acid detection
Aptometrists are biosensors whose biological component is aptamers. Aptamers are single-stranded molecules of DNA or RNA [106]. They have a very high temperature stability compared to antibodies that are made of proteins. These molecules of nucleic acid are obtained in vitro through a selective process called systematic evolution of ligands by exponential enrichment (SELEX) by being placed next to the target molecule [107]. SELEX is a common method currently used for isolating high-affinity single-stranded (ss) DNAs or RNAs from a large library with random sequences. Monitoring the enriching progress of candidate aptamers has always been a critical and constrained stage in the SELEX process. So far, numerous approaches for evaluating library enriching have been tested, including real-time quantitative PCR (qPCR), enzyme-linked oligonucleotide assay (ELONA), dot blotting, electrophoretic mobility shift assay (EMSA) and surface plasmon resonance (SPR) [108]. The steps of the SELEX process are summarized as follows: (1) nucleotide binding to target molecules, (2) separation of the aptamer-target complex from other unbound nucleotides, (3) separation of the aptamer from the target, (4) amplification of the desired aptamer by PCR to modify the properties of selected aptamers and (5) separate the two strands of DNA and reduce the concentration to increase the affinity of the selected aptamers [109].
Aptamers have the ability to bind to a wide range of molecules, containing mineral ions, small organic molecules, proteins, nucleic acids, viruses, bacteria and even cells and tissues [110–115]. Applications for these emerging molecules include use in bioimaging, therapeutic agents, drug delivery systems, disease diagnostics, dietary studies, new drugs and agents for the detection of hazardous substances [116–118]. Aptamers are often recognized as a substitute for antibodies because these molecules have overcome many of the weaknesses of antibodies [119]. Aptamers usually have much lower immunogenicity and toxicity than antibodies due to their acidic nucleic nature. In the case of toxins or molecules with very low immune responses, aptamers can be detected easily and with very high quality.
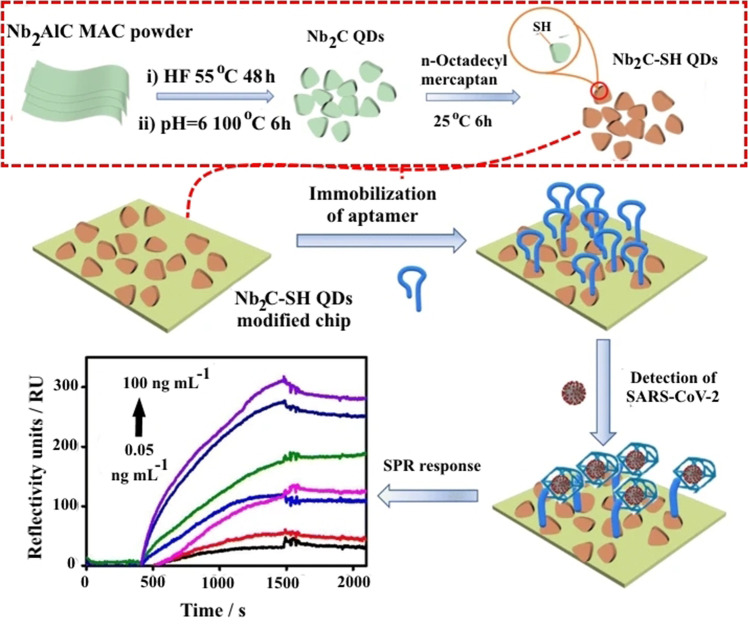
Using thiol-modified niobium carbide MXene quantum dots (Nb2C-SH QDs) as the bioplatform for anchoring N-gene-targeted aptamer, a new label-free surface plasmon resonance (SPR) aptasensor has been developed for the detection of N-gene of SARS-CoV-2. The immobilized aptamer strands altered configuration in the presence of SARS-CoV-2 N-gene to exclusively interact with N-gene. As a result of the increased contact area or increased distance between the aptamer and the SPR chip, the SPR signal irradiated by the laser (He–Ne) with the wavelength (λ) of 633 nm changed. Following that, Nb2C-SH QDs were added to the gold chip for SPR measurements via covalent binding of the Au–S linkage, as well as self-assembling interactions. Nb2C-SH QDs not only increased aptamer bioaffinity but also improved the SPR response. The production of Nb2C-SH QDs and the fabrication of an Nb2C-SH QD-based SPR aptasensor for detecting the SARS-CoV-2 N-gene are depicted in Fig. 9. Within the concentration range of 0.05–100 ng/mL, the Nb2C-SH QD-based SPR aptasensor had an LOD of 4.9 pg/mL for N-gene. The sensor also demonstrated great selectivity and stability in the presence of different respiratory viruses and proteins in human serum. Furthermore, the Nb2C-SH QD-based SPR aptasensor demonstrated a wide range of applications for qualitative N-gene analysis in a variety of samples, including seafood, seawater and human blood. As a result, our research can shed light on how the aptasensor for detecting SARS-CoV-2 in complicated settings is built [120].
Fig. 9.
The scheme of production of Nb2C-SH QDs and the fabrication of an Nb2C-SH QD-based SPR aptasensor for detecting the SARS-CoV-2 N-gene. Reproduced with permission from [120]
In 2021, a voltammetric genosensor has been constructed by Farzin et al. for the early detection of COVID-19 by identifying the RNA-dependent RNA polymerase (RdRP) sequence as a novel coronavirus target. For genome replication and gene transcription, the SARS-CoV-2 employs an RdRP. For the first time, silver ions (Ag+) in hexathia-18-crown-6 (HT18C6) were used as a redox probe. The HT18C6 (Ag) carbon paste electrode (CPE) was modified with chitosan and PAMAM dendrimer-coated silicon quantum dots (SiQDs@PAMAM) for probe sequence immobilization (aminated oligonucleotides). The current intensity of differential pulse voltammetry using the redox probe decreased as the concentration of target sequence increased. Based on this signal-off tendency, the suggested genosensor demonstrated a strong linear reaction to SARS-CoV-2 RdRP in the concentration range of 1.0 pM–8.0 nM, with a regression equation I (A) = 6.555 log [RdRP sequence] (pM) + 32.676 (R2 = 0.995) and a limit of detection (LOD) of 0.3 pM. The standard addition method in human sputum samples with various spike concentrations of the RdRP sequence demonstrated excellent recovery for real sample analysis (> 95%). As a result, the evolved voltammetric genosensor can be used to detect the RdRP sequence of SARS-CoV-2 in sputum samples [121].
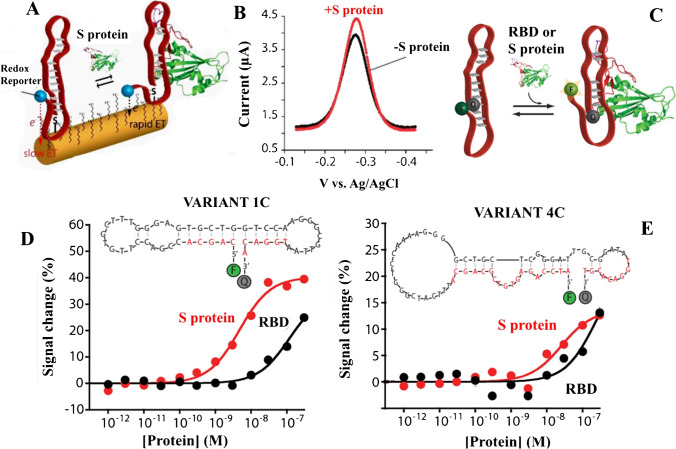
A novel electrochemical aptamer-based (EAB) sensor capable of detecting the SARS-CoV-2 S protein in bodily fluids quickly and effectively has been reported by Idili et al. [122]. EAB sensors are used to create a readily detectable electrochemical signal by binding-induced conformation changes in a covalently attached, redox reporting aptamer. In particular, conformation modification alterations affect the rate at which the redox reporter exchanges electrons with the electrode inquiring (here, we utilized Atto MB2, a derivative of methylene blue), as seen in Fig. 10A, B. Based on the binding-inducing conformational conversions of the recipient, this detection technique leads to directly diluted biological fluids with reagentless, fast and selective assays. In addition, fluorescence spectroscopy evaluated the binding activity of the chosen S-protein aptamers (Fig. 10C). Through the use of optically labelled versions, aptamers can support the conformation mechanism caused by binding, which makes them the ideal option for the EAB sensing platform. In contrast with previous techniques utilizing antibodies and aptamers for detection of SARS-CoV-2 antigens, the resultant EAB sensor exhibited equivalent bioanalytical efficiency. Picomolar concentrations of the S protein were identified in buffer, serum and 50% artificial saliva using the 1C variation (Fig. 10D, E). The results are also extremely encouraging, suggesting that the EAB sensor can distinguish between comparable targets (the receptor-binding domain (RBD) sections of different coronaviruses) and other proteins, in addition to detecting the target in undiluted blood and fake saliva. Moreover, fast binding kinetics and the one-stage operation of the aptamer enable the target to be detected in 15 s, making the EAB sensor perfect for supporting high-frequency tests at the care point. Without a doubt, the potential for EAB sensing to be a legitimate alternative to current POC testing is far ahead of other technologies used for decades (e.g. LFIA and ELISA) in terms of use and market availability. Furthermore, their abilities to support non-calibratory and dual-reporter methods (i.e. to allow the target to be quantified directly), as well as their adaptability, may also be combined with mobile phones or portable electrochemical installations.
Fig. 10.
A Mechanism of electron exchanging in the interrogating electrode. B Single-step detection of S-protein by DPV. C Scheme of fluorophore–quencher couple at the two ends of the aptamer for studying binding activities by fluorescence spectroscopy. D and E Binding activities of the 1C and 4C variants of aptamers labelled with a fluorophore–quencher couple at the two ends against the SARS-CoV-2 RBD (black curves) and the S protein (red curves) in solution. Reproduced with permission from [122]
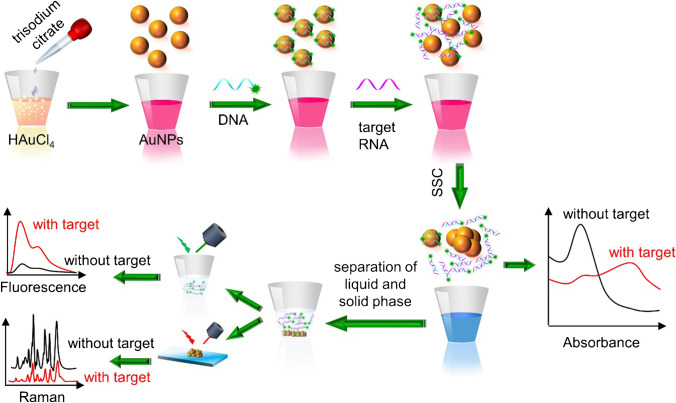
In order to detect special RNAs in SARS-CoV-2 using colorimetric, surface-enhanced Raman scattering (SERS), and fluorescence techniques, Gao et al. [123] developed a three-mode biosensor based on gold nanoparticles (AuNPs). According to their fundamental aggregation property and affinity energy to distinct biomolecules, AuNPs with an average size of 17 nm were produced, and colorimetric, SERS and fluorescence signals of sensors were continuously measured. Citrate-stabilized AuNPs are well distributed against aggregation, as seen in Fig. 11. This is owing to the negative capping agent’s electrostatic repulsion. Salt-induced self-aggregation of AuNPs causes a substantial reduction in absorbance and a noticeable colour shift when saline sodium citrate (SSC) is added. When AuNPs are combined with a certain number of DNA probes, the probes bind to the surface of the AuNPs and protect them against SSC-induced aggregation.
Fig. 11.
A schematic of the triple-mode biosensors used to detect COVID-19 viral RNA. Reproduced with permission from [123]
DNA binds selectively to target RNAs in the presence of target RNAs, producing a DNA-RNA complex that is then released from the surface of AuNPs, causing unprotected AuNPs to congregate. The quantity of target RNA is rendered into a shift and intensity change of the absorbance peak, a change in the Raman signal of AuNPs coupled with residue DNA probes and a variation in the fluorescence intensity of the supernatant. Each operating mode in the suggested biosensor may detect single-base mismatches in the target gene, reducing false positive/negative readings. The suggested biosensor does not need the extraction and purification of viral RNA. The suggested biosensor provides a relatively easy detection method when compared to PCR-based detection. Due to the long-term stability, the reaction solution conjugated to the DNA probe may be stored in the reaction chamber and ready for diagnosis. The operator only needs to load the detection sample and then centrifuge the supernatant to separate it from the aggregated AuNPs after adding the SSC buffer. A photoluminescence system is used to test the fluorescence intensity, and an automatic, portable microplate reader is used to test the absorption spectrum, while a micro-Raman spectrometer is used to record the Raman spectrum. Using 96 or 384 microplates, the proposed detection method can detect 96 or 384 samples simultaneously. In all triple modes, the sensor reaches a femtomole level detection limit of 160 fM in absorbance mode, 259 fM in fluorescence mode and 395 fM in SERS mode. The suggested sensing platform offers a novel method for detecting COVID-19 and other infections that is rapid, sensitive and selective [123].
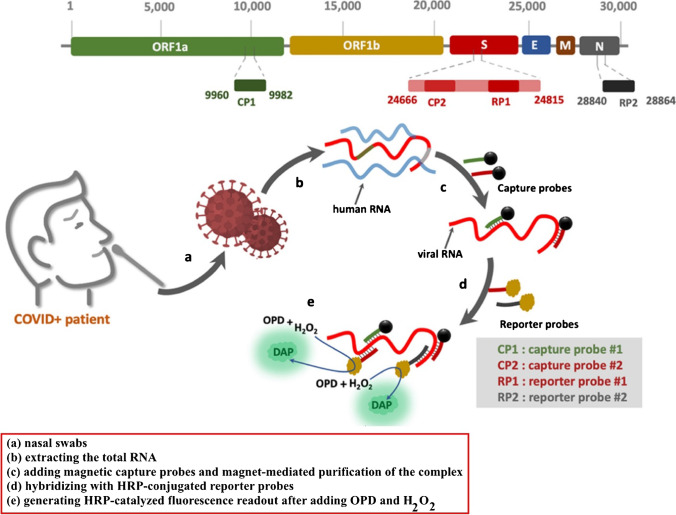
In another work, Zayani et al. described the development of a magnetofluorescent bioplatform to detect SARS-CoV-2 viral RNA directly in total RNA collected from COVID-19-positive patients’ nasopharyngeal swabs. Two capture probes attached to magnetic beads through a biotin/streptavidin linkage, targeting two particular locations in the ORF1a and S genes, yielded a greater fluorescence response (Fig. 12). Through the oxidation of o-phenylenediamine to fluorescent 2,3-diaminophenazine, two horseradish peroxidase (HRP)-conjugated reporter sequences, corresponding to the loci of the S and N genes, were utilized to detect the presence of viral RNA. The bioplatform possesses a linear dynamics range from 0.01 up to 3.0 ng (1 × 103 to 9 × 107 copies/μL) with a low LOD of 0.01 ng of viral RNA (1 × 103 copies/μL) under optimum conditions. This platform is highly selective and sensitive that can distinguish SARS-CoV-2 RNA from similar viruses such as West Nile, hepatitis C, measles and non-polio viruses. In addition, 46 clinical samples verified the proposed biosensor (36 COVID-19-positive and 10 COVID-19-negative samples, as assessed with the gold standard RT-qPCR method). The sensitivity and specificity of the proposed technique achieved 100%. Finally, having such a simple and specific technique available in the field, at a main point of care, can aid in the identification of SARS-CoV-2 infection in resource-limited situations [124].
Fig. 12.
The schematic depicts the step-by-step process for capturing and detecting viral RNA using magnetic probes and HRP-terminated reporters. Reproduced with permission from [124]
Pramanik et al. showed the ability to quickly diagnose, within 10 min, specific SARS-CoV-2 spike recombinant antigen or SARS-CoV-2 spike protein pseudo-type baculovirus using Rhodamine 6G (Rh-6G) dye-coupled DNA aptamer-adhering gold nanostar (GNS) spectroscopy. Because the Rh-6G-attached single-stand DNA aptamer enveloped the GNS, the NSET method quenched 99% of the dye’s fluorescence. The fluorescence signal remains in the presence of spike antigen or virus due to aptamer–spike protein binding. In particular, 130 fg/mL for antigen and 8 particles/mL for virus were established as a limit of detection of the NSET test. Finally, it was proven that GNSs with DNA aptamer may terminate the infection by inhibiting the receptor-binding capacity of angiotensin-converting enzyme 2 (ACE2) and dissolving the virus’ lipid membrane [125].
Conclusions and future prospects
COVID-19 is a serious and dangerous infectious disease with symptoms similar to SARS in the form of fever, cough and fatigue. The disease is mostly transmitted through respiratory droplets and close contact. This disease is a major threat to world health and safety. Bioanalytical methods designed to diagnose COVID-19 disease are superior to other diagnostic methods due to their lower cost, higher accuracy, better detection limit and lower error. The advantages and disadvantages of various biosensing methods used to detect the SARS-CoV-2 virus are given in Table S2. Electrochemical methods can be used in further studies of this disease and similar diseases, and even by simulating the disease using relevant biosensors due to their high response speed, which in addition to helping the advancement of science, also offers the possibility of rapid diagnosis and achievement. It provides the appropriate treatment method, so it is recommended to use the simulation and design of appropriate biosensors to make the necessary predictions to prevent or even diagnose and treat similar emerging diseases that may occur. Nanotechnology has the potential to accelerate the development of unique diagnostic sensors, the integration of novel devices, improved optimization/validation and improvements in sensing performance at the point of care. Future research should focus on developing novel and next-generation non-invasive, specific, inexpensive and quick biosensing techniques and technologies for diagnostic applications, particularly in the management of pandemics and life-threatening infectious illnesses. However, certain difficulties require further investigation and attention. For starters, the majority of these technologies and materials have been studied on a laboratory scale, implying that employing them in real-world circumstances may not be as precise as in the lab [126]. Furthermore, none of these biosensors has yet been developed for detecting the SARS-CoV-2 virus. As a result, the commercialization of numerous efficient biosensors should be hastened. Aside from the approaches and biosensors given, innovative methods such as AI-based technologies, wearable biosensors for continuous public monitoring and single-use disposable sensors for individual testing should be researched for SARS-CoV-2 mass screening [49].
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
Fariba Mollarasouli: writing—original draft, writing—review and editing, software.
Nader Zare-Shehneh: writing—original draft.
Mehrorang Ghaed: writing—review and editing.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fariba Mollarasouli, Email: f.mollarasouli@gmail.com.
Mehrorang Ghaedi, Email: m_ghaedi@yahoo.com.
References
- 1.Xu X, Chen P, Wang J, et al. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farnoosh G, Alishiri G, Zijoud SRH, et al (2020) Understanding the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease (COVID-19) based on available evidence - a narrative review. J Mil Med 22:1–11. 10.30491/JMM.22.1.1
- 3.Chan JF-W, To KK-W, Chen H, Yuen K-Y. Cross-species transmission and emergence of novel viruses from birds. Curr Opin Virol. 2015;10:63–69. doi: 10.1016/j.coviro.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He J, Tao H, Yan Y, et al. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heymann DL. Data sharing and outbreaks: best practice exemplified. Lancet. 2020;395:469–470. doi: 10.1016/S0140-6736(20)30184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Wang X-J. Potential inhibitors against 2019-nCoV coronavirus M protease from clinically approved medicines. J Genet Genomics. 2020;47:119–121. doi: 10.1016/j.jgg.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan R, Zhang Y, Li Y, et al (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science (80- ) 367:1444–1448. 10.1126/science.abb2762 [DOI] [PMC free article] [PubMed]
- 9.Ji W, Wang W, Zhao X, et al. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Read R (2020) Flawed methods in “COVID-19: attacks the 1-beta chain of hemoglobin and captures the porphyrin to inhibit human heme metabolism.” 10.26434/chemrxiv.11938173
- 12.Boccia S, Ricciardi W, Ioannidis JPA. What other countries can learn from Italy during the COVID-19 pandemic. JAMA Intern Med. 2020;180:927. doi: 10.1001/jamainternmed.2020.1447. [DOI] [PubMed] [Google Scholar]
- 13.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/nejmoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh GKM (2017) Viral shapeshifters: strange behaviors of HIV and other viruses. Simplicity Research Institute
- 16.Goh G, Dunker A, Foster J, Uversky V. HIV vaccine mystery and viral shell disorder. Biomolecules. 2019;9:178. doi: 10.3390/biom9050178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou P, Yang X Lou, Wang XG, et al (2020) Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv. 10.1101/2020.01.22.914952
- 19.He J, Tao H, Yan Y, et al (2020) Molecular mechanism of evolution and human infection with the novel coronavirus (2019-nCoV). bioRxiv 2020.02.17.952903. 10.1101/2020.02.17.952903
- 20.Ding N, Zhao K, Lan Y, et al. Induction of atypical autophagy by porcine hemagglutinating encephalomyelitis virus contributes to viral replication. Front Cell Infect Microbiol. 2017;7:56. doi: 10.3389/fcimb.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segars J, Katler Q, McQueen DB, et al. Prior and novel coronaviruses, coronavirus disease 2019 (COVID-19), and human reproduction: what is known? Fertil Steril. 2020;113:1140–1149. doi: 10.1016/j.fertnstert.2020.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kearney JE (2020) Chloroquine as a potential treatment and prevention measure for the 2019 novel coronavirus: a review. J Chem Inf Model 53:1689–1699. 10.20944/preprints202003.0275.v1
- 23.Li X, Geng M, Peng Y, et al. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gundlapally J, Kumar A, Kashyap A, et al (2020) In search of novel coronavirus 19 therapeutic targets. HELIX 10:01–08. 10.29042/2020-10-2-01-08
- 25.Chiu SS, Hung Chan K, Wing Chu K, et al. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin Infect Dis. 2005;40:1721–1729. doi: 10.1086/430301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 27.Zhang N, Wang L, Deng X, et al. Recent advances in the detection of respiratory virus infection in humans. J Med Virol. 2020;92:408–417. doi: 10.1002/jmv.25674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Tai W, Zhou Y, Jiang S. Vaccines for the prevention against the threat of MERS-CoV. Expert Rev Vaccines. 2016;15:1123–1134. doi: 10.1586/14760584.2016.1167603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo PCY, Huang Y, Lau SKP, Yuen K-Y. Coronavirus genomics and bioinformatics analysis Viruses. 2010;2:1804–1820. doi: 10.3390/v2081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson ME, Chen LH (2020) Travellers give wings to novel coronavirus (2019-nCoV). J Travel Med 27: 10.1093/jtm/taaa015 [DOI] [PMC free article] [PubMed]
- 31.Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China. JAMA. 2020;323:1239. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 32.Gralinski LE, Menachery VD. Return of the coronavirus: 2019-nCoV. Viruses. 2020;12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Lu Q, Liu M, et al (2020) Epidemiological and clinical features of the 2019 novel coronavirus outbreak in China. medrxiv. 10.1101/2020.02.10.20021675
- 34.Ye M, Ren Y, Lv T. Encephalitis as a clinical manifestation of COVID-19. Brain Behav Immun. 2020;88:945–946. doi: 10.1016/j.bbi.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Filatov A, Sharma P, Hindi F, Espinosa PS (2020) Neurological complications of coronavirus disease (COVID-19): encephalopathy. Cureus 12: 10.7759/cureus.7352 [DOI] [PMC free article] [PubMed]
- 36.Bikdeli B, Madhavan MV, Jimenez D, et al. COVID-19 and thrombotic or thromboembolic disease: implications for prevention, antithrombotic therapy, and follow-up. J Am Coll Cardiol. 2020;75:2950–2973. doi: 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nickbakhsh S, Ho A, Marques DFP, et al. Epidemiology of seasonal coronaviruses: establishing the context for the emergence of coronavirus disease 2019. J Infect Dis. 2020;222:17–25. doi: 10.1093/infdis/jiaa185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganji A, Farahani I, Khansarinejad B, et al. Increased expression of CD8 marker on T-cells in COVID-19 patients. Blood Cells, Mol Dis. 2020;83:102437. doi: 10.1016/j.bcmd.2020.102437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan S, Yi Q, Fan S, et al (2020) Characteristics of lymphocyte subsets and cytokines in peripheral blood of 123 hospitalized patients with 2019 novel coronavirus pneumonia (NCP). medRxiv 2020.02.10.20021832. 10.1101/2020.02.10.20021832
- 40.Ghazavi A, Ganji A, Khaki M, Mosayebi G (2018) Existential philosophy of the immune system: defense or homeostasis? HBI_Journals 21:110–120
- 41.Wan Y, Shang J, Sun S, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020;94:e02015–e2019. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khaki M, Ghazavi A, Ghasami K, et al (2011) Evaluation of viral antibodies in Iranian multiple sclerosis patients. Neurosci J 16:224 LP – 228 [PubMed]
- 43.Sun J, He W-T, Wang L, et al. COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 2020;26:483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou P, Yang X-L, Wang X-G, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai C-C, Liu YH, Wang C-Y, et al. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J Microbiol Immunol Infect. 2020;53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Organization WH. Water, sanitation, hygiene, and waste management for the COVID-19 virus: interim guidance, 23 April 2020. World Health Organization; 2020. [Google Scholar]
- 47.Tang Y-W, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID-19: current issues and challenges. J Clin Microbiol. 2020;58:e00512–e520. doi: 10.1128/JCM.00512-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Wang Y, Ye D, Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents. 2020;55:105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pishva P, Yüce M (2021) Nanomaterials to tackle the COVID-19 pandemic. Emergent Mater 1–19 [DOI] [PMC free article] [PubMed]
- 50.Eggins B (2002) Analytical techniques in the sciences. John Wiley & Sons, Ltd., Chichester, UK
- 51.Ronkainen NJ, Halsall HB, Heineman WR. Electrochemical biosensors. Chem Soc Rev. 2010;39:1747. doi: 10.1039/b714449k. [DOI] [PubMed] [Google Scholar]
- 52.Turner APF. Biosensors: sense and sensibility. Chem Soc Rev. 2013;42:3184. doi: 10.1039/c3cs35528d. [DOI] [PubMed] [Google Scholar]
- 53.Hashemi SA, Mousavi SM, Bahrani S, et al. Coupled graphene oxide with hybrid metallic nanoparticles as potential electrochemical biosensors for precise detection of ascorbic acid within blood. Anal Chim Acta. 2020;1107:183–192. doi: 10.1016/j.aca.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 54.Yadav N, Chhillar AK, Rana JS. Detection of pathogenic bacteria with special emphasis to biosensors integrated with AuNPs. Sensors Int. 2020;1:100028. doi: 10.1016/j.sintl.2020.100028. [DOI] [Google Scholar]
- 55.Luo X, Morrin A, Killard AJ, Smyth MR. Application of nanoparticles in electrochemical sensors and biosensors. Electroanalysis. 2006;18:319–326. doi: 10.1002/elan.200503415. [DOI] [Google Scholar]
- 56.Yu C-X (2020) Electrochemical biosensors with silver nanoparticles as signal labels. Int J Electrochem Sci 15:3869–3890. 10.20964/2020.05.53
- 57.Zhu C, Yang G, Li H, et al. Electrochemical sensors and biosensors based on nanomaterials and nanostructures. Anal Chem. 2015;87:230–249. doi: 10.1021/ac5039863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J. Carbon-nanotube based electrochemical biosensors: a review. Electroanalysis. 2005;17:7–14. doi: 10.1002/elan.200403113. [DOI] [Google Scholar]
- 59.Tyagi M, Tomar M, Gupta V. NiO nanoparticle-based urea biosensor. Biosens Bioelectron. 2013;41:110–115. doi: 10.1016/j.bios.2012.07.062. [DOI] [PubMed] [Google Scholar]
- 60.Narayanaswamy R, Wolfbeis OS. Optical sensors: industrial environmental and diagnostic applications. Springer; 2004. [Google Scholar]
- 61.Yakoh A, Pimpitak U, Rengpipat S, et al. Paper-based electrochemical biosensor for diagnosing COVID-19: detection of SARS-CoV-2 antibodies and antigen. Biosens Bioelectron. 2021;176:112912. doi: 10.1016/j.bios.2020.112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alafeef M, Dighe K, Moitra P, Pan D. Rapid, ultrasensitive, and quantitative detection of SARS-CoV-2 using antisense oligonucleotides directed electrochemical biosensor chip. ACS Nano. 2020;14:17028–17045. doi: 10.1021/acsnano.0c06392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fabiani L, Saroglia M, Galatà G, et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moerner WE. New directions in single-molecule imaging and analysis. Proc Natl Acad Sci. 2007;104:12596–12602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cox WG, Singer VL. Fluorescent DNA hybridization probe preparation using amine modification and reactive dye coupling. Biotechniques. 2004;36:114–122. doi: 10.2144/04361RR02. [DOI] [PubMed] [Google Scholar]
- 66.Velusamy V, Arshak K, Korostynska O, et al. An overview of foodborne pathogen detection: in the perspective of biosensors. Biotechnol Adv. 2010;28:232–254. doi: 10.1016/j.biotechadv.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 67.Janshoff A, Galla H-J, Steinem C. Piezoelectric mass-sensing devices as biosensors—an alternative to optical biosensors? Angew Chemie. 2000;39:4004–4032. doi: 10.1002/1521-3773(20001117)39:22<4004::AID-ANIE4004>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 68.Mayer KM, Hafner JH. Localized surface plasmon resonance sensors. Chem Rev. 2011;111:3828–3857. doi: 10.1021/cr100313v. [DOI] [PubMed] [Google Scholar]
- 69.Szunerits S, Boukherroub R. Sensing using localised surface plasmon resonance sensors. Chem Commun. 2012;48:8999. doi: 10.1039/c2cc33266c. [DOI] [PubMed] [Google Scholar]
- 70.Moitra P, Alafeef M, Dighe K, et al. Selective naked-eye detection of SARS-CoV-2 mediated by N gene targeted antisense oligonucleotide capped plasmonic nanoparticles. ACS Nano. 2020;14:7617–7627. doi: 10.1021/acsnano.0c03822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Funari R, Chu K-Y, Shen AQ. Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip. Biosens Bioelectron. 2020;169:112578. doi: 10.1016/j.bios.2020.112578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu G, Gai Z, Tao Y, et al. Dual-functional plasmonic photothermal biosensors for highly accurate severe acute respiratory syndrome coronavirus 2 detection. ACS Nano. 2020;14:5268–5277. doi: 10.1021/acsnano.0c02439. [DOI] [PubMed] [Google Scholar]
- 73.Qiu G, Gai Z, Saleh L, et al. Thermoplasmonic-assisted cyclic cleavage amplification for self-validating plasmonic detection of SARS-CoV-2. ACS Nano. 2021;15:7536–7546. doi: 10.1021/acsnano.1c00957. [DOI] [PubMed] [Google Scholar]
- 74.Peng X, Zhou Y, Nie K, et al. Promising near-infrared plasmonic biosensor employed for specific detection of SARS-CoV-2 and its spike glycoprotein. New J Phys. 2020;22:103046. doi: 10.1088/1367-2630/abbe53. [DOI] [Google Scholar]
- 75.Karakuş E, Erdemir E, Demirbilek N, Liv L. Colorimetric and electrochemical detection of SARS-CoV-2 spike antigen with a gold nanoparticle-based biosensor. Anal Chim Acta. 2021;1182:338939. doi: 10.1016/j.aca.2021.338939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.El-Said WA, Al-Bogami AS, Alshitari W. Synthesis of gold nanoparticles@reduced porous graphene-modified ITO electrode for spectroelectrochemical detection of SARS-CoV-2 spike protein. Spectrochim Acta Part A Mol Biomol Spectrosc. 2022;264:120237. doi: 10.1016/j.saa.2021.120237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosensors for detection of pathogenic bacteria. Biosens Bioelectron. 1999;14:599–624. doi: 10.1016/S0956-5663(99)00039-1. [DOI] [PubMed] [Google Scholar]
- 78.Touhami A. Biosensors and nanobiosensors: design and applications. Nanomedicine. 2015;15:374–400. [Google Scholar]
- 79.Erofeev AS, Gorelkin PV, Kolesov DV, et al. Label-free sensitive detection of influenza virus using PZT discs with a synthetic sialylglycopolymer receptor layer. R Soc Open Sci. 2019;6:190255. doi: 10.1098/rsos.190255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang R, Wang L, Callaway ZT, et al. A nanowell-based QCM aptasensor for rapid and sensitive detection of avian influenza virus. Sensors Actuators B Chem. 2017;240:934–940. doi: 10.1016/j.snb.2016.09.067. [DOI] [Google Scholar]
- 81.Alhalaili B, Popescu IN, Kamoun O, et al. Nanobiosensors for the detection of novel coronavirus 2019-nCoV and other pandemic/epidemic respiratory viruses: a review. Sensors. 2020;20:6591. doi: 10.3390/s20226591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kizek R, Krejcova L, Michalek P, et al. Nanoscale virus biosensors: state of the art. Nanobiosensors Dis Diagnosis. 2015;4:47. doi: 10.2147/NDD.S56771. [DOI] [Google Scholar]
- 83.Zuo B, Li S, Guo Z, et al. Piezoelectric immunosensor for SARS-associated coronavirus in sputum. Anal Chem. 2004;76:3536–3540. doi: 10.1021/ac035367b. [DOI] [PubMed] [Google Scholar]
- 84.Taha BA, Al Mashhadany Y, Hafiz Mokhtar MH, et al. An analysis review of detection coronavirus disease 2019 (COVID-19) based on biosensor application. Sensors. 2020;20:6764. doi: 10.3390/s20236764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y, Ren R, Pu H, et al. Field-effect transistor biosensor for rapid detection of Ebola antigen. Sci Rep. 2017;7:10974. doi: 10.1038/s41598-017-11387-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Seo G, Lee G, Kim MJ, et al. Correction to rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:12257–12258. doi: 10.1021/acsnano.0c06726. [DOI] [PubMed] [Google Scholar]
- 87.Kim J, Campbell AS, de Ávila BE-F, Wang J. Wearable biosensors for healthcare monitoring. Nat Biotechnol. 2019;37:389–406. doi: 10.1038/s41587-019-0045-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Purohit B, Kumar A, Mahato K, Chandra P. Smartphone-assisted personalized diagnostic devices and wearable sensors. Curr Opin Biomed Eng. 2020;13:42–50. doi: 10.1016/j.cobme.2019.08.015. [DOI] [Google Scholar]
- 89.Wei Q, Li T, Wang G, et al. Fe3O4 nanoparticles-loaded PEG–PLA polymeric vesicles as labels for ultrasensitive immunosensors. Biomaterials. 2010;31:7332–7339. doi: 10.1016/j.biomaterials.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 90.Xue Q, Kan X, Pan Z, et al. An intelligent face mask integrated with high density conductive nanowire array for directly exhaled coronavirus aerosols screening. Biosens Bioelectron. 2021;186:113286. doi: 10.1016/j.bios.2021.113286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eissa S, Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal Chem. 2021;93:1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 92.Zhao H, Liu F, Xie W, et al. Ultrasensitive supersandwich-type electrochemical sensor for SARS-CoV-2 from the infected COVID-19 patients using a smartphone. Sensors Actuators B Chem. 2021;327:128899. doi: 10.1016/j.snb.2020.128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Torrente-Rodríguez RM, Lukas H, Tu J, et al. SARS-CoV-2 RapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mavrikou S, Moschopoulou G, Tsekouras V, Kintzios S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Sensors. 2020;20:3121. doi: 10.3390/s20113121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mavrikou S, Tsekouras V, Hatziagapiou K, et al. Clinical application of the novel cell-based biosensor for the ultra-rapid detection of the SARS-CoV-2 S1 spike protein antigen: a practical approach. Biosensors. 2021;11:224. doi: 10.3390/bios11070224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim H, Abbas N, Shin S. A rapid diagnosis of SARS-CoV-2 using DNA hydrogel formation on microfluidic pores. Biosens Bioelectron. 2021;177:113005. doi: 10.1016/j.bios.2021.113005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zare-Shehneh N, Mollarasouli F, Ghaedi M (2021) Recent advances in carbon nanostructure-based electrochemical biosensors for environmental monitoring. Crit Rev Anal Chem 1–17 [DOI] [PubMed]
- 98.Mollarasouli F, Kurbanoglu S, Ozkan SA. The role of electrochemical immunosensors in clinical analysis. Biosensors. 2019;9:86. doi: 10.3390/bios9030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mojsoska B, Larsen S, Olsen DA, et al. Rapid SARS-CoV-2 detection using electrochemical immunosensor. Sensors. 2021;21:390. doi: 10.3390/s21020390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rahmati Z, Roushani M, Hosseini H, Choobin H. An electrochemical immunosensor using SARS-CoV-2 spike protein-nickel hydroxide nanoparticles bio-conjugate modified SPCE for ultrasensitive detection of SARS-CoV-2 antibodies. Microchem J. 2021;170:106718. doi: 10.1016/j.microc.2021.106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee J-H, Choi M, Jung Y, et al. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen Z, Zhang Z, Zhai X, et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG, using lanthanide-doped nanoparticles-based lateral flow immunoassay. Anal Chem. 2020;92:7226–7231. doi: 10.1021/acs.analchem.0c00784. [DOI] [PubMed] [Google Scholar]
- 103.Zeng L, Li Y, Liu J, et al. Rapid, ultrasensitive and highly specific biosensor for the diagnosis of SARS-CoV-2 in clinical blood samples. Mater Chem Front. 2020;4:2000–2005. doi: 10.1039/D0QM00294A. [DOI] [Google Scholar]
- 104.Rashed MZ, Kopechek JA, Priddy MC, et al. Rapid detection of SARS-CoV-2 antibodies using electrochemical impedance-based detector. Biosens Bioelectron. 2021;171:112709. doi: 10.1016/j.bios.2020.112709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Djaileb A, HojjatJodaylami M, Coutu J, et al. Cross-validation of ELISA and a portable surface plasmon resonance instrument for IgG antibody serology with SARS-CoV-2 positive individuals. Analyst. 2021;146:4905–4917. doi: 10.1039/D1AN00893E. [DOI] [PubMed] [Google Scholar]
- 106.Famulok M, Szostak JW. In vitro selection of specific ligand-binding nucleic acids. Angew Chemie Int Ed English. 1992;31:979–988. doi: 10.1002/anie.199209791. [DOI] [Google Scholar]
- 107.Stoltenburg R, Reinemann C, Strehlitz B. SELEX—a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 108.Liu Q, Zhang W, Chen S, et al. SELEX tool: a novel and convenient gel-based diffusion method for monitoring of aptamer-target binding. J Biol Eng. 2020;14:1–13. doi: 10.1186/s13036-019-0223-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Peyrin E. Nucleic acid aptamer molecular recognition principles and application in liquid chromatography and capillary electrophoresis. J Sep Sci. 2009;32:1531–1536. doi: 10.1002/jssc.200900061. [DOI] [PubMed] [Google Scholar]
- 110.Radi A-E, O’Sullivan CK (2006) Aptamer conformational switch as sensitive electrochemical biosensor for potassium ion recognition. Chem Commun 3432. 10.1039/b606804a [DOI] [PubMed]
- 111.Tang Z, Parekh P, Turner P, et al. Generating aptamers for recognition of virus-infected cells. Clin Chem. 2009;55:813–822. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen HW, Medley CD, Sefah K, et al. Molecular recognition of small-cell lung cancer cells using aptamers. ChemMedChem. 2008;3:991–1001. doi: 10.1002/cmdc.200800030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhen SJ, Huang CZ, Wang J, Li YF. End-to-end assembly of gold nanorods on the basis of aptamer−protein recognition. J Phys Chem C. 2009;113:21543–21547. doi: 10.1021/jp905084e. [DOI] [Google Scholar]
- 114.Li X, Zhang W, Liu L, et al. In vitro selection of DNA aptamers for metastatic breast cancer cell recognition and tissue imaging. Anal Chem. 2014;86:6596–6603. doi: 10.1021/ac501205q. [DOI] [PubMed] [Google Scholar]
- 115.Brody EN, Gold L. Aptamers as therapeutic and diagnostic agents. Rev Mol Biotechnol. 2000;74:5–13. doi: 10.1016/S1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 116.Wu X, Hu J, Zhu B, et al. Aptamer-targeted magnetic nanospheres as a solid-phase extraction sorbent for determination of ochratoxin A in food samples. J Chromatogr A. 2011;1218:7341–7346. doi: 10.1016/j.chroma.2011.08.045. [DOI] [PubMed] [Google Scholar]
- 117.Bouchard PR, Hutabarat RM, Thompson KM. Discovery and development of therapeutic aptamers. Annu Rev Pharmacol Toxicol. 2010;50:237–257. doi: 10.1146/annurev.pharmtox.010909.105547. [DOI] [PubMed] [Google Scholar]
- 118.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. doi: 10.1093/clinchem/45.9.1628. [DOI] [PubMed] [Google Scholar]
- 119.Priyanka SM, Bhalla V, et al. Nanobioprobe mediated DNA aptamers for explosive detection. Chem Commun. 2014;50:1080. doi: 10.1039/c3cc47562j. [DOI] [PubMed] [Google Scholar]
- 120.Chen R, Kan L, Duan F, et al. Surface plasmon resonance aptasensor based on niobium carbide MXene quantum dots for nucleocapsid of SARS-CoV-2 detection. Microchim Acta. 2021;188:316. doi: 10.1007/s00604-021-04974-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Farzin L, Sadjadi S, Sheini A, Mohagheghpour E. A nanoscale genosensor for early detection of COVID-19 by voltammetric determination of RNA-dependent RNA polymerase (RdRP) sequence of SARS-CoV-2 virus. Microchim Acta. 2021;188:121. doi: 10.1007/s00604-021-04773-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Idili A, Parolo C, Alvarez-Diduk R, Merkoçi A (2021) Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sensors acssensors.1c01222. 10.1021/acssensors.1c01222 [DOI] [PMC free article] [PubMed]
- 123.Gao Y, Han Y, Wang C, et al. Rapid and sensitive triple-mode detection of causative SARS-CoV-2 virus specific genes through interaction between genes and nanoparticles. Anal Chim Acta. 2021;1154:338330. doi: 10.1016/j.aca.2021.338330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zayani R, Rezig D, Fares W, et al. Multiplexed magnetofluorescent bioplatform for the sensitive detection of SARS-CoV-2 viral RNA without nucleic acid amplification. Anal Chem. 2021;93:11225–11232. doi: 10.1021/acs.analchem.1c01950. [DOI] [PubMed] [Google Scholar]
- 125.Pramanik A, Gao Y, Patibandla S, et al. Aptamer conjugated gold nanostar-based distance-dependent nanoparticle surface energy transfer spectroscopy for ultrasensitive detection and inactivation of corona virus. J Phys Chem Lett. 2021;12:2166–2171. doi: 10.1021/acs.jpclett.0c03570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Iravani S. Nano- and biosensors for the detection of SARS-CoV-2: challenges and opportunities. Mater Adv. 2020;1:3092–3103. doi: 10.1039/D0MA00702A. [DOI] [Google Scholar]
- 127.Wang N, Shang J, Jiang S, Du L. Subunit vaccines against emerging pathogenic human coronaviruses. Front Microbiol. 2020;11:298. doi: 10.3389/fmicb.2020.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Song Z, Xu Y, Bao L, et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.