Abstract
Coenzyme Q (CoQ) serves as an electron carrier in aerobic respiration and has become an interesting target for biotechnological production due to its antioxidative effect and benefits in supplementation to patients with various diseases. Here, we review discovery of the pathway with a particular focus on its superstructuration and regulation, and we summarize the metabolic engineering strategies for overproduction of CoQ by microorganisms. Studies in model microorganisms elucidated the details of CoQ biosynthesis and revealed the existence of multiprotein complexes composed of several enzymes that catalyze consecutive reactions in the CoQ pathways of Saccharomyces cerevisiae and Escherichia coli. Recent findings indicate that the identity and the total number of proteins involved in CoQ biosynthesis vary between species, which raises interesting questions about the evolution of the pathway and could provide opportunities for easier engineering of CoQ production. For the biotechnological production, so far only microorganisms have been used that naturally synthesize CoQ10 or a related CoQ species. CoQ biosynthesis requires the aromatic precursor 4-hydroxybenzoic acid and the prenyl side chain that defines the CoQ species. Up to now, metabolic engineering strategies concentrated on the overproduction of the prenyl side chain as well as fine-tuning the expression of ubi genes from the ubiquinone modification pathway, resulting in high CoQ yields. With expanding knowledge about CoQ biosynthesis and exploration of new strategies for strain engineering, microbial CoQ production is expected to improve.
Keywords: Coenzyme Q10 (CoQ10), Corynebacterium glutamicum, Escherichia coli, Metabolic engineering, Q complex, Ubi super complex, Yeast
Introduction
Coenzyme Q (CoQ), also called ubiquinone, plays an essential role in the respiratory chain of eukaryotes and many prokaryotes. CoQ is composed of a benzoquinone head group conjugated to a polyprenyl chain which length varies between organisms. Saccharomyces cerevisiae and Escherichia coli produce CoQ6 and CoQ8, respectively, whereas humans synthesize CoQ10 (Fig. 1). The most well-known function of CoQ is to transfer electrons and protons in respiratory chains that sustain bioenergetics. CoQ also acts as a cofactor in uridine biosynthesis, fatty acid oxidation, and for mitochondrial uncoupling proteins. Additionally, CoQ possesses antioxidant and lipid-solubility properties that protect lipids and lipoproteins from oxidative damage (Lee et al. 2012). The roles of CoQ are numerous and have been reviewed recently (Abby et al. 2020; Baschiera et al. 2021; Cirilli et al. 2021).
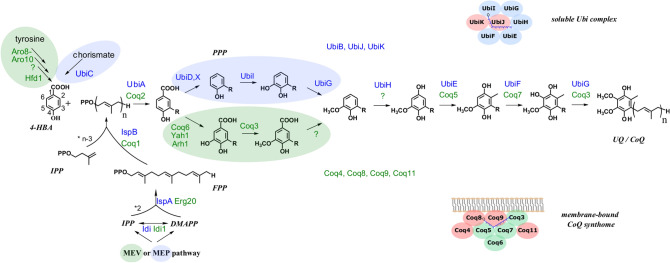
Fig. 1.
Comparative view of the eukaryotic (S. cerevisiae) and prokaryotic (E. coli) CoQ/UQ biosynthesis pathways. The proteins are in blue (E. coli) or green (S. cerevisiae), and the steps that differ between both organisms are highlighted. The numbering of the carbon atoms applied to all intermediates is given for 4-hydroxybenzoic acid (4-HBA) and the polyprenyl chain (n = 6 for S. cerevisiae, n = 8 for E. coli, n = 10 for CoQ10, the CoQ form found in humans) is depicted by R on all intermediates derived from 4-HBA. The Ubi complex and the CoQ synthome illustrate the supramolecular organization of some proteins of the pathways (enzymes in green/blue, accessory proteins in pink). Isopentenyl diphosphate (IPP), dimethylallyl diphosphate (DMAPP) and farnesyl diphosphate (FPP) are building blocks for the synthesis of the polyprenyl diphosphate tail which is added onto 4-HBA by UbiA/Coq2
Although CoQ10 is synthesized in human cells and taken up with food, age (Kalén et al. 1989), disease states and use of certain pharmacotherapeutic agents such as statins can lead to CoQ10 deficiency (Potgieter et al. 2013). Studies support that dietary supplementation of CoQ10, i.a., is beneficial for patients with cardiovascular and neurodegenerative diseases by modulating inflammatory and oxidative DNA damage responses (Yubero-Serrano et al. 2012; Gutierrez-Mariscal et al. 2012), improves symptoms of chronic heart failure, reduces cardiovascular mortality (Mortensen et al. 2014), decreases lead-acetate induced neurotoxicity (Yousef et al. 2019) and potentially slows the functional decline in early Parkinson Disease (Shults 2002). Primary CoQ10 deficiency is caused by mutations in genes of the synthetic pathway and may lead to, e.g., infantile encephalomyopathy and ataxia, which can be mitigated by CoQ10 supplementation (Quinzii et al. 2006). CoQ10 is a highly demanded food supplement, mainly in the form of softgels, capsules, and tablets. To increase its bioavailability, self-emulsified drug delivery systems, nanoemulsions, or cyclodextrin complexes have been developed (Arenas‐Jal et al. 2020). Due to its use as a food supplement and to reduce wrinkles (Žmitek et al. 2017), the industrial production of CoQ10 is desired. Although different chemical synthetic approaches have been described (Luo et al. 2017), they suffer from poor tautomer selectivity because they generally yield polyprenyl chains with cis and trans isomers, whereas the natural CoQ isoforms have an all-trans configuration. Thus, microbial bio-production of CoQ10 has been developed and in the following, we review CoQ biosynthesis in model microorganisms before focusing on CoQ10 production by different bacteria.
Overview of CoQ biosynthesis pathways in microorganisms
Whereas CoQ is found in almost all eukaryotes, its distribution in bacteria is much more narrow as CoQ is encountered only within the phylum Proteobacteria (Schoepp-Cothenet et al. 2013). The global architecture of CoQ biosynthesis is shared between bacteria and eukaryotes as exemplified by the prototypic pathways from the bacterium E. coli and the yeast S. cerevisiae (Fig. 1). Three stages can be distinguished: the synthesis of the precursor of the benzoquinone head group, the synthesis and conjugation of the polyprenyl side chain, and the sequential modifications of the head group on prenylated intermediates.
Synthesis of the precursors of the head group of CoQ
4-Hydroxybenzoic acid (4-HBA) serves as a precursor of CoQ in prokaryotes and eukaryotes but is produced differently (Fig. 1). Bacteria possess one of two non-orthologous enzymes (UbiC or XanB2) to catalyze the one-step conversion of chorismate to 4-HBA (Siebert et al. 1994; Zhou et al. 2013). The production of 4-HBA from l-tyrosine in eukaryotes is a multi-step process not fully elucidated, which in S. cerevisiae depends at least on aromatic aminotransferases I and II (Aro8, Aro9) and the aldehyde dehydrogenase Hfd1 (Payet et al. 2016; Robinson et al. 2021). 4-Aminobenzoic acid (4-ABA) is also a CoQ precursor in the yeasts S. cerevisiae and Schizosaccharomyces pombe (Pierrel et al. 2010; Marbois et al. 2010; Nishida et al. 2020), and additional molecules like para-coumarate and resveratrol were also identified as precursors (Xie et al. 2015), although they are likely converted into 4-HBA before entering the CoQ pathway. A recent review on precursors of the benzoquinone head group of CoQ is available for further details (Fernández-Del-Río and Clarke 2021).
Synthesis of the polyprenyl side chain
Isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) are precursors for the side chain of ubiquinone and are the end products of the mevalonate (MVA) pathway in eukaryotes, archaea and some eubacteria or of the methylerythritol phosphate (MEP) pathway in plants and most bacteria (Pérez-Gil and Rodríguez-Concepción 2013). The MVA pathway from S. cerevisiae and the MEP pathway from E. coli have been reviewed lately (Kawamukai 2018).
IPP and DMAPP are reversibly isomerized by isopentenyl diphosphate isomerase (Idi/Idi1), and two IPP molecules are added to one DMAPP by a farnesyl diphosphate synthase (IspA/Erg20) to generate farnesyl diphosphate (FPP) with 3 isoprenyl units (Fig. 1). FPP is then extended with sequential additions of IPP molecules by a trans-isoprenyl diphosphate synthase (IspB/Coq1). The length of the chain is determined by the size of the pocket which accommodates the growing polyprenyl diphosphate in the enzyme (Nagel et al. 2019).
Finally, the polyprenyl diphosphate chain is added onto 4-HBA by a membrane-bound 4-hydroxybenzoate 3-polyprenyl transferase, UbiA in E. coli and the related Coq2 in yeast (Li 2016). Interestingly, E. coli UbiA promiscuously accepts polyprenyl diphosphates of different lengths (Okada et al. 1998), as a result of polyprenyl diphosphates gaining access to the active site via an unrestricted lateral portal (Cheng and Li 2014). Catalysis occurs in lipid bilayers and the prenylated 4-HBA products are released into membranes (Cheng and Li 2014; Huang et al. 2014).
Functionalization of the head group
CoQ is obtained after functionalization of the phenyl ring of polyprenyl 4-HBA via one decarboxylation, three hydroxylation and three methylation steps (Fig. 1). Most steps are catalyzed by enzymes that share homology between eukaryotes and prokaryotes. The biochemistry of CoQ biosynthesis has been reviewed recently in bacteria (Aussel et al. 2014; Abby et al. 2020) and eukaryotes (Kawamukai 2016; Alcázar-Fabra et al. 2016; Stefely and Pagliarini 2017; Wang and Hekimi 2019; Fernández-del-Río and Clarke 2021), thus it will only be briefly discussed here.
Decarboxylation
In E. coli, prenyl 4-HBA is decarboxylated into octaprenylphenol by the UbiD-UbiX system that consists of the 3-octaprenyl-4-hydroxybenzoate decarboxylase UbiD and its associated flavin prenyltransferase UbiX (Fig. 1). UbiX produces the prenylated FMN used as a cofactor by UbiD (Marshall et al. 2017, 2019). Although widely conserved in many bacterial species, the UbiD-UbiX system is absent in some, suggesting that alternative systems exist, as recently proposed for Xanthomonas campestris and Francisella tularensis (Zhou et al. 2019; Kazemzadeh et al. 2021). In eukaryotes, the C1-decarboxylation step remains genetically and biochemically uncharacterized (Fernández-del-Río and Clarke 2021).
Hydroxylation reactions
The three hydroxylation reactions required for the biosynthesis of CoQ involve a large repertoire of O2-dependent hydroxylases in bacteria. In E. coli, three related class A flavoprotein monooxygenases (FMOs), UbiH (octaprenyl-methoxyphenol 1-hydroxylase), UbiI (octaprenylphenol 5-hydroxylase), and UbiF (demethoxyubiquinone 6-hydroxylase) hydroxylate the carbon atoms C1, C5, and C6, respectively (Fig. 1). Other bacterial species contain instead newly identified FMOs, like UbiM and UbiL, which are able to hydroxylate several positions of the head group (Pelosi et al. 2016). Some species contain a carboxylate-bridged diiron hydroxylase named Coq7 (demethoxyubiquinone 6-hydroxylase), which catalyzes a C6-hydroxylation (Stenmark et al. 2001). Overall, the number of CoQ hydroxylases present in bacterial genomes is highly variable (1–4), which suggests a complex evolutionary history (Abby et al. 2020). The situation is even more complex if we consider the newly identified O2-independent pathway, which is found in ~ 30% of CoQ synthesizing bacteria and involves two U32 proteins, UbiU and UbiV, as putative O2-independent hydroxylases (Pelosi et al. 2019). This pathway could be of interest for industrial production of CoQ under O2-limiting conditions.
The composition in CoQ hydroxylases seems more homogenous in eukaryotes with Coq6 (4-hydroxy-3-polyprenylbenzoate 5-hydroxylase), related to bacterial FMOs, catalyzing the C5-hydroxylation and Coq7 hydroxylating C6 (Fig. 1). However, some variation exists since a new FMO has recently been demonstrated to replace Coq7 in land plants, green algae and apicomplexans (Latimer et al. 2021; Xu et al. 2021a). The eukaryotic C1-hydroxylase is not yet known.
Methylation reactions
The three methylation reactions in the biosynthesis of CoQ are catalyzed by the S-adenosyl-l-methionine (SAM)-dependent UbiG (bifunctional 5-O- and 6-O-methyltransferase) and UbiE (C2-methyltransferase) proteins (Fig. 1), which are homologous to Coq3 and Coq5 in yeast, respectively (Kawamukai 2016). UbiG/Coq3 are needed for both O-methylation reactions of the pathway, while the C-methylation reaction is catalyzed by UbiE/Coq5. Note that UbiE is also involved in the biosynthesis of menaquinone in bacteria (Lee et al. 1997).
Supramolecular organization of the enzymes that modify the head group
After prenylation by UbiA/Coq2, all biosynthetic intermediates of the CoQ pathway are highly hydrophobic due to their polyprenyl tail, which may complicate substrate accessibility for head group-modifying enzymes. Interestingly, aforementioned hydroxylases and methyltransferases are known to be part of multiprotein complexes termed CoQ synthome in S. cerevisiae and Ubi complex in E. coli (He et al. 2014; Hajj Chehade et al. 2019). Accessory proteins, important for CoQ biosynthesis, but not involved in the catalysis of specific steps, are also found in those complexes (Fig. 1). The complexes have not been structurally characterized and even the stoichiometry of the proteins is unknown (Stefely and Pagliarini 2017; Wang and Hekimi 2019). So far, only a few direct interactions between specific Ubi or Coq proteins have been confirmed.
In S. cerevisiae, the CoQ synthome associates with the inner mitochondrial membrane (IMM) and includes Coq4, Coq8, Coq9 and Coq11, in addition to the Coq3, Coq4, Coq5 and Coq7 enzymes (Kawamukai 2016). The function of Coq4 remains elusive. Coq8 belongs to a family of atypical kinases, namely the UbiB family, and has been proposed to couple ATP hydrolysis to the extraction of CoQ precursors from the IMM and/or to the formation of the CoQ synthome (Reidenbach et al. 2018). Coq9 possesses an amphipathic helix that controls membrane association and the binding of lipids, including CoQ biosynthetic intermediates (Lohman et al. 2019). Moreover, Coq9 physically associates with Coq7 and was therefore suggested to present CoQ intermediates to the enzymes of the CoQ synthome (Lohman et al. 2019). At last, Coq11 is also part of the CoQ synthome and is required for efficient CoQ biosynthesis in yeast, but neither plant nor mammalian orthologs have been identified to date (Allan et al. 2015).
In contrast to the yeast CoQ synthome, the Ubi complex in E. coli is soluble and contains the five enzymes (UbiE to UbiI) that catalyze the last six reactions of the pathway (Fig. 1), transforming polyprenyl phenol into CoQ (Hajj Chehade et al. 2019). Two additional proteins, UbiJ and UbiK, are required for efficient CoQ biosynthesis, and are part of the Ubi complex (Fig. 1). UbiJ binds CoQ biosynthetic intermediates via its Sterol Carrier Protein 2 (SCP2) domain (Hajj Chehade et al. 2019) and interacts with UbiK (Loiseau et al. 2017), suggesting that UbiJ might present the head group of the hydrophobic intermediates to Ubi enzymes within the Ubi complex (Hajj Chehade et al. 2019). Interestingly, UbiJ is only required for the O2-dependent biosynthesis of CoQ, whereas UbiT participates only in the O2-independent biosynthesis of CoQ (Pelosi et al. 2019). Whether UbiT is part or not of the Ubi complex is currently unknown, but UbiT has been proposed to replace UbiJ under anaerobic conditions, since it contains an SCP2 domain and was recently shown to bind polyisoprenoid lipids in Pseudomonas aeruginosa (Vo et al. 2020). The UbiD/UbiX decarboxylation system is not part of the Ubi complex, but both proteins are soluble in E. coli cell extracts and co-migrate at around 700 kDa (Hajj Chehade et al. 2019), compatible with a UbiD6-UbiX12 association suggested by their individual 3D-multimeric structures (PDB IDs: 4RHE, 5M1D).
Overall, it appears that most head group-modifying steps of the CoQ biosynthesis pathways are taking place within multiprotein complexes composed of hydroxylases, methyltransferases and lipid-binding proteins that may serve in substrate presentation.
Regulation of CoQ biosynthesis
Besides CoQ, E. coli synthesizes two other isoprenoid quinones, demethyl-menaquinone 8 (DMK8) and menaquinone (MK8). Dioxygen availability has long been known to influence the composition of the quinone pool, high aeration favoring the accumulation of CoQ8 over (D)MK8, whereas microaerobic or anaerobic conditions increase the MK8 content and decrease CoQ8 (Nitzschke and Bettenbrock 2018). The biomass-specific CoQ content of aerobic glucose cultures was found to decrease throughout the exponential phase (Bekker et al. 2007). Consistent with early reports of catabolic repression affecting the CoQ pathway, a 2-fold increase in CoQ8 content was obtained by using glycerol instead of glucose as a carbon source (Martínez et al. 2019). This effect may be mediated at least in part by transcriptional regulation since the expression of several genes of the pathway (ubiA,C,D,X) was increased in glycerol medium compared to glucose (Martínez et al. 2019 and references therein). A previous report that exposure of E. coli to low osmotic pressure dramatically increased CoQ8 content has recently been disproven (Tempelhagen et al. 2020).
The regulation of CoQ biosynthesis in eukaryotes has been reviewed lately and is particularly complex in mammals (Villalba and Navas 2021). In S. cerevisiae, several mechanisms control CoQ production including the phosphorylation level of several Coq proteins, notably Coq7 (Martín-Montalvo et al. 2013), the regulation of the abundance of Coq5 via the Puf3 RNA-binding protein (Lapointe et al. 2018), the Snf2-dependent splicing of the PTC7 mRNA which encodes a phosphatase (Awad et al. 2017). Interestingly, increasing the mitochondrial methylation capacity by deleting the cho2 gene encoding a phosphatidylethanolamine N-methyltransferase resulted in a five-fold elevation of the cellular CoQ content (Ayer et al. 2021). This last study also identified several other mutants with increased CoQ levels (two- to twelve-fold), opening avenues to elucidate the various pathways and actors that control CoQ biosynthesis.
Strategies to improve ubiquinone-related production in microorganisms
Improving precursor supply of benzoquinone ring for ubiquinone production
As an alternative to chemical 4-HBA production from petroleum-derived phenol, bio-based production has been substantiated by extending the shikimate pathway or the tyrosine biosynthetic pathway (Lee and Wendisch 2017). To facilitate the 4-HBA production through the extended shikimate pathway, the gene ubiC encoding chorismate-pyruvate lyase (CPL) was expressed in Klebsiella pneumoniae (Müller et al. 1995), E. coli (Barker and Frost 2001), Pseudomonas putida (Yu et al. 2016), S. cerevisiae (Krömer et al. 2013), or Corynebacterium glutamicum (Kitade et al. 2018). In particular, elaborated strain development was extensively carried out in C. glutamicum, which features a high 4-HBA tolerance (Kitade et al. 2018; Kallscheuer and Marienhagen 2018; Purwanto et al. 2018). The engineering strategies are as follows; (1) introduction of feedback-resistant CPL from E. coli or Providencia rustigianii, (2) blocking of carbon flux to the final reactions of aromatic amino acid biosynthesis, TCA cycle, and/or the quinate/shikimate utilization (QSU) pathway, (3) overexpression of the shikimate pathway genes, (4) increased pools of the precursors phosphoenolpyruvate and erythrose-4-phosphate, (5) reduced accumulation of by-products, including lactate, dihydroxyacetone phosphate, and protocatechuate (PCA), and (6) reduced accumulation of shikimate pathway intermediates, including dehydroshikimate and shikimate. As a consequence, the highest 4-HBA product titer up to about 37 g/L was achieved in a two-stage bioprocess (Kitade et al. 2018). Thus, to provide aromatic precursor for CoQ10 biosynthesis, feedback-resistant CPL and AroG from E. coli were introduced into a mutant C. glutamicum, in which pobA, pcaHG, and qsuABD encoding 4-HBA hydroxylase, PCA dioxygenase, and QSU pathway enzymes (putative shikimate importer, 3-dehydroshikimate dehydratase, and quinate/shikimate dehydrogenase), respectively, were deleted (Burgardt et al. 2021). Meanwhile, P. taiwanensis was tailored to produce 4-HBA via l-tyrosine (Lenzen et al. 2019). This was enabled by expressing tyrAfbr, tal, fcs, ech, and vdh coding for a mutated prephenate dehydrogenase, tyrosine-ammonia lyase, feruloyl-CoA synthetase, enoyl-CoA hydratase, and vanillin dehydrogenase, together with blockage of carbon flux to l-tryptophan, homogentisate, and PCA. The resulting strain yielded about 10 g/L 4-HBA from glycerol in fed-batch cultivations. Besides 4-HBA, 4-ABA can also be used as aromatic precursor of CoQ to form 3-hexaprenyl-4-aminobenzoate by the action of 4-HBA-polyprenyl transferase in yeasts (Marbois et al. 2010; Nishida et al. 2020). A feasibility study to produce 4-ABA (around 0.25 mM) via chorismate was implemented in a mutant S. cerevisiae by expressing abz1 encoding 4-aminobenzoate synthase (Krömer et al. 2013). Several microbes such as E. coli (Huang and Gibson 1970; Koma et al. 2014), C. glutamicum (Kubota et al. 2016), and Bacillus subtilis (Averesch and Rothschild 2019) have been successfully engineered for high titer 4-ABA production. Of these, the highest titer of 4-ABA (about 43 g/L) was obtained by introduction of 4-ABA biosynthetic step from chorismate into the recombinant C. glutamicum overexpressing the shikimate pathway genes (Kubota et al. 2016).
Improving precursor supply of polyprenyl diphosphate for ubiquinone production
Since IPP and DMAPP are building blocks not only for the polyprenyl tail of CoQ, but also for a vast variety of natural terpenes and terpenoids like chlorophylls, carotenoids and various quinones, ways to increase their supply have been studied for many organisms and products.
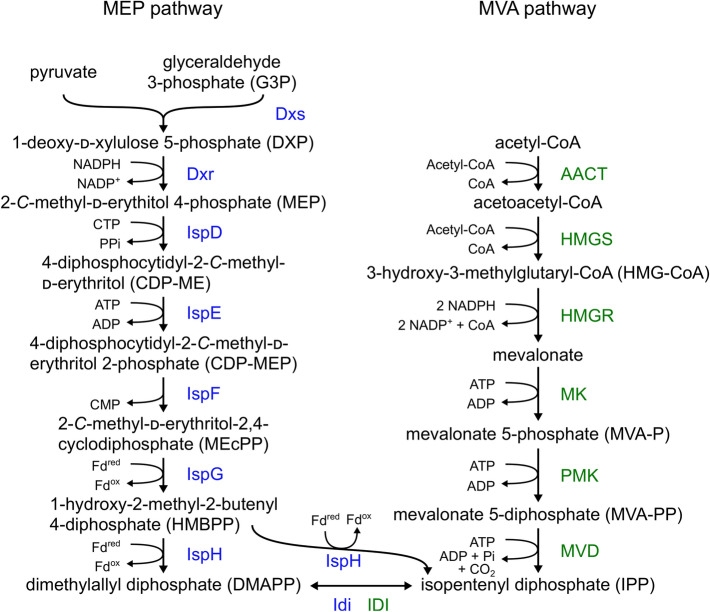
The entry point for the MEP pathway is the condensation of glyceraldehyde 3-phosphate (G3P) and pyruvate to 1-deoxy-d-xylulose 5-phosphate (DXP) by DXP synthase (Dxs), followed by reduction to MEP by DXP reductoisomerase (Dxr) (Fig. 2). MEP is converted to IPP in a series of reactions, catalyzed by 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase (IspD), 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase (IspE), 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase (IspF), 4-hydroxy-3-methylbut-2-enyl diphosphate synthase (IspG) and 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (IspH). IPP is isomerized to DMAPP by isopentenyl-diphosphate isomerase (Idi) (Rohmer 1999). Most metabolic engineering strategies to produce IPP-derived terpenes and terpenoids have followed the rationale of overexpressing genes that code for rate-limiting enzymes in the MEP pathway. The overexpression of dxs and idi has been established in different organisms like C. glutamicum and the cyanobacterial Synechocystis sp. for the production of patchoulol (Henke et al. 2018) and bisabolene (Rodrigues and Lindberg 2021), respectively. Overexpressing dxs, dxr and idi has improved production of isoprene in E. coli (Lv et al. 2013) and menaquinone-7 in B. subtilis (Ma et al. 2019; Liao et al. 2021). Volke et al. showed by metabolic control analysis in E. coli that indeed Dxs and Idi constitute major flux controlling steps and that upon dxs overexpression, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate accumulates intracellularly, while it is also exported outside the cells rather than being reduced to 4-hydroxy-3-methylbut-2-enyl diphosphate. Overexpression of ispG and ispH did not increase the flux, however (Volke et al. 2019).
Fig. 2.
Overview of the MEP and MVA pathways. E. coli gene product names in blue represent the reactions of the MEP pathway and enzyme names in green represent the reactions of the MVA pathway in S. cerevisiae
Besides the MEP pathway, IPP and DMAPP are synthesized via the MVA pathway that branches off the central carbon metabolism at acetyl-CoA. Two acetyl-CoA molecules are condensed to acetoacetyl-CoA by an acetoacetyl-CoA thiolase (AACT) (Fig. 2). This is followed by another condensation with acetyl-CoA to 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) via HMG-CoA synthase (HMGS) and subsequent reduction by HMG-CoA reductase (HMGR) to mevalonate. These reactions comprise the upper mevalonate pathway. The remaining reactions, referred to as the lower mevalonate pathway, contain two phosphorylation steps and a decarboxylation by mevalonate kinase (MK), phosphomevalonate kinase (PMK) and mevalonate diphosphate decarboxylase (MVD), respectively, yielding IPP (Miziorko 2011). The utilization of the MVA pathway in addition to the native MEP pathway in E. coli has been employed for a variety of products like isoprene (Liu et al. 2019), limonene (Wu et al. 2019), isoprenoid alcohols (Zada et al. 2018) as well as CoQ10 (Zahiri et al. 2006). The MVA pathway genes were heterologously expressed and originated from Streptococcus pneumoniae, Enterococcus faealis, S. cerevisiae and Methanosarcina mazei amongst others. A synthetic pathway was designed as alternative to the MEP and MVA pathways using a retrosynthetic approach with the goal to challenge the limitations in the natural pathways caused by carbon and energy inefficiencies, complex chemistry and regulatory mechanisms. This pathway centers on the production of isoprenoid alcohols, e.g. prenol or isoprenol, in order to diphosphorylate them to IPP and DMAPP and enabled E. coli to produce more than 2 g/L of prenol (Clomburg et al. 2019).
Besides engineering the direct precursor pathways, other approaches have successfully improved production of target compounds. Disruption or downregulation of pathways that compete for the common precursors IPP and DMAPP like carotenoid and hopanoid biosynthesis, has led to higher production of patchoulol by C. glutamicum (Henke et al. 2018) and CoQ10 by Rhodobacter sphaeroides (Zhu et al. 2017a) and Rhodopseudomonas palustris (Xu et al. 2021b). Another strategy to obtain higher titers would be to modify pathways of the central carbon metabolism. It is long known that optimizing distribution between G3P and pyruvate, the precursors of the MEP pathway, can increase flux towards the MEP pathway (Farmer and Liao 2001). In addition, combining the Entner–Doudoroff pathway with the MEP pathway increased isoprene titers in E. coli three-fold (Liu et al. 2013). With the advent of CRISPRi-mediated repression, fast screening of many target genes among different pathways allows to find suitable candidates to direct flux towards IPP and DMAPP in shorter time as it has been shown for E. coli (Tian et al. 2019) and C. glutamicum (Göttl et al. 2021). Lastly, cofactor economy plays an important role as some enzymes of the MEP and MVA pathways as well as of the CoQ10 synthesis are dependent on NAD(P)H, ATP or SAM. Zhou et al. have increased the NADPH pool by expressing a NADH kinase from S. cerevisiae, deleting the NADPH-dependent aldehyde reductase YjgB and overexpressing genes coding for flavodoxin I (fldA) and flavodoxin/ferredoxin NADP+ reductase (fpr) that are known to act as a NADPH-Fpr-FldA reducing system and to activate IspG and IspH (Zhou et al. 2017).
Taken together, the pathways and reactions leading up to IPP and DMAPP offer many possibilities for metabolic engineering approaches. But it is crucial to balance the modifications to avoid metabolic pitfalls that compromise the organisms’ vitality. Strategies like changing NAD(P)H/NAD(P)+ ratio or central metabolic pathways often are associated with reduced cell growth and overexpression or heterologous expression of genes in target pathways may perturb regulation to prevent buildup of toxic intermediates (George et al. 2018). Further metabolic engineering strategies to specifically produce CoQ10 will be addressed in another section.
CoQ10 production by bacteria natively synthesizing CoQ10
Since CoQ10 biosynthesis requires many enzymatic steps, and since their reaction mechanisms and regulation are still not fully elucidated, first production approaches were based on native CoQ10 producers, mainly Agrobacterium tumefaciens and R. sphaeroides (Table 1). Unlike secreted products of biotechnological interest such as amino acids (Wendisch 2020), CoQ10 is cell-bound, i.e., incorporated into cell membranes. Consequently, biomass production had to be maximized, e.g., by media optimization or mutagenesis, to achieve good CoQ10 titers (Yuan et al. 2012). Random mutagenesis and selection with menadione and sodium azide as inhibitors of the respiratory system generated mutants that overcame the growth inhibition with increased CoQ10 production. Thus, titers up to 350 mg/L were reached in pH–stat fed-batch fermentations using these classically obtained mutants (Kim et al. 2015). A. tumefaciens was used for two up-scaling steps (300 L and 5000 L) and produced CoQ10 to a cellular content of 8.54 mg/g DCW and a titer of 458 mg/L (Ha et al. 2007a). CoQ10 content and titer were elevated upon controlling the concentration of the carbon substrate sucrose and optimizing pH and dissolved oxygen levels (Ha et al. 2007b). R. sphaeroides has been employed for 100 L fermentation in which under phosphate limitation a titer of 1.95 g/L was reached, the highest reported in literature (Zhang et al. 2019). R. sphaeroides fermentation has been realized commercially as it also benefits from the fact that CoQ production can operate with non-toxic wastewater (He et al. 2021).
Table 1.
Representative examples of CoQ10 production strategies with natural, mutant, and metabolically engineered hosts
| Production host | Key strategies | Titer (mg/L) | Content (mg/g) | Volumetric productivity (mg/L/h) | References |
|---|---|---|---|---|---|
| Native CoQ producers and derived mutant strains | |||||
| A. tumefaciens KCCM 10413 | Controlling sucrose concentration, fed-batch cultivation | 627 | 9.25 | 5.23 | Ha et al. (2007a) |
| A. tumefaciens KCCM 10413 | Controlling pH and dissolved oxygen, 5000 L fed-batch cultivation | 458 | 8.54 | 3.82 | Ha et al. (2007b) |
| A. tumefaciens 1.2554 | Media optimization, mutagenesis, fed-batch cultivation | 120 | 3.86 | 1.25 | Yuan et al. (2012) |
| A. tumefaciens S02-13 | Adaptive laboratory evolution with menadione and sodium azide | 350 | ~ 4.2 | 3.89 | Kim et al. (2015) |
| R. sphaeroides KACC 91339P | Optimizing fermentation conditions, 150 L fed-batch cultivation | 55 | 8.12 | 0.78 | Kien et al. (2010) |
| R. sphaeroides Shenzhou6 | Mutagenesis using atmospheric and room temperature plasma treatment with vitamin K3 for selection pressure | 590 | – | 5.9 | Zou et al. (2019) |
| R. sphaeroides HY01 | Phosphate limitation, 100 L fed-batch cultivation | 1950 | ~ 24.4 | ~ 25.7 | Zhang et al. (2019) |
| Metabolically engineered native CoQ producers | |||||
| E. coli | Expression of ddsA from A. tumefaciens and MVA pathway genes from S. pneumoniae | – | 2.43 | – | Zahiri et al. (2006) |
| E. coli | Deletion of ispB, expression of ddsA from Sphingomonas baekryungensis, optimization of cultivation conditions | 0.70 | 0.43 | 0.10 | Martínez et al. (2015) |
| R. sphaeroides 2.4.1 | Overexpression of dxs, dxr, idi, ispD (MEP pathway); overexpression of fused ubiG and ubiE | 138 | 12.94 | 2.88 | Lu et al. (2015) |
| R. sphaeroides 2.4.1 | Overexpression of rate-limiting enzymes, increasing NADH/NAD+ ratio and oxygen uptake | 600 | 8.3 | 6.25 | Zhu et al. (2017b) |
| R. sphaeroides 2.4.1 | Overexpression of transcriptional repressor ppsR to decrease carotenoid synthesis and crtE to improve GGPP supply | 73 | 5.67 | – | Zhu et al. (2017a) |
| R. sphaeroides 2.4.1 | Modifying respiratory chain by deletion of sdhB, two-step oxygen supply culture strategy | 71 | 4.59 | 0.74 | Zhang et al. (2018) |
| R. sphaeroides ATCC 17023 | Deletion of fruA and fruB, increasing uptake of glucose via non-PTS by expression of galP | 78 | 4.53 | 1.08 | Yang et al. (2021) |
| R. palustris TIE-1 | Deletion of shc and crtB to disrupt carotenoid and hopanoid synthesis, overexpression of dxs, dps, ubiA | 3.6 | 8.2 | 0.05 | Xu et al. (2021b) |
| Metabolically engineered producers that do not natively synthesize CoQ | |||||
| C. glutamicum ATCC 13032 | Metabolic engineering to produce 4-HBA and DPP, expression of E. coli genes from ubiquinone pathway | 0.43 | 0.04 | 0.004 | Burgardt et al. (2021) |
Metabolic engineering strategies for the overproduction of CoQ10
Metabolic engineering allows for improving production rationally in native CoQ10 producers and for enabling CoQ10 production in microorganism that do not possess a native CoQ biosynthesis pathway (Lee et al. 2017). Strategies for (heterologous) overproduction involve the identification and elimination of bottlenecks and flux redistribution in the precursor pathways, use of the MVA pathway in addition to the MEP pathway and/or reducing competitive production of carotenoids as reviewed above. CoQ10 has been produced in metabolically engineered eukaryotes and prokaryotes, but as there are less studies about eukaryotic producers and their CoQ10 content is not competitive with most bacterial production hosts, the following sections will only focus on the latter.
Metabolic engineering of E. coli to produce CoQ10
E. coli is a natural CoQ8 producer and merely the expression of a heterologous decaprenyl diphosphate synthase is required for CoQ10 production since the polyprenyl transferase UbiA promiscuously accepts polyprenyl diphosphates of different lengths (Cheng and Li 2014), as was shown before (Martínez et al. 2015). E. coli synthesizes both, menaquinone and ubiquinone, with menaquinone biosynthesis being nonessential under aerobic conditions. Blocking the menaquinone pathway in addition to expression of dxs and ubiA and supplementation of pyruvate and 4-HBA boosted CoQ8 content 4-fold. Growth was not affected under aerobic conditions by the disruption of menaquinone biosynthesis (Xu et al. 2014). CoQ10 production by this industrially important organism has received attention some years ago (Table 1) (Zahiri et al. 2006; Cluis et al. 2011), but some natively CoQ10 producing bacteria like R. sphaeroides proved to be superior hosts for CoQ10 production. Nevertheless, a recent example of efficient menaquinone-7 (MK7) production to a titer of 1350 mg/L has shown that quinone production by E. coli should not be underestimated. This was achieved by optimized heterologous expression of MVA pathway genes and screening several heterologous Idi enzymes to improve IPP supply, overexpression of endogenous and exogenous MK pathway genes and enhancing the flux from chorismate to 1,4-dihydroxy-2-naphthoate, the direct precursor for demethylmenaquinone (Gao et al. 2021).
Genetic engineering of bacteria that natively produce CoQ10
Studies on native CoQ10 producers that have been genetically engineered for its overproduction are quite rare with exception of R. sphaeroides. This purple photosynthetic bacterium emerged as the most promising organism for CoQ10 production in recent years and will therefore be the focus here (Table 1). In one approach, genes that code for enzymes of the aerobic respiration chain were deleted due to relationship between CoQ10 synthesis and respiration chain activity. A R. sphaeroides mutant defective for succinate dehydrogenase (sdhB) overproduced CoQ10 under low oxygen conditions, which was exploited in a two-step oxygen supply culture strategy to increase the CoQ10 titer from 50 mg/L in the wild type to 71 mg/L in the recombinant strain (Zhang et al. 2018). In another study, deletion of the gene for the only known phosphotransferase system (PTS) in R. sphaeroides, PTSFru, combined with heterologous expression of a galactose:H+ symporter gene to improve provision with PEP as CoQ10 precursor increased the glucose consumption rate by 39% and the biomass concentration by 80% compared to the wild type and the CoQ10 titer to 78 mg/L, which was 50% higher than the wild type (Yang et al. 2021). Metabolic bottlenecks in the ubiquinone pathway of R. sphaeroides were identified to be UbiE, UbiH, and UbiG. A UbiG-UbiE fusion protein overcame this bottleneck (138 mg/L) despite slightly lower biomass concentration than the wild type (Lu et al. 2015). UbiA was not rate-limiting contrary to observations for E. coli and A. tumefaciens (Zhang et al. 2007; Cluis et al. 2011). Although not fully understood, heterologous expression of Vitreoscilla hemoglobin (vgb) slightly improved the titer in this R. sphaeroides strain (Lu et al. 2015) to 164 mg/L when the NADH/NAD+ ratio was modified as well. While increasing the NADH/NAD+ ratio influenced the biomass negatively, expression of vgb counteracted this and in combination, growth was superior to the parent strain. A fed-batch fermentation yielded 600 mg/L CoQ10 production (Zhu et al. 2017b).
CoQ10 production in bacteria that do not natively produce CoQ
A bacterium natively lacking CoQ biosynthesis has recently been engineered for CoQ10 production (Table 1) (Burgardt et al. 2021). Previously, C. glutamicum was engineered for high-level production of the aromatic CoQ10 precursor 4-HBA (Kitade et al. 2018; Purwanto et al. 2018). Two steps were required to enable CoQ10 production by the 4-HBA producing C. glutamicum strain. First, overproduction of the prenyl precursor of CoQ10, decaprenyl diphosphate (DPP), was achieved by heterologous expression of DPP synthase gene ddsA from Paracoccus denitrificans (Burgardt et al. 2021). Second, genes for the whole ubiquinone pathway from E. coli were expressed and the resulting strain produced 0.43 mg/L (Burgardt et al. 2021). Although the titer was low, this is the first proof-of-concept of producing CoQ10 by a microorganism lacking native CoQ biosynthesis. The fact that C. glutamicum has been used safely for more than 50 years in fermentative amino acid production, which is operated at a scale of 6 million tons per year (Wendisch 2020), forecasts that optimization of CoQ10 production by this bacterium holds large potential. Previous engineering of C. glutamicum for high-level production of aromatic compounds including the CoQ10 precursor 4-HBA (Lee and Wendisch 2017) as well as for products derived from the MEP pathway (Heider et al. 2014; Henke and Wendisch 2019; Li et al. 2021) provides a sound basis to de-bottleneck transfer of CoQ10 biosynthesis from native CoQ10 producing microbes to C. glutamicum and to gain an in-depth understanding of CoQ10 biosynthesis in the respective donor microbes.
Conclusions and future perspectives
CoQ is a key component in eukaryotic and bacterial cells as it is required for energy generation while also fulfilling numerous other functions. Future research has to fully elucidate CoQ biosynthesis since some parts of CoQ biosynthesis remain uncharacterized, e. g., the C1-decarboxylation and the C1-hydroxylation steps in the aromatic ring modification in eukaryotes. Recent advances, however, have been made in the understanding of the UbiD-UbiX system in bacteria, the diversity of CoQ hydroxylases, and especially, the supramolecular organization of enzymes that finalize the aromatic ring modification towards CoQ. Regarding the latter, the structural characterization and stoichiometry of the involved Ubi or Coq proteins are still missing, but hydroxylases and methyltransferases as well as associated lipid-binding proteins have been identified. In terms of microbial production of CoQ10, further research on the rational improvement of CoQ10 production is required. Although employment of mutagenized natural CoQ10 producers and process optimization led to impressive CoQ10 titers, the underlying mechanisms have not been understood. Metabolic engineering will not only enable the use of renewable resources for CoQ10 production and improve CoQ10 titers and productivities, but rational pathway reconstruction will help to expand the knowledge about the CoQ biosynthesis.
Author contributions
All authors wrote, revised, and edited the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. AB and VW acknowledge funding by the state of North Rhine Westphalia (NRW) and the “European Regional Development Fund (EFRE),” Project “Cluster Industrial Biotechnology (CLIB) Kompetenzzentrum Biotechnologie (CKB)” 34.EFRE0300095/1703FI04. VW and J-HL acknowledge support by the German-Korean MOBKOR program jointly funded by the National Research Foundation of Korea (NRF-2016K1A3A1A04940618) and the German Federal Ministry of Education and Research. JHL acknowledges support by Basic Science Research Program through the National Research Foundation of Korea (NRF-2018R1D1A1B07047207). LP and FP acknowledge financial support by ANR (project O2-taboo, ANR-19-CE44-0014), the Université Grenoble Alpes (UGA), and the French Centre National de la Recherche Scientifique (CNRS). The funders had no role in study design and data analysis and interpretation.
Declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fabien Pierrel and Volker F. Wendisch shared corresponding authors.
Fabien Pierrel and Arthur Burgardt shared first authors.
Contributor Information
Fabien Pierrel, Email: fabien.pierrel@univ-grenoble-alpes.fr.
Volker F. Wendisch, Email: volker.wendisch@uni-bielefeld.de
References
- Abby SS, Kazemzadeh K, Vragniau C, et al. Advances in bacterial pathways for the biosynthesis of ubiquinone. Biochim Biophys Acta Bioenerg. 2020;1861:148259. doi: 10.1016/j.bbabio.2020.148259. [DOI] [PubMed] [Google Scholar]
- Alcázar-Fabra M, Navas P, Brea-Calvo G. Coenzyme Q biosynthesis and its role in the respiratory chain structure. Biochim Biophys Acta BBA Bioenerg. 2016;1857:1073–1078. doi: 10.1016/j.bbabio.2016.03.010. [DOI] [PubMed] [Google Scholar]
- Allan CM, Awad AM, Johnson JS, et al. Identification of Coq11, a new coenzyme Q biosynthetic protein in the CoQ-synthome in Saccharomyces cerevisiae. J Biol Chem. 2015;290:7517–7534. doi: 10.1074/jbc.M114.633131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Jal M, Suñé-Negre JM, García-Montoya E. Coenzyme Q10 supplementation: efficacy, safety, and formulation challenges. Compr Rev Food Sci Food Saf. 2020;19:574–594. doi: 10.1111/1541-4337.12539. [DOI] [PubMed] [Google Scholar]
- Aussel L, Pierrel F, Loiseau L, et al. Biosynthesis and physiology of coenzyme Q in bacteria. Biochim Biophys Acta. 2014;1837:1004–1011. doi: 10.1016/j.bbabio.2014.01.015. [DOI] [PubMed] [Google Scholar]
- Averesch NJH, Rothschild LJ. Metabolic engineering of Bacillus subtilis for production of para-aminobenzoic acid—unexpected importance of carbon source is an advantage for space application. Microb Biotechnol. 2019;12:703–714. doi: 10.1111/1751-7915.13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad AM, Venkataramanan S, Nag A, et al. Chromatin-remodeling SWI/SNF complex regulates coenzyme Q6 synthesis and a metabolic shift to respiration in yeast. J Biol Chem. 2017;292:14851–14866. doi: 10.1074/jbc.M117.798397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayer A, Fazakerley DJ, Suarna C, et al. Genetic screening reveals phospholipid metabolism as a key regulator of the biosynthesis of the redox-active lipid coenzyme Q. Redox Biol. 2021;46:102127. doi: 10.1016/j.redox.2021.102127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker JL, Frost JW. Microbial synthesis of p-hydroxybenzoic acid from glucose. Biotechnol Bioeng. 2001;76:376–390. doi: 10.1002/bit.10160. [DOI] [PubMed] [Google Scholar]
- Baschiera E, Sorrentino U, Calderan C, et al. The multiple roles of coenzyme Q in cellular homeostasis and their relevance for the pathogenesis of coenzyme Q deficiency. Free Radic Biol Med. 2021;166:277–286. doi: 10.1016/j.freeradbiomed.2021.02.039. [DOI] [PubMed] [Google Scholar]
- Bekker M, Kramer G, Hartog AF, et al. Changes in the redox state and composition of the quinone pool of Escherichia coli during aerobic batch-culture growth. Microbiology. 2007;153:1974–1980. doi: 10.1099/mic.0.2007/006098-0. [DOI] [PubMed] [Google Scholar]
- Burgardt A, Moustafa A, Persicke M, et al. Coenzyme Q10 biosynthesis established in the non-ubiquinone containing Corynebacterium glutamicum by metabolic engineering. Front Bioeng Biotechnol. 2021 doi: 10.3389/fbioe.2021.650961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W, Li W. Structural insights into ubiquinone biosynthesis in membranes. Science. 2014;343:878–881. doi: 10.1126/science.1246774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirilli I, Damiani E, Dludla PV, et al. Role of coenzyme Q10 in health and disease: an update on the last 10 years (2010–2020) Antioxidants. 2021;10:1325. doi: 10.3390/antiox10081325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clomburg JM, Qian S, Tan Z, et al. The isoprenoid alcohol pathway, a synthetic route for isoprenoid biosynthesis. Proc Natl Acad Sci. 2019;116:12810–12815. doi: 10.1073/pnas.1821004116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Ekins A, Narcross L, et al. Identification of bottlenecks in Escherichia coli engineered for the production of CoQ10. Metab Eng. 2011;13:733–744. doi: 10.1016/j.ymben.2011.09.009. [DOI] [PubMed] [Google Scholar]
- Farmer WR, Liao JC. Precursor balancing for metabolic engineering of lycopene production in Escherichia coli. Biotechnol Prog. 2001;17:57–61. doi: 10.1021/bp000137t. [DOI] [PubMed] [Google Scholar]
- Fernández-Del-Río L, Clarke CF. Coenzyme Q biosynthesis: an update on the origins of the benzenoid ring and discovery of new ring precursors. Metabolites. 2021;11:385. doi: 10.3390/metabo11060385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chen H, Wang G, et al. Highly efficient production of menaquinone-7 from glucose by metabolically engineered Escherichia coli. ACS Synth Biol. 2021;10:756–765. doi: 10.1021/acssynbio.0c00568. [DOI] [PubMed] [Google Scholar]
- George KW, Thompson MG, Kim J, et al. Integrated analysis of isopentenyl pyrophosphate (IPP) toxicity in isoprenoid-producing Escherichia coli. Metab Eng. 2018;47:60–72. doi: 10.1016/j.ymben.2018.03.004. [DOI] [PubMed] [Google Scholar]
- Göttl VL, Schmitt I, Braun K, et al. CRISPRi-library-guided target identification for engineering carotenoid production by Corynebacterium glutamicum. Microorganisms. 2021;9:670. doi: 10.3390/microorganisms9040670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Mariscal FM, Perez-Martinez P, Delgado-Lista J, et al. Mediterranean diet supplemented with coenzyme Q10 induces postprandial changes in p53 in response to oxidative DNA damage in elderly subjects. Age. 2012;34:389–403. doi: 10.1007/s11357-011-9229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S, Kim S, Seo J, et al. Controlling the sucrose concentration increases coenzyme Q10 production in fed-batch culture of Agrobacterium tumefaciens. Appl Microbiol Biotechnol. 2007;76:109–116. doi: 10.1007/s00253-007-0995-8. [DOI] [PubMed] [Google Scholar]
- Ha S-J, Kim S-Y, Seo J-H, et al. Optimization of culture conditions and scale-up to pilot and plant scales for coenzyme Q10 production by Agrobacterium tumefaciens. Appl Microbiol Biotechnol. 2007;74:974–980. doi: 10.1007/s00253-006-0744-4. [DOI] [PubMed] [Google Scholar]
- Hajj Chehade M, Pelosi L, Fyfe CD, et al. A soluble metabolon synthesizes the isoprenoid lipid ubiquinone. Cell Chem Biol. 2019;26:482–492. doi: 10.1016/j.chembiol.2018.12.001. [DOI] [PubMed] [Google Scholar]
- He CH, Xie LX, Allan CM, et al. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim Biophys Acta BBA Mol Cell Biol Lipids. 2014;1841:630–644. doi: 10.1016/j.bbalip.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Lu H, Zhang G, Ren Z. Production of coenzyme Q10 by purple non-sulfur bacteria: current development and future prospect. J Clean Prod. 2021;307:127326. doi: 10.1016/j.jclepro.2021.127326. [DOI] [Google Scholar]
- Heider SAE, Peters-Wendisch P, Wendisch VF, et al. Metabolic engineering for the microbial production of carotenoids and related products with a focus on the rare C50 carotenoids. Appl Microbiol Biotechnol. 2014;98:4355–4368. doi: 10.1007/s00253-014-5693-8. [DOI] [PubMed] [Google Scholar]
- Henke NA, Wendisch VF. Improved astaxanthin production with Corynebacterium glutamicum by application of a membrane fusion protein. Mar Drugs. 2019;17:621. doi: 10.3390/md17110621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke NA, Wichmann J, Baier T, et al. Patchoulol production with metabolically engineered Corynebacterium glutamicum. Genes. 2018;9:1–15. doi: 10.3390/genes9040219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Gibson F. Biosynthesis of 4-aminobenzoate in Escherichia coli. J Bacteriol. 1970;102:767–773. doi: 10.1128/jb.102.3.767-773.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Levin EJ, Liu S, et al. Structure of a membrane-embedded prenyltransferase homologous to UBIAD1. PLoS Biol. 2014;12:e1001911. doi: 10.1371/journal.pbio.1001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalén A, Appelkvist E-L, Dallner G. Age-related changes in the lipid compositions of rat and human tissues. Lipids. 1989;24:579–584. doi: 10.1007/BF02535072. [DOI] [PubMed] [Google Scholar]
- Kallscheuer N, Marienhagen J. Corynebacterium glutamicum as platform for the production of hydroxybenzoic acids. Microb Cell Factories. 2018;17:70. doi: 10.1186/s12934-018-0923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamukai M. Biosynthesis of coenzyme Q in eukaryotes. Biosci Biotechnol Biochem. 2016;80:23–33. doi: 10.1080/09168451.2015.1065172. [DOI] [PubMed] [Google Scholar]
- Kawamukai M. Biosynthesis and applications of prenylquinones. Biosci Biotechnol Biochem. 2018;82:963–977. doi: 10.1080/09168451.2018.1433020. [DOI] [PubMed] [Google Scholar]
- Kazemzadeh K, Hajj Chehade M, Hourdoir G, et al. The biosynthetic pathway of ubiquinone contributes to pathogenicity of Francisella novicida. J Bacteriol. 2021;203:e00400–e421. doi: 10.1128/JB.00400-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kien NB, Kong I-S, Lee M-G, Kim JK. Coenzyme Q10 production in a 150-l reactor by a mutant strain of Rhodobacter sphaeroides. J Ind Microbiol Biotechnol. 2010;37:521–529. doi: 10.1007/s10295-010-0699-4. [DOI] [PubMed] [Google Scholar]
- Kim T-S, Yoo J-H, Kim S-Y, et al. Screening and characterization of an Agrobacterium tumefaciens mutant strain producing high level of coenzyme Q10. Process Biochem. 2015;50:33–39. doi: 10.1016/j.procbio.2014.10.024. [DOI] [Google Scholar]
- Kitade Y, Hashimoto R, Suda M, et al. Production of 4-hydroxybenzoic acid by an aerobic growth-arrested bioprocess using metabolically engineered Corynebacterium glutamicum. Appl Environ Microbiol. 2018 doi: 10.1128/AEM.02587-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koma D, Yamanaka H, Moriyoshi K, et al. Production of p-Aminobenzoic acid by metabolically engineered Escherichia coli. Biosci Biotechnol Biochem. 2014;78:350–357. doi: 10.1080/09168451.2014.878222. [DOI] [PubMed] [Google Scholar]
- Krömer JO, Nunez-Bernal D, Averesch NJH, et al. Production of aromatics in Saccharomyces cerevisiae—a feasibility study. J Biotechnol. 2013;163:184–193. doi: 10.1016/j.jbiotec.2012.04.014. [DOI] [PubMed] [Google Scholar]
- Kubota T, Watanabe A, Suda M, et al. Production of para-aminobenzoate by genetically engineered Corynebacterium glutamicum and non-biological formation of an N-glucosyl byproduct. Metab Eng. 2016;38:322–330. doi: 10.1016/j.ymben.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Lapointe CP, Stefely JA, Jochem A, et al. Multi-omics reveal specific targets of the RNA-binding protein Puf3p and its orchestration of mitochondrial biogenesis. Cell Syst. 2018;6:125–135.e6. doi: 10.1016/j.cels.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latimer S, Keene SA, Stutts LR, et al. A dedicated flavin-dependent monooxygenase catalyzes the hydroxylation of demethoxyubiquinone into ubiquinone (coenzyme Q) in Arabidopsis. J Biol Chem. 2021 doi: 10.1016/j.jbc.2021.101283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Wendisch VF. Biotechnological production of aromatic compounds of the extended shikimate pathway from renewable biomass. J Biotechnol. 2017;257:211–221. doi: 10.1016/j.jbiotec.2016.11.016. [DOI] [PubMed] [Google Scholar]
- Lee PT, Hsu AY, Ha HT, Clarke CF. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J Bacteriol. 1997;179:1748–1754. doi: 10.1128/jb.179.5.1748-1754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B-J, Lin Y-C, Huang Y-C, et al. The relationship between coenzyme Q10, oxidative stress, and antioxidant enzymes activities and coronary artery disease. Sci World J. 2012;2012:1–8. doi: 10.1100/2012/792756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SQE, Tan TS, Kawamukai M, Chen ES. Cellular factories for coenzyme Q10 production. Microb Cell Factories. 2017;16:1–16. doi: 10.1186/s12934-017-0646-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzen C, Wynands B, Otto M, et al. High-yield production of 4-hydroxybenzoate from glucose or glycerol by an engineered Pseudomonas taiwanensis VLB120. Front Bioeng Biotechnol. 2019;7:130. doi: 10.3389/fbioe.2019.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Bringing bioactive compounds into membranes: the UbiA superfamily of intramembrane aromatic prenyltransferases. Trends Biochem Sci. 2016;41:356–370. doi: 10.1016/j.tibs.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Swofford CA, Rückert C, et al. Heterologous production of α-Carotene in Corynebacterium glutamicum using a multi-copy chromosomal integration method. Bioresour Technol. 2021;341:125782. doi: 10.1016/j.biortech.2021.125782. [DOI] [PubMed] [Google Scholar]
- Liao C, Ayansola H, Ma Y, et al. Advances in enhanced menaquinone-7 production from Bacillus subtilis. Front Bioeng Biotechnol. 2021;9:656. doi: 10.3389/fbioe.2021.695526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sun Y, Ramos KRM, et al. Combination of Entner-Doudoroff pathway with MEP increases isoprene production in engineered Escherichia coli. PLoS ONE. 2013;8:e83290. doi: 10.1371/journal.pone.0083290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C-L, Bi H-R, Bai Z, et al. Engineering and manipulation of a mevalonate pathway in Escherichia coli for isoprene production. Appl Microbiol Biotechnol. 2019;103:239–250. doi: 10.1007/s00253-018-9472-9. [DOI] [PubMed] [Google Scholar]
- Lohman DC, Aydin D, Von Bank HC, et al. An isoprene lipid-binding protein promotes eukaryotic coenzyme Q biosynthesis. Mol Cell. 2019 doi: 10.1016/j.molcel.2018.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiseau L, Fyfe C, Aussel L, et al. The UbiK protein is an accessory factor necessary for bacterial ubiquinone (UQ) biosynthesis and forms a complex with the UQ biogenesis factor UbiJ. J Biol Chem. 2017;292:11937–11950. doi: 10.1074/jbc.M117.789164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Ye L, Lv X, et al. Identification and elimination of metabolic bottlenecks in the quinone modification pathway for enhanced coenzyme Q10 production in Rhodobacter sphaeroides. Metab Eng. 2015;29:208–216. doi: 10.1016/j.ymben.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Luo M, Yang X, Hu J, et al. The synthesis of coenzyme Q10. Curr Org Chem. 2017;21:489–502. doi: 10.2174/1385272820666160811123714. [DOI] [Google Scholar]
- Lv X, Xu H, Yu H. Significantly enhanced production of isoprene by ordered coexpression of genes dxs, dxr, and idi in Escherichia coli. Appl Microbiol Biotechnol. 2013;97:2357–2365. doi: 10.1007/s00253-012-4485-2. [DOI] [PubMed] [Google Scholar]
- Ma Y, McClure DD, Somerville MV, et al. Metabolic engineering of the MEP pathway in Bacillus subtilis for increased biosynthesis of menaquinone-7. ACS Synth Biol. 2019;8:1620–1630. doi: 10.1021/acssynbio.9b00077. [DOI] [PubMed] [Google Scholar]
- Marbois B, Xie LX, Choi S, et al. para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J Biol Chem. 2010;285:27827–27838. doi: 10.1074/jbc.M110.151894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall SA, Payne KAP, Leys D. The UbiX-UbiD system: the biosynthesis and use of prenylated flavin (prFMN) Arch Biochem Biophys. 2017;632:209–221. doi: 10.1016/j.abb.2017.07.014. [DOI] [PubMed] [Google Scholar]
- Marshall SA, Payne KAP, Fisher K, et al. The UbiX flavin prenyltransferase reaction mechanism resembles class I terpene cyclase chemistry. Nat Commun. 2019;10:2357. doi: 10.1038/s41467-019-10220-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez I, Méndez C, Berríos J, et al. Batch production of coenzyme Q10 by recombinant Escherichia coli containing the decaprenyl diphosphate synthase gene from Sphingomonas baekryungensis. J Ind Microbiol Biotechnol. 2015;42:1283–1289. doi: 10.1007/s10295-015-1652-3. [DOI] [PubMed] [Google Scholar]
- Martínez I, Zelada P, Guevara F, et al. Coenzyme Q production by metabolic engineered Escherichia coli strains in defined medium. Bioprocess Biosyst Eng. 2019;42:1143–1149. doi: 10.1007/s00449-019-02111-y. [DOI] [PubMed] [Google Scholar]
- Martín-Montalvo A, González-Mariscal I, Pomares-Viciana T, et al. The phosphatase Ptc7 induces coenzyme Q biosynthesis by activating the hydroxylase Coq7 in yeast. J Biol Chem. 2013;288:28126–28137. doi: 10.1074/jbc.M113.474494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miziorko HM. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch Biochem Biophys. 2011;505:131–143. doi: 10.1016/j.abb.2010.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen SA, Rosenfeldt F, Kumar A, et al. The effect of coenzyme Q10 on morbidity and mortality in chronic heart failure. JACC Heart Fail. 2014;2:641–649. doi: 10.1016/j.jchf.2014.06.008. [DOI] [PubMed] [Google Scholar]
- Müller R, Wagener A, Schmidt K, Leistner E. Microbial production of specifically ring-13C-labelled 4-hydroxybenzoic acid. Appl Microbiol Biotechnol. 1995;43:985–988. doi: 10.1007/BF00166913. [DOI] [PubMed] [Google Scholar]
- Nagel R, Schmidt A, Peters RJ. Isoprenyl diphosphate synthases: the chain length determining step in terpene biosynthesis. Planta. 2019;249:9–20. doi: 10.1007/s00425-018-3052-1. [DOI] [PubMed] [Google Scholar]
- Nishida I, Yanai R, Matsuo Y, et al. Benzoic acid inhibits coenzyme Q biosynthesis in Schizosaccharomyces pombe. PLoS ONE. 2020;15:e0242616. doi: 10.1371/journal.pone.0242616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzschke A, Bettenbrock K. All three quinone species play distinct roles in ensuring optimal growth under aerobic and fermentative conditions in E. coli K12. PLoS ONE. 2018;13:1–18. doi: 10.1371/journal.pone.0194699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Kainou T, Tanaka K, et al. Molecular cloning and mutational analysis of the ddsA gene encoding decaprenyl diphosphate synthase from Gluconobacter suboxydans. Eur J Biochem. 1998;255:52–59. doi: 10.1046/j.1432-1327.1998.2550052.x. [DOI] [PubMed] [Google Scholar]
- Payet LA, Leroux M, Willison JC, et al. Mechanistic details of early steps in coenzyme Q biosynthesis pathway in yeast. Cell Chem Biol. 2016;23:1241–1250. doi: 10.1016/j.chembiol.2016.08.008. [DOI] [PubMed] [Google Scholar]
- Pelosi L, Ducluzeau AL, Loiseau L, et al. Evolution of ubiquinone biosynthesis: multiple proteobacterial enzymes with various regioselectivities to catalyze three contiguous aromatic hydroxylation reactions. mSystems. 2016 doi: 10.1128/mSystems.00091-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi L, Vo C-D-T, Abby SS, et al. Ubiquinone biosynthesis over the entire O2 range: characterization of a conserved O2-independent pathway. Mbio. 2019;10:e01319-19. doi: 10.1128/mBio.01319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Gil J, Rodríguez-Concepción M. Metabolic plasticity for isoprenoid biosynthesis in bacteria. Biochem J. 2013;452:19–25. doi: 10.1042/BJ20121899. [DOI] [PubMed] [Google Scholar]
- Pierrel F, Hamelin O, Douki T, et al. Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem Biol. 2010;17:449–459. doi: 10.1016/j.chembiol.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Potgieter M, Pretorius E, Pepper MS. Primary and secondary coenzyme Q10 deficiency: the role of therapeutic supplementation. Nutr Rev. 2013;71:180–188. doi: 10.1111/nure.12011. [DOI] [PubMed] [Google Scholar]
- Purwanto HS, Kang M, Ferrer L, et al. Rational engineering of the shikimate and related pathways in Corynebacterium glutamicum for 4-hydroxybenzoate production. J Biotechnol. 2018;282:92–100. doi: 10.1016/j.jbiotec.2018.07.016. [DOI] [PubMed] [Google Scholar]
- Quinzii C, Naini A, Salviati L, et al. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidenbach AG, Kemmerer ZA, Aydin D, et al. Conserved lipid and small-molecule modulation of COQ8 reveals regulation of the ancient kinase-like UbiB family. Cell Chem Biol. 2018;25:154–165. doi: 10.1016/j.chembiol.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson KP, Jochem A, Johnson SE, et al. Defining intermediates and redundancies in coenzyme Q precursor biosynthesis. J Biol Chem. 2021 doi: 10.1016/j.jbc.2021.100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues JS, Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for improved bisabolene production. Metab Eng Commun. 2021;12:00159. doi: 10.1016/j.mec.2020.e00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohmer M. The discovery of a mevalonate-independent pathway for isoprenoid biosynthesis in bacteria, algae and higher plants. Nat Prod Rep. 1999;16:565–574. doi: 10.1039/a709175c. [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, van Lis R, Atteia A, et al. On the universal core of bioenergetics. Biochim Biophys Acta BBA - Bioenerg. 2013;1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- Shults CW. Effects of coenzyme Q10 in early parkinson disease: evidence of slowing of the functional decline. Arch Neurol. 2002;59:1541. doi: 10.1001/archneur.59.10.1541. [DOI] [PubMed] [Google Scholar]
- Siebert M, Severin K, Heide L. (1994) Formation of 4-hydroxybenzoate in Escherichia coli: characterization of the ubiC gene and its encoded enzyme chorismate pyruvate-lyase. Microbiology. 1994;140:897–904. doi: 10.1099/00221287-140-4-897. [DOI] [PubMed] [Google Scholar]
- Stefely JA, Pagliarini DJ. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem Sci. 2017;42:824–843. doi: 10.1016/j.tibs.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark P, Grünler J, Mattsson J, et al. A new member of the family of di-iron carboxylate proteins. J Biol Chem. 2001;276:33297–33300. doi: 10.1074/jbc.C100346200. [DOI] [PubMed] [Google Scholar]
- Tempelhagen L, Ayer A, Culham DE, et al. Cultivation at high osmotic pressure confers ubiquinone 8–independent protection of respiration on Escherichia coli. J Biol Chem. 2020;295:981–993. doi: 10.1016/S0021-9258(17)49909-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Kang JW, Kang A, Lee TS. Redirecting metabolic flux via combinatorial multiplex CRISPRi-mediated repression for isopentenol production in Escherichia coli. ACS Synth Biol. 2019;8:391–402. doi: 10.1021/acssynbio.8b00429. [DOI] [PubMed] [Google Scholar]
- Villalba JM, Navas P. Regulation of coenzyme Q biosynthesis pathway in eukaryotes. Free Radic Biol Med. 2021;165:312–323. doi: 10.1016/j.freeradbiomed.2021.01.055. [DOI] [PubMed] [Google Scholar]
- Vo C-D-T, Michaud J, Elsen S, et al. The O2-independent pathway of ubiquinone biosynthesis is essential for denitrification in Pseudomonas aeruginosa. J Biol Chem. 2020;295:9021–9032. doi: 10.1074/jbc.RA120.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volke DC, Rohwer J, Fischer R, Jennewein S. Investigation of the methylerythritol 4-phosphate pathway for microbial terpenoid production through metabolic control analysis. Microb Cell Factories. 2019;18:192. doi: 10.1186/s12934-019-1235-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hekimi S. The complexity of making ubiquinone. Trends Endocrinol Metab. 2019;30:929–943. doi: 10.1016/j.tem.2019.08.009. [DOI] [PubMed] [Google Scholar]
- Wendisch VF. Metabolic engineering advances and prospects for amino acid production. Metab Eng. 2020;58:17–34. doi: 10.1016/j.ymben.2019.03.008. [DOI] [PubMed] [Google Scholar]
- Wu J, Cheng S, Cao J, et al. Systematic optimization of limonene production in engineered Escherichia coli. J Agric Food Chem. 2019;67:7087–7097. doi: 10.1021/acs.jafc.9b01427. [DOI] [PubMed] [Google Scholar]
- Xie LX, Williams KJ, He CH, et al. Resveratrol and para-coumarate serve as ring precursors for coenzyme Q biosynthesis. J Lipid Res. 2015;56:909–919. doi: 10.1194/jlr.M057919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang S, Zhao J, et al. Improving coenzyme Q8 production in Escherichia coli employing multiple strategies. J Ind Microbiol Biotechnol. 2014;41:1297–1303. doi: 10.1007/s10295-014-1458-8. [DOI] [PubMed] [Google Scholar]
- Xu J-J, Zhang X-F, Jiang Y, et al. A unique flavoenzyme operates in ubiquinone biosynthesis in photosynthesis-related eukaryotes. Sci Adv. 2021;7:eabl3594. doi: 10.1126/sciadv.abl3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Ma X, Yao J, et al. Increasing coenzyme Q10 yield from Rhodopseudomonas palustris by expressing rate-limiting enzymes and blocking carotenoid and hopanoid pathways. Lett Appl Microbiol. 2021;73:88–95. doi: 10.1111/lam.13479. [DOI] [PubMed] [Google Scholar]
- Yang Y, Li L, Sun H, et al. Improving CoQ10 productivity by strengthening glucose transmembrane of Rhodobacter sphaeroides. Microb Cell Factories. 2021;20:207. doi: 10.1186/s12934-021-01695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef SAO, Fahad AA, Abdel Moneim AE, et al. The neuroprotective role of coenzyme Q10 against lead acetate-induced neurotoxicity is mediated by antioxidant, anti-inflammatory and anti-apoptotic activities. Int J Environ Res Public Health. 2019;16:2895. doi: 10.3390/ijerph16162895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Plan MR, Winter G, Krömer JO. Metabolic engineering of Pseudomonas putida KT2440 for the production of para-hydroxy benzoic acid. Front Bioeng Biotechnol. 2016;4:90. doi: 10.3389/fbioe.2016.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Tian Y, Yue T. Improvement of coenzyme Q10 production: mutagenesis induced by high hydrostatic pressure treatment and optimization of fermentation conditions. J Biomed Biotechnol. 2012;2012:1–8. doi: 10.1155/2012/607329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yubero-Serrano EM, Gonzalez-Guardia L, Rangel-Zuñiga O, et al. Mediterranean diet supplemented with coenzyme Q10 modifies the expression of proinflammatory and endoplasmic reticulum stress-related genes in elderly men and women. J Gerontol Ser A. 2012;67A:3–10. doi: 10.1093/gerona/glr167. [DOI] [PubMed] [Google Scholar]
- Zada B, Wang C, Park J-B, et al. Metabolic engineering of Escherichia coli for production of mixed isoprenoid alcohols and their derivatives. Biotechnol Biofuels. 2018;11:210. doi: 10.1186/s13068-018-1210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahiri HS, Yoon SH, Keasling JD, et al. Coenzyme Q10 production in recombinant Escherichia coli strains engineered with a heterologous decaprenyl diphosphate synthase gene and foreign mevalonate pathway. Metab Eng. 2006;8:406–416. doi: 10.1016/j.ymben.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Zhang D, Li Z, Wang F, et al. Expression of various genes to enhance ubiquinone metabolic pathway in Agrobacterium tumefaciens. Enzyme Microb Technol. 2007;41:772–779. doi: 10.1016/j.enzmictec.2007.06.014. [DOI] [Google Scholar]
- Zhang J, Gao D, Cai J, et al. Improving coenzyme Q10 yield of Rhodobacter sphaeroides via modifying redox respiration chain. Biochem Eng J. 2018;135:98–104. doi: 10.1016/j.bej.2018.04.006. [DOI] [Google Scholar]
- Zhang L, Liu L, Wang K-F, et al. Phosphate limitation increases coenzyme Q10 production in industrial Rhodobacter sphaeroides HY01. Synth Syst Biotechnol. 2019;4:212–219. doi: 10.1016/j.synbio.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang JY, Wu J, et al. The diffusible factor synthase XanB2 is a bifunctional chorismatase that links the shikimate pathway to ubiquinone and xanthomonadins biosynthetic pathways. Mol Microbiol. 2013;87:80–93. doi: 10.1111/mmi.12084. [DOI] [PubMed] [Google Scholar]
- Zhou J, Yang L, Wang C, et al. Enhanced performance of the methylerythritol phosphate pathway by manipulation of redox reactions relevant to IspC, IspG, and IspH. J Biotechnol. 2017;248:1–8. doi: 10.1016/j.jbiotec.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Zhou L, Li M, Wang X-Y, et al. Biosynthesis of Coenzyme Q in the Phytopathogen Xanthomonas campestris via a Yeast-Like Pathway. Mol Plant-Microbe Interact. 2019;32:217–226. doi: 10.1094/MPMI-07-18-0183-R. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Lu W, Ye L, et al. Enhanced synthesis of coenzyme Q10 by reducing the competitive production of carotenoids in Rhodobacter sphaeroides. Biochem Eng J. 2017 doi: 10.1016/j.bej.2017.03.019. [DOI] [Google Scholar]
- Zhu Y, Ye L, Chen Z, et al. Synergic regulation of redox potential and oxygen uptake to enhance production of coenzyme Q10 in Rhodobacter sphaeroides. Enzyme Microb Technol. 2017;101:36–43. doi: 10.1016/j.enzmictec.2017.03.005. [DOI] [PubMed] [Google Scholar]
- Žmitek K, Pogačnik T, Mervic L, et al. The effect of dietary intake of coenzyme Q10 on skin parameters and condition: results of a randomised, placebo-controlled, double-blind study: The effect of dietary intake of coenzyme Q10 on skin parameters and condition. BioFactors. 2017;43:132–140. doi: 10.1002/biof.1316. [DOI] [PubMed] [Google Scholar]
- Zou R-S, Li S, Zhang L-L, et al. Mutagenesis of Rhodobacter sphaeroides using atmospheric and room temperature plasma treatment for efficient production of coenzyme Q10. J Biosci Bioeng. 2019;127:698–702. doi: 10.1016/j.jbiosc.2018.12.005. [DOI] [PubMed] [Google Scholar]