ABSTRACT
Hypoxia-inducible transcription factor 1 (HIF-1) has been shown to enhance microbial killing and ameliorate the course of bacterial infections. While the impact of HIF-1 on inflammatory diseases of the gut has been studied intensively, its function in bacterial infections of the gastrointestinal tract remains largely elusive. With the help of a publicly available gene expression data set, we inferred significant activation of HIF-1 after oral infection of mice with Salmonella enterica serovar Typhimurium. Immunohistochemistry and Western blot analyses confirmed marked HIF-1α protein stabilization, especially in the intestinal epithelium. This prompted us to analyze conditional Hif1a-deficient mice to examine cell type-specific functions of HIF-1 in this model. Our results demonstrate enhanced noncanonical induction of HIF-1 activity upon Salmonella infection in the intestinal epithelium as well as in macrophages. Surprisingly, Hif1a deletion in intestinal epithelial cells did not impact inflammatory gene expression, bacterial spread, or disease outcomes. In contrast, Hif1a deletion in myeloid cells enhanced intestinal Cxcl2 expression and reduced the cecal Salmonella load. In vitro, HIF-1α-deficient macrophages showed overall impaired transcription of mRNA encoding proinflammatory factors; however, the intracellular survival of Salmonella was not impacted by HIF-1α deficiency.
KEYWORDS: HIF-1, Salmonella, gastrointestinal infection, host-pathogen interactions, innate immunity
INTRODUCTION
With drastically increasing numbers of antibiotic-resistant bacteria and few novel antibiotic discoveries, innovative strategies to combat acute and chronic infections are urgently needed. Altering the immune response rather than killing the pathogen directly has become a focus of investigation. The transcription factor hypoxia-inducible factor 1 (HIF-1) functions as a cellular energy switch orchestrating energy homeostasis, metabolism, angiogenesis, and evasion of apoptosis in response to low oxygen levels (hypoxia) in multiple cell types and tissues (1). Interestingly, HIF-1 further regulates the innate immune response to invading bacterial pathogens in ways that remain to be understood in greater detail to evaluate its potential as a therapeutic target (2, 3). HIF-1 is a heterodimer consisting of an α- and a β-subunit. HIF-1α is hydroxylated by oxygen-dependent prolyl hydroxylases (PHDs) and marked for proteasomal degradation by von Hippel-Lindau tumor suppressor protein (pVHL) under normoxic (or, rather, nonstressed) conditions. This prevents the α-subunit from translocating into the nucleus and binding to the HIF-1β subunit when oxygen levels are adequate (4, 5). The closely related HIF-2α isoform has been shown to exhibit overlapping transcriptional activity but is mainly expressed in endothelial and bone marrow cells (6, 7). Gram-positive and Gram-negative bacteria as well as LPS (lipopolysaccharide) potently stabilize HIF-1α (2, 8, 9), thereby promoting innate immune effector functions of phagocytes (10, 11). Pharmacological stabilization of HIF-1α by inhibition of iron- and oxygen-dependent PHDs utilizing iron chelators was shown to reduce bacterial burdens, suggesting that this approach can be an effective tool against bacterial infections in vivo and in vitro (12, 13). These findings clearly argue for the potential of HIF-1 targeting for the treatment of bacterial infections.
Hypoxia, physiologically defined as an oxygen concentration below 1% or partial pressure under 10 mm Hg, is a hallmark of tissue inflammation and is best explained by reduced tissue perfusion combined with increased oxygen uptake by immune cells (14). Luminal cells of the healthy gastrointestinal epithelium are marked by physiological hypoxia (15, 16), which overlaps HIF-1α stabilization. This is functionally linked to the production of short-chain fatty acids by the intestinal microbiota, which promotes oxygen consumption by intestinal epithelial cells (IECs), ultimately resulting in HIF-1 protein stabilization (17). HIF-1 guards intestinal barrier integrity in two principal ways: (i) by supporting the continuous proliferation of IECs via interaction with Wnt and Notch signaling (18, 19) and (ii) by direct barrier stabilization (20, 21) through the securing of tight junctions (22). While the function of HIF-1 has been extensively studied in intestinal inflammation and tumorigenesis, albeit with conflicting results demonstrating both pro- and anti-inflammatory effects (18, 20, 23), the impact of HIF-1 on bacterial infections of the intestinal tract remains largely elusive. In an elegant study, the group of Paul Beck reported the functional significance of HIF-1 for bacterium-induced intestinal injury. They showed that IEC-specific Hif1a knockout mice displayed more severe intestinal injury and inflammation in response to Clostridium difficile infection (24). Furthermore, pharmacological stabilization of HIF-1α attenuated C. difficile-induced injury, further supporting the functional importance of the HIF-1 pathway in this setting. In line with these results, Hartmann and colleagues showed that IEC-specific Hif1a knockout mice were highly susceptible to orogastric Yersinia enterocolitica infection, resulting in a drastic increase in mortality. Those authors linked Y. enterocolitica-induced HIF-1α stabilization to bacterial siderophore expression, including salmochelin, a siderophore expressed by Salmonella (25).
Nontyphoidal Salmonella represents an important cause of gastrointestinal infections throughout the world, mainly by the ingestion of contaminated food and water (26, 27). Salmonella initially colonizes the small intestine (SI) and invades IECs, overcoming the mucosal barrier and interfering with intracellular bactericidal responses through the expression of virulence factors, including type III secretion systems (T3SSs), allowing the translocation of effector molecules into the host cytosol on the one hand and the endomembrane system on the other (28–31). It can bypass humoral immunity by hijacking phagocytes to egress from the gut into the bloodstream or lymphatic system, potentially causing life-threatening systemic infections (32, 33). The above-described influence of HIF-1 on cellular immune functions as well as the tight sealing of the intestinal wall therefore led us to further investigate its role during gastrointestinal infections, using Salmonella enterica subsp. enterica serovar Typhimurium, a nontyphoidal strain that causes typhoid fever-like systemic infections in mice (33, 34).
(This work was partly presented in poster form at Novel Concepts in Innate Immunity 2019 in Tübingen, Germany.)
RESULTS
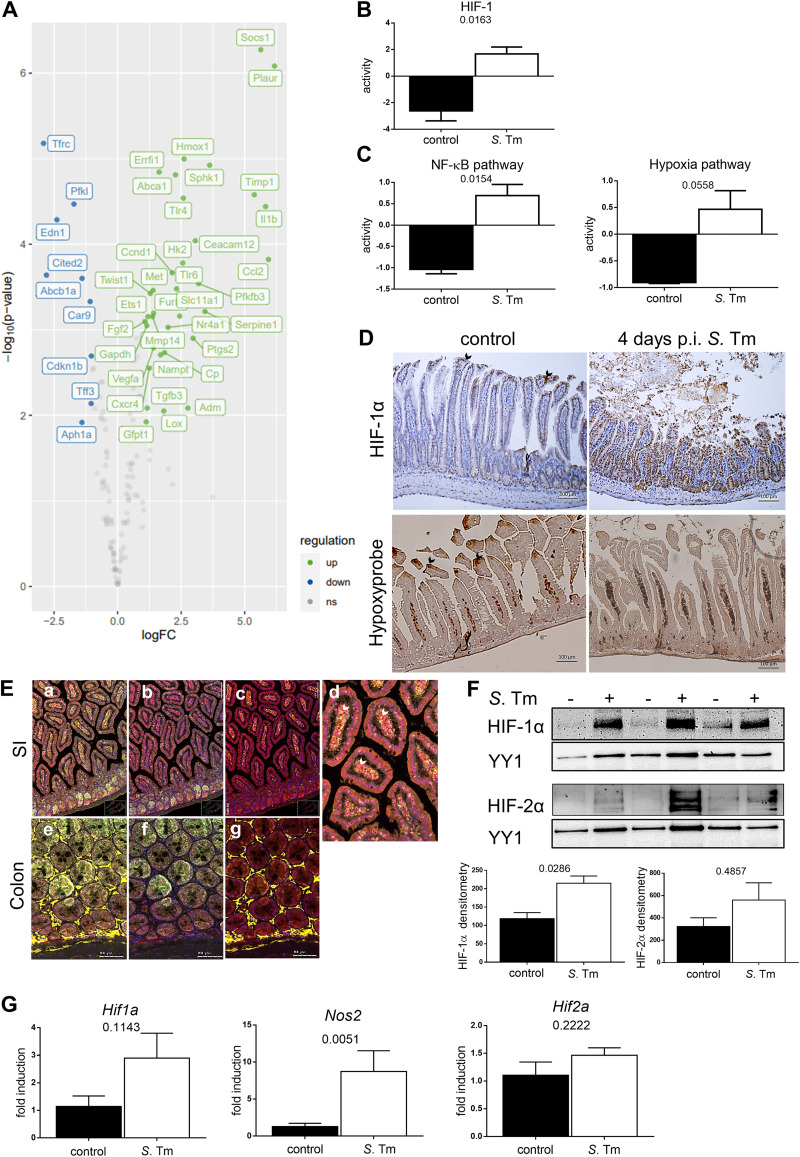
HIF-1 is highly stabilized in response to Salmonella in the small intestine.
As infections with other enteric pathogens such as Y. enterocolitica result in the stabilization of HIF-1α (2, 25), we sought to address the relevance of HIF-1 transcription factor activity in the setting of gastrointestinal Salmonella infection in an oral murine model (35). We performed a transcription factor analysis using DoRothEA (36) on a published gene expression data set (4 days after infection with Salmonella) (37). This analysis convincingly demonstrated the upregulation of a large subset of HIF-1 target genes (Fig. 1A). Based on the expression changes of these HIF-1 target genes, HIF-1 transcription factor activity was calculated, showing a significant increase in response to Salmonella in the gut (Fig. 1B). In order to understand the upstream pathways involved in HIF-1 transcriptional activation, we additionally applied the PROGENy pathway analysis tool to the published data set (38, 39). While HIF-1 and NF-κB share stimuli as well as target genes, the gene expression of Hif1a is directly regulated by NF-κB (8). Besides hypoxia-related gene activation, this analysis implicated increased hypoxia-independent (i.e., noncanonical) HIF-1α stabilization as the NF-κB pathway that was upregulated (Fig. 1C). In line with these in silico results, we demonstrated robust stabilization of intestinal epithelial HIF-1α accompanying mucosal inflammation 4 days after infection with Salmonella (Fig. 1D). Of note, the physiological villus tip-pronounced hypoxia pattern vanished upon Salmonella infection (Fig. 1D), while HIF-1 stabilization in intestinal epithelial cells (IECs) was observed. HIF-1α accumulation was further detected in the small intestinal lamina propria, arguing for a role of HIF-1α in immune cells in this model as well (Fig. 1E).
FIG 1.
Gene expression and pathway analysis in response to stabilization of HIF-1 upon Salmonella Typhimurium (S. Tm) infection in the gut. (A) Gene expression analysis of HIF-1 target genes after orogastric S. Typhimurium infection. ns, not significant. (B and C) Computation of HIF-1 transcriptional activity (B) and NF-κB and hypoxia pathway activities (C) upon orogastric Salmonella infection in comparison to uninfected controls. (D) Representative images of small intestines 4 days after oral PBS administration (control) or S. Typhimurium infection stained for HIF-1α (top) and Hypoxyprobe (bottom) with hematoxylin (blue) counterstaining. Arrows point toward luminal villus tips. (E) Multiplex immunofluorescence staining of HIF-1α (red) (a to g), Ly6G (neutrophils) (green) (a, b, and d to f), F4/80 (macrophages) (yellow) (a, d, e, and g), and DAPI (nuclei) (blue) (a to g) in the small intestine (SI) and colon. White arrows point toward HIF-1α-positive cells within the lamina propria. (F) HIF-1α and HIF-2α Western blots of nuclear extracts from IECs isolated from the control or at 4 days p.i. (n = 3), with YY1 as a loading control, and the corresponding densitometry. (G) qPCR analysis of Hif1a (n = 4), Nos2, and Hif2a (n = 5) mRNA expression in uninfected IECs and after infection relative to the reference gene β-actin and normalized to the control. Data represent means with standard errors of the means (SEM). *, P values according to a two-tailed t test (B and C) or a Mann-Whitney U test (F and G).
Small intestinal IECs were isolated for mRNA extraction and to obtain nuclear protein extracts (Fig. 1F and G). Since previous studies on chemically induced inflammation of the gut, such as dextran sulfate sodium (DSS)-induced colitis, hinted at an influence of HIF-2 on intestinal mucosal inflammatory processes (40), we examined HIF-1α as well as HIF-2α stabilization in IECs isolated from healthy and Salmonella-infected wild-type mice. While the HIF-2α protein level was enhanced in individual samples, only HIF-1α stabilization was consistently found upon infection in all infected animals (Fig. 1F). To analyze if the lack of HIF-1 accumulation could be compensated for by HIF-2, we compared the HIF-2α levels in HIF-1α-deficient IECs (Hif1aIEC) to those in wild-type (WT) controls and found no difference between uninfected controls and Salmonella-infected animals (see Fig. S3A in the supplemental material). Next, we sought to elucidate whether the observed HIF-1 activation in IECs was transcriptional or posttranslational and whether it resulted in the HIF-1-dependent upregulation of HIF-1 target genes. Quantitative PCR (qPCR) analysis (Fig. 1G) of isolated WT IECs showed a slight upregulation of Hif1a and a significant upregulation of the HIF-1 target gene Nos2 in response to Salmonella infection. Therefore, we conclude that epithelial HIF-1, rather than HIF-2, is stabilized and transcriptionally active upon Salmonella infection in a hypoxia-independent fashion.
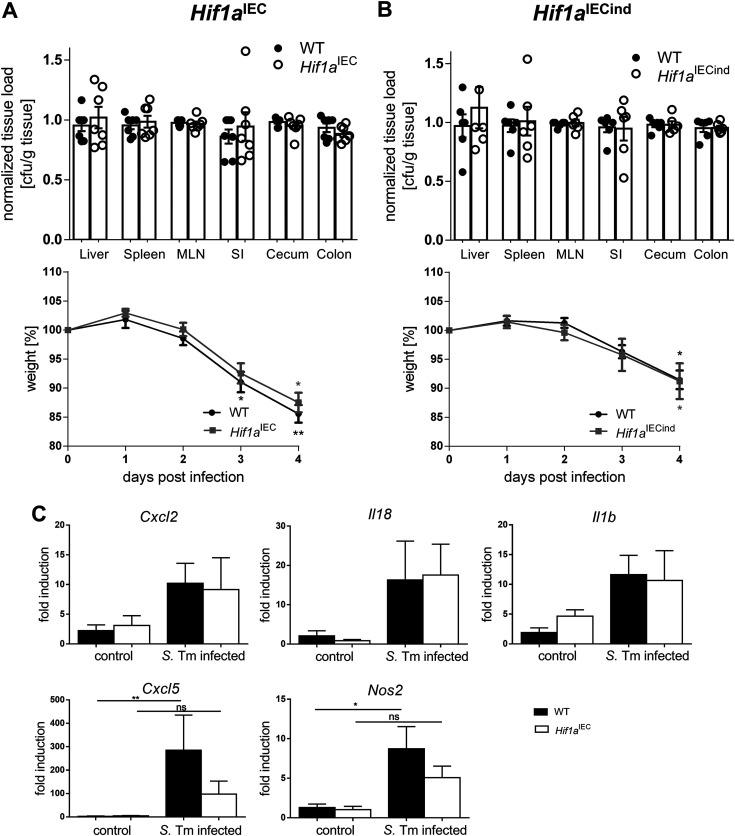
Epithelial lack of HIF-1α does not influence disease severity or systemic bacterial spread.
The above-described results suggested the functional relevance of HIF-1 for epithelial antimicrobial host defense against Salmonella. To evaluate this, we used mice with a constitutive Hif1a knockout in IECs (termed Hif1aIEC) and assayed disease severity, inflammatory activity in the gut, and bacterial spread to the mesenteric lymph nodes (MLNs), spleen, and liver tissue. Four days after Salmonella infection, the indicated organs were harvested, homogenized, diluted, and plated onto LB (lysogeny broth) agar with selective antibiotics for CFU counting (Fig. 2A). Furthermore, animals were weighed over the whole course of infection. Since differences in bacterial counts between individual experiments were observed and most likely caused by minor variations in the bacterial infection inocula, bacterial numbers were normalized to those found in the respective WT animals for a better comparison between organs. Both wild-type littermates and Hif1aIEC mice exhibited significant weight loss (almost 20%), illustrating the severity of the infection. Unexpectedly, Hif1aIEC mice showed no difference in bacterial organ counts or body weights compared to their WT littermates. To rule out the potential functional compensation for the constitutive loss of HIF-1 in Hif1aIEC mice, we additionally utilized a second IEC-specific knockout mouse line where the Hif1a deletion in IECs was induced prior to infection (termed Hif1aIECind). The gene deletion efficiency in both animal models, Hif1aIEC and Hif1aIECind mice, was confirmed beforehand (Fig. S1B to D). Again, no difference in the bacterial organ loads or weight loss could be observed (Fig. 2B). Finally, we compared the gene expression patterns of selected cytokines and chemokines in IECs isolated from WT and Hif1aIEC mice as well as healthy controls at 4 days postinfection (p.i.) (Fig. 2C). While a general upregulation was observed upon infection, only Cxcl5 and the HIF-1 target gene Nos2 showed slightly reduced upregulation in Hif1a-deficient IECs compared to the WT. The nonnormalized CFU relative to tissue weight are shown in Fig. S1.
FIG 2.
Intestinal epithelial HIF-1 loss does not affect systemic infection, weight loss, or inflammatory responses in a Salmonella infection model. (A and B) Quantification of systemic Salmonella spread (normalized CFU per gram of tissue) in the liver, spleen, mesenteric lymph nodes (MLN), small intestine (SI), cecum, and colon and weight loss (over the time course of infection) of WT littermates and Hif1aIEC mice harboring a constitutive HIF-1α knockout in IECs at 4 days postinfection (n = 7) (A) and WT littermates and tamoxifen-inducible HIF-1α-deficient (Hif1aIECind) mice (n = 6) (B). (C) mRNA expression of Cxcl2, Il18, Il1b, Cxcl5, and Nos2 relative to β-actin in WT and Hif1aIEC IECs of control mice (n = 5) and 4 days after infection with Salmonella Typhimurium (S. Tm) (n = 7) normalized to the WT control. Data represent means with SEM. *, P < 0.05; **, P < 0.01 (according to a Kruskal-Wallis test with Dunn’s posttest [A to C]).
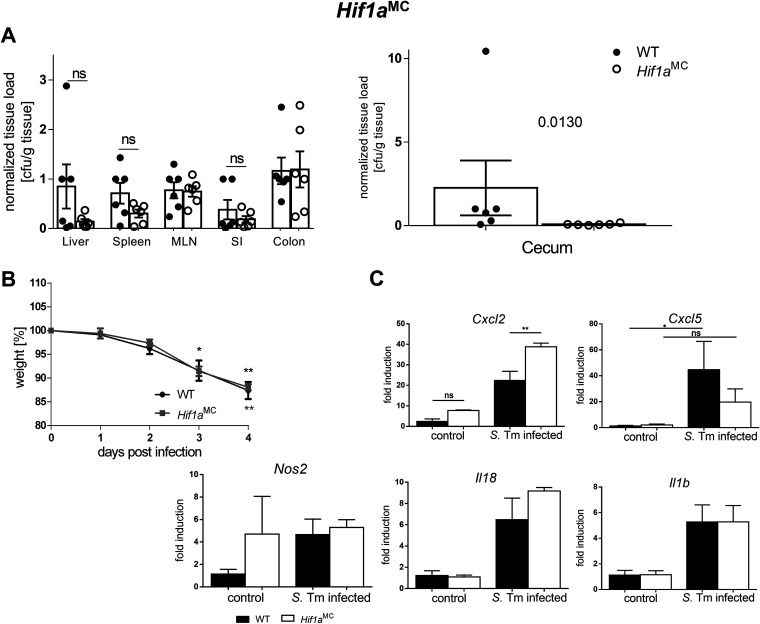
HIF-1 in myeloid cells influences the antimicrobial host response and systemic bacterial spread but not disease severity.
Macrophages and neutrophils play an essential role in the response to Salmonella by coordinating the innate and adaptive immune responses through the recognition of pathogen-associated molecular patterns (PAMPs), leading to an antimicrobial inflammatory immune response (41). Of note, Salmonella is a facultative intracellular pathogen and can survive within macrophages in so-called Salmonella-containing vacuoles (SCVs), bypassing the antimicrobial attack of the macrophage (41, 42). Since the influence of HIF-1 on the bactericidal functions of macrophages has been well described (10), we explored the effect of myeloid cell-specific Hif1a loss on systemic infection and disease outcomes as well as mucosal inflammation. Unexpectedly, the evaluation of weight changes and health status (Fig. 3B) did not show significant differences between wild-type and Hif1aMC mice. However, systemic Salmonella spread was impacted by HIF-1α deficiency. Bacterial numbers in the ceca of knockout animals were significantly reduced, while the same tendency was observed for the liver and spleen (Fig. 3A). Gene expression analysis of IECs isolated from infected wild-type and Hif1aMC animals showed significantly higher expression levels of the chemokine Cxcl2, while Cxcl5 expression was slightly reduced, in infected Hif1aMC mice (Fig. 3C). Since HIF-1α-dependent migratory defects of immune cells were reported previously (10), we further addressed potential preexisting infiltration differences of immune cells in the healthy mucosal tissues of Hif1aIEC and Hif1aMC mice by fluorescence-activated cell sorter (FACS) analysis of the lamina propria. However, no difference in the compositions of leukocytes in both mouse lines compared to their wild-type littermates was detected (Fig. S2). We addressed pathological changes and polymorphonuclear leukocyte (PMN) infiltration (Fig. S2A and S4) as well as macrophage infiltration (Fig. S2B and C) between these groups. While PMN infiltration upon infection in the small intestines of Hif1aMC mice was slightly reduced, the overall pathoscoring did not differ significantly between groups, nor was a difference in macrophage frequencies observed. The nonnormalized CFU relative to tissue weight are shown in Fig. S1.
FIG 3.
Myeloid cell-specific HIF-1α deletion affects Cxcl2 expression in IECs but not systemic Salmonella infection. (A and B) Quantification of systemic Salmonella spread/organ CFU after 4 days of infection (A) and quantification of weight loss during the time course of infection of WT littermates and Hif1aMC mice harboring an HIF-1α knockout in myeloid cells (n = 4). (C) mRNA expression of Cxcl2, Il18, Il1b, Cxcl5, and Nos2 relative to β-actin in IECs from WT and Hif1aMC uninfected controls (n = 3) and 4 days after infection with Salmonella Typhimurium (S. Tm) (n = 4) normalized to the WT control. Data represent means with SEM. *, P < 0.05; **, P < 0.01 (according to a Kruskal-Wallis test with Dunn’s posttest or Mann-Whitney U test [A {cecum}]).
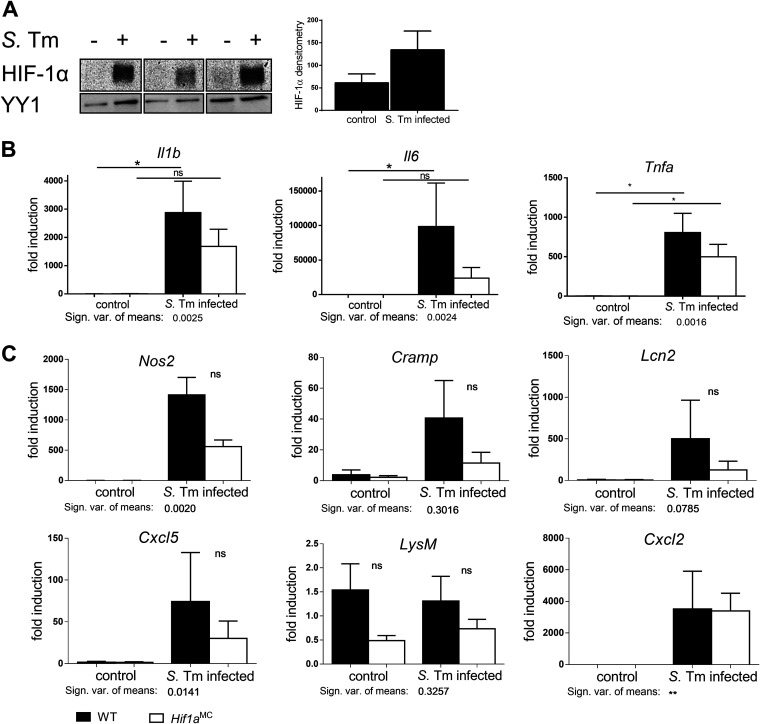
HIF-1 is highly stabilized in macrophages upon Salmonella infection and influences the inflammatory response.
Given the well-established supportive role of HIF-1 for microbial killing of macrophages, we were surprised that Hif1aMC mice by tendency exhibited a reduction in systemic bacterial spread after Salmonella infection compared to the wild-type. As macrophages represent one of the first lines of defense against Salmonella (32), we therefore sought to characterize the functional importance of HIF-1α for macrophages directly. Bone marrow-derived macrophages (BMDMs) were generated from wild-type mice and infected with Salmonella. This resulted in the robust stabilization of HIF-1α as shown by Western blotting (Fig. 4A). To further elucidate HIF-1’s impact on the transcriptional response to Salmonella, gene expression analyses in BMDMs of Hif1aMC mice and their wild-type littermates were performed. The expression levels of genes involved in direct bactericidal or inflammatory functions of phagocytes and in the response to Salmonella were compared (Fig. 4B and C). Of note, while the overall mRNA expression levels of proinflammatory mediators in HIF-1α-deficient macrophages upon infection were lower, the functional inactivation of HIF-1 had no significant influence on the Salmonella-induced regulation of the proinflammatory mediators Il1b, Il6, and Tnfa (Fig. 4B); Lcn2 (inhibiting intracellular Salmonella growth due to iron chelation [43]); Nos2; various chemokines; and the antimicrobial peptide cathelicidin-related antimicrobial peptide (CRAMP) (44) (Fig. 4C). In our hands, HIF-2α was not stabilized in response to Salmonella in wild-type or Hif1a-deficient macrophages, arguing against a compensatory effect of HIF-2 (Fig. S3E).
FIG 4.
Salmonella Typhimurium (S. Tm)-dependent HIF-1 stabilization alters the transcription of cytokines, AMPs, and chemokines in macrophages. (A) HIF-1α Western blotting and densitometry relative to the loading control YY1 (n = 3) of control and Salmonella-infected (4 h) bone marrow-derived macrophages (BMDMs), at an MOI of 10. (B and C) Relative mRNA expression of WT and Hif1aMC BMDMs in uninfected controls and 4 h after infection with Salmonella (MOI of 1) of the cytokines Il1b, Il6, and Tnfa (B) and the AMPs (antimicrobial peptides) and chemokines Nos2, Cramp, Cxcl2, Cxcl5, Lcn2, and LysM (lysozyme) (C) relative to β-actin (n = 5 [Nos2, n = 3]) and normalized to the WT control. Data represent means with SEM. *, P < 0.05; **, P < 0.01 (according to a Mann-Whitney U test [A] or a Kruskal-Wallis test with Dunn’s posttest [B and C]).
HIF-1 affects E. coli but not Salmonella killing by macrophages.
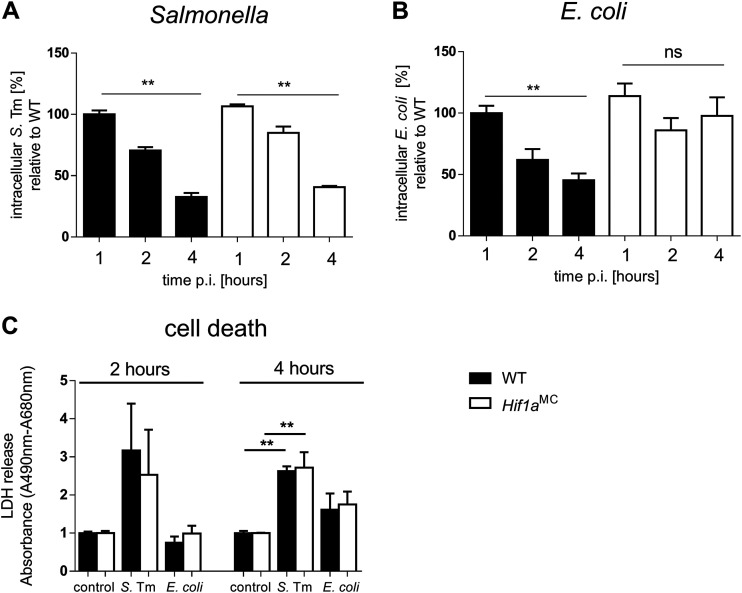
In order to reconcile our observation with the known role of HIF-1 in antimicrobial activity in macrophages (11, 13), we performed comparative intracellular killing assays using Salmonella and the nonpathogenic commensal bacterium Escherichia coli. Both WT and Hif1aMC BMDMs (bone marrow-derived macrophages) exhibited equally and significantly reduced intracellular Salmonella counts over the time course of 4 h of infection (Fig. 5A). In sharp contrast, while wild-type macrophages killed a significant percentage of intracellular E. coli bacteria, Hif1aMC cells failed to do so (Fig. 5B). To investigate whether macrophages were killing intracellular bacteria or were lysed by them in turn, we measured LDH (lactate dehydrogenase) activity in the cell culture supernatant as an indicator of cell death (Fig. 5C). LDH levels relative to the untreated control, in both WT and Hif1a-deficient BMDMs, were elevated upon Salmonella infection already after 2 h, while LDH levels upon E. coli infection were still as low as those in the control. This suggested that Salmonella induced cell death of a notable number of infected BMDMs in this setting. Intracellularly, macrophages can kill bacteria through an oxidative burst utilizing ROS (reactive oxygen species) and RNS (reactive nitrogen species). Unlike E. coli, Salmonella is able to evade and bypass this antimicrobial host defense mechanism (45). Salmonella converts the phagosome through the expression of a type III secretion system (T3SS) and the translocation of effector proteins encoded by Salmonella pathogenicity island 2 (SPI-2) into so-called Salmonella-containing vacuoles (SCVs) (45). Here, Salmonella survives and even replicates. We used an sseB mutant strain (ΔsseB) that lacks a functional SPI-2 T3SS and prevents SCV formation (Fig. S3) as well as the complemented ΔsseB psseB strain (46). However, no significant influence of HIF-1α on the number of intracellular SPI-2 T3SS-deficient Salmonella bacteria was noted within the 4-h time frame (Fig. S5). These results do not support a significant influence of the SPI-2 T3SS on resistance to HIF-1-induced bacterial killing in macrophages within the investigated time frame.
FIG 5.
E. coli but not Salmonella intracellular survival in macrophages is HIF-1 dependent. (A) Intracellular survival of Salmonella in bone marrow-derived macrophages (BMDMs) from WT littermates and HIF-1α-deficient (Hif1aMC) mice over a time course of 4 h (MOI of 10) (n = 3). Extracellular bacteria were killed by the addition of medium containing gentamicin after 30 min of incubation. (B) Intracellular survival of E. coli in WT and Hif1aMC BMDMs over the time course of infection (n = 4). (C) Cell death analysis measured by lactate dehydrogenase (LDH) release into the surrounding medium of the controls and Salmonella- and E. coli-infected BMDMs after 2 and 4 h (absorbance [A490 − A680]). Data represent means with SEM. *, P < 0.05; **, P < 0.01 (according to a Kruskal-Wallis test with Dunn’s posttest [A to C]).
DISCUSSION
Originally identified as a master regulator of the cellular response to hypoxia, HIF-1 emerged as a central regulator of immune cell functions (47). Utilizing the same mechanisms as those in the hypoxic setting, it shifts cellular metabolism, changes the transcription of a broad spectrum of genes, and therefore influences the immune functions of epithelial and endothelial cell types (13, 48) as well as phagocytes (10, 11), making it a potential tool to fight various bacterial and nonbacterial infections. The activation of macrophages with different inflammatory stimuli such as succinate (49) or bacteria (as shown for Mycobacterium tuberculosis) leads to a shift from the tricarboxylic acid (TCA) cycle to glycolysis, HIF-1 stabilization, and further macrophage activation (50–52). HIF-1 stabilization facilitates the induction of proinflammatory cytokines such as interleukin-1β and additional mechanisms controlling intracellular bacteria in particular (49, 53). In this study, we addressed the role of HIF-1 in intestinal infections with Salmonella, utilizing genetically modified mice harboring either a myeloid (Hif1aMC) or an intestinal epithelial (Hif1aIEC) cell-specific Hif1a knockout (54–56).
Although HIF-1α was robustly upregulated in IECs in response to Salmonella, its deletion did not change the overall disease outcome in the respective infection models. No difference between wild-type and knockout mice was observed regarding weight loss as well as systemic organ spread of Salmonella in both settings. Since mice with a deletion of Hif1a in IECs develop normally, we further analyzed whether these results could be due to a compensatory mechanism and utilized a tamoxifen-inducible IEC-specific Hif1a knockout mouse strain (Hif1aIECind) (55, 57). However, systemic bacterial spread and weight loss were comparable to those in the constitutive IEC-specific Hif1a knockout line. The deletion of Hif1a in myeloid cells, on the other hand, influenced systemic spread and the bacterial burden in the ceca.
Different research groups have reported that intestinal epithelial HIF-1 impacts enteric inflammatory diseases, especially inflammatory bowel disease (IBD) and DSS-induced colitis, and thereby plays a critical role in intestinal tumorigenesis and inflammation (18, 58). Surprisingly, Hif1a deletion in IECs did not alter the mRNA expression of genes encoding different cytokines, chemokines, and antimicrobial peptides (AMPs) potentially involved in the inflammatory response to bacterial pathogens. Only a slight decrease of Cxcl5 and Nos2 was observed upon Hif1a deletion. On the contrary, Hif1aMC mice showed increases in Cxcl2 mRNA expression upon Salmonella infection, hinting at a potential anti-inflammatory phenotype of HIF-1-possessing myeloid cells in this setting, a finding which could explain the reduced bacterial burden in these mice. Also, in vitro, Hif1a-deficient macrophages showed globally lower mRNA expression levels of multiple effectors involved in the specific response to Salmonella. Yet, intracellular Salmonella survival rates were comparable between Hif1aMC and wild-type cells, although it needs to be taken into account that in the Hif1aMC model (utilizing LysMcre), not only macrophages but also other cells of the myeloid lineage are targeted for Hif1a deletion (56). Neutrophils are known to impact systemic and, to a lesser extent, local inflammatory responses to Salmonella in the gut (59). Salmonella-caused enterocolitis is further characterized by a massive neutrophil infiltration of the colon and terminal ileum (60, 61). Therefore, neutrophil HIF-1α-dependent mechanisms toward Salmonella need to be addressed in more detail in the future.
Two macrophage mechanisms with central importance for the killing of intracellular pathogens are the production of ROS and RNS. The TCA cycle intermediates succinate and fumarate have been shown to inhibit PHD function and induce ROS formation, leading to robust HIF-1 stabilization (49, 62). NO (nitric oxide) species also inhibit PHDs and stabilize HIF-1 additionally through S-nitrosylation (63). However, a Salmonella mutant (ΔsseB) that is more susceptible to ROS due to its inability to form Salmonella-containing vacuoles (SCVs) intracellularly (46) again showed comparable intracellular survival rates in both wild-type and Hif1a-deficient macrophages, while, consistent with recent studies (13), E. coli survival was positively impacted by Hif1a deficiency. Therefore, we conclude that Salmonella potentially bypasses the ROS response in a different way. However, we were not able to prolong in vitro infection times due to high levels of cell death. It is therefore necessary to bear in mind that the strongest SPI-2 expression might not occur within the 4-h time frame (64).
Several studies have shown inducible NO synthase 2 (NOS2) and, therefore, NO to be of functional relevance for HIF-1-dependent immunity and the induction of a positive-feedback loop that drives the inflammatory macrophage response (51). However, chronologically, the induction of NOS2 and reactive nitrogen species follows the induction of ROS at late, severe stages of Salmonella infection (41). In all examined mouse lines, weight loss of almost 20% was observed, demonstrating a severe course of infection but potentially obscuring an HIF-1-dependent phenotype in the very final stages of a Salmonella infection model further portraying survival.
Salmonella has been shown to evade the RNS response by limiting NOS2 substrates (65). Salmonella also exhibits ROS/RNS detoxification enzymes such as SodC (superoxide dismutase) (66) on the one hand as well as PhoPQ (67), a factor crucial for the evasion of nitrosative stress and intracellular survival, on the other hand. They could therefore avoid or alter the main HIF-1 immune response of phagocytes in general. Salmonella, like other intracellular bacteria such as Chlamydophila pneumoniae (68) and Francisella tularensis (69), which actively avoid HIF-1 stabilization or possess HIF-1-degrading virulence factors, might therefore not be susceptible to HIF-1-dependent immunity and rather reprogram the ROS and RNS responses. Taken together, our findings indicate that the functional importance of HIF-1 for bacterial killing depends on the pathogen under investigation. For a diverse range of bacteria, from E. coli (13) to methicillin-resistant Staphylococcus aureus (MRSA) (12), HIF could present a therapeutic target but potentially not for all bacteria. It is important to note that our study did not address the effects of (pharmacological) stabilization of HIF-α. It surely is possible that this approach can result in a reduction of the bacterial burden in Salmonella infection, as has been shown for other infectious agents (12, 13), and also resulted in barrier protection in DSS-induced colitis (70). Evidently, a more detailed understanding of HIF-1 immunity and bacterial adaptation mechanisms of professional intracellular pathogens like Salmonella toward it is necessary to fully evaluate HIF-1’s potential as a therapeutic target.
MATERIALS AND METHODS
Mouse models.
For the constitutive deletion of Hif1a specifically in intestinal epithelial cells (IECs) (termed Hif1aIEC), Villin-Cre transgenic mice expressing the Cre recombinase under the control of the Villin1 promoter (54) were crossed with mice harboring a floxed Hif1a locus (55). For inducible IEC-specific Hif1a deletion (termed Hif1aIECind), VillinCre-ERT2 transgenic mice, which harbor Cre recombinase fused to the ligand binding domain of the human estrogen receptor under the control of Villin1 (57), were crossed with Hif1a-floxed mice. For constitutive myeloid cell-specific Hif1a deletion (termed Hif1aMC), LysMcre mice harboring Cre under the control of the murine M lysozyme promoter (56) were crossed with Hif1a-floxed mice.
Murine Salmonella infection.
For in vivo studies, 10-week-old mice from the above-described mouse lines and their cohoused littermates were used. Mice from the inducible Hif1a IEC knockout line were further administered tamoxifen (intraperitoneally [i.p.]) (100 mg/kg of body weight) for five consecutive days to induce Hif1a deletion. For comparable courses of infection among the experimental groups and between animals, this model requires oral streptomycin treatment of all mice before the oral administration of Salmonella to produce a niche for bacterial colonization and invasion in the gut. Accordingly, animals were treated with streptomycin via oral gavage (20 mg/50 μL H2O) 48 h after the last tamoxifen injection and/or 1 day before infection. The next day, animals received either 100 μL phosphate-buffered saline (PBS) (controls) or 108 CFU Salmonella Typhimurium in 100 μL PBS, also via oral gavage (35, 71). At 4 days postinfection (p.i.), animals were sacrificed for organ harvest. Along control treatment or infection, mice were examined and weighed at least once daily in accordance with the approved animal protocol. Then, liver, spleen, mesenteric lymph nodes (MLNs), small intestine (SI), cecum, and colon were collected and homogenized in sterile PBS. Counts of viable Salmonella CFU were obtained by serial dilutions and plating onto LB (lysogeny broth) agar plates supplemented with ampicillin. All animal experiments (and the choice of humane endpoints) were performed according to German animal protection law (TierSchG) and approved by the local animal welfare committee (Landesamt für Natur-, Umwelt-, und Verbraucherschutz Nordrhein-Westfalen, Recklinghausen) under code AZ84-02.04.2016.A491.
Bacterial strains.
For in vivo and in vitro infection experiments, a wild-type-like green fluorescent protein (GFP)-tagged ampicillin-resistant S. Typhimurium strain (ATCC 12048) was used. The isogenic ΔsseB S. Typhimurium strain ATCC 14028MvP643 and the constitutive ΔsseB psseB strain ATCC 14028 (Kanr) were kindly provided by Michael Hensel (University of Osnabrück, Germany) (46). E. coli K-12 strain D22 was utilized for in vitro use (72).
IEC isolation.
IECs were isolated by utilizing sections of all three parts of the small intestine. After sacrifice, intestinal sections were kept in PBS supplemented with 2% FBS (fetal bovine serum), cleaned from feces, and then turned inside out. Sections were incubated in 30 mM EDTA in PBS at 37°C. Next, IECs were detached in PBS with 2% FBS and isolated from other cell types by three sedimentation steps. The IEC pellets were snap-frozen and stored at −80°C until further preparation.
Western blotting.
IECs (snap-frozen pellets) or BMDMs (bone marrow-derived macrophages) were lysed in nuclear extraction buffer according to methods described previously by Carey et al. (73). Forty micrograms of nuclear extract was used for SDS-PAGE (reducing conditions) and wet-transfer blotted onto a nitrocellulose membrane. Immunoblotting was performed using primary antibodies against HIF-1α (catalog number 10006421; Cayman Chemical, Ann Arbor, MI, USA) (1:700), HIF-2α (catalog number NB100-122; Novus Biologicals) (1:1,000), and YY1 (catalog number 66281-1-Ig; Proteintech Europe) (1:1,000). Horseradish peroxidase (HRP)-labeled secondary antibodies were used (anti-mouse [catalog number 7076; Cell Signaling] [1:1,000] and anti-rabbit [catalog number 7074; Cell Signaling] [1:5,000]). For blocking and antibody dilution, 5% milk powder in Tris-buffered saline–Tween (TBS-T) buffer was used. Densitometry was calculated using ImageJ software (NIH, Bethesda, MD, USA) relative to YY1 and normalized to the values for the corresponding untreated wild-type controls.
RNA isolation and quantitative PCR.
Total RNA from snap-frozen IECs was isolated with peqGOLD RNAPure, and reverse transcription was performed using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA, USA). Next, quantitative real-time PCR (qPCR) was performed using the Applied Biosystems 7500 real-time PCR system in a 96-well format. Reaction mixtures contained 15 ng cDNA, 0.3 μM specific primer or primer mix according to the manufacturer’s instructions at 0.1 μM, and 1× Power SYBR green master mix (Applied Biosystems, Bleiswijk, The Netherlands). Primer mix for Il18 was obtained from Biomol GmbH (Hamburg, Germany). Primers (all from 5′ to 3′) against β-actin (forward [F], CAC TGT CGA GTC GCG TCC; reverse [R], TCA TCC ATG GCG AAC TGG TG), Hif1a (F, GCT TCT GTT ATG AGG CTC ACC; R, ATG TCG CCG TCA TCT GTT AG), Nos2 (F, AAG CCC CGC TAC TAC TCC AT; R, AAG CCA CTG ACA CTT CGC A), Cxcl2 (F, AAG TTT GCC TTG ACC CTG AA; R, AGG CAC ATC AGG TAC GAT CC), Cxcl5 (F, TGC CCT ACG GTG GAA GTC AT; R, AGC TTT CTT TTT GTC ACT GCC C), Il1b (F, CAA CCA ACA AGT GAT ATT CTC CAT G; R, GAT CCA CAC TCT CCA GCT GCA), Il6 (F, TGA GAA AAG AGT TGT GCA ATG GC; R, GCA TCC ATC ATT TCT TTG TAT CTC TGG), Tnfa (F, CCA TTC CTG AGT TCT GCA AAG G; R, AGG TAG GAA GGC CTG AGA TCT TAT C), Cramp (F, CAG CTG TAA CGA GCC TGG TG; R, CAC CTT TGC GGA GAA GTC CA), Lcn2 (F, ATGCACAGGTATCCTCAGGT; R, TGGCGAACTGGTTGTAGTCC), and LysM (F, ATG GAA TGG CTG GCT ACT ATG; R, ACC AGT ATC GGC TAT TGA TCT GA) were selected to span exon borders if possible and were validated according to MIQE guidelines (74). Relative mRNA expression was calculated using the comparative ΔCT method and normalized to β-actin expression using qbase+ 3.0 (Biogazelle, Zwijnaarde, Belgium).
Immunohistochemistry.
Immunohistochemical analysis was conducted on formalin-fixed and paraffin-embedded (FFPE) small intestines. Sections were stained with antibodies against HIF-1α (catalog number 10006421; Cayman Chemical), Ly6G (catalog number 12760; BioLegend), or F4/80 (catalog number 14-4801; eBioscience) and counterstained with hematoxylin. Hypoxic areas were visualized using the Hypoxyprobe-1 kit (Hypoxyprobe Inc., Burlington, MA, USA). Detailed staining procedures were previously described (75, 76).
Multispectral immunofluorescence staining.
FFPE sections of small and large intestines of infected wild-type animals (4 days p.i.) were deparaffinized and rehydrated according to standard procedures. This was followed by refixation in formalin and antigen retrieval. For triple staining, sections were subsequently incubated with the above-mentioned HIF-1α (1:10,000) and F4/80 (1:2,000) antibodies as well as Ly6G (1:4,000). For permanent labeling, HIF-1α was tagged with Opal 690, F4/80 was tagged with Opal 570, and Ly6G was tagged with Opal 520 (Opal 4-color immunohistochemistry [IHC] kit; PerkinElmer) according to the Opal kit protocol. Each staining was performed successively and followed by the detachment of the primary antibody, whereas the Opal label remained attached to the section. Finally, nuclei were stained with spectral 4′,6-diamidino-2-phenylindole (DAPI) (PerkinElmer). Fluorescence signals were detected using the TissueFAX Spectra multispectral cytometer and TissueFAXS Viewer software (Tissuegnostics, Romania).
Pathoscoring of small intestines and colons.
In accordance with the pathoscoring protocol established previously by Manja Barthel and colleagues (35), we addressed the development of submucosal edema, PMN infiltration, changes in goblet cell numbers, as well as epithelial damage upon Salmonella infection. In this study, we focused mainly on the small and large intestines instead of the cecum and therefore altered the criteria accordingly. We generally used a ×200 magnification for the determination of all parameters utilizing the Vectra 3.0 multiplex imaging system (PerkinElmer). The samples were further analyzed in a blind manner, and pseudonymization was removed after analysis. Since no differences between the wild types were observed, these groups were pooled, while three knockout animals each were analyzed from the Hif1aIEC and Hif1aMC lines. For small intestines, six microscopic pictures of the duodenum, jejunum, and ileum (two of each section) and three of colons (medial) were analyzed. In general, we compared each sample to the median of the wild-type control group. During the assessment of the alterations of the size of the submucosa/edema development, in the small intestine, no visible edema was detected; therefore, it was excluded from pathoscoring. The colon exhibited a measurable increase in submucosa size, and we employed the following units for detailed characterization (utilizing ImageJ) (measurement according to scale): 0 for no pathological changes, 1 for an increase of up to 60%, 2 for an increase of 61 to 100%, and 3 for an increase of 101% or more. PMN infiltration was evaluated by analysis of sections stained for neutrophils utilizing the above-mentioned Ly6G antibody. All Ly6G-positive cells were counted, and the total neutrophil number was then calculated relative to the tissue size, again by utilizing ImageJ software (calculation according to scale). An increase in neutrophil infiltration was observed, especially in the colon and less so in the small intestine. We therefore graduated the pathological units as 0 for 0 to 5 cells, 1 for 6 to 15 cells, 2 for 16 to 30 cells, and 3 for more than 30 cells, while a score of 3 was always accompanied by luminal infiltration of Ly6G-positive cells. The reduction in goblet cells was determined by calculating the goblet cell number relative to the tissue size as described above for PMN infiltration. The reduction in goblet cells was subdivided as follows: 0 for 0%, 1 for 1 to 41%, 2 for 42 to 96%, and 3 for more than 96%. The severity of epithelial damage was determined according to the following units: 0 for no changes, 1 for epithelial desquamation, 2 for erosion of up to 10 cells, and 3 for erosion of more than 10 cells, mostly also marked by ulceration. Again, these epithelial changes applied only partially to the SI. In cases where further morphological changes were observed for the SI, such as elongation of the small intestinal villi, we added +1 to the epithelial damage score of the corresponding sample.
Isolation and differentiation of bone marrow-derived macrophages.
Bone marrow from tibiae and femurs of 8- to 12-week-old WT and Hif1aMC mice was collected and seeded onto non-tissue-culture (TC)-treated cell culture plates in RPMI 1640 medium supplemented with 10% FBS overnight. Nonadherent cells were collected and cultured in RPMI 1640 (with penicillin and streptomycin [Pen/Strep]) supplemented with 20% FBS, 30% L929-conditioned medium, 100 U/mL penicillin, and 100 μg/mL streptomycin for 6 days for differentiation to naive macrophages (M0). Cells were then seeded overnight in RPMI 1640 (with Pen/Strep) supplemented with 10% FBS and 15% L929-conditioned medium. On the day of the assay, cells were washed twice with PBS, and the medium was changed to RPMI 1640 supplemented with 2% FBS without antibiotics 2 h before the assay.
Intracellular killing assay and in vitro infection.
Bacteria were grown overnight in LB medium supplemented with antibiotics as indicated. On the day of the assay, log-phase cultures were grown in LB (optical density [OD] of ∼0.55), washed with PBS twice, and then diluted in assay medium. For intracellular killing assays, BMDMs were infected with the indicated Salmonella and E. coli strains at an MOI (multiplicity of infection) of 10. Plates were then briefly centrifuged to ensure bacterium-cell contact. Thirty minutes after infection, gentamicin was added to cells (100-μg/mL final concentration) to kill extracellular bacteria. After 1, 2, and 4 h, cells were washed twice with PBS and then lysed with 0.0025% Triton X-100. Counts of viable bacteria/CFU were obtained by serial dilutions and plating onto LB agar plates. For RNA isolation, BMDMs were infected at an MOI of 1 instead to ensure cell viability, and no gentamicin was added to kill off excess extracellular bacteria. For lactate dehydrogenase (LDH) assays, the supernatants from intracellular killing assays were analyzed. The Pierce LDH cytotoxicity assay kit was used (Thermo Scientific, Waltham, MA), and the absorbance (A490 − A680) was measured.
Flow cytometry analyses of murine leukocyte populations in the lamina propria.
Peyer’s patches, mesenteric fat tissue, and feces were removed from the small intestines in HBSS (Hanks’ balanced salt solution) supplemented with 3% FBS. Mucus was removed using Nitex mesh to rinse. IECs were removed by two washes in HBSS-FBS containing 2 mM EDTA followed by collagenase VIII digestion (Sigma). Cells were then filtered using a 40-μm cell strainer and stained with the Live/Dead dye 7-amino-actinomycin D (7AAD; BioLegend) and antibodies against CD45.2 (clone 104; BioLegend), major histocompatibility complex class II (MHC-II) (clone M5.114.15.2; BioLegend), CD11c (clone N418; BioLegend), Ly6C (clone HK1.4; BioLegend), Ly6G (clone 1A8; BioLegend), CD103 (clone 2E7; BioLegend), CD11b (clone M1/70; BioLegend), Siglec F (clone E50-2440; BD), and CD64 (clone X54-5/7.1; BioLegend).
Bioinformatics analysis. (i) Data accessing and processing.
On the NCBI Gene Expression Omnibus (GEO), a publicly available transcriptomics data set by Altmeyer et al. studying the effect of Salmonella infection in a gastrointestinal infection (accession number GSE19174) was identified (37). Raw data were downloaded with the R package GEOquery (version 2.56.0) (77), focusing only on the wild-type samples. Samples associated with a PARP1 knockout were discarded. Subsequently, data were normalized first by removing lowly expressed genes, followed by background correction and between-array normalization using the R package vsn (version 3.56.0) (78). Finally, the probes were annotated with gene symbols, and expression was summarized in cases of duplicated gene symbols.
(ii) Differential gene expression analysis.
Differential gene expression analysis was performed using the R package limma (version 3.44.3) (79). A gene is considered significantly regulated with an absolute log fold change (logFC) of at least 1 and a false discovery rate (FDR) of ≤0.05.
(iii) Transcription factor analysis.
The activity of the transcription factor HIF-1 was inferred from gene expression data for each sample using the R/Bioconductor package DoRothEA (version 1.0.0 [https://saezlab.github.io/dorothea/]) (36). Activity is estimated by interrogating the expression of HIF-1 target genes. The difference in HIF-1 activities between control and infected samples was determined by a two-tailed t test.
Pathway analysis.
We inferred the activity of hypoxia and NF-κB for gene expression data for each sample using the R/Bioconductor package PROGENy (version 1.11.1 [https://saezlab.github.io/progeny/]) (38, 39). The activities are estimated by interrogating the expression of downstream affected genes instead of observing the expression of pathway members. Differences in pathway activities between control and infected samples were determined by a two-tailed t test.
Statistics.
Statistical analysis was performed using Prism 5.0 and 6.0 software (GraphPad Software, San Diego, CA, USA). Statistical significance was determined according to one-way analysis of variance followed by a Tukey post hoc test, a Kruskal-Wallis test with Dunn’s posttest, a two-tailed t test, or a Mann-Whitney U test, as indicated. Differences were considered statistically significant at a P value of <0.05. Asterisks in the graphs indicate statistically significant changes (*, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Data availability.
The code to perform the presented bioinformatics analysis is written in R and is freely available on GitHub at https://github.com/saezlab/HIF1A-activity-salmonella.
ACKNOWLEDGMENTS
Research in the Cramer lab was supported by grants from the Deutsche Forschungsgemeinschaft (CR 133/2-1 until 2-4 and CR 133/3-1). L.R. is supported by a START grant from the Medical Faculty of the RWTH Aachen University (691916).
M.H.W. and T.C. jointly supervised this study and provided funding. L.R., M.W.H., and T.C. conceptualized the study. L.R., A.D., S.J., K.Z., J.G., S.R., J.R., and V.C. performed experiments and analyzed data. C.H.H. and J.S.-R. predicted pathway activities based on microarray data. L.R. and T.C. wrote the original manuscript draft. L.R., A.D., S.J., K.Z., C.H.H., J.G., V.C., J.S.-R., M.W.H., and T.C. reviewed and edited the manuscript.
J.S.-R. has received funding from GSK and Sanofi and consultant fees from Travere Therapeutics. All other authors declare that no competing interests exist.
Footnotes
Supplemental material is available online only.
Contributor Information
Mathias W. Hornef, Email: mhornef@ukaachen.de.
Thorsten Cramer, Email: tcramer@ukaachen.de.
Manuela Raffatellu, University of California, San Diego, School of Medicine.
REFERENCES
- 1.Choudhry H, Harris AL. 2018. Advances in hypoxia-inducible factor biology. Cell Metab 27:281–298. 10.1016/j.cmet.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Werth N, Beerlage C, Rosenberger C, Yazdi AS, Edelmann M, Amr A, Bernhardt W, von Eiff C, Becker K, Schäfer A, Peschel A, Kempf VAJ. 2010. Activation of hypoxia inducible factor 1 is a general phenomenon in infections with human pathogens. PLoS One 5:e11576. 10.1371/journal.pone.0011576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nizet V, Johnson RS. 2009. Interdependence of hypoxic and innate immune responses. Nat Rev Immunol 9:609–617. 10.1038/nri2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL, Wang GL. 1992. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol 12:5447–5454. 10.1128/mcb.12.12.5447-5454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F, White SB, Zhao Q, Lee FS. 2001. HIF-1α binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci USA 98:9630–9635. 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. 1997. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1α regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA 94:4273–4278. 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, Harris AL. 2000. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol 157:411–421. 10.1016/s0002-9440(10)64554-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rius J, Guma M, Schachtrup C, Akassoglou K, Zinkernagel AS, Nizet V, Johnson RS, Haddad GG, Karin M. 2008. NF-κB links innate immunity to the hypoxic response through transcriptional regulation of HIF-1α. Nature 453:807–811. 10.1038/nature06905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brooks AC, Menzies-Gow N, Bailey SR, Cunningham FM, Elliott J. 2010. Endotoxin-induced HIF-1α stabilisation in equine endothelial cells: synergistic action with hypoxia. Inflamm Res 59:689–698. 10.1007/s00011-010-0180-x. [DOI] [PubMed] [Google Scholar]
- 10.Cramer T, Yamanishi Y, Clausen BE, Förster I, Pawlinski R, Mackman N, Haase VH, Jaenisch R, Corr M, Nizet V, Firestein GS, Gerber HP, Ferrara N, Johnson RS. 2003. HIF-1α is essential for myeloid cell-mediated inflammation. Cell 112:645–657. 10.1016/s0092-8674(03)00154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peyssonnaux C, Datta V, Cramer T, Doedens A, Theodorakis EA, Gallo RL, Hurtado-Ziola N, Nizet V, Johnson RS. 2005. HIF-1 alpha expression regulates the bacterial capacity of phagocytes. J Clin Invest 115:1806–1815. 10.1172/JCI23865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okumura CYM, Hollands A, Tran DN, Olson J, Dahesh S, Von Köckritz-Blickwede M, Thienphrapa W, Corle C, Jeung SN, Kotsakis A, Shalwitz RA, Johnson RS, Nizet V. 2012. A new pharmacological agent (AKB-4924) stabilizes hypoxia inducible factor-1 (HIF-1) and increases skin innate defenses against bacterial infection. J Mol Med (Berl) 90:1079–1089. 10.1007/s00109-012-0882-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin AE, Beasley FC, Olson J, Keller N, Shalwitz RA, Hannan TJ, Hultgren SJ, Nizet V. 2015. Role of hypoxia inducible factor-1α (HIF-1α) in innate defense against uropathogenic Escherichia coli infection. PLoS Pathog 11:e1004818. 10.1371/journal.ppat.1004818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Silver IA. 1977. Tissue PO2 changes in acute inflammation. Adv Exp Med Biol 94:769–774. 10.1007/978-1-4684-8890-6_106. [DOI] [PubMed] [Google Scholar]
- 15.He G, Shankar RA, Chzhan M, Samouilov A, Kuppusamy P, Zweier JL. 1999. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci USA 96:4586–4591. 10.1073/pnas.96.8.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, Grunberg S, Baldassano RN, Lewis JD, Li H, Thom SR, Bushman FD, Vinogradov SA, Wu GD. 2014. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147:1055–1063.e8. 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, Weir TL, Ehrentraut SF, Pickel C, Kuhn KA, Lanis JM, Nguyen V, Taylor CT, Colgan SP. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17:662–671. 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rohwer N, Jumpertz S, Erdem M, Egners A, Warzecha KT, Fragoulis A, Kühl AA, Kramann R, Neuss S, Rudolph I, Endermann T, Zasada C, Apostolova I, Gerling M, Kempa S, Hughes R, Lewis CE, Brenner W, Malinowski MB, Stockmann M, Schomburg L, Faller W, Sansom OJ, Tacke F, Morkel M, Cramer T. 2019. Non-canonical HIF-1 stabilization contributes to intestinal tumorigenesis. Oncogene 38:5670–5685. 10.1038/s41388-019-0816-4. [DOI] [PubMed] [Google Scholar]
- 19.Gustafsson MV, Zheng X, Pereira T, Gradin K, Jin S, Lundkvist J, Ruas JL, Poellinger L, Lendahl U, Bondesson M. 2005. Hypoxia requires Notch signaling to maintain the undifferentiated cell state. Dev Cell 9:617–628. 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Robinson A, Keely S, Karhausen J, Gerich ME, Furuta GT, Colgan SP. 2008. Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology 134:145–155. 10.1053/j.gastro.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tambuwala MM, Cummins EP, Lenihan CR, Kiss J, Stauch M, Scholz CC, Fraisl P, Lasitschka F, Mollenhauer M, Saunders SP, Maxwell PH, Carmeliet P, Fallon PG, Schneider M, Taylor CT. 2010. Loss of prolyl hydroxylase-1 protects against colitis through reduced epithelial cell apoptosis and increased barrier function. Gastroenterology 139:2093–2101. 10.1053/j.gastro.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 22.Saeedi BJ, Kao DJ, Kitzenberg DA, Dobrinskikh E, Schwisow KD, Masterson JC, Kendrick AA, Kelly CJ, Bayless AJ, Kominsky DJ, Campbell EL, Kuhn KA, Furuta GT, Colgan SP, Glover LE. 2015. HIF-dependent regulation of claudin-1 is central to intestinal epithelial tight junction integrity. Mol Biol Cell 26:2252–2262. 10.1091/mbc.E14-07-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cummins EP, Seeballuck F, Keely SJ, Mangan NE, Callanan JJ, Fallon PG, Taylor CT. 2008. The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology 134:156–165. 10.1053/j.gastro.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Hirota SA, Fines K, Ng J, Traboulsi D, Lee J, Ihara E, Li Y, Willmore WG, Chung D, Scully MM, Louie T, Medlicott S, Lejeune M, Chadee K, Armstrong G, Colgan SP, Muruve DA, MacDonald JA, Beck PL. 2010. Hypoxia-inducible factor signaling provides protection in Clostridium difficile-induced intestinal injury. Gastroenterology 139:259–269.e3. 10.1053/j.gastro.2010.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartmann H, Eltzschig HK, Wurz H, Hantke K, Rakin A, Yazdi AS, Matteoli G, Bohn E, Autenrieth IB, Karhausen J, Neumann D, Colgan SP, Kempf VAJ. 2008. Hypoxia-independent activation of HIF-1 by Enterobacteriaceae and their siderophores. Gastroenterology 134:756–767.e6. 10.1053/j.gastro.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Coburn B, Grassl GA, Finlay BB. 2007. Salmonella, the host and disease: a brief review. Immunol Cell Biol 85:112–118. 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- 27.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O’Brien SJ, Jones TF, Fazil A, Hoekstra RM, International Collaboration on Enteric Disease ‘Burden of Illness’ Studies. 2010. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis 50:882–889. 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 28.LaRock DL, Chaudhary A, Miller SI. 2015. Salmonellae interactions with host processes. Nat Rev Microbiol 13:191–205. 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Que F, Wu S, Huang R. 2013. Salmonella pathogenicity island 1 (SPI-1) at work. Curr Microbiol 66:582–587. 10.1007/s00284-013-0307-8. [DOI] [PubMed] [Google Scholar]
- 30.Zhang K, Riba A, Nietschke M, Torow N, Repnik U, Pütz A, Fulde M, Dupont A, Hensel M, Hornef M. 2018. Minimal SPI1-T3SS effector requirement for Salmonella enterocyte invasion and intracellular proliferation in vivo. PLoS Pathog 14:e1006925. 10.1371/journal.ppat.1006925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deiwick J, Nikolaus T, Shea JE, Gleeson C, Holden DW, Hensel M. 1998. Mutations in Salmonella pathogenicity island 2 (SPI2) genes affecting transcription of SPI1 genes and resistance to antimicrobial agents. J Bacteriol 180:4775–4780. 10.1128/JB.180.18.4775-4780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun Y, Reid B, Ferreira F, Luxardi G, Ma L, Lokken KL, Zhu K, Xu G, Sun Y, Ryzhuk V, Guo BP, Lebrilla CB, Maverakis E, Mogilner A, Zhao M. 2019. Infection-generated electric field in gut epithelium drives bidirectional migration of macrophages. PLoS Biol 17:e3000044. 10.1371/journal.pbio.3000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carter PB, Collins FM. 1974. The route of enteric infection in normal mice. J Exp Med 139:1189–1203. 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Bäumler AJ. 2001. Animal models of Salmonella infections: enteritis versus typhoid fever. Microbes Infect 3:1335–1344. 10.1016/S1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 35.Barthel M, Hapfelmeier S, Quintanilla-Martínez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Rüssmann H, Hardt W-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71:2839–2858. 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Alonso L, Holland CH, Ibrahim MM, Turei D, Saez-Rodriguez J. 2019. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res 29:1363–1375. 10.1101/gr.240663.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altmeyer M, Barthel M, Eberhard M, Rehrauer H, Hardt WD, Hottiger MO. 2010. Absence of poly(ADP-ribose) polymerase 1 delays the onset of Salmonella enterica serovar Typhimurium-induced gut inflammation. Infect Immun 78:3420–3431. 10.1128/IAI.00211-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert M, Klinger B, Klünemann M, Sieber A, Uhlitz F, Sauer S, Garnett MJ, Blüthgen N, Saez-Rodriguez J. 2018. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat Commun 9:20. 10.1038/s41467-017-02391-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holland CH, Szalai B, Saez-Rodriguez J. 2020. Transfer of regulatory knowledge from human to mouse for functional genomics analysis. Biochim Biophys Acta 1863:194431. 10.1016/j.bbagrm.2019.194431. [DOI] [PubMed] [Google Scholar]
- 40.Ramakrishnan SK, Shah YM. 2016. Role of intestinal HIF-2α in health and disease. Annu Rev Physiol 78:301–325. 10.1146/annurev-physiol-021115-105202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gogoi M, Shreenivas MM, Chakravortty D. 2019. Hoodwinking the big-eater to prosper: the Salmonella-macrophage paradigm. J Innate Immun 11:289–299. 10.1159/000490953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alpuche-Aranda CM, Racoosin EL, Swanson JA, Miller SI. 1994. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J Exp Med 179:601–608. 10.1084/jem.179.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaible UE, Kaufmann SHE. 2004. Iron and microbial infection. Nat Rev Microbiol 2:946–953. 10.1038/nrmicro1046. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberger CM, Gallo RL, Finlay BB. 2004. Interplay between antibacterial effectors: a macrophage antimicrobial peptide impairs intracellular Salmonella replication. Proc Natl Acad Sci USA 101:2422–2427. 10.1073/pnas.0304455101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Figueira R, Holden DW. 2012. Functions of the Salmonella pathogenicity island 2 (SPI-2) type III secretion system effectors. Microbiology (Reading) 158:1147–1161. 10.1099/mic.0.058115-0. [DOI] [PubMed] [Google Scholar]
- 46.Beuzon CR, Banks G, Deiwick J, Hensel M, Holden DW. 1999. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol 33:806–816. 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 47.Palazon A, Goldrath AW, Nizet V, Johnson RS. 2014. HIF transcription factors, inflammation, and immunity. Immunity 41:518–528. 10.1016/j.immuni.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leire E, Olson J, Isaacs H, Nizet V, Hollands A. 2013. Role of hypoxia inducible factor-1 in keratinocyte inflammatory response and neutrophil recruitment. J Inflamm (Lond) 10:28. 10.1186/1476-9255-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, Zheng L, Gardet A, Tong Z, Jany SS, Corr SC, Haneklaus M, Caffrey BE, Pierce K, Walmsley S, Beasley FC, Cummins E, Nizet V, Whyte M, Taylor CT, Lin H, Masters SL, Gottlieb E, Kelly VP, Clish C, Auron PE, Xavier RJ, O’Neill LAJ. 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 496:238–242. 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Prados J-C, Través PG, Cuenca J, Rico D, Aragonés J, Martín-Sanz P, Cascante M, Boscá L. 2010. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol 185:605–614. 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 51.Braverman J, Sogi KM, Benjamin D, Nomura DK, Stanley SA. 2016. HIF-1α is an essential mediator of IFN-γ-dependent immunity to Mycobacterium tuberculosis. J Immunol 197:1287–1297. 10.4049/jimmunol.1600266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gleeson LE, Sheedy FJ, Palsson-McDermott EM, Triglia D, O’Leary SM, O’Sullivan MP, O’Neill LAJ, Keane J. 2016. Cutting edge: Mycobacterium tuberculosis induces aerobic glycolysis in human alveolar macrophages that is required for control of intracellular bacillary replication. J Immunol 196:2444–2449. 10.4049/jimmunol.1501612. [DOI] [PubMed] [Google Scholar]
- 53.Knight M, Stanley S. 2019. HIF-1α as a central mediator of cellular resistance to intracellular pathogens. Curr Opin Immunol 60:111–116. 10.1016/j.coi.2019.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Madison BB, Dunbar L, Qiao XT, Braunstein K, Braunstein E, Gumucio DL. 2002. cis elements of the villin gene control expression in restricted domains of the vertical (crypt) and horizontal (duodenum, cecum) axes of the intestine. J Biol Chem 277:33275–33283. 10.1074/jbc.M204935200. [DOI] [PubMed] [Google Scholar]
- 55.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. 2000. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res 60:4010–4015. [PubMed] [Google Scholar]
- 56.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. 1999. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res 8:265–277. 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 57.el Marjou F, Janssen K-P, Chang BH-J, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39:186–193. 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 58.Robrahn L, Jiao L, Cramer T. 2020. Barrier integrity and chronic inflammation mediated by HIF-1 impact on intestinal tumorigenesis. Cancer Lett 490:186–192. 10.1016/j.canlet.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Cheminay C, Chakravortty D, Hensel M. 2004. Role of neutrophils in murine salmonellosis. Infect Immun 72:468–477. 10.1128/IAI.72.1.468-477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Day DW, Mandal BK, Morson BC. 1978. The rectal biopsy appearances in Salmonella colitis. Histopathology 2:117–131. 10.1111/j.1365-2559.1978.tb01700.x. [DOI] [PubMed] [Google Scholar]
- 61.Lamps LW. 2003. Pathology of food-borne infectious diseases of the gastrointestinal tract: an update. Adv Anat Pathol 10:319–327. 10.1097/00125480-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 62.Sudarshan S, Sourbier C, Kong H-S, Block K, Romero VAV, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, Lee M-J, Gourlay CW, Trepel J, Linehan WM, Neckers L. 2009. Fumarate hydratase deficiency in renal cancer induces glycolytic addiction and hypoxia-inducible transcription factor 1α stabilization by glucose-dependent generation of reactive oxygen species. Mol Cell Biol 29:4080–4090. 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst MW, Li C-Y. 2007. Regulation of HIF-1α stability through S-nitrosylation. Mol Cell 26:63–74. 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brown NF, Vallance BA, Coombes BK, Valdez Y, Coburn BA, Finlay BB. 2005. Salmonella pathogenicity island 2 is expressed prior to penetrating the intestine. PLoS Pathog 1:e32. 10.1371/journal.ppat.0010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lahiri A, Das P, Chakravortty D. 2008. Arginase modulates Salmonella induced nitric oxide production in RAW264.7 macrophages and is required for Salmonella pathogenesis in mice model of infection. Microbes Infect 10:1166–1174. 10.1016/j.micinf.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 66.Uzzau S, Bossi L, Figueroa-Bossi N. 2002. Differential accumulation of Salmonella[Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol Microbiol 46:147–156. 10.1046/j.1365-2958.2002.03145.x. [DOI] [PubMed] [Google Scholar]
- 67.Bourret TJ, Liu L, Shaw JA, Husain M, Vázquez-Torres A. 2017. Magnesium homeostasis protects Salmonella against nitrooxidative stress. Sci Rep 7:15083. 10.1038/s41598-017-15445-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rupp J, Gieffers J, Klinger M, van Zandbergen G, Wrase R, Maass M, Solbach W, Deiwick J, Hellwig-Burgel T. 2007. Chlamydia pneumoniae directly interferes with HIF-1α stabilization in human host cells. Cell Microbiol 9:2181–2191. 10.1111/j.1462-5822.2007.00948.x. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt EV, Diaz K, Griffin AJ, Rasmussen JA, Crane DD, Jones BD, Bosio CM. 2016. Metabolic reprogramming of host cells by virulent Francisella tularensis for optimal replication and modulation of inflammation. J Immunol 196:4227–4236. 10.4049/jimmunol.1502456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manresa MC, Taylor CT. 2017. Hypoxia inducible factor (HIF) hydroxylases as regulators of intestinal epithelial barrier function. Cell Mol Gastroenterol Hepatol 3:303–315. 10.1016/j.jcmgh.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Meynell GG, Subbaiah TV. 1963. Antibacterial mechanisms of the mouse gut. I. Kinetics of infection by Salmonella typhimurium in normal and streptomycin-treated mice studied with abortive transductants. Br J Exp Pathol 44:197–208. [PMC free article] [PubMed] [Google Scholar]
- 72.Normark S, Boman HG, Matsson E. 1969. Mutant of Escherichia coli with anomalous cell division and ability to decrease episomally and chromosomally mediated resistance to ampicillin and several other antibiotics. J Bacteriol 97:1334–1342. 10.1128/jb.97.3.1334-1342.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carey MF, Peterson CL, Smale ST. 2009. Dignam and Roeder nuclear extract preparation. Cold Spring Harb Protoc 2009:pdb.prot5330. 10.1101/pdb.prot5330. [DOI] [PubMed] [Google Scholar]
- 74.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 75.Rohwer N, Lobitz S, Daskalow K, Jöns T, Vieth M, Schlag PM, Kemmner W, Wiedenmann B, Cramer T, Höcker M. 2009. HIF-1α determines the metastatic potential of gastric cancer cells. Br J Cancer 100:772–781. 10.1038/sj.bjc.6604919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Daskalow K, Rohwer N, Raskopf E, Dupuy E, Kühl A, Loddenkemper C, Wiedenmann B, Schmitz V, Cramer T. 2010. Role of hypoxia-inducible transcription factor 1α for progression and chemosensitivity of murine hepatocellular carcinoma. J Mol Med (Berl) 88:817–827. 10.1007/s00109-010-0623-4. [DOI] [PubMed] [Google Scholar]
- 77.Sean D, Meltzer PS. 2007. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23:1846–1847. 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 78.Huber W, von Heydebreck A, Sültmann H, Poustka A, Vingron M. 2002. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics 18(Suppl 1):S96–S104. 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 79.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00222-21-s0001.pdf, PDF file, 2.1 MB (2.1MB, pdf)
Data Availability Statement
The code to perform the presented bioinformatics analysis is written in R and is freely available on GitHub at https://github.com/saezlab/HIF1A-activity-salmonella.