Abstract
PCR amplification of the recA gene followed by restriction fragment length polymorphism (RFLP) analysis was investigated for the rapid detection and identification of Burkholderia cepacia complex genomovars directly from sputum. Successful amplification of the B. cepacia complex recA gene from cystic fibrosis (CF) patient sputum samples containing B. cepacia genomovar I, Burkholderia multivorans, B. cepacia genomovar III, Burkholderia stabilis, and Burkholderia vietnamiensis was demonstrated. In addition, the genomovar identifications determined directly from sputum were the same as those obtained after selective culturing. Sensitivity experiments revealed that recA-based PCR could reliably detect B. cepacia complex organisms to concentrations of 106 CFU g of sputum−1. To fully assess the diagnostic value of the method, sputum samples from 100 CF patients were screened for B. cepacia complex infection by selective culturing and recA-based PCR. Selective culturing identified 19 samples with presumptive B. cepacia complex infection, which was corroborated by phenotypic analyses. Of the culture-positive sputum samples, 17 were also detected directly by recA-based PCR, while 2 samples were negative. The isolates cultured from both recA-negative sputum samples were subsequently identified as Burkholderia gladioli. RFLP analysis of the recA amplicons revealed 2 patients (12%) infected with B. multivorans, 11 patients (65%) infected with B. cepacia genomovar III-A, and 4 patients (23%) infected with B. cepacia genomovar III-B. These results demonstrate the potential of recA-based PCR-RFLP analysis for the rapid detection and identification of B. cepacia complex genomovars directly from sputum. Where the sensitivity of the assay proves a limitation, sputum samples can be analyzed by selective culturing followed by recA-based analysis of the isolate.
Patients with cystic fibrosis (CF) are extremely susceptible to pulmonary infection with a range of bacterial flora (29). Over the last 20 years, Burkholderia cepacia has emerged as an opportunistic microbial pathogen in patients with CF as well as immunocompromised patients without CF (13, 25). B. cepacia can be transmitted between patients, is frequently resistant to a wide range of antimicrobial treatments, and produces an increase in pulmonary symptoms and a decrease in long-term survival (9, 10, 27, 28). In addition, approximately 20% of all CF patients infected with B. cepacia succumb to cepacia syndrome, a necrotizing pneumonia with bacteremia which leads to an acute and frequently fatal clinical decline (17). In response to these serious problems, CF centers now segregate patients so that cross-infection with the organism is reduced (12, 20).
The taxonomy of B. cepacia has proved to be very complex. Initial phylogenetic investigations demonstrated that isolates previously classified as B. cepacia comprised at least five genotypically distinct genomovars, collectively referred to as the B. cepacia complex (30). B. cepacia genomovar V has been identified as Burkholderia vietnamiensis, a nitrogen-fixing organism associated with rice roots (11), while B. cepacia genomovar II and genomovar IV have been proposed as the new species Burkholderia multivorans and Burkholderia stabilis, respectively (30, 31). At present, B. cepacia genomovar III awaits assignment of a binomial species name pending the availability of suitable phenotypic identification criteria. Strains of B. cepacia genomovar I (which contains the type strain) will be known as B. cepacia when taxonomic reappraisal is complete. Very recently, the description of B. cepacia genomovar VI and genomovar VII (also known as Burkholderia ambifaria sp. nov.) as members of the B. cepacia complex has further demonstrated the extraordinary diversity of this group of organisms (6, 7, 16).
Ribotyping of serial B. cepacia complex strains has revealed that CF patients are infected and colonized with a single genomovar strain (3, 22). Although all species of the B. cepacia complex have been cultured from CF patients, the majority of infections result from strains of B. cepacia genomovar III and B. multivorans (21, 30). B. cepacia complex organisms also show differences with respect to transmissibility and virulence, with strains of B. cepacia genomovar III being responsible for most epidemic outbreaks as well as cases of cepacia syndrome (8, 30).
Knowledge of the B. cepacia complex genomovar species responsible for pulmonary infections is extremely important for appropriate segregation and grouping of CF patients into cohorts. We routinely use the recA-based diagnostic scheme recently described by Mahenthiralingam et al. (23) to identify B. cepacia complex isolates. Particular advantages of this multifaceted approach are its capacity to identify all genomovars of the B. cepacia complex and to differentiate B. cepacia genomovar III isolates into the two distinct recA cluster groups, known as III-A and III-B. This diagnostic approach provides the clinical microbiologist with a variety of experimental methods to identify genomovar-specific polymorphisms within B. cepacia complex isolates. These include restriction fragment length polymorphism (RFLP) analysis of the PCR-amplified recA gene, the use of genomovar-specific recA primers, and direct nucleotide sequencing of recA. Due to the flexibility of the approach, these tests can be used individually or, if desired, can be applied for multiple complementary analyses. We now describe the novel application of recA-based PCR-RFLP analysis for the rapid detection and identification of B. cepacia complex genomovars directly from CF sputum samples. The ability, using a single PCR, to both detect and differentiate all members of the B. cepacia complex in sputum may prove particularly valuable for diagnostic laboratories.
MATERIALS AND METHODS
Bacterial strains.
Table 1 lists the 25 B. cepacia complex reference strains used for this study, as well as their original sources of isolation. The organisms were obtained from the Belgium Coordinated Collections of Microorganisms/Laboratorium voor Microbiologie Ghent located at the University of Ghent (http://www.belspo.be/bccm/), the Canadian B. cepacia Strain Repository located at the University of British Columbia (16), and the American Type Culture Collection (Manassas, Va.) (http://www.atcc.org/home.cfm). The strains were selected to represent the different genomovar species of the B. cepacia complex, as previously determined by genotypic and phenotypic analyses (23, 30). Archived bacterial strains of other organisms found in CF patients, namely, Burkholderia gladioli, Ralstonia pickettii, Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia, and Haemophilus influenzae, were isolated from the sputum of adult patients attending the CF clinic at Belfast City Hospital. All organisms were stored at −70°C in defibrinated horse blood (E & O Laboratories, Bonnybridge, Scotland).
TABLE 1.
B. cepacia complex strains used to establish genomovar-specific RFLP patterns
| Reference straina | B. cepacia complex genomovar (species) | Original source of isolateb | Result of:
|
||
|---|---|---|---|---|---|
| recA-based PCR with primers BCR1 and BCR2c |
recA-based RFLP
|
||||
| HaeIIId | MnlIe | ||||
| LMG1222 | I | Allium cepa I | + | D | d |
| LMG17997 | I | Urinary tract | + | E | e |
| CEP509 | I | CF (Australia) | + | E | e |
| C5393 | II (B. multivorans) | CF (Canada) | + | F | a |
| C1576 | II (B. multivorans) | CF-e (UK) | + | C | a |
| LMG13010 | II (B. multivorans) | CF (Belgium) | + | F | a |
| LMG14273 | II (B. multivorans) | CF (Belgium) | + | F | j |
| ATCC 17616 | II (B. multivorans) | Soil | + | F | a |
| LMG12614 | III-A | CF-e (UK) | + | G | f |
| LMG12615 | III-A | CF-e (UK) | + | G | f |
| C5424 | III-A | CF-e (Canada) | + | G | f |
| C6433 | III-A | CF-e (Canada) | + | G | f |
| C1394 | III-B | CF-e (UK) | + | H | h |
| PC184 | III-B | CF-e (US) | + | J | i |
| CEP511 | III-B | CF-e (Australia) | + | I | h |
| C7322 | IV (B. stabilis) | CF (Canada) | + | J | b |
| C6061 | IV (B. stabilis) | CF (Canada) | + | J | b |
| LMG14294 | IV (B. stabilis) | CF (Belgium) | + | J | b |
| LMG14940 | IV (B. stabilis) | CF (Belgium) | + | J | b |
| LMG18888 | IV (B. stabilis) | Human blood | + | J | b |
| LMG16230 | V (B. vietnamiensis) | CF (Sweden) | + | A | c |
| LMG16232 | V (B. vietnamiensis) | CF (Sweden) | + | A | c |
| LMG10929 | V (B. vietnamiensis) | Rice rhizosphere | + | B | c |
| FC441 | V (B. vietnamiensis) | CGD (Canada) | + | A | c |
| C2822 | V (B. vietnamiensis) | CF (Canada) | + | B | c |
LMG, culture collection from Laboratorium voor Microbiologie, University of Ghent, Ghent, Belgium; ATCC, American Type Culture Collection; all other strains were from the Canadian B. cepacia Strain Repository.
CF, patient with CF; CF-e, patient with CF epidemic infection; CGD, patient with chronic granulomatous disease. UK, United Kingdom; US, United States.
+, positive result.
Letters correspond to alphabetical RFLP types as shown in Fig. 1A.
Letters correspond to alphabetical RFLP types as shown in Fig. 1B.
Study population.
For our clinical study, sputum samples were collected from patients attending the Bradbury Adult Cystic Fibrosis Center, Wythenshawe Hospital, Manchester, England. This group consisted of 100 adults (53 males; 47 females) with an age range of 17 to 50 years and a mean age of 25 years.
Processing of sputum samples.
Expectorated sputum samples were collected postphysiotherapy and physically mixed with an equal amount of fresh Sputolysin (Calbiochem, La Jolla, Calif.) before incubation at 37°C for 30 min.
Culturing of organisms.
Before analysis, B. cepacia complex reference strains were removed from storage and grown to confluence at 37°C for 48 h on Columbia agar (Oxoid Ltd., Basingstoke, England) supplemented with 5% (vol/vol) horse blood (E & O Laboratories) (blood agar). B. cepacia complex organisms in patient sputum samples were isolated by culturing at 37°C for 48 h on MAST selective agar (MAST Diagnostics, Liverpool, England). Culturing of organisms in nutrient broth (Oxoid) was performed overnight at 37°C.
Phenotypic analysis.
Phenotypic analysis was performed using the multitest API 20NE identification system (bioMèrieux, Marcy l'Etoile, France) in accordance with the manufacturer's instructions.
Preparation of template DNA from bacterial cultures.
Fresh cultures of the bacterial strains were suspended in 1 ml of 10 mM Tris-HCl buffer (pH 8.0) containing 1 mM EDTA (TE buffer) and centrifuged at 10,000 × g for 10 min. Supernatants were removed, and the resulting pellets of bacteria were resuspended in 0.2 ml of TE buffer. Genomic DNA was prepared using a high-purity PCR template kit (Roche Molecular Biochemicals, Lewes, England). In brief, samples were treated with 150 μg of lysozyme (Sigma-Aldrich) and incubated at 37°C for 30 min followed by incubation with 1 mg of proteinase K at 72°C for 10 min. Template DNA was precipitated with 100 μl of isopropanol and recovered from the samples by centrifugation in a membrane filter attached to an underlying collection tube. The DNA was then washed in 2 mM Tris-HCl (pH 7.5) containing 20 mM NaCl and 80% (vol/vol) ethanol before elution in 10 mM Tris (pH 8.5). Control samples consisting of 0.2 ml of sterile water (Biowhittaker, Walkersville, Md.) in place of the DNA samples were run in parallel. Successful isolation of bacterial genomic DNA was confirmed by electrophoresis in 0.7% (wt/vol) agarose gels (Life Technologies GIBCO BRL Products, Paisley, Scotland). The quantity and purity of the bacterial genomic DNA were assessed by measuring the absorbances at 260 and 280 nm.
Preparation of template DNA from CF patient sputum samples.
Liquefied (Sputolysin-treated) sputum (1 ml) was centrifuged at 10,000 × g for 10 min. The resulting pellet was resuspended in 0.2 ml of TE buffer. Bacterial cells were fractured by snap-freezing in liquid nitrogen for 3 min followed by heating at 100°C for 1 min (34). This step was repeated three more times. Samples were also treated with 150 μg of lysozyme at 37°C for 30 min to ensure complete bacterial cell lysis. After treatment with proteinase K and precipitation with isopropanol, DNA was purified as described for bacterial cultures.
PCR analysis.
PCR analysis was performed with a DNA thermal cycler (Cetus GeneAmp 9600; Perkin-Elmer Applied Biosystems, Foster City, Calif.). The B. cepacia complex recA gene (1,040 bp) was amplified using primers BCR1 and BCR2 (Table 2), which target the 5′ and 3′ ends of the recA gene locus, respectively (23). PCRs were performed with a total volume of 50 μl. Samples contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 200 μM each deoxynucleoside triphosphate (Amersham-Pharmacia Biotech, Little Chalfont, England), 150 nM each BCR1 and BCR2, 1.5 mM MgCl2, 5% (vol/vol) dimethyl sulfoxide (DMSO) (Sigma-Aldrich), 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer), and 100 ng of pure genomic DNA or 5 μl of sputum DNA. Samples were initially heated at 96°C for 3 min before amplification of the recA sequence using 35 cycles consisting of 1 min of denaturation at 96°C, 1 min of annealing at 56°C, and 1.5 min of extension at 72°C. The PCR was completed with a final extension step at 72°C for 10 min.
TABLE 2.
Primers used for the detection and identification of B. cepacia complex genomovars in CF sputum samples
| Primer | Sequence (5′ to 3′) | Specificity |
|---|---|---|
| BCR1a | TGA CCG CCG AGA AGA GCA A | recA gene—B. cepacia complex |
| BCR2a | CTC TTC TTC GTC CAT CGC CTC | recA gene—B. cepacia complex |
| PSL1b | AAC TAG TTG TTG GGG ATT CAT TTC | 16S rRNA—B. cepacia |
| PSR1b | TTT CGA GCA CTC CCG CCT CTC AG | 16S rRNA—B. cepacia |
| G1c | TCG GAA TCC TGC TGA GAG GC | 16S rRNA—genomovars I, III, and IV |
| G2c | GCC ATG GAT ACT CCA AAA GGA | 23S rRNA—genomovars I, III, and IV |
| BC-GIId | AGG CGG TCT GTT AAG ACA | 16S rRNA—genomovar II |
| BC-GVd | TAA TAC CGC ATA CGA TCT AT | 16S rRNA—genomovar V |
| BC-Rd | AGC ACT CCC GAA TCT CTT | 16S rRNA—genomovars II and V |
| PSLb | AGG ATT AGA TAC CCT GGT AGT CCA | 16S rRNA—all bacteria |
| PSRb | ACT TAA CCC AAC ATC TCA CGA CAC | 16S rRNA—all bacteria |
Primers described by Mahenthiralingam et al. (23).
Primers described by Campbell et al. (4).
Primers described by Whitby et al. (34); also known as PC-SSF and PC-SSR (21).
Primers described by LiPuma et al. (21).
The recA-based detection of B. cepacia complex organisms in sputum was also compared with rRNA-based PCR assays previously described by Campbell et al. (4) (primers PSL1 and PSR1, originally designed for B. cepacia), Whitby et al. (34) (primers G1 and G2 for B. cepacia genomovars I and III and B. stabilis as a group), and LiPuma et al. (21) (B. multivorans primers BC-GII and BC-R and B. vietnamiensis primers BC-GV and BC-R). To confirm successful extraction of bacterial DNA from sputum, primers PSL and PSR (which target 16S ribosomal DNA [rDNA] sequences of all bacteria) were used (4). See Table 2 for all 16S and 16S-23S rDNA primer sequences.
RFLP analysis of the B. cepacia complex recA gene.
For RFLP analysis, B. cepacia complex recA amplicons were digested with HaeIII (Amersham-Pharmacia Biotech, St. Albans, England) and MnlI (New England Biolabs Inc., Hitchin, England) restriction endonucleases (23). Amplicons (5 to 15 μl) were added to the endonuclease along with the appropriate enzyme buffer, in accordance with the manufacturer's instructions, before incubation at 37°C for 2 h. When required, amplicons were concentrated using a QIAquick PCR purification kit (Qiagen Inc.; http://www.qiagen.com) to help increase the intensity of the digested recA DNA fragments. RFLP patterns were analyzed as described previously (23).
Detection of PCR and RFLP products.
PCR-amplified products were routinely analyzed by electrophoresis in 2% (wt/vol) agarose gels (Life Technologies GIBCO BRL Products) containing 40 mM Tris buffer (pH 8.0) and 20 mM acetate. Restriction fragments were resolved in 3% (wt/vol) high-resolution Multiphore agarose gels (Flowgen, Lichfield, England). Molecular size markers (100-bp ladder; Life Technologies GIBCO BRL Products) were run in parallel on all gels. Resolved DNA products were stained with ethidium bromide and viewed under UV light. For clarity, the specific RFLP patterns obtained for strains of the different genomovars were classified as previously reported (23).
DNA sequence analysis.
PCR amplicons were sequenced on an ABI PRISM apparatus (Perkin-Elmer) using DyeDeoxy Terminator chemistry and AmpliTaq FS DNA polymerase in accordance with the manufacturer's instructions. Nucleotide sequences were compared with previously published sequences using a basic local alignment sequence tool (BLAST) (1).
Detection limits.
To determine the minimum number of B. cepacia complex organisms detectable in sputum using recA-based PCR amplification, Sputolysin-treated sputum from a CF patient without B. cepacia complex infection was inoculated with known concentrations of selected reference strains serially diluted in one-quarter-strength Ringer's solution (Oxoid). Experiments were performed using B. cepacia genomovar III-A strain C5424, B. cepacia genomovar III-B strain CEP511, and B. multivorans strains C5393 and LMG13010. These strains were selected because they represent the most prevalent genomovars recovered from patients with CF (21, 30). The number of organisms added to each sputum sample was determined by colony counts on blood agar plates inoculated in parallel. Cultures were incubated for 48 h at 37°C before enumeration. DNA was extracted from the inoculated sputum samples and analyzed by PCR as described earlier. Results were expressed as the number of CFU gram of sputum−1.
RESULTS
recA RFLP reference panel for B. cepacia complex genomovar identification.
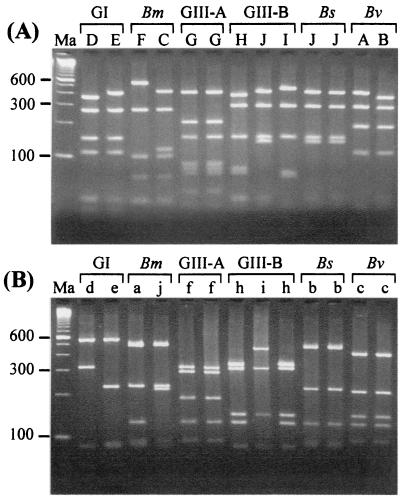
With primers BCR1 and BCR2, the 1,040-bp recA gene product was successfully amplified from the genomic DNAs of all 25 control organisms (Table 1). The recA amplicons from a sample of eight strains, selected to represent all genomovars of our reference panel, were sequenced, and their identities were confirmed by BLAST analysis (data not shown). A reference panel of genomovar-specific RFLP patterns was created using restriction endonucleases HaeIII and MnlI (Fig. 1A and B, respectively, and Table 1) as previously described (23).
FIG. 1.
RFLP analysis of the 1-kb recA gene amplified from strains of B. cepacia genomovar I (GI), B. multivorans (Bm), B. cepacia genomovar III (GIII), B. stabilis (Bs), and B. vietnamiensis (Bv). (A) HaeIII RFLP analysis. The alphabetical RFLP types, along with their genomovar status, are shown above the lanes. Lanes (left to right): Ma, molecular size markers (100-bp ladder); D, LMG1222; E, CEP509; F, C5393; C, C1576; G, LMG12614; G, C5424; H, C1394; J, PC184; I, CEP511; J, C7322; J, LMG14294; A, LMG16230; and B, C2822. (B) MnlI RFLP analysis. The alphabetical RFLP types, along with their genomovar status, are shown above the lanes. Lanes are as described for panel A, except for C1576, which was replaced with LMG14273.
Specificity of primers BCR1 and BCR2.
Since samples of sputum from CF patients contain a range of different bacterial flora, it was important to demonstrate that primers BCR1 and BCR2 did not react with other organisms commonly found in such samples. No recA amplicons were produced when the primers were tested against genomic DNAs prepared from B. gladioli, R. pickettii, P. aeruginosa, S. aureus, S. maltophilia, and H. influenzae isolates or sputum samples from 10 adult CF patients without B. cepacia complex infection.
recA-based PCR-RFLP detection and identification of B. cepacia complex genomovars in sputum.
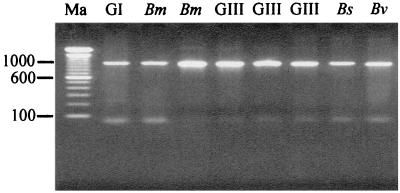
To investigate if the B. cepacia complex recA gene could be successfully amplified from sputum DNA preparations, 15 samples from CF patients infected with organisms of the B. cepacia complex were analyzed. These sputum samples contained B. cepacia genomovar I (n = 1), B. multivorans (n = 4), B. cepacia genomovar III (n = 7), B. stabilis (n = 1), and B. vietnamiensis (n = 2). Upon PCR analysis with recA primers BCR1 and BCR2, the 1,040-bp recA gene was successfully amplified from all the samples, demonstrating that recA-based detection of B. cepacia complex organisms directly from sputum DNA was possible. Figure 2 shows the recA gene product amplified from the sputum of patients infected with different B. cepacia complex genomovars. The amplified bands were clear and sharp, with no background reaction due to nonspecific binding of the primers. PCR amplification of the recA gene from sputum DNA also proved very reproducible.
FIG. 2.
PCR amplification of the 1-kb B. cepacia complex recA gene from the sputum of CF patients infected with genomovar I (GI), B. multivorans (Bm), genomovar III (GIII), B. stabilis (Bs), and B. vietnamiensis (Bv). The genomovar status of each sample is shown above each lane. Molecular size markers (100-bp ladder) were run in lane Ma.
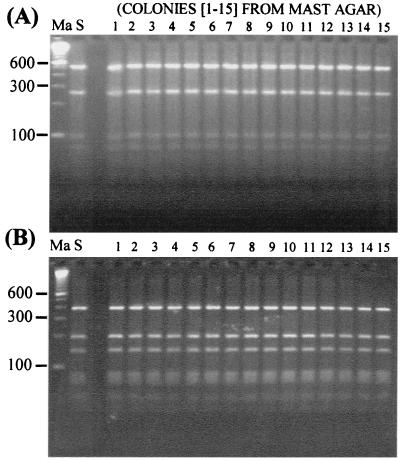
We compared the recA-based PCR-RFLP identification results obtained directly from the 15 sputum samples with those observed upon analysis of the B. cepacia complex organisms isolated from the sputum samples by selective culturing. A representative sample of 15 colonies, selected from different regions of each MAST agar plate, was picked and subcultured onto blood agar before DNA isolation and PCR-RFLP analysis as described in Materials and Methods. For all the patients, the genomovar results obtained directly from sputum samples were identical to those determined for all 15 colonies isolated from the samples by culturing. Figure 3A and B show the results obtained from two patients with B. multivorans and B. cepacia genomovar III (recA group III-A) infections upon HaeIII analysis of the recA gene amplified from their sputum samples and 15 colonies isolated by selective culturing. The identical genomovar-specific RFLP patterns observed between the sputum and culture samples can be clearly seen. Similar results were obtained with sputum samples containing B. cepacia genomovar I, B. stabilis, and B. vietnamiensis (data not shown).
FIG. 3.
Comparison of recA-based PCR-RFLP identification results (with HaeIII) produced directly from CF sputum and after selective culturing. (A) Patient infected with B. multivorans. The RFLP type produced directly from analysis of the patient's sputum is shown in lane S. The RFLP types produced upon analysis of 15 colonies isolated by selective culturing are shown in lanes 1 to 15. Molecular size markers (100-bp ladder) were run in lane Ma. (B) Patient infected with genomovar III (recA group III-A). The RFLP type obtained directly from analysis of the patient's sputum is shown in lane S. The RFLP types produced upon analysis of 15 colonies isolated by selective culturing are shown in lanes 1 to 15. Molecular size markers (100-bp ladder) were run in lane Ma.
Detection limit of the recA-based PCR assay.
Based on the success of these experiments, we examined the sensitivity of recA-based PCR for the detection of B. cepacia complex organisms in sputum. The detection limit of the PCR assay was investigated with four different B. cepacia complex genomovar strains, representing B. cepacia genomovars III-A and III-B and B. multivorans. In the presence of DMSO, the method could reliably detect 106 CFU g of sputum−1 for all four strains. The DMSO adjuvant significantly enhanced amplification of the recA gene compared to other additives examined, including bovine serum albumin, glycerol, Taq Extender (Stratagene, La Jolla, Calif.), and DyNAzyme EXT (Finnzymes, Espoo, Finland).
Clinical evaluation of recA-based PCR-RFLP analysis for detection and identification of B. cepacia complex genomovars in sputum.
To fully assess the diagnostic potential of the recA-based PCR-RFLP method, we screened 100 CF sputum samples for the presence of B. cepacia complex infection. Sputum samples were initially screened for the presence of B. cepacia complex organisms by culturing on MAST selective agar followed by phenotypic analysis of the isolates. Upon examination, growth was observed on 19 plates (Table 3). All 19 isolates were identified as B. cepacia upon phenotypic analysis.
TABLE 3.
Results of PCR analysis of culture-positive CF sputum samplesa
| Sample | Growth on MAST agar | Result obtained by method indicated
|
Genomovar determined by recA-based RFLP analysis | Results of 16S or 16S-23S rDNA PCR with primer pair:
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| recA-based PCR with primers BCR1 and BCR2 |
recA-based RFLP with:
|
||||||||
| HaeIIIb | MnlIc | PSL1-PSR1 | BC-GII–BC-R | G1-G2d | BC-GV–BC-R | ||||
| M1 | + | + | G | f | III-A | + | − | + | − |
| M2 | + | + | G | f | III-A | + | − | + | − |
| M9 | + | + | G | f | III-A | + | − | + | − |
| M16 | + | + | G | f | III-A | + | − | + | − |
| M18 | + | + | G | f | III-A | + | − | + | − |
| M20 | + | + | F | a | B. multivorans | + | + | − | + |
| M32 | + | + | G | f | III-A | + | − | + | − |
| M37 | + | + | F | a | B. multivorans | + | + | − | − |
| M48 | + | + | I | h | III-B | + | − | + | − |
| M54 | + | + | G | f | III-A | + | − | + | − |
| M55 | + | + | H | h | III-B | + | − | + | − |
| M64 | + | + | G | f | III-A | + | − | + | − |
| M69 | + | + | I | h | III-B | + | − | + | − |
| M71e | + | − | + | − | − | − | |||
| M84 | + | + | G | f | III-A | + | − | + | − |
| M85 | + | + | H | h | III-B | + | − | − | − |
| M87e | + | − | + | − | − | − | |||
| M89 | + | + | G | f | III-A | + | − | + | − |
| M94 | + | + | G | f | III-A | + | − | + | − |
+, positive result; −, negative result.
Primers also known as PC-SSF and PC-SSR (21).
Isolates grown on MAST selective agar were identified as B. gladioli.
Bacterial genomic DNA was successfully prepared from all patient sputum samples and confirmed by PCR with primers PSL and PSR, which amplified a 313-bp region of 16S rDNA from all bacteria (data not shown). All 100 samples were analyzed for the presence of B. cepacia complex genomovars by recA-based PCR. Of the 19 culture-positive sputum samples, 17 were identified by recA-based PCR, while 2 samples (M71 and M87) remained undetected (Table 3). RFLP analysis of the recA amplicons with HaeIII and MnlI revealed that 2 patients had B. multivorans infections (12%), while the remaining 15 patients were infected with B. cepacia genomovar III (88%). However, the B. cepacia genomovar III-infected patients did differ with respect to recA phylogenetic subcluster (23). A total of 11 patients (73%) were found to harbor strains of B. cepacia genomovar III recA group III-A, while the remaining 4 patients (27%) were infected with strains of B. cepacia genomovar III recA group III-B.
For comparison, all of the sputum samples were examined with oligonucleotide primers directed to the rRNA operon of the B. cepacia complex genomovars. The characteristic 209-bp PCR product produced with primers PSL1 and PSR1 was amplified from all 19 culture-positive sputum samples. However, a further 10 sputum samples from patients with no history of culturable B. cepacia complex infection also produced a positive reaction with these primers.
Samples were also analyzed with primers G1 and G2 (specific for B. cepacia genomovars I and III and B. stabilis) as well as B. multivorans primers BC-GII and BC-R and B. vietnamiensis primers BC-GV and BC-R (Table 3). Of the 15 sputum samples identified as containing genomovar III by recA-based PCR-RFLP analysis, 14 were found positive with primers G1 and G2, which produced an approximately 1,300-bp 16S-23S rDNA amplicon (Table 3). One sputum sample, from a patient with a B. cepacia genomovar III-B infection (M85), did not produce any amplicon with these primers. In addition, the culture-positive, recA-based PCR-negative sputum samples M71 and M87 were not detected with G1 and G2 (Table 3). All other sputum samples showed no reaction with these primers. With the 16S rDNA-specific B. multivorans primers BC-GII and BC-R, only two samples (M20 and M37) were positive. These results concurred with those of the recA-based PCR-RFLP analysis. Sputum sample M20 also reacted with B. vietnamiensis primers BC-GV and BC-R. This result reflected cross-reactivity with 16S rDNA from B. multivorans, a characteristic previously described for these primers (21). However, all other sputum samples (including M71 and M87) showed no reaction with the B. vietnamiensis primers. It was interesting that the 10 culture-negative sputum samples found positive with PSL1 and PSR1 were found negative when analyzed with G1 and G2, BC-GII and BC-R, and BC-GV and BC-R.
Due to their negative reaction with the recA primers BCR1 and BCR2 as well as the rDNA-based primers G1 and G2, BC-GII and BC-R, and BC-GV and BC-R, culture-positive sputum samples M71 and M87 were examined further. The isolates grown from both samples, which were also recA-based PCR negative, were sent to the Public Health Laboratory Service, Colindale, London, England, for identification. These investigations revealed that both isolates were not B. cepacia complex genomovars but were the closely related species B. gladioli.
DISCUSSION
Due to their taxonomic complexity, identification of B. cepacia complex pathogens has proved a challenging task for the clinical microbiologist. Currently, commercial phenotypic identification systems display significant variations in their capacity to accurately identify B. cepacia complex isolates and do not differentiate between individual genomovars (16, 19, 32). Not surprisingly, investigations have found that CF treatment centers frequently misidentify B. cepacia complex genomovars recovered from patients' sputum (24). Although phenotypic tests have now been described for analysis of the B. cepacia complex, it is still not possible to accurately differentiate all genomovars based on this approach (16).
In an attempt to resolve these problems, molecular biological approaches have been developed to facilitate accurate detection and identification of B. cepacia complex isolates (2, 21, 23, 26, 33). In addition, to aid laboratory diagnosis, a number of studies have also investigated the utility of PCR for the rapid detection of B. cepacia complex pathogens directly from CF sputum. For example, Campbell et al. (4) described a PCR assay for the detection of B. cepacia in sputum using primers PSL1 and PSR1. However, these primers were designed from published 16S rDNA sequences of B. cepacia strain ATCC 25416 (now known to be B. cepacia genomovar I) before taxonomic reappraisal initially identified five distinct genomovar species. Recent studies have now highlighted potential caveats associated with the use of PSL1 and PSR1, primarily their poor specificity for B. multivorans and B. vietnamiensis as well as the capacity to cross-react with other non-B. cepacia complex organisms (2, 5, 21). Karpati and Jonasson (18) similarly described a PCR assay for the detection of B. cepacia in sputum (also before taxonomic reappraisal) using primers directed to 16S rDNA sequences. However, 20% of B. cepacia strains analyzed with these primers failed to yield any amplified product, while other Burkholderia species, such as B. gladioli, Burkholderia caryophylli, and Burkholderia solanacearum, cross-reacted with the primers. More recently, Whitby et al. (34) described a PCR assay for the detection of B. cepacia complex pathogens in sputum based on primers G1 and G2 (also known as PC-SSF and PC-SSR), specific for the 16S-23S spacer region of the rRNA operon. However, since these primers reacted with B. cepacia genomovars I and III and B. stabilis only, they could not detect and identify all B. cepacia complex genomovars in sputum. We investigated whether it was possible to rapidly detect and identify B. cepacia complex organisms directly from crude sputum based on analysis of the recA gene. Our diagnostic algorithm used a single PCR step, with the B. cepacia complex recA primers BCR1 and BCR2, to detect the presence of B. cepacia complex species in sputum and, upon RFLP analysis of the amplicon, to identify the genomovar.
A number of different methods have been described for the preparation of bacterial DNA from sputum. In particular, Campbell et al. (4) and Whitby et al. (34) described the lysis of bacterial cells in sputum by freeze fracturing in liquid nitrogen followed by heating to 100°C. Upon centrifugation, the resulting supernatant containing the released DNA was used directly for PCR. We have used a modification of this method for the preparation of bacterial DNA from sputum. Rather than add a crude supernatant to the PCR (which contains cytoplasmic and proteinaceous debris released during cell lysis), we included an additional step in which the DNA is precipitated with isopropanol and then purified before analysis. This step served to remove any material that could potentially interfere with or inhibit the PCR assay and also produced a high-quality DNA template for amplification. Initial experiments demonstrated that amplification of the B. cepacia complex recA gene from genomovars in CF sputum was possible due to the high specificity of the BCR1 and BCR2 primers for the B. cepacia complex. In addition, the genomovar results obtained upon RFLP analysis of the recA gene amplified from sputum were identical to those determined after culturing of the organisms. These experiments highlighted the value of direct sputum detection and identification of B. cepacia complex organisms, which could be achieved within 1 day (if desired), as opposed to 3 to 4 days when a conventional selective culture step, which normally takes between 48 and 72 h for good-quality growth (15), was added. We further investigated the recA-based PCR-RFLP assay by assessing the sensitivity of the method. Our results revealed primers BCR1 and BCR2 could reliably detect B. cepacia complex organisms to concentrations of 106 CFU g of sputum−1. The detection limit observed with the recA-based PCR was higher than that reported with PCR assays based on analysis of 16S or 16S-23S rDNA (4, 34). This reduced sensitivity likely reflects the large size of the recA amplicon, in combination with only one copy of the gene (23) compared to the multiple copies of the rRNA operon that exist within bacterial cells.
Within a clinical setting, the diagnostic potential of the recA-based PCR-RFLP method for the detection and identification of B. cepacia complex organisms directly from sputum was impressive. When applied to sputum from 100 CF patients, the recA-based PCR successfully detected 17 of 19 culture-positive sputum samples that were identified as containing B. cepacia by phenotypic analysis. Sputum samples from two culture-positive patients (M71 and M87) did not produce any reaction when analyzed with the recA primers. In addition, both samples were negative with all 16S or 16S-23S rDNA primers (except for PSL1 and PSR1), providing further evidence that these patients may not have been infected with a member of the B. cepacia complex but rather a closely related species which was biochemically indistinguishable on selective agar (16, 32). To date, the recA primers BCR1 and BCR2 remain highly specific for members of the B. cepacia complex only (7, 16, 23). Also, recent studies have shown that suspected B. cepacia complex isolates that are recA-based PCR negative belong to other closely related species that are not members of the current complex (16; E. Mahenthiralingam, unpublished data). Our studies confirmed this view, since further investigations did indeed reveal that the culture-positive, recA-negative sputum samples M71 and M87 were infected with B. gladioli and not organisms of the B. cepacia complex. Phenotypic misidentification of B. gladioli as B. cepacia is common with the API 20NE strip, which does not contain the taxon B. gladioli in its database.
The 16S rDNA primers PSL1 and PSR1 detected all 17 sputum samples containing members of the B. cepacia complex, including both samples with B. multivorans. It was interesting that both culture-positive sputum samples infected with B. gladioli also produced a very strong positive reaction when analyzed with these primers, although previous studies have not observed any cross-reactivity between PSL1 and PSR1 and B. gladioli isolates (4, 5). In addition, we found that a further 10 sputum samples from patients without any history of culturable B. cepacia infection similarly produced an amplicon of the correct molecular weight when analyzed with the primers. These samples did not show any reaction with primers G1 and G2, BC-GII and BC-R, or BC-GV and BC-R. These results would therefore appear to confirm recent studies that demonstrated the reaction of PSL1 and PSR1 with non-B. cepacia complex organisms (5, 21). On the basis of these data, we recommend that results obtained using these primers, especially from sputum, be interpreted with caution.
RFLP analysis of our B. cepacia complex recA amplicons identified the genomovar responsible for infection. Two patients were infected with B. multivorans, while the remaining patients were infected with B. cepacia genomovar III. Analysis of these samples with the rDNA primers BC-GII and BC-R (for B. multivorans) and G1 and G2 (for B. cepacia genomovars I and III and B. stabilis) confirmed these results, although one B. cepacia genomovar III sample did not react with G1 and G2. These data therefore provide further evidence for the prevalence of both B. multivorans and B. cepacia genomovar III strains within the CF community. RFLP analysis of our recA amplicons also revealed that the B. cepacia genomovar III-infected patients differed with respect to the recA cluster group to which their genomovar belonged. The taxonomic significance of recA groups III-A and III-B is, at the moment, not clear. Recent studies have not found any significant biochemical differences between these two recA subgroups, except for the ability to reduce nitrate (16). Also, it has not been established whether B. cepacia genomovars III-A and III-B have different effects on CF patient morbidity and mortality. However, after retrospective review of our patients' records, we have established that the B. cepacia genomovar III-A and III-B strains identified in our clinical study correspond to the epidemic B. cepacia genomovar III strains 1 (Edinburgh-Toronto epidemic) and 2 (Manchester epidemic), respectively, which were previously reported as the cause of epidemic infections in CF patients attending the Bradbury Adult Cystic Fibrosis Center, Wythenshawe Hospital (14). More interestingly, the patients infected with these two different strains appeared to show no difference in clinical outcome, suggesting that infection with B. cepacia genomovar III-A or III-B has no significant effect on prognosis. Further investigations will be required to confirm this observation.
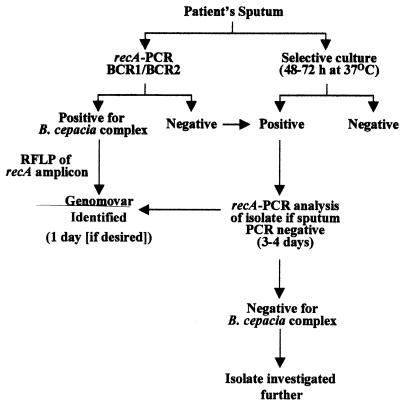
Although the detection limit of the recA-based PCR was higher than that of PCR assays based on the rRNA operon, the clinical results obtained with this method were excellent and comparable to those obtained by conventional culturing. This finding may reflect the fact that patients with B. cepacia complex infection frequently have high genomovar concentrations in their sputum and saliva (13, 15). For such patients, the recA-based PCR-RFLP method is an excellent way to rapidly detect and identify the B. cepacia complex genomovar present. When levels of infection are lower and the sensitivity of the assay proves a limitation, sputum samples can be analyzed by selective culturing followed by recA-based confirmation and identification of the isolate. Indeed, we normally perform a parallel culture step so that further analyses, such as continued diagnostic investigation (if required), molecular typing experiments, and susceptibility testing, can be carried out on the isolate. Figure 4 illustrates the experimental approach that we have adopted for routine recA-based detection and identification of B. cepacia complex genomovars in CF sputum.
FIG. 4.
Experimental approach for recA-based detection and identification of B. cepacia complex genomovars in CF sputum.
In summary, rapid detection and identification of B. cepacia complex pathogens from CF sputum based on recA is possible. In addition, since the very recently described B. cepacia genomovar VI and genomovar VII (B. ambifaria) species of the B. cepacia complex also show reaction with the recA primers BCR1 and BCR2 and can be distinguished by RFLP analysis (7, 16; Mahenthiralingam, unpublished), it should be possible to detect and identify these organism in CF sputum also. At present, we are examining the diagnostic value of genomovar-specific recA primers for the detection and identification of individual B. cepacia complex organisms in sputum.
ACKNOWLEDGMENTS
This work was supported by grants (PJ470 and PJ472) from the Cystic Fibrosis Trust, Bromley, United Kingdom.
We thank Tyrone Pitt and his staff at the Public Health Laboratory Service for assistance in identifying B. gladioli strains isolated from the sputum of our patients.
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D L. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind A, Schneider I, Jungwirth R, Roller C. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J Clin Microbiol. 1999;37:1335–1339. doi: 10.1128/jcm.37.5.1335-1339.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brisse S, Verduin C M, Milatovic D, Fluit A, Verhoef J, Laevens S, Vandamme P, Tümmler B, Verbrugh H A, van Belkum A. Distinguishing species of the Burkholderia cepacia complex and Burkholderia gladioli by automated ribotyping. J Clin Microbiol. 2000;38:1876–1884. doi: 10.1128/jcm.38.5.1876-1884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell P, Phillips J A, Heidecker G J, Krishnamani M R S, Zahorchak R, Stull T L. Detection of Pseudomonas (Burkholderia) cepacia using PCR. Pediatr Pulmonol. 1995;20:44–49. doi: 10.1002/ppul.1950200109. [DOI] [PubMed] [Google Scholar]
- 5.Clode F E, Kaufmann M E, Malnick H, Pitt T L. Evaluation of three oligonucleotide primer sets in PCR for the identification of Burkholderia cepacia and their differentiation from Burkholderia gladioli. J Clin Pathol. 1999;52:173–176. doi: 10.1136/jcp.52.3.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coenye T, LiPuma J J, Henry D, Hoste B, Vandemeulebroecke K, Gillis M, Speert D P, Vandamme P. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int J Syst Bacteriol. 2001;51:271–279. doi: 10.1099/00207713-51-2-271. [DOI] [PubMed] [Google Scholar]
- 7.Coenye T, Mahenthiralingam E, Henry D, LiPuma J J, Laevens S, Gillis M, Speet D P, Vandamme P. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex comprising biocontrol and cystic fibrosis-related isolates. Int J Syst Evol Microbiol. 2001;51:1481–1490. doi: 10.1099/00207713-51-4-1481. [DOI] [PubMed] [Google Scholar]
- 8.Coenye T, Schouls L M, Govan J R W, Kersters K, Vandamme P. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int J Syst Bacteriol. 1999;49:1657–1666. doi: 10.1099/00207713-49-4-1657. [DOI] [PubMed] [Google Scholar]
- 9.Corey M, Farewell V. Determinants of mortality from cystic fibrosis in Canada. Am J Epidemiol. 1996;143:1007–1017. doi: 10.1093/oxfordjournals.aje.a008664. [DOI] [PubMed] [Google Scholar]
- 10.Frangolias D D, Mahenthiralingam E, Rae S, Raboud J M, Davidson A G F, Wittmann R, Wilcox P G. Burkholderia cepacia in cystic fibrosis: variable disease course. Am J Respir Crit Care Med. 1999;160:1572–1577. doi: 10.1164/ajrccm.160.5.9805046. [DOI] [PubMed] [Google Scholar]
- 11.Gillis M, Van T V, Bardin R, Goor M, Hebbar P, Willems A, Segers P, Kersters K, Heulin T, Fernandez M P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int J Syst Bacteriol. 1995;45:274–289. [Google Scholar]
- 12.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 14.Haworth C S, Dodd M E, Doherty C, Super M, Hambleton G, Vandamme P, Govan J R W, Webb A K. The morbidity and mortality associated with two epidemic strains of Burkholderia cepacia with genomovar III status in cystic fibrosis patients. Pediatr Pulmonol. 1997;14(Suppl.):290. [Google Scholar]
- 15.Henry D, Campbell M, McGimpsey C, Clarke A, Louden L, Burns J L, Roe M H, Vandamme P, Speert D. Comparison of isolation media for recovery of Burkholderia cepacia complex from respiratory secretions of patients with cystic fibrosis. J Clin Microbiol. 1999;37:1004–1007. doi: 10.1128/jcm.37.4.1004-1007.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henry D A, Mahenthiralingam E, Vandamme P, Coenye T, Speert D P. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J Clin Microbiol. 2001;39:1073–1078. doi: 10.1128/JCM.39.3.1073-1078.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isles A, Macluskey I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 18.Karpati F, Jonasson J. Polymerase chain reaction for the detection of Pseudomonas aeruginosa, Stenotrophomonas maltophilia and Burkholderia cepacia in sputum of patients with cystic fibrosis. Mol Cell Probes. 1996;10:397–403. doi: 10.1006/mcpr.1996.0055. [DOI] [PubMed] [Google Scholar]
- 19.Kiska D L, Kerr A, Jones M C, Caracciolo J A, Eskridge B, Jordan M, Miller S, Hughes D, King N, Gilligan P H. Accuracy of four commercial systems for identification of Burkholderia cepacia and other gram-negative nonfermenting bacilli recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:886–891. doi: 10.1128/jcm.34.4.886-891.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LiPuma J J. Burkholderia cepacia: management issues and new insights. Clin Chest Med. 1998;19:473–486. doi: 10.1016/s0272-5231(05)70094-0. [DOI] [PubMed] [Google Scholar]
- 21.LiPuma J J, Dulaney B J, McMenamin J D, Whitby P W, Stull T L, Coenye T, Vandamme P. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J Clin Microbiol. 1999;37:3167–3170. doi: 10.1128/jcm.37.10.3167-3170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LiPuma J J, Fischer M C, Dasen S E, Mortensen J E, Terrence I S. Ribotype stability of serial pulmonary isolates of Pseudomonas cepacia. J Infect Dis. 1991;164:133–136. doi: 10.1093/infdis/164.1.133. [DOI] [PubMed] [Google Scholar]
- 23.Mahenthiralingam E, Bischof J, Byrne S K, Radomski C, Davies J E, Av-Gay Y, Vandamme P. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38:3165–3173. doi: 10.1128/jcm.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMenamin J D, Zaccone T M, Coeyne T, Vandamme P, LiPuma J J. Misidentification of Burkholderia cepacia in US cystic fibrosis treatment centers. Chest. 2000;117:1661–1665. doi: 10.1378/chest.117.6.1661. [DOI] [PubMed] [Google Scholar]
- 25.O'Neil K M, Herman J H, Modlin J F, Moxon E R, Winkelstein J A. Pseudomonas cepacia: an emerging pathogen in chronic granulomatous disease. J Pediatr. 1986;108:940–942. doi: 10.1016/s0022-3476(86)80934-9. [DOI] [PubMed] [Google Scholar]
- 26.Segonds C, Heulin T, Marty N, Chabanon G. Differentiation of Burkholderia species by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene and application to cystic fibrosis isolates. J Clin Microbiol. 1999;37:2201–2208. doi: 10.1128/jcm.37.7.2201-2208.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson I N, Finlay J, Winstanley D J, Dewhurst N, Nelson J, Butler S, Govan J R W. Multi-resistance isolates possessing characteristics of both Burkholderia (Pseudomonas) cepacia and Burkholderia gladioli from patients with cystic fibrosis. J Antimicrob Chemother. 1994;34:353–361. doi: 10.1093/jac/34.3.353. [DOI] [PubMed] [Google Scholar]
- 28.Sun L, Jiang R-Z, Steinbach S, Holmes A, Campanelli C, Forester J, Tan Y, Riley M, Goldstein R. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF center epidemics in North America and Britain. Nat Med. 1995;1:661–666. doi: 10.1038/nm0795-661. [DOI] [PubMed] [Google Scholar]
- 29.Tümmler B, Kiewitz C. Cystic fibrosis: an inherited susceptibility to bacterial respiratory infections. Mol Med Today. 1999;5:351–358. doi: 10.1016/s1357-4310(99)01506-3. [DOI] [PubMed] [Google Scholar]
- 30.Vandamme P, Holmes B, Vancanneyt M, Coenye T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 31.Vandamme P, Mahenthiralingam E, Holmes B, Coenye T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Pelt C, Verduin C M, Goessens W H F, Vos M C, Tümmler B, Segonds C, Reubsaet F, Verbrugh H, van Belkum A. Identification of Burkholderia spp. in the clinical microbiology laboratory: comparison of conventional and molecular methods. J Clin Microbiol. 1999;37:2158–2164. doi: 10.1128/jcm.37.7.2158-2164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitby P W, Carter K B, Hatter K L, LiPuma J J, Stull T L. Identification of members of the Burkholderia cepacia complex by species-specific PCR. J Clin Microbiol. 2000;38:2962–2965. doi: 10.1128/jcm.38.8.2962-2965.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitby P W, Dick H L N, Campbell P W, Tullis D E, Matlow A, Stull T L. Comparison of culture and PCR for detection of Burkholderia cepacia in sputum samples of patients with cystic fibrosis. J Clin Microbiol. 1998;36:1642–1645. doi: 10.1128/jcm.36.6.1642-1645.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]