Abstract
Simple PCR and sequencing assays that utilize a single pair of degenerate primers were used to characterize a 429-bp-long DNA fragment internal (sodAint) to the sodA gene encoding the manganese-dependent superoxide dismutase in 40 coagulase-negative staphylococcal (CNS) type strains. The topology of the phylogenetic tree obtained was in general agreement with that which was inferred from an analysis of their 16S rRNA or hsp60 gene sequences. Sequence analysis revealed that the staphylococcal sodA genes exhibit a higher divergence than does the corresponding 16S ribosomal DNA. These results confirm that the sodA gene constitutes a highly discriminative target sequence for differentiating closely related bacterial species. Clinical isolates that could not be identified at the species level by phenotypical tests were identified by use of this database. These results demonstrate the usefulness of this method for rapid and accurate species identification of CNS isolates, although it does not allow discrimination of subspecies. The sodA sequence polymorphisms observed with staphylococcal species offer good opportunities for the development of assays based on DNA chip technologies.
Coagulase-negative staphylococci (CNS), which are part of the normal skin flora, have emerged as predominant pathogens in hospital-acquired infections (8, 15). They are associated with the presence of foreign bodies, such as prosthetic valves, cerebrospinal fluid shunts, and orthopedic prostheses, as well as intravascular, urinary, and dialysis catheters. Therefore, it has become increasingly important to accurately identify these isolates to the species level in order to define the clinical significance of these bacteria, to carry out a proper epidemiologic surveillance, and to manage patients infected with CNS in case of relapse. A variety of manual and automated methods have been developed for the identification of CNS that are important in human medicine (6, 7, 19, 24). These methods, based on phenotypic characteristics, include conventional identifications and several commercial kits. Unfortunately, the overall accuracy of these systems is low, ranging from 50 to 70% (6, 7, 19, 24). Several genotypic methods based on the analysis of PCR products derived from selected DNA targets have thus been developed for species-level identification of CNS, including electrophoretic analysis (2) and determination (12) of the 16S ribosomal DNA (rDNA) sequence. In the latter case, however, the interpretation of these data may be complicated by the fact that closely related species may have identical 16S rDNA sequences or, alternatively, that divergent 16S rDNA sequences may exist within a single organism (26). To solve this problem, it is possible to use alternative monocopy target sequences which exhibit a higher divergence than those of the 16S rDNA. Recently, partial sequencing of the highly conserved and ubiquitous hsp60 and tuf genes have been found to be useful for identification and taxonomic classification of species of the genus Staphylococcus (5, 17, 18). It was previously reported that PCR and sequencing of the sodA gene of the gram-positive cocci which encodes the manganese-dependent superoxide dismutase (Mn-SOD), with the use of a single pair of degenerate primers, constitute a valuable approach to the genotypic identification of streptococcal (22) and enterococcal (23) species. In the present study, we report the use of the same universal primers (21) to construct a sodA database of 40 staphylococcal type species and we demonstrate the usefulness of this library for a rapid sequence-based identification method for CNS isolates.
(This study was partially presented at the 100th General Meeting of the American Society for Microbiology, Los Angeles, Calif., 21 to 25 May 2000).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The main characteristics of the staphylococcal strains used in this study, including the type strains, are listed in Tables 1 and 2. Type strains were obtained from the Collection de l'Institut Pasteur (CIP). All cultures were grown at 37°C in brain heart infusion broth and subcultured on brain heart infusion agar for examination of the purity and the colony characteristics. Clinical isolates of CNS were identified by the ID 32 Staph system (API-bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions and by use of APILAB ID 32 software.
TABLE 1.
Staphylococcal type strains used in this study
| Strain | Other designationa | sodAint accession no. |
|---|---|---|
| S. aureus subsp. anaerobius CIP 103780 T | ATCC 35844 | Not determined |
| S. aureus subsp. aureus CIP 65.8 T | ATCC 12600 | Not determined |
| S. arlettae CIP 103501 T | ATCC 43959 | AJ34894 |
| S. auricularis CIP 103587 T | ATCC 33753 | AJ34895 |
| S. capitis subsp. capitis CIP 81.53 T | ATCC 27840 | AJ34896 |
| S. capitis subsp. ureolyticus CIP 104192 T | ATCC 49326 | AJ34897 |
| S. caprae CIP 104000 T | ATCC 35538 | AJ34898 |
| S. carnosus subsp. carnosus CIP 103274 T | ATCC 51365 | AJ34899 |
| S. carnosus subsp. utilis CIP 105758 T | DSM 11676 | AJ34900 |
| S. chromogenes CIP 81.59 T | DSM 20454 | AJ34901 |
| S. cohnii subsp. cohnii CIP 81.54 T | ATCC 29974 | AJ34902 |
| S. cohnii subsp. urealyticum CIP 104024 T | ATCC 49330 | AJ34903 |
| S. condimenti CIP 105760 T | DSM 11674 | AJ34904 |
| S. delphini CIP 103732 T | ATCC 49171 | AJ34905 |
| S. epidermidis CIP 81.55 T | ATCC 14990 | AJ34906 |
| S. equorum CIP 103502 T | ATCC 43958 | AJ34907 |
| S. felis CIP 103366 T | ATCC 49168 | AJ34908 |
| S. gallinarum CIP 103504 T | ATCC 35539 | AJ34909 |
| S. haemolyticus CIP 81.56 T | ATCC 29970 | AJ34910 |
| S. hominis subsp. hominis CIP 81.57 T | ATCC 27844 | AJ34911 |
| S. hominis subsp. novobiosepticus CIP 105719 T | ATCC 700236 | AJ34912 |
| S. hyicus CIP 81.58 T | ATCC 11249 | AJ34913 |
| S. intermedius CIP 81.60 T | ATCC 29663 | AJ34914 |
| S. kloosii CIP 103503 T | ATCC 43959 | AJ34915 |
| S. lentus CIP 8163 T | ATCC 29070 | AJ34916 |
| S. lugdunensis CIP 103642 T | ATCC 43809 | AJ34917 |
| S. lutrae CIP 105399 T | ATCC 700373 | AJ34918 |
| S. muscae CIP 103641 T | ATCC 49910 | AJ34919 |
| S. pasteuri CIP 103540 T | ATCC 51129 | AJ34920 |
| S. piscifermentans CIP 103958 T | DSM 7373 | AJ34921 |
| S. pulvereri CIP 104364 T | DSM 9930 | AJ34922 |
| S. saccharolyticus CIP 103275 T | ATCC 14953 | AJ34923 |
| S. saprophyticus subsp. bovis CIP 105260 T | AJ34924 | |
| S. saprophyticus subsp. saprophyticus CIP 76.125 T | ATCC 15305 | AJ34925 |
| S. schleiferi subsp. coagulans CIP 104370 T | ATCC 49545 | AJ34926 |
| S. schleiferi subsp. schleiferi CIP 103643 T | ATCC 43808 | AJ34927 |
| S. sciuri subsp. carnaticus CIP 105826 T | ATCC 700058 | AJ34928 |
| S. sciuri subsp. sciuri CIP 81.62 T | ATCC 29062 | AJ34929 |
| S. simulans CIP 81.64 T | ATCC 27848 | AJ34930 |
| S. vitulus CIP 104850 T | ATCC 51145 | AJ34931 |
| S. warneri CIP 81.65 T | ATCC 27836 | AJ34932 |
| S. xylosus CIP 81.66 T | ATCC 29971 | AJ34933 |
| Macrococcus caseolyticusb CIP 100755 T | ATCC 13548 | AJ34934 |
ATCC, American Type Culture Collection; DSM, Deutsche Sammlung Von Mikrooganismen.
Formerly designated Staphylococcus caseolyticus.
TABLE 2.
Identification of various staphylococcal strains by sequencing the sodAint fragment
| Strain | Origin | Bacterial species (% identity)a | sodAint accession no. |
|---|---|---|---|
| NEM1997 | Cheese | M. caseolyticus (100) | AJ34935 |
| NEM1998 | Cheese | M. caseolyticus (100) | AJ34936 |
| NEM1999 | Middle ear fluid | S. auricularis (99.8) | AJ34937 |
| NEM2000 | Middle ear fluid | S. auricularis (99.8) | AJ34938 |
| NEM2001 | Urine | S. capitis (99.8) | AJ34939 |
| NEM2002 | Blood | S. capitis (99.8) | AJ34940 |
| NEM2003 | Blood | S. capitis (100) | AJ34941 |
| NEM2004 | Urine | S. caprae (100) | AJ34942 |
| NEM2005 | Blood | S. caprae (100) | AJ34943 |
| NEM2006 | Cheese | S. chromogenes (99.8) | AJ34944 |
| NEM2007 | Cheese | S. chromogenes (99.8) | AJ34945 |
| NEM2008 | Blood | S. epidermidis (100) | AJ34946 |
| NEM2009 | Catheter | S. epidermidis (100) | AJ34947 |
| NEM2010 | Blood | S. epidermidis (99.5) | AJ34948 |
| NEM2011 | Catheter | S. haemolyticus (100) | AJ34949 |
| NEM2012 | Blood | S. haemolyticus (99.5) | AJ34950 |
| NEM2013 | Cutaneous abcess | S. lugdunensis (99.8) | AJ34951 |
| NEM2014 | Synovial fluid | S. lugdunensis (99.8) | AJ34952 |
| NEM2015 | Nasal swab | S. pasteuri (100) | AJ34953 |
| NEM2016 | Urine | S. saprophyticus (99.8) | AJ34954 |
| NEM2017 | Blood | S. schleiferi (99.8) | AJ34955 |
| NEM2018 | Vagina | S. simulans (100) | AJ34956 |
| NEM2019 | Cheese | S. vitulus (99.8) | AJ34957 |
| NEM2020 | Blood | S. warneri (99.8) | AJ34958 |
| NEM2021 | Blood | S. xylosus (99.3) | AJ34959 |
| NEM2022 | Cheese | S. xylosus (99.5) | AJ34960 |
The species identification was based on the phylogenetic position of the sodAint fragment of the strain studied relative to those of the type strains, as shown in Fig. 1. The numbers in parentheses indicate the percentages of identity of the sodAint fragments with that of the corresponding type strains.
DNA manipulations.
Rapid extraction of bacterial genomic DNA collected from 2 ml of an overnight culture was performed with the InstaGen Matrix (Bio-Rad) according to the manufacturer's instructions. The sodA degenerate primers d1 (5′-CCITAYICITAYGAYGCIYTIGARCC-3′) and d2 (5′-ARRTARTAIGCRTGYTCCCAIACRTC-3′) were used to amplify an internal fragment, designated sodAint, representing approximately 83% of the sodA gene. PCRs were performed on a Gene Amp System 2400 thermal cycler (Perkin-Elmer Cetus, Courtaboeuf, France) in a final volume of 50 μl containing 150 ng of DNA as the template, 0.5 μM each primer, a 200 μM concentration of each deoxynucleoside triphosphate, and 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer) in a 1× amplification buffer (10 mM Tris-HCl [pH 8.3], 50 mM KCl, 1.5 mM MgCl2). The PCR mixtures were denatured (3 min at 95°C) and then subjected to 30 cycles of amplification (60 s of annealing at 37°C, 45 s of elongation at 72°C, and 30 s of denaturation at 95°C). PCR products were resolved by electrophoresis on a 1% agarose gel stained with ethidium bromide. PCR products were purified on an S-400 Sephadex (Pharmacia, Uppsala, Sweden) column, and both strands were directly sequenced with the oligonucleotides d1 and d2 by using an ABI-PRISM Big Dye terminator sequencing kit on an ABI-PRISM 310 Genetic Analyzer (Perkin-Elmer) as previously described (23). All precautions to prevent carryover of amplified DNA were used.
Sequence analysis.
Nucleotide sequences were analyzed with Perkin-Elmer software programs (Sequence Analysis, Sequence Navigator, and Autoassembler). Multiple alignment of sod genes was carried out by the CLUSTAL X program (9). The construction of the unrooted phylogenetic tree was performed by the neighbor-joining method (25). The topology of the phylogenetic tree was evaluated by bootstrap analyses to give the degree of confidence intervals for each node on the phylogenetic tree. The confidence values were determined for branches which showed possible monophyletic clades of related organisms separated at each node. It is generally accepted that the monophyly of a clade can be accepted if the clade occurs in more than 95% of the bootstrapped trees (4).
Protein extraction and SOD activity assay.
Crude cell lysates of staphylococcal strains were prepared as follows. Cells from 10 ml of overnight cultures were harvested by centrifugation, washed with an equal volume of Tris-EDTA buffer (50 mM Tris HCl [pH 7.6], 50 mM EDTA), suspended in 2 ml of TELL lysis buffer (50 mM Tris HCl [pH 7.6], 50 mM EDTA, 200 mg of lysozyme/liter and 30 mg of lysostaphin/liter), and incubated for 1 h at 37°C. After ultrasonic disruption for 3 min in 30-s pulses at 4°C, the lysates were cleared by centrifugation (16, 170 × g, for 10 min at 4°C). The supernatants were recovered and stored at −20°C until needed. Fifty micrograms of total proteins was electrophoresed through a 10% nondenaturing polyacrylamide gel which was stained for SOD activity by the method of Beauchamp and Fridovich (1).
Nucleotide sequence accession numbers.
The sequences determined were submitted to the EMBL gene bank and assigned the accession numbers listed in Tables 1 and 2.
RESULTS AND DISCUSSION
CNS express a single Mn-SOD.
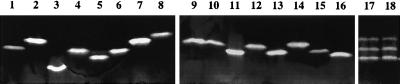
It has recently been reported that Staphylococcus aureus possesses two genes encoding Mn-SOD, designated sodA and sodM (3, 21, 28). In order to confirm these results, we analyzed the SOD activities of the type strains S. aureus subsp. aureus and S. aureus subsp. anaerobius and 25 unrelated clinical isolates of S. aureus following electrophoresis of protein extracts in a nondenaturing polyacrylamide gel. All strains of S. aureus produce three closely migrating bands of SOD activity (Fig. 1 and data not shown). Previous work demonstrated that the upper band of activity corresponds to SodM, the lowest band corresponds to SodA, and the middle band was proposed to result from the formation of a hybrid protein composed of SodM and SodA (28). This analysis indicates that, unlike all the low-GC-content, gram-positive bacilli and cocci described so far, S. aureus carries genes encoding two unrelated SODs. The fact that S. aureus strains possess two different sod genes impedes direct sequencing of PCR products with the primers d1 and d2, since both genes coamplify with this pair of oligonucleotides. Therefore, to determine if CNS possess one gene encoding Mn-SOD, the SOD activities of crude bacterial extracts of the 40 type strains of CNS were analyzed following electrophoresis in nondenaturing polyacrylamide gels. This analysis revealed that all the strains studied produce a single band of SOD activity (Fig. 1 and data not shown), which suggests that CNS, as opposed to S. aureus, express a single type of Mn-SOD.
FIG. 1.
SOD activity gel. Crude extracts (50 μg) of various staphylococcal type strains were loaded onto a nondenaturing 10% polyacrylamide gel stained for SOD activity. Lanes 1 to 18, S. capitis subsp. capitis, S. chromogenes, S. cohnii subsp. cohnii, S. epidermidis, S. haemolyticus, S. hominis subsp. hominis, S. hyicus; S. intermedius, S. lentus, S. lugdunensis, S. saprophyticus subsp. saprophyticus, S. schleiferi subsp. schleiferi, S. simulans, S. vitulus, S. warneri, S. xylosus, S. aureus subsp. aureus, and S. aureus subsp. anaerobius, respectively.
Amplification and sequencing of the sodAint gene from various CNS type strains.
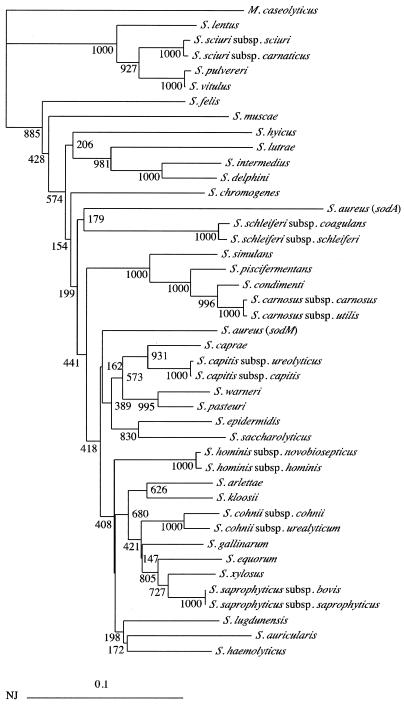
By using the primers d1 and d2 in a PCR assay, we amplified an internal fragment representing approximately 85% of the sodA gene in 40 type strains of CNS (Staphylococcus arlettae, Staphylococcus auricularis, Staphylococcus capitis subsp. capitis, Staphylococcus capitis subsp. urealyticus, Staphylococcus caprae, Staphylococcus carnosus subsp. carnosus, Staphylococcus carnosus subsp. utilis, Staphylococcus chromogenes, Staphylococcus cohnii subsp. cohnii, Staphylococcus cohnii subsp. urealyticus, Staphylococcus condimenti, Staphylococcus delphini, Staphylococcus epidermidis, Staphylococcus equorum, Staphylococcus felis, Staphylococcus gallinarum, Staphylococcus haemolyticus, Staphylococcus hominis subsp. hominis, Staphylococcus hominis subsp. novobiosepticus, Staphylococcus hyicus, Staphylococcus intermedius, Staphylococcus kloosii, Staphylococcus lentus, Staphylococcus lugdunensis, Staphylococcus lutrae, Staphylococcus muscae, Staphylococcus pasteuri, Staphylococcus piscifermentans, Staphylococcus pulvereri, Staphylococcus saccharolyticus, Staphylococcus saprophyticus subsp. bovis, Staphylococcus saprophyticus subsp. saprophyticus, Staphylococcus schleiferi subsp. coagulans, Staphylococcus schleiferi subsp. schleiferi, Staphylococcus sciuri subsp. carnaticus, Staphylococcus sciuri subsp. sciuri, Staphylococcus simulans, Staphylococcus vitulus, Staphylococcus warneri, Staphylococcus xylosus). We also included in this study Macrococcus caseolyticus, which was formerly designated Staphylococcus caseolyticus (13). A single amplification product having the expected size of 480 bp was observed with all staphylococcal species (data not shown). Direct sequencing of these amplicons gave rise to electropherograms devoid of overlapping peaks, which confirms that these strains contain a single type of sod gene. Sequence analysis of these amplicons revealed that they were actual sodAint fragments, since the corresponding deduced polypeptides all possessed the amino acids characteristic of the Mn-SOD at the expected positions (data not shown). Multiple alignment of the staphylococcal sodAint DNA sequences was carried out by the CLUSTAL X program. The sequences of the degenerated primers d1 and d2 and alignment gaps were not taken into consideration for calculations. Although some differences could be observed, the topology of the phylogenetic tree obtained (Fig. 2) was in general agreement with that which was inferred from an analysis of their 16S rRNA or hsp60 gene sequences (17, 27). The phylogenetic position of M. caseolyticus is the most distant from any other CNS species (Fig. 2 and data not shown), an observation consistent with the decision to remove this species from the genus Staphylococcus (13). It is worth noting, however, that most staphylococcal species groups are not supported by significant bootstrap values (i.e., ≥95%). In fact, if this critical value is used, only three major clusters corresponding to species groups S. sciuri, S. intermedius, and S. simulans are defined (Fig. 2).
FIG. 2.
Phylogenetic unrooted tree showing relationships among the sodAint fragments from various staphylococcal type strains. The tree was established from an analysis of the sequences listed in Table 1 by using the neighbor-joining method. The sodAint sequence of M. caseolyticus type strains included in this work was used as an outgroup sequence to root the tree. The value on each branch is the estimated confidence limit (expressed as a percentage) for the position of the branch as determined by bootstrap analysis. Only the bootstrap values superior to 95% were considered significant (4). The scale bar (neighbor-joining distance) represents 10% differences in nucleotide sequences. The accession numbers of sodA and sodM were AF121672 and Z49245, respectively.
The S. sciuri group includes S. sciuri, S. lentus, S. pulvereri, and S. vitulus. These four species differ from the other Staphylococcus species by several remarkable features. They, are novobiocin resistant and oxidase positive, they are the sole species possessing cytochrome c in their electron transport systems, and they all share the same characteristic pattern of amino acid substitution in their HSP60 proteins (14, 17). Interestingly, we observed that their sodAint sequences, when compared to other CNS sequences, contained an additional codon which codes for a prolyl residue at position 78 of the corresponding 143-amino-acid-long partial SodA protein (data not shown). It is notable that the sodAint sequences of S. pulvereri and S. vitulus display 99.5% identity, which confirms that they constitute a single species (20).
An S. intermedius group consisting of S. intermedius, S. delphini, and S. lutrae was recovered in 98.1% of the bootstrap trees. The fact that S. schleiferi was not included in this cluster is in disagreement with the results from DNA annealing studies or from a phylogenetic analysis of their 16S rDNA or hsp60 gene sequences (17, 27). Moreover, the related species S. hyicus, S. muscae, and S. chromogenes did not cluster to form a S. hyicus species subgroup (Fig. 2).
The S. simulans group species, defined in 100% of the bootstrap trees (Fig. 2), consists of S. simulans, S. piscifermentans, S. condimenti, and S. carnosus.
The S. saprophyticus group, as defined by 16S rDNA sequence analysis (91% of bootstrap value), includes the novobiocin-resistant and oxidase-negative species S. saprophyticus, S. arlettae, S. kloosi, S. cohnii, S. gallinarum, S. equorum, and S. xylosus (27). In our analysis, however, the monophyly of this clade is uncertain since it is associated with a bootstrap value of 68% (Fig. 2). Similarly, the S. epidermidis group (S. epidermidis, S. capitis, S. caprae, and S. saccharolyticus), which constitutes a monophyletic clade supported by a high bootstrap value (97%) on the basis of 16S rDNA sequence analysis (27), did not form a clearly distinct lineage in our study (38.9% of bootstrap value). On the other hand, association of S. pasteuri and S. warneri to the S. epidermidis group was inferred from our treeing analysis (Fig. 2).
As reported in an analysis of their 16S rDNA sequences (27), we found that the branching order of the species S. auricularis, S. haemolyticus, S. hominis, and S. lugdunensis was uncertain in our sodAint-based phylogenetic analysis.
Evidence for horizontal transfer of a sod gene from CNS to S. aureus.
The sodA and sodM genes from S. aureus display 77.5% sequence identity, which indicates that the presence of these two isofunctional genes in this bacterium is not due to a recent duplication event. In a sodAint phylogenetic tree that includes the sequences of Bacillus subtilis, Clostridium perfringens, Enterococcus faecalis, Enterococus faecium, Lactococcus lactis, Streptococcus pyogenes, Streptococcus agalactiae, and Streptococcus pneumoniae, both the sodM and sodA genes from S. aureus were clearly positioned within the staphylococcal lineage (data not shown). However, it is worth noting that the phylogenetic position of the sodM gene in the sod tree (Fig. 2) is similar to that of S. aureus in 16S rDNA or hsp60 gene trees (17, 27). This might indicate that sodM is the indigenous S. aureus sod gene whereas sodA was acquired by horizontal gene transfer from an as-yet-uncharacterized CNS. This hypothesis is based on the fact that S. aureus possesses a remarkable ability to acquire useful genes from various bacteria by lateral gene transfer, as revealed by genome sequence analysis (16). Accordingly, it has been proposed that the mecA homologue present in S. sciuri is the evolutionary precursor of the S. aureus methicillin resistance gene mecA (29). Efforts are currently being made to track the original host of the S. aureus sodA gene.
Species identification of clinical and environmental isolates of staphylococci by sequencing the sodAint gene.
Pairwise comparison of the staphylococcal sodAint sequences revealed that their mean identity (81.5%) is inferior to that calculated from a comparison of their 16S rDNA genes (mean identity, 98%) but is similar to that computed from a comparison of their hsp60 genes (mean identity, 82%) (17). These results confirm that sodA might constitute a more discriminative target sequence than does the 16S RNA to differentiate closely related bacterial species, as already demonstrated for differentiating closely related species belonging to the genera Streptococcus and Enterococcus (11, 22, 23). The sodAint fragments of S. cohnii subsp. cohnii and S. cohnii subsp. urealyticum display 4% sequence divergence, enabling the distinction between these two subspecies (Table 3). However, the sodAint fragments of the remaining pairs of type strain subspecies (S. capitis subsp. capitis and S. capitis subsp. ureolyticus, S. carnosus subsp. carnosus and S. carnosus subsp. utilis, S. hominis subsp. hominis and S. hominis subsp. novobiosepticus, S. saprophyticus subsp. bovis and S. saprophyticus subsp. saprophyticus, S. schleiferi subsp. coagulans and S. schleiferi subsp. schleiferi, S. sciuri subsp. carnaticus and S. sciuri subsp. sciuri) display more than 99.3% sequence identity. This finding is consistent with the observation that the hsp60 genes of S. schleiferi subsp. coagulans and S. schleiferi subsp. schleiferi display 98% sequence identity (17). We therefore concluded that the sodA gene, like the hsp60 gene, does not allow discrimination at the subspecies level.
TABLE 3.
Identity matrix based on pairwise comparisons of sodAint fragments of CNS type strains
| Straina | % Identity with the following strainb:
|
||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | |
| 1. S. arlettae | 82.3 | 86.5 | 86.2 | 84.6 | 80.7 | 80.4 | 79.3 | 86.2 | 88.1 | 81.4 | 76.5 | 84.6 | 88.1 | 77.4 | 89.7 | 86.9 | 87.6 | 87.9 | 74.8 | 77.6 | 91.1 | 71.8 | 85.8 | 76.9 | 73.9 | 85.3 | 82.1 | 71.1 | 83.7 | 88.8 | 88.8 | 77.6 | 77.9 | 71.8 | 71.8 | 81.6 | 71.6 | 85.1 | 89.0 |
| 2. S. auricularis | 83.2 | 83.0 | 83.0 | 79.9 | 79.9 | 81.1 | 83.0 | 82.8 | 80.6 | 74.6 | 80.9 | 82.8 | 75.1 | 86.0 | 84.1 | 83.0 | 83.0 | 73.2 | 74.4 | 80.4 | 65.7 | 83.7 | 72.3 | 72.3 | 83.0 | 79.6 | 69.2 | 78.6 | 83.7 | 83.7 | 75.8 | 76.2 | 69.2 | 69.2 | 79.6 | 69.2 | 82.8 | 82.1 | |
| 3. S. capitis subsp. capitis | 99.8 | 93.7 | 79.0 | 79.0 | 84.8 | 84.1 | 84.8 | 79.3 | 78.8 | 89.3 | 85.3 | 78.3 | 85.1 | 87.2 | 86.9 | 86.5 | 79.3 | 77.9 | 85.1 | 72.5 | 84.4 | 74.4 | 75.1 | 90.2 | 81.4 | 74.4 | 87.2 | 86.2 | 86.2 | 81.1 | 81.8 | 73.0 | 73.2 | 83.9 | 74.8 | 89.5 | 85.3 | ||
| 4. S. capitis subsp. ureolyticus | 93.9 | 78.8 | 78.8 | 84.6 | 84.4 | 84.6 | 79.0 | 79.0 | 89.0 | 85.1 | 78.1 | 84.6 | 86.9 | 86.7 | 86.2 | 79.0 | 78.1 | 85.3 | 72.3 | 84.1 | 74.8 | 74.8 | 90.0 | 81.1 | 74.1 | 86.9 | 86.0 | 86.0 | 80.9 | 81.6 | 72.7 | 73.0 | 83.7 | 74.6 | 89.3 | 84.8 | |||
| 5. S. caprae | 76.5 | 76.5 | 83.0 | 83.0 | 83.0 | 77.9 | 79.7 | 88.1 | 86.0 | 77.2 | 82.5 | 89.7 | 86.5 | 86.0 | 77.2 | 79.7 | 85.5 | 71.1 | 84.6 | 75.5 | 76.9 | 90.4 | 79.0 | 72.5 | 87.4 | 87.2 | 87.2 | 80.0 | 80.9 | 71.3 | 71.6 | 81.4 | 73.0 | 89.7 | 84.8 | ||||
| 6. S. carnosus subsp. carnosus | 99.5 | 73.9 | 78.3 | 78.8 | 96.8 | 73.7 | 77.6 | 79.0 | 74.8 | 80.7 | 75.1 | 78.8 | 79.7 | 69.7 | 71.8 | 79.0 | 69.9 | 79.5 | 70.9 | 71.8 | 79.0 | 93.8 | 69.0 | 75.8 | 78.1 | 78.1 | 76.5 | 76.9 | 70.8 | 70.6 | 88.0 | 69.4 | 77.6 | 78.8 | |||||
| 7. S. carnosus subsp. utilis | 73.7 | 78.1 | 78.6 | 96.8 | 73.7 | 77.4 | 79.0 | 74.6 | 80.7 | 74.8 | 78.6 | 79.5 | 69.7 | 71.8 | 79.0 | 69.9 | 78.8 | 70.6 | 71.6 | 79.0 | 93.8 | 69.0 | 75.8 | 78.1 | 78.1 | 76.0 | 76.5 | 70.8 | 70.6 | 87.7 | 69.4 | 77.6 | 78.8 | ||||||
| 8. S. chromogenes | 80.0 | 79.3 | 74.4 | 77.9 | 80.4 | 80.0 | 75.1 | 82.8 | 80.4 | 80.4 | 80.2 | 77.6 | 78.6 | 78.3 | 69.5 | 80.4 | 77.2 | 76.2 | 83.7 | 77.9 | 70.2 | 78.6 | 81.6 | 81.6 | 78.8 | 79.3 | 70.9 | 70.9 | 80.0 | 70.6 | 81.6 | 80.9 | |||||||
| 9. S. cohnii subsp. cohnii | 95.6 | 80.2 | 78.3 | 85.5 | 88.3 | 75.1 | 90.2 | 85.3 | 87.9 | 88.3 | 76.7 | 77.2 | 86.2 | 70.2 | 85.3 | 75.5 | 74.4 | 85.3 | 80.9 | 72.7 | 82.3 | 90.4 | 90.4 | 79.0 | 79.0 | 73.4 | 73.4 | 80.4 | 73.2 | 85.1 | 89.5 | ||||||||
| 10. S. cohnii subsp. urealyticum | 80.2 | 78.8 | 85.1 | 90.4 | 75.1 | 91.6 | 86.5 | 87.9 | 88.3 | 76.5 | 78.1 | 87.6 | 70.9 | 86.7 | 76.5 | 74.1 | 87.6 | 80.2 | 71.8 | 83.7 | 91.8 | 91.8 | 77.6 | 77.6 | 72.7 | 72.7 | 80.0 | 72.3 | 86.7 | 90.0 | |||||||||
| 11. S. condimenti | 74.8 | 78.8 | 79.3 | 75.3 | 81.8 | 76.2 | 80.0 | 80.9 | 69.7 | 72.3 | 79.3 | 69.9 | 79.3 | 71.3 | 72.0 | 79.3 | 94.7 | 69.7 | 75.1 | 78.6 | 78.6 | 75.5 | 76.0 | 71.5 | 71.3 | 89.1 | 70.1 | 78.6 | 79.5 | ||||||||||
| 12. S. delphini | 77.4 | 78.6 | 75.1 | 78.1 | 78.3 | 78.1 | 78.3 | 75.8 | 91.8 | 79.5 | 69.7 | 78.1 | 81.8 | 73.0 | 80.4 | 75.1 | 70.2 | 75.5 | 80.9 | 80.9 | 80.9 | 80.9 | 69.5 | 69.5 | 76.7 | 70.2 | 79.3 | 78.3 | |||||||||||
| 13. S. epidermidis | 83.4 | 79.5 | 84.1 | 84.6 | 87.9 | 88.1 | 76.2 | 78.1 | 84.1 | 69.5 | 82.8 | 74.4 | 75.8 | 86.5 | 79.0 | 72.5 | 88.6 | 83.7 | 83.7 | 80.0 | 79.7 | 73.7 | 73.7 | 80.2 | 73.0 | 85.8 | 84.1 | ||||||||||||
| 14. S. equorum | 75.5 | 89.3 | 86.9 | 85.8 | 85.8 | 74.1 | 76.9 | 89.7 | 70.2 | 85.1 | 73.2 | 73.9 | 86.0 | 81.1 | 70.6 | 83.9 | 93.0 | 93.0 | 76.9 | 77.2 | 70.6 | 70.6 | 83.0 | 71.1 | 85.8 | 91.4 | |||||||||||||
| 15. S. felis | 75.1 | 76.9 | 76.5 | 76.5 | 76.7 | 74.4 | 79.7 | 69.9 | 79.0 | 75.1 | 76.5 | 76.9 | 74.1 | 71.8 | 75.1 | 76.9 | 76.9 | 76.7 | 76.7 | 72.3 | 71.8 | 73.4 | 72.3 | 76.5 | 76.7 | ||||||||||||||
| 16. S. gallinarum | 86.7 | 88.1 | 88.6 | 74.8 | 79.5 | 90.0 | 71.3 | 87.2 | 75.8 | 74.6 | 87.4 | 83.7 | 72.5 | 82.8 | 91.8 | 91.8 | 77.4 | 76.9 | 72.3 | 72.3 | 81.6 | 73.0 | 86.0 | 91.4 | |||||||||||||||
| 17. S. haemolyticus | 87.6 | 87.9 | 74.6 | 78.1 | 86.0 | 69.0 | 87.6 | 73.9 | 78.1 | 86.7 | 78.8 | 70.2 | 83.7 | 88.6 | 88.6 | 78.6 | 78.8 | 70.4 | 70.6 | 77.4 | 70.2 | 86.0 | 86.9 | ||||||||||||||||
| 18. S. hominis subsp. hominis | 99.3 | 75.8 | 79.0 | 86.7 | 70.9 | 87.4 | 75.5 | 79.0 | 86.0 | 81.4 | 73.4 | 83.7 | 85.5 | 85.5 | 80.0 | 80.0 | 73.2 | 73.0 | 81.1 | 73.4 | 85.5 | 86.7 | |||||||||||||||||
| 19. S. hominis subsp. novobiosepticus | 75.5 | 79.0 | 86.2 | 71.1 | 87.6 | 75.5 | 78.8 | 85.5 | 82.3 | 73.4 | 83.4 | 86.0 | 86.0 | 79.5 | 79.5 | 73.2 | 73.0 | 81.1 | 73.4 | 85.5 | 86.7 | ||||||||||||||||||
| 20. S. hyicus | 75.3 | 73.9 | 66.4 | 73.9 | 73.7 | 70.4 | 76.7 | 69.5 | 68.1 | 75.3 | 74.8 | 74.8 | 74.8 | 74.8 | 68.8 | 69.0 | 71.1 | 68.5 | 76.0 | 74.4 | |||||||||||||||||||
| 21. S. intermedius | 80.2 | 68.3 | 79.5 | 81.1 | 74.1 | 79.5 | 72.5 | 68.5 | 76.9 | 80.4 | 80.4 | 77.4 | 77.9 | 67.4 | 67.4 | 74.8 | 68.5 | 79.3 | 78.3 | ||||||||||||||||||||
| 22. S. kloosii | 72.0 | 87.6 | 76.9 | 75.1 | 87.4 | 80.0 | 73.4 | 84.8 | 90.9 | 90.9 | 78.1 | 77.9 | 73.0 | 73.0 | 81.6 | 73.9 | 85.3 | 90.9 | |||||||||||||||||||||
| 23. S. lentus | 69.7 | 68.3 | 69.5 | 73.9 | 68.5 | 90.0 | 66.9 | 72.0 | 72.0 | 66.9 | 67.6 | 88.9 | 88.7 | 69.0 | 90.0 | 73.4 | 70.6 | ||||||||||||||||||||||
| 24. S. lugdunensis | 76.5 | 74.8 | 84.4 | 79.5 | 70.4 | 82.3 | 86.9 | 86.9 | 78.6 | 79.0 | 70.2 | 69.9 | 77.6 | 70.9 | 84.4 | 84.6 | |||||||||||||||||||||||
| 25. S. lutrae | 71.3 | 75.8 | 72.0 | 69.7 | 73.7 | 74.4 | 74.4 | 77.9 | 78.1 | 68.1 | 68.1 | 70.6 | 69.7 | 72.7 | 74.6 | ||||||||||||||||||||||||
| 26. S. muscae | 76.0 | 71.3 | 70.4 | 74.4 | 74.4 | 74.4 | 74.6 | 74.4 | 71.6 | 71.1 | 72.3 | 70.9 | 74.4 | 74.4 | |||||||||||||||||||||||||
| 27. S. pasteuri | 80.7 | 73.7 | 86.2 | 88.1 | 88.1 | 81.6 | 82.1 | 75.5 | 75.8 | 83.7 | 74.1 | 94.4 | 86.5 | ||||||||||||||||||||||||||
| 28. S. piscifermentans | 70.4 | 75.8 | 80.2 | 80.2 | 78.1 | 78.1 | 70.8 | 70.6 | 90.7 | 70.8 | 79.5 | 80.0 | |||||||||||||||||||||||||||
| 29. S. pulvereri | 69.9 | 73.2 | 73.2 | 69.5 | 69.9 | 93.5 | 93.3 | 68.3 | 99.5 | 74.1 | 73.0 | ||||||||||||||||||||||||||||
| 30. S. saccharolyticus | 85.1 | 85.1 | 81.1 | 80.9 | 71.6 | 71.8 | 78.1 | 70.4 | 84.4 | 84.8 | |||||||||||||||||||||||||||||
| 31. S. saprophyticus subsp. bovis | 100.0 | 79.0 | 79.0 | 72.7 | 72.7 | 80.7 | 73.7 | 87.6 | 94.4 | ||||||||||||||||||||||||||||||
| 32. S. saprophyticus subsp. saprophyticus | 79.0 | 79.0 | 72.7 | 72.7 | 80.7 | 73.7 | 87.6 | 94.4 | |||||||||||||||||||||||||||||||
| 33. S. schleiferi subsp. coagulans | 98.8 | 69.5 | 69.7 | 78.3 | 69.9 | 79.7 | 78.6 | ||||||||||||||||||||||||||||||||
| 34. S. schleiferi subsp. schleiferi | 69.0 | 69.2 | 79.0 | 70.4 | 80.4 | 78.6 | |||||||||||||||||||||||||||||||||
| 35. S. sciuri subsp. carnaticus | 99.3 | 69.0 | 93.5 | 74.4 | 72.0 | ||||||||||||||||||||||||||||||||||
| 36. S. sciuri subsp. sciuri | 69.0 | 93.3 | 74.4 | 72.0 | |||||||||||||||||||||||||||||||||||
| 37. S. simulans | 68.8 | 82.5 | 81.1 | ||||||||||||||||||||||||||||||||||||
| 38. S. vitulus | 74.6 | 73.4 | |||||||||||||||||||||||||||||||||||||
| 39. S. warneri | 84.4 | ||||||||||||||||||||||||||||||||||||||
| 40. S. xylosus | |||||||||||||||||||||||||||||||||||||||
The main characteristics of the strains are listed in Table 1.
The strain numbers correspond to the strains identified by the numbers on the left.
Twenty-six unrelated CNS isolates were identified by conventional microbiological tests, the ID 32 Staph system, and the sodAint system (Table 2). In all cases, the sodAint sequences of the isolates displayed less than 1.5% divergence with that of the corresponding type strain (Table 2). For 14 strains (NEM1999, NEM2000, NEM2002, NEM2004, NEM2006, NEM2008, NEM2009, NEM2010, NEM2011, NEM2013, NEM2014, NEM2016, NEM2021, and NEM2022), the two methods gave the same results. The remaining 12 isolates were not identified at the species level, or were misidentified, by the conventional microbiological phenotypic tests. These strains were correctly identified by the sodAint system. For example, NEM1997 and NEM1998 were identified by the ID 32 Staph system as S. capitis although they were novobiocin resistant and oxidase positive. Both strains were identified as M. caseolyticus following sodAint-based sequence analysis. NEM2001, NEM2003, NEM2004, NEM2015, and NEM2018 were formerly misidentified by conventional methods as S. hominis, S. epidermidis, S. haemolyticus, S. warneri, and S. epidermidis, respectively. They were subsequently unambiguously identified with the sodA system as S. capitis, S. caprae, S. pasteuri, and S. simulans, respectively. These latter species are often misidentified by conventional methods (10), and genotypic methods are often necessary to identify these uncommon species. Lastly, NEM2005, NEM2012, NEM2017, NEM2019, and NEM2020, which were not identified by the ID 32 Staph system, were identified by the sodAint method as S. caprae, S. haemolyticus, S. schleiferi, S. vitulus, and S. warneri, respectively.
Conclusions.
We have determined the sodAint sequences of 40 type strains of CNS and demonstrated the usefulness of this database for species-level identification of staphylococcal isolates. This method consists of a PCR carried out with a single pair of degenerate oligonucleotides for amplification of a staphylococcal sodAint fragment and direct sequencing of the PCR product with the same degenerate primers. Under these conditions, the delay required for bacterial identification is less than 24 h. This method might be useful in reference laboratories for characterization of strains that could not be assigned to a species on the basis of their conventional phenotypic reaction. Furthermore, the sodA sequence polymorphisms observed with staphylococcal species offer good opportunities for the development of assays based on DNA chip technologies.
ACKNOWLEDGMENTS
We thank C. Bizet for the gift of staphylococcal type strains (CIP) and S. Nair for critical reading of the manuscript.
This work was supported by the Institut Pasteur and by the University Paris V.
REFERENCES
- 1.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 2.Chesneau O, Morvan A, Aubert S, el Solh N. The value of rRNA gene restriction site polymorphism analysis for delineating taxa in the genus Staphylococcus. Int J Syst Evol Microbiol. 2000;50:689–697. doi: 10.1099/00207713-50-2-689. [DOI] [PubMed] [Google Scholar]
- 3.Clements M O, Watson S P, Foster S J. Characterization of the major superoxide dismutase of Staphylococcus aureus and its role in starvation survival, stress resistance, and pathogenicity. J Bacteriol. 1999;181:3898–3903. doi: 10.1128/jb.181.13.3898-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Felsenstein J. Confidence limits on phylogeny and approach using the boostrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 5.Goh S H, Potter S, Wood J O, Hemmingsen S M, Reynolds R P, Chow A W. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J Clin Microbiol. 1996;34:818–823. doi: 10.1128/jcm.34.4.818-823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grant C E, Sewell D L, Pfaller M, Bumgardner R V, Williams J A. Evaluation of two commercial systems for identification of coagulase-negative staphylococci to species level. Diagn Microbiol Infect Dis. 1994;18:1–5. doi: 10.1016/0732-8893(94)90126-0. [DOI] [PubMed] [Google Scholar]
- 7.Ieven M, Verhoeven J, Pattyn S R, Goossens H. Rapid and economical method for species identification of clinically significant coagulase-negative staphylococci. J Clin Microbiol. 1995;33:1060–1063. doi: 10.1128/jcm.33.5.1060-1063.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarvis W R, Martone W J. Predominant pathogens in hospital infections. J Antimicrob Chemother. 1992;29:19–24. doi: 10.1093/jac/29.suppl_a.19. [DOI] [PubMed] [Google Scholar]
- 9.Jeanmougin F, Thompson J D, Gouy M, Higgins D G, Gibson T J. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- 10.Kawamura Y, Hou X-G, Sultana F, Hirose K, Miyake M, Shu S-E, Ezaki T. Distribution of Staphylococcus species among human clinical specimens and emended description of Staphylococcus caprae. J Clin Microbiol. 1998;36:2038–2042. doi: 10.1128/jcm.36.7.2038-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamura Y, Whiley R A, Shu S E, Ezaki T, Hardie J M. Genetic approaches to the identification of the mitis group within the genus Streptococcus. Microbiology. 1999;145:2605–2613. doi: 10.1099/00221287-145-9-2605. [DOI] [PubMed] [Google Scholar]
- 12.Kloos W E. Taxonomy and systematics of staphylococci indigenous to humans. In: Croosley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingston; 1997. pp. 113–137. [Google Scholar]
- 13.Kloos W E, Ballard D N, George C G, Webster J A, Hubner R J, Ludwig W, Schleifer K H, Schubert K. Delimiting the genus Staphylococcus through description of Macrococcus caseolyticus gen. nov., comb. nov. and Macrococcus equipercicus sp. nov., Macrococcus bovicus sp. nov. and Macrococcus carouselicus sp. nov. Int J Syst Bacteriol. 1998;48:859–877. doi: 10.1099/00207713-48-3-859. [DOI] [PubMed] [Google Scholar]
- 14.Kloos W E, Bannerman T L. Staphylococcus and Micrococcus. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C.: American Society for Microbiology; 1995. pp. 282–298. [Google Scholar]
- 15.Kloos W E, Bannerman T L. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7:117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y, Lian J, Ito T, Kanamori M, Matsumaru H, Maruyama A, Murakami H, Hosoyama A, Mizutani-Ui Y, Takahashi N K, Sawano T, Inoue R, Kaito C, Sekimizu K, Hirakawa H, Kuhara S, Goto S, Yabuzaki J, Kanehisa M, Yamashita A, Oshima K, Furuya K, Yoshino C, Shiba T, Hattori M, Ogasawara N, Hayashi H, Hiramatsu K. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/s0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 17.Kwok A Y, Su S C, Reynolds R P, Bay S J, Av-Gay Y, Dovichi N J, Chow A W. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int J Syst Bacteriol. 1999;49:1181–1192. doi: 10.1099/00207713-49-3-1181. [DOI] [PubMed] [Google Scholar]
- 18.Martineau F, Picard F J, Ke D, Paradis S, Roy P H, Ouellette M, Bergeron M G. Development of a PCR assay for identification of staphylococci at genus and species levels. J Clin Microbiol. 2001;39:2541–2547. doi: 10.1128/JCM.39.7.2541-2547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perl T M, Rhomberg P R, Bale M J, Fuchs P C, Jones R N, Koontz F P, Pfaller M A. Comparison of identification systems for Staphylococcus epidermidis and other coagulase-negative Staphylococcus species. Diagn Microbiol Infect Dis. 1994;18:151–155. doi: 10.1016/0732-8893(94)90084-1. [DOI] [PubMed] [Google Scholar]
- 20.Petras P. Staphylococcus pulvereri = Staphylococcus vitulus? Int J Syst Bacteriol. 1998;48:617–618. doi: 10.1099/00207713-48-2-617. [DOI] [PubMed] [Google Scholar]
- 21.Poyart C, Berche P, Trieu-Cuot P. Characterization of superoxide dismutase genes from Gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 22.Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J Clin Microbiol. 1998;36:41–47. doi: 10.1128/jcm.36.1.41-47.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poyart C, Quesnes G, Trieu-Cuot P. Sequencing the gene encoding manganese-dependent superoxide dismutase for rapid species identification of enterococci. J Clin Microbiol. 2000;38:415–418. doi: 10.1128/jcm.38.1.415-418.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Renneberg J, Rieneck K, Gutschik E. Evaluation of Staph ID 32 system and Staph-Zym system for identification of coagulase-negative staphylococci. J Clin Microbiol. 1995;33:1150–1153. doi: 10.1128/jcm.33.5.1150-1153.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Stackebrandt E, Goebel B M. A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 27.Takahashi T, Satoh I, Kikuchi N. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol. 1999;49:725–728. doi: 10.1099/00207713-49-2-725. [DOI] [PubMed] [Google Scholar]
- 28.Wright Valderas M, Hart M E. Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus. J Bacteriol. 2001;183:3399–3407. doi: 10.1128/JB.183.11.3399-3407.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu S W, de Lencastre H, Tomasz A. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J Bacteriol. 2001;183:2417–2424. doi: 10.1128/JB.183.8.2417-2424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]