Abstract
Biomedical researchers have subsequently been inspired the development of new approaches for precisely changing an organism’s genomic DNA in order to investigate customized diagnostics and therapeutics utilizing genetic engineering techniques. Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR) is one such technique that has emerged as a safe, targeted, and effective pharmaceutical treatment against a wide range of disease-causing organisms, including bacteria, fungi, parasites, and viruses, as well as genetic abnormalities. The recent discovery of very flexible engineered nucleic acid binding proteins has changed the scientific area of genome editing in a revolutionary way. Since current genetic engineering technique relies on viral vectors, issues about immunogenicity, insertional oncogenesis, retention, and targeted delivery remain unanswered. The use of nanotechnology has the potential to improve the safety and efficacy of CRISPR/Cas9 component distribution by employing tailored polymeric nanoparticles. The combination of two (CRISPR/Cas9 and nanotechnology) offers the potential to open new therapeutic paths. Considering the benefits, demand, and constraints, the goal of this research is to acquire more about the biology of CRISPR technology, as well as aspects of selective and effective diagnostics and therapies for infectious illnesses and other metabolic disorders. This review advocated combining nanomedicine (nanomedicine) with a CRISPR/Cas enabled sensing system to perform early-stage diagnostics and selective therapy of specific infectious disorders. Such a Nano-CRISPR-powered nanomedicine and sensing system would allow for successful infectious illness control, even on a personal level. This comprehensive study also discusses the current obstacles and potential of the predicted technology.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s40097-022-00472-7.
Keywords: Gene editing, CRISPR/Cas, Infectious diseases, Nanomedicine, Biosensor, Diseases management, Personalized healthcare
Introduction
Every year, thousands of people are afflicted with potentially fatal chronic illnesses, and many infectious diseases are incurable due to the frequency and specificity of outbreaks [1]. Infectious diseases are the second leading cause of mortality worldwide and the third leading cause of death in India. Acute lower respiratory tract infections, viral hepatitis, tuberculosis, diarrheal disorders, HIV/AIDS, and malaria are all infectious diseases that kill people all over the world [2]. The worldwide burden of infectious diseases in the contemporary environment is dictated by the incidence and prevalence of active cases, notably the appearance of new and re-emerging disorders [3]. Due to its exceptional characteristics and a lack of sufficient medical facilities, many infectious illnesses still lack acceptable diagnostic and treatment techniques and constitute a severe concern to the vulnerable community. As a result, significant improvements in existing and alternative diagnosis and therapy techniques for the prevention of these illnesses are required [4].
The ideal diagnostic test, according to the World Health Organization (WHO), is affordable and trustworthy for any pathogen, provides rapid findings, can be used in point-of-care settings, and requires little to no sophisticated equipment or expert assistance. No test has yet satisfied all of these conditions. As a result, finding new and more accurate diagnostic tools for certain infectious diseases is crucial right now [5]. Methods that include amplification of nucleic acid sequences (molecular method) with detection and visualization of antigens (immunological approach) provide a more precise diagnosis [6]. The extraction of pathogenic nucleic acids from biological materials and identification by polymerase chain reaction (PCR) is a traditional and successful way to diagnose infectious illnesses [7].
Based on the concept that nucleic acids are desirable biomarkers for illnesses, diagnostic procedures rely primarily on finding the disease’s target sequence and cleaving it to generate a readable signal [8]. In recent years, advances in genome manipulation and editing techniques have been considered as a significant advancement in the field of genetic engineering, providing biologists with methods to precisely influence nearly any organism’s genomic DNA to incorporate desired mutations, such as extension, correction, replacement, and excision, contributing to the speed of research for discovering the basis for unusual genetic disorders and developing therapeutics [9, 10]. This allows researchers to investigate gene expression and control by developing in vitro (cellular) or in vivo (animal) disease models, which might aid in the development of regenerative medicine for genetic and infectious disorders [11].
Over time, advances in next-generation biomedical technology have accelerated research and translation to previously astounding peaks. Genetic modification utilizing programmable nucleases is one of the key technologies that might be used to generate novel treatment techniques for infectious diseases. Examples of molecular scissors include homing endonucleases (HEs), transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 (Cas9) [12]. CRISPR/Cas systems are unique in that they provide prokaryotes with heritable adaptive tolerance to invading genomic material. CRISPR’s genomic locus serves as a memory store device, storing sequences of nucleic acid spacers originating from invading genetic elements. The sequences are later recovered to assist the Cas enzyme’s selective elimination of foreign invaders. CRISPR/Cas systems function at the molecular level, spanning adaptive mechanisms, crRNA maturation, and system-level intervention with significant biological variety [13]. The CRISPR/Cas technique aimed at identifying nuclear acids for molecular diagnostics was originally described in 2016. CRISPR/Cas has previously been used to develop various effective methods for detecting and diagnosing infectious and non-infectious illnesses [14]. Genetic disorders, like infectious illnesses, have congenital traits. Biochemical, physiological, or genetic factors such as bacteria, viruses, or fungus induce mutations in the DNA, resulting in such conditions. Treatments for these genetic diseases have traditionally relied on viral-mediated transgenic expression and RNA interference techniques [15].
Antimicrobial medications are the most common form of prevention for infectious infections. Pathogens may develop immune in circumstances where antimicrobial therapy is insufficient for therapeutic purposes [16]. Antibiotic therapy can harm the human symbiotic microbiome; however, a component of the pathogen population usually survives owing to drug tolerance or persistence. The patient (side effects and contraindications) as well as the infection might be blamed for pharmacological treatment issues (drug-resistance). Another hurdle to drug usage for possible pathogen specificity in novel drugs is the amount and length of administration, which results in increased side effects and toxicity [17]. The most challenging challenge thus far has been delivering the CRISPR technology to the target cells. The non-specific targeting of nucleic acids by the CRISPR system has been a subject of concern because it is designed for therapeutic and diagnostic reasons [18]. The CRISPR/Cas enzyme approach must be delivered successfully in order to avoid off-target effects inside the gene and to ensure that the tool reaches the desired cell or tissue. Off-target difficulties are being investigated by several researchers, either through the development of off-target detection methods or the development of CRISPR tools [19].
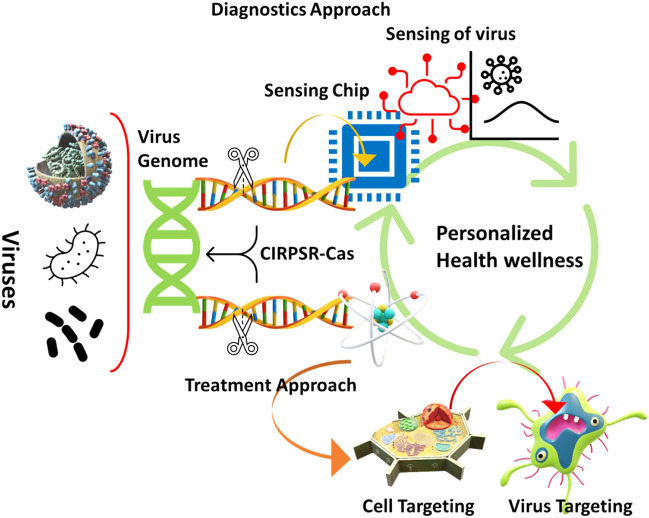
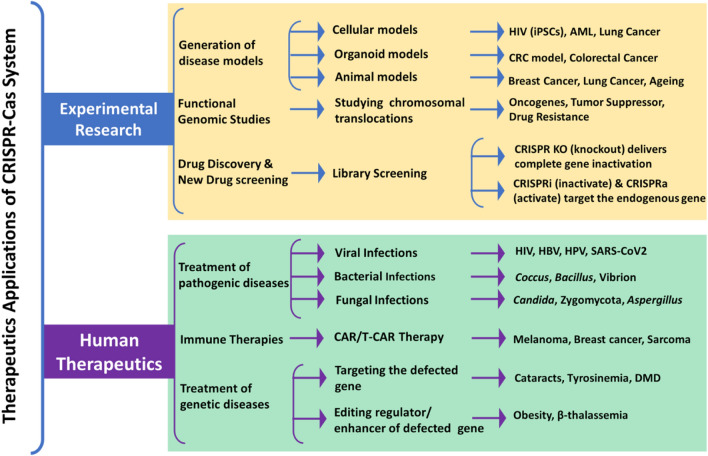
Despite the fact that the CRISPR/Cas mechanism may be involved in the development of drug resistance, it is not a promising drug target because, despite their important role in bacterial immunity, they are not required for cell survival; additionally, structural variants of CRISPR/Cas exist within the same species and vary between strains. Such limitations can be overcome thanks to the usage of nano-mediated technologies. Nanotechnology formulations for different pathogens, such as bacteria, viruses, fungi, or parasites, combined with CRISPR/Cas, are now becoming increasingly relevant, and can pave the way for the creation of viable diagnosis and therapy techniques (Fig. 1). This review aims to outline the machineries of CRISPR/Cas and their applications in infectious illness diagnosis and treatment techniques, as well as present a review of therapeutic nanoparticles and their targeted delivery applications in disease diagnostics.
Fig. 1.
An overview of the CRISPR Cas-based efficient diagnostics and in vivo delivery approaches and genetic engineering mechanisms of nanotechnology-supported CRISPR/Cas9 payload in various forms of nanomedicine and drug delivery systems
Introduction to the CRISPR/Cas9
Different mechanisms as a defense are utilized by the microorganisms to avoid damage to their genome from the bacteriophage infections and transfer of plasmids. Eubacteria and archaea have an RNA-based defense system that adapts to the recognition and destruction of external DNA and RNA. This offers acquired immunity against plasmids and viruses’ attacks. The use of RNA driven endonucleases named the CRISPR-associated (Cas) 9 enzymes, is one such defensive technique [20–22]. The CRISPR/Cas9 system is an adaptive immune system in prokaryotes that aids microorganisms to react to foreign genetic material and destroy it. It is present in a greater diversity of prokaryotic genomes, reported in 1987 in Escherichia coli as irregular genomic arrays in which different interspersed “spacer” sequences distinguish a replicated similar sequence [23]. Microbes exposed to foreign genetic material by transformation, conjugation and transduction are stimulated towards defense mechanisms that detect foreign DNA and protect themselves from genomic intruders [24].
Structure of CRISPR/Cas
The key components of CRISPR/Cas systems comprise CRISPR RNAs (crRNAs) besides Cas enzyme. The accompanying nucleic acid chain demonstrates the complementarity with the crRNAs. Therefore, the binding of crRNA to the unique invasive pathogen genome sequence results in the shredding of the target DNA/RNA sequence by Cas enzyme [15]. CRISPR loci are composed of 20–40 bp length repeat sequences, which are interspaced by unique 20–58 bp sequences called spacers resulting from small segments of external genetic material (protospacers) [25, 26]. The CRISPR locus has an AT-rich leader sequence and is frequently followed by a downstream succession of sequences known as cas genes, which code for nucleolytic Cas proteins [27]. Streptococcus pyogenes (SpCas9) Cas9 is the best characterized and most used for the modification of the DNA chain. SpCas9 consists of 1368 amino acids and is classified into various regions, each with a different function [28]. In the CRISPR/Cas system, the Cas9 protein serves as a facilitator for site-specific DNA binding and infectious genetic material cleavage [15].
Composition of CRISPR/Cas
The CRISPR/Cas structures can be classified into two classes which include multisubunit crRNAs receptors complexes and single protein receptor, respectively, six types and multiple subtypes depending upon locus organization and gene conservation [29, 30]. The CRISPR class 1 is characterized by multisubunit effector nucleases and comprises the types (I, III, and IV). Type I is characterized by the signature cas3 gene and is currently classified into seven subtypes (I-A, I-B, I-C, I-D, I-E, I-F, and I-U). Type III utilizes a multiprotein complex with the signature gene cas10 and includes four subtypes (III-A, III-B, III-C, III-D) [31]. Type IV loci, which share common protein sets such as Cas7 (Csf2), Cas5 (Csf3), and a smaller variant of Cas 8 (Csf1), are classified into two subtypes (IV-A and IV-B) [32]. Due to a lack of expertise, despite the fact that several Classes 1 CRISPR systems have been reported of the intracellular process underlying the defence mechanisms, regular implementations of the class as a genome-editing tool have been restricted [33, 34].
The CRISPR class 2 system is distinguished by a single effector protein, Cas9, which is subclassified into types II, V, and VI based on the components required for pre-crRNA processing and the variety of the effector protein’s domains [35]. The type II CRISPR locus employs a single effector protein (Cas9) that is driven by a dual-RNA heteroduplex (crRNA-tracrRNA) (trans-activating crRNA) and is divided into three subtypes (II-A, II-B, and II-C) [36]. crRNA and Cas12a (also known as Cpf1) protein are components of the type V CRISPR system. A RuvC endonuclease domain in the Cas12a protein cleaves the non-targeting strand and the targeted strand sequentially to produce DSBs. The type V exemplified by Cas12a is broken by ten recognized subtypes ranging from A-I to U based on domain organisation similarities [35, 37]. The type VI CRISPR systems consist of a single RNA-guided Cas13 effector protein primarily aimed at defending RNA from bacteria from genetic mobility. Type VI CRISPR/Cas structures based on Cas13 phylogeny can be classified into four subtypes (VI-A, VI-B, VI-C, VI-D) [38, 39].
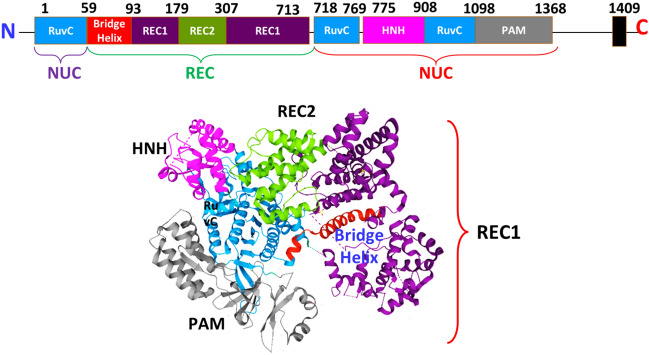
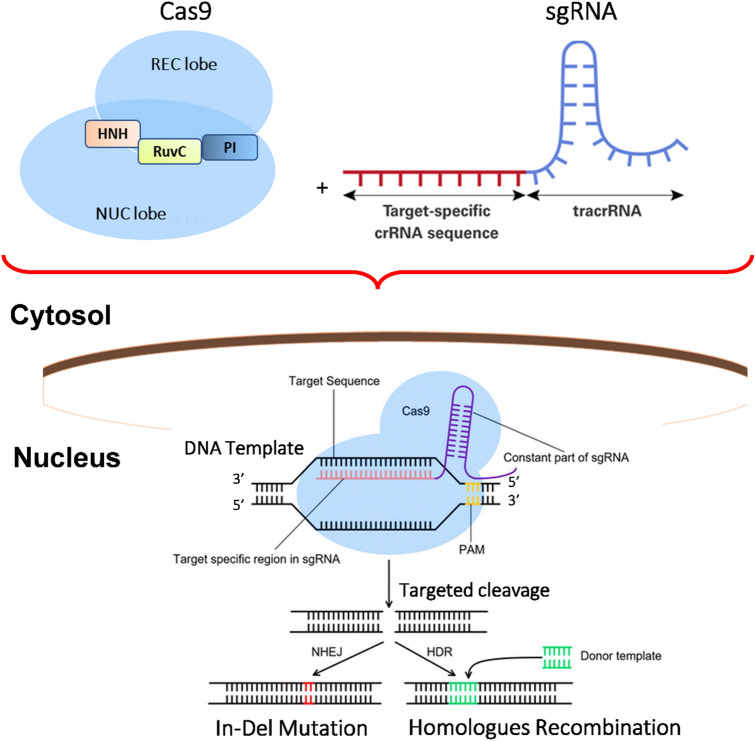
Cas9 is a big multimeric polypeptide with 1409 amino acid residues in its crystal structure. The Recognition unit (REC) is split into three facets: the bridge helix (residues 60–93), the region REC1 (residues 94–179 and 308–713), as well as the region REC2 (residues 180–307), and the Nuclease unit (NUC) is made up of the RuvC (residues 1–59, 718–769, and 909–1098) (Fig. 2). Mutational research has shown that intervention generated in vivo by Cas9 includes both the RuvC and the HNH patterns [25, 40].
Fig. 2.
Domain organization and crystal structure of CRISPR/Cas9 from S. pyogenes (PDB ID: 4CMP) displaying various domains and protein regions in the structure. The interdomain borders are labelled with residue numbers. RuvC Resolvase, NUC Nuclease Unit, REC Recognition Unit, PAM Protospacer Adjacent Motif
Functioning mechanism of CRISPR/Cas
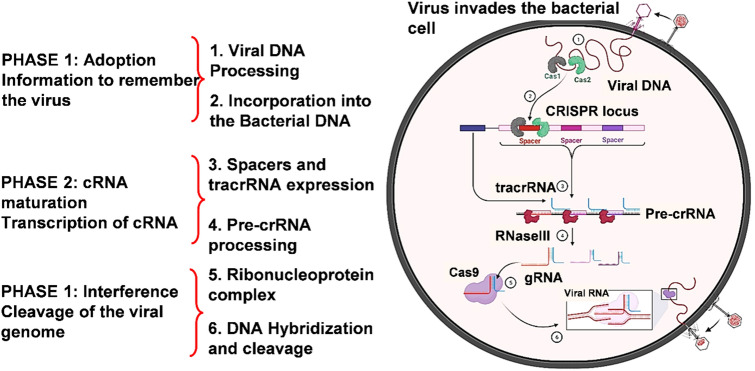
The CRISPR/Cas system is a sequence-specific defensive mechanism that identifies and hydrolyses invading genetic material [27]. The mechanism can be divided into the following three stages: (1) adaptation or spacer acquisition, (2) crRNA maturation, and (3) target interference.
Adaptation of CRISPR/Cas
In this stage, the spacer is incorporated into the CRISPR array after foreign genetic elements are identified, protospacer sequences are selected and analyzed, helping the host organism to remember the intruder’s genetic code, showing the immune system's adaptive existence [13]. This phase provides the genetic memory that is required to neutralize re-invading nucleic acids via the subsequent expression and interruption processes [41]. In several CRISPR/Cas types, the collection of protospacers and their processing prior to integration is still unidentified. Recent discoveries, on the other hand, have shed light on the biochemistry of the spacer integration mechanism [27]. After the spacer has been modified in size, the availability of the integrated host factor (IHF) protein is responsible for the spacers integration into the host genome. Cas1 and Cas2 are nuclease proteins that are important components of CRISPR/Cas complexes of three different kinds (I, II, and III). These two proteins form dimer that effort together to get foreign DNA. Cas1 is a nuclear integrase that really can cleave the bacterial genome into a DNA spacer, whereas Cas2 is an endoribonuclease that primarily cleaves RNAs [26, 27].
crRNA maturation of CRISPR/Cas
The CRISPR series is transcribed into a long precursor crRNA (pre-crRNA), which is then processed into mature reference crRNAs, which are used to decipher invader sequences [42]. Although there are significant type-specific modifications, often these organisms' CRISPR/Cas loci transcription to form an RNA–protein guiding complexes includes a systematic pattern. Across both forms, the CRISPR locus is transcribed, and the complex CRISPR ribonucleoprotein (crRNP) is generated from the RNA processed by Cas ribonucleases [43]. The pre-crRNA is cleaved into its constituent parts, each containing a single spacer and partial repeats (Fig. 3). In this case pre-crRNA cleavage is catalyzed by a ubiquitous bacterial enzyme, the RNase III encoded outside the local CRISPR, which includes tracrRNA, a distinct CRISPR-complementary RNA species [44]. The Cas protein accountable for processing differs by subtypes, and they do not process the pre-crRNA of each other as the three CRISPR/Cas forms coexist in nature [45]. In terms of pre-crRNA processing and the configurations of the crRNP complexes produced, Type I and III CRISPR/Cas systems are identical. Except for Type I-C, which uses Cas5d, all Type I and III systems use the Cas6 protein to process pre-crRNA [46].
Fig. 3.
CRISPR/Cas-based immunity's fundamental steps. Each of the three execution phases of the CRISPR-mediated intervention mechanism comprises an information processing component (CRISPR adaptation) and a two-part executive component (CRISPR expression and CRISPR-based interference). The introduction of extra spacers into the CRISPR locus, which could be naive or primed acquisitions, with both the latter leading in the acquirement of spacers from the very same genetic elements such as plasmids, is the first stage of the adaptation mechanism to identify the virus. CRISPR region transcription and CRISPR RNA processing are mostly the final two processes, which are preceded by pathogen or transportable genetic material identification and CRISPR RNA destruction
Interference of CRISPR/Cas
The effector component recognizes the gene encoding in the invading nucleic acid via complementary base pairing and triggers sequential cleavage, preventing the reproduction and proliferation of exogenous genetic elements [47]. CRISPR/Cas systems functions on the notion that crRNA coupled Cas protein identifies the corresponding protospacer, causing the target gene to be degraded by nuclease [41]. The mature crRNA activates the interference cascade complex, which recognizes target nucleotide sequences similar to the packed crRNA that facilitates intervention by cleaving each strand of the intruding genomic DNA following R-loop formation [48]. Cas3 cleaves the target DNA and subsequently destroys it with the help of a 3′-5′ endonuclease. A 7- to 8-ntd sequence at the spacer’s 5′ end directs target sequences homology coupling and initiates R-loop formation, which necessitates the identification of a protospacer adjacent motif (PAM) ahead of the target protospacer [49, 50].
Role of CRISPR/Cas9 technology in infectious diseases
Due to the overwhelming effectiveness of RNA-guided nucleic acid degradation, the CRISPR/Cas system (as summarized in Table 1) has emerged as a viable contender for the creation of next-generation antimicrobial drugs to combat contagious infections, particularly those caused by antimicrobial resistant (AMR) microorganisms. Additionally, the varied adaptability of the CRISPR/Cas system enables it to specifically destroy a bacterial isolate species among a vast population, allowing CRISPR/Cas bacteriocins to precisely change the design of a diverse bacterial species [51]. This makes CRISPR/Cas antimicrobials particularly useful for treating infections in natural complex microbial consortia, such as the intestinal microbiota. CRISPR technology has demonstrated therapeutic potential as an antibacterial agent, with the ability to target antibiotic-resistant and/or very pathogenic microorganisms [41, 52]. CRISPR-mediated antimicrobials have a distinct advantage over other antimicrobial methods in that they can destroy bacteria based on their sequence. This may be useful in situations where only a small number of bacteria within a genus must be eliminated, which is difficult to do with current strategies [53]. It elucidates basic host-microbe relationships, advancement of fast and reliable diagnostics methods, and improved prevention and treatment of infectious diseases [54]. The transmission of drug resistance and pathogenicity by foreign DNA is a major contributor to the spread of more aggressive and dangerous bacterial strains. In plasmids and phages, CRISPR/Cas mechanisms have been shown to induce toxicity and antimicrobial resistance [55]. Mechanisms to grasp the causes of human infection by pathogens (bacteria, fungi, and viruses) are necessary to optimize therapeutic intervention and the rational development of targeted treatment and vaccines. In this regard, diverse pathogens are utilized to report the gene and protein contributions to molecular pathogens in CRISPR/Cas9-based gene editing [56] (Fig. 4).
Table 1.
Application of CRISPR systems in disease diagnostics and therapeutics
| Name | Enzyme | Preamplification | Applications |
|---|---|---|---|
| NASBACC | Cas9 | NASBA | Insight amongst African and American ZIKV |
| CRISPR–Chip | Cas9 | – | gDNA identification in cells in vitro and DMD individuals |
| CRISDA | Cas9 nickase | SDA | gDNA Identification; SNPs related with breast cancer in cell cultures |
| FLASH | Cas9 | PCR | Antibiotic resistant genes in clinical specimens; gDNA identification |
| CAS-EXPAR | Cas9 | EXPAR | Methylated DNA detection in L. monocytogenes mRNA |
| Cas9nAR | Cas9 nickase | Strand-displacing DNA polymerase | Bacterial detection (S. typhimurium, E. coli, M. smegmatis, S. erythraea); KRAS SNP detection in cell lines |
| DETECTR | Cas12a | RPA | HPV16 and HPV18 identification in clinical specimens |
| Cas14-DETECTR | Cas14 (Cas12f) | PCR | HERC2 SNPs in clinical isolates identified |
| HOLMES | Cas12a | PCR | Screening of viral infections (PRV, JEV); virus-strain differentiation in cell cultures and clinical specimens; SNP classification |
| CRISPR-materials | Cas12a | RPA | EBOV synthetic RNA detection |
| CDetection | Cas12b | RPA | HPV16 screening, Human Blood groups genotyping (ABO), and BRCA1 and TP53 SNPs |
| HOLMESv2 | Cas12b | LAMP | SNP distinction in cell cultures; identification of RNA viruses (JEV); screening of human mRNA and circular RNA; Methylation |
| E-CRISPR | Cas12a | – | Virus (HPV16, PB19) and polypeptide (TGF-ß1) identification |
| – | Cas13 | – | Human mRNA and bacteriophage λ-RNA detection |
| SHERLOCK | Cas13 | NASBA or RPA | Viral (DENV, ZIKV) and bacterium (E. coli, K. pneumoniae, M. tuberculosis, P. aeruginosa, S. aureus) detection; viral strain differentiation; SNP identification |
| SHERLOCKv2 | Cas13 | RPA | Virus (DENV, ZIKV) and bacterium (P. aeruginosa, S. aureus) detection; viral strain differentiation; SNP identification |
Fig. 4.
Application of CRISPR/Cas technology in therapeutics. HIV (iPSCs) Human Immunodeficiency Virus (Induced pluripotent stem cells), HBV Hepatitis B Virus, AML Acute Myeloid Leukemia, CRC Colorectal Cells, HPV Human Papillomavirus, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2, CAR/T-CAR Chimeric antigen receptor T cells, DMD Duchenne muscular dystrophy
Viral infections and CRISPR/Cas
Since CRISPR/Cas9 evolved in bacteria to directly target viruses’ invaders and foreign DNA, it is apparent that it may be used to treat viral infections [56]. After initial infection, several viruses retain their genome or inject in the human chromosomal DNA episomally inside host genome, resulting in long-term infection. Viruses that induce severe infection include HIV, hepatitis viruses, papillomaviruses, herpesviruses, and coronaviruses. CRISPR technology has been utilised to suppress or eliminate recurring viral infections in vitro and in animal models in recent years, providing promise for therapies for infectious diseases that are dormant and chronic [13].
AIDS continues to be a global public health problem affected by HIV a retrovirus, which results in the loss of the immune system. Initial CRISPR–Cas9 replication-competent HIV studies have shown successful viral replication suppression in relatively brief cultured cells assays [57, 58]. Virus escape, however, was not addressed in the experiments, despite HIV-1's well-known tendency to build resistance to inhibitors. More research has found that CRISPR/Cas9 may effectively restrict HIV replication, but that the virus frequently evades this inhibition due to acquired mutations clustered around at the Cas9 catalytic domain [59, 60].
According to the WHO, about 250 million individuals are found to be infected with the hepatitis B virus (HBV), a DNA virus that multiplies by reverse transcriptase and is associated with an increased risk of liver failure and hepatorenal syndrome [61–63]. CRISPR systems can be used to combat viral illnesses by preventing viral reproduction while maintaining the normalcy of the affected cell. Cas9 employs conserved HBV areas that are crucial for virus survivability and reproduction and are not specifically targeted by traditional antiretroviral drugs [64]. The potential to destroy viruses is the most persistent issue. However, Ramanan et al. obtained the highest outcome of HBV cleavage using CRISPR/Cas9, which is a 92% reduction of covalently closed circular DNA (cccDNA) in in-vitro studies [65, 66].
Although there is an effective HBV vaccine, a cure is still unclear. Li et al. utilized CRISPR/Cas9 to delete a full-length 3,175-bp HBV DNA fragment from chronically infected cells that were chromosomally incorporated and episomally found, raising the possibility of absolute HBV prevention [67]. Unlike HBV pathophysiology, which is caused by elevated levels of viral infection, Human Papillomavirus (HPV) pathogenesis is caused by an ill-fated infection that multiplies in basal keratinocytes of the skin or mucosal epithelium [68]. Targeting the E7 gene using the CRISPR/Cas system promoted apoptosis, reduced proliferation and differentiation, and restored production of the pRb protein in HPV16 positive cervical cancer SiHa and Caski cell lines. As a consequence, exploiting the HPV16-E7 gene using the CRISPR/Cas technology might be a potential therapeutic tool for treating HPV infection and HPV-related cervical cancer [69, 70].
CRISPR/Cas9 controlled cleavage allows the treatment of disease-associated viral strains with exceptional reliability and accuracy [71]. Advances in genome editing using CRISPR/Cas9 support virological research and could provide a treatment for chronic herpesvirus infections by treating these viruses directly inside infected cells. Nucleoside analogues like Foscarnet, Valganciclovir, Ganciclovir, and are currently used to treat herpes virus infection [72]. CRISPR technologies are now being applied to diagnose and treat the new coronavirus (SARS-CoV-2), an emerging pathogen that has infected more than 25.5 million people and killed more than 850,000 people, which is of considerable urgent importance to scientists. SARS-CoV-2 is a Coronaviridae virus with an enveloped RNA genome that causes moderate to extreme symptoms in people of all ages [73, 74]. A novel approach to reducing SARS-CoV-2 replication to fight viral attack appears to be using the CRISPR/Cas system by targeting the positive-sense genome and viral mRNAs for endogenous closing viral genome templates and switching off their gene expression [75–77].
Bacterial infections and CRISPR/Cas
Though CRISPR technologies originated in bacteria, antibacterial therapies can be produced that can be provided via the cell wall. Nonetheless, it has recently been proven that it may be used in clinical studies to make bacteria susceptible to medicines and minimise horizontal transfer of resistant strains [78]. In Streptococcus thermophilus strains, scientists have identified two related CRISPR loci. The CRISPR system’s spacer sequences were discovered to somehow be analogous to several phage and chromosomal sequences, lending credence to the theory that CRISPR is a bacterial defensive mechanism against external elements [79]. Streptococcus pneumoniae possess the ability to inject the genetic material into the host through horizontal gene transfer or direct uptake [80]. In response to an immune system attack, S. pneumoniae changes the surface polysaccharide capsule to withstand antibodies and develops new capsule-encoding genes. According to research, the S. pneumoniae capsule gene seems to be the genesis of the CRISPR1 locus and therefore can prohibit rodents from surviving contamination by preventing the transition of these genes to non-capsulated S. pneumoniae. CRISPR/Cas interference can help to avoid the formation of new harmful strains since it provides a significant survival advantage for antibiotic resistance or pathogenicity [81, 82].
With about 10 million infections and 1.5 million fatalities per year, Mycobacterium tuberculosis (M. tb) is considered the world's most powerful disease. In this context, genetic alteration is important to better understand genetic mutations as well as the discovery of anti-M. tb medications and vaccinations in M. tb key genes [83]. Sequence-specific regulatory suppression in M. tb was achieved using CRISPR/Cas9 systems from S. pyogenes and S. thermophilus [84, 85]. Antibiotic-resistant Pseudomonas aeruginosa has emerged as a paradigm for new or previously overlooked antibacterial medicines [86].
P. aeruginosa CRISPR/Cas systems have previously been shown to be correlated with minimal genome sizes and restriction in sulphonamide tolerance of mobile genes, and P. aeruginosa has been indicated to be an example of a pathogenic bacteria where CRISPR/Cas is evident in the case of HGT [87]. Citorik et al. employed CRISPR/Cas9 to target sequences in Escherichia coli variants that allowed resistance to antibiotics and pathogenicity [88]. Studies predicted that impact can be produced using native or import systems with the type I-E CRISPR/Cas system in E. coli and is equally potent irrespective of the genomic site, strand or transcription of the objective sequence. Furthermore, the specificity of CRISPR RNA targeting enabled researchers to discern even highly related strains in pure or mixed cultures [89].
Fungal infections and CRISPR/Cas
For fungal infections, diverse variations of CRISPR/Cas9 methods have been developed, and each of these techniques could be the best option for a specific fungus. Saccharomyces cerevisiae, the yeast, was the first fungus for which CRISPR/Cas9 was used to manipulate the genome, and the fungus' ability to preserve plasmids aided the invention of CRISPR/Cas9 methods [90, 91]. Based on the original CRISPR/Cas9 studies in yeasts, the genetic manipulation method was altered for both model and non-model filamentous fungi. A CRISPR/Cas9 system had first been developed into the genome of filamentous fungus Trichoderma reesei by introducing a codon-optimized CAS9 gene through Agrobacterium tumefaciens-mediated transformation [92].
Other human fungal diseases, such as Malassezia, Fusarium, Blastomyces dermatitidis, and others, have been studied using CRISPR/Cas9 technologies; however, this work concentrated on major fungal pathogens seen in clinical settings. The efficacy and versatility of CRISPR/Cas9 have the potential to have a significant impact on the medical microbiology, both within the research facility enhancing genetic manipulation for investigating bacteria and fungi physiology and pathophysiology, deconstructing the function of virulence genes, and discovering different potential therapeutic targets and host–pathogen interaction—and at point-of-care enhancing the prognosis of fungal infected individuals [93].
The CRISPR/Cas9 genome editing has been used to effectively treat Candida albicans and Aspergillus spp., including the opportunistic pathogen A. fumigatus, which are both human-pathogenic fungi [94, 95]. Drug-resistant diploid Candida clinical isolate strains may be targeted using a codon-optimized Cas9 technology, according to research. This opens the door to investigating mechanisms that render Candida susceptible to antifungals utilizing CRISPR technology [96]. Arras et al. proved the adaptability of the CRISPR paradigm by disrupting the ADE2 gene in Cryptococcus neoformae. In a mouse inhalation model, Cas9-bearing C. neoformans strains were demonstrated to have no influence over virulence, making it a critical element in studying C. neoformans pathogenesis. [97, 98].
Furthermore, the CRISPR/Cas9 approach may well be utilised without modification in Myceliophthora thermophilic bacteria, such as M. heterothallica, implying that it might be employed in a wide range of thermophilic fungus. This widespread adaptability enables for in-depth examination of these fungi, some of which have uncommon properties such as a reproductive reproduction, both for fundamental research and the development of new hosts for commercial biotechnological applications [99, 100]. Mucormycosis is fungal infections caused by filamentous fungi Mucor circinelloides, belonging to the Mucorales order. Infection happens as spores are inhaled, ingested, or traumatically implanted into a vulnerable (i.e., immunocompromised) human [101]. The resultant disease mechanisms are marked by a high mortality rate and sometimes rapid clinical development. Mucorales infections are uncommon in immunocompetent people, but they can be lethal in immunocompromised patients [102]. CRISPR/Cas9 technology's potential to rapidly implement specific and accurate DNA sequence editing, as well as its effective adaptation in a large variety of fungal organisms indicates that it may be a promising advanced method for Rhizopus delemar research. A recent study found that CRISPR/Cas9 technology was active in Mucor circinelloides, a similar but distinct Mucormycoses pathogen [103, 104].
Potential applications of CRISPR/Cas9 system in diagnostics
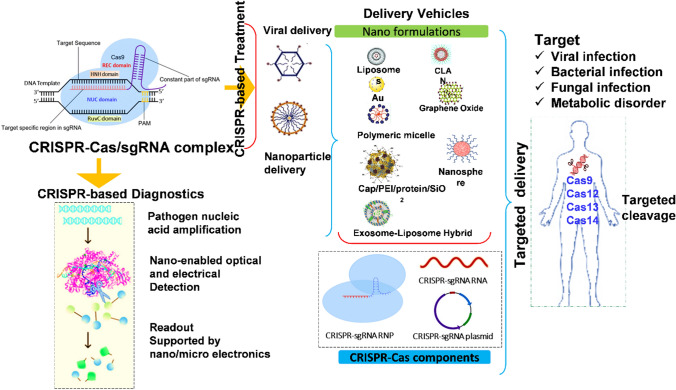
Infectious pathogen diagnostics depend heavily on an effective, quick, and low-cost nucleic acid detection system. The importance of early and reliable disease diagnosis in the implementation of early disease care, accompanied by effective adjustment of treatment measures, if possible, through easy monitoring, is critical. Point of Care (POC) diagnostic systems are effective tools for minimizing clinical delays, which is critical because delays or ineffective therapies will result in high mortality and the spread of infectious agents [105]. The most widely used tool, PCR-based diagnostics, has high sensitivity and accuracy, but it requires specialized equipment and skilled expertise, which restricts its usage. In the meantime, sequencing has become increasingly important in nucleic acid identification, but its high difficulty and expense preclude it from being used in fast on-site infectious diagnostics. As opposed to PCR, the nucleic acid isothermal amplification has emerged as a potential option for fast and compact identification, but its sensitivity and specificity must be enhanced [106, 107].
CRISPR technologies, which use specially engineered synthetic sgRNA, can detect nucleic acids implicated in both infectious and non-infectious diseases, as well as in the advancement of mobile screening procedures to advance the diagnosis, management, and prevention of infectious disease [26]. It has been demonstrated that the SARS-CoV-2 virus has mutated and produced variant in different countries among other factors such as age, ethnicity, location, area, etc. Exploring facets of nanobiotechnology to explore nanoscale diagnostic systems of selective SARS-CoV-2 detection is therefore a critical component of managing COVID-19 [108–110].
Methods that focus on CRISPR/Cas show considerable potential for identifying viruses including SARS-CoV-2, Zika virus (ZIKV), Dengue virus (DENV), Ebola, HPV, and M. tuberculosis because of their high accuracy and sensitivity, rapid, profitable, and ease of use [111, 112]. Importantly, the CRISPR/Cas-based identification system’s simplicity allows for the fast implementation of screening approaches in the event of an infectious disease epidemic. Further enhancements and changes are being made to the techniques to make them more flexible in the detection of bacterial and viral nucleic acids [113, 114]. For detection purposes, CRISPR/Cas utilize either the Cas13 enzyme in the indiscriminate cleaving of ssRNA or the Cas12a enzyme in the cleaving of single-beached DNA as “collateral action” [115]. The CRISPR/Cas technology relies on the following two methods for the detection of pathogens: specific cleavage of DNA and collateral cleavage.
Specific cleavage: Cas9
Several researchers working on infectious disease diagnostics have used CRISPR/Cas9. Researchers employed a hybrid strategy based on nucleic acid sequence-based amplification (NASBA) and the CRISPR/Cas9 technology to consistently identify between African and American Zika virus strains in in-vitro studies and a macaque model [116]. The researchers used (ds)DNA as a substrate for the Cas9 endonuclease, which is an intermediate in the NASBA amplification process. Following Cas9 cleavage along with a strain-specific PAM, the sgRNA-Cas9 complex cleaves the resulting dsDNA, resulting in curtailed or complete DNA fragments (Fig. 5). The ledge transition caused by full-length DNA fragments results in a colour shift that allowed the strains to be distinguished [117].
Fig. 5.
The mechanism of Cas9 cleavage of crRNA-tracrRNA target DNA. When the Cas9-crRNA-tracrRNA complex attaches to PAM-containing foreign DNA, Cas9 releases the double strands of genetic Material, allowing crRNA and foreign DNA to form a duplex. The REC lobe as well as the NUC lobe constitutes two separate components of Cas9. The REC lobe senses nucleic acid. The HNH, RuvC, and C-terminal regions of the NUC lobe are connected by a PAM (PI) interaction domain. The HNH and RuvC domains cleave the DNA strand to form a duplex of crRNA and other DNA, resulting in a double-stranded break in the target DNA
Because CRISPR is found in nearly all bacterial defence systems, detecting the CRISPR mechanism in microbial pathogens became a promising technique. Some diagnostic techniques based on these principles include CRISPR serotyping/subtyping, diagnostic assays classified as single guide RNA (sgRNA), and yet another diagnostic test based on CRISPR interference (dCas9) [118]. However, recent research indicated that CRISPR/Cas9 can be used for HIV infection therapy, as the DNA double-strand repair triggered by NHEJ upon Cas9 cleavage of HIV1 genomic regions produces a genetic variation at the cleavage site, facilitating viral immunity [56, 119].
The CRISPR/Cas9 method has been recognized as a valuable tool for detecting oncogenes and other cancer factors as well, and it has been incorporated into medical science and currently used to explore the genetic pathways in nearly all fields of cancer [120]. The strategy of CRISPR-Chip was another significant step forward in CRISPR diagnostics. CRISPR-Chip incorporates dCas9 protein with a graphene-based electronic transistor which binds to target DNA and used in identifying genetic alterations in clinical specimens from Duchenne muscular dystrophy patients [121]. Alternatively, CRISPR-Chip sensor rendered by immobilizing Cas9 with a sgRNA specific to the SARS COV-2 target sequence on a graphene-based field-effect transistor (GFET) produced electrical signal by complexing Cas9–sgRNA linking the target nucleic acid system without amplification [122, 123]. Quan and colleagues identified a tool for detecting pathogens resistant to antimicrobial therapy, finding low abundance sequences by hybridization (FLASH) in gram-positive bacteria such as S. aureus and malaria-causing Plasmodium falciparum. It incorporates a group of sgRNAs and Cas9 proteins to break the gene of interest into smaller fragments appropriate for more next-generation Illumina sequencing. In FLASH analysis, 90.1% of reads corresponded to targeted genes, contrasted to 0.3% in NGS alone. This accounted to a 293-fold increase in the average reads per million (rpM) of gene products. A sequencing resolution of 500 to 5000 reads was adequate for each FLASH-NGS sample to retrieve 10 or more reads per gene for 100% of gene products. Despite the fact that there were lesser on-target hits, at 100 fg (35 S. aureus genome copies in 30 l, or 1.9 aM), all intended regions were captured with at least 10 rpM, and more than 50% were enclosed at 10 fg (0.19 aM). Down to 0.4 pmol, which corresponds to 50 copies of each Cas9-guide RNA complex per S. aureus genomic copy, the quantity of Cas9 protein used seems to have little effect on template strand concentration [124].
CRISPR/Cas9-triggered nicking endonuclease-mediated strand displacement amplification (CRISDA) was developed in addition to the fluorescence technique for susceptible multiplication and identification of double-stranded DNA (dsDNA), and this method takes complete privilege of the increased selectivity, restrictive precision, and distinct morphological rearrangements of CRISPR signaling pathways in recognizing the target DNA. This employs Cas9 protein to sense a relatively specific target and create unique nicks in quasi-DNA that are subsequently replicated in vitro by DNA polymerases [125, 126]. Cas9 induced exponentially amplified response (Cas-EXPAR), a novel site-specific Cas9-based nucleotide detection method, was developed in 2018, based on nicking action of target-specific Cas9's nucleases [127]. CAS-EXPAR, when combined with an exponentially amplification process, can capture the target at relatively low concentrations as 0.82 amol (0.82 × 10−18 mol/L). This technology was then refined to check the methylation status of DNA fragments in Listeria monocytogenes RNA. Changes in DNA methylation were shown to have a significant association with diseases such as cancer. Cas-EXPAR offers a flexible and alternative for detecting basic or methylated DNA molecules, making it an ideal tool for early tumour diagnosis [128, 129].
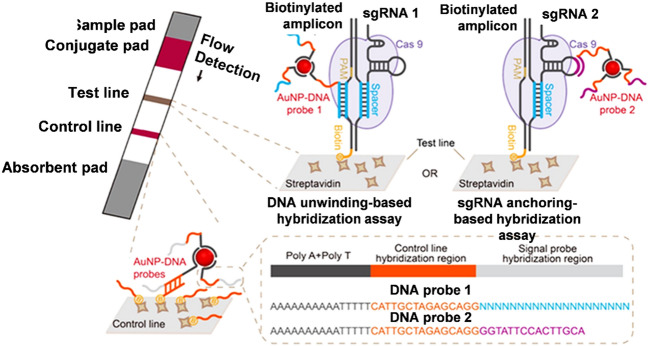
In a CRISPR/Cas9-mediated lateral flow nucleic acid assay (CASLFA), the amplified target sequence is combined with sgRNA, Cas9, and the Au-NP probe to identify African swine fever virus (ASFV). The sgRNA stabilisation technique is used to create an AuNP-DNA probe that can be used with a poly adenine (A)-Au affinity labelled probe. Genomic samples were effectively amplified utilizing isothermal amplification with biotinylated primers in the DNA unwinding-based hybridized studies (Fig. 6). The biotinylated amplicons trickled onto the sample pad and formed AuNP-DNA probe complexes that continued to cascade down the test line and were collected by the precoated streptavidin after a brief incubation with the designed Cas9/sgRNA1 for interaction [130]. CASLFA tests were performed on chromosomal DNA samples from ASFV-infected specimens in varied replicate ratios. Analytical sensitivity was assessed using both naked eye observations and grayscale analysis, yielding 200 copies of viral genomic DNA as the threshold limit that may be detected [130]. The main benefit of LFAs is that their findings are simple to comprehend because they are generally basic and can be seen with the naked eye. LFAs, on the other hand, lack precision and efficiency, making these traditional paper-based systems unsuitable for the more quantifiable evaluates required in clinical applications [131].
Fig. 6.
ASFV detection in suspected swine serum samples using the CASLFA technique.
(Copyright Permission American Chemical Society 2020, Ref. 129)
The FNCas9 Editor-Linked Uniform Detection Assay (FELUDA), a COVID-19 diagnostic platform, was developed by Azhar et al., and it employs FnCas9, fluorescein amidites (FaM) tracrRNA-sgRNA, and anti-FAM antibody conjugated with metal (gold) nanoparticles for rapid detection of infectious infections. They used RT-PCR amplification and accurate dFaCas9 binding to construct a lateral flow method for COVID-19 viral detection. Signaling augmentation by collateral fragmentation of reporter proteins is employed in several CRISPRDx systems to identify low copy counts of nucleic acids; however, FELUDA is quite efficient in detecting SNVs without it though. Its strength comes from its ability to accurately detect nucleotide bases and their variants. The affinity-based FELUDA tests could be used to create panels of mutation-scanning sgRNAs on a microchip for rapid monitoring of many targets from clinical material [132].
Collateral cleavage: orthologues CRISPR/Cas systems
In many Cas family orthologues, including Cas12, Cas13, and Cas14 effectors, cutting the targeted genetic material might result in disintegration of unwanted single-strand DNA (ssDNA) or single-strand RNA (ssRNA) [132]. Cas12a and Cas13 of the CRISPR/Cas orthologues family have been shown to have a remarkable potential to cause indiscriminate cleavage of ssDNA (single-stranded DNA) and RNA upon target identification. Because arbitrary cleavage activity may readily correlate detection of certain nucleic acid sequences with the database, this technology is used in diagnostics [133, 134].
CRISPR/Cas 12 system
Cas 12 is a CRISPR-associated effector protein (also known as Cpf1) from Class 2 that is utilised in CRISPR-based diagnostics [135]. For target cleavage, it comprises of a single RuvC endonuclease domain and T-enriched PAM sequence recognition. Its functioning in the ssDNase on a coupled dsDNA substrate is reflected in its dsDNA cleavage [136]. Doudna et al. proposed the DNA endonuclease-target CRISPR transporter (DETECTR), a CRISPR/Cas diagnostic platform for detecting high-risk, invasive genital tumours associated within human papillomaviruses HPV16 and HPV18, with a maximum precision of 96%. This successful presentation increases the future use of CRISPR/Cas technology in other areas from infectious diseases [133].
In a three-stage approach, DETECTR utilizes the Type V Cas12a enzyme for selective binding to Genomic substrates. First, a guide RNA within a particular viral genome directs theCas12a enzyme to a double-stranded DNA sequence. The Cas12a enzyme acts on the single-stranded DNA molecule through cleavage of the linking between quencher molecule and a reporter fluorophore arbitrarily until it binds to its viral genetic target (Fig. 7A). The fluorophore and quencher emit a fluorescent light, which is observed as “collateral” cleavage [26, 137]. In Lachnospiraceae bacterium ND2006, crystallography studies reported that Cas12a (LbCas12a) protein displays non-specific collateral involvement and degrades all the neighboring DNA molecules after target RNA is recognized. The researchers observed that LbCas12a-crRNA, coupled to a ssDNA activator molecule, facilitated trans-ssDNA cleavage at a frequency of 250 every sec, with a catalytic efficiency (kcat/Km) of 5.1108 s−1 M−1, using the fluorescence quencher (FQ)-labeled reporter experiment. LbCas12a-crRNA was able to achieve 1250 spins every second while being connected to a dsDNA activator, with a catalytic efficiency comparable to diffusion and a kcat/Km of 1.7109 s−1 M−1 [133, 138].
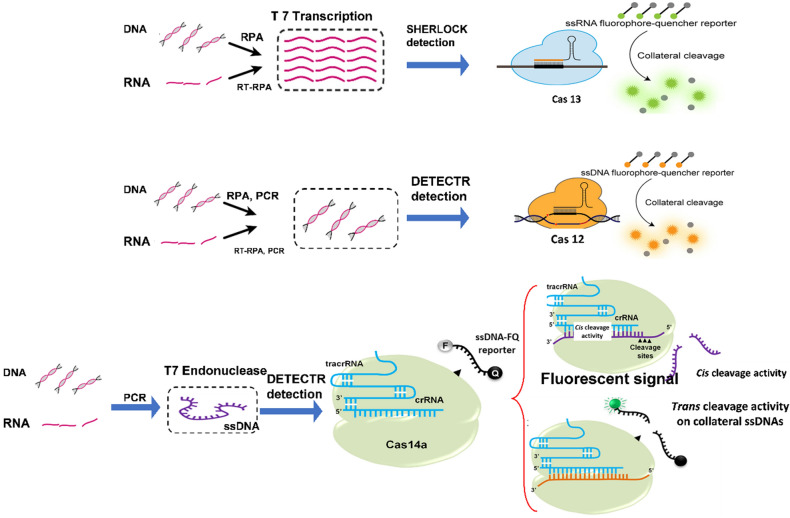
Fig. 7.
Overview of endonuclease involvement and nucleic acid detection mechanisms (collateral cleavage). A Cas12 Cleavage Activity: Binding of the Cas12-crRNA complex to the target (dsDNA), in the DETECTR technology, after the Cas12's collateral nuclease action activity allows non-specific cleavage of the reporter protein, enabling the excitation wavelength to be captured. B Cas13 Cleavage Activity: Cas13 is led by a single CRISPR RNA (crRNA) in the SHERLOCK technology for cleaving ssRNA or mRNA, and the similar mechanism continues. Target binding enables a single-beam (ssDNA; Cas12a) or single-beamed RNA (ssRNA; Cas13) collateral cleavage, given in trans for Cas12a and Cas13. C The Cas14a ribonucleoprotein single-stranded complex executing cis-cleavage on ssDNA substrates. A Cas14-gRNA-ssDNA tri-complex adjuvant is necessary for the cleavage of a ssDNA-fluorophore-quencher (FQ) reporter molecule and the emission of a fluorophore
The CRISPR-based Fluorescent Diagnostic System (CRISPR-FDS) was termed after studies demonstrated enhanced process parameters for the SARS-CoV2 detection assay by combining RT-PCR with a Cas12a-based nucleic acid detection technique. All experiments spiked with 108 copies of the target amplicon revealed a substantial CRISPR-FDS signal after a 20-min readout interval, whereas samples spiked with 109 copies indicated complete substrate conversion. Because single copy RNAs should be identified as replication frequencies above 0.69, the reported detection limits of 108 amplicons every CRISPR-FDS output specimen indicates that this method ought to be extremely tolerant of RT-RPA or RT-PCR pre-amplification effectiveness [139]. The DETECTR method pre-amplifies conserved gene regions in the N and E genes of SARS-CoV-2 using reverse transcription–loop-mediated isothermal amplification (RT-LAMP), and the amplicons are detected using Cas12a's collateral ssDNA cleavage activity [140].
Another study uses an ultra-sensitive and robust method to identify pulmonary tuberculosis using Cas12a/potential gRNAs. Researchers have developed a CRISPR/Cas12a-powered luminescence diagnostic test that includes isothermal recombinase polymerase amplification with Cas12a trans-cleavage activity and can be stimulated by target-specific DNA, allowing for accurate and sensitive quantification of the bacterium M. tb from clinical samples [141]. In addition, development of another Cas 12a protein HOLMES (one-hour low-cost multipurpose highly effective system) which uses PCR to enrich target nucleic acids instead of the DETECTR isothermal amplification method was applicated to diagnose DNA viruses, such as Japanese encephalitis virus and Aujeszky's disease virus [134].
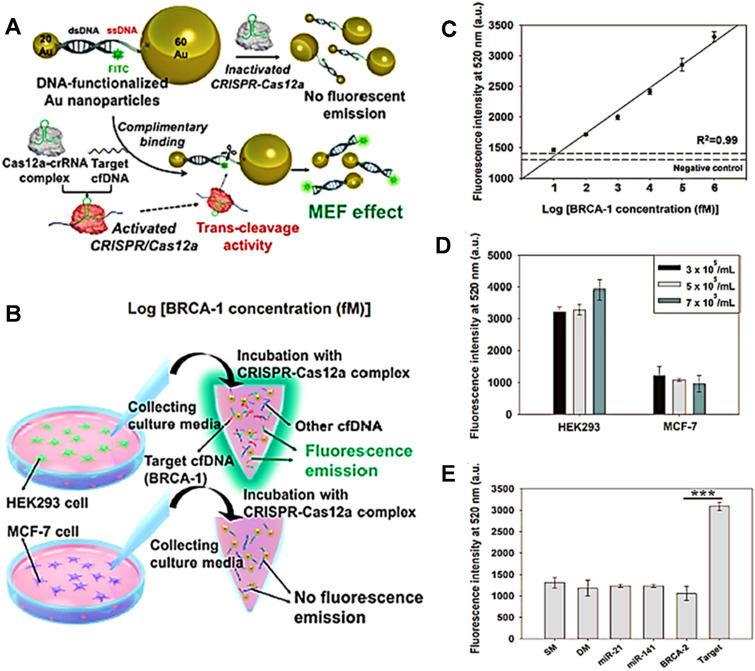
Further research in medical diagnostics reported the All-In-One Dual CRISPR/Cas12a (AIOD-CRISPR) detection tool for highly precise, ultrasensitive, and quicker visual identification of viral nucleic acids such as HIV and SARS-CoV2 genomic RNA with high sensitivity [142]. When incubated at 37 °C, the RT-AIOD-CRISPR had the maximum efficiency with 0.32 U/L AMV. Researchers further tested the sensitivities of the RT-AIOD-CRISPR assay using varying copy numbers of HIV-1 gag RNA templates [143]. Another technique for CRISPR/Cas12a diagnosis relies on Au nanoparticle (AuNP)-assisted metal-enhanced fluorescence (MEF) detection in breast cancer which does not necessitate nucleic acid amplification. Researchers tested a biomarker for sensitive and specific of intracellular caspase-3 based on MEF of pre-apoptotic cells, implying that the developed CRISPR/Cas12a nanosensor may be used to detect additional DNA targets such as other cfDNAs (cell-free DNA), genomic DNA, and viral DNA (Fig. 8). They created a CRISPRCas12a-assisted AuNP nanosensor with great sensitivity (as low as 0.34 fM) and speed for detecting cfDNA (less than 30 min). In comparison to earlier CRISPR-Cas-based detection systems, the suggested technique eliminated the obvious limitations of preamplifier approaches, such as time, cost, and the creation of false-positive signals. The suggested method effectively replaced targeted amplification with a signal intensification approach based on DNA complexed AuNP-based MEF. The approach not only demonstrated strong fluorescence enhancement mediated by the active CRISPR-Cas12a complex in association with target DNA, but also allowed for sensible target DNA concentration evaluation by colour change. Visual detection of viral nucleic acid using a combination of Au nanoparticles (AuNPs)-based colorimetry and CRISPR/Cas12a had been a success [144, 145].
Fig. 8.
Determination of cfDNA in buffer solution and human serum, both quantitatively and qualitatively. a Diagrammatic representation of MEF-based precise targeted cfDNA assessment without nucleic acid multiplication using CRISPR/Cas12a. The trans-cleavage (collateral effect) effect had been used to activate the CRISPR/Cas12a complex, which degraded the ssDNA between both the 20- and 60-AuNPs. The fluorescence intensity of dsDNA-functionalized 20-AuNP was higher than those of 60-AuNP. b An illustration depicting the steps how cfDNA is measured in cell culture medium. c Fluorescence intensities of Au nanosensor with activated CRISPR/Cas12a detection indicate that the intensity of the Au nanosensor was substantially related to the concentration of BRCA-1 d Comparative measurement of cfDNA fluorescent intensities with active CRISPR/Cas12a in HEK293 and MCF-7 culture medium from various cell numbers. e The fluorescence intensities of several nucleic acids (single-mismatched (SM), double-mismatched (DM), miR-21, miR-141, and BRCA-1 target DNA) were compared to validate selectivity sensing. (
Copyright permission American Chemical Society 2019 Ref. 145)
Study reports from the endpoint identification of Plasmodium species with programmed Cas12a detection and Au-NP, gold nanoparticles via lateral flow strip, resulted in cleavage of target sequences and collateral cleavage of spiked fluorophore-labelled ssDNA reporter detectable by fluorescent measurement or lateral flow data [146]. Studies demonstrated the ability of solid-state CRISPR/Cas12a-assisted nanopores (SCAN) to detect HIV-1 DNA with high sensitivity and specificity. SCAN was reported to be capable of detecting target DNA amounts greater than 10 nM in less than one hour. Preamplification procedures would likely be required at concentrations less than 10 nM, as in prior Cas12a-based experiments. The SCAN's specificity for identifying two separate sections of the HIV-1 gene has also been confirmed. The concept was applied to glass nanopores and HIV-1 DNAs in research and may be applied to other nanopore forms and DNA targets that could open new possibilities for molecular diagnostic applications [147]. Quantitative reporters enabling nucleic acid measurement include CRISPR/Cas12a combined with metallic nanoparticles connected to magnetic beads via single stranded DNAs. This technology can assess fake cell-free DNA samples in blood as well as numerous cancer abnormalities in pure DNA samples with allelic fractions as low as 0.01 percent [148].
CRISPR/Cas 13 system
Unlike Cas12, Cas13 is a single-unit enzyme in Class 2 type VI CRISPR-effector protein (also called C2c2) that targets single-stranded RNA with a high degree of precision [149] Cas13 enzymes also have collateral cleavage activity, like their type V complements, but most Cas13 orthologues need a protospacer flanking site (PFS) rather than a PAM to activate cleavage. The synthesis of these two features makes them appealing for diagnostic applications [150]. Zhang et al. designed Specific High Sensitivity Enzymatic Reporter Unlocking (SHERLOCK), an in-vitro nucleic acid detection tool, based on Cas13’s “collateral cleavage” action [151].
Cas13a is an RNA-binding protein, and, therefore, SHERLOCK relies on T7 RNA polymerase and reverse transcription (RT)-RPA for amplifying DNA to RNA for detection (Fig. 8B). However, the use of multiple procedures can result in cost and analytical variations. Furthermore, since RNA is less stable in human specimens than DNA, SHERLOCK can produce inaccurate results, posing challenges for clinical use [152]. Importantly, the studies revealed that SHERLOCK could differentiate between very closely related sequences with as little as one nucleotide difference by carefully designing the Cas13a guide RNA and thus be used to distinguish between distinct variants of the same virus, such as the American and African Zika virus strains, as well as genotypic SNPs in the human genome, such as cancer-causing mutations in the BRAF and EGFR genomes [153]. SHERLOCK uses a commercialized laterally flowing strips with a fluorescein (FAM) biotin RNA sensor for single-objective identification. Antibodies conjugated to AntiFAM AuNPs are used in the detection response. The existence of a second band indicates that the RNA sensor has been cleaved, enabling the FAM antibodies to run down the strip and attach to the second protein. The full-length RNA sensor accumulates at the first streptavidin line in a negative test [154].
Another study from Liu et al. reported enhanced detection method based on CRISPR/Cas13a nanosensor capable of directly detecting H7N9 virus and achieving high sensitivity. As a result, the point-of-care monitoring for the flu outbreak became a reality and this H7N9 diagnostic approach based on CRISPR/Cas13a will aid in the potential monitoring of the H7N9 influenza A virus epidemic [155]. For early disease detection and progression avoidance, still clinicians are using biomarkers for disease diagnosis. Studies reported CRISPR/Cas13a electrochemical microfluid biosensors for on-site microRNAs identification for brain cancer in the adolescent blood serum without the amplification micro-RNAs [156–158]. Furthermore, researchers developed Heating Unextracted Diagnostic Samples to Obliterate Nucleases (HUDSON), a tool which utilizes thermal and chemical reduction to hydrolyze viral proteins and deactivates the elevated amounts of proteolytic enzymes present in bodily secretions, designed to check viral nucleic acid effectively using SHERLOCK [159].
Researchers used HUDSON in conjunction with SHERLOCK to build a biosensor-based viral monitoring system for ZIKV and DENV, with exceptional productivity and little apparatus or sample processing requirements. This approach is as sensitive and specific as amplification-based nucleic acid diagnostics, with performance and apparatus requirements equivalent to quick antigen testing [160–162]. Using 16S rRNA CRISPR/Cas13-based biosensor, it is now possible to detect M. tb following reverse transcription is prior to and after amplification. In the meantime, preventing RNA depletion should be prioritized, which could restrict the CRISPR/Cas13 system’s use in tuberculosis molecular diagnosis [163]. Hu et al. (2020) outlined the prototype of DNA-AuNPs bioprobes using a freezing-based approach that was salt-free and did not require thiol manipulation. By combining the CRISPR/Cas13a method, these AuNPs bioprobes were used to detect RNA. The bioprobes described above were used to investigate Listeria monocytogenes, Pseudomonas aeruginosa, Staphylococcus aureus and Salmonella typhimurium [164].
CRISPR/Cas 14 system
Cas14 (also known as Cas12f) is a class 2 type V protein that is partial the size of other Cas enzymes and comprises 24 variations divided into three subgroups (Cas14a-c). It recognizes and cleaves ssDNA without the use of a protospacer adjacent motif (PAM) (Fig. 8C). This capability, together with Cas14's modest size, makes the CRISPR/Cas14-based technology a good choice for identifying DNA SNPs with high specificity [165, 166]. Cas14a is integrated into the DETECTR platform to create a new ssDNA detection system known as Cas14a-DETECTR, a virus detection system used for nasopharyngeal swabs and washings of infected children suffering from inadequately treated acute respiratory infections caused by another one of four human bocavirus genotypes (HBoV1) [167]. In owing to increased genotyping of SNPs, Cas14-DETECTR’s development offers potential for utility in identifying ssDNA viruses that are therapeutically, economically, and environmentally relevant. Cas12b-based DNA detection (CD detection) has been used to directly identify plasma HPV cell less tumour DNA in Cas14-DETECTR, demonstrating that Cas12b-based DNA detection (CD detection) might assist in quickly determining viral infectiousness [168].
Therapeutic applications of CRISPR/Cas technology
According to WHO’s 2019 Global Health Estimates, the global mortality rate from infectious illnesses is increasing significantly, accounting for up to seven of the world’s ten leading causes of death [169]. Diseases like polio, tetanus, measles, and so on can be avoided by vaccines, whereas the most common cure for infectious diseases is the use of pathogen-specific medicines. Bacteriophages (phage therapy), blood preparations (hemotherapy), gamma globulins, or immunoglobulins (serotherapy) are often used in addition to medications that specifically attack the pathogen [170]. Recent medicinal research explorations have led to the effective improvement of therapeutic agent functionality in disease management. However, there is a significant limitation to the effective treatment of multiple diseases, and that limitation is the distribution of the therapeutic drug to the target region. Targeted therapy is an approach to disease management that involves administering sufficient doses of drug carrier to the affected region of the body for an extended period. To address this, one of the ultimate aims of nanomedicine is the invention of safer and more efficient therapeutic nanoparticles [16, 171].
One of the therapeutic uses of CRISPR technology is the introduction of different tools into target cells under in vivo settings. The far more common carrier for administering such techniques are viral vectors, and numerous researchers have chosen adeno-associated virus (AAV) as their preferred vector; nevertheless, the number of exogenous genes that may be transported by these transgenic viruses is limited. Because most Cas enzymes have a large molecular weight, creating recombinant AAVs using Cas proteins and crRNA/sgRNA can be difficult [142, 172]. Drug-resistant bacteria and persistent viral diseases like HIV and hepatitis B virus (HBV) are the principal targets of novel CRISPR-based infectious disease treatments [13]. Several biomaterials, including lipid, polymeric, and inorganic nanoparticles, have recently been developed enabling transitory expression of CRISPR systems in vivo (NPs).
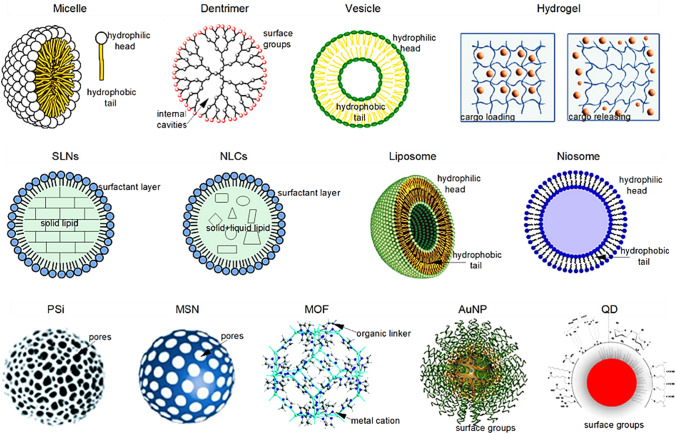
Nano-formulations and drug delivery approaches
Despite breakthroughs in CRISPR/Cas9 technology, significant therapeutic applications are unlikely in the future due to a lack of appropriate delivery technology. The scarcity of effective distribution mechanisms remains its most major constraint to genetic engineering's near-limitless possibilities. Conventional viral delivery systems based on retrovirus or adenoviruses have a high transfection effectiveness, but their intrinsic toxicity, which includes immune system response initiation and viral DNA invasion into recipient chromosomal, has limited their usage [173]. Adeno-associated viruses are considered safer, but their limited packing ability poses a restriction in the delivery of components of CRISPR/Cas9. Nonviral delivery systems are, instead, better than genome-editing components and are not limited by their size, but their distribution performance is comparatively poor [174]. A range of nonviral delivery mechanisms for genome editing has been developed because of recent developments in nanotechnology (Fig. 9).
Fig. 9.
Various nano system suitable to deliver CRISPR/Cas for therapeutics. The different approach combined with CRISPR/Cas aids in targeting various genetic and metabolic disorders. Lipid-based delivery systems investigate for lung cancer, heredity diseases, transthyretin amyloidosis, Alzheimer’s diseases, diabetes, leukemia, tyrosinemia, and hurler syndrome. Metal nanoparticle-based delivery systems investigate for Duchenne muscular dystrophy, X-fragile syndrome, Human Immunodeficiency Virus (HIV) infection, Human papillomavirus (HPV), autism spectrum disorders (ADS), central serous chorioretinopathy (CSCR-4), WHIM syndrome. Polymeric peptides-based delivery systems investigate for cancer, monosodium urate crystal, antibacterial resistance in MRSA, CML, hypercholesterolemia, intercranial tumor, osteosarcoma, septic shock, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Cell-fusion peptide-based delivery methods have been investigated for murine leukemia virus (MLV), Lesch-Nyhan syndrome and hyperuricemia, kidney, and urinary tract congenital abnormalities, and Enhanced Green Fluorescence Protein (E-GFP) tumors.
(Copyright Permission Elsevier 2021, Ref. 70)
Furthermore, several techniques for improving delivery effectiveness have been explored, including promoting endosomal escape, active targeting, and cargo release in response to stimuli. Natural extracellular vesicles, as well as synthetic nanoparticles based on liposomes, polymers, or polypeptides, have been used to deliver CRISPR/Cas9 [175]. Targetable unit manipulation on nanocarriers to accomplish specific gene delivery is a promising technique that is being extensively used in gene therapy to reduce risk factors on body cells [176]. Encapsulated nanocarrier protects susceptible CRISPR materials from degradation in biological fluids by nucleases and proteases. Since nucleic acids possess negative charges and Cas9 nanocarriers have net positive charge, the overall positive charge on the nanoparticle's surface helps facilitate cellular uptake by interacting with negatively charged cell membrane surface to increase load capacity and by contacting cells or cell-penetrating peptides (CPPs) to allow cell wall permeation [177]. In addition, physicochemical properties of nano-formulation are critical in targeting and release. Modifying the physicochemical properties of the nano-formulation in a particular way to target disease cells can enhance distribute drugs more effectively [178–180]. However, the activation of the delivered CRISPR device at disease sites has been difficult to monitor, rising the chance of nano-carrying or non-targeting the gene editing in healthy tissues which is inevitable if the CRISPR-based drug with high dosages is used [181].
Lipid-based targeted delivery of CRISPR/Cas9
Lipid NPs are spherical three-dimensional vesicles formed by a lipid molecule assembling. For the development of nanocarrier systems, lipid materials are well characterized because of the tertiary amines in their composition. Lipid nanoparticles (LNPs) predicated on ionizable cationic lipids have indeed been developed to stimulate endosomal egress by exhibiting a cationic charge in the decreased pH of late endosomes. Although these LNPs were designed to work with RNA interference (RNAi), they can also be used in the delivery of CRISPR/Cas systems [181–183]. A large variety of independently tested lipid nanocarriers for their capabilities to supply CRISPR/Cas9 devices have been studied. Research was conducted using a lipid-gold NP formulation to target Polo-like kinase-1, a mitosis regulator, overexpressed in many forms of cancer. This research modified HIV-1 Tat peptide cationic gold nanocluster to increase nuclear targeting and complexed it using electrostatic attraction with Cas9 protein and pDNA encoding sgRNA (Plk1 targets) [184].
In vivo and in vitro studies revealed a unique polyethylene glycol phospholipid-modified cationic lipid nanoparticle (PLNP)-based approach for efficiently delivering CRISPR/Cas9 to treated melanoma cells that was considerably more efficient than conventional transfection kits. Because of the CRISPR/Cas9 system’s effective gene-editing capacity and the excellent gene delivery capability of PLNPs, this approach is more accessible and secure than viral and/or hydrodynamic injection-based gene delivery approaches and has the potential to be probable in the treatment of a variety of gene-related illnesses, including tumours, infectious diseases, and heritable diseases [185]. One of the first promising Cas9 NP approaches employed viral and nonviral NP administration in hepatocytes to enable homology-directed regeneration in a murine model of hereditary tyrosinemia type 1. This method obtained genetic manipulation in 6 percent of hepatocytes with a 24.1-percent indel rate calculated by high-throughput sequencing of the desired target in total liver genomic DNA after adjusting the length of time of Lipid NP and Adeno-Associated Virus (AAV) administration to increase the traversal between peak Cas9 and sgRNA expression [186].
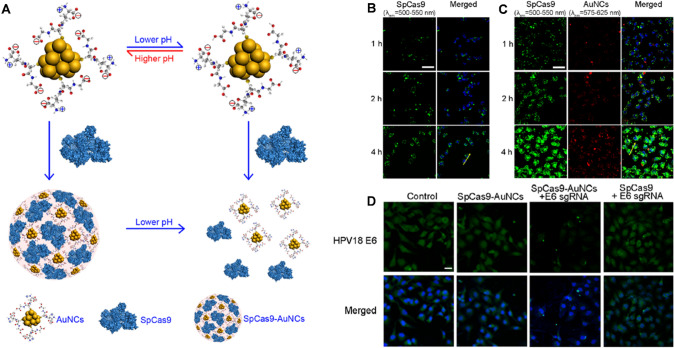
CRISPR/Cas9 nano complexes consisting of R7L10 peptides with Cas9-sgRNA ribonucleoprotein were shown to have the lowest off-target efficacy in adult mouse brains [187]. A single administration of CRISPR–Cas9 delivered by liver-targeting lipid nanoparticles (LNP), resulted in a 60% downregulation of the target gene (ATTR) and a 70–80% corresponding decrease in serum concentration of the dysfunctional transthyretin protein in a preclinical mouse model of transthyretin amyloidosis [188, 189]. Park et al. studied CRISPR/Cas9-loaded nanocomplexes that successfully targeted beta-secretase 1 “BACE1” in post-mitotic nerves in adult mouse brains and revealed therapeutic benefits in five familial Alzheimer’s disease (5XFAD) and APP knock-in models [190]. Other nanoscale combinations relying on electromagnetic contacts have been developed to transport CRISPR/Cas9. Cas9 and sgRNA encoding plasmid DNA that inactivates E6 and E7 HPVs in cervical cancer cells, leading to cell cycle inhibition and probable death of cells [191].
The therapeutic method was tested by interrupting the DPP-4 gene in Type 2 Diabetes Mellitus utilizing a lecithin-based liposomal nanocarrier particle (NL) to deliver Cas9-RNPs. Hepatocytes can effectively ingest lecithin-based NL particles during vivo tests in mice, suggesting that the greater potency of the gene disruption impact may be attributable to longer NL buildup in the liver. However, in a similar manner to incretins, downregulation of DPP-4 by NL Cas9-RNP treatment resulted in glucose tolerance and insulin resistance reconciliation [192]. Xu et al. developed a cationic lipid-assisted polymeric nanoparticle (CLANs), a flexible mechanism for delivering pCas9 into chronic myeloid leukaemia (CML) cells, B cells, neutrophils, and macrophages for in vivo gene editing [193].
Yin et al. demonstrated a method for delivering CRISPR/Cas9 components that employed two different vehicles: lipid nanoparticles provided Cas9 mRNA as well as an AAV administered the sgRNA/HDR sequence. They were able to repair the Fah (fumarylacetoacetate hydrolase) gene in the murine model of inherited tyrosinemia using the same strategy [194]. Other researchers reported a study that used a lipid nanoparticle carrier to encapsulate modified sgRNA and Cas9-encoding mRNA. After a single administration, the mouse Ttr (transthyretin) gene was reduced by more than 97 percent in serum protein levels in the liver [195]. Another study discovered that delivering Cas9 mRNA and sgRNA in vivo using a lipid nanoparticle containing disulphide bonds (BAMEA-O16B) had a significant tailoring effectiveness (80%) [196, 197].
Metal nanoparticles for efficient CRISPR/Cas9 delivery
Widder, Senyei, and others proposed magnetic micro- and nanoparticles for medication delivery in the late 1970s. Pharmacological substances are coupled to or enclosed within magnetic micro- or nanoparticles in this approach. Magnetic centers with protective coatings or functionalized polymers, as well as porous polymers containing magnetic particles, can be used to produce such nanostructures [198–200]. Because of their remarkable physicochemical stability and biocompatibility, gold-based nanoparticles (AuNC and AuNP) are increasingly being investigated as non-viral protein and DNA delivery methods. The AuNC is a gold-based nanostructure composed of golden molecule clusters that are smaller than 3 nm range in size. Several studies have documented the capacity of AuNC and AuNP endorsed multi-layer lipid NPs to deliver various CRISPR/Cas9 processes utilizing gold-based nanoparticles and lipid NPs’ delivery capabilities [201].
Since AuNPs facilitate the delivery of CRISPR/Cas content in various forms via endocytosis, they appear promising in the treatment of DMD since they are safe in terms of eliciting an immune response, providing better dose control, and allow multiple injections of the treatment. The effectiveness of dystrophin rescue and muscle strength enhancement of several CRISPR/Cas9 systems can be boosted by editing further pluripotent of the stem cells found in muscle tissues at different times. When used in combination with an NHEJ technique, gold nanoparticles may be more effectively in gene editing [202, 203].
In Thy1-YFP and Ai9 animals with X-fragile syndrome, CRISPR-Gold (CRISPR designed with gold nanoparticles) were utilised to transport Cas9 or Cpf1 to the brain via localized intracranial infusion, resulting in a significant reduction in recurring X-fragile syndrome symptoms (XFS) [204]. CRISPR Gold Cas9 RNP with prototype DNA was infused intramuscularly and repaired 5.4% of dystrophin genetic mutations. In a mouse model of autism spectrum disorder (ASD), intracranial infusion of CRISPR Gold Cas9 RNP caused targeted gene editing of 14.5% of the GRM5 gene and regained 40–50% of protein synthesis in the brain and other organs, reversing the consequences of ASD [205, 206].
The use of graphene oxide (GO)-based carriers in a genetic manipulation strategy based on CRISPR/Cas 9 technology has demonstrated to be a successful technique. Macromolecules in Cas9/single-guide RNA (sgRNA) combinations electrostatically loaded with PEI/PEG have been reported to transport CRISPR/Cas9 complexes in their genetic engineering process, where Cas9 retains endonuclease behaviour equivalent to much like its free form. GFP-expressing human AGS cells treated with GO-mediated Cas9/sgRNA demonstrated a 39 percent reduction in gene disruption while maintaining high cytocompatibility and low cell toxicity [207–209].
Studies have indicated that the magnetofection of viral and non-viral methods to the delivery CRISPR/Cas9 system in swine fibroblasts is the alternative approach. In combination with a non-uniform magnetic field, the use of PEI-Mag 2 iron oxide nanoparticles proved to be a successful, rapid, and non-toxic strategy to boost the particular action for CRISPR/Cas9 therapeutics. However, further experiments are required to applicate the use in in-vivo conditions towards the utilization of magnetic nanoparticles adapted to deliver the gene-editing mechanism of CRISPR/Cas9 into porcine fibroblast cell lines [210]. Kaushik et al. developed a delivery mechanism for the eradication of HIV infection, based on magnetic nanoparticles. Reports stated that the resulting nanoconjugates could cross an in vitro blood–brain barrier (BBB) model that demonstrated their ability in vivo to enter the central nervous system providing an important boost to CRISPR antiviral therapy (Fig. 10) [211–217].
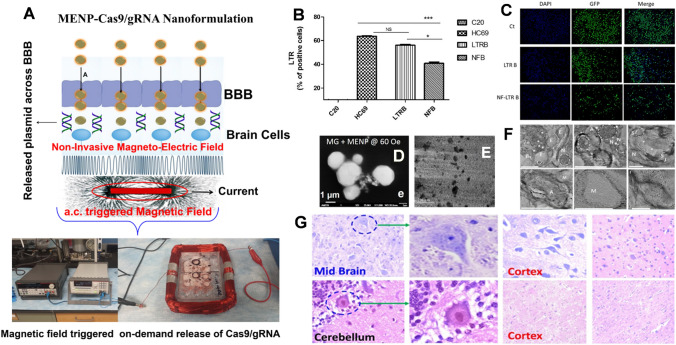
Fig. 10.
A The CRISPR/Cas9/gRNA bind with MENPs electrostatic interaction and their delivery across the BBB under the influence by static magnetic field. Set-up of ac-magnetic field stimulation (60 Oe for 30 min) were applied to release CRISPR/Cas9/gRNA (98%) from the nanomedicine MENPs-CRISPR/Cas9 nanomedicine. The efficacy of released CRISPT-Cas9 against eradicating HIV latent virus confirms using LTR expression (B) and imaging (C),
Copyright permission Springer Nature-2019 [210]. The cell-uptake evaluation of MENS as confirmed by SEM (D) and TEM (E), Copyright permission Springer Nature-2017 [214]. These MENPs were delivered to the brain of (F) mice, TEM imaging of post-MENPs injection mice brain tissue shows the uniform distribution of particle in all the brain cell types (Copyright permission Springer Nature-2016 [215] and G baboon, the histology studies confirmed that MENPs did not affect integrity of major brain (a) and peripheral (b) tissues. (Copyright permission American Chemical Society 2019 [216])
Magneto-electric nanoparticle (MENPs) are the core–shell nano-architecture and have been establish as an efficient drug nano-carries to deliver drug across the BBB, a non-invasive approach (Fig. 10). These MENPs due to desired surface charge bind with CRISPR/Cas9/gRNA (designed specifically to target latent and activated HIV infection) via electrostatic interaction [211]. A magnetically guided (0.3 T for 3 h) approach was optimized to deliver MENPs-CRISPT-Cas9 nanomedicine across the BBB without affecting the cellular junction of the BBB (Fig. 10A). After delivery confirmation, an externally controlled ac-magnetic field (60 Oe, pulse-to-pulse, for 30 min) to achieve 98% release of CRISPR/Cas9 from the nanomedicine. The efficacy of the released drug was evaluated by LTR expression assay (Fig. 10B) and further validated using fluorescent imaging (Fig. 10C). The results of the studies confirmed that release CRISPR/Cas9 is more efficient eradicate HIV then the pure. This might be due to easy cell-uptake of MENPs [215] under the influence of ac-magnetic field which is known to produce localized electric field and acoustics at the MENPS-cell interface (Fig. 10D, E). To establish MENP-CRISPR/Cas9/gRNA nanomedicine to treat neuroHIV, the magnetically guided delivery of MENPs have been demonstrated in mice [216] and non-human primate i.e., baboon [216]. The static magnetic field of 0.3 Tesla for mice and 3 T for baboon was applied from 3 h. The post-injection brain tissue TEM imaging confirmed [216] that MENPs are uniformly distributed in the all the cell type without any agglomeration (Fig. 10F). The effect of MENPs on mice brain tissue integrity, blood-toxicity profiling, and motor coordination function we also evaluated using appropriate techniques and outcomes project MENPs as a safe nano-system. In case of baboon, MRI was utilized to deliver MENPs to the brain and pre and post imaging analysis confirmed the successful delivery [216]. The MENPs are distributed in major region of interest and did not affect the integrity of brain and peripheral tissue without affecting toxicity profiling as confirmed by blood analysis up to 4 weeks (Fig. 10G). The results of these studies proposed MENPs as a potential nano-system for delivering a drug to the brain across the BBB and on-demand controlled release of the drug on ac-magnetic field. Investigation also suggests that MENPs are safe are suitable for clinical application where developing efficient therapies for brain diseases is the focus.
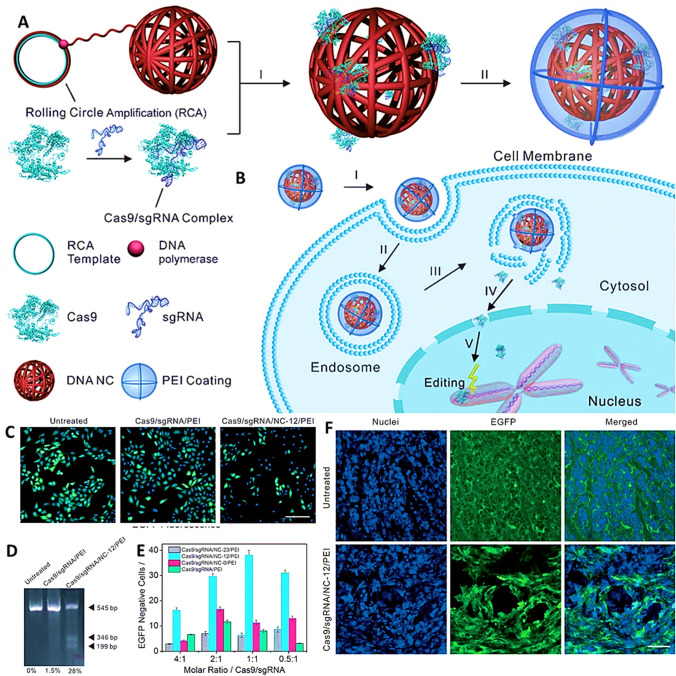
Polymeric peptide based CRISPR/Cas9 delivery
Polymeric nanoparticles obtained from polymerized or monomeric components have intensified their importance, over the years through natural biopolymers by developing new biomedical tools due to their compatibility controlled released and biodegradability [218]. Due to the diverse range of polymers available for biological transfection, encapsulating the Cas9 protein, DNA, or RNA inside polymeric carriers could be an approach for the delivery of Cas enzyme. Polymeric carriers, for example poly-lactic/glycolic acid copolymers (PLGA) or polycaprolactone (PCL), can be generated using natural monomers as coating materials on the magnetic nanoparticles because of their hydrophilic nature and reduced di-pole attractions between the magnetic nanoparticles. This is due to their hydrophilic atmosphere and thus to the fact that their di-polymeric attractions are reduced, and the results of this can be well dispersed aqueous nanoparticles [219].
Although polymeric carriers are often used to transport other proteins, there are only a few instances of polymeric Cas9 delivery in the literature. These may be due to the fact of high molecular weight dependence, multiple optimizations, difficult surface homogenous modifications and plus the ability for a surface or a carrier [220]. Wang et al. created a potent gene-editing method with increased structural penetrability for administering Cas9 and sgRNA plasmids for treatment of cancer. The polypeptide (poly(4((2(piperidin1yl) ethyl) aminomethyl) benzyl glutamate) PEGylated nanoparticles vector displayed 100% deletion effectiveness of polo-like kinase 1 (Plk1) in HeLa malignant cells, which decreased Plk1 expression levels and significantly suppressed tumor cell proliferation, increasing the survival period of tumour harboring rodents [221].
Liu et al. experimentally demonstrated delivery of expression vector employing cationic lipid aided PEG-PLGA nanoparticles (CLANs), encrypting Cas9 and sgRNA, relying on a repository of poly (ethylene glycol)-b-poly (lactide-co-glycolide) (PEG-b-PLGA) cationic lipoproteins, and it has been confirmed that such framework could well suppress the expression of pro – inflammatory elements [222]. Recently, researchers have demonstrated that plasmids encoding Cas9 and sgRNA delivered by poly (beta-amino ester) (PBAE) nanoparticles can accomplish 1-cut knockout and 2-cut deletion edits. The PBAE/DNA nanoparticles developed here to hold the promise for non-viral gene editing utilizing DNA and these findings further has implications for the creation and testing of next-generation non-viral delivery vehicles for CRISPR/Cas9 gene editing in general [223].
Researchers investigated the in vitro transfection of PLGA nanoparticles identified with fluorescent labelled encapsulating high molecular weight CRISPR/Cas9 plasmid DNA in murine macrophages derived from bone-marrow using the nanoprecipitation process, which forms particulate matter by incorporating a hydrogen-based organic polymer and medicinal solution in an aqueous solution with a polymer surfactant [224]. Timin and colleagues used polymeric and hybrid poly-l-arginine hydrochloride/dextran sulphate (PARG/DEXS)3 capsules to deliver CRISPR/Cas9 components for safe and effective gene editing. In the HEK293T cells, the dTomato gene was effectively knocked off, indicating that providing CRISPR/Cas9 mRNA results in better transfection than plasmid DNA transfection [225].
Sun et al. reported an in vivo delivery of polymeric core–shell nanoparticle loaded with Cas9-sgRNA complex through a PEI coating on a DNA nanoclew using U2OS-E-GFP tumour bearing mice and demonstrated E-GFP disruption by 25% [226]. The Cas9/sgRNA/NC-12/PEI nanoparticles carrying the AF647-labeled Cas9 were first assessed through confocal laser scanning microscopy (CLSM), a depth-selective approach for determining the position of given therapeutics inside the cells. Over the span of 6 h, the tagged Cas9 connects to the cell membrane, subsequently penetrates the cytosol, and finally localises to the nucleus, as demonstrated by the red fluorescence signal from AF647-Cas9 colocalizing with the blue fluorophore of labelled nuclei.Based on signs that Cas9/sgRNA would penetrate the nuclear membrane, researchers assessed the degree to what targeted DNA cleavage and repair via the endogenous NHEJ pathway could promote the creation of insertions and deletions. When designed to target the E-GFP coding region, most in-dels might alter the base sequence, preventing proper E-GFP production [226]. They investigated at how nanoparticles carrying either as the E-GFP-targeting sgRNA (Cas9/sgRNA/NC-12/PEI) or the non-targeting cgRNA (Cas9/cgRNA/NC-12/PEI) affected E-GFP expression. Confocal microscopy and flow cytometry studies revealed that the sgRNA suppressed fluorescence in 36% of the cells, while the cgRNA had no impact when compared with untreated cells (Fig. 11). Interestingly, when DNA nanoclew and sgRNA guide sequence were partly complementary, the editing efficiency was increased, which suggests that a modified DNA nanoclew may be used to include many multi-editing sgRNAs [227]. ZIFs are zeolitic imidazolate frameworks with imidazolate (C3H3N2) linkers and metal ions. These are metal-organic frameworks composites with very porosity architectures capable of encapsulating a wide range of organic compounds, minimal cytotoxicity, and pH sensitivity. These ZIF characteristics, according to a recent study, might be exploited to help in the spread of Cas9 RNP. When ZIF-Cas9 Ribonucleoprotein nanoparticles were injected into CHO cells, they enhanced endosomal escape and reduced target gene expression more effectively [228, 229].
Fig. 11.
Design of the DNA NC based CRISPR–Cas9 delivery system. A Cas9/sgRNA/NC/PEI preparation. I RCA generated the NC, which was then integrated with the Cas9/sgRNA complex. II Regarding improved endosome egress, PEI was coated onto Cas9/sgRNA/NC. B Cas9/sgRNA transport to the cell nucleus using a DNA NC-based transporter for genetic manipulation. I Adherence to the cell surface; II localization; III escape from endosome; IV nuclear transport; V chromosomal search and double-strand break introduction for genetic manipulation. C U2OS flow cytometry analysis and fluorescent microscopy depictions. Cas9/sgRNA/PEI and Cas9/sgRNA/NC-12/PEI treated E-GFP cells. Green indicates E-GFP, while blue represents Hoechst 33,342-stained nuclei. D Cas9/sgRNA genetic manipulation delivered via DNA NCs (8 mg mL−1) covered with PEI (10 mg mL−1). U2OS assay using T7EI. Cas9/gRNA/NC-12/PEI and Cas9/gRNA/PEI were used to treat E-GFP cells. E An E-GFP disruption test using Cas9/gRNA supplied via various DNA NCs. After treatment with Cas9/sgRNA/NC-23/PEI, Cas9/sgRNA/NC-12/PEI, Cas9/sgRNA/PEI at various Cas9/sgRNA molar ratios, the percentages of E-GFP negative cells are shown. F Cas9/sgRNA delivery into U2OS in vivo. Nude mice with E-GFP xenograft tumours. Tumor slices were taken ten days after Cas9/sgRNA/NC-12/PEI was injected intratumorally. An FITC-conjugated GFP antibody was used to label the E-GFPs, and Hoechst 33,342 was used to stain the nuclei. (
Copyright Permission Willey 2015, Ref. 226)
According to the studies, gold nanocrystals (AuNCs) can self-assemble alongside SpCas9 protein, and the resultant complex (SpCas9 AuNCs) effectively transfer SpCas9 protein into the nucleus of the cell. SpCas9-AuNCs are stable over a wide range pH level but disintegrate at lower pH levels (Fig. 12A). SpCas9 is transported inside cells via the assembling breakdown process, in which it conducts its fragmentation activity. Additionally, after transfecting HPV18 E6 sgRNA into cervical cancer cells, the researchers used self-assembled SpCas9AuNCs nanoparticles to successfully eliminate the E6 oncogene, reinstating the functioning of the tumor-suppressive protein p53 and causing death. Since successful intracellular uptake and transportation of SpCas9 into the nucleus of the cell is necessary for genetic disruptions, the assimilation of SpCas9AuNCs into HeLa cells were investigated by treating cells with SpCas9AuNCs for 1, 2, and 4 h (Fig. 12B). SpCas9 and AuNCs were detected in green and red fluorescences, respectively. Indigenous SpCas9 incubated alone were unable to penetrate the nuclear membrane even after 4 h of incubation. SpCas9-AuNCs-incubated cells, on the other hand, had a greater green fluorescence intensity over native SpCas9-incubated cells, indicating that perhaps the nanoparticles produced assisted the process of cellular uptake (Fig. 12C). Additionally, as the incubation time were prolonged, increased SpCas9 was found in the nucleus of the cell, where cleavage of target DNA occurs mediated by endonucleases. This finding was quantitatively supported by line scanning patterns of absorption spectra. The excellent cellular uptake capacity was further confirmed by flow cytometry (Fig. 12D). SpCas9AuNCs have also been found to be easily absorbed by BCP1 cells, which are extremely difficult to transfect, suggesting how this technology might be utilised to transfect a wide range of cell cultures which are difficult to transfect [230].
Fig. 12.
A pH-induced assembly disassembly of SpCas9AuNCs is depicted schematically. Electrostatic interaction enables AuNCs to self-assemble with SpCas9 at a higher pH, whilst SpCas9AuNCs deconstruct at the lower pH, weakening its contact with AuNCs. B Confocal laser scanning microscopy (CLSM) pictures of SpCas9 and SpCas9-AuNCs C treated HeLa cells at 1, 2, and 4 h, correspondingly. To monitor the SpCas9 dispersion within the cell, an anti-Cas9 antibody and an antimouse secondary antibody have been used in combination using Alexa Fluor 488 to stain the SpCas9 protein, while AuNCs was displayed in red fluorescence. D Western blot examination and immunofluorescence test of the HPV18 E6 protein in HeLa cells treated with SpCas9AuNCs, SpCas9AuNCs with E6 sgRNA, or SpCas9 with E6 sgRNA. (Copyright permission American Chemical Society 2019 Ref. 230)
Cell-fusion peptides for intracellular delivery of CRISPR/Cas9
CPPs (cell penetrating peptides) are an approach to evaluate while designing a delivery vector because they may make it much easier for cells to access and leave endosomes. Though cell penetrating peptides have indeed been utilised to deliver CRISPR–Cas9, when it is used alone, these are ineffective in protecting the elements from breakdown. CPPs, on the other hand, might be utilised as a supplement to numerous nanostructures [229]. The very first work to employ CPPs for cellular uptake of CRISPR/Cas9 into mammalian cells was reported by Ramakrishna et al. Traditionally, physical delivery techniques like as electroporation and microinjection were utilised, but the researchers herein co-produced Cas9 with CPP, with sgRNA targeting the C–C chemokine receptor type 5 (CCR5) in combination with a similar CPR which does not contain maleimide activity. Genetic disrupting efficiency (percent indels) just at CCR5 gene locus were tested using HeLa, HEK293T, NCCIT (mouse pluripotent embryonal carcinoma cell line), H9 human embryonic stem cells, and human dermal fibroblasts [177, 184].
Using dormant HIV-1-infected human 293 T cells, calcium phosphate transfection was utilised to transfer plasmid DNA containing Cas9 and sgRNA, which was engineered to assault the provirus genome, prohibiting viral proteins from being produced and removing inner genetic material from recipient cell nucleus [231, 232]. The complex molecular aggregation of NPs made from an amphiphilic peptide with Cas9 RNPs was reported by Lostale-Seijo et al. Using hydrazone binding, a cationic cell-penetrating peptide (P) was covalently and individually attached to a spectrum of lipophilic aldehyde tails (Tn) to generate the amphiphilic protein (PTn). The researchers scanned a repertoire of NPs synthesized with lipotropic aldehydes of varied carbon chains to determine the deletion proportion of the E-GFP gene in E-GFP-expressing HeLa cells [184, 233].
The CRISPR/Cas9 technique was effectively administered in mesenchymal stem cells (MSCs) for the first time utilizing hybridized extracellular vesicles, which were designed by merely incubating exosomes with liposomes. CRISPR/Cas9 has been a significant step forward in gene therapy, which has proven tough over decades. Nevertheless, the resultant hybrid nanoparticles could be released into the cytoplasm of MSCs and transcribed by incapsulated genes. Exosome–liposome hybrid nanoparticles in MSCs can supply the CRISPR–Cas9 mechanism, making them potential for in vivo gene manipulation [234, 235]. To facilitate distribution by cationic lipids-based transfection ingredients like RNAiMAX, a composite molecule comprising Cas9 and negatively supercharged proteins was designed. Injection of the Cas9 RNP/RNAiMAX combination into the cochlear of transgene Atoh1 (atonal bHLH transcription factor 1)-GFP mice led in a 13% drop in GFP in the transgene mice [236]. On the other hand, Mangeot and co-workers developed nano-blades, a vector based on the murine leukaemia virus (MLV), to transport Cas9 RNPs for in vivo gene editing [237].
Challenges associated with nano-CRISPR/Cas Systems
Despite its great specificity and sensitivity, CRISPR/Cas-based fluorescent nucleic acid detection has numerous drawbacks, including the need for pre-amplification techniques like LAMP and RPA, the high cost of fluorescent signal readout equipment, and scientific knowledge. However, while CRISPR/Cas-based lateral flow colorimetric detection only gives qualitative detection findings, it has been considered as a quick, reliable, and cost-effective technique for on-site diagnosis by the naked eye. Furthermore, the electrochemical biosensors based on CRISPR/Cas provide a very desired free pre-amplification approach for fast nucleic acid diagnostics. However, problems persist in miRNA detection due to their short size of 19-23 nucleotides, which clearly impedes the creation of sgRNAs [238]. Although SHERLOCK has demonstrated excellence in detecting pathogens, on no account efforts have been made to identify miRNA since its smaller size makes the amplification difficulty [239]. Nevertheless, this method has shown great potential in diagnostics and other site-specific applications, many factors such as immunogenicity, off-target site delivery methods and toxicity must be addressed to affect its effectiveness, particularly if being used for in vivo human gene therapy.
Compatibility with the host immune system
Targeted delivery of the CRISPR/Cas protein is necessary for the proper mechanism and expression of the Cas protein, hence, the immunogenicity risk of gene editing through CRISPR/Cas9 depends greatly on the path of the delivery. While Cas9 in vivo treatments pose distinct problems to vector immunogenicity, in-vitro cell delivery of Cas9 permits better inspection of residues in the product. Most therapies in CRISPR/Cas either attempt to express Cas9 by nucleic acids in the target cells or to put the protein into a viral capsid or nanoparticles. Anti-Cas9 neutralizing antibodies can therefore only affect success when Cas9 is exposed extracellularly and outside of the vector hull, for example, after tissue injection [240, 241].
One approach to this issue is the usage of a CRISPR/Cas System protein delivery that could have less immuno-genesis since the Cas protein is only available for a limited duration in the target cell [242]. The extent of immunogenicity, when combined with the immunogenic effects caused by some delivery vehicles, could render the CRISPR/Cas system ineffective. Exogenous RNA transmitted by lipid nanoparticles has been shown to stimulate Toll-like receptors and cause immune responses [243]. Lipid-based vectors can offer as a viable alternative to viral vectors for in vivo applications since lipids are less immunogenic than viruses [244].
The Cas9 protein (SpCas9) expression in mice as a cell and humoral immune response raises concerns about its safety and efficacy as gene therapy or epi-gene therapy in humans. Immune responses to other exogenous pathways of gene transfer, as well as preclinical models, show that Cas9 infections are immune to humans [245]. The CRISPR mechanism is usually packed into AAV vectors for gene therapy and does not come into close contact with the antibody, while the main immune response is the humoral immune response of AAVs, and the cellular immune response mediated by Cas enzyme. Cellular immunity to Cas enzyme has been reported, but all of these findings are focused on in vitro studies. It is still unclear if the Cas enzyme used in vivo can cause cellular immunity in these CRISPR-edited cells [246, 247].
To minimize off-targeting of CRISPR system.
Several reports have described that Cas9 binds to unpredicted genomic sites for cleavage, a phenomenon known as off-target effects. CRISPR/Cas mediated genome therapy off target results would seriously hinder its application, as it can cause genetic instability and increase the risk of some diseases by inserting undesirable mutations at non-target sites [248]. However, several scientists are trying to understand the factors affecting the variables that influence them and produced a variety of useful approaches to minimize off-target effects [249]. Even though the sgRNA is programmed to target a single gene of interest, the same Cas9/sgRNA also targets a large range of non-specific genes. Long-term constitutive expression of Cas9/sgRNA in gene-based CRISPR transmission exacerbates the problem: repetitive exposure of Cas9/sgRNA to non-specific genes will result in significant off-target results. Off-target effects have been reduced using a variety of approaches, including designing a high-specificity Cas9 protein [250, 251].
Novel delivery method for retention and higher efficacy
CRISPR/Cas components delivery is still also a significant challenge in gene therapy in the efficient delivery of individual cells, tissues and organs for accurate genome editing. An ideal distribution vector should have several advantages including non-toxicity, efficient delivery, low costs, high performance, and biodegradability [252]. As a result, the type of delivery vector should be selected with caution to understand the side effects of viral vectors. AAVs and adenoviruses, unlike lentiviruses, minimize the risk of unwanted DNA integration into the host genome, which unlike may pose a concern for biomedical studies [253].
However, in pre-clinical trials, the safety concern of viral vectors remains a big problem. For this purpose, researchers have focused on non-viral vectors, such as liposomes, polymers, and nanoparticles [254]. Nonviral vectors, such as cell penetrating peptides, lipid nanoparticles, lipoplexes/polyplexes, and gold nanoparticles, have a number of advantages over viral vectors, including ease of scale-up, lack of immunogenicity, and distribution of the CRISPR/Cas mechanism as RNPs, which reduces off-target effects [181]. Inorganic nanoparticles on the other hand, are easier to manufacture as compared to viral and lipid/polymer-based vectors, as they have a more predictable composition, size, and dispersion, are easier to describe and structurally flocculate, and are much more consistent over time. As a result, future investigations are directed on the in vitro and in vivo use of inorganic nanoparticles as CRISPR/Cas9 delivery vehicles [206].
In an AAV vector genome, for example, nucleic acid sequences cause TLR9 and induce interferon type I which increases the MHC expression on immune cells and targets epithelial cells. Hence, the adaptive immune response is initiated and the risk of Cas9 by pre-existing memory T cells is increased. Usually, LNPs include cationic lipids that aid in endocytosis, releases to cytoplasm and PEGylated lipids that help stabilize and deter unspecified interactions. PEGylation decreases immunogenicity and improves blood flow. An LNP-based lipid and PEG-DMG aid method for the enveloping of Cas9 mRNA and modified sgRNA has recently been able to develop a long-lasting liver editing of around 70% and benefiting from the multi-dose system [195, 206, 255].
Toxicity assessment with reference to the disease models
The Cas enzyme are derived from prokaryotic (bacterial) sources, hence in vivo administration of such proteins and the production of targeted antibodies into body tissues harboring these proteins is likely to cause cellular damage [256]. Since the development of this technology, the cause of Cas9/dCas9 toxicity in bacteria has alternated between evident and enigmatic. Early findings have shown the potential for non-specific nuclease activity toxicity of the Cas9 protein. However, the idea must be amended in comparable toxicities with dCas9 [257]. On the other hand, a low dose of NCs including metals, oxides of metals, polymers, and composites is usually non-toxic, prolonged delivery of such agents can lead to long-term aggregation and potential toxicity. As a result, more extensive research using in vivo subjects are required to determine the short- and long-term toxicity (small animals to non-human primates) [258].
Towards point of care testing for personalized health management
Despite its potential, CRISPR diagnosis address several problems, which most commonly occur in other point of care diagnoses, before to the widespread availability of dispersed testing [259]. Biosensing is a technology that identifies potential biological and (bio)chemical agents by using a biobased or biomimetic recognition element that either undergoes a (bio)chemical reaction (for instance, enzyme-based biosensors) or binds the protein of interest (i.e., analyte) in an extremely precise manner [260]. Post-service testing usually cost more per-test due to increasing complexity in the production process and the lack of automation prevalent in a laboratory environment. For example, the typical cost of a point-of-care influenza test is estimated at two to five times that of a central laboratory RT-PCR test. However other factors, such as sequence limitations, quantitative analysis, standardization, target amplification, multiplexing etc. are some of the challenges coming up in the way that could impact the point of care sensing systems.
Conclusion and future directions
CRISPR/Cas mechanisms in prokaryotes have heritable adaptive immunity. Clinical applications of CRISPR technologies are continuing to expand as new insights into the composition and operation of these structures are getting discovered. Initial research on CRISPR related treatments for infectious diseases have, to date, focused on detection and care of diseases resistant to pathogenic drug-resistant bacteria and of chronic viral infections, both of which contribute greatly to the global burden of disease, and to modern tools are needed to investigate underlying pathways and reliably detect and cure infections in all environments. While the first-generation gene therapy Zolgensma is labelled the “most expensive drug ever”, for the treatment of spinal muscular atrophy (SMA) in pediatric patients with a price of more than $2.1 million, still it cannot be considered as a permanent feature in the class of medicines. In the light of the boom of CRISPR-based research and gene therapy advancement and the spread of its use from rare genetic diseases to widespread disorders of ubiquity like chronic pain, cardiovascular diseases and Alzheimer's, the hope for a few thousand dollars in gene therapy is not completely unacceptable nor has it been uncommon in decades [206, 261]. The perspectives of Nano-CRISPR/Cas system to manage a targeted infectious disease in a personalized manner are illustrated in Fig. 13.
Fig. 13.
The perspective of nanotechnology assisted CRISPR/Cas system for efficient diagnostics and treatment of a viral infection/diseases, overall, towards personalized health management
Despite reliable and safe application, CRISPR-based infectious disease therapy continues to be a challenge. The CRISPR/Cas system has enormous potential for gene editing, but it has a substantial influence on CRISPR/Cas distribution to cells. However, there are still many areas of delivery that may be enhanced to increase the possibility for applicability:
Immunity to the CRISPR/Cas system and the vehicle used to administer it which can be resolved using a CRISPR/Cas system protein supply that is likely to have less immunogenicity, as the Cas protein is only present in the target cell for a limited time.
Biomaterials developed to improve delivery effectiveness, a high-capability and non-immunogenic cargo is needed for successful genetic modifying in a biocompatible, excellently, high-capacity freight and these characteristics are important for non-virus delivery design
Temporally and spatially regulation of Cas9 activity, the CRISPR/Cas system's unexpected off-target impact is a big concern and it's important to regulate CRISPR/Cas system components to particular target locations before Cas9 is turned on, as well as the distribution of certain factors that turn on this machinery at a specified time.
The efficiency at which antimicrobial resistance (AMR) is accomplished can be vastly improved if CRISPR/Cas constructs can be reprogrammed to address certain genes of interest. Such progress could have implications for combating reservoirs of resistance and could help antibiotics retain or regain their antimicrobial activity.
This quick screening of genetic material would aid in the discovery of genes involved in drug resistance to neuro-disintegration. As a result, researchers will be able to create successful therapeutics towards fighting both genetic and infectious diseases. Thorough genetic testing can help identify suppression variants such loss-of-function mutations in CCR5 and PCSK9, which safeguard against HIV infection and hypertriglyceridemia, respectively.
CRISPR-based diagnostics can detect antibiotic resistance mutations in bacterial/viral strains allowing us to quickly establish the proper treatment strategy, reducing overtreatment while also allowing us to gain a better understanding of the resistance.
If a mutated gene is hard to repair owing to its genomic context, a pseudogene that can be triggered to substitute the mutant gene may exist. However, if the illness is caused by a protein with abnormal properties (such as misfolding and aggregation in a tissue) (e.g., amyloidosis), the protein’s production might be suppressed at multiple sites along its expression pathway.
The ability of viral vectors to load CRISPR/Cas9 elements is typically inadequate. In this regard, nonviral vectors can be rationally engineered in terms of packaging CRISPR/Cas9 cargo effectively and are simpler to use for therapeutic purposes.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Authors do acknowledge their respective departments and institutions for providing support and facilities.
Author contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. In brief specifically, AKD visualized and articulated the article, VG, MK, and GO introduced the biological aspects with reference to various diseases, N-YK and C-ZL introduced engineering aspects, YM evaluated the aspects of materials science, AK edited and evaluated aspects of biosensing and drug delivery. All authors read and approved the final manuscript. All authors agree to take responsibility and be accountable for the contents of the manuscript.
Funding
YKM thanks funding by Interreg Deutschland–Denmark with money from the European Regional Development Fund, project number 096-1.1-18 (Access and Acceleration).
Declarations
Conflict of interest
Author declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fauci AS. Infectious diseases: considerations for the 21st Century. Clin. Infect. Dis. 2001;32:675–685. doi: 10.1086/319235. [DOI] [PubMed] [Google Scholar]
- 2.Menon GR, Singh L, Sharma P, Yadav P, Sharma S, Kalaskar S, Singh H, Adinarayanan S, Joshua V, Kulothungan V, Yadav J, Watson LK, Fadel SA, Suraweera W, Rao MV, Dhaliwal RS, Begum R, Sati P, Jamison DT, Jha P. National burden estimates of Healthy Life Lost in India, 2017: an analysis using direct mortality data and indirect disability data. Lancet Glob Health. 2019;7:E1675–E1684. doi: 10.1016/S2214-109X(19)30451-6. [DOI] [PubMed] [Google Scholar]
- 3.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakhrebah MA, Nassar MS, Alsuabeyl MS, Zaher WA, Meo SA. CRISPR technology: new paradigm to target the infectious disease pathogens. Eur. Rev. Med. Pharmacol. Sci. 2018;22:3448–3452. doi: 10.26355/eurrev_201806_15169. [DOI] [PubMed] [Google Scholar]
- 5.Caliendo AM, Gilbert DN, Ginocchio CC, et al. Better tests, Better Care: Improved diagnostics for infectious diseases. Clin. Infect. Dis. 2013;57:S139–170. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker DH. Principles of diagnosis of infectious diseases. Pathobiol. Hum. Dis. 222–225 (2014)
- 7.Watzinger F, Ebner K, Lion T. Detection and monitoring of virus infections by real-time PCR. Mol. Aspects. Med. 2006;27:254–298. doi: 10.1016/j.mam.2005.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foss DV, Hochstrasser ML, Wilson RC. Clinical applications of CRISPR-based genome editing and diagnostics. Transfusion. 2019;59:1389–1399. doi: 10.1111/trf.15126. [DOI] [PubMed] [Google Scholar]
- 9.Verma R, Sahu R, Singh DD, Egbo TE. A CRISPR/Cas9 based polymeric nanoparticles to treat/inhibit microbial infections. Semin. Cell. Dev. Biol. 2019;96:44–52. doi: 10.1016/j.semcdb.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 10.Rocha LFM, Braga LAM, Mota FB. Gene editing for treatment and prevention of human diseases: a global survey of gene editing-related researchers. Hum. Gene Ther. 2020;31:852–862. doi: 10.1089/hum.2020.136. [DOI] [PubMed] [Google Scholar]
- 11.Torres-Ruiz R, Rodriguez-Perales S. CRISPR/Cas9 technology: applications and human disease modelling. Brief Funct. Genomics. 2017;16:4–12. doi: 10.1093/bfgp/elw025. [DOI] [PubMed] [Google Scholar]
- 12.Trevisan M, Palù G, Barzon L. Genome editing technologies to fight infectious diseases. Exp. Rev Anti Infect Ther. 2017;15:1001–1013. doi: 10.1080/14787210.2017.1400379. [DOI] [PubMed] [Google Scholar]
- 13.Strich JR, Chertow DS. CRISPR/Cas biology and its application to infectious diseases. J. Clin. Microbiol. 2019;57:1307–1318. doi: 10.1128/JCM.01307-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mustafa MI, Makhawi AM. SHERLOCK and DETECTR: CRISPR/Cas systems as potential rapid diagnostic tools for emerging infectious diseases. J. Clin. Microbiol. 2021;59:e00745–e820. doi: 10.1128/JCM.00745-20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Yadav N, Narang J, Chhillar AK, Rana JS. CRISPR: A new paradigm of theranostics. Nanomedicine. 2021;33:102350. doi: 10.1016/j.nano.2020.102350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yetisgin AA, Cetinel S, Zuvin M, Kosar A, Kutlu O. Therapeutic nanoparticles and their targeted delivery applications. Molecules. 2020;25:2193. doi: 10.3390/molecules25092193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillaireau H, Couvreur P. Nanocarriers’ entry into the cell: relevance to drug delivery. Cell Mol Life Sci. 2009;66:2873–2896. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar P, Malik YS, Ganesh B, et al. CRISPR/Cas system: an approach with potentials for COVID-19 diagnosis and therapeutics. Front. Cell. Infect. Microbiol. 2020;10:576875. doi: 10.3389/fcimb.2020.576875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D, Bae S, Park J, et al. Digenome-seq: genome-wide profiling of CRISPR/Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. [DOI] [PubMed] [Google Scholar]
- 20.Lone BA, Karna SKL, Ahmad F, Shahi N, Pokharel YR. CRISPR/Cas9 system: a bacterial tailor for genomic engineering. Genet. Res. Int. 2018;2018:3797214. doi: 10.1155/2018/3797214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Rodríguez DR, Ramírez-Solís R, Garza-Elizondo MA, Garza-Rodríguez ML, Barrera-Saldaña HA. Genome editing: a perspective on the application of CRISPR/Cas9 to study human diseases (Review) Int. J. Mol. Med. 2019;43:1559–1574. doi: 10.3892/ijmm.2019.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang F, Doudna JA. CRISPR/Cas9 structures and mechanisms. Annu. Rev. Biophys. 2017;46:505–529. doi: 10.1146/annurev-biophys-062215-010822. [DOI] [PubMed] [Google Scholar]
- 23.Kennedy EM, Cullen BR. Gene Editing: a new tool for viral disease. Annu. Rev. Med. 2017;68:401–411. doi: 10.1146/annurev-med-051215-031129. [DOI] [PubMed] [Google Scholar]
- 24.Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 25.Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jolany Vangah S, Katalani C, Booneh HA, Hajizade A, Sijercic A, Ahmadian G. CRISPR-based diagnosis of infectious and noninfectious diseases. Biol. Proc. Online. 2020;22:22. doi: 10.1186/s12575-020-00135-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hille F, Charpentier E. CRISPR/Cas: biology, mechanisms and relevance. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2016;371:20150496. doi: 10.1098/rstb.2015.0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro LF, Ribeiro LFC, Barreto MQ, Ward RJ. Protein engineering strategies to expand CRISPR/Cas9 applications. Int. J. Genomics. 2018;2018:1652567. doi: 10.1155/2018/1652567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Wan T, Xin H, et al. Delivery of CRISPR/Cas9 for therapeutic genome editing. J. Gene Med. 2019;21:3107. doi: 10.1002/jgm.3107. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Huang Z. Recent advances in structural studies of the CRISPR/Cas-mediated genome editing tools. Natl. Sci. Rev. 2018;6:438–451. doi: 10.1093/nsr/nwy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR/Cas systems. Curr. Opin. Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinilla-Redondo R, Mayo-Muñoz D, Russel J, et al. Type IV CRISPR/Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res. 2020;48:2000–2012. doi: 10.1093/nar/gkz1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Makarova KS, Koonin EV. Annotation and Classification of CRISPR/Cas Systems. Methods Mol Biol. 2015;1311:47–75. doi: 10.1007/978-1-4939-2687-9_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, van der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR/Cas systems. Science. 2016;353:6299. doi: 10.1126/science.aad5147. [DOI] [PubMed] [Google Scholar]
- 35.Moon SB, Kim DY, Ko JH, Kim YS. Recent advances in the CRISPR genome editing tool set. Exp. Mo.l Med. 51 .1–11 (2019). [DOI] [PMC free article] [PubMed]
- 36.Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR/Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z, Dong H, Cui Y, Cong L, Zhang D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb. Cell Fact. 2020;19:172. doi: 10.1186/s12934-020-01431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell MR. Molecular Mechanisms of RNA Targeting by Cas13-containing Type VI CRISPR/Cas Systems. J. Mol. Biol. 2019;431:66–87. doi: 10.1016/j.jmb.2018.06.029. [DOI] [PubMed] [Google Scholar]
- 39.Toro N, Mestre MR, Martínez-Abarca F, González-Delgado A. Recruitment of Reverse Transcriptase-Cas1 Fusion Proteins by Type VI-A CRISPR/Cas Systems. Front. Microbiol. 2019;10:2160. doi: 10.3389/fmicb.2019.02160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rath D, Amlinger L, Rath A, Lundgren M. The CRISPR/Cas immune system: biology, mechanisms and applications. Biochimie. 2015;117:119–128. doi: 10.1016/j.biochi.2015.03.025. [DOI] [PubMed] [Google Scholar]
- 42.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pougach K, Semenova E, Bogdanova E, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol. Microbiol. 2010;77:1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Charpentier E, Richter H, van der Oost J, White MF. Biogenesis pathways of RNA guides in archaeal and bacterial CRISPR/Cas adaptive immunity. FEMS Microbiol Rev. 2015;39:428–441. doi: 10.1093/femsre/fuv023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carte J, Christopher RT, Smith JT, et al. The three major types of CRISPR/Cas systems function independently in CRISPR RNA biogenesis in Streptococcus thermophilus. Mol. Microbiol. 2014;93(1):98–112. doi: 10.1111/mmi.12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nam KH, Haitjema C, Liu X, et al. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR/Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol. Life Sci. 2014;71:449–465. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiedenheft B, van Duijn E, Bultema JB, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc. Natl. Acad. Sci. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barrangou R. CRISPR/Cas systems and RNA-guided interference. Wiley Interdiscip. Rev. RNA. 2013;4:267–278. doi: 10.1002/wrna.1159. [DOI] [PubMed] [Google Scholar]
- 50.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat. Rev. Genet. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duan C, Cao H, Zhang LH, Xu Z. Harnessing the CRISPR/Cas systems to combat antimicrobial resistance. Front. Microbiol. 2021;12:716064. doi: 10.3389/fmicb.2021.716064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uribe RV, Rathmer C, Jahn LJ, Ellabaan M, Li SS, Sommer M. Bacterial resistance to CRISPR/Cas antimicrobials. Sci. Rep. 2021;11:17267. doi: 10.1038/s41598-021-96735-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bikard D, Barrangou R. Using CRISPR/Cas systems as antimicrobials. Curr. Opin. Microbiol. 2017;37:155–160. doi: 10.1016/j.mib.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 54.Ramalingam S, Thangavel S. CRISPR/Cas9 probing of infectious diseases and genetic disorders. Indian J. Pediatr. 2019;86:1131–1135. doi: 10.1007/s12098-019-03037-9. [DOI] [PubMed] [Google Scholar]
- 55.Gholizadeh P, Aghazadeh M, Asgharzadeh M, Kafil HS. Suppressing the CRISPR/Cas adaptive immune system in bacterial infections. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:2043–2051. doi: 10.1007/s10096-017-3036-2. [DOI] [PubMed] [Google Scholar]
- 56.Doerflinger M, Forsyth W, Ebert G, Pellegrini M, Herold MJ. CRISPR/Cas9-the ultimate weapon to battle infectious diseases? Cell Microbiol. 2017;19:e12693. doi: 10.1111/cmi.12693. [DOI] [PubMed] [Google Scholar]
- 57.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao Q, Guo D, Chen S. Application of CRISPR/Cas9-based gene editing in HIV-1/AIDS therapy. Front Cell Infect Microbiol. 2019;9:69. doi: 10.3389/fcimb.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang G, Zhao N, Berkhout B, Das AT. CRISPR/Cas9 can inhibit HIV-1 replication but NHEJ repair facilitates virus escape. Mol. Ther. 2016;24:522–526. doi: 10.1038/mt.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das AT, Binda CS, Berkhout B. Elimination of infectious HIV DNA by CRISPR/Cas9. Curr. Opin. Virol. 2019;38:81–88. doi: 10.1016/j.coviro.2019.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin G, Zhang K, Li J. Application of CRISPR/Cas9 technology to HBV. Int J. Mol Sci. 2015;16:26077–26086. doi: 10.3390/ijms161125950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Block TM, Mehta AS, Fimmel CJ, Jordan R. Molecular viral oncology of hepatocellular carcinoma. Oncogene. 2003;22:5093–5107. doi: 10.1038/sj.onc.1206557. [DOI] [PubMed] [Google Scholar]
- 63.Lee WM. Hepatitis B virus infection. NEJM. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 64.Zeisel MB, Lucifora J, Mason WS, et al. Towards an HBV cure: state-of-the-art and unresolved questions–report of the ANRS workshop on HBV cure. Gut. 2015;64:1314–1326. doi: 10.1136/gutjnl-2014-308943. [DOI] [PubMed] [Google Scholar]
- 65.Ramanan V, Shlomai A, Cox DB, et al. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhen S, Hua L, Liu YH, et al. Harnessing the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 67.Li H, Sheng C, Wang S, et al. Removal of integrated hepatitis B virus DNA using CRISPR/Cas9. Front. Cell Infect. Microbiol. 2017;7:91. doi: 10.3389/fcimb.2017.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McLaughlin-Drubin ME, Münger K. Oncogenic activities of human papillomaviruses. Virus Res. 2009;143:195–208. doi: 10.1016/j.virusres.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hu Z, Yu L, Zhu D, et al. Disruption of HPV16-E7 by CRISPR/Cas system induces apoptosis and growth inhibition in HPV16 positive human cervical cancer cells. Biomed Res Int. 612823, (2014) [DOI] [PMC free article] [PubMed]
- 70.Xu X, Liu C, Wang Y, Koivisto O, Zhou J, Shu Y, Zhang H. Nanotechnology-based delivery of CRISPR/Cas9 for cancer treatment. Adv. Drug Deliv. Rev. 2021;176:113891. doi: 10.1016/j.addr.2021.113891. [DOI] [PubMed] [Google Scholar]
- 71.van Diemen, F.R., Lebbink, R.J.: CRISPR/Cas9, a powerful tool to target human herpesviruses. Cell Microbiol.19 (2017) [DOI] [PubMed]
- 72.Chen YC, Sheng J, Trang P, Liu F. Potential application of the CRISPR/Cas9 system against herpesvirus infections. Viruses. 2018;10:291. doi: 10.3390/v10060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nalawansha DA, Samarasinghe KTG. Double-barreled CRISPR technology as a novel treatment strategy for COVID-19. ACS Pharmacol. Transl. Sci. 2020;3:790–800. doi: 10.1021/acsptsci.0c00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR/Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Freije CA, Myhrvold C, Boehm CK, et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Konwarh R. Can CRISPR/Cas technology be a felicitous stratagem against the COVID-19 fiasco? Prospects and Hitches. Front. Mol. Biosci. 2020;7:557377. doi: 10.3389/fmolb.2020.557377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tiwari S, Juneja S, Ghosal A, Bandara N, Khan R, Wallen SL, Ramakrishna S, Kaushik A. Antibacterial and antiviral high-performance nanosystems to mitigate new SARS-CoV-2 variants of concern. Curr. Opin. Biomed. Eng. 2022;21:100363. doi: 10.1016/j.cobme.2021.100363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl. Acad. Sci. 2015;112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Loureiro A, da Silva GJ. CRISPR/Cas: converting a bacterial defence mechanism into a state-of-the-art genetic manipulation tool. Antibiotics. 2019;8:18. doi: 10.3390/antibiotics8010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Griffith F. The significance of pneumococcal types. J. Hyg. 1928;27:113–159. doi: 10.1017/S0022172400031879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Avery OT, Macleod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 83.Zumla A, Raviglione M, Hafner R. Fordham von Reyn C Tuberculosis. New Engl. J. Med. 2013;368:745–755. doi: 10.1056/NEJMra1200894. [DOI] [PubMed] [Google Scholar]
- 84.Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- 85.Singh AK, Carette X, Potluri LP, et al. Investigating essential gene function in Mycobacterium tuberculosis using an efficient CRISPR interference system. Nucleic Acids Res. 2016;44:143. doi: 10.1093/nar/gkw625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cady KC, Bondy-Denomy J, Heussler GE, Davidson AR, O'Toole GA. The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J. Bacteriol. 2012;194:5728–5738. doi: 10.1128/JB.01184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wheatley RM, MacLean RC. CRISPR/Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2021;15:1420–1433. doi: 10.1038/s41396-020-00860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR/Cas systems. MBio. 2014;5:e00928–e1013. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR/Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Q, Coleman JJ. Progress and challenges: development and implementation of CRISPR/Cas9 technology in filamentous fungi. Comput. Struct. Biotechnol. J. 2019;17:761–769. doi: 10.1016/j.csbj.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu R, Chen L, Jiang Y, Zhou Z, Zou G. Efficient genome editing in filamentous fungus Trichoderma reesei using the CRISPR/Cas9 system. Cell Discov. 2015;1:15007. doi: 10.1038/celldisc.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Morio F, Lombardi L, Butler G. The CRISPR toolbox in medical mycology: state of the art and perspectives. PLoS Pathog. 2020;16:1008201. doi: 10.1371/journal.ppat.1008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fuller KK, Chen S, Loros JJ, Dunlap JC. Development of the CRISPR/Cas9 system for targeted gene disruption in Aspergillus fumigatus. Eukaryot. Cell. 2015;14:1073–1080. doi: 10.1128/EC.00107-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Krappmann S. CRISPR/Cas9, the new kid on the block of fungal molecular biology. Med. Mycol. 2017;55:16–23. doi: 10.1093/mmy/myw097. [DOI] [PubMed] [Google Scholar]
- 96.Vyas VK, Barrasa MI, Fink GR. A Candida albicans CRISPR system permits genetic engineering of essential genes and gene families. Sci. Adv. 2015;1:1500248. doi: 10.1126/sciadv.1500248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shanmugam K, Ramalingam S, Venkataraman G, Hariharan GN. The CRISPR/Cas9 system for targeted genome engineering in free-living fungi: advances and opportunities for lichenized fungi. Front Microbiol. 2019;10:62. doi: 10.3389/fmicb.2019.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arras SD, Chua SM, Wizrah MS, Faint JA, Yap AS, Fraser JA. Targeted genome editing via CRISPR in the pathogen Cryptococcus neoformans. PLoS ONE. 2016;11:164322. doi: 10.1371/journal.pone.0164322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van den Brink J, van Muiswinkel GC, Theelen B, Hinz SW, de Vries RP. Efficient plant biomass degradation by thermophilic fungus Myceliophthora heterothallica. Appl. Environ. Microbiol. 2013;79:1316–1324. doi: 10.1128/AEM.02865-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu Q, Gao R, Li J, et al. Development of a genome-editing CRISPR/Cas9 system in thermophilic fungal Myceliophthora species and its application to hyper-cellulase production strain engineering. Biotechnol. Biofuels. 2017;10:1. doi: 10.1186/s13068-016-0693-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ribes JA, Vanover-Sams CL, Baker DJ. Zygomycetes in human disease. Clin Microbiol Rev. 2000;13:236–301. doi: 10.1128/CMR.13.2.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arroyo MA, Schmitt BH, Davis TE, Relich RF. Detection of the dimorphic phases of mucor circinelloides in blood cultures from an immunosuppressed female. Case Rep. Infect. Dis. 3720549 (2016) [DOI] [PMC free article] [PubMed]
- 103.Bruni GO, Zhong K, Lee SC, Wang P. CRISPR/Cas9 induces point mutation in the mucormycosis fungus Rhizopus delemar. Fungal Genet Biol. 2019;124:1–7. doi: 10.1016/j.fgb.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nagy G, Szebenyi C, Csernetics Á, et al. Development of a plasmid free CRISPR/Cas9 system for the genetic modification of Mucor circinelloides. Sci Rep. 2017;7:16800. doi: 10.1038/s41598-017-17118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumar S, Nehra M, Khurana S, et al. Aspects of point-of-care diagnostics for personalized health wellness. Int. J. Nanomed. 2021;16:383–402. doi: 10.2147/IJN.S267212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Shang X, Huang X. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg. Microbes Infect. 2020;9:1682–1691. doi: 10.1080/22221751.2020.1793689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem. Rev. 2015;115:12491–12545. doi: 10.1021/acs.chemrev.5b00428. [DOI] [PubMed] [Google Scholar]
- 108.Kaushik AK, Dhau JS, Gohel H, et al. Electrochemical sars-cov-2 sensing at point-of-care and artificial intelligence for intelligent covid-19 management. ACS Appl. Bio Mater. 2020;3:7306–7325. doi: 10.1021/acsabm.0c01004. [DOI] [PubMed] [Google Scholar]
- 109.Chen FE, Lee PW, Trick AY, et al. Point-of-care CRISPR/Cas-assisted SARS-CoV-2 detection in an automated and portable droplet magnetofluidic device. Biosens. Bioelectron. 2021;190:113390. doi: 10.1016/j.bios.2021.113390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Paliwal, P., Sargolzaei, S., Bhardwaj, S.K., Bhardwaj, V., Dixit, C., Kaushik, A.: Grand challenges in bio-nanotechnology to manage the covid-19 pandemic. Front. Nanotech. 2 (2020)
- 111.Jain S, Nehra M, Kumar R, et al. Internet of medical things (IoMT)-integrated biosensors for point-of-care testing of infectious diseases. Biosens Bioelectron. 2021;179:113074. doi: 10.1016/j.bios.2021.113074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaushik A. Manipulative magnetic nanomedicine: the future of COVID-19 pandemic/endemic therapy. Expert Opin Drug Deliv. 2021;18:531–534. doi: 10.1080/17425247.2021.1860938. [DOI] [PubMed] [Google Scholar]
- 113.Yuen KS, Wang ZM, Wong NM, et al. Suppression of Epstein-Barr virus DNA load in latently infected nasopharyngeal carcinoma cells by CRISPR/Cas9. Virus Res. 2018;244:296–303. doi: 10.1016/j.virusres.2017.04.019. [DOI] [PubMed] [Google Scholar]
- 114.Chertow DS. Next-generation diagnostics with CRISPR. Science. 2018;360:381–382. doi: 10.1126/science.aat4982. [DOI] [PubMed] [Google Scholar]
- 115.Chiu C. Cutting-edge infectious disease diagnostics with CRISPR. Cell Host Microbe. 2018;23:702–704. doi: 10.1016/j.chom.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pardee K, Green AA, Takahashi MK, et al. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165:1255–1266. doi: 10.1016/j.cell.2016.04.059. [DOI] [PubMed] [Google Scholar]
- 117.Xiang X, Qian K, Zhang Z, et al. CRISPR/Cas systems based molecular diagnostic tool for infectious diseases and emerging 2019 novel coronavirus (COVID-19) pneumonia. J. Drug Target. 2020;28:727–731. doi: 10.1080/1061186X.2020.1769637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Uppada V, Gokara M, Rasineni GK. Diagnosis and therapy with CRISPR advanced CRISPR based tools for point of care diagnostics and early therapies. Gene. 2018;656:22–29. doi: 10.1016/j.gene.2018.02.066. [DOI] [PubMed] [Google Scholar]
- 119.Wang Z, Pan Q, Gendron P, et al. CRISPR/Cas9-derived mutations both inhibit HIV-1 replication and accelerate viral escape. Cell. Rep. 2016;15:481–489. doi: 10.1016/j.celrep.2016.03.042. [DOI] [PubMed] [Google Scholar]
- 120.Tian X, Gu T, Patel S, Bode AM, Lee MH, Dong Z. CRISPR/Cas9—an evolving biological tool kit for cancer biology and oncology. NPJ Precis Oncol. 2019;3:8. doi: 10.1038/s41698-019-0080-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hajian R, Balderston S, Tran T, et al. Detection of unamplified target genes via CRISPR/Cas9 immobilized on a graphene field-effect transistor. Nat. Biomed. Eng. 2019;3:427–437. doi: 10.1038/s41551-019-0371-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bruch R, Urban GA, Dincer C. Unamplified gene sensing via Cas9 on graphene. Nat. Biomed. Eng. 2019;3:419–420. doi: 10.1038/s41551-019-0413-4. [DOI] [PubMed] [Google Scholar]
- 123.Cordaro A, Neri G, Sciortino MT, Scala A, Piperno A. Graphene-based strategies in liquid biopsy and in viral diseases diagnosis. Nanomaterials. 2020;10:1014. doi: 10.3390/nano10061014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Quan J, Langelier C, Kuchta A, et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019;47:83. doi: 10.1093/nar/gkz418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Srivastava S, Upadhyay DJ, Srivastava A. Next-generation molecular diagnostics development by CRISPR/Cas tool: rapid detection and surveillance of viral disease outbreaks. Front. Mol. Biosci. 2020;7:582499. doi: 10.3389/fmolb.2020.582499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhou W, Hu L, Ying L, Zhao Z, Chu PK, Yu XF. A CRISPR/Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat Commun. 2018;9:5012. doi: 10.1038/s41467-018-07324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Huang M, Zhou X, Wang H, Xing D. Clustered regularly interspaced short palindromic repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal Chem. 2018;90:2193–2200. doi: 10.1021/acs.analchem.7b04542. [DOI] [PubMed] [Google Scholar]
- 128.Jia F, Li X, Zhang C, Tang X. The expanded development and application of CRISPR system for sensitive nucleotide detection. Protein Cell. 2020;11:624–629. doi: 10.1007/s13238-020-00708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Field AE, Robertson NA, Wang T, Havas A, Ideker T, Adams PD. DNA methylation clocks in aging: categories, causes, and consequences. Mol. Cell. 2018;71:882–895. doi: 10.1016/j.molcel.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wang X, Xiong E, Tian T, et al. Clustered regularly interspaced short palindromic repeats/Cas9-mediated lateral flow nucleic acid assay. ACS Nano. 2020;14:2497–2508. doi: 10.1021/acsnano.0c00022. [DOI] [PubMed] [Google Scholar]
- 131.van Dongen JE, Berendsen JTW, Steenbergen RDM, Wolthuis RMF, Eijkel JCT, Segerink LI. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens. Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Azhar M, Phutela R, Ansari AH, et al. Rapid, field-deployable nucleobase detection and identification using FnCas9. biorxiv. 2020.
- 133.Chen JS, Ma E, Harrington LB, et al. CRISPR/Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li SY, Cheng QX, Wang JM, et al. CRISPR/Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rusk N. Spotlight on Cas12. Nat Methods. 2019;16:215. doi: 10.1038/s41592-019-0347-5. [DOI] [PubMed] [Google Scholar]
- 136.Paul B, Montoya G. CRISPR/Cas12a: Functional overview and applications. Biomed. J. 2020;43:8–17. doi: 10.1016/j.bj.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kocak DD, Gersbach CA. From CRISPR scissors to virus sensors. Nature. 2018;557:168–169. doi: 10.1038/d41586-018-04975-8. [DOI] [PubMed] [Google Scholar]
- 138.Murugan K, Seetharam AS, Severin AJ, Sashital DG. CRISPR/Cas12a has widespread off-target and dsDNA-nicking effects. J. Biol. Chem. 2020;295:5538–5553. doi: 10.1074/jbc.RA120.012933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Huang Z, Tian D, Liu Y, et al. Ultra-sensitive and high-throughput CRISPR-powered COVID-19 diagnosis. Biosens. Bioelectron. 2020;164:112316. doi: 10.1016/j.bios.2020.112316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Palaz F, Kalkan AK, Tozluyurt A, Ozsoz M. CRISPR-based tools: alternative methods for the diagnosis of COVID-19. Clin. Biochem. 2021;89:1–13. doi: 10.1016/j.clinbiochem.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xu H, Zhang X, Cai Z, et al. An isothermal method for sensitive detection of mycobacterium tuberculosis complex using clustered regularly interspaced short palindromic repeats/Cas12a Cis and trans cleavage. J. Mol. Diagn. 2020;22:1020–1029. doi: 10.1016/j.jmoldx.2020.04.212. [DOI] [PubMed] [Google Scholar]
- 142.Kumar P, Malik YS, Ganesh B, et al. CRISPR/Cas system: an approach with potentials for COVID-19 diagnosis and therapeutics. Front. Cell Infect. Microbiol. 2020;10:576875. doi: 10.3389/fcimb.2020.576875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ding, X, Yin, K, Li, Z, Liu, C.: All-in-One Dual CRISPR/Cas12a (AIOD-CRISPR) assay: a case for rapid, ultrasensitive and visual detection of novel coronavirus SARS-CoV-2 and HIV virus. BioRxiv. 2020.03.19.998724 (2020).
- 144.Phan QA, Truong LB, Medina-Cruz D, Dincer C, Mostafavi E. CRISPR/Cas-powered nanobiosensors for diagnostics. Biosens. Bioelectr. 2022;197:113732. doi: 10.1016/j.bios.2021.113732. [DOI] [PubMed] [Google Scholar]
- 145.Choi JH, Lim J, Shin M, Paek SH, Choi JW. CRISPR/Cas12a-based nucleic acid amplification-free DNA biosensor via Au nanoparticle-assisted metal-enhanced fluorescence and colorimetric analysis. Nano Lett. 2021;21:693–699. doi: 10.1021/acs.nanolett.0c04303. [DOI] [PubMed] [Google Scholar]
- 146.Lee RA, Puig H, Nguyen PQ, et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc. Natl. Acad. Sci. 2020;117:25722–25731. doi: 10.1073/pnas.2010196117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nouri R, Jiang Y, Lian XL, Guan W. Sequence-specific recognition of HIV-1 DNA with solid-state CRISPR/Cas12a-assisted nanopores (SCAN) ACS Sens. 2020;5:1273–1280. doi: 10.1021/acssensors.0c00497. [DOI] [PubMed] [Google Scholar]
- 148.Shao N, Han X, Song Y, Zhang P, Qin L. CRISPR/Cas12a coupled with platinum nanoreporter for visual quantification of SNVs on a volumetric bar-chart chip. Anal. Chem. 2019;91:12384–12391. doi: 10.1021/acs.analchem.9b02925. [DOI] [PubMed] [Google Scholar]
- 149.Abudayyeh OO, Gootenberg JS, Konermann S, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gootenberg JS, Abudayyeh OO, Lee JW, et al. Nucleic acid detection with CRISPR/Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tsou JH, Leng Q, Jiang F. A CRISPR test for detection of circulating nuclei acids. Transl. Oncol. 2019;12:1566–1573. doi: 10.1016/j.tranon.2019.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Sashital DG. Pathogen detection in the CRISPR/Cas era. Genome Med. 2018;10:32. doi: 10.1186/s13073-018-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Shihong Gao D, Zhu X, Lu B. Development and application of sensitive, specific, and rapid CRISPR/Cas13-based diagnosis. J Med Virol. 2021;93:4198–4204. doi: 10.1002/jmv.26889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu Y, Xu H, Liu C, et al. CRISPR/Cas13a nanomachine based simple technology for avian influenza A (H7N9) virus on-site detection. J. Biomed Nanotechnol. 2019;15:790–798. doi: 10.1166/jbn.2019.2742. [DOI] [PubMed] [Google Scholar]
- 156.Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, Dincer C, Urban GA. CRISPR/cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free MIRNA diagnostics. Adv. Mater. 2019;31:1905311. doi: 10.1002/adma.201905311. [DOI] [PubMed] [Google Scholar]
- 157.Ibrahim AU, Al-Turjman F, Sa'id Z, Ozsoz M. Futuristic CRISPR-based biosensing in the cloud and internet of things era: an overview. Multimed Tools Appl. 2020;8:1–29. doi: 10.1007/s11042-020-09010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sheng Y, Zhang T, Zhang S, Johnston M, Zheng X, Shan Y, Liu T, Huang Z, Qian F, Xie Z, Ai Y, Zhong H, Kuang T, Dincer C, Urban GA, Hu J. A CRISPR/Cas13a-powered catalytic electrochemical biosensor for successive and highly sensitive RNA diagnostics. Biosens. Bioelectr. 2021;178:113027. doi: 10.1016/j.bios.2021.113027. [DOI] [PubMed] [Google Scholar]
- 159.Myhrvold C, Freije CA, Gootenberg JS, et al. Field-deployable viral diagnostics using CRISPR/Cas13. Science. 2018;360:444–448. doi: 10.1126/science.aas8836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Chotiwan N, Brewster CD, Magalhaes T, et al. Rapid and specific detection of Asian- and African-lineage Zika viruses. Sci. Transl. Med. 2017;9:538. doi: 10.1126/scitranslmed.aag0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Chen W, Huang Z, Hu S, et al. Invited review: Advancements in lateral flow immunoassays for screening hazardous substances in milk and milk powder. J. Dairy Sci. 2019;102:1887–1900. doi: 10.3168/jds.2018-15462. [DOI] [PubMed] [Google Scholar]
- 162.Khambhati K, Bhattacharjee G, Singh V. Current progress in CRISPR-based diagnostic platforms. J. Cell Biochem. 2019;120:2721–2725. doi: 10.1002/jcb.27690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lyu C, Shi H, Cui Y, et al. CRISPR-based biosensing is prospective for rapid and sensitive diagnosis of pediatric tuberculosis. Int. J. Infect. Dis. 2020;101:183–187. doi: 10.1016/j.ijid.2020.09.1428. [DOI] [PubMed] [Google Scholar]
- 164.Hu M, Yuan C, Tian T, et al. Single-step, salt-aging-free, and thiol-free freezing construction of AuNP-based bioprobes for advancing CRISPR-based diagnostics. J. Am. Chem. Soc. 2020;142:7506–7513. doi: 10.1021/jacs.0c00217. [DOI] [PubMed] [Google Scholar]
- 165.Karvelis T, Bigelyte G, Young JK, et al. PAM recognition by miniature CRISPR/Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020;48:5016–5023. doi: 10.1093/nar/gkaa208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Harrington LB, Burstein D, Chen JS, et al. Programmed DNA destruction by miniature CRISPR/Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Aquino-Jarquin G. CRISPR/Cas14 is now part of the artillery for gene editing and molecular diagnostic. Nanomedicine. 2019;18:428–431. doi: 10.1016/j.nano.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 168.Vatankhah M, Azizi A, Sanajouyan Langeroudi A, et al. CRISPR-based biosensing systems: a way to rapidly diagnose COVID-19. Crit. Rev. Clin. Lab. Sci. 2021;58:225–241. doi: 10.1080/10408363.2020.1849010. [DOI] [PubMed] [Google Scholar]
- 169.Global health estimates. World Health Organization. https://www.who.int/data/global-health-estimates. Accessed 10 Sept (2021).
- 170.Zaychikova MV, Danilenko VN, Maslov DA. CRISPR/Cas systems: prospects for use in medicine. Appl. Sci. 2020;10:9001. doi: 10.3390/app10249001. [DOI] [Google Scholar]
- 171.Vashist A, Kaushik A, Vashist A, et al. Recent trends on hydrogel-based drug delivery systems for infectious diseases. Biomater Sci. 2016;4:1535–1553. doi: 10.1039/C6BM00276E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yin LJ, Hu SQ, Guo F. The application of CRISPR-Cas9 gene editing technology in viral infection diseases. Yi Chuan Hereditas. 2015;37:412–418. doi: 10.16288/j.yczz.14-460. [DOI] [PubMed] [Google Scholar]
- 173.Kim D, Le QV, Wu Y, Park J, Oh YK. Nanovesicle-mediated delivery systems for CRISPR/Cas genome editing. Pharmaceutics. 2020;12:1233. doi: 10.3390/pharmaceutics12121233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wu Z, Yang H, Colosi P. Effect of genome size on AAV vector packaging. Mol Ther. 2010;18:80–86. doi: 10.1038/mt.2009.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Elmowafy EM, Tiboni M, Soliman ME. Biocompatibility, biodegradation and biomedical applications of poly (lactic acid)/poly (lactic-co-glycolic acid) micro and nanoparticles. J. Pharm. Investig. 2019;49:347–380. doi: 10.1007/s40005-019-00439-x. [DOI] [Google Scholar]
- 176.Lyu Y, Yang C, Lyu X, Pu K. Active delivery of CRISPR system using targetable or controllable nanocarriers. Small. 2021;17:2005222. doi: 10.1002/smll.202005222. [DOI] [PubMed] [Google Scholar]
- 177.Ramakrishna S, Kwaku Dad AB, Beloor J, Gopalappa R, Lee SK, Kim H. Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. 2014;24:1020–1027. doi: 10.1101/gr.171264.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gage A, Brunson K, Morris K, et al. Perspectives of manipulative and high-performance nanosystems to manage consequences of emerging new severe acute respiratory syndrome coronavirus 2 variants. Front. Nanotechnol. 3. (2021)
- 179.Bhardwaj V, Kaushik A, Khatib ZM, Nair M, McGoron AJ. Recalcitrant issues and new frontiers in nano-pharmacology. Front. Pharmacol. 2019;10:1369. doi: 10.3389/fphar.2019.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ortiz-Casas B, Galdámez-Martínez A, Gutiérrez-Flores J, et al. Bio-acceptable 0d and 1d ZnO nanostructures for cancer diagnostics and treatment. Mater. Today. 2021;50:533–569. doi: 10.1016/j.mattod.2021.07.025. [DOI] [Google Scholar]
- 181.Wilbie D, Walther J, Mastrobattista E. Delivery aspects of CRISPR/Cas for in vivo genome editing. Acc Chem Res. 2019;52:1555–1564. doi: 10.1021/acs.accounts.9b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Thi EP, Mire CE, Lee AC, et al. siRNA rescues nonhuman primates from advanced Marburg and Ravn virus disease. J. Clin. Invest. 2017;127:4437–4448. doi: 10.1172/JCI96185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jayant RD, Sosa D, Kaushik A, et al. Current status of Non-viral gene therapy for CNS disorders. Expert Opin Drug Deliv. 2016;13:1433–1445. doi: 10.1080/17425247.2016.1188802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Givens BE, Naguib YW, Geary SM, Devor EJ, Salem AK. Nanoparticle-based delivery of CRISPR/Cas9 genome-editing therapeutics. AAPS J. 2018;20:108. doi: 10.1208/s12248-018-0267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang L, Wang P, Feng Q, et al. Lipid nanoparticle-mediated efficient delivery of Crispr/cas9 for tumor therapy. NPG Asia Mater. 9 (2017).
- 186.Mukalel AJ, Riley RS, Zhang R, Mitchell MJ. Nanoparticles for nucleic acid delivery: applications in cancer immunotherapy. Cancer Lett. 2019;458:102–112. doi: 10.1016/j.canlet.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Barman NC, Khan NM, Islam M, et al. CRISPR/Cas9: a promising genome editing therapeutic tool for Alzheimer's disease-a narrative review. Neurol Ther. 2020;9:419–434. doi: 10.1007/s40120-020-00218-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sheridan C. CRISPR therapeutics push into human testing. Nat Biotechnol. 2017;35:3–5. doi: 10.1038/nbt0117-3. [DOI] [PubMed] [Google Scholar]
- 189.Norgren N, Olsson M, Nyström H, et al. Gene expression profile in hereditary transthyretin amyloidosis: differences in targeted and source organs. Amyloid. 2014;21:113–119. doi: 10.3109/13506129.2014.894908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Park H, Oh J, Shim G, et al. In vivo neuronal gene editing via CRISPR/Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer's disease. Nat. Neurosci. 2019;22:524–528. doi: 10.1038/s41593-019-0352-0. [DOI] [PubMed] [Google Scholar]
- 191.Kennedy EM, Kornepati AV, Goldstein M, et al. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J. Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Cho EY, Ryu JY, Lee HAR, et al. Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes. J. Nanobiotechnol. 2019;17:19. doi: 10.1186/s12951-019-0452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Xu CF, Iqbal S, Shen S, Luo YL, Yang X, Wang J. Development of “CLAN” nanomedicine for nucleic acid therapeutics. Small. 2019;15:1900055. doi: 10.1002/smll.201900055. [DOI] [PubMed] [Google Scholar]
- 194.Yin H, Song CQ, Dorkin JR, et al. Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 2016;34:328–333. doi: 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Finn JD, Smith AR, Patel MC, et al. A single administration of CRISPR/Cas9 lipid nanoparticles achieves robust and persistent in vivo genome editing. Cell Rep. 2018;22:2227–2235. doi: 10.1016/j.celrep.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 196.Chuang YF, Phipps AJ, Lin FL, et al. Approach for in vivo delivery of CRISPR/Cas system: a recent update and future prospect. Cell Mol. Life Sci. 2021;78:2683–2708. doi: 10.1007/s00018-020-03725-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Liu J, Chang J, Jiang Y, et al. Fast and efficient CRISPR/Cas9 genome editing in vivo enabled by bioreducible lipid and messenger RNA nanoparticles. Adv Mater. 2019;31:e1902575. doi: 10.1002/adma.201902575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Senyei A, Widder K, Czerlinski C. Magnetic guidance of drug carrying microspheres. J. Appl. Phys. 1978;49:3578–3583. doi: 10.1063/1.325219. [DOI] [Google Scholar]
- 199.Widder KJ, Senyel AE, Scarpelli GD. Magnetic microspheres: a model system of site-specific drug delivery in vivo. Proc. Soc. Exp. Biol. Med. 1978;158:141–146. doi: 10.3181/00379727-158-40158. [DOI] [PubMed] [Google Scholar]
- 200.McBain SC, Yiu HH, Dobson J. Magnetic nanoparticles for gene and drug delivery. Int. J. Nanomed. 2008;3:169–180. doi: 10.2147/ijn.s1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tang H, Zhao X, Jiang X. Synthetic multi-layer nanoparticles for CRISPR/Cas9 genome editing. Adv Drug Deliv. Rev. 2021;168:55–78. doi: 10.1016/j.addr.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 202.Babačić H, Mehta A, Merkel O, Schoser B. CRISPR/Cas gene-editing as plausible treatment of neuromuscular and nucleotide-repeat-expansion diseases: a systematic review. PLoS ONE. 2019;14:e0212198. doi: 10.1371/journal.pone.0212198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shahbazi R, Sghia-Hughes G, Reid JL, et al. Targeted homology-directed repair in blood stem and progenitor cells with CRISPR nanoformulations. Nat. Mater. 2019;18:1124–1132. doi: 10.1038/s41563-019-0385-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Alagoz M, Kherad N. Advance genome editing technologies in the treatment of human diseases: CRISPR therapy (Review) Int. J. Mol. Med. 2020;46:521–534. doi: 10.3892/ijmm.2020.4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lee B, Lee K, Panda S, et al. Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nat. Biomed. Eng. 2018;2:497–507. doi: 10.1038/s41551-018-0252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Tay LS, Palmer N, Panwala R, Chew WL, Mali P. Translating CRISPR/Cas therapeutics: approaches and challenges. CRISPR J. 2020;3:253–275. doi: 10.1089/crispr.2020.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yue H, Zhou X, Cheng M, Xing D. Graphene oxide-mediated Cas9/sgRNA delivery for efficient genome editing. Nanoscale. 2018;10:1063–1071. doi: 10.1039/C7NR07999K. [DOI] [PubMed] [Google Scholar]
- 208.Liu J, Cui L, Losic D. Graphene and graphene oxide as new nanocarriers for drug delivery applications. Acta Biomater. 2013;9:9243–9257. doi: 10.1016/j.actbio.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 209.Imani R, Mohabatpour F, Mostafavi F. Graphene-based Nano-Carrier modifications for gene delivery applications. Carbon. 2018;140:569–591. doi: 10.1016/j.carbon.2018.09.019. [DOI] [Google Scholar]
- 210.Hryhorowicz M, Grześkowiak B, Mazurkiewicz N, Śledziński P, Lipiński D, Słomski R. Improved delivery of CRISPR/Cas9 system using magnetic nanoparticles into porcine fibroblast. Mol. Biotechnol. 2019;61:173–180. doi: 10.1007/s12033-018-0145-9. [DOI] [PubMed] [Google Scholar]
- 211.Kaushik A, Yndart A, Atluri V, et al. Magnetically guided non-invasive CRISPR/Cas9/gRNA delivery across blood-brain barrier to eradicate latent HIV-1 infection. Sci. Rep. 2019;9:3928. doi: 10.1038/s41598-019-40222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Nehra M, Uthappa UT, Kumar V, et al. Nanobiotechnology-assisted therapies to manage brain cancer in personalized manner. J. Control Release. 2021;338:224–243. doi: 10.1016/j.jconrel.2021.08.027. [DOI] [PubMed] [Google Scholar]
- 213.Sadique MA, Yadav S, Ranjan P, et al. High-performance antiviral nano-systems as a shield to inhibit viral infections: SARS-CoV-2 as a model case study. J. Mater. Chem. B. 2021;9:4620–4642. doi: 10.1039/D1TB00472G. [DOI] [PubMed] [Google Scholar]
- 214.Varahachalam SP, Lahooti B, Chamaneh M, et al. Nanomedicine for the SARS-CoV-2: state-of-the-art and future prospects. Int. J. Nanomed. 2021;16:539–560. doi: 10.2147/IJN.S283686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kaushik, A., Nikkhah-Moshaie, R., Sinha, R., Bhardwaj, V., Atluri, V., Jayant, R.D., Yndart, A., Kateb, B., Pala, N., Nair, M.: Investigation of AC-magnetic field stimulated nanoelectroporation of magneto-electric nano-drug-carrier inside CNS cells. Sci. Rep. 7, (2017). [DOI] [PMC free article] [PubMed]
- 216.Kaushik, A., Jayant, R.D., Nikkhah-Moshaie, R., Bhardwaj, V., Roy, U., Huang, Z., Ruiz, A., Yndart, A., Atluri, V., El-Hage, N., Khalili, K., Nair, M.: Magnetically guided central nervous system delivery and toxicity evaluation of magneto-electric nanocarriers. Sci. Rep. 6, (2016). [DOI] [PMC free article] [PubMed]
- 217.Kaushik A, Rodriguez J, Rothen D, Bhardwaj V, Jayant RD, Pattany P, Fuentes B, Chand H, Kolishetti N, El-Hage N, Khalili K, Kenyon NS, Nair M. MRI-guided, noninvasive delivery of magneto-electric drug nanocarriers to the brain in a nonhuman primate. ACS Appl. Bio Mater. 2019;2:4826–4836. doi: 10.1021/acsabm.9b00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Dizaj SM, Jafari S, Khosroushahi AY. A sight on the current nanoparticle-based gene delivery vectors. Nanoscale Res. Lett. 2014;9:252. doi: 10.1186/1556-276X-9-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kumar R, Mondal K, Panda PK, et al. Core-shell nanostructures: perspectives towards drug delivery applications. J. Mater. Chem. B. 2020;8:8992–9027. doi: 10.1039/D0TB01559H. [DOI] [PubMed] [Google Scholar]
- 220.Glass Z, Lee M, Li Y, Xu Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2018;36:173–185. doi: 10.1016/j.tibtech.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wang HX, Song Z, Lao YH, et al. Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proc. Natl. Acad. Sci. 2018;115:4903–4908. doi: 10.1073/pnas.1712963115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Liu Y, Cao ZT, Xu CF, Lu ZD, Luo YL, Wang J. Optimization of lipid-assisted nanoparticle for disturbing neutrophils-related inflammation. Biomaterials. 2018;172:92–104. doi: 10.1016/j.biomaterials.2018.04.052. [DOI] [PubMed] [Google Scholar]
- 223.Rui Y, Varanasi M, Mendes S, Yamagata HM, Wilson DR, Green JJ. Poly (Beta-Amino Ester) nanoparticles enable nonviral delivery of CRISPR/Cas9 plasmids for gene knockout and gene deletion. Mol. Ther. Nucleic Acids. 2020;20:661–672. doi: 10.1016/j.omtn.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Jo A, Ringel-Scaia VM, McDaniel DK, et al. Fabrication and characterization of PLGA nanoparticles encapsulating large CRISPR/Cas9 plasmid. J. Nanobiotechnol. 2020;18:16. doi: 10.1186/s12951-019-0564-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Timin AS, Muslimov AR, Lepik KV, et al. Efficient gene editing via non-viral delivery of CRISPR/Cas9 system using polymeric and hybrid microcarriers. Nanomedicine. 2018;14:97–108. doi: 10.1016/j.nano.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 226.Sun W, Ji W, Hall JM, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR/Cas9 for genome editing. Angew Chem. Int. Ed. Engl. 2015;54:12029–12033. doi: 10.1002/anie.201506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Luther DC, Lee YW, Nagaraj H, Scaletti F, Rotello VM. Delivery approaches for CRISPR/Cas9 therapeutics in vivo: advances and challenges. Expert Opin. Drug Deliv. 2018;15:905–913. doi: 10.1080/17425247.2018.1517746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chen B, Yang Z, Zhu Y, Xia Y. Zeolitic imidazolate framework materials: recent progress in synthesis and applications. J. Mater. Chem. A. 2014;2:16811–16831. doi: 10.1039/C4TA02984D. [DOI] [Google Scholar]
- 229.Eoh J, Gu L. Biomaterials as vectors for the delivery of CRISPR/Cas9. Biomater. Sci. 2019;7:1240–1261. doi: 10.1039/C8BM01310A. [DOI] [PubMed] [Google Scholar]
- 230.Ju E, Li T, Ramos da Silva S, Gao SJ. Gold nanocluster-mediated efficient delivery of Cas9 protein through pH-induced assembly-disassembly for inactivation of virus oncogenes. ACS Appl. Mater. Interfaces. 2019;11:34717–34724. doi: 10.1021/acsami.9b12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci. Rep. 2013;3:2510. doi: 10.1038/srep02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rabiee, N., Bagherzadeh, M., Ghadiri, A. M., Kiani, M., Ahmadi, S., Jajarmi, V., Fatahi, Y., Aldhaher, A., Tahriri, M., Webster, T. J., Mostafavi, E.: Calcium-based nanomaterials and their interrelation with chitosan: optimization for pCRISPR delivery. J. Nanostruct. Chem., 1–14 (2021). [DOI] [PMC free article] [PubMed]
- 233.Lostalé-Seijo I, Louzao I, Juanes M, Montenegro J. Peptide/Cas9 nanostructures for ribonucleoprotein cell membrane transport and gene edition. Chem. Sci. 2017;8:7923–7931. doi: 10.1039/C7SC03918B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lin Y, Wu J, Gu W, et al. Exosome-liposome hybrid nanoparticles deliver CRISPR/Cas9 system in MSCs. Adv Sci. 2018;5:1700611. doi: 10.1002/advs.201700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Khunger A, Kaur N, Mishra YK, Ram Chaudhary G, Kaushik A. Perspective and prospects of 2D MXenes for smart biosensing. Mater. Lett. 2021;304:130656. doi: 10.1016/j.matlet.2021.130656. [DOI] [Google Scholar]
- 236.Zuris JA, Thompson DB, Shu Y, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2015;33:73–80. doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Mangeot PE, Risson V, Fusil F, et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019;10:45. doi: 10.1038/s41467-018-07845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. MicroRNA: function, detection, and bioanalysis. Chem. Rev. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 239.Wang M, Zhang R, Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens. Bioelectron. 2020;165:112430. doi: 10.1016/j.bios.2020.112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Staahl BT, Benekareddy M, Coulon-Bainier C, et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017;35:431–434. doi: 10.1038/nbt.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wagner DL, Peter L, Schmueck-Henneresse M. Cas9-directed immune tolerance in humans-a model to evaluate regulatory T cells in gene therapy? Gene Ther. 2021;28:549–559. doi: 10.1038/s41434-021-00232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Mout R, Ray M, Yesilbag Tonga G, et al. Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS Nano. 2017;11:2452–2458. doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kedmi R, Ben-Arie N, Peer D. The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation. Biomaterials. 2010;31:6867–6875. doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 244.Reichmuth AM, Oberli MA, Jaklenec A, Langer R, Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ferdosi SR, Ewaisha R, Moghadam F, et al. Multifunctional CRISPR/Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019;10:1842. doi: 10.1038/s41467-019-09693-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wagner DL, Amini L, Wendering DJ, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat. Med. 2019;25:242–248. doi: 10.1038/s41591-018-0204-6. [DOI] [PubMed] [Google Scholar]
- 247.Xiao P, Bai R, Zhang T, et al. Extensive adaptive immune response of Aavs and Cas proteins in non-human primates. Sci. Bull. 2021;66:2061–2064. doi: 10.1016/j.scib.2021.02.009. [DOI] [PubMed] [Google Scholar]
- 248.Fu Y, Foden JA, Khayter C, et al. High-frequency off-target mutagenesis induced by CRISPR/Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Xu Y, Li Z. CRISPR/Cas systems: overview, innovations and applications in human disease research and gene therapy. Comput. Struct. Biotechnol. J. 2020;18:2401–2415. doi: 10.1016/j.csbj.2020.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR/Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529:490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Sahel DK, Mittal A, Chitkara D. CRISPR/Cas system for genome editing: progress and prospects as a therapeutic tool. J. Pharmacol. Exp. Ther. 2019;370:725–735. doi: 10.1124/jpet.119.257287. [DOI] [PubMed] [Google Scholar]
- 253.Nayak S, Herzog RW. Progress and prospects: immune responses to viral vectors. Gene Ther. 2010;17:295–304. doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Mout R, Ray M, Lee YW, Scaletti F, Rotello VM. In vivo delivery of CRISPR/Cas9 for therapeutic gene editing: progress and challenges. Bioconjug. Chem. 2017;28:880–884. doi: 10.1021/acs.bioconjchem.7b00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Mishra S, Webster P, Davis ME. PEGylation significantly affects cellular uptake and intracellular trafficking of non-viral gene delivery particles. Eur. J. Cell Biol. 2004;83:97–111. doi: 10.1078/0171-9335-00363. [DOI] [PubMed] [Google Scholar]
- 256.Charlesworth CT, Deshpande PS, Dever DP, et al. Identification of pre-existing adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019;25:249–254. doi: 10.1038/s41591-018-0326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Misra, C.S., Bindal, G., Sodani, M., et al.: Determination of cas9/dcas9 associated toxicity in microbes. BioRxiv. 848135 (2019).
- 258.Nair M, Jayant RD, Kaushik A, Sagar V. Getting into the brain: potential of nanotechnology in the management of NeuroAIDS. Adv Drug Deliv Rev. 2016;103:202–217. doi: 10.1016/j.addr.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Sharma PK, Kim ES, Mishra S, et al. Ultrasensitive and reusable graphene oxide-modified double-interdigitated capacitive (DIDC) sensing chip for detecting SARS-CoV-2. ACS Sens. 2021;6:3468–3476. doi: 10.1021/acssensors.1c01437. [DOI] [PubMed] [Google Scholar]
- 260.Morales-Narváez E, Dincer C. The impact of biosensing in a pandemic outbreak: Covid-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.