ABSTRACT
Uropathogenic Escherichia coli (UPEC) causes the majority of uncomplicated urinary tract infections (UTI), which affect nearly half of women worldwide. Many UPEC strains carry an annotated intimin-like adhesin (ila) locus in their genome related to a well-characterized virulence factor in diarrheagenic E. coli pathotypes. Its role in UPEC uropathogenesis, however, remains unknown. In prototype UPEC strain CFT073, there is an ila locus that contains three predicted intimin-like genes, sinH, sinI, and ratA. We used in silico approaches to determine the phylogeny and genomic distribution of this locus among uropathogens. We found that the currently annotated intimin locus-encoded proteins in CFT073 are more closely related to invasin proteins found in Salmonella. Deletion of the individual sinH, sinI, and ratA genes did not result in measurable effects on growth, biofilm formation, or motility in vitro. On average, sinH was more highly expressed in clinical strains during active human UTI than in human urine ex vivo. Unexpectedly, we found that strains lacking this ila locus had increased adherence to bladder cells in vitro, coupled with a decrease in bladder cell invasion and death. The sinH mutant displayed a significant fitness defect in the murine model of ascending UTI, including reduced inflammation in the bladder. These data confirmed an inhibitory role in bladder cell adherence to facilitate invasion and inflammation; therefore, the ila locus should be termed invasin-like rather than intimin-like. Collectively, our data suggest that loss of this locus mediates measurable interactions with bladder cells in vitro and contributes to fitness during UTI.
KEYWORDS: Escherichia coli, adhesins, intimin, invasin, urinary tract infection, virulence
INTRODUCTION
Urinary tract infections (UTI) pose a significant health threat worldwide. Nearly one-third of women will experience at least one UTI and receive antibiotic treatment by age 24, with a cumulative lifetime risk of 60.4% (1, 2). Recurrent infections, defined as having more than one UTI within a 6-month period, are extremely common and account for over 25% of infections (3, 4). Due to their prevalence and recurrence, UTI have a significant impact on the U.S. health care system. UTI-associated costs in the United States totaled more than $3.5 billion in 2015, a steep increase from $1.6 billion in 1995 (1, 5). The overwhelming majority of uncomplicated UTI are caused by uropathogenic Escherichia coli (UPEC), which is becoming increasingly antibiotic resistant (4, 6), driving an essential need for improved treatments and therapeutics (6, 7).
UPEC normally resides in the gut as a part of the commensal microbiota without causing disease (8, 9); however, when exposed to the urethral opening, bacteria can ascend into the bladder and bind host cells utilizing type 1 fimbriae and other adhesins to colonize the bladder (5, 10). If the infection is not treated in a timely manner, UPEC can potentially utilize other fimbrial adhesins, as well as flagella, to ascend the ureters and colonize the kidneys (11, 12), causing acute pyelonephritis (5). Adherence is a crucial virulence property essential in almost every step of pathogenesis, which allows for the colonization, ascension, and persistence of the infection throughout the urinary tract. For this reason, critical UPEC adhesins are often targeted for novel therapeutics and the development of vaccines to prevent UTI (13–15). We sought to investigate the role of the intimin-like adhesin (ila) locus in prototype strain CFT073, which carries the sinH-, sinI-, and ratA-like genes.
Intimin is an adhesin that functions with its cognate receptor, Tir (translocated intimin receptor), to tightly adhere to host epithelial cells during pedestal formation in a type III secretion system (T3SS)-dependent manner (16, 17). The most well-studied examples of intimin are found in enteropathogenic E. coli (EPEC) and enterohemorrhagic E. coli (EHEC) (18). These pathogens cause diarrhea when they colonize the intestine and adhere to the host using intimin to form pedestals, ultimately resulting in their characteristic attaching and effacing (A/E) lesions (19, 20). In EHEC and EPEC, intimin is encoded by eaeA on the locus of enterocyte effacement (LEE) pathogenicity island, which also encodes the T3SS and the Tir protein (20, 21). Bacteria secrete Tir directly into the host epithelial cell cytoplasm using the T3SS, where Tir ultimately becomes translocated to the host plasma membrane and acts as a receptor to bind intimin expressed on the bacterial surface (16). In this way, the bacteria produce both the ligand and receptor for close or tight adhesion to the host cells. The binding of intimin to the Tir protein in the host membrane triggers cytoskeletal changes that are attributed to the symptoms of EPEC and EHEC infection (22).
However, Tir-independent host binding cannot be ruled out due to the diversity of polymorphic sites detected in the C-terminal domain, which have been designated α, β, γ, δ, and ε intimin subtypes (23, 24). The diversity of the extracellular C-terminal domain may contribute to tissue tropism, as the ability of intimin to bind β-1 integrins was detected in T cells in vitro (25). However, the relevancy of non-Tir intimin binding in vivo is unclear. A related protein, invasin found in Yersinia spp., is known to bind β-1 integrins during infection and shares strong homology with intimin (26). Specifically, the N-terminal domains of intimin have high homology with invasin proteins, suggesting that the two may share secretion and membrane insertion mechanisms. Unlike intimins, which promote extracellular attachment, invasins mediate bacterial entry into eukaryotic cells (27).
The virulence-associated genetic island CS54 in Salmonella enterica consists of the intimin- and invasin-related genes (shdA, ratB, ratA, sivH, and sivI) (28). Although many Shiga toxin-producing E. coli strains also harbor the CS54 genomic island (29), it is generally not found intact in commensal strains of E. coli. In contrast, many UPEC strains have inherited a three-gene portion of the CS54 genomic island, including sinH (sivH), sinI (sivI), and ratA. Interestingly, Salmonella enterica subsp. II and Salmonella bongori, in addition to S. enterica subsp. I, have also inherited these same three loci, indicating a pattern of distinct phylogenetic inheritance (28). The Salmonella invasin homolog (sivH) was determined to have a role in intestinal colonization, but not fecal shedding, while both sivI and ratA produced no phenotypes in the murine gut (28).
Although the role of intimin has been well studied in the context of enteric E. coli strains, the roles of intimin in UPEC have yet to be fully characterized. By deleting the genes in the ila locus in prototype UPEC strain CFT073, we identified a role for these adhesins in host bladder cell interactions. We observed decreased bladder cell death and reduced populations of intracellular bacteria in vitro when we deleted each gene in the ila locus. Furthermore, the sinH mutant was outperformed by the wild type (WT) in the murine ascending model of UTI during both cochallenge and independent infections. Concordant with our in vitro findings, we also observed reduced inflammation in the bladders, but not kidneys, of mice infected with a ΔsinH mutant compared to those with WT infection. Considering the significant but modest decreases in bladder colonization in vivo, we must acknowledge that SinH could have originally been acquired for the gut niche of the UPEC life cycle and that the interactions in the urinary tract are coincidental.
RESULTS
The intimin-like locus in E. coli CFT073 has predicted host cell interaction domains and was likely acquired from other enteric pathogens.
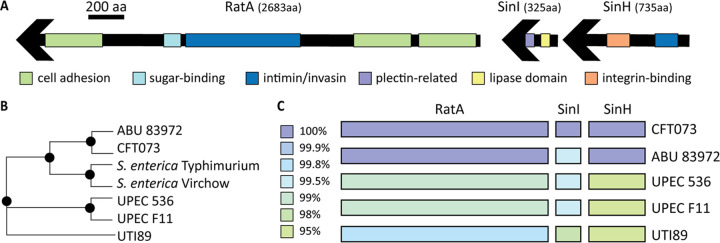
Because these intimin-like proteins have not been previously characterized in UPEC and cognate genes typically associated with this island are missing in prototype UPEC strain CFT073, we conducted domain analysis to predict their potential origin and function. To predict the function of these intimin-like proteins encoded in the CFT073 genome, we utilized protein prediction software (Phyre 2) to identify known protein domains (30). One region of SinH was predicted with 47% identity (100% confidence) to resemble the transmembrane beta-domain of invasin from Yersinia pseudotuberculosis (Fig. 1A; Table 1): invasin is an intimin homolog used for cell adhesion and invasion by Yersinia spp. (21). This region was also predicted with 45% identity (100% confidence) to resemble the transmembrane beta-domain of intimin from an EHEC O157:H7 strain (Fig. 1A, noted in dark blue; Table 1). The predicted beta-barrel structure of this region is an important structure of intimin and critical for surface expression (18, 21). Indeed, by utilizing I-TASSER software (31), predicted models detected a beta-barrel, and the top cellular component gene ontology (GO) score (GO:0043231) predicts a membrane-bound structure. Additional regions of CFT073 SinH also displayed structural homology to integrin-binding domains with high confidence (Table 1). These structural similarities provide further evidence that SinH is likely to play a role in host interactions, similar to other members of the intimin and invasin families.
FIG 1.
The intimin-like adhesin locus contains predicted host cell interaction domains and is highly conserved among UPEC strains. (A) Phyre2-predicted structures of various regions of SinH, SinI, and RatA putative proteins are represented. Only domains predicted with greater than 70% confidence are shown. Green-highlighted regions indicate homology to known cell adhesion domains, light blue indicates sugar-binding domains, dark blue indicates intimin/invasin domains, purple indicates plectin-related domains, yellow indicates lipase domains, and orange indicates integrin-binding domains. Exact parameters for predicted regions are given in Table 1. (B) A phylogenetic tree was created in PATRIC to display the homology of SinH to other bacterial strains. CFT073 SinH is more similar to S. enterica (SivH) than are other UPEC strains, UTI89, F11, and UPEC536. (C) PATRIC protein blast tool was used to compare the SinH-RatA CFT073 locus to other prototype UPEC strains. The colors indicate the percent amino acid homology in each strain, ranging from 95% to 100% identity.
TABLE 1.
PHYRE 2 predicted domains of the intimin-like putative proteinsa
| Locus | Amino acids | % Coverage | Confidence | % Identity | PDB molecule | PDB title |
|---|---|---|---|---|---|---|
| SinH | 109–353 | 33 | 100 | 47 | Invasin | X-ray crystal structure of the transmembrane beta-domain from invasin2 from Yersinia pseudotuberculosis |
| 111–353 | 32 | 100 | 45 | Intimin | X-ray crystal structure of the transmembrane beta-domain from intimin2 from EHEC strain O157:H7 | |
| 497–612 | 15 | 97.4 | 22 | Invasin/intimin cell adhesion | Invasin/intimin cell adhesion fragments; immunoglobulin-like beta-sandwich | |
| 504–735 | 31 | 96.8 | 18 | Invasin | Crystal structure of invasin: a bacterial integrin-binding protein | |
| SinI | 68–173 | 32 | 90.5 | 15 | Cellulose-binding protein | Nucleoside hydrolase-related hypothetical protein from Saccharophagus degradans that is associated with carbohydrate metabolism |
| 85–158 | 22 | 96 | 21 | Plectin-related protein | Crystal structure of the Ig-pH domain of actin-binding protein SCAB1 | |
| 192–256 | 19 | 81.6 | 24 | Colipase-binding domain | Lipase/lipooxygenase domain (PLAT/LH2 domain) | |
| 197–253 | 17 | 72.4 | 32 | Serine-rich repeat protein 1 | Crystal structure of keratin 4 binding domain of surface adhesin in Streptococcus agalactiae | |
| 214–312 | 30 | 73.7 | 20 | CC chemokine receptor 7 | Human CC chemokine receptor 7 in complex with the intracellular allosteric antagonist Cmp2105 | |
| RatA | 182–427 | 9 | 100 | 36 | Putative invasin | Structure of Yersinia pseudotuberculosis adhesin |
| 1217–1385 | 6 | 79 | 15 | Surface layer protein | Crystal structure of the S-layer protein SbsC domains 4 and 5 | |
| 1290–1388 | 3 | 96.9 | 9 | Invasin/intimin cell adhesion fragments | Immunoglobulin-like beta-sandwich | |
| 1476–1754 | 10 | 97.9 | 6 | Ig domain protein group 1 domain protein | Structure of Yersinia pseudotuberculosis invasin | |
| 1945–2252 | 11 | 92.7 | 13 | Attaching and effacing protein, pathogenesis factor | FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections |
For each locus listed in the first column, the unique amino acid regions for each recognized domain are listed in the second column. The third column indicates the percent coverage of the entire predicted protein product that the recognized region spans. The fourth and fifth columns indicate the confidence and percent identity of the Protein Database (PDB) molecule listed in the sixth column. The last column gives a brief description of the PDB molecules as listed by PHYRE 2.0.
SinI demonstrated less structural resemblance to intimin and invasin than SinH: however, they may be interacting with one another, working as a two-partner system to initiate contact with the host cell. To this point, unlike SinH, SinI was not predicted to have a beta-barrel structure; however, the top structural analog predicted by I-TASSER was the Yersinia invasin InvD (PDB 5LDYA) (31). By utilization of PYHRE, one region was predicted with 96.2% confidence to be a plectin-related domain (cytoskeleton scaffolding), indicated in purple in Fig. 1A, resembling the actin-binding host protein SCAB1 (Fig. 1A; Table 1). It is possible that SinH-SinI plays a role similar to the effects of intimin-Tir binding on host actin polymerization and microfilament aggregation upon adhesion (32). Additionally, SinI was predicted with 90.5% confidence to resemble a carbohydrate metabolism protein involved in cellulose binding (Table 1), also a predicted function of intimin (21). Therefore, the sequence and functional similarities of these binding domains of SinI strengthen the putative relationship between intimin and CFT073’s operon (sinH-sinI). Although intimin and invasin are related proteins, intimin is used for external cell pedestal formation and invasin is used to promote bacterial penetration or invasion of host cells. SinH and SinI could potentially contribute to these functions in UPEC, because both extracellular and intracellular colonization and replication have been observed for this pathogen (33–35).
The third and largest gene in this CFT073 genomic island is annotated as ratA-like based on homology with RatA genes of Salmonella and other pathogenic E. coli species and was discovered prior to the ribosome-associated toxin RatA in E. coli K-12 (36). The previously annotated toxin-antitoxin system ratAB locus in CFT073 was renamed pasTI to avoid confusion with the ratA-like locus described here (37). We found that many domains of this putative protein product displayed homology to invasin in Yersinia spp. and A/E proteins (Fig. 1A; Table 1). Many of the functions ascribed to the recognized domains of RatA involve interactions with host cells. Additionally, the SignalP database detected a signal sequence for secretion (38), and STRING software predicted strong gene association between sinH, sinI, and ratA in CFT073 (39). We therefore propose that the ratA-like locus in CFT073 be considered part of the putative intimin-like adhesin (ila) locus along with sinH-sinI, as others have suggested in Yersinia spp. (18).
When using cluster analyses on PATRIC (40), we found that the SinH-like protein in CFT073 was more similar to that of SivH in Salmonella enterica (72.6% to 78.6%) than are other UPEC strains, such as UTI89 and 536 (71% and 72%, respectively, to S. enterica) (Fig. 1B). The amino acid sequence similarity between CFT073 SinH and homologous proteins in closely related strains are visually represented in Fig. S1A in the supplemental material. These differences are most evident in areas with repeated domains (Fig. S1A). The amino acid sequence of SinI in CFT073 has only a 45% to 66.9% similarity to Salmonella enterica serovars. CFT073 RatA interestingly displayed homology to both RatA and RatB proteins in Salmonella enterica serovar Typhimurium (42% and 43%, respectively). This may suggest that CFT073 originally acquired RatB as well, during horizontal gene transfer.
To further investigate the potential genetic lineage of the ila locus among UPEC strains, we utilized PATRIC to perform amino acid comparisons of UPEC type strains 536, F11, ABU 83972, UTI89, and CFT073 (40). All UPEC strains exhibited >95% similarity throughout the intimin-like locus (Fig. 1C). ABU 83972, a strain that causes asymptomatic bacteriuria, was most similar to CFT073, with >99% similarity to all three predicted proteins (Fig. 1C). We also compared sequences of known uropathogens representing other bacterial species. The SinI and RatA predicted proteins were only identified in a single species, Citrobacter freundii, with low (<40%) similarity; however, SinH was found in three other uropathogens, again with low similarity (Fig. S1B). Thus, the genes of this ila genetic island do not seem to be highly conserved among most UTI pathogens.
We found that sinH is predicted to function in an operon with downstream gene sinI, but ratA may be transcribed separately according to predictive promoter analyses that detected two independent transcription start sites upstream of sinH and ratA independently. To understand the operon structure within the ila locus, we used reverse transcription PCR (RT-PCR) on CFT073 cDNA with primers spanning the junctions between loci. As predicted, we found that sinI and sinH were cotranscribed, as demonstrated by the expected size product amplified by primer set A on CFT073 cDNA (Fig. S2). Primer set B, however, spanning the sinI-ratA junction, did not amplify from a cDNA template, indicating that these two genes are not cotranscribed under these conditions (Fig. S2). Although ratA is not transcribed in an operon with sinH and sinI, the locus seems to have been horizontally acquired as a genomic island, as seen in Salmonella and Shiga toxin-producing E. coli (29).
The intimin-like locus is present in recently isolated UPEC and is expressed during late-stationary-phase growth.
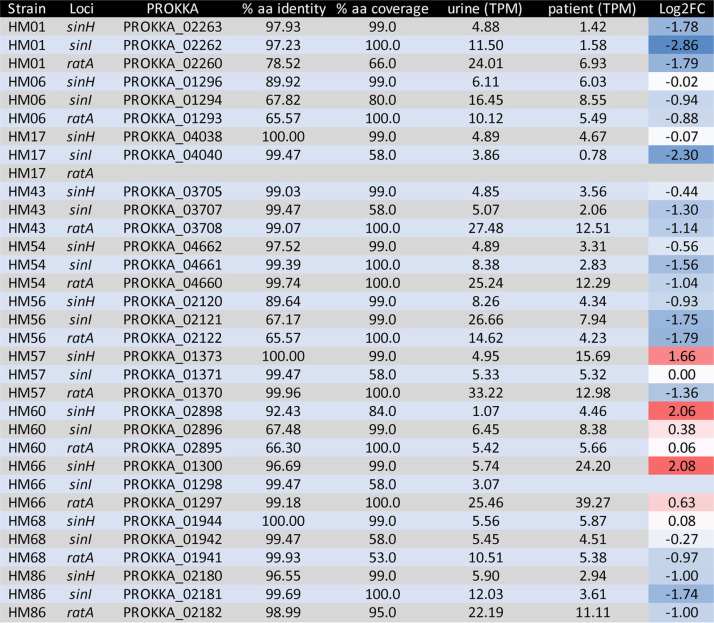
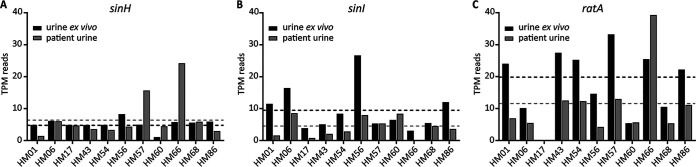
Previously, our laboratory analyzed gene expression data from RNA isolated from 14 UPEC isolates collected and immediately stabilized in the urine of patients with active UTI (41). To better understand the temporal and environmental cues triggering expression of the ila locus, we mined this expression database. The ila locus was found in 11/14 (79%) of the clinical isolates; among these, ratA was missing from strain HM17 (Table 2). Overall, SinH was the most conserved and SinI was the most variable in amino acid identity and coverage among the collection of clinical UPEC isolates. Of the three genes in this locus, sinH was the only gene more highly expressed in the patient-collected sample, as a mean of all isolates, in comparison to expression when cultured in human urine ex vivo (Fig. 2A). The mean is skewed by two strains that had increased expression: isolates HM66 and HM57 had the most transcripts per million (TPM) from patient isolate samples (Table 2). HM60 and HM66 also had high log2 fold change (FC) expression when patient samples were compared to urine, suggesting upregulation during human UTI (Table 2). Interestingly, the entire ila locus of HM60 also displayed very low amino acid identity to SinH, SinI, and RatA of CFT073, with 92.4%, 62.5%, and 66.3% identity, respectively (Table 2). As an average, sinI expression was higher during growth in pooled human urine ex vivo than in vivo (Fig. 2B). The only clinical isolate to have higher expression of sinI in patient samples was HM60 (Fig. 2B). ratA was collectively, in both patient and urine samples, the most highly expressed gene of the ila locus (Fig. 2C). This could be due to a lack of translation efficiency observed with large transcripts and may not necessarily indicate importance during infection. HM66 had both the highest raw TPM reads and the largest log2 FC (Table 2). In general, the expression of the ila locus was relatively low under both conditions, considering that the average gene TPM fell between 5 and 20 reads. This is not surprising, considering that previous studies have shown that the majority of bacterial transcripts during infection are associated with ribosomal and protein synthesis machinery to promote rapid growth of UPEC in vivo (41).
TABLE 2.
In vitro and in vivo expression of sinH, sinI, and ratA in strains isolated from human UTIa
Expression of sinH, sinI, and ratA in 11 UPEC clinical isolates was measured in transcripts per million (TPM), with a higher value indicating greater expression. The table is sorted by genome and then gene locus, given in columns 1 and 2, respectively. Columns 4 and 5 show the percent amino acid similarity of the clinical strain compared to the CFT073 sequence, while columns 6 and 7 indicate the gene expression in vitro (6) and in vivo (7). Column 8 provides the log2 fold change (FC) of expression between in vivo and in vitro (patient/urine), with positive values indicating greater expression in human patients (red) and negative values indicating greater expression in human urine ex vivo (blue). The values are color coded via gradient scale; thus, the darker the red, the more intense the upregulation in patient samples, and the red transitions toward white as the values approach zero, indicative of equal expression in vivo and in vitro.
FIG 2.
In clinical UPEC isolates, sinH has higher expression in patient samples, while ratA is increased in human urine ex vivo. Clinical UPEC isolates were collected from patients with active UTI. Patients urinated directly into specimen jars containing RNAProtect to preserve bacterial transcripts. These same UPEC isolates were cultured in human urine ex vivo for RNA isolation. Transcripts per million (TPM) are plotted for both in vivo (gray bars) and ex vivo (black bars) samples collected for each UPEC clinical isolate. The mean values are indicated by similarly colored dashed lines. The reads aligning to sinH (A), sinI (B), and ratA (C) are graphed.
To further assess the role of the ila locus in CFT073, we generated three mutants (ΔsinH, ΔsinI, ΔratA) as well as an operon deletion mutant (ΔsinH-sinI). We began by testing the ability of the mutant constructs to grow in vitro at 37°C under aerated conditions. No growth defect was observed for any of the mutants compared to WT CFT073 in either LB medium or filter-sterilized pooled human urine (Fig. S3A and B). The lack of phenotype by the mutant constructs demonstrates that these genes are not required for in vitro growth in either LB medium or human urine. In fact, intimin-like mutants outperformed WT CFT073 during growth in human urine (Fig. S3B).
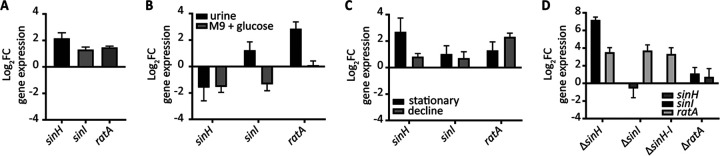
Virulence factor expression can have a large energetic cost, and, as a consequence, these genes are generally tightly regulated and expressed only under specific conditions. For example, surface adhesins and pili are typically optimally expressed under static culture conditions (42). To this point, we sought to investigate the optimal expression conditions of the ila locus in CFT073 via quantitative PCR (qPCR). We first cultured the WT strain under both static and aerated conditions in LB medium at 37°C to mid-log phase and found that all three genes were upregulated during static culture conditions (Fig. 3A). Next, we sought to determine the optimal growth medium by comparing LB medium, pooled human urine, and M9 minimal medium containing 0.4% glucose. We measured the relative expression of sinH, sinI, and ratA in these different growth media during mid-log phase under static culture conditions. When normalized to growth in LB medium, the expression of sinH and sinI was decreased in M9 minimal medium containing glucose and either increased (sinI) or decreased (sinH) in human urine (Fig. 3B). Expression of ratA was upregulated in human urine and unchanged in M9 (Fig. 3B). Similar trends were observed under aerated conditions (Fig. S4A). To quantify the temporal expression of the ila locus during bacterial growth phases, we measured gene expression during mid-log- and stationary-phase aerated cultures and static decline-phase cultures (43). Decline phase was achieved when stationary cultures (overnight with aeration at 37°C) were transferred to room temperature on the benchtop for 24 h before sampling. All samples were cultured in LB medium and normalized to mid-log expression in CFT073 cultured statically (Fig. 3C) or with shaking (Fig. S4B). When compared to static cultures, the entire ila locus was upregulated in both stationary- and decline-phase cultures (Fig. 3C). sinH expression was highest during stationary phase, and ratA was highest during decline-phase culture (Fig. 3C). Similar trends were observed in aerated LB cultures, apart from sinH, which was downregulated during stationary growth (Fig. S4B). Finally, we measured the expression of ila genes in the mutant constructs, to determine any potential compensatory regulatory mechanisms. Interestingly, both sinH and sinI single mutants and the sinH-sinI double mutant demonstrated upregulation of the ratA locus when normalized to WT CFT073 expression (Fig. 3D; Fig. S4C). However, the inverse was not true; the ratA mutant did not have altered expression of the sinH-sinI locus (Fig. 3D; Fig. S4C). Since it was previously determined that sinI and sinH are cotranscribed (Fig. S2), it is important to note that the ΔsinH strain had increased sinI expression; in contrast, the ΔsinI strain did not have altered sinH expression compared to the WT (Fig. 3D; Fig. S4C). Upregulation of sinI in the absence of sinH could be due simply to polar effects from the mutation on downstream genes, or it could indicate a regulatory mechanism suggesting cooperation within genes and gene products in the intimin-like locus, as STRING software predicted (39). Taken together, the results demonstrate that the ila locus is most highly expressed in stationary decline phase, under static culture conditions with amino acid carbon sources (LB medium or human urine). To assess the function of these gene products under optimal expression, we employed the decline-phase culture model (overnight at 37°C with aeration and then transferred to room temperature on the benchtop for 24 h) in many subsequent experiments in this study.
FIG 3.
Coordinated upregulation of the intimin-like locus in static- and late-stationary-phase cultures. WT CFT073 was cultured under specific in vitro conditions to isolate RNA for the resulting qPCR to determine expression levels of sinH, sinI, and ratA. The comparative threshold cycle (CT) method was used to determine the relative log2 fold change of each strain under specified conditions. (A) Bacteria were cultured to mid-log phase in LB medium either statically or with aeration at 37°C. Static conditions were normalized to shaking aeration; bars represent the mean of 10 biological replicates. Error bars represent the standard error of the mean (SEM). (B) Growth of CFT073 in human urine (black bars) or M9 medium plus 0.4% glucose (gray bars) was normalized to that in LB. Growth was conducted to mid-log phase under static conditions at 37°C. Bars represent the mean of five biological replicates, and error bars represent the SEM. (C) Growth under either stationary conditions (black bars) or cultured at 37°C with aeration overnight and then another overnight incubation on the benchtop (gray bars) was normalized to mid-log cultures under static conditions. Bars represent the mean of five biological replicates, and error bars represent the SEM. (D) Mutant strains were cultured in the 48-h culture model described previously and normalized to WT CFT073 under static conditions. The expression levels of sinH (dark gray), sinI (black), and ratA (light gray) are indicated in each strain, with bar height representing the mean and error bars representing the SEM.
Deletion of intimin-like genes has no effect on biofilm formation or motility but may influence type 1 fimbria expression.
We hypothesized that these genes could function in biofilm formation in UPEC because the intimin-like proteins are predicted to function as adhesins (18). The ability to form biofilms has been proposed to be advantageous for UPEC during UTI, especially in the context of cystitis and catheter-associated infections (44). To determine if intimin-like genes play a role in bacterium-bacterium adhesion or adherence to abiotic surfaces, we measured the ability of ila mutants to form biofilms in LB medium and human urine using a static microtiter plate assay. The minimal biofilms formed by CFT073 in human urine were extremely consistent, displaying no discrepancies in biofilm formation for any mutant strains at either 24 h or 48 h (Fig. S5A). Similarly, in LB medium, there were no differences observed in biofilm formation (Fig. S5B). Since CFT073 is a relatively poor biofilm former, we also assessed potential surface changes by measuring curli expression, utilizing agar containing Congo red. Strains were incubated at 30°C for 72 h and analyzed for either a red or white appearance indicating a curli-positive or curli-negative phenotype, respectively (45). The ΔsinI strain was observed to have a red ring just inside the leading colony ruffled edge (positive for curli expression), while all other mutants looked very similar to the WT strain (Fig. S5C). Thus, while we expect that intimin-like proteins are surface expressed, the loss of intimin-like genes do not appear to alter to biofilm formation in CFT073.
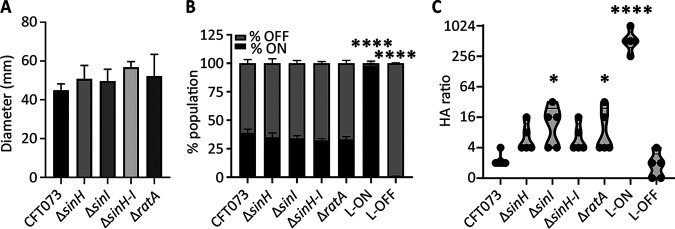
Motility and adhesion typically have an inverse relationship due to the presence of type 1 fimbriae on the surface of the cell, which antagonize the function and expression of flagella used for motility (46, 47). We suspected that the presence of SinH, SinI, and RatA may also have an inverse relationship with motility because of their putative role in intimate interactions with host cells. To test the swimming ability of the mutant strains, we stabbed bacteria into the center of 0.25% soft agar plates and measured the diameters of their motility after incubation at 30°C. The mutants displayed swimming motility very similar to that of WT CFT073, with the operon mutant (ΔsinH-sinI) demonstrating a nonsignificant increase in motility (Fig. 4A).
FIG 4.
Intimin-like genes may have cross talk with type 1 fimbriae. (A) CFT073 and intimin-like adhesin mutants were stabbed into motility agar and then incubated at 30°C for 16 h. The average diameter (mm) of the swim radius from three independent trials is represented by bars, and the error bars represent the SEM. (B) The invertible element assay was performed on WT, intimin-like mutants, CFT Fim L-ON, and Fim L-OFF cultured statically for 18 h at 37°C. ImageJ software was used to quantify the intensity of Fim-ON and Fim-OFF sized bands. The mean of three independent trials of both the percentage ON (black) and the percentage OFF (gray) of the total population are displayed with the SEM. (C) Bacterial strains were cultured for 72 h statically at 37°C. Serial 1:2 dilutions of culture were coincubated with erythrocytes, mixed, and observed after 1 h. The last titer detected with positive hemagglutination is recorded for each strain, and each biological replicate is indicated by a point. The violin plot width indicates the distribution of data, and the horizontal line indicates the median. If significant, P values determined by one-way analysis (ANOVA) with Dunnett’s multiple-comparison test, compared to WT CFT073, are indicated (*, P < 0.05; ****, P < 0.0001).
Considering previous studies describing the competition of surface-expressed proteins with one another (46), we hypothesized that the type 1 fimbriae may be differentially expressed in these strains. We utilized both invertible element and hemagglutination assays to assess the contributions of type 1 fimbriae in an intimin-null background. In E. coli, expression of type 1 fimbriae is controlled by an invertible element in the fim promoter, with the orientation controlled by several recombinases. The ON/OFF orientation can be detected by a PCR assay (48). Bacterial cultures were incubated statically in LB medium overnight, and the orientation of the fim invertible element was determined utilizing PCR. All intimin-like mutants displayed ratios of Fim-ON to -OFF populations similar to those of the WT strain CFT073 (Fig. 4B). Interestingly, fimS was in the OFF orientation for both the WT and the ila mutants following growth for 48 h at 37°C with aeration and subsequent static growth on the benchtop for 24 h (Fig. S6A).
The ability of CFT073 to agglutinate erythrocytes is largely dependent on the type 1 fimbriae, which are mannose-sensitive adhesins, although there are other surface structures that are mannose resistant (e.g., P fimbriae) and can also contribute to hemagglutination (49). To determine whether the ila locus is associated with hemagglutination (HA) ability, we tested our mutant strains with guinea pig erythrocytes that lack the P fimbria receptor. These assays were conducted using decline-phase cultures, when ila genes were optimally expressed. The exception to this is sinH expression, which was highest during stationary phase and could therefore produce the weaker phenotypes observed with ΔsinH and ΔsinH-sinI strains. The bacterial cultures were serially diluted in phosphate-buffered saline (PBS) and then mixed with erythrocytes; thus, the higher the HA titer (up to 1:1,024), the lower the bacterial CFU required to produce the phenotype. The CFT073 fim L-ON and -OFF constructs are positive and negative controls, respectively (48). WT CFT073 demonstrated similar levels of HA as L-OFF negative controls (Fig. 4C). The intimin-like mutants all had a slightly higher HA titer, which was significant in ΔsinI and ΔratA mutants compared to the WT (Fig. 4C). Additionally, we constructed ila mutants in both CFT073 fim L-ON and L-OFF backgrounds. In these strains, the mutants no longer behave significantly differently than the respective parental strains (Fig. S6B and C). However, more variation is observed in the L-OFF background (Fig. S6C), which could suggest that the loss of ila has effects on other, nonmajor hemagglutinins, which would explain the very subtle phenotypes observed in WT assays (Fig. 4C), as the type 1 fimbriae are known to be the dominant feature contributing to this phenotype.
Intimin-like adhesins mediate host bladder cell interactions in vitro.
The two best characterized fimbriae in UPEC are type 1 (fim) and P (pap) fimbriae, which are utilized for colonizing the bladder and kidneys, respectively (50). Interactions between bacterial adhesins and host cells facilitate phenotypes such as adherence and attachment, invasion of host cells, immune cell interactions, and bacterially mediated cell death. To assess the role of intimin-like adhesins with host epithelial cells, we performed a series of assays to quantify interactions with both bladder and kidney cell lines in vitro. Compared to WT CFT073, all intimin-like mutants displayed significantly increased adherence, ranging from 185% to 255% of the level of wild-type adherence to bladder cells in vitro (Fig. 5A). Although a slight increase in adherence to kidney cells was also observed, these differences were not significant (Fig. 5B). When investigating the ability of UPEC to internalize into T24 bladder cells in culture using the gentamicin protection assay, we observed that the absence of intimin-like genes resulted in a significant loss of intracellular CFU (Fig. 5C). This is in direct contrast to an observed increase in cell association (Fig. 5A). We did test the sensitivity of the ila mutants to gentamicin and found no differences from that of the WT under assay conditions, all strains being fully sensitive. However, no decrease in intracellular CFU was observed using the kidney cell line (Fig. 5D). Taken together, these results suggest that the ila may hinder cell adherence mediated by type 1 or P fimbriae but ultimately contribute to the invasion of cultured bladder cells after adherence. In fact, we were able to increase the invasion ability of both WT CFT073 and the ΔsinH strain by providing a copy of sinH on the arabinose-inducible pBAD vector (Fig. S7A). Additionally, when nonpathogenic E. coli K-12 MG1655 was given a copy of CFT073 sinH, intracellular invasion into bladder cells was increased 287% to 300% compared to empty vector using two independent constructs (Fig. S7B).
FIG 5.
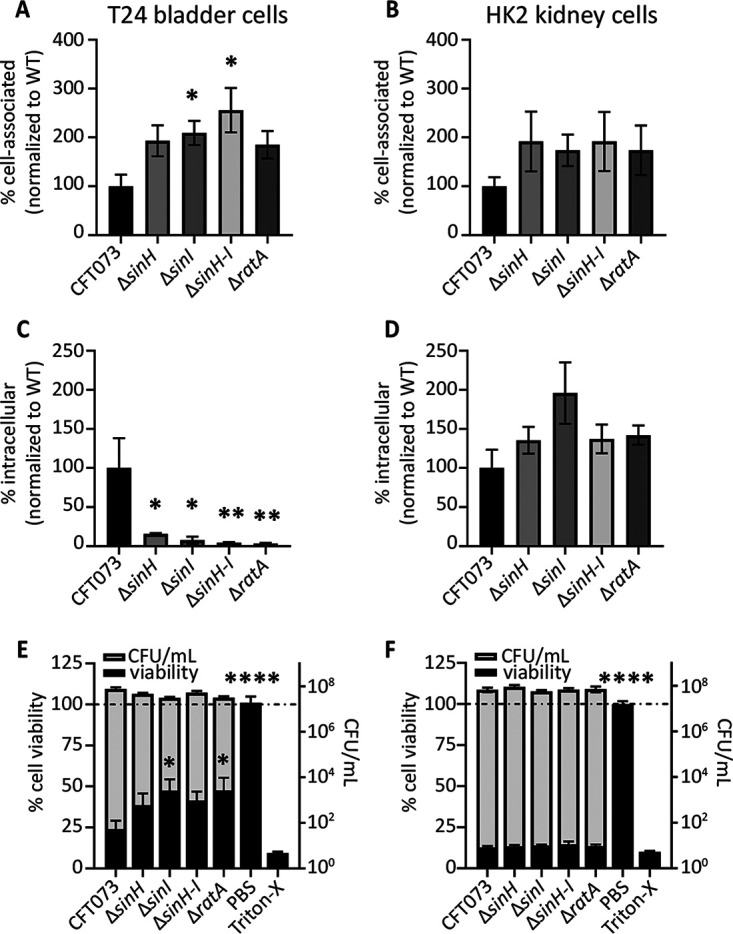
Intimin-like genes play a role in host bladder cell interactions in vitro but not with kidney cells. UPEC host cell association was determined in both T24 bladder (A) and HK2 kidney (B) cell lines. UPEC was added to cells at an MOI of 1:50 and incubated for 1 h before enumeration of the attached CFU. Data are the mean of five biological replicates; all data are normalized to WT adherence. Error bars represent the SEM. (C and D) UPEC cell internalization was determined in both T24 bladder (C) and HK2 kidney (D) cell lines. After 1 h of coculture of bacteria with host cells, the media were supplemented with gentamicin to kill extracellular bacteria. Bars represent the mean of internalized UPEC CFU burden from five biological replicates, with the SEM shown. (E and F) UPEC host cell-killing ability was determined in both T24 bladder (E) and HK2 kidney (F) cell lines. UPEC was cocultured with host cells for 5 h before determination of the number of UPEC CFU from cell supernatants and host viability as measured by the MTT assay. The left y axis plots the mean cell viability as determined by A570 values via MTT, normalized to RPMI 1640 only as an internal control (black bars). The right y axis plots mean CFU/mL as determined by the collected cell supernatants (gray bars). Both data sets display the SEM. If significant, P values determined by one-way ANOVA with Dunnett’s multiple-comparison test, compared to WT CFT073, are indicated (*, P < 0.05; **, P < 0.01; ****, P < 0.0001).
To further assess the effect of the ila locus on host cell populations, we performed an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] cell death assay to quantify the number of viable cells after 5 h of coculture with UPEC strains. Cell culture medium with no bacteria (only PBS) served as a negative control, and 0.4% Triton-X was used as a positive cell death control in these assays. Bladder cells had an average of 76% cell death (24% viability) after treatment with WT CFT073; however, cell death was significantly reduced in the absence of the intimin-like genes sinI and ratA, ranging from 52.5% to 61.5% (Fig. 5E). UPEC infection of kidney cell lines induced maximum detectable cell death in this assay determined by positive controls, and no differences were observed between the WT and mutant constructs (Fig. 5F). It is known that CFT073 is readily able to kill kidney cells in culture, mainly through hemolysin-mediated mechanisms (34), and this could explain the lack of differences observed. Collectively, the observed differences demonstrated cell type-specific interactions of the intimin-like adhesins of CFT073. Our findings indicate that the ila protein products decrease eukaryotic cell viability in bladder cells but not in kidney cells.
SinH is a CFT073 fitness factor in the murine model of UTI.
CFT073 sinH was previously identified as a potential fitness factor during bladder colonization in our transposon insertion sequencing (Tn-seq) studies (51). To verify these results, we conducted a cochallenge experiment of WT CFT073 against the ΔsinH mutant in CBA/J mice at three time points: 24 h, 48 h, and 72 h (Fig. 6A). Mice were transurethrally inoculated with 108 CFU containing equal numbers of the ΔsinH mutant and the WT strain. The outputs of each strain were compared by calculating a competitive index (CI). A negative log10 CI indicates that the mutant strain was outcompeted by the WT strain during infection. At 24 h, 48 h, and 72 h postinoculation, the ΔsinH mutant had a significant colonization defect during infection in the urine and bladder (Fig. 6A). Additionally, the 48-h time point showed a significant defect of the mutant in kidney colonization (Fig. 6A). The output CFU for each organ site at each time point is comparable to that of other studies conducted in this mouse model; clearance of infection is indicated by points on the dashed line at the limit of detection in Fig. S8.
FIG 6.
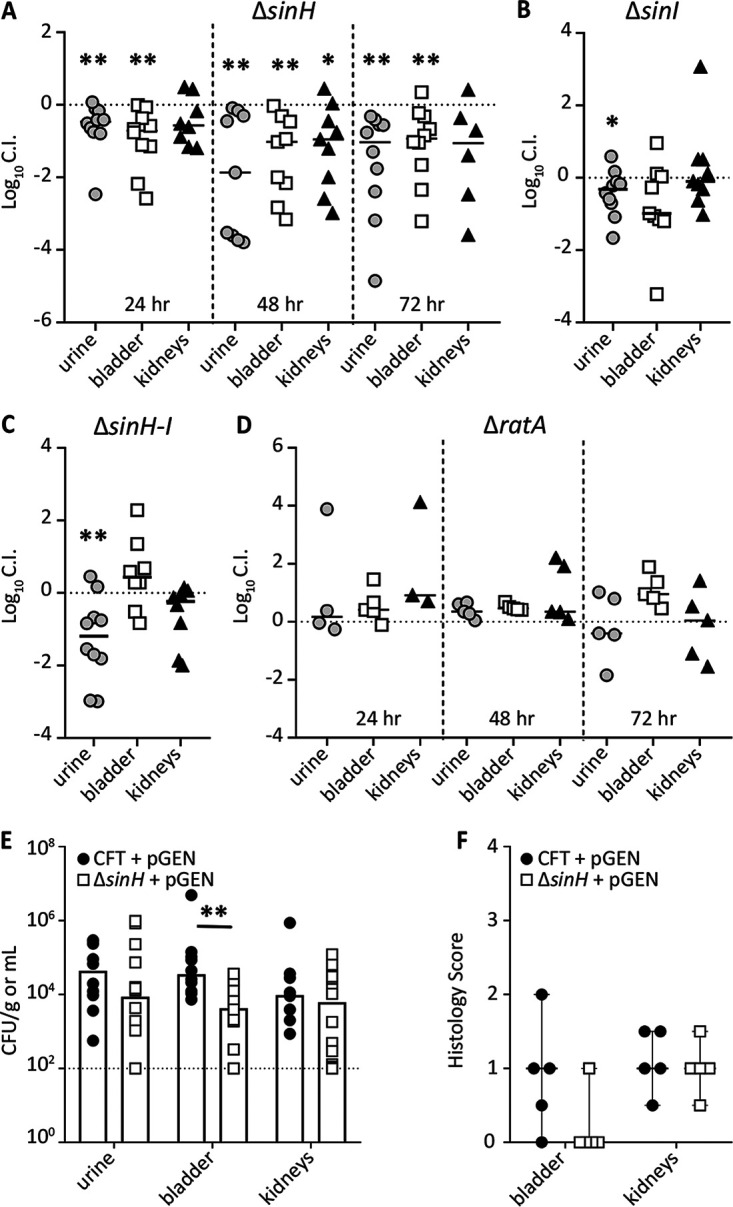
sinH-like adhesin serves as a fitness factor during bladder infection. (A to D) CFT073 and ΔsinH (A), ΔsinI (B), ΔsinH-sinI (C), and ΔratA (D) isogenic mutants were transurethrally inoculated at 2 × 108 CFU, which consisted of a 1:1 mixture of WT CFT073 and mutant, into CBA/J mice. Urine, bladder, and kidneys were collected, and homogenates were plated on LB agar with and without kanamycin to determine each contributing bacterial burden of the mutant and WT, respectively. Each data point represents the log10 competitive index (CI) calculated for individual mouse organs. Solid horizontal black lines represent the median. Horizontal dashed lines represent a competitive index of 0, which indicates that the WT and mutant have equal fitness. If significant, P values determined by the Wilcoxon signed rank test are indicated by asterisks (*, P < 0.05; **, P < 0.01). Infections were terminated at 48 h unless noted otherwise. (E) Five mice were transurethrally inoculated with either the WT or the ΔsinH mutant carrying the empty pGEN-MCS plasmid for 48 h. The total CFU output per gram of tissue (or milliliter of urine) is plotted for each individual, with the bar indicating the median bacterial burden. If significant, P values determined by the Mann-Whitney test are indicated by asterisks (**, P < 0.01). (F) Corresponding histopathology inflammation scores were determined for bladders and kidneys of individually challenged mice (n = 5). Each dot represents a single organ, and the horizontal line represents the median with range.
We also measured the contribution of SinI, SinHI, and RatA in colonization of the urinary tract by testing these mutants for fitness at 48 h. Compared to WT CFT073, the ΔsinI mutant displayed a slight defect in the urine and bladder (Fig. 6B), although this defect was statistically significant only in the urine. Deletion of the entire operon resulted in the mutant strain displaying a significant fitness defect in the urine, but no defect was observed in the bladder (Fig. 6C). The kidney showed no competitive advantage for either the mutant strain or WT CFT073 (Fig. 6C). The individual outputs in CFU/gram of tissue are shown in Fig. S8. Since ratA produces a large protein that is not encoded by the sinH-sinI operon, we conducted a similar time point cochallenge experiment as done with the ΔsinH mutant. At both 24 h and 48 h, the ΔratA strain had no fitness defect in any of the three organ sites (Fig. 6D). However, at 72 h, a fitness advantage in the bladder was observed but was not significant (Fig. 6D).
Because the 48-h infection model showed a significant defect during the ΔsinH mutant cochallenge in all three organ sites, we conducted an additional in vivo experiment using this time point to assess an independent model of infection. CBA/J mice were transurethrally inoculated with either WT CFT073 or the ΔsinH mutant, both of which expressed the empty vector pGEN plasmid. The median CFU value of the ΔsinH mutant plus pGEN in the urine was 5-fold lower than that of the WT, but this difference was not significant (Fig. 6E). However, in the bladders, the mutant bacterial burden was nearly a log lower (P = 0.0307) (Fig. 6E). The kidneys had no difference in colonization. Collectively, these data demonstrate that loss of SinH leads to bladder colonization defects, as detected by a reduced CFU burden, during murine UTI. We then generated a pGEN vector containing a WT copy of sinH under the control of its native promoter and provided it to the ΔsinH mutant. While bladder colonization was partially restored with the complemented mutant strain, these results were not significant (Fig. S9A).
We also performed histopathology of bladder and kidney tissue of these mice, because the cell culture experiments showed that ila gene products affect cell viability (Fig. 5) and it is known that EHEC eaeA contributes to inflammation (52). Blinded histopathology scoring of hematoxylin and eosin (H&E)-stained sections demonstrated that inflammation scores in the bladders of mice infected with the ΔsinH mutant were lower than those for WT infection (Fig. 6F). No difference in immune recruitment was observed in the kidneys (Fig. 6F). These data match closely to the host cell interactions observed in bladder, but not kidney, cell lines in vitro (Fig. 5). Unfortunately, the histopathology scores did not improve in the bladders of mice infected with the complemented strain (Fig. S9B). Collectively, we determined that loss of SinH leads to a measurable colonization defect in the bladders of mice, most likely due to interactions with host cells and inflammation.
DISCUSSION
Adherence to host epithelial cells using fimbrial and nonfimbrial adhesins is critical for bacterial pathogenesis during urinary tract infection. Intimin is an adhesin used by intestinal pathogens to bind the Tir receptor expressed on epithelial cells and cause cytoskeletal changes, resulting in A/E lesions and diarrhea (16, 19, 20). Several UPEC strains contain genes annotated as “intimin-like” within their genome (53), but these have not been thoroughly investigated in the context of urinary tract infection and may play a similar role in interactions with host uroepithelial cells. Classical intimin interacts with the bacterially secreted Tir ligand (21). Although UPEC has a Tir domain containing protein TcpC, which is known to interact with host macrophages (54), there is no predicted interaction between TcpC and the ila locus. When examining the predicted protein structures of SinH, SinI, and RatA, we could not find the known sequence that facilitates the interaction between Tir and intimin (21). We therefore solely investigated the ila locus in the prototype UPEC strain CFT073.
We found that loss of sinH, sinI, and ratA had no impact on in vitro CFT073 growth in LB medium or human urine. This indicates that our observed phenotypes were likely not due to a growth defect. Interestingly, sinH and sinI were not consistently upregulated or downregulated among clinical isolates when a collection of recent UPEC strains was compared directly from patient samples with culture in urine ex vivo (41). The finding that they are not expressed equally indicates that they may have different posttranscriptional modifications, a second transcriptional start site, or the action of regulatory small RNAs or that the mRNA of sinI is less stable during active infection, among other plausible mechanisms. This contrasts with our in vitro experimental conditions that indicate that sinH and sinI are cotranscribed. Additionally, SinI is not well conserved across bacterial species, while SinH is, potentially indicating that sinH is a gene more necessary for bacterial infection than sinI. Likewise, sinH on an inducible vector alone was able to promote invasion ability in E. coli K-12, which lacks sinI. However, a strong consideration in the transcriptome data set is that only gene expression from bacteria eliminated in urine was measured (41). Bacteria eliminated in the urine may represent the majority of sequenced transcriptomes with decreased expression of these putative adhesins, since they are largely planktonic. Those attached to or intracellular in exfoliated epithelial cells could represent populations with increased expression of the putative adhesins, which would make the amount of cell shedding directly related to detectable ila transcripts in any given isolate sample. While there does seem to be a pattern surrounding sinH and sinI expression in vitro, their lack of equal expression ex vivo seems to indicate that other transcriptional modifications occur in this operon during active human UTI.
Biofilms are utilized by UPEC during infection to adhere to one another in the bladder and are a part of the intracellular life cycle with the formation of intracellular bacterial communities (5, 55), so we hypothesized that the defect of the ΔsinH mutant observed in the bladder could be due to an inability to form biofilms. However, we found that the absence of ila genes had no effect on biofilm formation in urine, and when cultured in LB medium, the genes proved to be a mild hindrance to biofilm formation. sinI acts as a biofilm regulator in Bacillus subtilis (56), but sinI in CFT073 has very little homology and appears not to behave in this manner in vitro. However, when using Congo red agar to assess curli expression, we noticed a red ring just inside the leading edge of the ΔsinI colony, which could indicate cell surface changes in this mutant. Additionally, we found that the ΔsinI and ΔratA mutants both had a significantly increased ability to hemagglutinate cells. These data together suggest that deletion of these surface-expressed structures may allow for other UPEC adhesins to decorate the outer membrane.
In concordance with this hypothesis, ila mutants displayed increased cell association coupled with reduced intracellular populations and decreased cytotoxicity, specifically in bladder cell lines. When considering that cell adherence is increased while invasion is decreased, this may suggest a stepwise role of intimin-like adhesins in the later stages of host cell interactions after type 1 fimbriae have initiated attachment to mannosylated residues on uroplakins (10, 57). Interestingly, intimin, invasins, and the type 1 fimbriae have all been determined to bind β-1 integrins (25, 26, 58). The finding of decreased intracellular populations in ila-null strains may be directly linked to decreased cell death, as intracellular bacterial communities (IBC) have been documented to egress from bladder cells in vitro, causing host cell death (59, 60). Only a very small percentage of the total UPEC population actively invades the host bladder tissue during UTI, especially in comparison to rates of invasion by gut pathogens (61), and this may explain only the moderate fitness defects seen in our ascending model of UTI. It remains possible that the function of the ila locus in CFT073 may contribute primarily to colonization in the intestine and is beneficial for UPEC bladder cell interactions during extraintestinal colonization, consequentially. Survival and persistence in the intestinal reservoir are important features of UPEC that facilitate their ability to access the urinary tract (5, 8).
In Salmonella enterica serovar Typhimurium, sivH and sivI are carried along with ratA (28), providing additional evidence for the potential relationship of these genes in CFT073 due to their homology to a preestablished genetic island. Using PCR on cDNA, we confirmed that sinH and sinI are cotranscribed as a single operon, separate from ratA. In silico analysis predicted that many domains of CFT073 RatA resemble intimin and invasins from other bacterial species. To this point, we demonstrated that all ila-null strains, including the ΔratA mutant, had increased cell association but decreased intracellular bladder cell populations in vitro following invasion assays. These data indicate that perhaps the functional role of the ila locus during UTI is more similar to that of invasin than intimin.
SinH has been proposed to facilitate secretion of other intimins via its transmembrane β-barrel structure, which is absent in other intimin subtypes (21). Thus, without SinH, other intimins would potentially have no way of accessing the host cell. This could account for the colonization defect of the ΔsinH mutant, because without SinH, SinI, RatA, and other intimin-like proteins may be detrimentally accumulating inside the cell, conferring an energetic cost to the bacterium. This is especially important when taking into consideration that CFT073 and most UPEC strains do not possess a type III secretion system (T3SS), the mechanism utilized by enteric E. coli for intimin secretion (21). EPEC and EHEC use T3SS to deliver their receptor (Tir) to allow for a contact point with the host. In contrast, UPEC can intimately adhere to the host cells (via type 1 or P fimbriae) using native epithelial host receptors. Therefore, UPEC does not require a T3SS to inject the receptor, as it already has the capability to bind intimately to the urinary epithelial cells and subsequently allow the ila gene products to function.
While eaeA encodes the most classic example of intimin, a homolog is also commonly encoded by the gene fdeC, named for factor adherence E. coli (18). A gene within the CFT073 genome was identified as fdeC and was subsequently shown to be essential for infection in the mouse bladder and kidneys, potentially due to the inability of a fdeC mutant strain to resist flushing mechanisms in the bladder (53, 62). Many uropathogenic strains carry fdeC; in fact, 99% of extraintestinal pathogenic E. coli strains carry fdeC (53), which displays similarity to both intimin and invasin genes. Mice immunized with the FdeC antigen were protected against infection in the kidneys (53). Due to the established importance of fdeC during infection, we suggest that the ila locus may have some functional redundancy masking in vivo phenotypes, since CFT073 carries both fdeC and ila. In addition to the role SinH has as a potential adhesin, it may also be involved in the host immune response through antibody-mediated immunity. Humans infected with EPEC lacking eaeA developed significantly less-robust adaptive immune responses with less antibody development (63). Considering the potential role of intimin-like adhesins during the immune response, they could be a desirable target for vaccine development.
SinH was found to be important for bladder colonization in a murine model of ascending UTI, originally detected through Tn-seq screening of a transposon ordered library (51). The ΔsinH mutant in competition with WT CFT073 displayed a significant defect in colonization of all three organ sites at 48 h, as well as a subtle but significant defect in the urine and bladder at 24 h and 72 h of infection. The ΔsinH mutant was also less able to colonize murine bladders during independent infection, suggesting an even stronger role within the niche in the absence of competition with the WT. Given intimin’s putative role in the immune response (63, 64), it was interesting that we observed reduced inflammation in the bladders of ΔsinH mutant-infected mice. Intimin also contains a C-type lectin domain, which is typically used for carbohydrate binding (21) and may correspond to the predicted cellulose-binding domain of SinI. C-type lectins are also used by phagocytes to engulf bacteria (65), which could be partially responsible for the intimin-mediated immune response during Salmonella enterica infection and similarly true for UPEC.
Collectively, these findings support that SinH, SinI, and RatA are structural invasin-like, rather than intimin-like, homologs. SinH is highly conserved across UPEC strains, and the amino acid sequence and protein structural similarities are strong in comparison to those of invasins from enteric pathogens. Our data demonstrate that this ila locus contains genes coding for surface-expressed proteins that modulate the intracellular and immune interactions of UPEC during UTI. These functions were perhaps coincidentally inherited and therefore subtle, due to their enteric origin. However, the widespread presence of these genes in other UPEC strains suggests that they are evolutionarily beneficial to uropathogens and the many stages of their infection life cycle. Future studies will provide more mechanistic insight into the molecular interactions of these bacterial proteins with host cells.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli CFT073 was isolated from the blood and urine of a patient with acute pyelonephritis (34). E. coli strains and plasmids utilized in this work are listed in Table S1 in the supplemental material. Lysogeny broth (LB), which contains 0.5 g NaCl, 5 g yeast extract, and 10 g tryptone/L, was used to routinely culture bacteria at 37°C under aerated conditions and was inoculated from single colonies. Overnight LB bacterial cultures were washed once with phosphate-buffered saline (PBS) and then inoculated 1:100 into 3 mL LB or filter-sterilized pooled human urine collected from at least four female donors. These cultures were incubated with aeration at 37°C in a Bioscreen-C automated growth curve machine (Growth Curves USA), with collection of optical density at 600 nm (OD600) readings every 15 min for 15 h.
Bioinformatics analysis.
The PATRIC database (40) was used to determine amino acid homology of the predicted SinH, SinI, and RatA proteins by utilizing the protein BLAST tool. Comparative sequence alignment and phylogenetic trees were esthetic outputs of this analysis. PHYRE2 (30) and I-TASSER (31) online softwares were used to determine protein domain homology. SignalP-5.0 (38) and STRING software (39) online interface were also used for analyses.
Construction of bacterial mutants and complementation vectors.
E. coli CFT073 mutants were generated using the lambda red recombinase system (66). Briefly, primers were designed to be homologous to regions on the 5′ and 3′ ends of sinH, sinI, ratA, or the operon (sinH-sinI) and used to replace the gene(s) with a kanamycin resistance cassette amplified from the pKD4 template plasmid (66). Primers used for the generation and confirmation of mutants can be found in Table S2. The lambda red fragments were generated with ∼35 bp of homology and confirmed by PCR. The PCR product was digested with DpnI and then electroporated into CFT073 containing the pKD46 recombinase-containing plasmid. Potential mutant colonies were screened using primers flanking the gene(s) of interest by PCR amplification and comparing the gene product size to the fragment generated by WT CFT073. Mutant strains were maintained in 25 μg/mL kanamycin.
To perform in vitro and in vivo complementation experiments, plasmids were designed by PCR amplifying sinH from WT CFT073 genomic DNA with flanking ends for backbone overlap (pGEN_MCS or pBAD), using Easy-A high-fidelity polymerase (Agilent). Vectors were generated using the NEBuilder HiFi DNA assembly cloning kit (New England BioLabs). Oligonucleotides used for complementation construct design are listed in Table S2. Gel-purified PCR products were assembled according to the manufacturer’s instructions, and the enzymatic reaction product was incubated at 50°C for 1 h. The NEBuilder reaction product was dialyzed for 30 min using a VSWF 0.025-μm filter disk. The cleaned-up DNA was electroporated into E. coli TOP10 cells. The resulting plasmids were verified by restriction digest, and then 1 μL of DNA was transformed into electrocompetent CFT073, K-12 MG1655, or ila mutants.
RNA isolation and quantitative RT-PCR.
Overnight bacterial cultures incubated in LB broth were back-diluted 1:100 into the appropriate medium type and cultured under static or aerated conditions to an OD600 of 0.5 to 0.6 (mid-log) or of 0.1 to 0.2 for human urine samples. Stationary cultures were collected at either an OD600 of 1.0 or after overnight incubation if an OD600 of 1.0 could not be achieved. A 2:1 (vol/vol) ratio of RNA Protect (Qiagen) was added to 300 to 500 μL of bacterial culture and incubated at room temperature for 5 min. Samples were then pelleted and frozen at −80°C.
RNA isolation was performed with the RNeasy minikit (Qiagen) after treatment with lysozyme and proteinase K, and RNA samples were then DNase treated using a Turbo DNA-free kit (Invitrogen). The absence of DNA contamination was confirmed via PCR with CFT073 genomic DNA (gDNA) primers. cDNA synthesis was performed with an iScript kit (Bio-Rad) on an equal amount of RNA for each sample, roughly 1 μg total. The resulting cDNA was diluted 1:50 in water and utilized for real-time quantitative reverse transcription PCR (qRT-PCR) in a QuantStudio 3 PCR system (Applied Biosystem) with SYBRGreen PowerUp reagent (Invitrogen). Primers designed to amplify internal regions for either sinH, sinI, ratA, or control gene gapA were utilized. Samples were normalized to gapA transcript levels by subtracting the comparative threshold cycle (CT) values of gapA from the values of monitored genes normalized to conditions indicated by the ΔΔCT method. Finally, data were presented as the log2 fold change using the formulation log2(2−ΔΔCT).
Biofilm assays.
The ability of WT CFT073 and mutant strains to form biofilms was assessed by adherence to 24-well plates. Bacteria were cultured from a single colony at 37°C under aerated conditions, and then 108 CFU were inoculated into 1 mL LB or filter-sterilized pooled human urine in 24-well plates. Cultures used to inoculate urine were pelleted, washed, and resuspended in urine before inoculation. Bacteria were allowed to form biofilms for 24 h under 37°C static conditions. Then, all wells were aspirated, washed twice with 1 mL PBS, and stained with 500 μL 0.1% crystal violet for 10 min. Biofilms were gently washed with PBS. The crystal violet was dissolved in 500 μL 100% ethanol. Biofilm formation was assessed by measuring the A570 of each well. Experiments were repeated in biological triplicates, with technical duplicates within each trial.
Congo red agar was made as previously described (67). Five microliters of overnight culture was spotted on Congo red agar and incubated at 30°C for 72 h before images were taken.
Motility assays.
The swimming motility of WT CFT073 and mutant strains was measured using semisoft agar containing 10 g tryptone, 5 g NaCl, and 2.5 g agar/L. Bacterial cultures were incubated with aeration at 37°C overnight in LB and then normalized to an OD600 of 1.0 and resuspended in 10 mM HEPES (pH 7.4). Resuspended cultures were then stabbed into semisoft agar using a sterile inoculating needle and incubated for 16 h at 30°C. Experiments were repeated in biological triplicates, each with technical duplicates.
Quantification of type 1 fimbriae.
For the invertible element assay (48), bacteria were cultured statically overnight at 37°C and normalized to an OD600 of 0.5 in water. PCR was performed to amplify the invertible region of fimS using primers listed in Table S2. Five hundred nanograms of PCR product was digested with SnaB1 enzyme (NEB) overnight at 37°C. Digestions were run on 3% agarose and electrophoresed at 100 V to visualize separation of digested bands. fimS in the ON orientation yields bands at 403 bp and 198 bp, while the OFF orientation yields 440-bp and 161-bp bands. Bands were visualized and quantified using the Bio-Rad software.
For the hemagglutination assay, bacteria were cultured statically in LB at 37°C for 72 h (68). Cells were washed and resuspended in 1× PBS to be serially diluted 1:2 throughout the rows of a round-bottom 96-well plate. Guinea pig erythrocytes (catalog no. IGPRBC10ML-33782) were washed, resuspended at 3% (vol/vol), diluted in PBS, and then added to each well of the plate. Bacteria-and-erythrocyte coculture was gently mixed and allowed to settle for 1 h. To assess the role of the type 1 fimbriae, 50 mM mannose was added as a negative control as previously described (69).
Host cell interaction assays.
T24 bladder cells (ATCC HTB-4) and HK2 kidney cells (ATCC CRL-2190) were maintained in RPMI 1640 with l-glutamine and 25 mM HEPES (Corning) with 10% fetal bovine serum (FBS) (Corning) and antibiotics (Sigma 10 mg/mL) at 37°C with 5% CO2. Cells were seeded into treated 24-well or 96-well plates and allowed to grow to confluence for assays.
Cell interactions were assessed by adding a multiplicity of infection (MOI) of 100 CFU per host cell. Cell association was determined after 1 h of incubation with UPEC and host cells, in which cells were washed and then lysed in 0.4% Triton-X in PBS. CFU were enumerated by drip plating on LB agar. Similarly, internalization assays were conducted after 1 h of incubation with UPEC, followed by 1 h of incubation with RPMI 1640 supplemented with 100 μg/mL gentamicin to kill extracellular bacteria. Cells were then washed and lysed, and lysates were serially diluted to enumerate bacterial CFU. The procedure was modified from reference 70.
Cell viability after coincubation with CFT073 was determined utilizing the cell proliferation kit I (Millipore Sigma catalog no. 11465007001). A total of 107 CFU of CFT073 strains was added to confluent monolayers of host cells and incubated at 37°C with 5% CO2 for 5 h. Then, medium was replaced with RPMI 1640 containing penicillin (100 μg/mL), streptomycin (100 μg/mL), and gentamicin (100 μg/mL) for 2 h. Cells were washed, and 100 μL of MTT-containing RPMI 1640 was added to each well, according to the kit protocol. After 2 to 4 h of incubation, 100 μL of solubilization reagent was added, and ultimately, the A570 was measured to determine cellular respiration. RPMI 1640 with PBS alone served as the positive control, and 0.4% Triton-X served as the negative control for cell viability.
Murine model of ascending UTI.
The individual fitness of each mutant was assessed in the CBA/J mouse model of ascending UTI (71). WT CFT073 and mutant strains were cultured overnight in LB broth under 37°C aerated conditions from a single colony. A bacterial suspension for inoculation was created with a 1:1 ratio of WT CFT073 to kanamycin-resistant mutants (ΔsinH, ΔsinI, ΔratA, or ΔsinH-sinI) in PBS. CBA/J female mice that were 6 to 8 weeks old and 20 to 22 g (Jackson Laboratories) were anesthetized with ketamine/xylazine and then infected transurethrally with 50 μL containing 2 × 108 CFU per mouse using a sterile polyethylene catheter (inside diameter of 0.28 mm by outside diameter of 0.61 mm) connected to an infusion pump (Harvard Apparatus). Input CFU/mL was confirmed by plating spiral dilutions (Spiral Biotech) onto LB agar plates with and without antibiotics. Bladders and kidneys were aseptically removed for each mouse, weighed, and then homogenized (OMNI International) in 3 mL of 1× PBS. The homogenates were diluted and spiral plated onto LB agar plates with and without kanamycin (25 μg/mL) to determine the CFU output/g tissue for each strain per mouse. Counts were determined using QCount software (Spiral Biotech), and then the number of mutant CFU (antibiotic plates) was subtracted from the total number of CFU (plain plates) to determine the number of WT CFT073 bacteria present in the input and output. The following formula was used to calculate log10 competitive indices (CI) for each mouse:
ACKNOWLEDGMENTS
We thank Anna Sintsova for providing raw gene expression data files from her study (41). We thank Arwen Frick-Cheng, Chris Alteri, and Melanie Pearson for their critical edits and feedback on the manuscript. We acknowledge Kate Eaton for her blinded histology scoring.
This work was funded by the National Institutes of Health Public Health Service grant no. R01AI059722 and F32AI147527.
Footnotes
Supplemental material is available online only.
Contributor Information
Harry L. T. Mobley, Email: hmobley@umich.edu.
Igor E. Brodsky, University of Pennsylvania
REFERENCES
- 1.Foxman B, Barlow R, D'Arcy H, Gillespie B, Sobel JD. 2000. Urinary tract infection: self-reported incidence and associated costs. Ann Epidemiol 10:509–515. 10.1016/S1047-2797(00)00072-7. [DOI] [PubMed] [Google Scholar]
- 2.Foxman B. 2003. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Dis Mon 49:53–70. 10.1067/mda.2003.7. [DOI] [PubMed] [Google Scholar]
- 3.Stamm WE, Norrby SR. 2001. Urinary tract infections: disease panorama and challenges. J Infect Dis 183(Suppl 1):S1–S4. 10.1086/318850. [DOI] [PubMed] [Google Scholar]
- 4.Foxman B. 1990. Recurring urinary tract infection: incidence and risk factors. Am J Public Health 80:331–333. 10.2105/ajph.80.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kot B. 2019. Antibiotic resistance among uropathogenic Escherichia coli. Pol J Microbiol 68:403–415. 10.33073/pjm-2019-048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta K, Scholes D, Stamm WE. 1999. Increasing prevalence of antimicrobial resistance among uropathogens causing acute uncomplicated cystitis in women. JAMA 281:736–738. 10.1001/jama.281.8.736. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 157:1127–1129. 10.1016/S0022-5347(01)65154-1. [DOI] [PubMed] [Google Scholar]
- 9.Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. 2008. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol 46:2529–2534. 10.1128/JCM.00813-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou G, Mo WJ, Sebbel P, Min G, Neubert TA, Glockshuber R, Wu XR, Sun TT, Kong XP. 2001. Uroplakin Ia is the urothelial receptor for uropathogenic Escherichia coli: evidence from in vitro FimH binding. J Cell Sci 114:4095–4103. 10.1242/jcs.114.22.4095. [DOI] [PubMed] [Google Scholar]
- 11.Lane MC, Alteri CJ, Smith SN, Mobley HL. 2007. Expression of flagella is coincident with uropathogenic Escherichia coli ascension to the upper urinary tract. Proc Natl Acad Sci USA 104:16669–16674. 10.1073/pnas.0607898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73:7657–7668. 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brumbaugh AR, Mobley HL. 2012. Preventing urinary tract infection: progress toward an effective Escherichia coli vaccine. Expert Rev Vaccines 11:663–676. 10.1586/erv.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langermann S, Ballou WR. 2001. Vaccination utilizing the FimCH complex as a strategy to prevent Escherichia coli urinary tract infections. J Infect Dis 183(Suppl 1):S84–S86. 10.1086/318857. [DOI] [PubMed] [Google Scholar]
- 15.Spaulding CN, Hultgren SJ. 2016. Adhesive pili in UTI pathogenesis and drug development. Pathogens 5:30. 10.3390/pathogens5010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. 1997. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell 91:511–520. 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 17.Jarvis KG, Girón JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA 92:7996–8000. 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heinz E, Stubenrauch CJ, Grinter R, Croft NP, Purcell AW, Strugnell RA, Dougan G, Lithgow T. 2016. Conserved features in the structure, mechanism, and biogenesis of the inverse autotransporter protein family. Genome Biol Evol 8:1690–1705. 10.1093/gbe/evw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothbaum R, McAdams AJ, Giannella R, Partin JC. 1982. A clinicopathologic study of enterocyte-adherent Escherichia coli: a cause of protracted diarrhea in infants. Gastroenterology 83:441–454. 10.1016/S0016-5085(82)80342-9. [DOI] [PubMed] [Google Scholar]
- 20.McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA 92:1664–1668. 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Batchelor M, Prasannan S, Daniell S, Reece S, Connerton I, Bloomberg G, Dougan G, Frankel G, Matthews S. 2000. Structural basis for recognition of the translocated intimin receptor (Tir) by intimin from enteropathogenic Escherichia coli. EMBO J 19:2452–2464. 10.1093/emboj/19.11.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knutton S, Lloyd DR, McNeish AS. 1987. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect Immun 55:69–77. 10.1128/iai.55.1.69-77.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adu-Bobie J, Frankel G, Bain C, Goncalves AG, Trabulsi LR, Douce G, Knutton S, Dougan G. 1998. Detection of intimins alpha, beta, gamma, and delta, four intimin derivatives expressed by attaching and effacing microbial pathogens. J Clin Microbiol 36:662–668. 10.1128/JCM.36.3.662-668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGraw EA, Li J, Selander RK, Whittam TS. 1999. Molecular evolution and mosaic structure of alpha, beta, and gamma intimins of pathogenic Escherichia coli. Mol Biol Evol 16:12–22. 10.1093/oxfordjournals.molbev.a026032. [DOI] [PubMed] [Google Scholar]
- 25.Frankel G, Lider O, Hershkoviz R, Mould AP, Kachalsky SG, Candy DC, Cahalon L, Humphries MJ, Dougan G. 1996. The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to beta1 integrins. J Biol Chem 271:20359–20364. 10.1074/jbc.271.34.20359. [DOI] [PubMed] [Google Scholar]
- 26.Isberg RR, Leong JM. 1990. Multiple beta 1 chain integrins are receptors for invasin, a protein that promotes bacterial penetration into mammalian cells. Cell 60:861–871. 10.1016/0092-8674(90)90099-Z. [DOI] [PubMed] [Google Scholar]
- 27.Adams TM, Wentzel A, Kolmar H. 2005. Intimin-mediated export of passenger proteins requires maintenance of a translocation-competent conformation. J Bacteriol 187:522–533. 10.1128/JB.187.2.522-533.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Baumler AJ. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infect Immun 71:629–640. 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dallman T, Cross L, Bishop C, Perry N, Olesen B, Grant KA, Jenkins C. 2013. Whole genome sequencing of an unusual serotype of Shiga toxin-producing Escherichia coli. Emerg Infect Dis 19:1302–1304. 10.3201/eid1908.130016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knutton S, Baldwin T, Williams PH, McNeish AS. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 57:1290–1298. 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulett GC, Totsika M, Schaale K, Carey AJ, Sweet MJ, Schembri MA. 2013. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr Opin Microbiol 16:100–107. 10.1016/j.mib.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. 10.1128/iai.58.5.1281-1289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garofalo CK, Hooton TM, Martin SM, Stamm WE, Palermo JJ, Gordon JI, Hultgren SJ. 2007. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect Immun 75:52–60. 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamaguchi Y, Park JH, Inouye M. 2011. Toxin-antitoxin systems in bacteria and archaea. Annu Rev Genet 45:61–79. 10.1146/annurev-genet-110410-132412. [DOI] [PubMed] [Google Scholar]
- 37.Norton JP, Mulvey MA. 2012. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathog 8:e1002954. 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 39.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. 2019. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sintsova A, Frick-Cheng AE, Smith S, Pirani A, Subashchandrabose S, Snitkin ES, Mobley H. 2019. Genetically diverse uropathogenic Escherichia coli adopt a common transcriptional program in patients with UTIs. Elife 8:e49748. 10.7554/eLife.49748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas WE, Nilsson LM, Forero M, Sokurenko EV, Vogel V. 2004. Shear-dependent 'stick-and-roll' adhesion of type 1 fimbriated Escherichia coli. Mol Microbiol 53:1545–1557. 10.1111/j.1365-2958.2004.04226.x. [DOI] [PubMed] [Google Scholar]
- 43.Bertrand RL. 2019. Lag phase is a dynamic, organized, adaptive, and evolvable period that prepares bacteria for cell division. J Bacteriol 201:e00697-18. 10.1128/JB.00697-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klemm P, Schembri M. 2004. Type 1 fimbriae, curli, and antigen 43: adhesion, colonization, and biofilm formation. EcoSal Plus 1:ecosalplus.8.3.2.6. 10.1128/ecosalplus.8.3.2.6. [DOI] [PubMed] [Google Scholar]
- 45.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lane MC, Simms AN, Mobley HL. 2007. Complex interplay between type 1 fimbrial expression and flagellum-mediated motility of uropathogenic Escherichia coli. J Bacteriol 189:5523–5533. 10.1128/JB.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simms AN, Mobley HL. 2008. Multiple genes repress motility in uropathogenic Escherichia coli constitutively expressing type 1 fimbriae. J Bacteriol 190:3747–3756. 10.1128/JB.01870-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim JK, Gunther NWt, Zhao H, Johnson DE, Keay SK, Mobley HL. 1998. In vivo phase variation of Escherichia coli type 1 fimbrial genes in women with urinary tract infection. Infect Immun 66:3303–3310. 10.1128/IAI.66.7.3303-3310.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hagberg L, Jodal U, Korhonen TK, Lidin-Janson G, Lindberg U, Svanborg EC. 1981. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect Immun 31:564–570. 10.1128/iai.31.2.564-570.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subashchandrabose S, Mobley HLT. 2015. Virulence and fitness determinants of uropathogenic Escherichia coli. Microbiol Spectr 3:UTI-0015-2012. 10.1128/microbiolspec.UTI-0015-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shea AE, Marzoa J, Himpsl SD, Smith SN, Zhao L, Tran L, Mobley HLT. 2020. Escherichia coli CFT073 fitness factors during urinary tract infection: identification using an ordered transposon library. Appl Environ Microbiol 86:e00691-20. 10.1128/AEM.00691-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Zhai D, Fan Z, Qu D, Chen G, Su S, Meng J, Jia M, Luo X, Li M. 2020. PAMP protects intestine from Enterohemorrhagic Escherichia coli infection through destroying cell membrane and inhibiting inflammatory response. Biochem Biophys Res Commun 523:939–946. 10.1016/j.bbrc.2020.01.035. [DOI] [PubMed] [Google Scholar]
- 53.Nesta B, Spraggon G, Alteri C, Moriel DG, Rosini R, Veggi D, Smith S, Bertoldi I, Pastorello I, Ferlenghi I, Fontana MR, Frankel G, Mobley HL, Rappuoli R, Pizza M, Serino L, Soriani M. 2012. FdeC, a novel broadly conserved Escherichia coli adhesin eliciting protection against urinary tract infections. mBio 3:e00010-12. 10.1128/mBio.00010-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Waldhuber A, Puthia M, Wieser A, Cirl C, Dürr S, Neumann-Pfeifer S, Albrecht S, Römmler F, Müller T, Zheng Y, Schubert S, Groß O, Svanborg C, Miethke T. 2016. Uropathogenic Escherichia coli strain CFT073 disrupts NLRP3 inflammasome activation. J Clin Invest 126:2425–2436. 10.1172/JCI81916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderson GG, Palermo JJ, Schilling JD, Roth R, Heuser J, Hultgren SJ. 2003. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301:105–107. 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 56.Milton ME, Draughn GL, Bobay BG, Stowe SD, Olson AL, Feldmann EA, Thompson RJ, Myers KH, Santoro MT, Kearns DB, Cavanagh J. 2019. The solution structures and interaction of SinR and SinI: elucidating the mechanism of action of the master regulator switch for biofilm formation in Bacillus subtilis. J Mol Biol 432:343–357. 10.1016/j.jmb.2019.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mulvey MA, Lopez-Boado YS, Wilson CL, Roth R, Parks WC, Heuser J, Hultgren SJ. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494–1497. 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 58.Eto DS, Jones TA, Sundsbak JL, Mulvey MA. 2007. Integrin-mediated host cell invasion by type 1-piliated uropathogenic Escherichia coli. PLoS Pathog 3:e100. 10.1371/journal.ppat.0030100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klumpp DJ, Rycyk MT, Chen MC, Thumbikat P, Sengupta S, Schaeffer AJ. 2006. Uropathogenic Escherichia coli induces extrinsic and intrinsic cascades to initiate urothelial apoptosis. Infect Immun 74:5106–5113. 10.1128/IAI.00376-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Justice SS, Hunstad DA, Seed PC, Hultgren SJ. 2006. Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc Natl Acad Sci USA 103:19884–19889. 10.1073/pnas.0606329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bishop BL, Duncan MJ, Song J, Li G, Zaas D, Abraham SN. 2007. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat Med 13:625–630. 10.1038/nm1572. [DOI] [PubMed] [Google Scholar]
- 62.Easton DM, Allsopp LP, Phan MD, Moriel DG, Goh GK, Beatson SA, Mahony TJ, Cobbold RN, Schembri MA. 2014. The intimin-like protein FdeC is regulated by H-NS and temperature in enterohemorrhagic Escherichia coli. Appl Environ Microbiol 80:7337–7347. 10.1128/AEM.02114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, Kaper JB, Levine MM. 1993. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest 92:1412–1417. 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Higgins LM, Frankel G, Connerton I, Goncalves NS, Dougan G, MacDonald TT. 1999. Role of bacterial intimin in colonic hyperplasia and inflammation. Science 285:588–591. 10.1126/science.285.5427.588. [DOI] [PubMed] [Google Scholar]
- 65.Kerrigan AM, Brown GD. 2009. C-type lectins and phagocytosis. Immunobiology 214:562–575. 10.1016/j.imbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nhu NTK, Phan MD, Peters KM, Lo AW, Forde BM, Min Chong T, Yin WF, Chan KG, Chromek M, Brauner A, Chapman MR, Beatson SA, Schembri MA. 2018. Discovery of new genes involved in curli production by a uropathogenic Escherichia coli strain from the highly virulent O45:K1:H7 lineage. mBio 9:01462-18. 10.1128/mBio.01462-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hultgren SJ, Schwan WR, Schaeffer AJ, Duncan JL. 1986. Regulation of production of type 1 pili among urinary tract isolates of Escherichia coli. Infect Immun 54:613–620. 10.1128/iai.54.3.613-620.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Himpsl SD, Pearson MM, Mobley HLT. 2019. Using hemagglutination, surface shearing, and acid treatment to study fimbriae in Proteus mirabilis. Methods Mol Biol 2021:109–120. 10.1007/978-1-4939-9601-8_11. [DOI] [PubMed] [Google Scholar]
- 70.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. 2000. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J 19:2803–2812. 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagberg L, Engberg I, Freter R, Lam J, Olling S, Svanborg EC. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect Immun 40:273–283. 10.1128/iai.40.1.273-283.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00275-21-s0001.pdf, PDF file, 5.9 MB (5.9MB, pdf)