Abstract
The Applied Biosystems ViroSeq HIV-1 Genotyping System is a commercially available, integrated system for sequence-based analysis of drug resistance mutations in human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase (RT). We evaluated the performance of this system for analysis of non-subtype B HIV-1 by analyzing plasma samples from Ugandan women and infants. Plasma samples were obtained from 105 women and 25 infants enrolled in a Ugandan clinical trial. HIV-1 analysis was performed with the ViroSeq system according to the manufacturer's instructions, except that the volume of plasma used for analysis was less than the recommended 0.5 ml for some samples. Viral loads ranged from 2,313 to 2,336,400 copies/ml. PCR products suitable for sequencing were amplified from all samples tested. Complete sequences for protease (amino acids 1 to 99) and RT (amino acids 1 to 320) were obtained for 102 of 105 (97%) of the maternal samples tested and all 25 of the infant samples tested. Complete double-stranded sequences were obtained for 90 of 105 (86%) of the maternal samples tested and 22 of 25 (88%) of the infant samples tested. The sequences obtained with this system were used for HIV-1 subtyping. The subtypes identified were A, C, D, and A/D recombinant HIV-1. The performances of the seven sequencing primers were similar for the subtypes examined. The ViroSeq system performs well for analysis of Ugandan plasma samples with subtypes A, C, D, and A/D recombinant HIV-1. The availability of this genotyping system should facilitate studies of HIV-1 drug resistance in countries where these subtypes are prevalent.
Antiretroviral drugs can improve the health and extend the lives of patients with human immunodeficiency virus (HIV) type 1 (HIV-1) infection and can be used to prevent HIV-1 vertical transmission, the major cause of pediatric HIV infection. There are three major classes of antiretroviral drugs approved in the United States for clinical use: protease inhibitors, nucleoside reverse transcriptase (RT) inhibitors, and nonnucleoside RT inhibitors (NNRTIs). Unfortunately, the efficacies of these drugs are often limited by HIV-1 drug resistance, which is usually caused by mutations in the protease and RT enzymes. Sequence-based genotyping assays can be used to detect these mutations.
Most HIV-1 isolates can be categorized into subtypes (clades) on the basis of genetic differences. To date, almost all of the information characterizing HIV-1 drug resistance mutations and their effects on drug susceptibility comes from studies of subtype B, which is the most common subtype in the United States and Europe. However, the majority of HIV-1 infections worldwide are caused by other subtypes. Non-subtype B has been reported in the United States (5, 13, 16, 21; Beatrice, S. T., W. R. Oleszko, A. Punsalang, M. A. Chaisson, L. V. Torian, C. A. Schable, and I. B. Weisfuse, 12th World AIDS Conf., abstr. 42116, 1998; P. J. Weidle, C. E. Ganea, D. Pienniazek, A. Ramos, C. A. Schable, J. Enst, and J. McGowan, 12th World AIDS Conf., abstr. 13225, 1998) and accounts for a growing percentage of infections in Europe (7, 10; C. Loveday, H. Devereux, A. Burke, L. Dann, A. Phillips, and M. Johnson, 12th World AIDS Conf., abstr. 42167, 1998). Research on drug resistance in non-subtype B HIV-1 is becoming increasingly important for two reasons: (i) the prevalence of non-subtype B is increasing in the United States and other regions where antiretroviral drugs are widely used, and (ii) the availability and use of antiretroviral drugs are growing throughout the world, where most infections are caused by non-subtype B HIV-1.
An integrated system for HIV-1 genotyping is available from Applied Biosystems (Foster City, Calif.). This system uses a total of 10 DNA primers for analysis (1 primer for reverse transcription, 2 primers for PCR amplification, and 7 primers for cycle sequencing). This assay was designed and optimized for analysis of subtype B HIV-1. However, the nucleotide sequences in the protease and RT coding regions are sufficiently different to allow these sequences to be used for subtype determination (2, 18, 22). Sequence differences in and around these regions among subtypes can complicate genotypic analysis of HIV-1 since the primers used in the analysis may not bind to target sequences.
Previous studies have demonstrated the successful use of the Applied Biosystems ViroSeq HIV-1 Genotyping System for analysis of subtype B HIV-1 in plasma (6) and of cultured isolates of different non-subtype B subtypes (J. Dileanis, N. Marlowe, B. Hoo, R. C. Brown, M. Bulmer, D. Huang, P. Palumbo, R. Schuurman, K. Van-Laethem, A.-M. Vandamme, and T. Elbeik, 5th Int. Cong. Drug Ther. HIV Infect., abstr. P338, 2000). In the study described in this report, we analyzed the performance of the ViroSeq system for analysis of plasma samples from Ugandan women and infants with non-subtype B HIV-1 infection. In Uganda, most HIV-1 infections are caused by subtypes A and D, which are prevalent in approximately equal proportions (15, 20); other subtypes (e.g., subtypes C and G) have also been reported (3, 4, 11, 17, 20). Analysis of subtype A and D HIV-1 is relevant to the worldwide AIDS epidemic, since those subtypes have also been found in other African countries, as well as in the United States, South America, Asia, Europe, the former Soviet Union, and other regions. In the present study, we focused our analysis on whether the ViroSeq system could successfully amplify DNA for sequencing and provide complete DNA sequences for genotypic analysis.
MATERIALS AND METHODS
Samples used for analysis.
Samples were obtained from Ugandan women and infants enrolled in the HIV Network for Prevention Trials (HIVNET) 012 clinical trial, which compared the efficacies of two different antiretroviral regimens for prevention of HIV-1 vertical transmission (14; M. Owor, M. Deseyve, C. Duefield, M. Musisi, T. Fleming, P. Musoke, L. Guay, F. Mmiro, and J. B. Jackson, XIII Int. AIDS Conf., abstr. LbOr1, 2000). The samples analyzed in the present study were collected 6 to 8 weeks after delivery from a subset of women and infants enrolled in the nevirapine (NVP) arm of the HIVNET 012 clinical trial. This included 33 women whose infants were HIV-1 infected by age 6 to 8 weeks despite NVP prophylaxis, 25 of the HIV-1-infected infants, and 72 women whose infants were alive and uninfected at 6 to 8 weeks of age. Viral load data were obtained in the HIVNET 012 clinical trial with the Amplicor MONITOR test kit (Roche, Branchburg, N.J.).
HIV-1 genotyping.
HIV-1 genotyping was performed with the Applied Biosystems ViroSeq HIV-1 Genotyping System (Applied Biosystems). Analysis in this system begins with HIV-1 RNA isolation, reverse transcription with Moloney murine leukemia virus RT, and a single 40-cycle PCR with AmpliTaq Gold. The PCR yields a 1.8-kb DNA product. PCR amplifications are performed with a uracil N-glycosylase contamination control system to reduce the risk of contamination of the PCR mixtures with products from previous amplification reactions. PCR products are purified with spin columns and analyzed by agarose gel electrophoresis prior to sequencing. The intensity of ethidium bromide staining of the products from each PCR is compared to that of a DNA molecular weight size standard included with the ViroSeq system; the manufacturer provides instructions to determine whether each PCR produced sufficient DNA for sequencing and whether the PCR products should be diluted prior to sequencing. DNA sequence analysis is performed with premixed BigDye sequencing reagents with seven different primers. BigDye terminator chemistry provides 98% accuracy at 550 bases for the ABI PRISM 377 DNA sequencer, which was used for analysis. Software provided with the ViroSeq system is used to assemble sequence data from the different primers into a contiguous sequence that can be inspected for identification of drug resistance mutations. The complete sequence includes the region that encodes protease amino acids 1 to 99 and RT amino acids 1 to 324. In the present study, genotyping was performed as recommended by the manufacturer, with the following exception. The plasma volume recommended for analysis is 0.5 ml. In the present study, all samples from infants were 0.1 ml and some samples from women were <0.5 ml, as indicated. Sequencing reactions were analyzed with an ABI PRISM 377 automated sequencer. Protease and RT nucleotide sequences were aligned by the Clustal method, and phylogenetic reconstructions were performed (DNASTAR; Lasergene, Madison, Wis.).
RESULTS
We evaluated the performance of the Applied Biosystems Viroseq HIV-1 Genotyping System for analysis of Ugandan plasma samples. Applied Biosystems recommends the use of 0.5-ml plasma samples with viral loads that were >2,000 copies/ml for analysis in the ViroSeq system. In our sample set, the volume of plasma available for analysis was limited. The samples used for analysis were <0.5 ml for 6 of 105 women and all 25 infants (Table 1). The viral loads of these samples were relatively high, although some samples had viral loads <10,000 copies/ml (Table 1). In the present study, PCR provided sufficient DNA for sequencing for all samples tested (Table 2).
TABLE 1.
Samples used for analysis
| Subject | No. of samples | Vol (ml)
|
No. of samples <0.5 ml | No. of samples for which viral load data were available | Viral load (no. of copies/ml)
|
No. of samples with <10,000 copies/ml | ||
|---|---|---|---|---|---|---|---|---|
| Range | Mean | Range | Mean | |||||
| Mother | 105 | 0.1–0.5 | 0.48 | 6 | 103 | 2,313–666,883 | 114,943 | 18 |
| Infant | 25 | 0.1 | 0.1 | 25 | 13 | 44,454–2,336,400 | 606,738 | 0 |
| Total | 130 | 0.1–0.5 | 0.41 | 31 | 116 | 2,313–2,366,400 | 170,058 | 18 |
TABLE 2.
Summary of assay performance
| Subject | No. of samples analyzed | No. of samples with PCR product sufficient for sequencing | No. of primer reactions performeda | No. (%) of primer reactions successful on first attempta | No. of primer reactions repeateda | No. (%) of repeat primer reactions successfula | No. (%) of genotypes obtained | No. (%) of genotypes with full double-stranded sequences |
|---|---|---|---|---|---|---|---|---|
| Mother | 105 | 105 | 630 | 603 (96) | 27 | 6 (22) | 102 (97) | 90 (86) |
| Infant | 25 | 25 | 150 | 144 (96) | 6 | 2 (33) | 22 (88) | 22 (88) |
| Total | 130 | 130 | 780 | 747 (96) | 33 | 8 (24) | 124 (95) | 112 (86) |
The analysis did not include the alternate primer D.
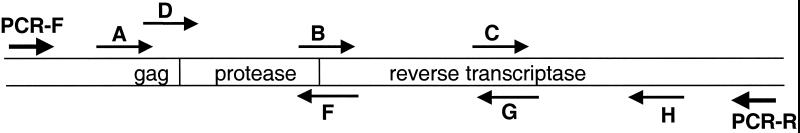
The locations and positions of the sequencing primers in the ViroSeq system are shown in Fig. 1. Primers A, B, C, and D sequence the sense strand of the PCR product. Primers A and D are alternate primers that bind to the heterogeneous gag region. Primers F, G, and H sequence the antisense strand of the PCR product. In this analysis, a total of 848 primer reactions were performed. This included analysis of all 130 samples with primers A, B, C, F, G, and H and analysis of the first 68 samples with the alternate primer, primer D (Table 3). Only 24 (35%) of the 68 reactions with primer D were successful. In contrast, the reactions with primer A were successful for most of those samples. Therefore, primer D was not included for routine analysis of the remaining 62 samples. An advantage of including only six primers for each sample (either primer A or D, but not both) is that 16 samples can be analyzed with a single 96-well plate and a single 96-lane sequencing gel. For primers A, B, C, F, G, and H, 96% of the reactions were successful on the first sequencing attempt. Sequencing reactions were repeated for 33 primers that initially failed. Eight (24%) of the repeat sequencing reactions were successful (Table 2). Note that, for one sample, reactions with all primers failed, accounting for 6 of 25 (24%) of the primer failures. Also, 2 of 7 primer A failures were for samples from a mother-infant pair, and 3 of 13 primer F failures were for samples from a mother and her twins. The alternate primer, primer D, was tested with four of the seven samples for which reactions with primer A failed, and reactions with primer D were successful for three of the four samples. Full-length sequences were obtained for 102 of 105 (97%) of the samples from women tested and 22 of 25 (88%) of the samples from infants tested. Complete double-stranded sequences were obtained for 112 of 130 (86%) of the samples tested (Table 2).
FIG. 1.
PCR and sequencing primers. The orientation and position of the PCR primers (PCR-F and PCR-R) and the seven sequencing primers in the ViroSeq HIV-1 Genotyping System are shown with respect to the protease and RT coding regions.
TABLE 3.
Performance of sequencing primers
| Sequencing primer | No. of reactions performed | No. (%) of reactions successful |
|---|---|---|
| A | 130 | 123 (95) |
| B | 130 | 129 (99) |
| C | 130 | 129 (99) |
| F | 130 | 117 (90) |
| G | 130 | 128 (99) |
| H | 130 | 129 (99) |
| D | 68 | 24 (35) |
To confirm the absence of sample cross-contamination, protease and RT nucleotide sequences from each sample were aligned and phylogenetic reconstructions were performed. No two sequences in the data set were identical, and the sequence obtained from each infant most closely resembled the sequence obtained from the corresponding mother.
HIV-1 subtyping was performed for the 102 women and 22 infants whose samples generated full-length sequences. Subtyping methods and the subtype analysis of the samples from the women are described in a separate report (11); those subtypes were 50A, 35D, 12 A/D recombinant, and 4C; the subtype of one sequence could not be determined. The subtypes identified in the infants were 10A, 9D, 2 A/D recombinant, and 1C. We compared the subtypes of samples for which reactions with one or more of the sequencing primers failed. No subtype was identified for the sample for which reactions with all seven primers failed. Among the remaining 6 samples for which reactions with primer A failed, 3 had subtype D and 3 had subtype A; and among the remaining 12 samples for which reactions with primer F failed, 4 had subtype A, 6 had subtype D, and 2 had A/D recombinant HIV-1. Samples for which reactions with primer D failed included those with subtype A, C, D, and A/D recombinant HIV-1. We noted no association between subtype and sequencing primer performance among the subtypes tested.
DISCUSSION
The study described in this report tested the performance of the Applied Biosystems ViroSeq HIV-1 Genotyping System for analysis of non-subtype B HIV-1 in Ugandan plasma samples. The performance of the ViroSeq system with non-subtype B samples, including subtype A, C, and D, and A/D recombinant HIV-1, was comparable to that reported for analysis of subtype B HIV-1 (6). The major difference in the performance of this system for analysis of subtype B and Ugandan samples was the performance of the alternate sequencing primer, primer D. Reactions with this primer were successful for only 35% of the Ugandan samples tested. In contrast, reactions with primer D were successful for 176 (92%) of 192 samples from a large cohort of pediatric patients in which all but two patients had subtype B infection (6). The failure of primer D did not pose a problem with analysis of the Ugandan samples, since reactions with primer A were successful for almost all of the samples tested. These results demonstrate the advantage of having alternate sequencing primers for the heterogeneous gag region.
Analysis of drug resistance in non-subtype B HIV-1 is becoming increasingly important as the availability and use of antiretroviral drugs increase in regions where non-subtype B is prevalent. Some group O isolates are naturally resistant to NNRTIs such as NVP, reflecting the presence of cysteine at position 181 in RT (Y181C) (9, 19). Subtype F isolates with decreased susceptibilities to NNRTIs and subtype G isolates with decreased susceptibilities to protease inhibitors have also been described (1, 8). Our recent analysis of NVP resistance in women in the HIVNET 012 clinical trial also suggests that the HIV-1 subtype may influence the emergence of NVP-resistant HIV-1 following single-dose NVP prophylaxis (11). Further studies are needed to examine the genotypic correlates of drug resistance in different HIV-1 subtypes and to examine the emergence of drug resistance in individuals infected with non-subtype B HIV-1.
Analysis of drug resistance in infants is becoming increasingly important, since antiretroviral prophylaxis is now recommended for prevention of HIV-1 vertical transmission. Furthermore, drug resistance may be most likely to arise in infants in resource-poor settings where non-subtype B is prevalent, since pregnant women and infants in those settings are more likely to receive shorter regimens of only one or two antiretroviral drugs. Those regimens are less likely to fully suppress HIV-1 replication. Our analysis of samples from infants involved in the HIVNET 012 clinical trial provided complete genotypes (protease and RT) for 22 of 25 infants. For two of the three remaining samples, partial genotypes were obtained which were sufficient for analysis of NVP resistance mutations. NVP-resistant HIV-1 was detected 6 to 8 weeks after delivery in 11 of 24 (46%) of these infants following the administration of a single dose of NVP (12). This analysis was performed with 0.1-ml plasma samples (one-fifth of the recommended sample volume). In many cases, it may be difficult to obtain larger plasma samples from infants. The ability to use the ViroSeq system for analysis of low-volume samples makes it attractive for analysis of antiretroviral drug resistance in infants.
This report demonstrates the successful use of the ViroSeq HIV-1 Genotyping System for analysis of A, C, D, and A/D recombinant HIV-1 in clinical plasma samples, including low-volume samples from infants. Further studies are needed to extend this analysis to include samples with other subtypes and with low viral loads.
ACKNOWLEDGMENTS
This work was supported by (i) the Elizabeth Glaser Pediatric AIDS Foundation; (ii) HIVNET, sponsored by the National Institute of Allergy and Infectious Diseases (NIAID), the National Institutes of Health (NIH), and the U.S. Department of Health and Human Services (DHHS), through contract N01-AI-35173 with Family Health International, contract N01-AI-45200 with the Fred Hutchinson Cancer Research Center, and subcontracts with JHU/Markerere University (Kampala, Uganda) (contract NO1-AI-35173-417); (iii) the HIV Prevention Trials Network (HPTN), sponsored by NIAID, National Institute of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse, National Institute of Mental Health, and the Office of AIDS Research of NIH, DHHS (contracts U01-AI-46745 and U01-AI-48054); (iv) the Pediatric and Adult AIDS Clinical Trials Groups Division of AIDS, NIAID, NIH); and (v) R29 34348 (NICH/HD, NIH). Reagents for HIV genotyping were provided by Applied Biosystems.
We thank Philippa Musoke and Francis Mmiro (Makerere University) for providing the plasma samples used for analysis. We acknowledge the assistance of Melissa Allen (Protocol Specialist, Family Health International). We thank Estelle Piwowar-Manning, Constance Ducar, and the laboratory staff in Uganda for assistance with sample processing. We also thank Eric Shulse and the Applied Biosystems Genotyping Team for helpful discussions and for providing the reagents used in the study.
REFERENCES
- 1.Apetrei C, Descamps D, Collin G, Loussert-Ajaka I, Damond F, Duca M, Simon F, Brun-Vezinet F. Human immunodeficiency virus type 1 subtype F reverse transcriptase sequence and drug susceptibility. J Virol. 1998;72:3534–3538. doi: 10.1128/jvi.72.5.3534-3538.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker-Pergola G, Kataaha P, Johnston-Dow L, Fung S, Jackson J B, Eshleman S H. Analysis of HIV-1 protease and reverse transcriptase in antiretroviral drug naïve Ugandan adults. AIDS Res Hum Retrovir. 2000;16:807–813. doi: 10.1089/088922200308800. [DOI] [PubMed] [Google Scholar]
- 3.Becker-Pergola G, Mellquist J L, Guay L, Mmiro F, Ndugwa C, Kataaha P, Jackson J B, Eshleman S H. Identification of diverse HIV-1 subtypes and dual HIV-1 infection in pregnant Ugandan women. AIDS Res Hum Retrovir. 2000;16:1099–1104. doi: 10.1089/088922200414938. [DOI] [PubMed] [Google Scholar]
- 4.Brennan C A, Lund J K, Golden A, Yamaguchi J, Vallari A S, Phillips J F, Kataaha P K, Jackson J B, Devare S G. Serologic and phylogenetic characterization of HIV-1 subtypes in Uganda. AIDS. 1997;11:1823–1832. doi: 10.1097/00002030-199715000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Brodine S K, Mascola J R, Weiss P J, Ito S I, Porter K R, Artenstein A W, Garland F C, McCutchan F E, Burke D S. Detection of diverse HIV-1 genetic subtypes in the USA. Lancet. 1995;346:1198–1199. doi: 10.1016/s0140-6736(95)92901-0. [DOI] [PubMed] [Google Scholar]
- 6.Cunningham S, Ank B, Lewis D, Wei L, Dileanis J, Jackson J B, Palumbo P, Krogstad P, Eshleman S H. Performance of the Applied Biosystems ViroSeq HIV-1 Genotyping System for analysis of HIV-1 in pediatric plasma samples. J Clin Microbiol. 2001;39:1254–1257. doi: 10.1128/JCM.39.4.1254-1257.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damond F, Apetrei E, Couturier E, Descamps D, Collin G, Brun-Vezinet F, Simon F. Prevalence of HIV-1 subtypes in France, 1996–1997, consequences for viral monitoring. AIDS. 1998;12:S73. [Google Scholar]
- 8.Descamps D, Apetrei C, Collin G, Damond F, Simon F, Brun-Vezinet F. Naturally occurring decreased susceptibility of HIV-1 subtype G to protease inhibitors. AIDS. 1998;12:1109–1111. [PubMed] [Google Scholar]
- 9.Descamps D, Collin G, Letourneur F, Apetrei C, Damond F, Loussert-Ajaka I, Simon F, Saragosti S, Brun-Vezinet F. Susceptibility of human immunodeficiency virus type 1 group O isolates to antiretroviral agents: in vitro phenotypic and genotypic analyses. J Virol. 1997;71:8893–8898. doi: 10.1128/jvi.71.11.8893-8898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich U, Ruppach H, Gehring S, Knechten H, Knickmann M, Jager H, Wolf E, Husak R, Orfanos C E, Brede H D, Rubsamen-Waigmann H, von Briesen H. Large proportion of non-B HIV-1 subtypes and presence of zidovudine resistance mutations among German seroconvertors. AIDS. 1997;11:1532–1533. [PubMed] [Google Scholar]
- 11.Eshleman S H, Becker-Pergola G, Deseyve M, Guay L A, Mracna M, Fleming T, Cunningham S, Musoke P, Mmiro F, Jackson J B. Impact of HIV-1 subtype on women receiving single dose NVP prophylaxis to prevent HIV-1 vertical transmission (HIVNET 012) J Infect Dis. 2001;184:914–917. doi: 10.1086/323153. [DOI] [PubMed] [Google Scholar]
- 12.Eshleman, S. H., M. Mracna, L. A. Guay, M. Deseyve, C. Cunningham, M. Mirochnick, P. Musoke, T. Fleming, M. G. Fowler, L. M. Mofenson, F. Mmiro, and J. B. Jackson. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012). AIDS, in press. [DOI] [PubMed]
- 13.Gao F, Yue L, Hill S C, Robertson D L, Graves A H, Saag M S, Shaw G M, Sharp P M, Hahn B H. HIV-1 sequence subtype D in the United States. AIDS Res Hum Retrovir. 1994;10:625–627. doi: 10.1089/aid.1994.10.625. [DOI] [PubMed] [Google Scholar]
- 14.Guay L A, Musoke P, Fleming T, Bagenda D, Allen M, Nakabiito C, Sherman J, Bakaki P, Ducar C, Deseyve M, Emel L, Mirochnick M, Fowler M G, Mofenson L, Miotti P, Dransfield K, Bray D, Mmiro F, Jackson J B. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-infant transmission of HIV-1 in Kampala, Uganda: HIVNET-012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 15.Hu D J, Baggs J, Downing R G, Pieniazek D, Dorn J, Fridlund C, Biryahwaho B, Sempala S D, Rayfield M A, Dondero T J, Lal R. Predominance of HIV-1 subtype A and D infections in Uganda. Emerg Infect Dis. 2000;6:609–615. doi: 10.3201/eid0606.000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irwin K L, Pau C P, Lupo D, Pienazek D, Luo C C, Olivo N, Rayfield M, Hu D J, Weber J T, Respess R A, Janssen R, Minor P, Ernst J. Presence of human immunodeficiency virus (HIV) type 1 subtype A infection in a New York community with high HIV prevalence: a sentinel site for monitoring HIV genetic diversity in North America. Centers for Disease Control and Prevention-Bronx Lebanon HIV Serosurvey Team. J Infect Dis. 1997;176:1629–1633. doi: 10.1086/517343. [DOI] [PubMed] [Google Scholar]
- 17.Kaleebu P, Bobkov A, Cheingsong-Popov R, Bieniasz P, Garaev M, Weber J. Identification of HIV-1 subtype G from Uganda. AIDS Res Hum Retrovir. 1995;11:657. doi: 10.1089/aid.1995.11.657. [DOI] [PubMed] [Google Scholar]
- 18.Pieniazek D, Rayfield M, Hu D J, Nkengasong J, Wiktor S Z, Downing R, Biryahwaho B, Mastro T, Tanuri A, Soriano V, Lal R, Dondero T. Protease sequences from HIV-1 group M subtypes A-H reveal distinct amino acid mutation patterns associated with protease resistance in protease inhibitor-naive individuals worldwide. HIV Variant Working Group. AIDS. 2000;14:1489–1495. doi: 10.1097/00002030-200007280-00004. [DOI] [PubMed] [Google Scholar]
- 19.Quinones-Mateu M E, Soriano V, Domingo E, Menendez-Arias L. Characterization of the reverse transcriptase of a human immunodeficiency virus type 1 group O isolate. Virology. 1997;236:364–373. doi: 10.1006/viro.1997.8748. [DOI] [PubMed] [Google Scholar]
- 20.Rayfield M A, Downing R G, Baggs J, Hu D J, Pieniazek D, Luo C C, Biryahwaho B, Otten R A, Sempala S D K, Dondero T J. A molecular epidemiologic survey of HIV in Uganda. HIV Variant Working Group. AIDS. 1998;12:521–527. doi: 10.1097/00002030-199805000-00014. [DOI] [PubMed] [Google Scholar]
- 21.Rowe P M. HIV-1 group O infection identified in USA. Lancet. 1996;348:116. doi: 10.1016/s0140-6736(05)64613-2. [DOI] [PubMed] [Google Scholar]
- 22.Vergne L, Peeters M, Mpoudi-Ngole E, Bourgeois A, Liegeois F, Toure-Kane C, Mboup S, Mulanga-Kabeya C, Saman E, Jourdan J, Reynes J, Delaporte E. Genetic diversity of protease and reverse transcriptase sequences in non-subtype-B human immunodeficiency virus type 1 strains: evidence of many minor drug resistance mutations in treatment-naive patients. J Clin Microbiol. 2000;38:3919–3925. doi: 10.1128/jcm.38.11.3919-3925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]