Abstract
Vancomycin-resistant enterococci (VRE) have recently become an increasing problem in hospitals in Poland, being responsible for a growing number of nosocomial outbreaks. In this work, we have analyzed the second outbreak of VRE with the VanB phenotype to be identified in the country. It was caused by clonal dissemination of a single strain of vancomycin-resistant Enterococcus faecalis (VRES) and horizontal transmission of vancomycin resistance genes among several vancomycin-resistant Enterococcus faecium (VREM) strains. Two similar restriction fragment length polymorphism types of the vanB gene cluster characterized VRES and VREM isolates, and they both contained the same vanB2 variant of the vanB gene. Two vancomycin-susceptible E. faecium (VSEM) isolates, recovered from the same wards during the outbreak, proved to be related to certain VREM isolates and could represent endemic strains that had acquired vancomycin resistance. One VSEM and four VREM isolates, all identified in the same patient, belonged to a single clone, although they revealed remarkable diversity in terms of susceptibility, PFGE patterns, plasmid content, and number of vanB gene cluster copies. Most probably they reflected the dynamic evolution of an E. faecium strain in the course of infection of a single patient. One of the VREM isolates turned out to be resistant to teicoplanin, which coincided with the use of this antibiotic in the patient's therapy. Its vanB gene variant differed by a single mutation from that found in other isolates; however, it also lacked a large part of the vanB gene cluster, including the regulatory genes vanRB and -SB, and the vancomycin-inducible promoter PYB. Expression of the resistance genes vanHB, -B, and -XB was constitutive in the mutant, and this phenomenon was responsible for its unusual phenotype.
Vancomycin-resistant enterococci (VRE) usually cause infections in severely debilitated, immunocompromised patients who undergo prolonged antimicrobial therapy (9, 12, 34, 36). Of the six different VRE phenotypes known to date (23, 31, 32, 35, 40, 44), phenotypes VanA and VanB are of the highest clinical importance as they are most frequently observed in two predominant enterococcal species, Enterococcus faecalis and Enterococcus faecium (12, 36, 37). Although the majority of nosocomial VRE outbreaks have been attributed to VanA organisms, outbreaks caused by VanB VRE have also been reported several times. They were found to result from either the clonal spread of resistant strains (13, 18, 47, 49) or the horizontal transfer of resistance determinants (10, 11, 42, 51) or both (28).
The VanB phenotype originally described in 1989 (45) is characterized by its high-level resistance to vancomycin and susceptibility to teicoplanin (44). It is determined by a cluster of genes—vanRB, -SB, -YB, -W, -HB, -B, and -XB—which reside within composite transposons such as Tn1547 or similar elements (21, 43). These transposons are found in chromosomal or plasmid DNA (42), and they may be horizontally transmitted either by themselves, together with other mobile elements (42, 43), or by plasmid conjugation (10). Resistance to vancomycin depends upon the products of vanHB, -B, and -XB genes, and their expression is regulated by a two-component system, which consists of proteins encoded by vanRB and -SB genes (VanRB-VanSB) (1, 4, 20). Genes vanYB, -W, -HB, -B, and -XB are transcribed together from promoter PYB, which is located upstream of vanYB, and this process is activated by VanRB in its phosphorylated form. VanRB phosphorylation is catalyzed by the VanSB sensor histidine kinase in response to its stimulation by vancomycin; switching off the resistance genes is, on the other hand, due to phosphatase activity of VanSB, which dephosphorylates VanRB in the absence of vancomycin. Since teicoplanin fails to interact with VanSB, VRE strains with the phenotype VanB demonstrate susceptibility to this glycopeptide (1, 4, 6, 7, 20).
This work presents an analysis of a VRE outbreak which occurred in 2000 in a Polish hospital, during which a VanB teicoplanin-resistant E. faecium strain was selected.
MATERIALS AND METHODS
Clinical isolates.
Thirteen vancomycin-resistant enterococcal isolates (six E. faecalis isolates [VRES] and seven E. faecium isolates [VREM]) were recovered from infected or colonized patients in two hematological wards of the University Hospital in Cracow between March and September 2000 (Table 1). All of the VRES isolates were identified in blood, urine, or sputum samples collected from different patients. Four out of the seven VREM isolates (3123, 3124A, 3124B, and 3128) were cultured from the clinical specimens (blood and urine) of a single patient, and three of them (3123, 3124A, and 3124B) were discerned in the same blood sample based on differences in their colony morphology or susceptibility. The remaining three VREM isolates were identified in the urine of another infected patient and in stool samples from two other patients. Carriage testing was performed as described previously (28).
TABLE 1.
Selected clinical data for isolates in this study
| Isolate no. | Phenotype | Sourceb | Date of isolation (mo/yr) | Matingc | PFGE | L-PCRd | MIC (μg/ml) ofe:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PEN | AMP | GEN | STR | VAN | TEC | CHL | TET | CIP | LNZ | Q-D | |||||||
| 2496 | VRES | i (blood) | 03/00 | − | A | RFLP-4 | 32 | 2 | >1,024 | >2,048 | 256 | 0.25 | 32 | 32 | 128 | 2 | ND |
| 2492 | VRES | i (urine) | 06/00 | − | A | RFLP-4 | 16 | 4 | >1,024 | >2,048 | 256 | 0.25 | 32 | 32 | 256 | 2 | ND |
| 2493 | VRES | i (blood) | 07/00 | − | A | RFLP-4 | 2 | 0.5 | >1,024 | >2,048 | 256 | 0.25 | 32 | 32 | 256 | 2 | ND |
| 3127 | VRES | i (blood) | 08/00 | − | A | RFLP-4 | 32 | 2 | >1,024 | >2,048 | 256 | 0.25 | 32 | 64 | 128 | 2 | ND |
| 3130 | VRES | i (sputum) | 09/00 | − | A | RFLP-4 | 32 | 2 | >1,024 | >2,048 | 256 | 0.25 | 32 | 64 | 128 | 2 | ND |
| 3135 | VRES | i (sputum) | 09/00 | − | A | RFLP-4 | 32 | 8 | >1,024 | >2,048 | 256 | 0.25 | 64 | 64 | 128 | 2 | ND |
| 2499 | VREM | c | 05/00 | − | a1 | RFLP-2 | >128 | 64 | >1,024 | >2,048 | 512 | 0.25 | 16 | 0.5 | 128 | 2 | 0.5 |
| 2498 | VREM | i (urine) | 07/00 | − | b | RFLP-2 | >128 | 64 | >1,024 | >2,048 | 256 | 0.5 | 8 | 1 | 256 | 2 | 1 |
| 2497 | VREM | c | 07/00 | − | a2 | — | >128 | 64 | >1,024 | >2,048 | 512 | 32 | 32 | 0.5 | 128 | 2 | 1 |
| 3123a | VREM | i (blood) | 08/00 | − | c1 | RFLP-2 | 128 | 128 | >1,024 | >2,048 | 256 | 0.5 | 64 | 64 | 64 | 2 | 0.5 |
| 3124Aa | VREM | i (blood) | 08/00 | + | c2 | RFLP-2 | >128 | 128 | 128 | >2,048 | 256 | 0.5 | 8 | 64 | 64 | 2 | 1 |
| 3124Ba | VREM | i (blood) | 08/00 | + | c3 | RFLP-2 | 128 | 128 | 128 | >2,048 | 128 | 0.25 | 8 | 64 | 64 | 2 | 1 |
| 3128a | VREM | i (urine) | 08/00 | + | c4 | RFLP-2 | >128 | 64 | 64 | >2,048 | 512 | 0.25 | 8 | 64 | 64 | 2 | 1 |
| 2494 | VSEM | i (blood) | 05/00 | − | a3 | ND | >128 | 128 | 128 | 2,048 | 2 | 0.25 | 8 | 1 | 256 | 2 | 1 |
| 3122a | VSEM | i (blood) | 08/00 | − | c5 | ND | >128 | 128 | 64 | >2,048 | 2 | 0.25 | 8 | 64 | 64 | 2 | 0.5 |
Isolates recovered from a single patient.
i, infection; c, carriage.
+ and − indicate isolates that produced and did not produce transconjugants, respectively.
RFLP types of vanB gene clusters amplified by L-PCR; —, lack of L-PCR product; ND, not determined.
Abbreviations: PEN, penicillin; AMP, ampicillin; GEN, gentamicin; STR, streptomycin; VAN, vancomycin; TEC, teicoplanin; CHL, chloramphenicol; TET, tetracycline; CIP, ciprofloxacin; LNZ, linezolid; Q-D, quinupristin-dalfopristin.
Two vancomycin-susceptible E. faecium (VSEM) isolates from different patients were involved in the study, and these were all vancomycin-susceptible enterococci identified as infection agents in the wards at that time. One of the patients was that from whom multiple VREM isolates were recovered; moreover, the VSEM isolate was cultured from the same blood sample as the three VREM isolates mentioned above (isolate 3122).
Genus identification of the isolates was performed according to the method of Facklam and Collins (22), and species were identified using the API ID32 STREP test (bioMérieux, Charbonnieres-les-Bains, France), supplemented by potassium tellurite reduction, motility, and pigment production tests (22).
Antimicrobial susceptibility testing.
MICs of different antimicrobial agents were evaluated by the agar dilution method according to NCCLS guidelines (38). The following agents were tested: penicillin, ampicillin, streptomycin, gentamicin, tetracycline, and chloramphenicol (Polfa Tarchomin, Warsaw, Poland); vancomycin (Eli Lilly, Indianapolis, Ind.); teicoplanin (Marion Merrell, Denham, United Kingdom); ciprofloxacin (Bayer, Wuppertal, Germany); linezolid (Pharmacia & Upjohn, Kalamazoo, Mich.); and quinupristin-dalfopristin (Rhone Poulenc Rorer, Paris, France). E. faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and the E. faecalis V583 VanB standard strain (21, 45) were used as reference strains.
Resistance transfer (mating).
The vancomycin resistance transfer experiment was performed using the filter-mating procedure (30), with E. faecalis FA2-2 (15) or E. faecium 64/3 (50) strains as recipients. Transconjugants were selected on BHI agar (Oxoid, Basingstoke, United Kingdom) plates supplemented with rifampin (64 μg/ml; Polfa Tarchomin), fusidic acid (64 μg/ml; Leo Pharmaceutical Products, Ballerup, Denmark), and vancomycin (32 μg/ml).
PFGE typing.
For the pulsed-field gel electrophoresis (PFGE) typing, total DNA of the isolates was purified according to the procedure of Clark et al. (14) and digested with the SmaI restriction enzyme (MBI Fermentas, Vilnius, Lithuania). PFGE was performed in a CHEF DRII system (Bio-Rad, Hercules, Calif.), under conditions described by de Lencastre et al. (17). The results were interpreted in accordance with the criteria set by Tenover et al. (48).
Detection of the vanB gene and the vanSB-vanYBregion.
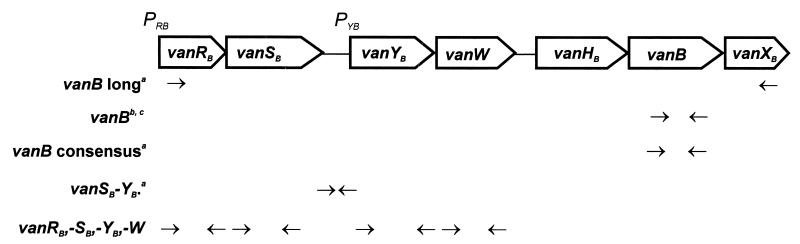
Total DNA of the isolates was purified with the use of a Genomic DNA Prep Plus kit (A&A Biotechnology, Gdańsk, Poland). The vanB gene was detected by specific PCR with two different pairs of primers, primers vanB (14) and primers vanB consensus (16), and the vanSB-vanYB region was amplified using primers proposed by Dahl et al. (16) (Fig. 1). Genomic DNA of the E. faecalis V583 VanB standard strain (21, 45) was used as a positive control.
FIG. 1.
Dislocation of PCR primers used in the study with respect to the scheme of the Tn1547 vanB gene cluster present in the E. faecalis V583 VanB reference strain (20, 21, 43, 45). Superscript letters: a, reference 16; b, reference 14; c, in study isolates the upstream vanB primer annealed within the vanHB gene and not in the vanB gene.
Sequencing of vanB gene-specific PCR products.
vanB gene-containing PCR products of approximately 1.1 kb, obtained for selected VRE isolates, were sequenced as previously described (29), with the addition of two internal primers: 5′-CGATCCGCACTACATCGG-3′ and 5′-AACGGCGATGCCCGCAT-3′.
RFLP analysis of the vanB gene cluster.
Restriction fragment length polymorphism (RFLP) of long PCR (L-PCR) products containing clusters of vanRB, -SB, -YB, -W, -HB, -B, and -XB genes was studied as reported previously (29) with the use of primers vanB long for L-PCR (16) (Fig. 1). DraI and PagI (isoschizomer of BspHI) restriction enzymes (MBI Fermentas) were used in the analysis and genomic DNA of the E. faecalis V583 VanB standard strain (21, 45) was included as a positive control.
Analysis of the vanB gene cluster location.
Undigested total DNAs of the isolates were separated by PFGE and blotted onto a Hybond-N+ membrane (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) for hybridization with the vanB gene cluster probe. The L-PCR amplicon of the vanB cluster of the E. faecalis V583 VanB reference strain (16, 21, 45) was used as the probe. Probe labeling, hybridization, and signal detection were performed with the ECL Random-Prime labeling and detection system (Amersham Pharmacia Biotech). DNA of E. faecalis V583 (21, 45) was used as a positive control.
Detection of vanA and vanRB, -SB, -YB, and -W genes.
Total DNAs of selected isolates were tested for the presence of vanA and vanRB, -SB, -YB, and -W genes. The vanA gene was detected by PCR with two different pairs of primers (14, 19), whereas the remaining genes were amplified with primers designed according to the Tn1547 sequence (reference 20; GenBank accession no. U35369) (Fig. 1). Sequences of these primers were as follows: vanRB1, 5′-CTTGTCGAGGATGATG-3′; vanRB2, 5′-CCTCCAATCGGTAACC-3′; vanSB1, 5′-GTCGGTGTAACGGCAAC-3′; vanSB2, 5′-GCTGGTTGTTTGCCTC-3′; vanYB1, 5′-GAATCATCACAAACGGC-3′; vanYB2, 5′-CTCTGTCTTGTCTGGC-3′; vanW1, 5′-GATTGACACAGCGCTTC-3′; vanW2, 5′-CTCCTGAATATCCACAC-3′. Amplicons of vanRB, -SB, -YB and -W genes, obtained for the E. faecalis V583 VanB reference strain (21, 45), were subsequently used as probes in the dot blot hybridization (46) with DNAs of the analyzed isolates. Probe labeling, hybridization and signal detection were performed as described above. DNAs isolated from the E. faecium BM4147 VanA standard strain (3) and the E. faecalis V583 VanB standard strain (21, 45) were used as controls.
Analysis of vanB gene expression by RT-PCR.
Expression of the vanB gene was studied in selected VREM isolates. Total RNA was purified from 10-ml cultures of the isolates grown overnight in Todd-Hewitt broth (THB) (Oxoid), THB with vancomycin (8 μg/ml), and THB with teicoplanin (8 μg/ml). RNA was obtained by using a Total RNA Prep Plus kit (A&A Biotechnology), treated with DNase I (amplification grade; Sigma Chemical Company, St. Louis, Mo.), and subjected to reverse transcription (RT) with the vanB long downstream primer (16) (Fig. 1). The SuperScript II reverse transcriptase (Gibco BRL Life Technologies Inc., Karlsruhe, Germany) was used in the reaction according to the manufacturer's protocol, and this was subsequently followed by specific PCR with the vanB consensus pair of primers (16). Total RNAs extracted from the E. faecalis V583 VanB standard strain (21, 45) and the VSEM 2494 clinical isolate were used as controls. RNA preparations were tested for the lack of DNA contamination by PCR that had not been preceded by RT. RT-PCR products were analyzed by agarose gel electrophoresis.
Nucleotide sequence accession numbers.
Nucleotide sequences of vanHB-vanB regions of VREM 2497 and 3123 isolates will appear in the EMBL database under accession numbers AJ306726 and AJ306727, respectively.
RESULTS
Antimicrobial susceptibility testing of VRE isolates.
MICs of various antimicrobials evaluated for VRE isolates are presented in Table 1. All the isolates demonstrated high-level resistance to vancomycin (MICs, 128 to 512 μg/ml) and susceptibility to teicoplanin (MICs, 0.25 to 0.5 μg/ml), except for a single VREM isolate (isolate 2497) which showed resistance to both glycopeptides (vancomycin MIC, 512 μg/ml; teicoplanin MIC, 32 μg/ml). Isolates were found to be uniformly resistant to ciprofloxacin and to high concentrations of at least streptomycin out of the two aminoglycosides tested (streptomycin and gentamicin). Resistance to other antimicrobials was also widely spread in the studied VRE population. All VREM isolates were resistant to penicillin and ampicillin, and the majority of VRES isolates demonstrated resistance to penicillin, chloramphenicol, and tetracycline. All VRE isolates revealed susceptibility to linezolid, and all VREM isolates were susceptible to quinupristin-dalfopristin. The four VREM isolates recovered from a single patient (3123, 3124A, 3124B, and 3128) demonstrated similar MIC patterns; however, significant differences were observed in the case of isolate 3123, which was characterized by clearly increased MICs of gentamicin and chloramphenicol compared to the other isolates in this group.
PFGE typing of VRE isolates.
Results of the analysis are shown in Table 1. All the VRES isolates produced identical PFGE banding patterns (PFGE type A) in contrast to the VREM isolates, which could be classified into three distinct types with subtypes (48) (PFGE types a1 and a2, b, and c1 to c4). Two isolates of PFGE subtypes a1 and a2 (2499 and 2497, respectively) were those recovered from different carriers, whereas all four VREM isolates of PFGE subtypes c1 to c4 were those collected from a single patient (3123, 3124A, 3124B, and 3128).
Susceptibility and PFGE typing of VSEM isolates.
Susceptibility testing and PFGE typing of the two VSEM isolates (2494 and 3122) were performed along with those of the VRE isolates, and the results are shown in Table 1. The VSEM isolates also revealed a multidrug resistance phenotype with resistance to penicillin, ampicillin, and ciprofloxacin and to high concentrations of streptomycin. Isolate 3122 was additionally resistant to tetracycline, similarly to the only tetracycline-resistant VREM isolates, all collected from the same patient (3123, 3124A, 3124B, and 3128). Moreover, when compared to isolates 3124A, 3124B, and 3128, isolate 3122 differed only with respect to the vancomycin MIC.
The VSEM isolates produced PFGE patterns that were similar to those of the VREM isolates. Isolate 2494 could be classified into PFGE type a (subtype a3), together with VREM isolates 2499 and 2497. Isolate 3122 represented another subtype of PFGE type c (subtype c5), which also included all the VREM isolates recovered from the same patient (3123, 3124A, 3124B, and 3128).
Vancomycin resistance transfer and susceptibility of transconjugants.
Results of mating are presented in Table 1. Only three VREM isolates (3124A, 3124B, and 3128) produced vancomycin-resistant transconjugants, and the efficiency of conjugation was low, ranging from 10−7 to 10−9 recombinants per donor cell. Susceptibility testing revealed that no other resistance determinants were cotransferred with those of vancomycin resistance.
Detection of the vanB gene and the vanSB-vanYBregion.
Presence of the vanB gene was checked in the VRE isolates by PCR with two different pairs of primers, vanB (14) and vanB consensus (Fig. 1) (16). In all VRE isolates, PCR with primers vanB, which are specific for the vanB1 gene variant (16), produced amplicons of about 1.1 kb instead of the 433 bp expected for vanB1 and observed in the case of vanB1-containing E. faecalis V583 (14, 21, 45). On the other hand, primers vanB consensus, which amplify a 484-bp fragment of all vanB gene variants known to date (16), yielded a product of about 500 bp (results not shown). The same pattern of vanB gene PCR products obtained with the two pairs of primers had previously been observed with VREM isolates from a hospital in Warsaw (29).
For all but one of the VRE isolates, PCR of the vanSB-vanYB region (which includes the PYB promoter [Fig. 1]) produced amplicons of approximately 300 bp that corresponded well to the size of 309 bp expected for the Tn1547 transposon present in the E. faecalis V583 strain (16, 21, 45). The only exception was the VREM 2497 isolate, for which no vanSB-vanYB-specific PCR product was observed (results not shown).
Sequencing of vanB gene-containing PCR products.
The 1.1-kb vanB gene-containing amplicons, obtained in PCR with vanB primers (14) for isolates VRES 2496, VREM 2497, and VREM 3123, were subjected to direct DNA sequencing. As had been found previously in a VREM isolate from a hospital in Warsaw (29), PCR products encompassed the 896-bp fragment of the vanB coding region starting from its 5′ end and the 201-bp fragment of the vanHB gene that is located directly upstream of vanB. This indicated that the forward primer of the vanB pair (14) annealed to a sequence present within the vanHB gene, instead of vanB. In the VRES 2496 and VREM 3123 isolates the analyzed sequence of 1,045 bp was identical to the corresponding region of the VREM strain from Warsaw (29) and contained the vanB2 (24) variant of the vanB gene originally identified in the United States (GenBank accession no. U94526) (39). The sequence found in the VREM 2497 isolate revealed a single base pair difference when compared to the others. An A-to-G mutation in position 388 of the vanB gene causes a Met-to-Val substitution at position 130 of the protein sequence with respect to E. faecalis V583 (21).
RFLP analysis of vanB gene clusters.
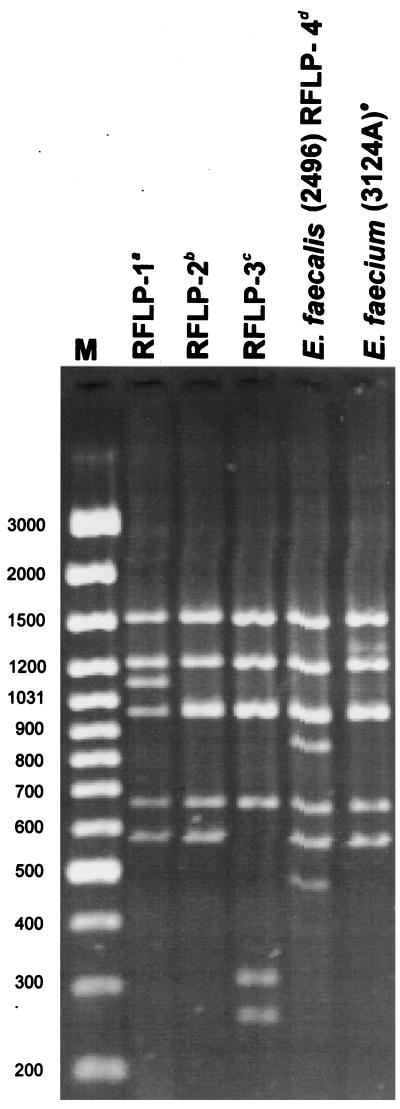
DNA molecules encompassing the vanB gene cluster region (Fig. 1) were amplified by L-PCR, and products of the expected size of about 6 kb were obtained for all but one of the VRE isolates. Results of the DraI/PagI (BspHI) RFLP study of these amplicons are shown in Table 1; Fig. 2 presents RFLP patterns characteristic for representative VRES and VREM isolates together with those specific for E. faecalis V583, RFLP-1 (16, 21, 45), and all VREM VanB strains identified in Poland previously, RFLP-2 (16, 29) and a novel one, here designated RFLP-3 (28). All the VRES isolates contained vanB gene clusters of the same RFLP type, which turned out to be unique when compared to the types described previously (16, 28) and was designated RFLP-4. On the contrary, the VREM isolates possessed a cluster variant of the RFLP-2 type, observed before in VRE isolates from several countries (16), including the VREM strain from Warsaw (29). RFLP-2 and RFLP-4 differed from each other by the presence of two additional DNA bands in RFLP-4. The only isolate that failed to produce the 6-kb amplicon in L-PCR was VREM 2497, for which an approximately 1.3-kb product was observed. This product, however, was also generated in the presence of the upstream vanB long primer alone and did not contain the vanB gene as revealed by hybridization (results not shown).
FIG. 2.
RFLP analysis of the vanB gene cluster with the use of DraI and PagI (BspHI) restriction enzymes. Lane M, GeneRuler 100-bp DNA Ladder Plus (MBI Fermentas). Superscript letters: a, RFLP-1 is a polymorph type of the original Tn1547 vanB cluster present in E. faecalis V583 (16, 20, 45); b, RFLP-2 was originally described by Dahl et al. (16) and here is represented by the cluster present in the VREM 8533 isolate from a hospital in Warsaw (29); c, RFLP-3 was identified by Kawalec et al. in another Warsaw hospital (28) and is represented here by the cluster of the VREM isolate 8284; d, E. faecalis 2496 represents all study VRES isolates; e, E. faecium 3124A is a representative of all study VREM isolates.
Location of vanB gene clusters.
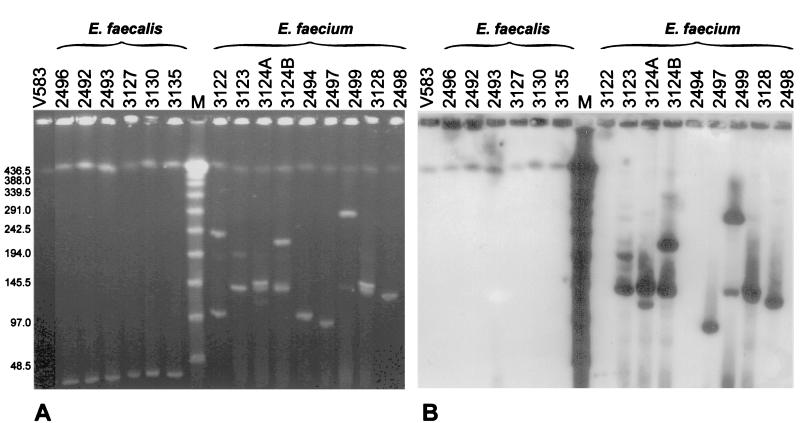
The isolates' undigested total DNA was separated by PFGE and hybridized with the vanB gene cluster probe. Results of the analysis are shown in Fig. 3. In all isolates PFGE visualized a DNA band of the same migration and the biggest size observed, which most probably represented their chromosomal DNA. It is also highly likely that plasmid molecules formed other bands seen in the gel, which in the case of E. faecium isolates demonstrated a remarkable diversity in size. In all the VRES isolates the vanB cluster probe hybridized with the putative chromosomal band, whereas in all the VREM isolates hybridization occurred with plasmid DNA. The hybridizing plasmids were of various sizes; however, all PFGE type c isolates from a single patient (3123, 3124A, 3124B, and 3128) possessed one of the vanB cluster-containing plasmids of the same migration rate (similar to the λ ladder 150-kb band). Several of the VREM isolates were characterized by two (or more) hybridizing plasmid DNA bands.
FIG. 3.
Analysis of location of vanB gene clusters by hybridization of the vanB cluster probe with undigested and PFGE-separated DNA of study isolates. (A) PFGE gel; (B) hybridization. Lanes contain DNA of the indicated isolates, DNA of the E. faecalis V583 VanB reference strain (lanes V583) (45), or a λ ladder PFGE marker (lanes M) (New England BioLabs, Beverly, Mass.).
Detection of vanA, and vanRB, -SB, -YB, and -W genes in the teicoplanin-resistant VREM isolate.
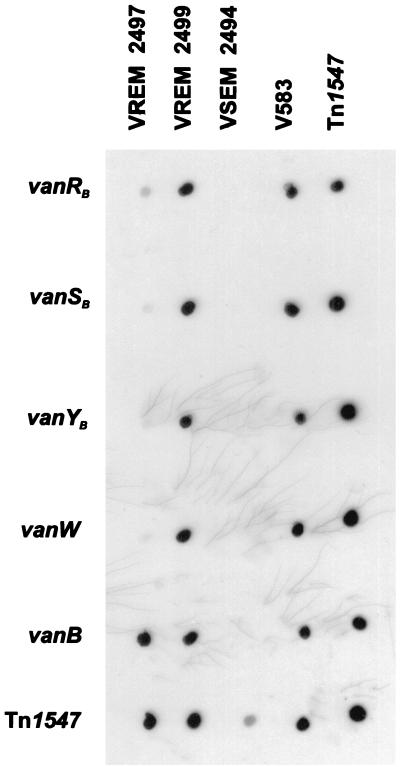
The only teicoplanin-resistant VREM isolate, 2497, which also turned out to be negative in the vanB gene cluster L-PCR and the vanSB-vanYB region PCR was subjected to a more detailed analysis. It was checked for the presence of the vanA gene; however, specific PCR did not yield any product (result not shown). Subsequently the isolate was tested for a set of genes of the vanB cluster (Fig. 1), including the regulatory genes vanRB and vanSB. PCR failed to amplify any products specific for vanRB (expected size, 634 bp), vanSB (expected size, 898 bp), vanYB (expected size, 664 bp), and vanW (expected size, 803 bp) genes that were obtained with DNA from a related VREM isolate with a clear VanB phenotype, isolate 2499, and the E. faecalis V583 VanB reference strain (21, 45) (results not shown). In order to confirm these results, dot blot hybridization was performed, in which amplicons of vanRB, -SB, -YB, -W, and -B genes of E. faecalis V583 were used as probes. DNA of isolate 2497 hybridized only with the vanB probe, whereas all the probes interacted with DNA of isolate 2499 (Fig. 4).
FIG. 4.
Analysis of presence of vanRB, -SB, -YB, and -W genes in the teicoplanin-resistant VREM 2497 isolate by dot blot hybridization with a set of specific probes. DNAs of particular isolates are dislocated in columns: VREM 2497, the teicoplanin-resistant isolate; VREM 2499, the isolate related to 2497 with the typical VanB phenotype; VSEM 2494, the VSEM isolate related to VREM 2497 and 2499; V583, the E. faecalis V583 VanB standard strain (45); Tn1547, L-PCR product encompassing the vanB gene cluster from the E. faecalis V583 strain. Rows represent hybridization with probes specific for corresponding genes; the Tn1547 probe was the labeled L-PCR product described above.
Expression analysis of the vanB gene in the teicoplanin-resistant VREM isolate.
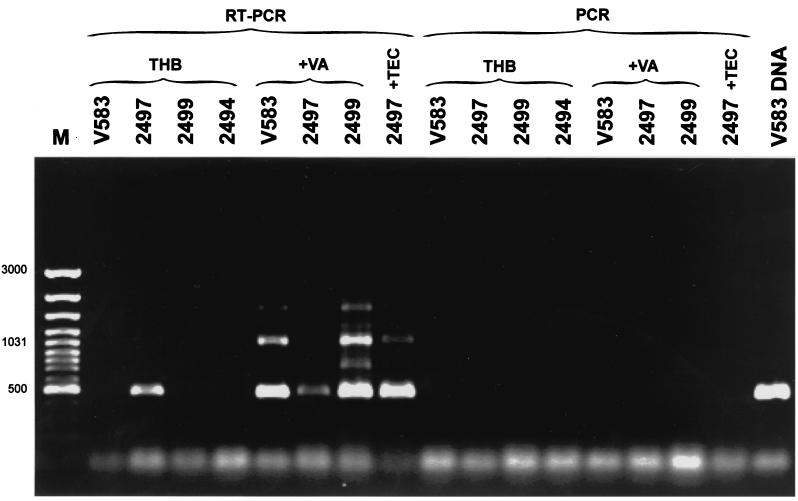
The absence of the regulatory genes vanRB and vanSB indicated the possibility of constitutive expression of the resistance genes vanHB, vanB, and vanXB in VREM isolate 2497. In order to check this hypothesis, VREM isolates 2497 (teicoplanin resistant) and 2499 (with the typical VanB phenotype) and the E. faecalis V583 VanB standard strain (21, 45) were grown in the absence and presence of vancomycin. Isolate 2497 was also grown with teicoplanin, and the VSEM 2494 isolate was grown in a nonsupplemented medium. Total RNA preparations of the cultures were subjected to RT-PCR, in which the downstream vanB long primer complementary to the vanXB gene (16) (Fig. 1) was used for RT, and vanB consensus primers, annealing to the vanB gene (16), were utilized in the subsequent PCR step. Results are shown in Fig. 5. The only non-glycopeptide-supplemented culture, in which the RT-PCR product of the expected size of approximately 500 bp was observed, was that of isolate 2497. It was also obtained in cultures of isolates 2497, 2499, and E. faecalis V583 grown with vancomycin and of isolate 2497 which was grown in the presence of teicoplanin. The length of the other major RT-PCR band of about 1 kb corresponded to the expected product size (1,069 bp), which could be amplified from the upstream vanB consensus and the downstream vanB long primer. Lack of any PCR product in reactions that were not preceded by RT excluded the possibility of amplification as a result of DNA contamination of RNA preparations, and specificity of the reaction was confirmed by PCR failure in the culture of VSEM isolate 2494.
FIG. 5.
RT-PCR analysis of vanB gene expression in the teicoplanin-resistant VREM 2497 isolate. M, GeneRuler 100-bp DNA Ladder Plus (MBI Fermentas); V583 DNA, product of PCR with vanB consensus primers (16) performed with DNA of the E. faecalis V583 VanB reference strain (45). RT-PCR lanes refer to RNA preparations that were subjected to both RT and PCR; PCR lanes refer to control reactions in which the RT step was omitted. Abbreviations: THB, refers to reactions performed on RNAs extracted from cultures grown in the absence of glycopeptides; +VA and +TEC, refer to cultures supplemented with vancomycin and teicoplanin, respectively. Strain designations: V583, the E. faecalis V583 VanB standard strain (45); 2497, the teicoplanin-resistant VREM isolate; 2499, isolate related to 2497 VREM with the typical VanB phenotype; 2494, isolate related to 2497 and 2499 VSEM isolate.
DISCUSSION
In mid-1999, the first VRE isolates with the VanB phenotype were identified in Poland, which resulted from independent selection events in two separate hospitals in Warsaw (28, 29). The data presented in this work document the third incidence of these microorganisms being reported in the country and describe the second Polish VanB VRE outbreak that occurred in 2000 in two hematology units of the University Hospital in Cracow (located about 300 km south of Warsaw). In contrast to the previously analyzed VRE epidemics, caused by VanA VREM in Gdańsk (27) and by VanB VREM strains in Warsaw (28), this outbreak was due to both E. faecium and E. faecalis. The isolates collected during the investigation demonstrated resistance to multiple antimicrobials, which is a common characteristic of VRE (8, 10, 12, 36), including all VRE isolates collected before in Poland (27, 28, 29). Except for a single VREM isolate, isolate 2497 (discussed below in detail), they all revealed the typical VanB phenotype with resistance to vancomycin and susceptibility to teicoplanin (1, 44). They were also frequently resistant to penicillins, aminoglycosides at high concentrations, ciprofloxacin, tetracycline, and chloramphenicol, and the only drugs with in vitro activity against all the isolates were quinupristin-dalfopristin (E. faecium) and linezolid.
Identification of the VanB phenotype was confirmed by PCR detection of the vanB gene with primers vanB consensus (16) and amplification of the unusually sized product (approximately 1.1 kb) with primers vanB (14) suggested that the gene variant present in the outbreak isolates was not vanB1 (16). Such a vanB gene amplicon has already been observed in a VREM strain from a Warsaw hospital (29) and was revealed to originate from the annealing of the upstream vanB primer within the adjacent vanHB gene. Sequence analysis of the amplified fragment demonstrated that selected VRES (isolate 2496) and VREM (isolate 3123) isolates contained a vanB2 gene variant identical to that identified in the Warsaw strain (29) and in an isolate from the United States, in which it was originally described (39). The corresponding region in VREM isolate 2947 differed from the remaining ones only by a single mutation, and it represented a novel, though closely related, vanB2 variant. These data, together with the results of the RFLP analysis of vanB gene clusters (discussed below), suggested that there may have been an epidemiological link between the VRE isolates analyzed here and the VREM strain identified in Warsaw about 9 months earlier. The direct transmission of the Warsaw strain to the hospital in Cracow seems rather unlikely, as indicated by the different PFGE patterns produced by VREM isolates from the two institutions (results not shown). However, it is possible that transmission of the particular variant of VanB phenotype determinants was mediated by another strain or, more probably, that their reservoir is widely spread in Poland.
The RFLP analysis of vanB gene clusters present in outbreak isolates demonstrated that all the VREM isolates contained the same cluster RFLP type, which had been identified before as RFLP-2 in VRE from Norway, Sweden, the United Kingdom, Germany, and the United States (16) and was also observed in the VREM strain from the Warsaw hospital (29). On the other hand, the VRES isolates were found to contain another polymorph of the region, which seems to represent a novel type (RFLP-4); however, it differed from RFLP-2 only by the presence of two additional restriction fragments. This observation, together with the vanB PCR and sequencing data, suggested that one of the vanB gene cluster variants evolved from the other due to DNA recombination (insertion or deletion of a DNA fragment). It is, however, impossible to reveal whether the two gene cluster variants were introduced independently into E. faecalis and E. faecium populations in the hospital or whether any of these was transmitted from one species to the other, which was then followed by its modification.
All the VRES isolates were found to be indistinguishable by PFGE, which indicated that the infections of six patients with this microorganism were due to clonal dissemination of a single strain. Contrarily, the VREM isolates collected from two infected patients represented two distinct PFGE types (types b and c); moreover, other isolates from two colonized patients, though related to each other, were classified as another type (type a). These data revealed that the spread of VREM in the hospital was mostly nonclonal and could, therefore, be mediated by horizontal transfer of VanB resistance genes among nonrelated strains. The hypothesis was supported by the fact that some VREM isolates (PFGE type c isolates 3124A, 3124B, and 3128) produced vancomycin-resistant transconjugants in mating experiments. The hybridization study of the vanB gene cluster probe with the undigested total DNA of isolates separated by PFGE demonstrated that in VRES, van genes were located in chromosomal DNA, whereas in VREM isolates they resided within plasmid molecules. These plasmids were of different sizes in isolates belonging to different PFGE types; moreover, some VREM isolates contained more than one vanB gene cluster-carrying plasmid. (In some isolates the multiple hybridizing bands could also represent different forms of the same plasmid.) These data suggested that the vanB gene cluster was located within an active transposon, which could be inserted into various DNA replicons. It was probably spread among E. faecium strains by horizontal transfer mediated by the element itself or, less likely, by conjugation of plasmids, followed by their multiple rearrangements.
Two VSEM isolates identified as etiologic agents of infections during the time of the VRE outbreak in the same wards were found by PFGE to be related to VREM isolates, and they also demonstrated susceptibility patterns similar to those of their VREM counterparts. They belonged to two different PFGE types discerned among E. faecium isolates: type a (VSEM isolate 2494), which included two VREM isolates from two other colonized patients (isolates 2497 and 2499), and type c (VSEM isolate 3122), which also grouped four VREM isolates from the same patient (3123, 3124A, 3124B, and 3128). It may be suggested that, as with previous reports (28, 41, 47), the VSEM isolates analyzed here represented endemic hospital strains which had acquired vancomycin resistance genes. However, a small number of VSEM isolates identified in the wards at the time did not allow us to study in detail the endemic vancomycin-susceptible enterococcus background of the outbreak, and it cannot be ruled out that the two VSEM isolates appeared due to a loss of VanB resistance determinants by VREM strains.
An interesting group of isolates was formed by five closely related PFGE type c E. faecium isolates collected from a single patient, four of which (VSEM 3122 and VREM 3123, 3124A, and 3124B) were identified in the same blood sample, with the remaining one (VREM 3128) being recovered from urine. Apart from the difference in vancomycin susceptibility (VSEM versus VREM) and variations in their PFGE patterns (five subtypes), these isolates also revealed some heterogeneity in susceptibility to other antimicrobials, plasmid profile, and number of vanB gene cluster copies. It is very likely that all these observations document evolutionary changes which occurred in the originally homogeneous E. faecium strain in the course of infection of a single patient. Similar data were previously obtained in the study of VREM incidence in the Warsaw hospital (29). Interestingly, all the isolates were recovered 23 days after the end of the weeklong vancomycin therapy of the patient, who was also treated with piperacillin-tazobactam and ceftazidime along with vancomycin. At the time of the E. faecium isolation, the patient was on prolonged therapy with cefepime and clindamycin. Such a profile of antibiotic treatment could create favorable conditions for the selection and evolution of enterococcal strains.
The most striking part of the outbreak analysis was the identification of a teicoplanin-resistant VREM isolate, isolate 2497, of PFGE type a that was related to two other isolates, VREM 2499 of the typical VanB phenotype and VSEM 2494. Isolate 2497 was recovered from a patient carrier who had been hospitalized for 6 months before the strain's isolation and had undergone prolonged antimicrobial therapy. The patient, among others, was treated twice with vancomycin and 4 days prior to identification of the isolate teicoplanin was introduced into therapy (together with amikacin and colistin). PCR analysis excluded the VanA phenotype in the isolate, whereas the vanB gene was amplified with primers vanB consensus (16) and vanB (14) as in all other isolates. The unusual PCR product of about 1.1 kb containing parts of vanHB and vanB genes (amplified with primers vanB) was found to differ only by a single nucleotide substitution compared to those obtained for isolates VREM 3123 and VRES 2496. However, the isolate failed to amplify the entire vanB gene cluster and the spacer region located between the vanSB and vanYB genes (which includes the vancomycin-inducible PYB promoter), which prompted us to investigate in greater detail the organization and expression of its glycopeptide resistance determinant.
PCR and hybridization revealed that isolate 2497 did not contain the four genes that are located upstream of the vanHB gene within the wild-type vanB gene cluster, namely, vanRB, -SB, -YB, and -W (16, 20, 43). The most important finding was the lack of genes vanRB and -SB, which code for the two-component VanRB-VanSB system that activates transcription of vanYB, -W, -HB, -B, and -XB genes from the PYB promoter in the presence of vancomycin but not teicoplanin (20). Therefore, it was hypothesized that the deletion of a large part of the vanB gene cluster, including vanRB and -SB genes and the PYB promoter, resulted in constitutive expression of resistance genes vanHB, -B, and -XB, and, subsequently, in teicoplanin resistance. In order to check this hypothesis the RT-PCR experiment was carried out, and it indeed demonstrated that the genes were transcribed in this isolate in the absence of glycopeptides. The promoter sequence responsible for the effect remains to be determined. In the work of Evers and Courvalin, S1 nuclease mapping suggested that a weak promoter might be present immediately upstream of the vanHB gene, but this result was poorly reproducible and was not confirmed by other approaches (20).
Teicoplanin-resistant VanB mutants have been rarely observed among clinical isolates of enterococci. Hayden et al. described a VanB VREM isolate for which the MIC of teicoplanin was 64 μg/ml and which constitutively expressed a 41-kDa membrane protein that was vancomycin inducible in the E. faecalis V583 reference strain (26). However, the protein was not studied in detail, and it is not known what modifications of the vanB gene cluster were responsible for the phenotype. On the other hand such mutants have been often obtained in the laboratory, including selection in an animal model, and all those studied down to the molecular level were found to result from point mutations in the vanSB gene (2, 4, 5, 6, 7, 25, 33). Some of these changes caused the VanSB ability to recognize teicoplanin as the expression inducer, whereas others abolished its phosphatase activity, which led to the permanent phosphorylation of VanRB and constitutive transcription of the resistance genes. Finally, VanSB null mutants were revealed to express the heterogeneous inducible phenotype that probably resulted from taking over the VanSB functions by a host kinase (2, 4, 5, 6, 7). Deletion mutants lacking the whole VanRB-VanSB regulatory system and the PYB promoter have not been, however, observed to date.
The data presented in this work document a VanB VRE outbreak, which even though it did not involve many patients (10 patients in all) turned out to be very complex and so reflected well the dynamics of VRE epidemiology. It was caused by VRES as well as VREM which could exchange vancomycin resistance determinants or acquire them independently. The VRE spread was both clonal (VRES) and nonclonal (VREM), and horizontal transmission of vanB gene clusters probably occurred due to the conjugative functions of the transposable elements containing the clusters. The VREM strains were undergoing diversification on the level of chromosomal and plasmid DNA, and evolution of one of them could be observed in the course of infection of a single patient. Treatment of one of the patients with teicoplanin was probably responsible for the selection of a VREM strain variant, which due to the deletion of a large part of the vanB gene cluster was also resistant to teicoplanin. This finding, together with data from other laboratories, indicates that various genetic changes may determine resistance to teicoplanin in VanB enterococci and thus limit its use against these microorganisms (2, 6, 33, 36).
ACKNOWLEDGMENTS
We thank Ewa Sadowy and Andrew Hazlewood for critical reading of the manuscript and Andrzej Pałucha for very helpful discussions and the SuperScript II enzyme. Also we are very thankful to Patrice Courvalin who kindly provided E. faecium BM4147 and E. faecalis V583, Wolfgang Witte for the E. faecium 64/3 strain, and Wolfgang Haas for E. faecalis FA2-2.
This work was partially financed by the U.S.-Poland Maria Skłodowska-Curie Joint Fund II (MZ/NIH 98-324) and the program SPUB-M-INCO-COPERNICUS (4 PR UE/P-05/DZ 112/2000) of the Polish Committee for Scientific Research (KBN).
REFERENCES
- 1.Arthur M, Courvalin P. Genetics and mechanisms of glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 1993;37:1563–1571. doi: 10.1128/aac.37.8.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur M, Depardieu F, Courvalin P. Regulated interactions between partner and non-partner sensors and response regulators that control glycopeptide resistance gene expression in enterococci. Microbiology. 1999;145:1849–1858. doi: 10.1099/13500872-145-8-1849. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Molinas C, Depardieu F, Courvalin P. Characterization of Tn1546, a Tn3-related transposon conferring glycopeptide resistance by synthesis of depsipeptide peptidoglycan precursors in Enterococcus faecium BM4147. J Bacteriol. 1993;175:117–127. doi: 10.1128/jb.175.1.117-127.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arthur M, Quintiliani R., Jr Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrob Agents Chemother. 2001;45:375–381. doi: 10.1128/AAC.45.2.375-381.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aslangul E, Baptista M, Fantin B, Depardieu F, Arthur M, Courvalin P, et al. Selection of glycopeptide-resistant mutants of vanB-type Enterococcus faecalis BM4281 in vitro and in experimental endocarditis. J Infect Dis. 1997;175:598–605. doi: 10.1093/infdis/175.3.598. [DOI] [PubMed] [Google Scholar]
- 6.Baptista M, Depardieu F, Courvalin P, Arthur M. Specificity of induction of glycopeptide resistance genes in Enterococcus faecalis. Antimicrob Agents Chemother. 1996;40:2291–2295. doi: 10.1128/aac.40.10.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baptista M, Rodrigues P, Depardieu F, Courvalin P, Arthur M. Single-cell analysis of glycopeptide resistance gene expression in teicoplanin-resistant mutants of VanB-type Enterococcus faecalis. Mol Microbiol. 1999;32:17–28. doi: 10.1046/j.1365-2958.1999.01308.x. [DOI] [PubMed] [Google Scholar]
- 8.Bell J M, Paton J C, Turnidge J. Emergence of vancomycin-resistant enterococci in Australia: phenotypic and genotypic characteristics of isolates. J Clin Microbiol. 1998;36:2187–2190. doi: 10.1128/jcm.36.8.2187-2190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce J M, Favero M S, Gaynes R P, Goldmann D A, Jarvis W R, Pugliese G, Weinstein R A. Populations at risk and routes of transmission. In: Pugliese G, Weinstein R A, editors. Issues and controversies in prevention and control of VRE. Chicago, Ill: Etna Communications; 1998. pp. 15–16. [Google Scholar]
- 10.Boyce J M, Opal S M, Chow J W, Zervos M J, Potter-Bynoe G, Sherman C B, Romulo R L C, Fortna S, Medeiros A A. Outbreak of multidrug-resistant Enterococcus faecium with transferable vanB class vancomycin resistance. J Clin Microbiol. 1994;32:1148–1153. doi: 10.1128/jcm.32.5.1148-1153.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carias L L, Rudin S D, Donskey C J, Rice L B. Genetic linkage and cotransfer of a novel vanB-containing transposon (Tn5382) and a low-affinity penicillin-binding protein 5 gene in a clinical vancomycin-resistant Enterococcus faecium isolate. J Bacteriol. 1998;180:4426–4434. doi: 10.1128/jb.180.17.4426-4434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cetnikaya Y, Falk P, Mayhall C G. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow J W, Kutitza A, Shlaes D M, Green M, Sahm D F, Zervos M J. Clonal spread of vancomycin-resistant Enterococcus faecium between patients in three hospitals in two states. J Clin Microbiol. 1993;31:1609–1611. doi: 10.1128/jcm.31.6.1609-1611.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clark N, Cooksey R, Hill B, Swenson J, Tenover F C. Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob Agents Chemother. 1993;37:2311–2317. doi: 10.1128/aac.37.11.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clewell D B, Tomich P K, Gawron-Burke M C, Franke A E, Yagi Y, An F Y. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J Bacteriol. 1982;152:1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl K H, Skov Simonsen G, Olsvik Ø, Sundsfjord A. Heterogeneity in vanB gene cluster of genetically diverse clinical strains of vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1999;43:1105–1110. doi: 10.1128/aac.43.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Lencastre H, Severina E P, Roberts R B, Kreiswirth B N, Tomasz A the BARG Initiative Pilot Study Group. Testing the efficacy of a molecular surveillance network: methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus faecium (VREF) genotypes in six hospitals in the metropolitan New York City area. Microb Drug Resist. 1996;2:343–351. doi: 10.1089/mdr.1996.2.343. [DOI] [PubMed] [Google Scholar]
- 18.Donskey C J, Schreiber J R, Jacobs M R, Shekar R, Salata R A, Gordon S, Whalen C C, Smith F, Rice L B the Northeast Ohio Vancomycin-Resistant Enterococcus Surveillance Program. A polyclonal outbreak of predominantly VanB vancomycin-resistant enterococci in northeast Ohio. Clin Infect Dis. 1999;29:573–579. doi: 10.1086/598636. [DOI] [PubMed] [Google Scholar]
- 19.Dutka-Malen S, Evers S, Courvalin P. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J Clin Microbiol. 1995;33:24–27. doi: 10.1128/jcm.33.1.24-27.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evers S, Courvalin P. Regulation of VanB-type resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. 1996;178:1302–1309. doi: 10.1128/jb.178.5.1302-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evers S, Reynolds P E, Courvalin P. Sequence of the vanB and ddl genes encoding D-alanine:D-lactate and D-alanine:D-alanine ligases in vancomycin-resistant Enterococcus faecalis V583. Gene. 1994;140:97–102. doi: 10.1016/0378-1119(94)90737-4. [DOI] [PubMed] [Google Scholar]
- 22.Facklam R, Collins M D. Identification of Enterococcus species isolated from human infections by a conventional test scheme. J Clin Microbiol. 1989;27:731–734. doi: 10.1128/jcm.27.4.731-734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fines M, Perichon B, Reynolds P, Sahm D F, Courvalin P. VanE, a new type of acquired glycopeptide resistance in Enterococcus faecalis BM4405. Antimicrob Agents Chemother. 1999;43:2161–2164. doi: 10.1128/aac.43.9.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gold H S, Ünal S, Cercenado E, Thauvin-Eliopulos C, Eliopulos G M, Wennerstein C B, Moellering R C., Jr A gene conferring resistance to vancomycin but not teicoplanin in isolates of Enterococcus faecalis and Enterococcus faecium demonstrates homology with vanB, vanA, and vanC genes of enterococci. Antimicrob Agents Chemother. 1993;37:1604–1609. doi: 10.1128/aac.37.8.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutmann L, Billot-Klein D, Al-Obeid S, Klare I, Francoual S, Collatz E, Van Heijenoort J. Inducible carboxypeptidase activity in vancomycin-resistant enterococci. Antimicrob Agents Chemother. 1992;36:77–80. doi: 10.1128/aac.36.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayden M K, Trenholme G M, Schultz J E, Sahm D F. In vivo development of teicoplanin resistance in a VanB Enterococcus faecium isolate. J Infect Dis. 1993;167:1224–1227. doi: 10.1093/infdis/167.5.1224. [DOI] [PubMed] [Google Scholar]
- 27.Kawalec M, Gniadkowski M, Hryniewicz W. Outbreak of vancomycin-resistant enterococci in a hospital in Gdańsk, Poland, due to horizontal transfer of different Tn1546-like transposon variants and clonal spread of several strains. J Clin Microbiol. 2000;38:3317–3322. doi: 10.1128/jcm.38.9.3317-3322.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawalec M, Gniadkowski M, Zaleska M, Ozorowski T, Konopka L, Hryniewicz W. Outbreak of vancomycin-resistant Enterococcus faecium of the phenotype VanB in a hospital in Warsaw, Poland: probable transmission of the resistance determinants into an endemic vancomycin-susceptible strain. J Clin Microbiol. 2001;39:1781–1787. doi: 10.1128/JCM.39.5.1781-1787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawalec M, Gniadkowski M, Zielińska U, Kłos W, Hryniewicz W. A vancomycin-resistant Enterococcus faecium strain carrying the vanB2 gene variant in a Polish hospital. J Clin Microbiol. 2001;39:811–815. doi: 10.1128/JCM.39.2.811-815.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klare I, Collatz E, Al.-Obeid S, Wagner J, Rodloff A C, Witte W. Glykopeptidresistenz bei Enterococcus faecium aus Besiedlungen und Infectionen von Patienten aus Intensivstationen Berliner Kliniken und einem Transplantationszentrum. Z Antimikrob Antineoplast Chemother. 1992;10:45–53. [Google Scholar]
- 31.Leclercq R, Derlot E, Duval J, Courvalin P. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N Engl J Med. 1988;319:157–161. doi: 10.1056/NEJM198807213190307. [DOI] [PubMed] [Google Scholar]
- 32.Leclercq R, Dutka-Malen S, Duval J, Courvalin P. Vancomycin resistance gene vanC is specific to Enterococcus gallinarum. Antimicrob Agents Chemother. 1992;36:2005–2008. doi: 10.1128/aac.36.9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lefort A, Baptista M, Fantin B, Depardieu F, Arthur M, Carbon C, Courvalin P. Two-step acquisition of resistance to the teicoplanin-gentamicin combination by VanB-type Enterococcus faecalis in vitro and in experimental endocarditis. Antimicrob Agents Chemother. 1999;43:476–482. doi: 10.1128/aac.43.3.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maki D G, Agger W A. Enterococcal bacteremia: clinical features, the risk of endocarditis and management. Medicine. 1988;67:248–269. [PubMed] [Google Scholar]
- 35.McKessar S J, Berry A M, Bell J M, Turnidge J D, Paton J C. Genetic characterization of vanG, a novel vancomycin resistance locus of Enterococcus faecalis. Antimicrob Agents Chemother. 2000;44:3224–3228. doi: 10.1128/aac.44.11.3224-3228.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray B E. Vancomycin-resistant enterococci. Am J Med. 1997;102:284–293. doi: 10.1016/S0002-9343(99)80270-8. [DOI] [PubMed] [Google Scholar]
- 37.Murray B E. Diversity among multidrug-resistant enterococci. Emerg Infect Dis. 1998;4:37–47. doi: 10.3201/eid0401.980106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, M7–A5. Wayne, Pa: NCCLS; 2000. [Google Scholar]
- 39.Patel R, Uhl J R, Kohner P, Hopkins M K, Steckelberg J M, Kline B, Cockerill F R I. DNA sequence variation within vanA, vanB, vanC-1, and vanC-2/3 genes of clinical Enterococcus isolates. Antimicrob Agents Chemother. 1988;42:202–205. doi: 10.1128/aac.42.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perichon B, Reynolds P E, Courvalin P. VanD-type glycopeptide-resistant Enterococcus faecium BM4339. Antimicrob Agents Chemother. 1997;41:2016–2018. doi: 10.1128/aac.41.9.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perlada D E, Smulian A G, Cushion M T. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J Clin Microbiol. 1997;35:2342–2347. doi: 10.1128/jcm.35.9.2342-2347.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quintiliani R, Jr, Courvalin P. Conjugal transfer of the vancomycin-resistance determinant vanB between enterococci involves the movement of large genetic elements from chromosome to chromosome. FEMS Microbiol Lett. 1994;119:359–364. doi: 10.1111/j.1574-6968.1994.tb06913.x. [DOI] [PubMed] [Google Scholar]
- 43.Quintiliani R, Jr, Courvalin P. Characterization of Tn1547, composite transposon flanked by the IS16 and IS256-like elements, that confers vancomycin resistance in Enterococcus faecalis BM4281. Gene. 1996;172:1–8. doi: 10.1016/0378-1119(96)00110-2. [DOI] [PubMed] [Google Scholar]
- 44.Quintiliani R, Jr, Evers S, Courvalin P. The vanB gene confers various levels of self-transferable resistance to vancomycin in enterococci. J Infect Dis. 1993;167:1220–1223. doi: 10.1093/infdis/167.5.1220. [DOI] [PubMed] [Google Scholar]
- 45.Sahm D F, Kissinger J, Gilmore M S, Murray B E, Mulder R, Solliday J, Clarke B. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 1989;33:1588–1591. doi: 10.1128/aac.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 47.Suppola J P, Kolho E, Salmenlinna S, Tarkka E, Vuopio-Varkila J, Vaara M. VanA and vanB incorporate into an endemic ampicillin-resistant vancomycin-sensitive Enterococcus faecium strain: effect on interpretation of clonality. J Clin Microbiol. 1999;37:3934–3939. doi: 10.1128/jcm.37.12.3934-3939.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tenover F C, Arbeit R, Goering V, Mickelsen P, Murray B M, Pershing D, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thal L, Donabedian S, Robinson-Dunn B, Chow J W, Dembry L, Clewell D B, Alshab D, Zervos M J. Molecular analysis of glycopeptide-resistant Enterococcus faecium isolates collected from Michigan Hospitals over a 6-year period. J Clin Microbiol. 1998;36:3303–3308. doi: 10.1128/jcm.36.11.3303-3308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Werner G, Klare I, Witte W. Arrangement of the vanA gene cluster in enterococci of different ecological origin. FEMS Microbiol Lett. 1997;155:55–61. doi: 10.1111/j.1574-6968.1997.tb12685.x. [DOI] [PubMed] [Google Scholar]
- 51.Woodford N, Jones B L, Baccus Z, Ludlam H A, Brown D F J. Linkage of vancomycin and high-level gentamicin resistance genes on the same plasmid in a clinical isolate of Enterococcus faecalis. J Antimicrob Chemother. 1995;35:179–184. doi: 10.1093/jac/35.1.179. [DOI] [PubMed] [Google Scholar]