Abstract
Introduction: Renal cancer is one of the most common cancers in the world, but the effect of therapies on advanced renal cancer has not improved for decades. Ferroptosis is an emerging type of programmed cell death and has been proved to play a vital role in many kinds of cancers. However, the mechanisms of ferroptosis regulated by long noncoding RNA (lncRNA) in the context of renal cancer was still unknown. Methods: We used bioinformation analysis to identify SLC16A1-AS1 as a survival-related lncRNA in renal cancer. The expression levels of SLC16A1-AS1 and microRNA-143-3p (miR-143-3p) were detected by quantitative reverse transcription–polymerase chain reaction. Cell counting kit-8 assay, 5-bromo-2′-deoxyuridine proliferation assay, and colony-formation assay were performed to evaluate cell viability and proliferation. Wound-healing assay and transwell assay were used to examine cell invasive and migration capacity. Dual-luciferase reporter assay and RNA-binding protein immunoprecipitation were used to identify the interaction among SLC16A1-AS1, miR-143-3p, and the target protein solute carrier family 7 membrane 11 (SLC7A11). Reduced glutathione and glutathione and lipid peroxidation measurements were carried out to evaluate the level of ferroptosis, and the expression levels of ferroptosis-related proteins were analyzed by western blot. Results: Our study revealed that SLC16A1-AS1 has high expression and was associated with overall survival in renal cancer. Knockdown SLC16A1-AS1 inhibited cell viability, proliferation, and migration of renal cancer cells. Furthermore, it was demonstrated that SLC16A1-AS1 served as a sponge of miR-143-3p, and knockdown SLC16A1-AS1 significantly increased the enrichment of miR-143-3p. And then, SLC7A11 was identified as the target protein of miR-143-3p, and overexpression miR-143-3p remarkably inhibited the expression of SLC7A11. Moreover, knockdown SLC16A1-AS1 could aggravate this effect. Finally, through inhibiting SLC7A11 expression, silencing SLC16A1-AS1 induced ferroptosis via increasing miR-143-3p. Conclusion: The present results suggest that silencing lncRNA SLC16A1-AS1 can induce ferroptosis through miR-143-3p/SLC7A11 signaling in renal cancer. Our study provided a novel view into the pathogenesis and treatment strategy of RCC.
Keywords: SLC16A1-AS1, miR-143-3p, SLC7A11, renal cell carcinoma, ferroptosis
Introduction
Renal cancer was estimated to be the sixth most new case in males and the ninth in females. 1 Clear cell carcinoma is the most common histologic type of renal cancer, accounting for over 80%. To date, the standard therapeutic strategy of most renal cancer cases is surgery with regular follow-up. As for advanced or metastatic renal cancer, over the last 2 decades, there are some therapy options emerging continuously, from inhibitors of the vascular endothelial growth factor to tyrosine kinase inhibitor and most of them have been demonstrated to be successful in therapeutic effect. Recently, an immune checkpoint inhibitor has gradually shown its efficacy and safety in clinical trials. 2 Despite the advances in therapeutic strategies, the overall prognosis of renal cancer patients has not significantly improved yet.
Ferroptosis is a novel type of programmed cell death, based mainly on iron accumulation and lipid peroxidation. Although no exclusive evidence reveals the normal physiological function for ferroptosis in the cellular process, emerging evidence suggests specific connections between ferroptosis and many pathological conditions, such as degenerative diseases, cardiovascular diseases, and renal ischemia/reperfusion injury. Ferroptosis has also shown its antitumor function in many kinds of cancers, indicating a potential role in cancer therapy.3–5 Ferroptosis can be triggered mainly through the transporter-dependent pathway (eg, inactivation of system xc−) or enzyme-dependent pathway (eg, inhibition of glutathione peroxidase 4 [GPX4]). System xc−, the cystine/glutamate antiporter, can import cystine, which is then reduced to cysteine (an ingredient of glutathione synthesis), and export glutamate. 6 System xc− is constituted of 2 subunits, solute carrier family 7 membrane 11 (SLC7A11) and solute carrier family 3 membrane 2 (SLC3A2), and SLC7A11 is the major subunit to function. Inhibition of SLC7A11 directly inactivates system xc−, leading to depletion of cellular cysteine and then lipid peroxidation, and finally triggers ferroptosis. Thus, targeting SLC7A11 could be a hopeful strategy in cancer therapy.7,8
Long noncoding RNA (lncRNA), the length of which is longer than 200 nucleotides, is a class of RNAs that is not translated into proteins. Because of distinct functions, lncRNAs play more and more important roles in biological processes, such as epigenetic regulation, cell cycle, and cell proliferation regulation.9,10 Emerging evidence has shown that lncRNAs have a tumor-promoting function or tumor-suppressing function via dysregulation of expression. It has been reported that lncRNAs have various regulatory mechanisms and sponge function is an important one of them. lncRNA sponges microRNA (miRNA) via combining with miRNA, acting as a competing-endogenous RNA (ceRNA), and thus decreases miRNA activity and increases miRNA targeted gene expression. 11 SLC16A1-AS1 gene is located on chromosome 1, at 1p13.2-p12, in antisense direction, SLC16A1-AS1 is a kind of lncRNA, which has been reported to be associated with bladder cancer and nonsmall cell lung cancer. However, the physiological function of SLC16A1-AS1 in renal cancer remains unknown.12,13
In the present study, we discovered that SLC16A1-AS1 was highly expressed in RCC cells. Notably, downregulation of SLC16A1-AS1 inhibited cell apoptosis and ferroptosis. Further, we discovered that SLC16A1-AS1 serves as a miRNA sponge for miR-143-3p and silencing SLC16A1-AS1 increased miR-143-3p and decreased ferroptosis key factor SLC7A11, and thus induced ferroptosis. Our study provided a novel view into the pathogenesis and treatment strategy of RCC.
Materials and Methods
Cell Culture and Treatment
All cell lines were purchased from ATCC. HK-2 cell line was cultured with Dulbecco’s modified eagle medium/10% fetal bovine serum media and renal cell carcinoma cell lines, 786-O, A498, and Caki-1, were all cultured with RPMI 1640/10% fetal bovine serum media. To ensure authenticity, all cell lines were verified through short tandem repeat authentication.
Quantitative Reverse Transcription–Polymerase Chain Reaction (qRT–PCR)
The RNA extraction was constructed by the TRIzol method. The RNAs in cytoplasmic and nuclear were isolated using nuclear and cytoplasmic extraction reagents. The complementary DNA was synthesized using PrimeScript™ RT Reagent Kit. Subsequently, qRT–PCR was performed in a Bio-Rad Real-time PCR system with SYBR Green PCR Master Mix.
Transfection
Cell transfection was carried out using Lipofectamine 3000. MiR-143-3p mimics, pcDNA3.1/SLC7A11 plasmid, pcDNA3.1/SLC16A1-AS1 plasmid, siRNA-SLC16A1-AS1, and negative control miRNA were synthesized by Shanghai Integrated Ribo Co., Ltd and transfected into the cells using Lipofectamine RNAiMAX Transfection Reagent.
Cell Counting Kit-8 (CCK-8) Assay
CCK-8 assay was used to evaluated cell viability. Cells were seeded in 96-well plates at a concentration of 2×103 /well. Before detecting, 10 μL of CCK-8 solution were added to each well. After 2 h of incubation, the absorbance of each well was detected using a Perkin Elmer microplate reader.
5-Bromo-2′-Deoxyuridine Proliferation Assay
To measure cell proliferation, a 5-bromo-2′-deoxyuridine (BrdU) proliferation assay was constructed. Then 5 × 10 4 cells/well were seeded in 24-well plates and 24 h later, cells were incubated with 10 μM BrdU for 40 min. Then, cells were fixed with 4% cold paraformaldehyde and incubated with hydrochloric acid to break the DNA structure. After incubating with anti-BrdU overnight and secondary antibody for 1 h, labeled cells were observed using a fluorescence microscope.
Colony-Formation Assays
Firstly, cells were seeded in 6-well plates evenly at a concentration of about 200 cells per well. Before detecting, cells were fixed with methanol and stained with 0.1% crystal violet. Colonies with >0.05 mm diameter were counted and colony number was analyzed using ImageJ software.
Transwell Invasion Assay
A Transwell invasion assay was performed to evaluate the invasive capacity of renal cancer cells with Millipore transwell chambers. Cells were seeded in the top chambers at a density of 2 × 104 cells/well in 100 μL RPMI 1640 medium without serum and the lower chambers were filled with 600 μL complete RPMI 1640 medium as mentioned before. After 24 h incubation, the cells were stained with 0.1% crystal violet dye and the images were analyzed through ImageJ software.
Wound-Healing Assay
A wound-healing assay was carried out to evaluate migration in renal cancer cells. Cells were seeded in 6-well plates at a density of 1 × 105 cells/well. After overnight incubation, the surface of cells was scratched with a straight gap, and 24-h and 48-h later, the width of the gap was recorded, respectively.
RNA-Binding Protein Immunoprecipitation (RIP)
RIP assay was used to discover the interactions among lncRNA, miRNA, and target messenger RNA (mRNA). Cells were co-transfected with MS2bs-SLC16A1-AS1, MS2bs-SLC16A1-AS1-mut, and MS2bs-Rluc by Lipofectamine 3000, and then, RIP assay was performed using a protein immunoprecipitation kit according to the manufacturer’s instruction. The anti-Ago2 antibodies were utilized to pulldown RNA-induced silencing complex, which binds mRNA, miRNA, and lncRNA, and the abundance of SLC16A1-AS1, miR-143-3p, and SLC7A11 was detected by qRT–PCR.
Dual-Luciferase Reporter Assay
Dual-luciferase reporter assay was used to examine the binding between miRNA and its target mRNA. The 3′-untranslated region (3′-UTR) fragment with and without the potential miR-143-3p binding site of SLC7A11 mRNA was inserted into luciferase reporter vectors named SLC7A11-WT and SLC7A11-MUT. Cells were co-transfected with SLC7A11-WT or SLC7A11-MUT and miR-143-3p mimics, and then, the luciferase activity was measured using the dual-luciferase reporter assay system according to the manufacturer’s instruction.
Reduced Glutathione and Glutathione Assay
Reduced glutathione and glutathione (GSH/GSSG) ratio in renal cancer cells was detected using Glutathione Detection Kit (Beyotime Biotechnology) according to the manufacturer’s instruction.
Lipid Peroxidation Measurements
After different treatments, cells were stained with 5 μM Liperfluo (Dojindo) at 37°C for 30 min in the dark and then cells were stained with Hoechst nuclear stain (Dojindo) for 10 min. Finally, the cells were washed with phosphate-buffer saline twice and detected under an Olympus FV1200 confocal microscopy.
Western Blot
Total proteins of renal cancer cells were extracted by radioimmunoprecipitation assay buffer (RIPA) lysis buffer, separated by SDS–polyacrylamide gel electrophoresis, and then transferred onto a piece of polyvinylidene difluoride transfer membrane. The membranes were incubated with antibodies against GPX4 (ab125066, Abcam), cyclooxygenase 2 (COX2; ab62331, Abcam), ACSL4 (ab155282, Abcam), and glyceraldehyde 3-phosphate dehydrogenase (ab8245, Abcam), and after incubation with a secondary antibody, the proteins were visualized by chemiluminescence on Bio-Rad ChemiDoc XRS+.
Statistical Analysis
GraphPad Prism (Version 8.0) and SPSS 25.0 were used to analyze all the data. The differences between groups were analyzed through Student’s t-test and one-way analysis of variance. P-value <.05 was considered to be statistically significant.
Results
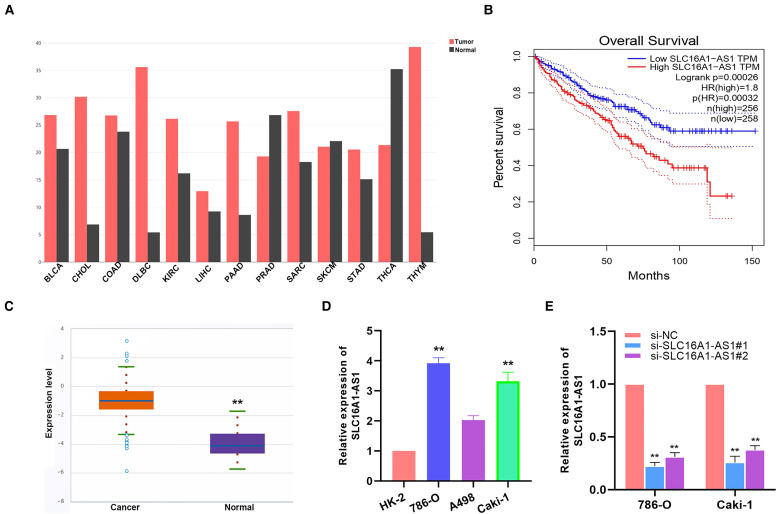
SLC16A1-AS1 is highly expressed in renal cancer cells. Through bioinformatic analysis, it was revealed that the expression level of SLC16A1-AS1 in tumor cases was higher than that in normal cases (Figure 1A and C). And the patients with high expression of SLC16A1-AS1 had poorer overall survival than the patients with low expression (Figure 1B). In vitro, the expressions of SLC16A1-AS1 in renal cancer cell lines were all significantly higher than that in a normal renal tubular epithelial cell line (Figure 1D). For further research, we designed and synthesized siRNA for SLC16A1-AS1 in 786-O cells and verified the expression level by PCR. (Figure 1E).
Figure 1.
SLC16A1-AS1 is highly expressed in renal cancer cells. (A) Analysis from the TCGA database shows the expression of SLC16A1-AS1 in various malignant tumors. (B) A Kaplan–Meyer survival analysis shows patients with high expression of SLC16A1-AS1 had poorer overall survival than the patients with low expression. (C) Analysis from Starbase 3.0 shows the expression of SLC16A1-AS1 in RCC samples and normal tissues. (D) SLC16A1-AS1 expression in normal renal cell (HK-2) and 3 RCC cell lines (786-O, A498, and Caki-1). (E) Knockdown efficacy of 2 distinct si-SLC16A1-AS1 evaluated by qRT–PCR. *p<.05, **p<.01 compared with the NC group.
Abbreviations: RCC, renal cell carcinoma; qRT–PCR, quantitative reverse transcription–polymerase chain reaction; NC, negative control.
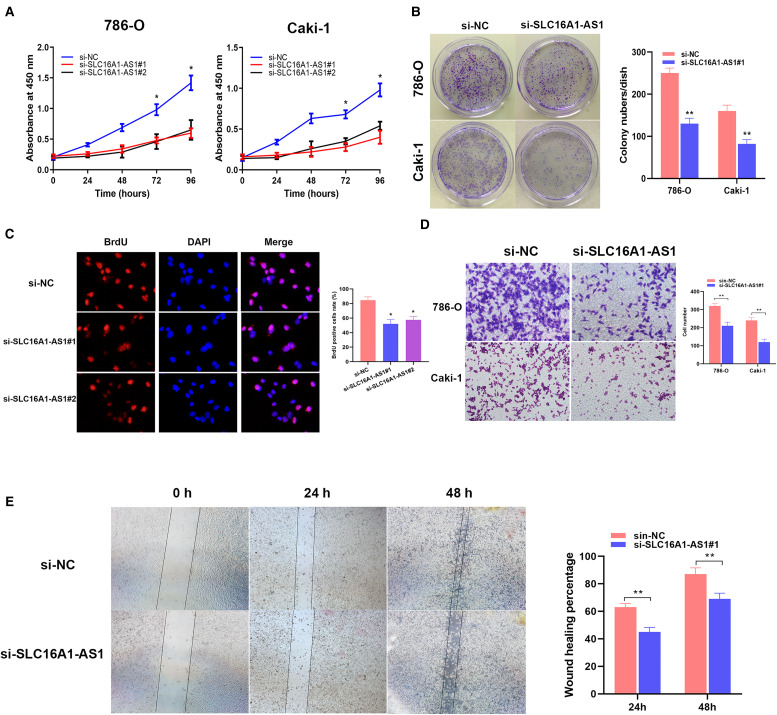
SLC16A1-AS1 promotes renal cancer proliferation and migration.
CCK-8 assays showed that silencing of SLC16A1-AS1 efficiently inhibited cell viability in renal cancer cells (Figure 2A). Colony-formation assays and BrdU assays were performed to evaluate the effect on cell proliferation after silencing SLC16A1-AS1, and as expected, the number of the colony and BrdU-positive cells was decreased, suggesting that the proliferation in renal cancer cells was significantly suppressed (Figure 2B and C). Transwell invasion assay showed a decrease in cell invasive capacity after transfection with si-SLC16A1-AS1 (Figure 2D). As for migration ability, the results of wound-healing assays demonstrated that the deficiency of SLC16A1-AS1 prolonged wound-healing time indicating a poor migration ability in renal cancer (Figure 2E). All these trends were reversed by overexpression of SLC16A1-AS1(Figure 2A to E). These results uncovered that SLC16A1-AS1 was probably a pro-cancer lncRNA.
Figure 2.
SLC16A1-AS1 promotes renal cancer proliferation and migration. (A) The relative proliferation levels of 786-O and Caki-1 cells were checked by CCK-8 assays. (B) Colony-formation assays were performed to evaluate cell proliferation after transfection. (C) BrdU assay was performed to assess cell proliferation. (D) A Transwell invasion assay was performed to detect cell invasive capacity. (E) Wound-healing assays were performed to evaluate cell migration. *p<.05, **p<.01 compared with the NC group.
Abbreviations: CCK-8: cell counting kit-8; NC: negative control; OV: overexpression; BrdU, 5-bromo-2′-deoxyuridine.
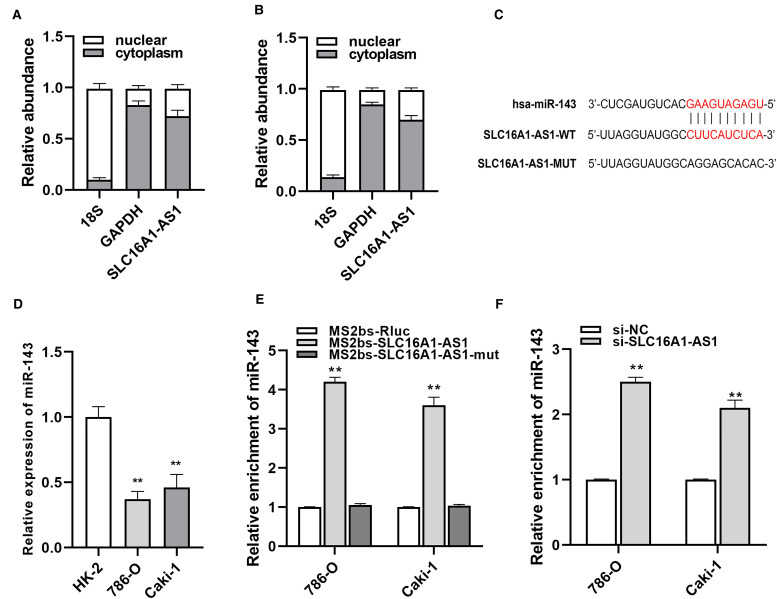
SLC16A1-AS1 acts as a sponge of miR-143-3p. We found that SLC16A1-AS1 was mainly located in the cytoplasm indicating a potential interaction with miRNAs (Figure 3A and B). To further explore the mechanism of SLC16A1-AS1 in antitumor progression, the database was applied to figure out the potential interaction between SLC16A1-AS1 and miRNA, and finally, miR-143-3p was predicted to be the best-matched one (Figure 3C). Then, the expression of miR-143-3p was evaluated between normal cells and renal cancer cells, and as a result, cancer cells all had relatively lower expression of miR-143-3p than normal cells (Figure 3D). MS2-RIP assays uncovered that miR-143-3p was enriched in the MS2bs-SLC16A1-AS1 group while knockdown SLC16A1-AS1 significantly upregulated miR-143-3p expression in renal cancer cells (Figure 3E and F). These results indicated that SLC16A1-AS1 inhibits renal cancer growth mainly through sponging miR-143-3p.
Figure 3.
SLC16A1-AS1 acts as a sponge of miR-143-3p. (A, B) The localization of SLC16A1-AS1 was examined in 786-O and Caki-1 cells. (C) The putative binding site of miR-143-3p on SLC16A1-AS1. (D) The expression level of miR-143-3p in RCC cells was determined by qRT–PCR. (E) MS2-based RIP assay in RCC cells. (F) The expression level of miR-143-3p in RCC cells after transfection with si-SLC16A1-AS1. *p<.05, **p<.01 compared with the NC group.
Abbreviations: miR-143-3p, microRNA-143-3p; RCC, renal cell carcinoma; qRT–PCR, quantitative reverse transcription–polymerase chain reaction; RIP, RNA-binding protein immunoprecipitation; NC, negative control.
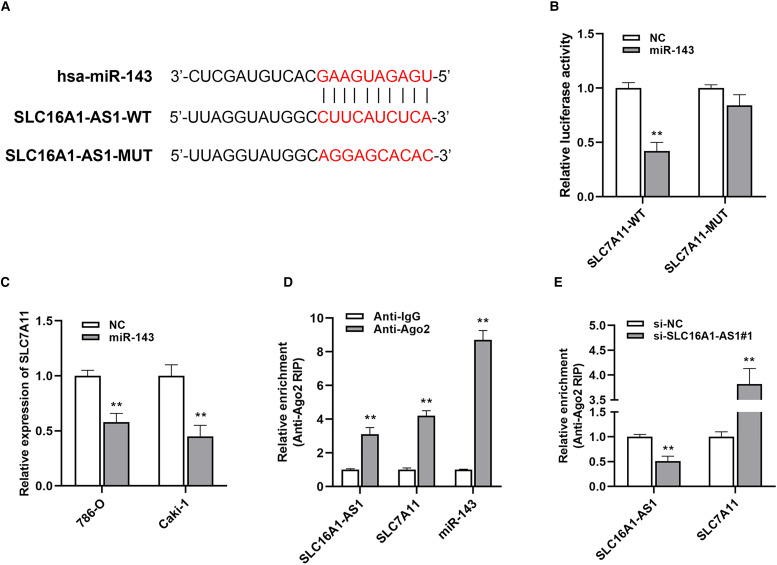
SLC7A11 serves as the target of miR-143-3p and is downregulated via SLC16A1-AS1 knockdown.
SLC7A11 was identified as the potential target of miR-143-3p based on the TargetScan online database (Figure 4A). Luciferase reporter assays revealed that the luciferase activity was significantly decreased in 293T cells after transfected with miR-143-3p and wildtype 3′-UTR-SLC7A11 reporter, while was not different statistically after transfected with a mutant 3′-UTR-SLC7A11 reporter (Figure 4B). Additionally, the expression of SLC7A11 was decreased after being transfected with a miR-143-3p mimic in renal cancer cells (Figure 4C). Ago2-related RIP assay demonstrated that SLC16A1-AS1, SLC7A11, and miR-143-3p were all enriched in renal cancer cells, and moreover, knockdown SLC16A1-AS1 remarkably increased SLC7A11 enrichment to Ago2 (Figure 4D and E). These results suggested that SLC16A1-AS1 knockdown inhibits the expression of SLC7A11 via serving as the ceRNA for miR-143-3p.
Figure 4.
SLC7A11 serves as the target of miR-143-3p and is downregulated via SLC16A1-AS1 knockdown. (A) The putative binding site of miR-143-3p within SLC7A11. (B) A dual-luciferase reporter assay was performed to verify the correlation between miR-143-3p and SLC7A11. (C) SLC7A11 expression in RCC cells was detected by qRT–PCR analysis. (D, E) RIP assay was performed to assess the enrichment of SLC16A1-AS1, SLC7A11, and miR-143-3p on Ago2 relative IgG. *p<.05, **p<.01 compared with the NC group.
Abbreviations: miR-143-3p, microRNA-143-3p; RCC, renal cell carcinoma; qRT–PCR, quantitative reverse transcription–polymerase chain reaction; RIP, RNA-binding protein immunoprecipitation.
Silencing SLC16A1-AS1 induces ferroptosis through downregulating SLC7A11.
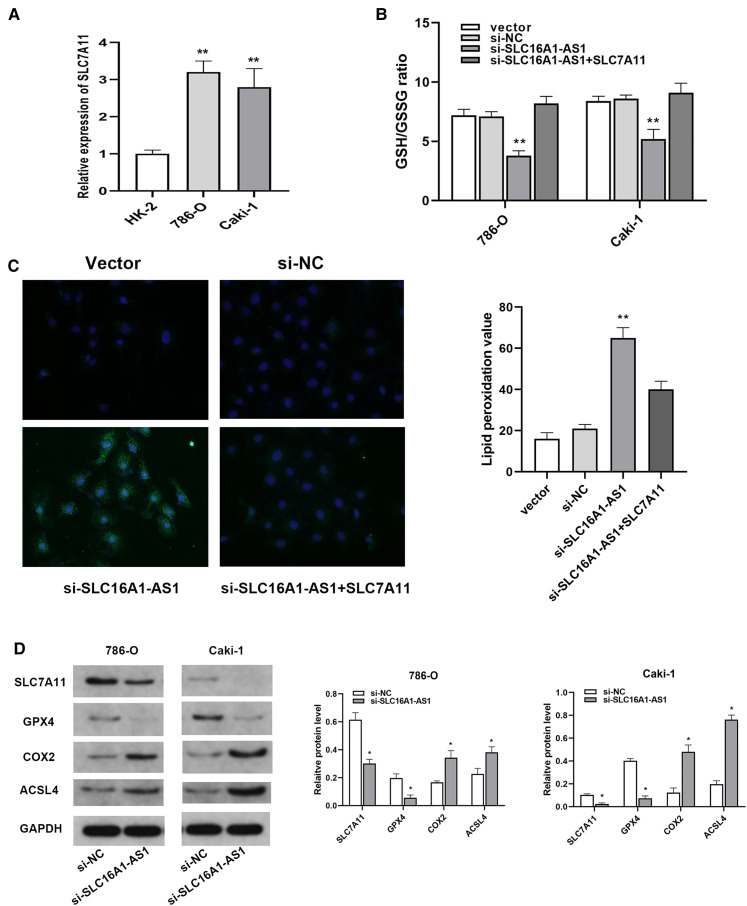
Compared with HK-2 cells, the levels of SLC7A11 expression in 786-O and Caki-1 cells were higher (Figure 5A). After detecting reduced glutathione and oxidized glutathione, we discovered that silencing SLC16A1-AS1 remarkably decreased GSH/GSSG ratio in renal cancer cells, while overexpression of SLC7A11 totally reversed the effect (Figure 5B). The level of lipid peroxidation was detected by liperfluo staining and the results have shown that silencing SLC16A1-AS1 significantly increased the level of lipid peroxidation in cell membranes, however, SLC7A11 overexpression notably reduced the amount of lipid peroxidation (Figure 5C). As for ferroptosis-related markers, western blot analysis revealed that silencing SLC16A1-AS1 upregulated ferroptosis promoters (COX2 and ACSL4) and downregulated ferroptosis inhibitors (GPX4) in renal cancer cells (Figure 5D).
Figure 5.
Silencing SLC16A1-AS1 induces ferroptosis through downregulating SLC7A11. (A) The expression level of SLC7A11 in RCC cells and normal cells. (B) GSH/GSSG assay was conducted in RCC cells after silencing SLC16A1-AS1. (C) Liperfluo level was examined using immunofluorescence staining. (D) Western blot analyses of SLC7A11 and ferroptosis-related proteins (GPX4, COX2, and ACSL4) in RCC cells after silencing SLC16A1-AS1. *p<.05, **p<.01 compared with the NC group.
Abbreviations: GSH/GSSG, reduced glutathione and glutathione; RCC, renal cell carcinoma; GPX4, glutathione peroxidase 4; COX2, cyclooxygenase 2.
Discussion
In this study, silencing SLC16A1-AS1 induced ferroptosis in RCC cells, which was identified with ferroptosis-specific characteristics, such as increased iron concentration and lipid peroxidation level and decreased ratio of GSH/GSSG. Naturally, a ferroptosis inhibitor could alleviate the above changes. Ferroptosis, a novel nonapoptosis regulated cell death, has become a hot point in the last few years, especially in cancer research studies. The fast-growing studies have reported that the proliferation of cancer cells could be significantly controlled by inducing ferroptosis, such as in hematological malignancies, hepatocellular carcinoma (HCC), lung cancer, gastroenteric cancer, and renal cancer.3,8,14–17 Ferroptosis can be triggered through 2 main mechanisms: Fenton reaction mediated by iron accumulation and antioxidant deregulation. 18 There are 2 pathways to cause antioxidant deregulation, transport-dependent pathway and enzyme-dependent pathway. 6 And the final step of these 2 mechanisms to induce ferroptosis is the increase of lipid peroxidation, especially polyunsaturated fatty acids peroxidation. 5 For example, SLC7A11 is the key subunit of system xc−, which is the main transporter of cystine/glutamate. Inhibition of SLC7A11 can induce ferroptosis by limiting the biosynthesis of glutathione which is the main antioxidant in cells. 19 In our study, the SLC7A11 expression level was decreased in RCC cells with SLC16A1-AS1 silence and increased in cells with SLC16A1-AS1 overexpression. Therefore, ferroptosis in RCC was induced through inhibition of SLC7A11 mediated by SLC16A1-AS1.
Recently, lncRNA SLC16A1-AS1 has been reported to be associated with various kinds of cancers. Liu et al 12 identified SLC16A1-AS1 as an independent prognostic biomarker in nonsmall cell lung cancer through microarray analysis. Logotheti et al 13 revealed a new regulatory mechanism of SLC16A1-AS1 in bladder cancer through which SLC16A1-AS1 induced by E2F1 forms a complex with E2F1 promoter and enhances SLC16A1/MCT1 expression, which promotes metabolic reprogramming and invasiveness of bladder cancer. As for HCC, bioinformation analysis was carried out by Song et al 20 to identify a cluster of lncRNAs as a potential diagnostic and prognostic biomarker of HCC and SLC16A1-AS1 was one of them. Tian et al 21 reported that SLC16A1-AS1 was upregulated in HCC and clarified it as a poor-survival predictive factor. Besides, SLC16A1-AS1 has also shown its distinct function in some other cancers, such as glioblastoma, oral squamous cell carcinoma, tongue squamous cell carcinoma, and cervical squamous cell carcinoma.22–25 In the present study, the expression of SLC16A1-AS1 was significantly increased, and meanwhile, knockdown of SLC16A1-AS1 decreased the level of apoptosis and ferroptosis in RCC cells. This indicated that SLC16A1-AS1 might play a vital role in regulating RCC cell survival.
Increasing evidence has uncovered that miRNA sponge function is the main mechanism of lncRNAs to perform a physiological function, and naturally, lncRNA SLC16A1-AS1 can also act as a ceRNA to sponge miRNA. Pei et al 24 discovered that SLC16A1-AS1 served as the sponge of miR-310b-3p and then upregulated the expression of CHD5, the target of miR-310b-3p. Through SLC16A1-AS1/miR-310b-3p/CHD5 axis, the proliferation and invasiveness of HCC cells were suppressed and the radiosensitivity of HCC cells was enhanced. 24 Our results unveiled that SLC16A1-AS1 served as the sponge of miR-143-3p and upregulated the expression of SLC7A11, which was clarified as the downstream target of miR-143-3p. miR-143-3p, known as a tumor suppressor, is located at chromosome 5q32. MiR-143-3p is downregulated in many kinds of cancers and its expression is regulated by p53 and hypoxia-inducible factor-1α. 26 In colorectal cancer, miR-143-3p inhibited cancer cell growth and proliferation and enhanced the cancer cell sensitivity to chemotherapeutic agents, and miR-143-3p showed the same effects in bladder cancer and prostate cancer.27–30 As for renal cancer, Takai et al 31 reported that miR-143-3p suppressed tumor growth through downregulating KRAS networks. Notably, the KRAS network has been demonstrated to be the main target gene of miR-143-3p in the previous studies, while whether SLC7A11 has the link to miR-143-3p is still unknown yet. SLC7A11 is the main subunit of system xc− and targeting SLC7A11 can regulate ferroptosis. Thus, in our present study, we revealed that SLC7A11 was the target of miR-143-3p and could be regulated by SLC16A1-AS1–miR-143-3p ceRNA pair to play its role in ferroptosis. Due to the limitation of condition, in vivo assay and the in-depth study of the mechanism will be continued in the follow-up study.
Conclusions
In summary, our results demonstrated that lncRNA might play vital roles in renal cancer acting as miRNA sponges. We found that the SLC16A1-AS1/miR-143-3p/SLC7A11 axis promoted RCC cells apoptosis and ferroptosis through a mechanism involving ceRNA. Our findings proved that SLC16A1-AS1 was associated with ferroptosis in renal cancer, indicating that SLC16A1-AS1 may be a treatment target in renal cancer.
Abbreviations
- BrdU
5-bromo-2′-deoxyuridine
- CCK-8
cell counting kit-8
- ceRNA
competing-endogenous RNA
- COX2
cyclooxygenase 2
- GPX4
glutathione peroxidase 4
- GSH/GSSG
reduced glutathione and glutathione
- HCC
hepatocellular carcinoma
- lncRNA
long noncoding RNA
- miR-143-3p
microRNA-143-3p
- miRNA
microRNA
- mRNA
messenger RNA
- qRT–PCR
quantitative reverse transcription–polymerase chain reaction
- RIP
RNA-binding protein immunoprecipitation
- SLC3A2
solute carrier family 3 membrane 2
- SLC7A11
solute carrier family 7 membrane 11
- 3′-UTR
3′-untranslated region.
Footnotes
Ethics Approval and Consent to Participate: Our study was approved and checked by The Ethics Committee of Renmin Hospital of Wuhan University.
Availability of Data and Material: The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors’ Contributions: Yang Du contributed to the conception and design of the work. Yan ze Li and Heng cheng Zhu conducted the experiments. Yan ze Li analyzed data and wrote this manuscript. Hong chao Zhao and Lei Wang revised and reviewed the manuscript. All authors contributed to final approval of the version to be published and agree to be accountable for all aspects of the work.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Yang Du https://orcid.org/0000-0002-3803-8245
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics. CA Cancer J Clin. 2021;71(1):7‐33. [DOI] [PubMed] [Google Scholar]
- 2.Riaz IB, He H, Ryu AJ, et al. A living, interactive systematic review and network meta-analysis of first-line treatment of metastatic renal cell carcinoma. Eur Urol. 2021;80(6):712–723. [DOI] [PubMed] [Google Scholar]
- 3.Bebber CM, Muller F, Prieto CL, Weber J, von Karstedt S. Ferroptosis in cancer cell biology. Cancers. 2020;12(1). 10.3390/cancers12010164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han C, Liu Y, Dai R, Ismail N, Su W, Li B. Ferroptosis and its potential role in human diseases. Front Pharmacol. 2020;11:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stockwell BR, Friedmann AJ, Bayir H, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171(2):273‐285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Yu C, Kang R, Kroemer G, Tang D. Cellular degradation systems in ferroptosis. Cell Death Differ. 2021;28(4):1135‐1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato M, Onuma K, Domon M, et al. Loss of the cystine/glutamate antiporter in melanoma abrogates tumor metastasis and markedly increases survival rates of mice. Int J Cancer. 2020;147(11):3224–3235. [DOI] [PubMed] [Google Scholar]
- 8.Yang WS, SriRamaratnam R, Welsch ME, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1-2):317‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965‐3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393‐407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wan P, Su W, Zhang Y, et al. LncRNA H19 initiates microglial pyroptosis and neuronal death in retinal ischemia/reperfusion injury. Cell Death Differ. 2020;27(1):176‐191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu HY, Lu SR, Guo ZH, et al. lncRNA SLC16A1-AS1 as a novel prognostic biomarker in non-small cell lung cancer. J Investig Med. 2020;68(1):52‐59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logotheti S, Marquardt S, Gupta SK, et al. LncRNA-SLC16A1-AS1 induces metabolic reprogramming during bladder cancer progression as target and co-activator of E2F1. Theranostics. 2020;10(21):9620‐9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J, Chen X, Jiang L, et al. DJ-1 suppresses ferroptosis through preserving the activity of S-adenosyl homocysteine hydrolase. Nat Commun. 2020;11(1):1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Zandkarimi F, Zhang Y, et al. Energy-stress-mediated AMPK activation inhibits ferroptosis. Nat Cell Biol. 2020;22(2):225‐234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin-Sanchez D, Fontecha-Barriuso M, Martinez-Moreno JM, et al. Ferroptosis and kidney disease. Nefrologia. 2020;40(4):384–394. [DOI] [PubMed] [Google Scholar]
- 17.Linkermann A, Skouta R, Himmerkus N, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci USA. 2014;111(47):16836‐16841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26(3):165‐176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang L, Liu W, Liu F, et al. IMCA Induces ferroptosis mediated by SLC7A11 through the AMPK/mTOR pathway in colorectal cancer. Oxid Med Cell Longev. 2020;2020:1675613. 10.1155/2020/1675613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song M, Zhong A, Yang J, et al. Large-scale analyses identify a cluster of novel long noncoding RNAs as potential competitive endogenous RNAs in progression of hepatocellular carcinoma. Aging. 2019;11(22):10422‐10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tian J, Hu D. LncRNA SLC16A1-AS1 is upregulated in hepatocellular carcinoma and predicts poor survival. Clin Res Hepatol Gastroenterol. 2021;45(2):101490. [DOI] [PubMed] [Google Scholar]
- 22.Feng H, Zhang X, Lai W, Wang J. Long non-coding RNA SLC16A1-AS1: its multiple tumorigenesis features and regulatory role in cell cycle in oral squamous cell carcinoma. Cell Cycle. 2020;19(13):1641‐1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long Y, Li H, Jin Z, Zhang X. LncRNA SLC16A1-AS1 is upregulated in glioblastoma and promotes cancer cell proliferation by regulating miR-149 methylation. Cancer Manag Res. 2021;13:1215‐1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei S, Chen Z, Tan H, Fan L, Zhang B, Zhao C. SLC16A1-AS1 Enhances radiosensitivity and represses cell proliferation and invasion by regulating the miR-301b-3p/CHD5 axis in hepatocellular carcinoma. Environ Sci Pollut Res Int. 2020;27(34):42778‐42790. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H, Jin S, Ji A, et al. LncRNA SLC16A1-AS1 suppresses cell proliferation in cervical squamous cell carcinoma (CSCC) through the miR-194/SOCS2 axis. Cancer Manag Res. 2021;13:1299‐1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokumaru Y, Takabe K, Yoshida K, Akao Y. Effects of MIR143 on rat sarcoma signaling networks in solid tumors: a brief overview. Cancer Sci. 2020;111(4):1076‐1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28(10):1385‐1392. [DOI] [PubMed] [Google Scholar]
- 28.Noguchi S, Yasui Y, Iwasaki J, et al. Replacement treatment with microRNA-143 and -145 induces synergistic inhibition of the growth of human bladder cancer cells by regulating PI3K/Akt and MAPK signaling pathways. Cancer Lett. 2013;328(2):353‐361. [DOI] [PubMed] [Google Scholar]
- 29.Xu B, Niu X, Zhang X, et al. miR-143 decreases prostate cancer cells proliferation and migration and enhances their sensitivity to docetaxel through suppression of KRAS. Mol Cell Biochem. 2011;350(1-2):207‐213. [DOI] [PubMed] [Google Scholar]
- 30.Yoshikawa Y, Taniguchi K, Tsujino T, et al. Anti-cancer effects of a chemically modified mir-143 on bladder cancer by either systemic or intravesical treatment. Mol Ther Methods Clin Dev. 2019;13:290‐302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takai T, Tsujino T, Yoshikawa Y, et al. Synthetic miR-143 exhibited an anti-cancer effect via the downregulation of K-RAS networks of renal cell cancer cells in vitro and in vivo. Mol Ther. 2019;27(5):1017‐1027. [DOI] [PMC free article] [PubMed] [Google Scholar]