ABSTRACT
Patients with cystic fibrosis (CF) experience lifelong respiratory infections, which are a significant cause of morbidity and death. These infections are polymicrobial in nature, and the predominant bacterial species undergo a predictable series of changes as patients age. Young patients have populations dominated by opportunists that are typically found within the microbiome of the human nasopharynx, such as nontypeable Haemophilus influenzae (NTHi); these are eventually supplanted, and the population within the CF lung is later dominated by pathogens such as Pseudomonas aeruginosa. In this study, we investigated how initial colonization with NTHi impacts colonization and persistence of P. aeruginosa in the respiratory tract. Analysis of polymicrobial biofilms in vitro by confocal microscopy revealed that NTHi promoted greater P. aeruginosa biofilm volume and diffusion. However, sequential respiratory infection of mice with NTHi followed by P. aeruginosa resulted in significantly lower levels of P. aeruginosa, compared to infection with P. aeruginosa alone. Coinfected mice also had reduced airway tissue damage and lower levels of inflammatory cytokines, compared with P. aeruginosa-infected mice. Similar results were observed after instillation of heat-inactivated NTHi bacteria or purified NTHi lipooligosaccharide endotoxin prior to P. aeruginosa introduction. Based on these results, we conclude that NTHi significantly reduces susceptibility to subsequent P. aeruginosa infection, most likely due to priming of host innate immunity rather than a direct competitive interaction between species. These findings have potential significance with regard to therapeutic management of early-life infections in patients with CF.
KEYWORDS: cystic fibrosis, bacteria, Haemophilus influenzae, Pseudomonas aeruginosa, biofilm, lung infection
INTRODUCTION
Cystic fibrosis (CF) is an autosomal recessive genetic illness that causes dysfunctional ionic transport at epithelial surfaces, with concomitant impacts on mucociliary defenses and clearance in the lungs. As a consequence, patients with CF experience lifelong infections of the airway mucosa, which cause chronic inflammation and epithelial damage, among other sequelae (1–3). Despite significant therapeutic gains that have significantly increased the average life span for CF patients, airway infections and associated respiratory complications remain a leading cause of morbidity and death in patients with CF disease (4). The microbial populations within the CF lung are complex, polymicrobial communities that undergo dynamic changes that correlate with the patient’s age and overall health and can be a determinant of disease severity (3, 5–7). Despite years of study of opportunistic infections in the context of CF, there remains much to be learned about how specific pathogens and opportunists impact the course and severity of disease (6–9). Nontypeable Haemophilus influenzae (NTHi) is a nonencapsulated, Gram-negative opportunist that normally resides in the human nasopharynx and upper airways as part of the normal flora (10–12). Although it is typically benign in healthy individuals, NTHi is among the most common bacterial species isolated from CF patients during the first year of life and is thus considered a common early-stage pathogen (13–15). As with other mucosal airway infections, NTHi persists in the lung within biofilm communities (16, 17). Like other mucosal pathogens, NTHi produces lipooligosaccharide (LOS) endotoxins with truncated carbohydrate regions (18). Like most endotoxins, the NTHi LOS elicits host inflammation via Toll-like receptor 4 and is likely an integral component for symptomatic responses to NTHi infection (19, 20).
As CF patients age, the bacterial population diversity typically declines, and the microbial population in the lung becomes dominated by late-stage pathogens, such as Pseudomonas aeruginosa (8, 9, 21, 22). P. aeruginosa is a Gram-negative opportunistic pathogen that has a high level of inherent resistance to antimicrobials and host immune effectors and thus is difficult to treat (23). To adapt to the CF airway environment, P. aeruginosa populations typically undergo mucoid conversion to overexpress the exopolysaccharide alginate, which is a component of P. aeruginosa biofilms (24, 25). Mucoid conversion of P. aeruginosa is associated with increased antibiotic resistance, persistent inflammation, and overall worse clinical outcomes (26). Initial P. aeruginosa infections, which may take place within the first year of life, frequently become chronic after mucoid conversion (19, 20). While the role of P. aeruginosa in end-stage CF disease has been well established, less has been done to establish the role of P. aeruginosa in early-stage CF disease, particularly the ages at which NTHi colonization peaks in frequency (4, 9).
Bacteria reside in the respiratory system as a complex, evolving, polymicrobial community in which individual organisms may interact with one another in a synergistic, antagonistic, or null fashion (11, 27–29). Changes in these communities as different species, or strains within species, interact are thought to greatly influence antibiotic susceptibility, immune evasion, and mutations for chronic colonization, all influencing patient outcomes (8, 30). To fully understand how specific species contribute to disease pathogenesis, we must characterize community members in concert, modeling their interactions after the environment in which they naturally reside, rather than as individual species (5, 8, 31). In this study, we aimed to assess the polymicrobial interaction between NTHi and P. aeruginosa both in vitro and in vivo, using clinically isolated bacterial strains from patients with CF. Mice were intratracheally infected with NTHi and/or mucoid P. aeruginosa (mPA) to produce localized, acute respiratory infections. To model the temporal acquisition of NTHi and P. aeruginosa in CF patients, we tested whether precolonization with NTHi influences the colonization of P. aeruginosa and we began to characterize how this interaction may influence the innate immune response.
RESULTS
NTHi and P. aeruginosa form polymicrobial biofilms in vitro that support P. aeruginosa growth.
In any environment that contains more than one bacterial species, including the CF lungs, organisms may interact with one another synergistically, competitively, or in a null fashion (4, 5). To begin to characterize the polymicrobial interaction between NTHi and P. aeruginosa, we first wanted to evaluate how these two organisms form a polymicrobial biofilm in vitro. Since NTHi and P. aeruginosa are typically acquired sequentially over a patient’s life span, dual-species static biofilms were sequentially seeded with NTHi followed by P. aeruginosa 12 h later. These polymicrobial biofilms were grown for a total of 18 h, while single-species biofilms of NTHi and P. aeruginosa were grown for 12 h and 6 h, respectively, at 37°C in 5% CO2. Time points for growth were selected based on the peak viability of NTHi and P. aeruginosa, as determined by counts of viability over time (see Fig. S1A in the supplemental material). Utilizing various strains of NTHi, we assessed viable colony counts of P. aeruginosa from sequential dual biofilms, compared to single-species P. aeruginosa biofilms. In vitro, precolonization with NTHi resulted in significant increases in P. aeruginosa growth when the two organisms were inoculated in roughly the same amounts, suggesting a synergistic relationship (**, P < 0.01) (see Fig. S1B to D).
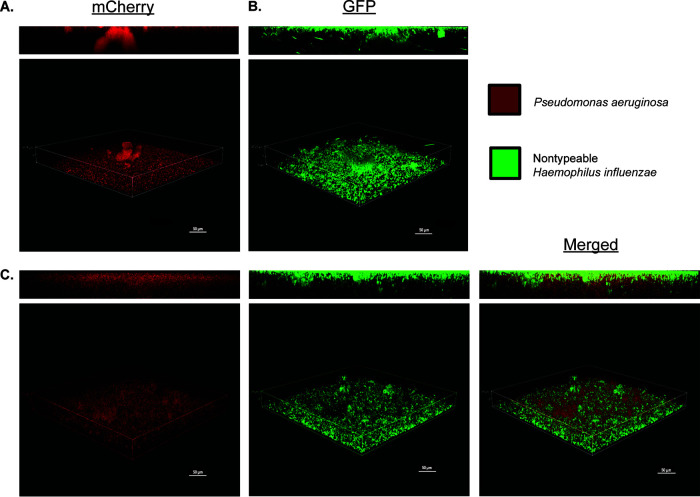
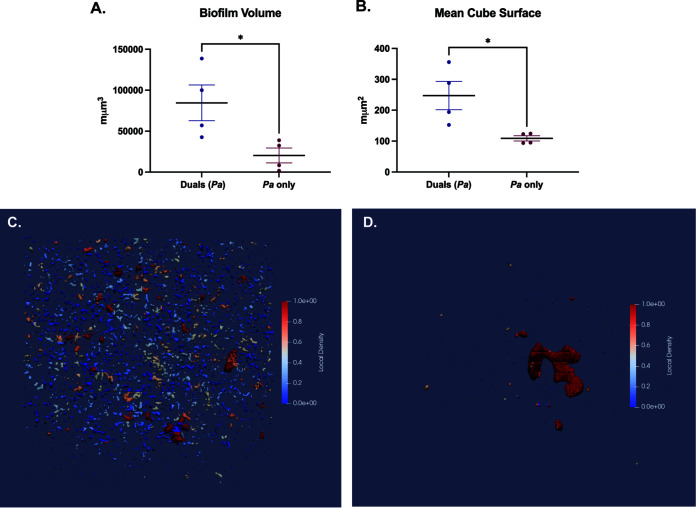
We then visualized NTHi and P. aeruginosa single- and dual-species fluorescent biofilms with confocal laser scanning microscopy (CLSM) after 18 h of growth using strains NTHi 86028NP-gfp+ and mPA 08-31-mCherry+. In single-species biofilms, both organisms formed lawn structures with some towers (Fig. 1A and B). Sequentially seeded dual-species biofilms showed a complex lawn with more diffuse P. aeruginosa colony formation, which appeared to support P. aeruginosa growth. As expected, the spatial orientation of dual biofilms was stratified, reflecting the sequential introduction of NTHi followed by P. aeruginosa, with a significant lawn structure with NTHi on the bottom (Fig. 1C). Quantification of P. aeruginosa in both single- and dual-species biofilms with BiofilmQ software revealed that the total volume of P. aeruginosa biofilm was greater in dual-species biofilms than in single-species P. aeruginosa biofilms (Fig. 2A). Segmentation within BiofilmQ software allowed fluorescent images to be divided into 1-μm cubes, with each cube being individually tracked. Following segmentation, the mean cube surface was calculated as the number of points between occupied volume and unoccupied volume in the cube. The calculated mean cube surface showed that dual-species biofilms had more P. aeruginosa than single-species P. aeruginosa biofilms, recapitulating the results seen with total biofilm volume measurements and verifying the accuracy of cubing (Fig. 2B). This segmentation technique was used for further three-dimensional visualization of single- and dual-species biofilms of P. aeruginosa. Segmentation showed that there was more diffuse local density of P. aeruginosa associated with dual-species growth (Fig. 2C), rather than single-species growth (Fig. 2D). Overall, BiofilmQ quantification and viable colony counts from coculture suggest that, in vitro, NTHi positively influences P. aeruginosa growth, producing greater biofilm volume and more dispersed spatial distribution when the species are grown together, compared to a single-species P. aeruginosa biofilm.
FIG 1.
NTHi and P. aeruginosa form polymicrobial biofilms in vitro. Biofilms of P. aeruginosa mPA 08-31 (pUCP19+mCherry+) (A), NTHi 86-028NP (PGM1.1+gfp) (B), and NTHi and P. aeruginosa (C) on abiotic glass surfaces are shown. Inoculation of NTHi preceded P. aeruginosa by 12 h, and both organisms grew for an additional 6 h at 37°C in 5% CO2 (18 h total). Imaged at ×40 magnification; the scale bars represent 50 μm. GFP, green fluorescent protein.
FIG 2.
Quantification of NTHi and P. aeruginosa (Pa) polymicrobial biofilms. (A and B) Bacterial density within fluorescent biofilms was quantified using BiofilmQ software to measure biofilm volume of P. aeruginosa in dual- and single-species biofilms (A) and the mean cube surface of P. aeruginosa (quantified as 1 cube equals 1 μm) (B). (C and D) Cubing can be visualized as local density of P. aeruginosa in both polymicrobial NTHi/P. aeruginosa biofilms (C) and P. aeruginosa biofilms (D). Values are mean ± SEM (n = 4). *, P < 0.05, Mann-Whitney test, two-tailed.
NTHi infection decreases the bacterial burden of P. aeruginosa in mouse lung.
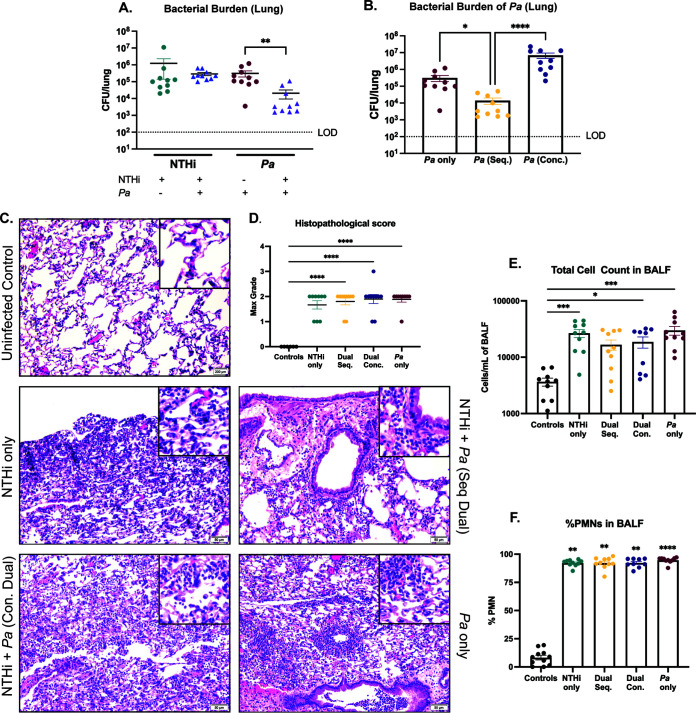
While our in vitro results suggest that NTHi and P. aeruginosa may positively interact in polymicrobial biofilms, we aimed to evaluate how each species is impacted during polymicrobial infection within the host. Through serial passage, we induced antibiotic resistance in a CF sputum sample-derived clinical isolate of NTHi, HI-1, developing a model strain for acute infection (NTHi HI-1r) that can be differentially plated in the presence of P. aeruginosa. To model the sequential acquisition of an early-life pathogen (NTHi) followed by a later-stage pathogen (P. aeruginosa) that typically ensues as CF patients age, BALB/cJ mice were intratracheally infected with NTHi HI-1r (inoculum of ∼108 CFU/mouse) followed by an mPA isolate, mPA 08-31 (inoculum of ∼107 CFU/mouse), 24 h later (see Fig. S2A). Both organisms colonized the lungs and were detectable by viable plate counting of lung homogenates 48 h postinfection. However, colonization of P. aeruginosa was significantly impeded after sequential introduction with NTHi, compared to P. aeruginosa single infection (P < 0.01) (Fig. 3A). Because this contrasts with the in vitro findings, this result supports the importance of studying polymicrobial interactions within a host to accurately model human diseases. To determine the importance of temporal spacing in this interaction, we also infected mice with both organisms concurrently (inoculum of ∼107 CFU/ml of each species) (see Fig. S2B). When we compared the bacterial burdens of P. aeruginosa in the lungs after a single infection, a sequential dual infection, and a concurrent dual infection, we saw that mice sequentially infected with NTHi followed by P. aeruginosa showed significant reduction in P. aeruginosa abundance, compared to a P. aeruginosa-only infection (P < 0.05). Additionally, sequential dual infection resulted in less P. aeruginosa in lung homogenates than concurrent dual infection (P < 0.0001) (Fig. 3B). This suggests that the competitive interaction between NTHi and P. aeruginosa is dependent on the time of introduction and P. aeruginosa colonization is impeded only following sequential infection. Histopathological analysis of hematoxylin and eosin (H&E)-stained lung sections indicated immune cell infiltration and inflammation 48 h after infection with both NTHi and P. aeruginosa (Fig. 3C). H&E-stained slides were graded for the severity of lung injury, as indicated by neutrophil infiltration of alveoli, perivascular inflammation, and peribronchial inflammation. At 48 h postinfection, when large burdens of both NTHi and P. aeruginosa were present, there was moderate tissue damage for all infection groups, with no significant difference between groups. All infections resulted in significantly more severe tissue damage than a phosphate-buffered saline (PBS) vehicle control (P < 0.0001) (Fig. 3D). Immune cell infiltration was expressed by total cell infiltrates counted in the bronchoalveolar lavage fluid (BALF) samples after cytospin preparation and differential staining. As expected from histological results, all infection groups showed significantly more total cell counts, as well as polymorphonuclear leukocyte (PMN) infiltrates in those counts, than did uninfected controls, with no significant difference between infection groups (Fig. 3E and F). Weight loss per animal was evaluated over the course of infection, and weight loss increased, compared to uninfected controls (see Fig. S3A). Taken together, these results suggest that in vivo NTHi inhibits the bacterial burden of P. aeruginosa in the airways when introduced sequentially but not concurrently and infection with either organism results in respiratory tissue damage with a significant inflammatory response.
FIG 3.
NTHi infection significantly decreases P. aeruginosa (Pa) colonization/persistence in mouse lung. Either BALBc/J mice were intratracheally infected with NTHi (NTHi Hi-1r) 24 h prior to introduction of mPA (mPA 08-31) in a sequential fashion or the species were introduced at the same time (concurrently) and assessed 48 h postinfection. (A) Bacterial counts of NTHi and P. aeruginosa in the lung homogenates enumerated by plate counting. (B) Bacterial counts of P. aeruginosa in the lung homogenates, comparing sequential infection and concurrent infection. (C and D) Severity of disease indicated by H&E-stained lung sections (×200 magnification) (C) and semiquantitative grading by a pathologist (L.N.) (D). (E) Total cell counts in BALF samples quantified by differential cell counts. (F) Percentage of PMNs in the BALF samples. Values are mean ± SEM (n = 10). LOD, limit of detection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Kruskal-Wallis test with Dunn’s multiple-comparison test.
Impacts of NTHi on P. aeruginosa are not strain specific.
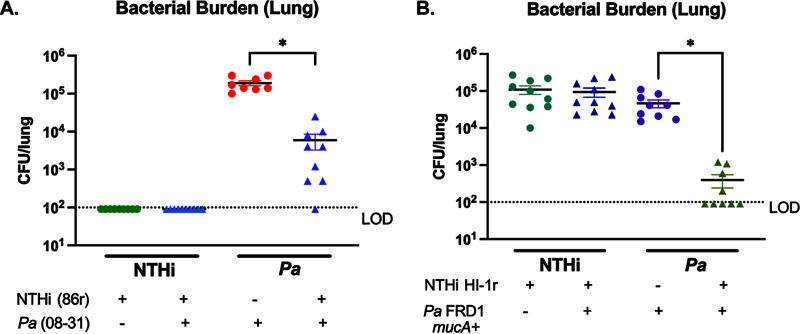
Our coinfection results indicate that a clinical isolate of NTHi, HI-1r, significantly inhibits the bacterial burden of mPA isolate mPA 08-31 in the airways. To determine whether the impact of NTHi on P. aeruginosa in vivo is strain specific, we induced antibiotic resistance in laboratory strain NTHi 86-028NP to develop strain NTHi 86r, allowing for differential plating of NTHi and P. aeruginosa. Following the sequential infection scheme described previously, we intratracheally introduced NTHi 86-028NPr (inoculum of ∼108 CFU/mouse) into the airways of BALBc/J mice, followed by the introduction of mPA 08-31 (inoculum of ∼107 CFU/mouse) 24 h later (see Fig. S2C). Unlike the positive interaction between NTHi and P. aeruginosa seen in our polymicrobial biofilms with the parent strain of NTHi 86r (NTHi 86-028NP), in the airways viable colony counts of P. aeruginosa were significantly decreased after preintroduction of NTHi 86r, compared to a P. aeruginosa single infection (P < 0.05) (Fig. 4A). This finding confirms that the contrasting, synergistic interaction between NTHi 86-028NP and mPA 08-31 seen in vitro (Fig. 1 and 2) is dependent on the lack of host response, rather than strain specificity of NTHi. Mucoid conversion of P. aeruginosa is a common consequence of long-term colonization (22, 27, 32). For young patients, the initial colonizing P. aeruginosa strain is nonmucoid for a variable length of time, not yet producing the alginate component that is strongly associated with persistent infection (22). To extend the impact of this phenotype to the minority of patients colonized with P. aeruginosa before mucoid conversion, we tested whether preintroduction of NTHi inhibits the abundance of nonmucoid P. aeruginosa. To do this, we performed sequential infections with the clinical strain NTHi HI-1r followed by a nonmucoid P. aeruginosa strain (FRD1mucA+). Viable colony counts from lung homogenates revealed that nonmucoid isogenic mutants of P. aeruginosa, in addition to the mucoid strain evaluated previously, were significantly diminished after preintroduction of NTHi, compared to P. aeruginosa alone (P < 0.05) (Fig. 4B). Weight loss per animal was evaluated over the course of infection, and weight loss increased, compared to uninfected controls (see Fig. S3B and C). These results show that NTHi infection significantly reduces the colonization/persistence of P. aeruginosa, regardless of the specific strain of NTHi or the mucoidy status of P. aeruginosa.
FIG 4.
NTHi impact on subsequent P. aeruginosa (Pa) infection is strain independent. BALBc/J mice were intratracheally infected with spectinomycin-resistant NTHi 86-028NP (NTHi 86r) followed by mPA (mPA 08-31). (A) Bacterial counts in the lung homogenates were enumerated by viable colony counting 72 h postinfection. Similarly, intratracheal infections were also performed with spectinomycin-resistant NTHi HI-1 (NTHi HI-1r) and nonmucoid P. aeruginosa (FRD1mucA+). (B) Bacterial counts in the lung homogenates were enumerated by viable colony counting 72 h postinfection. Values are mean ± SEM (n = 10). LOD, limit of detection. *, P < 0.05, Kruskal-Wallis test with Dunn’s multiple-comparison test.
Infection with NTHi followed by P. aeruginosa results in less severe airway tissue damage and less inflammatory cytokine infiltration, compared with P. aeruginosa alone.
While the clinical isolate NTHi Hi-1r persists longer than previously established NTHi strains, such as NTHi 86-028NP (data not shown), this strain still begins to be cleared from the airways of mice around 72 h postinfection, with viable colony counts below the limit of detection for some animals. We wanted to test whether the duration of P. aeruginosa inhibition in the lungs extended past the optimal time point for NTHi viability, when fewer viable bacteria would be present for interspecies interaction. BALBc/J mice were intratracheally infected with NTHi HI-1r, followed 24 h later by mPA 08-31, and viable colony counts from lung homogenates were assessed 72 h after the dual infection, extending the previously established 48-h time period (see Fig. S2C). As expected, sequential introduction of NTHi prior to P. aeruginosa significantly reduced colonization of P. aeruginosa, compared to P. aeruginosa alone (P < 0.001) (Fig. 5A). Reflecting the inhibition of P. aeruginosa abundance in viable colony counts during dual-species infection, histological grading of tissue damage revealed a significantly decreased damage score in dual-species-infected animals, compared to a P. aeruginosa single infection (P < 0.01) (Fig. 5B). H&E staining of lung sections of each infection group revealed more severe immune cell infiltration, pleuritis, perivascular inflammation, and peribronchial inflammation 72 h postinfection than seen at the 48-h postinfection time point (Fig. 5C). Finally, total cell counts in BALF samples and the percentages of PMN infiltrates in those counts were not different among single- or dual infection groups but were significantly increased in comparison to uninfected controls (Fig. 5D and E). Weight loss per animal was evaluated over the course of infection, and weight loss increased, compared to uninfected controls (see Fig. S3D).
FIG 5.
NTHi and P. aeruginosa (Pa) dual infection reduces the severity of airway tissue damage. BALBc/J mice were intratracheally infected with NTHi (NTHi Hi-1r) 24 h prior to introduction of mPA (mPA 08-31) and assessed 72 h postinfection. (A) Bacterial counts of NTHi and P. aeruginosa in the lung homogenates enumerated by viable colony counting (n = 13 to 15). (B and C) Severity of disease indicated by semiquantitative grading by a pathologist (L.N.) (B) and H&E-stained lung sections (×200 magnification) (C) (n = 6 to 10). (D) Total cell counts in BALF samples quantified by differential counting (n = 6 to 8). (E) Percentage of PMNs in the BALF samples (n = 6 to 9). Values are mean ± SEM. LOD, limit of detection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Kruskal-Wallis test with Dunn’s multiple-comparison test.
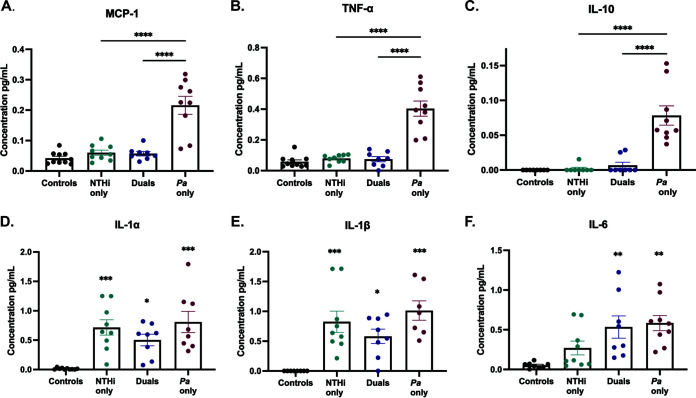
To evaluate the host immune response following NTHi and P. aeruginosa single or dual infections, we measured cytokine and chemokine levels in BALF samples with DuoSet enzyme-linked immunosorbent assay (ELISA) kits designed to detect both proinflammatory and anti-inflammatory markers, including monocyte chemoattractant protein 1 (MCP-1), tumor necrosis factor alpha (TNF-α), interleukin 10 (IL-10), IL-1α, IL-1β, and IL-6. Levels of several of these well-established proinflammatory markers, including MCP-1 and TNF-α, and the anti-inflammatory marker IL-10 were significantly higher after P. aeruginosa single infection, compared to a dual infection with NTHi and P. aeruginosa (P < 0.0001) (Fig. 6A to C). While IL-1α, IL-1β, and IL-6 levels were not different between single-species-infected and dual-species-infected groups at 72 h postinfection, dual-species infection resulted in smaller increases in IL-1α and IL-β, compared to uninfected controls, than did single-species infection (Fig. 6D to F). Taken together, these data suggest that, at 72 h postinfection, NTHi still inhibits P. aeruginosa bacterial burden in the lungs. In addition to inhibiting the abundance of P. aeruginosa, sequential infection reduces the severity of damage to airway tissues, as measured by histological scoring, and reduces inflammatory cytokine levels in BALF samples.
FIG 6.
Inflammatory cytokine levels after dual infection are reduced, compared to P. aeruginosa (Pa) infection alone. Cytokine and chemokine analyses were performed on BALF supernatants from infected or mock-infected mice 72 h postinfection using DuoSet ELISA kits for MCP-1 (A), TNF-α (B), IL-10 (C), IL-1α (D), IL-1β (E), and IL-6 (F). Values are mean ± SEM (n = 7 to 10). Dual infection indicates sequential introduction. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, one-way ANOVA with Tukey’s multiple-comparison test for post hoc analysis.
Inhibitory effect of NTHi on P. aeruginosa infection is independent of bacterial viability.
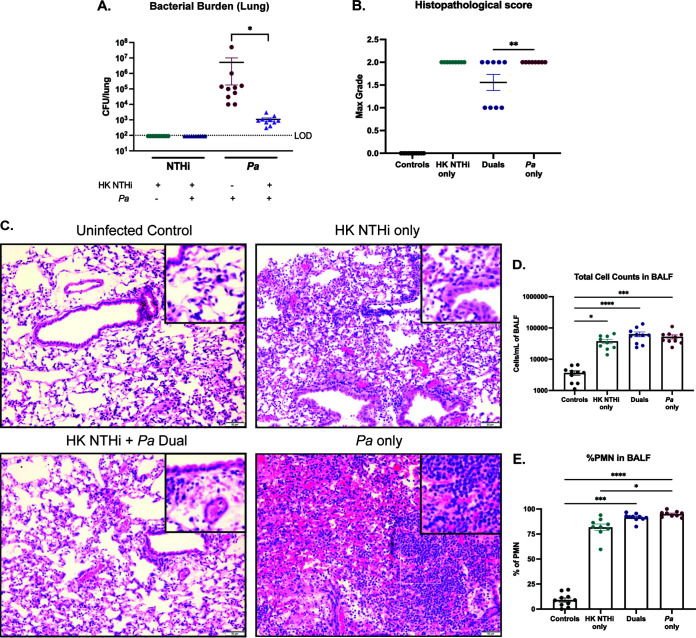
Previous polymicrobial studies involving NTHi suggest that this pathogen either may directly engage in interspecies interactions by active processes or may be passively involved in priming the host for further immune stimuli (33–36). To determine whether live NTHi cells are required to diminish P. aeruginosa establishment in the lungs, we sequentially infected mice with heat-killed (HK) NTHi HI-1r (inoculum equivalent to ∼108 CFU/ml) followed by the mPA strain mPA 08-31 (inoculum of ∼107 CFU/ml) 24 h later (see Fig. S2C). The total infection time for this experiment was 72 h, repeating our previously established time point at which live NTHi infection reduced P. aeruginosa infection. Viable colony counts from lung homogenates showed that P. aeruginosa counts were still significantly reduced by pretreatment with HK NTHi (P < 0.05) (Fig. 7A). Additionally, P. aeruginosa infection resulted in a significantly higher histopathological score than HK NTHi/P. aeruginosa dual infection, suggesting that nonviable NTHi is sufficient to protect the lung parenchyma from the severe effects of P. aeruginosa infection and indicating that the protection from tissue damage previously seen at this time point during dual infection is independent of NTHi viability (P < 0.01) (Fig. 7B). H&E-stained lung sections harvested from this experiment showed more neutrophilic infiltration and more severe tissue damage after P. aeruginosa single infection, compared to dual infection (Fig. 7C). As seen previously, the total cell counts in BALF samples, the percentage of PMN infiltration, and the weight loss per animal were not different among single or dual infection groups but were significantly increased in comparison to uninfected controls (Fig. 5D and E; also see Fig. S3E). Overall, we found that HK NTHi is sufficient to significantly impede the abundance of P. aeruginosa in the lungs at 72 h postinfection, suggesting that the interaction between NTHi and P. aeruginosa may be entirely mediated by the immune response of the host.
FIG 7.
Impact of NTHi on P. aeruginosa (Pa) infection is independent of bacterial viability. BALBc/J mice were intratracheally infected with HK NTHi (NTHi Hi-1r) 24 h prior to introduction of mPA (mPA 08-31) and assessed 72 h after sequential infection. (A) Bacterial counts of NTHi and P. aeruginosa in the lung homogenates enumerated by viable colony counting (n = 10). (B and C) Severity of disease indicated by semiquantitative grading by a pathologist (L.N.) (B) and H&E-stained lung sections (×200 magnification) (C) (n = 8 to 10). (D) Total cell counts in BALF samples quantified by differential counting. (E) Percentage of PMNs in the BALF samples (n = 9 or 10). Values are mean ± SEM. LOD, limit of detection. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Kruskal-Wallis test with Dunn’s multiple-comparison test.
Sequential introduction with LOS alone reduces the bacterial burden of P. aeruginosa in the lungs.
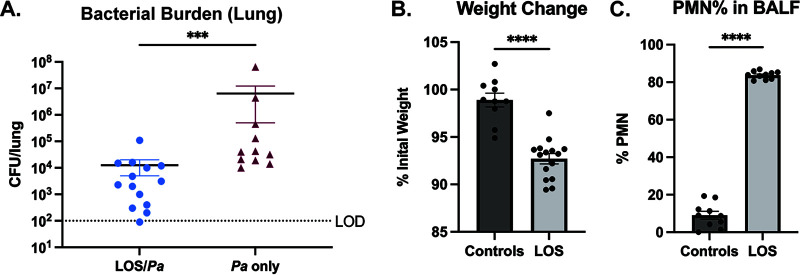
Like many mucosal pathogens, NTHi expresses “rough” LOS endotoxins with short nonrepeating saccharide chains (18). As with other Gram-negative endotoxins, NTHi LOS features hexaacylated lipid A, which elicits host inflammatory responses via Toll-like receptor 4 (19, 20). To test the impact of NTHi endotoxin on P. aeruginosa infection, mice were intratracheally dosed with purified LOS (7 μg/g) 24 h prior to infection with mPA 08-31, and P. aeruginosa bacterial loads from the lung homogenates were assessed 72 h postinfection (see Fig. S2C). Bacterial counts revealed that pretreatment with LOS significantly reduced subsequent colonization by P. aeruginosa (P < 0.001) (Fig. 8A). Compared to uninfected controls, purified LOS caused significant weight loss and PMN infiltration in BALF samples (P < 0.0001) (Fig. 8B and C), which is consistent with a lung inflammatory response. Based on these findings, we conclude that innate immune priming via NTHi endotoxin is a likely mechanism for the inhibitory effect of NTHi on subsequent P. aeruginosa infection.
FIG 8.
Challenge with NTHi LOS endotoxin elicits host inflammation and reduces colonization of P. aeruginosa (Pa). BALBc/J mice were intratracheally dosed with purified LOS (∼7 μg/g body weight) 24 h prior to introduction of mPA (mPA 08-31) and assessed 72 h after dual challenge. (A) Bacterial burden of P. aeruginosa in the lung homogenates with or without prior challenge with LOS (n = 11 to 15). (B and C) LOS alone was sufficient to initiate the response to infection, as measured by percentage of initial weight (B) and percentage of PMNs in BALF samples (C), compared to uninfected controls (n = 10 to 15). Values are mean ± SEM. LOD, limit of detection. ***, P < 0.001; ****, P < 0.0001, Mann-Whitney test, two-tailed.
DISCUSSION
With the advent and widespread use of culture-independent technologies for profiling microbial consortia, it has become clear that the populations within the lungs of patients with CF are diverse and subject to dynamic change. These shifts are likely of importance in determining whether species will persist or be cleared, as well as in the initiation of the inflammatory events that are a hallmark of CF disease (6–8, 27, 37, 38). The consequences of respiratory exacerbations include an intense neutrophilic inflammatory response that can cause significant lung damage and decreased respiratory function, contributing significantly to the morbidity and death associated with CF disease (8, 38). There is still much to learn regarding how specific organisms interact within the compromised respiratory tract of CF airways and how these interactions influence disease progression (8).
The complex microbial communities in the lung often change in dominant species and overall population diversity as patients age (22, 27). Decreased microbial diversity and the emergence of mPA as the dominant microbial species are significantly associated with detrimental host immune responses, lung function decline, and eventual patient death (22, 27). Previous studies have focused on characterizing key CF pathogens individually; however, these infections are undoubtedly polymicrobial in nature, influencing the pathogenesis of disease as organisms interact (3, 19). NTHi is commonly carried within the microbiome of the nasopharynx and upper airways in healthy individuals and has long been recognized as an opportunistic pathogen in patients with impaired mucociliary defenses, including young children with CF (10, 39). In this work, we addressed the role of preceding NTHi infection on susceptibility to respiratory infection with P. aeruginosa; the results indicate that NTHi primes an innate host response that impedes P. aeruginosa colonization.
Polymicrobial interactions in static biofilms provide basic information about interspecies bacterial interactions. Our results indicate that NTHi and P. aeruginosa form polymicrobial biofilms that support P. aeruginosa growth (Fig. 1 and 2; also see Fig. S1 in the supplemental material). However, to understand polymicrobial infections in patients, we must determine how these two organisms interact within the lung environment. In a mouse model, we tested how NTHi impacted P. aeruginosa colonization following sequential introduction, representing how patients typically acquire these organisms over time, in contrast to concurrent acquisition.
Bacterial infections in the CF lungs result in chronic inflammation, as microbial and phagocytic debris builds up in the airways, contributing to the reduced lung function and overactive immune response associated with exacerbations (10). The inherently diminished airway defenses associated with CF transmembrane conductance regulator (CFTR) dysfunction are thought to predispose CF patients to frequent microbial infections, followed by an inadequate, yet overactive, innate immune response that further initiates tissue damage by airway remodeling (12, 29). Histopathological analysis of lung sections can evaluate immune cell infiltration within alveoli, pleura, perivascular spaces, and peribronchial spaces and grade the severity of tissue damage. In CF, the ensuing tissue damage following chronic bacterial infection is a strong indicator of respiratory failure, the leading cause of morbidity and death (12). Preintroduction of NTHi not only reduces the bacterial burden of P. aeruginosa but also significantly reduces tissue damage, compared to a P. aeruginosa single infection, an effect that may reduce airway remodeling following infection (Fig. 5). While dual infection reduces the severity of airway parenchymal damage, we wanted to know whether inflammatory cytokine levels were reduced after dual infection as well. The BALF samples collected from single- and dual-infected mice showed trending decreases in early response cytokines, such as IL-1-α, IL-1-β, and IL-6, in dual-infected mice, compared to P. aeruginosa single infection (Fig. 6A to C). Since the ILs are some of the first cytokines to respond to bacterial stimuli, we suspect that significant differences between dual- and single-infected groups may appear at earlier time points, prior to 72 h postinfection (11, 30). We saw significant decreases in levels of inflammatory cytokines MCP-1, TNF-α, and IL-10 in sequential dual infection, compared to P. aeruginosa single infection, suggesting that the reduction in bacterial burden also diminishes a potentially harmful innate immune response (Fig. 6D to F). Most notably, this reduction in TNF-α levels could extend beneficial effects for CF patients, as the continual production of this early response cytokine indicates an ongoing and overactive innate immune response in the CF lung (11).
Defining how different microbes interact within the CF lung to impact bacterial colonization and persistence and host response has the potential to guide rational treatment strategies to lengthen periods of asymptomatic or mildly symptomatic infection that are typically associated with younger patients. The results of our infection studies indicate that colonization of the lung with NTHi may provide inherent resistance to subsequent colonization with P. aeruginosa. A likely mechanism for such an effect would be stimulation of innate immune defenses, which can prime host phagocyte responses to respond to incoming pathogens (34, 36, 40, 41). We next addressed the hypothesis that NTHi initiates a host inflammatory response that impedes subsequent P. aeruginosa colonization by testing the impact of pretreatment with purified NTHi endotoxin. We intratracheally administered purified LOS to mice prior to P. aeruginosa and found that LOS alone promoted the reduction in P. aeruginosa colonization (Fig. 8). These results raise interesting possibilities regarding whether aggressive treatment against early-stage opportunists, such as NTHi, may be contraindicated or even harmful by providing a window of susceptibility to other infections. It is also notable that, for opportunistic airway infections associated with chronic obstructive pulmonary disease (COPD), an extensive body of recent work indicates that treatment with inflammatory agonists that evoke innate responses can confer protection, and such agonists are under active investigation as therapeutics (42–46). The concept of therapeutically inducing a low level of airway inflammation to enhance resistance may merit further study in CF-related infections.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
NTHi 86-028NP is a well characterized patient isolate for which a fully annotated genome sequence is available (47). NTHi HI-1 is a CF patient isolate that was kindly provided by Timothy Starner (University of Iowa Children’s Hospital). NTHi 86-028NPr and NTHi HI-1r were derived from NTHi 86-028NP and NTHi HI-1, respectively, by serial passage on plates containing spectinomycin (16). NTHi 86-028NP-gfp+ was constructed by electroporation with plasmid PGM1.1+gfp (provided by K. Mason, Nationwide Children’s Research Hospital) (48, 49). For HK NTHi experiments, bacteria were scraped from a plate into sterile PBS, resuspended to ∼109 CFU/ml, and incubated at 65°C for 1 h; bacterial killing was confirmed by lack of growth. All NTHi strains were routinely cultured on supplemented brain heart infusion (sBHI) agar (Difco) containing 10 μg/ml hemin (ICN Biochemical) and 1 μg/ml NAD (Sigma). NTHi LOS (a gift from Michael Apicella, University of Iowa) was purified from strain NTHi 2019 according to standard methodology (50) and was intratracheally administered as described previously (51, 52).
P. aeruginosa mPA08-31 is a mucoid clinical isolate from a CF patient at the University of Alabama at Birmingham (UAB) Hospital and was provided by S. Birket (UAB) (53, 54). A nonmucoid derivative of P. aeruginosa (FRD1mucA+) was provided by J. Scoffield (UAB) (55). P. aeruginosa mPA08-31-mCherry+ was derived by electroporation with plasmid pUCP19+mCherry (provided by D. Wozniak, Ohio State University). All P. aeruginosa strains were routinely cultured on Luria-Bertani (LB) agar (Difco). Strains were streaked for colony isolation before inoculation into LB broth and shaking overnight at 37°C and 200 rpm.
CLSM of static biofilms.
In vitro biofilms were prepared by resuspending NTHi cells from an overnight plate culture to∼108 CFU/ml in 2× sBHI or diluting an overnight broth culture of P. aeruginosa to ∼108 CFU/ml in LB broth. Dual-species biofilms were sequentially seeded with NTHi preceding P. aeruginosa by 12 h in a 35-mm glass-bottom confocal dish (MatTek) in 2-ml increments, such that the final density in each well was ∼108 CFU of one or both organisms. NTHi biofilms were incubated for 12 h, and P. aeruginosa biofilms were incubated for 6 h. Sequentially seeded polymicrobial biofilms were incubated for 18 h. Viable colony counts of NTHi or P. aeruginosa from biofilms were obtained by plating on sBHI agar with spectinomycin or LB agar, respectively. For fluorescence imaging of NTHi 86028NP-gfp+ and mPA08-31-mCherry+, all dishes were incubated at 37°C in 5% CO2 before being washed with sterile PBS and fixed with 4% paraformaldehyde (Alfa Aesar, Tewksbury, MA). CLSM was performed using an A1R HD25 confocal laser microscope (Nikon, Tokyo, Japan) at UAB. All images were processed using NIS-Elements v5.0 software.
Fluorescent biofilm quantification.
Fluorescent biofilms were quantified using BiofilmQ software (56). Single fluorescence channels were automatically segmented using the Otsu algorithm. Background was removed with semimanual thresholding denoising. Global biofilm parameters for the fluorescence channel representing P. aeruginosa were quantified for both single-species and dual-species biofilms to assess biovolume. Three-dimensional spatial representation of cubing (representing 1 μm per cube) was calculated by BiofilmQ and visualized with ParaView according to the manufacturer’s instructions.
Mouse model of respiratory infections.
BALB/cJ mice (8 to 10 weeks of age) were obtained from the Jackson Laboratory (Bar Harbor, ME). For single-species infections, mice were anesthetized with isoflurane and intratracheally infected with NTHi (∼109 CFU in 100 μl PBS) or P. aeruginosa (∼108 CFU in 100 μl PBS). For dual infections, mice were anesthetized with isoflurane and infected intratracheally with NTHi and P. aeruginosa (∼109 CFU NTHi and ∼108 CFU P. aeruginosa in 100 μl PBS) either concurrently or sequentially, with infection with NTHi occurring 24 h prior to P. aeruginosa introduction. For viable plate counting, the left lung of each mouse was harvested and homogenized in 500 μl of sterile PBS at 30 Hz/s. Lung homogenates were serially diluted in PBS and plated on LB agar to obtain viable colony counts of P. aeruginosa and on sBHI agar containing spectinomycin (2 mg/ml) for selection of NTHi HI-1r. All samples from polymicrobial infections were also plated on sBHI agar without antibiotic for total bacterial counts of both organisms. For histological analysis, the right lung of each animal was inflated with 10% buffered formalin and stored at 4°C until processing. All mouse infection protocols were approved by the UAB Institutional Animal Care and Use Committee.
BALF sample collection.
Bronchoalveolar lavage was performed after euthanasia by flushing mouse lungs with 6 ml of cold PBS in 1-ml increments as described previously (18). Collected BALF samples were stored on ice until processing, and the first 1 ml isolated was centrifuged (1,350 × g for 5 min) to separate the supernatant from immune cells. The cell pellet was used for total and differential cell counts. The remaining 5 ml of collected BALF was stored at −20°C until use for cytokine analyses.
Cytokine analyses and differential cell counting.
Cytokine analyses were performed on the 5 ml of collected BALF via DuoSet ELISA kits (R&D Systems, Minnesota, MN). For total and differential cell counts, cell pellets from BALF samples were resuspended in fresh PBS and collected by cytospin at 500 rpm for 5 min. Cells were stained with a Kwik-Diff differential cell stain (Thermo Fisher Scientific, Waltham, MA). Three representative fields from each spot were counted to determine the composition of immune cells.
Histological analysis.
Inflated right lungs from infected animals were stored in 10% neutral buffered formalin (Thermo Fisher Scientific) at 4°C until processing. Sections from each lobe of the right lung were trimmed and sent to the UAB Comparative Pathology Laboratory to be processed, embedded in paraffin, sectioned, and stained with H&E. Images of lung sections (×200 magnification) were taken with an Olympus DP25 camera (Tokyo, Japan) using an Olympus BX40 microscope. Semiquantitative grading of all lung sections was performed by a board-certified surgical pathologist (L.N.). Semiquantitative histopathological scores were assigned using a scoring matrix based primarily on neutrophilic influx. Severity was rated on a scale of 0 to 3, where 0 represents no observable neutrophils, 1 represents dispersed acute inflammation (mild damage), 2 represents dense infiltration (moderate damage), and 3 represents solid infiltration with necrosis (severe damage). The final histopathological score represents the maximum grade assigned to each replicate.
Statistical analyses.
All bar graphs represent sample mean ± standard error of the mean (SEM). All mouse experiments were repeated at least two times, and data from independent experiments were combined for analyses. For nonparametric analyses, differences between groups were analyzed by the Kruskal-Wallis test with the uncorrected Dunn’s test for multiple comparisons. For normally distributed data sets, as determined by the Shapiro-Wilk normality test, one-way analysis of variance (ANOVA) was used with Tukey’s multiple-comparison test. Outliers were detected via the ROUT method (Q value of 1%) and excluded from analysis. All statistical tests were performed using GraphPad Prism 9 (San Diego, CA).
ACKNOWLEDGMENTS
We gratefully acknowledge helpful input and discussions with colleagues in the UAB Center for Cystic Fibrosis Research. We thank Michael Apicella (University of Iowa) for providing purified LOS and Timothy Starner (University of Iowa Children’s Hospital), Daniel Wozniak (Ohio State University), Jessica Scoffield (UAB), and Susan Birket (UAB) for providing bacterial strains.
This work was supported by grants to W.E.S. from the Cystic Fibrosis Foundation (grants CFFSWORDS1810 and CFFSWORDS20G0). N.R.L. and M.S.M were supported by predoctoral fellowships from the Cystic Fibrosis Foundation (RDP Rowe19RO). B.C.H. was supported by a fellowship from the UAB Predoctoral Training Program in Lung Biology (grant T32 HL13640).
Footnotes
Supplemental material is available online only.
Contributor Information
W. Edward Swords, Email: wswords@uabmc.edu.
Marvin Whiteley, Georgia Institute of Technology School of Biological Sciences.
REFERENCES
- 1.Lyczak JB, Cannon CL, Pier GB. 2002. Lung infections associated with cystic fibrosis. Clin Microbiol Rev 15:194–222. 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ratjen F, Bell SC, Rowe SM, Goss CH, Quittner AL, Bush A. 2015. Cystic fibrosis. Nat Rev Dis Primers 1:15010. 10.1038/nrdp.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magalhaes AP, Lopes SP, Pereira MO. 2016. Insights into cystic fibrosis polymicrobial consortia: the role of species interactions in biofilm development, phenotype, and response to in-use antibiotics. Front Microbiol 7:2146. 10.3389/fmicb.2016.02146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cystic Fibrosis Foundation. 2019. Cystic fibrosis patient registry annual data report. Cystic Fibrosis Foundation, Bethesda, MD. https://www.cff.org/medical-professionals/patient-registry. [Google Scholar]
- 5.O'Toole GA. 2018. Cystic fibrosis airway microbiome: overturning the old, opening the way for the new. J Bacteriol 200:e00561-17. 10.1128/JB.00561-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YJ, LiPuma JJ. 2016. The microbiome in cystic fibrosis. Clin Chest Med 37:59–67. 10.1016/j.ccm.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caverly LJ, Zhao J, LiPuma JJ. 2015. Cystic fibrosis lung microbiome: opportunities to reconsider management of airway infection. Pediatr Pulmonol 50(Suppl 40):S31–S38. 10.1002/ppul.23243. [DOI] [PubMed] [Google Scholar]
- 8.Filkins LM, O'Toole GA. 2015. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog 11:e1005258. 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zemanick ET, Sagel SD, Harris JK. 2011. The airway microbiome in cystic fibrosis and implications for treatment. Curr Opin Pediatr 23:319–324. 10.1097/MOP.0b013e32834604f2. [DOI] [PubMed] [Google Scholar]
- 10.Erwin AL, Smith AL. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol 15:355–362. 10.1016/j.tim.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 11.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. 2016. The microbiome and the respiratory tract. Annu Rev Physiol 78:481–504. 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg F, Dickson RP, Nagrath D, Ramnath N. 2020. The lung microbiome: a central mediator of host inflammation and metabolism in lung cancer patients? Cancers (Basel) 13:13. 10.3390/cancers13010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardines R, Giufre M, Pompilio A, Fiscarelli E, Ricciotti G, Di Bonaventura G, Cerquetti M. 2012. Haemophilus influenzae in children with cystic fibrosis: antimicrobial susceptibility, molecular epidemiology, distribution of adhesins and biofilm formation. Int J Med Microbiol 302:45–52. 10.1016/j.ijmm.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 14.Rayner RJ, Hiller EJ, Ispahani P, Baker M. 1990. Haemophilus infection in cystic fibrosis. Arch Dis Child 65:255–258. 10.1136/adc.65.3.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saliu F, Rizzo G, Bragonzi A, Cariani L, Cirillo DM, Colombo C, Dacco V, Girelli D, Rizzetto S, Sipione B, Cigana C, Lore NI. 2021. Chronic infection by nontypeable Haemophilus influenzae fuels airway inflammation. ERJ Open Res 7:00614-2020. 10.1183/23120541.00614-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starner TD, Zhang N, Kim G, Apicella MA, McCray PB. Jr.. 2006. Haemophilus influenzae forms biofilms on airway epithelia: implications in cystic fibrosis. Am J Respir Crit Care Med 174:213–220. 10.1164/rccm.200509-1459OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swords WE. 2012. Nontypeable Haemophilus influenzae biofilms: role in chronic airway infections. Front Cell Infect Microbiol 2:97. 10.3389/fcimb.2012.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preston A, Mandrell RE, Gibson BW, Apicella MA. 1996. The lipooligosaccharides of pathogenic Gram-negative bacteria. Crit Rev Microbiol 22:139–180. 10.3109/10408419609106458. [DOI] [PubMed] [Google Scholar]
- 19.Choi J, Cox AD, Li J, McCready W, Ulanova M. 2014. Activation of innate immune responses by Haemophilus influenzae lipooligosaccharide. Clin Vaccine Immunol 21:769–776. 10.1128/CVI.00063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAleer JP, Vella AT. 2008. Understanding how lipopolysaccharide impacts CD4 T-cell immunity. Crit Rev Immunol 28:281–299. 10.1615/critrevimmunol.v28.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grahl N, Dolben EL, Filkins LM, Crocker AW, Willger SD, Morrison HG, Sogin ML, Ashare A, Gifford AH, Jacobs NJ, Schwartzman JD, Hogan DA. 2018. Profiling of bacterial and fungal microbial communities in cystic fibrosis sputum using RNA. mSphere 3:e00292-18. 10.1128/mSphere.00292-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns JL, Gibson RL, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith AL, Ramsey BW. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis 183:444–452. 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 23.Gellatly SL, Hancock RE. 2013. Pseudomonas aeruginosa: new insights into pathogenesis and host defenses. Pathog Dis 67:159–173. 10.1111/2049-632X.12033. [DOI] [PubMed] [Google Scholar]
- 24.Rossi E, La Rosa R, Bartell JA, Marvig RL, Haagensen JAJ, Sommer LM, Molin S, Johansen HK. 2021. Pseudomonas aeruginosa adaptation and evolution in patients with cystic fibrosis. Nat Rev Microbiol 19:331–342. 10.1038/s41579-020-00477-5. [DOI] [PubMed] [Google Scholar]
- 25.Gill JF, Deretic V, Chakrabarty AM. 1987. Alginate production by the mucoid Pseudomonas aeruginosa associated with cystic fibrosis. Microbiol Sci 4:296–299. [PubMed] [Google Scholar]
- 26.Mayer-Hamblett N, Aitken ML, Accurso FJ, Kronmal RA, Konstan MW, Burns JL, Sagel SD, Ramsey BW. 2007. Association between pulmonary function and sputum biomarkers in cystic fibrosis. Am J Respir Crit Care Med 175:822–828. 10.1164/rccm.200609-1354OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns JL, Emerson J, Stapp JR, Yim DL, Krzewinski J, Louden L, Ramsey BW, Clausen CR. 1998. Microbiology of sputum from patients at cystic fibrosis centers in the United States. Clin Infect Dis 27:158–163. 10.1086/514631. [DOI] [PubMed] [Google Scholar]
- 28.Dickson RP, Erb-Downward JR, Huffnagle GB. 2013. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med 7:245–257. 10.1586/ers.13.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dickson RP, Erb-Downward JR, Prescott HC, Martinez FJ, Curtis JL, Lama VN, Huffnagle GB. 2014. Cell-associated bacteria in the human lung microbiome. Microbiome 2:28. 10.1186/2049-2618-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson RP, Huffnagle GB. 2015. The lung microbiome: new principles for respiratory bacteriology in health and disease. PLoS Pathog 11:e1004923. 10.1371/journal.ppat.1004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welp AL, Bomberger JM. 2020. Bacterial community interactions during chronic respiratory disease. Front Cell Infect Microbiol 10:213. 10.3389/fcimb.2020.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burns JL, Ramsey BW, Smith AL. 1993. Clinical manifestations and treatment of pulmonary infections in cystic fibrosis. Adv Pediatr Infect Dis 8:53–66. [PubMed] [Google Scholar]
- 33.Lysenko ES, Clarke TB, Shchepetov M, Ratner AJ, Roper DI, Dowson CG, Weiser JN. 2007. Nod1 signaling overcomes resistance of S. pneumoniae to opsonophagocytic killing. PLoS Pathog 3:e118. 10.1371/journal.ppat.0030118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. 2005. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog 1:e1. 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Armbruster CE, Hong W, Pang B, Weimer KE, Juneau RA, Turner J, Swords WE. 2010. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio 1:e00102-10. 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weimer KE, Armbruster CE, Juneau RA, Hong W, Pang B, Swords WE. 2010. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis 202:1068–1075. 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hampton TH, Green DM, Cutting GR, Morrison HG, Sogin ML, Gifford AH, Stanton BA, O'Toole GA. 2014. The microbiome in pediatric cystic fibrosis patients: the role of shared environment suggests a window of intervention. Microbiome 2:14. 10.1186/2049-2618-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zemanick ET, Harris JK, Wagner BD, Robertson CE, Sagel SD, Stevens MJ, Accurso FJ, Laguna TA. 2013. Inflammation and airway microbiota during cystic fibrosis pulmonary exacerbations. PLoS One 8:e62917. 10.1371/journal.pone.0062917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Starner TD, McCray PB. Jr, American College of Physicians, American Physiological Society. 2005. Pathogenesis of early lung disease in cystic fibrosis: a window of opportunity to eradicate bacteria. Ann Intern Med 143:816–822. 10.7326/0003-4819-143-11-200512060-00010. [DOI] [PubMed] [Google Scholar]
- 40.Ratner AJ, Lysenko ES, Paul MN, Weiser JN. 2005. Synergistic proinflammatory responses induced by polymicrobial colonization of epithelial surfaces. Proc Natl Acad Sci USA 102:3429–3434. 10.1073/pnas.0500599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mukherjee S, Weimer KE, Seok SC, Ray WC, Jayaprakash C, Vieland VJ, Swords WE, Das J. 2014. Host-to-host variation of ecological interactions in polymicrobial infections. Phys Biol 12:016003. 10.1088/1478-3975/12/1/016003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cleaver JO, You D, Michaud DR, Pruneda FA, Juarez MM, Zhang J, Weill PM, Adachi R, Gong L, Moghaddam SJ, Poynter ME, Tuvim MJ, Evans SE. 2014. Lung epithelial cells are essential effectors of inducible resistance to pneumonia. Mucosal Immunol 7:78–88. 10.1038/mi.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duggan JM, You D, Cleaver JO, Larson DT, Garza RJ, Guzman Pruneda FA, Tuvim MJ, Zhang J, Dickey BF, Evans SE. 2011. Synergistic interactions of TLR2/6 and TLR9 induce a high level of resistance to lung infection in mice. J Immunol 186:5916–5926. 10.4049/jimmunol.1002122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans SE, Tuvim MJ, Fox CJ, Sachdev N, Gibiansky L, Dickey BF. 2011. Inhaled innate immune ligands to prevent pneumonia. Br J Pharmacol 163:195–206. 10.1111/j.1476-5381.2011.01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans SE, Xu Y, Tuvim MJ, Dickey BF. 2010. Inducible innate resistance of lung epithelium to infection. Annu Rev Physiol 72:413–435. 10.1146/annurev-physiol-021909-135909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moghaddam SJ, Clement CG, De la Garza MM, Zou X, Travis EL, Young HW, Evans CM, Tuvim MJ, Dickey BF. 2008. Haemophilus influenzae lysate induces aspects of the chronic obstructive pulmonary disease phenotype. Am J Respir Cell Mol Biol 38:629–638. 10.1165/rcmb.2007-0366OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrison A, Dyer DW, Gillaspy A, Ray WC, Mungur R, Carson MB, Zhong H, Gipson J, Gipson M, Johnson LS, Lewis L, Bakaletz LO, Munson RS, Jr.. 2005. Genomic sequence of an otitis media isolate of nontypeable Haemophilus influenzae: comparative study with H. influenzae serotype d, strain KW20. J Bacteriol 187:4627–4636. 10.1128/JB.187.13.4627-4636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vogel AR, Szelestey BR, Raffel FK, Sharpe SW, Gearinger RL, Justice SS, Mason KM. 2012. SapF-mediated heme-iron utilization enhances persistence and coordinates biofilm architecture of Haemophilus. Front Cell Infect Microbiol 2:42. 10.3389/fcimb.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitchell MA, Skowronek K, Kauc L, Goodgal SH. 1991. Electroporation of Haemophilus influenzae is effective for transformation of plasmid but not chromosomal DNA. Nucleic Acids Res 19:3625–3628. 10.1093/nar/19.13.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apicella MA, Griffiss JM, Schneider H. 1994. Isolation and characterization of lipopolysaccharides, lipooligosaccharides and lipid A. Methods Enzymol 235:242–252. 10.1016/0076-6879(94)35145-7. [DOI] [PubMed] [Google Scholar]
- 51.Vernooy JH, Dentener MA, van Suylen RJ, Buurman WA, Wouters EF. 2001. Intratracheal instillation of lipopolysaccharide in mice induces apoptosis in bronchial epithelial cells: no role for tumor necrosis factor-α and infiltrating neutrophils. Am J Respir Cell Mol Biol 24:569–576. 10.1165/ajrcmb.24.5.4156. [DOI] [PubMed] [Google Scholar]
- 52.Ehrentraut H, Weisheit CK, Frede S, Hilbert T. 2019. Inducing acute lung injury in mice by direct intratracheal lipopolysaccharide instillation. J Vis Exp 10.3791/59999. [DOI] [PubMed] [Google Scholar]
- 53.Das S, Howlader DR, Zheng Q, Ratnakaram SSK, Whittier SK, Lu T, Keith JD, Picking WD, Birket SE, Picking WL. 2020. Development of a broadly protective, self-adjuvanting subunit vaccine to prevent infections by Pseudomonas aeruginosa. Front Immunol 11:583008. 10.3389/fimmu.2020.583008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDaniel MS, Schoeb T, Swords WE. 2020. Cooperativity between Stenotrophomonas maltophilia and Pseudomonas aeruginosa during polymicrobial airway infections. Infect Immun 88:e00855-19. 10.1128/IAI.00855-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Silo-Suh LA, Suh SJ, Ohman DE, Wozniak DJ, Pridgeon JW. 2015. Complete genome sequence of Pseudomonas aeruginosa mucoid strain FRD1, isolated from a cystic fibrosis patient. Genome Announc 3:e00153-15. 10.1128/genomeA.00153-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hartmann R, Jeckel H, Jelli E, Singh PK, Vaidya S, Bayer M, Rode DKH, Vidakovic L, Diaz-Pascual F, Fong JCN, Dragos A, Lamprecht O, Thoming JG, Netter N, Haussler S, Nadell CD, Sourjik V, Kovacs AT, Yildiz FH, Drescher K. 2021. Quantitative image analysis of microbial communities with BiofilmQ. Nat Microbiol 6:151–156. 10.1038/s41564-020-00817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00568-21-s0001.pdf, PDF file, 2.3 MB (2.3MB, pdf)