Abstract
The transmission of SARS-CoV-2 coronavirus has led to the COVID-19 pandemic. Nucleic acid testing while specific has limitations for mass surveillance. One alternative is the main protease (Mpro) due to its functional importance in mediating the viral life cycle. Here, we describe a combination of modular substrate and gold colloids to detect Mpro via visual readout. The strategy involves zwitterionic peptide that carries opposite charges at the C-/N-terminus to exploit the specific recognition by Mpro. Autolytic cleavage releases positively-charged moiety that assembles the nanoparticles with rapid color changes (t <10 min). We determine a limit of detection for Mpro in breath condensate matrices <10 nM. We further assayed ten COVID-negative subjects and found no false-positive result. In the light of simplicity, our test for viral protease is not limited to an equipped laboratory, but also is amenable to integrating as portable point-of-care devices including those on face-coverings.
Keywords: covid-19, mpro/3clpro/nsp5, colorimetric analysis, zwitterionic peptide, smart mask
Graphical Abstract
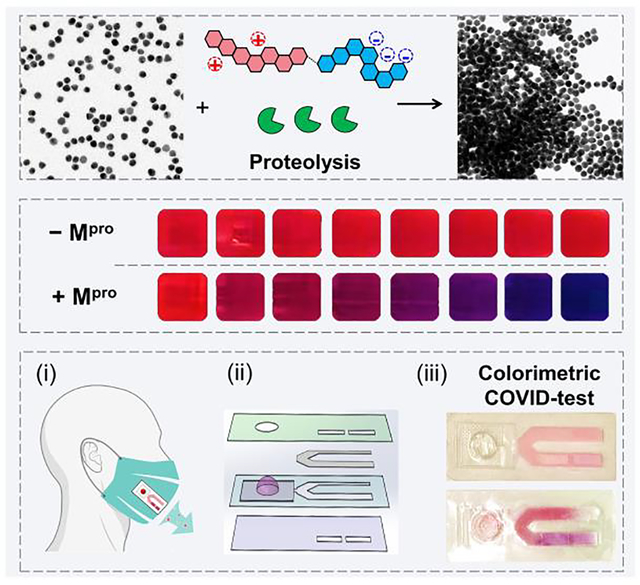
We reported a rapid colorimetric detection of SARS-CoV-2 protease using a zwitterionic peptide and gold colloids. Our sensor showed good performance in exhaled breath condensate with a limit of detection in the low nanomolar range. This technology can be integrated into regular face coverings for COVID-detection.
Introduction
Proteolytic post-processing is a critical step in the lifecycle of many proteins from infectious microorganisms (e.g., virus).[1, 2] For example, the functional polypeptides required for SARS-CoV-2 replication and transcription are released from the proteins by extensive proteolytic processing predominately by the main protease (Mpro, also known as 3CLpro/nsp5).[3] Analytical detection of these proteolytic events could have value in diagnosis and therapeutic screening development.[4] Established protocols for probing proteases include the use of optically-addressed substrates, enzyme-linked immunosorbent assay, and gel electrophoresis.[5, 6] However, these techniques require labor-intensive chromophore labelling, multi-step detection procedures, and/or specific instrumentation. A cost-effective, easy, and colorimetric sensing platform could thus solve these major limitations analogous to the impact of lateral flow assays.
Colorimetric sensors based on gold nanoparticles (AuNPs) are popular because of their unique photophysical properties including a surface plasmon resonance (SPR) band with a high molar absorption coefficient. Aggregation enhances the near-field interactions of proximal nanoparticles and leads to a bathochromic shift in the SPR band; this in turn leads to a pronounced color change.[7] Strategies for plasmonic coupling can be grouped into chemical linking and physical factors.[8, 9] For example, dithiothreitol is a simple molecule to cluster AuNPs due to the strong thiol-to-gold bond.[10, 11, 12] Nanoplasmonic sensing based on this covalent bond have been reported for thrombin, trypsin, furin, etc.[10, 13]
Alternatively, controlled physical factors (e.g., ionic strength, solvent polarity, and ligand hydrophilicity) can also impact colloidal dispersion.[9, 14, 15] In particular, nanoparticles primarily stabilized through electrostatic double layer repulsions tend to aggregate via the addition of excess salt due to charge screening.[15] Tuning non-specific electrostatic interactions has also led to several simple and rapid colorimetric assays for heavy metal ions.[16] In principle, these electrostatic processes could be repurposed for sensing other targets (here, proteases) by rationally designing the electrophoretic property of a specific substrate where the selectivity is based on molecular recognition between the enzyme and the substrate.
Here we report on a general formula of modular zwitterionic peptides, (Asp)n−(AA)x−(Arg)m, that induce AuNP aggregation in response to proteases. We then apply this system to colorimetrically measure Mpro. Unlike nucleic acids testing, protease detection uses autolytic cleavage of the substrates that amplifies signals without the polymerase chain reaction. The key role of Mpro in the propagation of infectious SARS-CoV-2—in addition to a lack of human homologues—make this protease an ideal maker for COVID-19 diagnosis.[2, 4]
Results and Discussion
In our system, the modular zwitterionic peptide has oppositely charged termini, i.e., (Asp)n and (Arg)m, flanking a specific Mpro cleavage site (AA)x. Because n ≥ m, the intact peptide carries a net neutral charge. Accordingly, the interactions between the intact peptide and sulfonated ligand-coated AuNPs are minimized given the negatively charged sulfonate groups. Proteolytic segmentation of the peptide would switch the electrophoretic property of the corresponding fragment, thus triggering the collapse of gold suspension via electrostatic attractions. This induces a visible color change. By using a combination of the zwitterionic peptide and AuNPs in label-free conditions, we showed that this approach can rapidly detect Mpro at approximately 3 to 10 nM in buffer and exhaled breath condensate in 10 min. Several mutated peptides were built to investigate the sensing mechanism and to optimize the sensor performance. Finally, we reframed the sensor configuration by building it into a lateral flow strip to create portable sensing devices as a point-of-care surveillance tool for COVID disease. Overall, the novel peptide-enabled colorimetric assay does not require interfacial bioconjugations or any specific instrumentations, and thus may have value in surveillance of COVID.
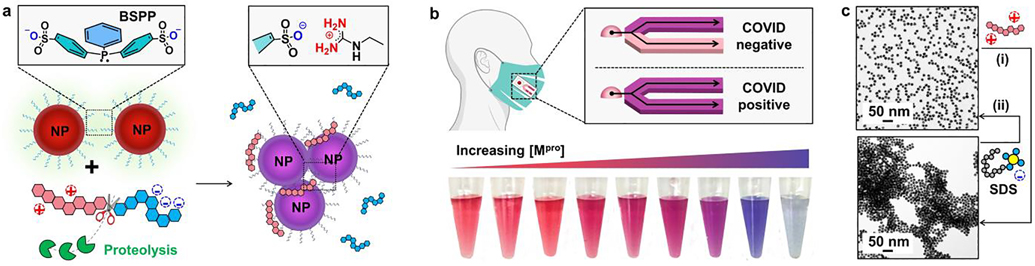
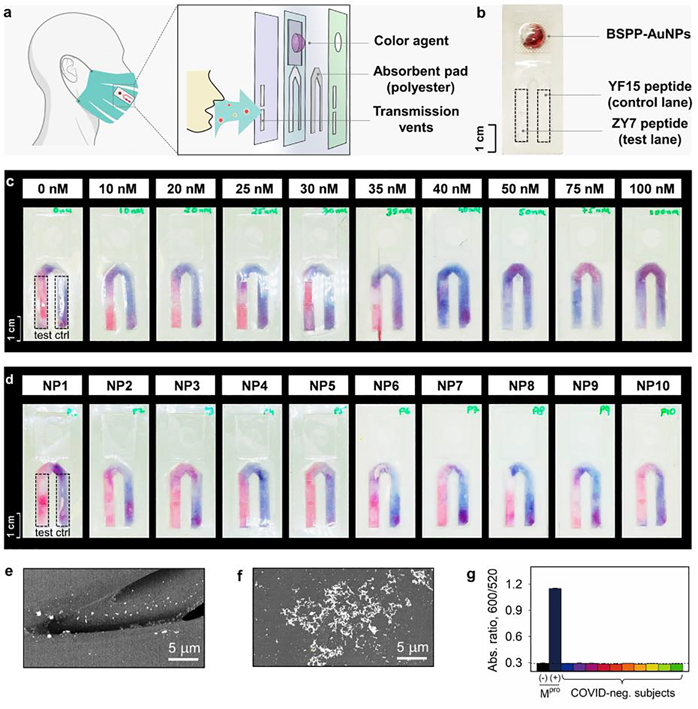
Figure 1a schematically shows the intact peptide and its proteolytic fragments with distinct physicochemical properties (e.g., net charge, interfacial affinity) to modulate the dispersity of a gold suspension. We hypothesize that successful proteolysis of the substrate will induce clustering of the monodispersed particles, which changes the color from ruby red to violet-blue by naked eye. In addition, this color change reporter when integrated with routinely-used mask could allow one to easily monitor the COVID-infectious status (Figure 1b,c).
Figure 1.
The surveillance approach uses a color change based on peptide-gold assembly. (a) Schematic illustration of the aggregation of bis(p-sulfonatophenyl)phenylphosphine-coated gold nanoparticles (BSPP-AuNPs) caused by the proteolytic hydrolysis of the intact peptide where the net charge of intact peptide and its fragment is reversed. The green cartoon designates proteases, i.e., Mpro; the tandem hexagon represents a modular zwitterionic peptide with a Mpro recognition site. (b) The colorimetric test could be coupled with face coverings with a lateral flow strip to indicate COVID infection in-situ (top). White-light image of BSPP-AuNPs after incubating with the proteolytic products. The visual color shifts from ruby red to violet-blue with increasing amount of Mpro from 0−200 nM (bottom). (c) TEM images of the dispersed BSPP-AuNPs (top) and proteolysis-induced gold aggregates [bottom, (i)]. The flocculated AuNPs can be redispersed using ionic surfactant additives, such as sodium dodecyl sulfate [SDS, (ii)], due to the restored electrostatic repulsion.
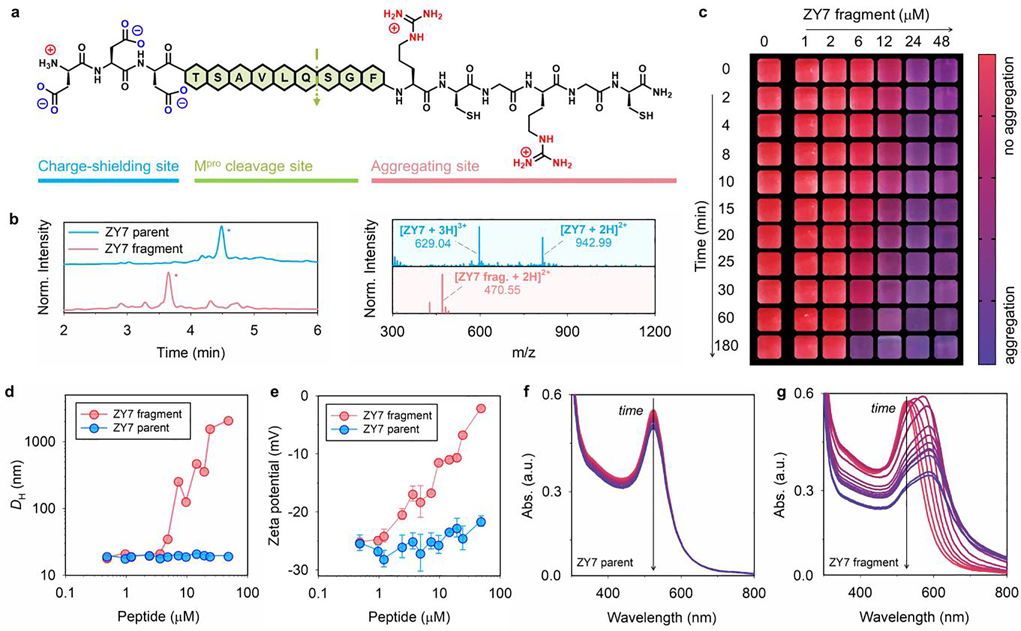
The zwitterionic peptides were rationally designed to encompass arranging three functional domains in a linear N-to-C-terminal configuration (Figure 2a), including: (i) an N-terminal charge shielding domain made of repeating aspartic acid (Asp, D) for neutralizing the charge of the intact peptide, i.e., part iii; (ii) an exposed protease-recognition sequence; and (iii) a C-terminal aggregating site for clustering gold colloids. The aggregating domain (part iii) has a repeating arginine (Arg, R) and a cysteine (Cys, C) bridge with high affinity to AuNPs via electrostatic and coordinating interactions.[17] Overall, the peptide design meets two key requirements. The first is little or no attraction between the intact peptide and AuNPs. This is achieved by matching the charge state of intact peptide to that of particle surfaces, which typically originates from the terminal groups of the surface ligand.[18] Second, peptide fragmentation by the protease exposes the aggregating domain that favors tight self-assembly of particles. This modular design is also intended to obviate the need for major revisions when targeting a new protease. That is, the specificity of the sensor is tuned by changing only the cleavage site while keep the other components the same. In addition, the inclusion of aspartic acid and arginine residue improves the hydrophilicity of the oligopeptide and ensures the sensor performance in aqueous media. Figure 2a shows a representative peptide ZY7 synthesized based on the above criteria: DDDTSAVLQSGFRCGRGC (net charge = 0). Specifically, ZY7 contains a Mpro cleavage sequence LQ↓SG [i.e., at C-terminus of glutamine (Q)] surrounded by a negatively and positively charged moiety (i.e., DDD vs. RCGR).[19] To the best of our knowledge, no mammalian protease cleaves after a C-terminal glutamine. Hence this cleavage site is unique for viral proteases only. Proteolysis of ZY7 peptide by Mpro has been confirmed by HPLC and ESI-MS and results in the formation of two fragments: DDDTSAVLQ (net charge = −3) and SGFRCGRGC (net charge = +3, with a dithiol bridge); see Figure 2b and Table 1.
Figure 2.
Modular zwitterionic peptide and protease-induced gold aggregation. (a) Peptide ZY7 has three domains, including a charge-shielding (DDD), a Mpro recognition (LQ↓SG), and an aggregating site (RCGRGC). The net charge of ZY7 and its aggregating fragment is 0 and +3, respectively. (b) HPLC and ESI-MS data show that Mpro selectively cleaves the ZY7 peptide at the C-terminus of glutamine (Q). Peak with * is the product. (c) The color evolution of BSPP-AuNPs (3.8 nM, 50 μL) in the presence of pre-cleaved ZY7 fragments. Shown are the cropped images with a color bar where purple represents more aggregation. The particle aggregation kinetic is concentration- and time-dependent, as shown in horizontal and vertical directions. (d) DLS profiles of BSPP-AuNPs (3.8 nM, 100 μL) incubated with increasing concentrations of ZY7 parent peptide (blue) and ZY7 fragments (red). No change of the hydrodynamic diameter (DH) for BSPP-AuNPs was observed when incubated with ZY7 peptide, while such a change became sizable in the presence of ZY7 fragments of more than ~3 μM. (e) Zeta potential measurements of AuNPs (3.8 nM, 100 μL) incubated with increasing concentrations of ZY7 parent peptide (blue) and ZY7 fragments (red). The ZY7 fragments adsorbed to the particle and altered the surface charges from −26.5 to −2.5 mV. Error bars represent triplicate measurements for one sample. (f-g) The time progression of optical absorption of AuNPs (3.8 nM, 50 μL) when incubated with ZY7 parent peptide and its fragments (c = 6.0 μM). The curves from red to purple were recorded every 10 min for 2 h. Noticeable peak changes were observed at 520 and 600 nm for a fragment-AuNP mixture.
Table 1.
Peptide information and limit of detection (LoD) for Mpro.
| Name | Peptide sequence | M. W. (g mol−1) | Net charge [a] | Operation window [b] (μM) | LoD, Mpro (nM) | Description | ||
|---|---|---|---|---|---|---|---|---|
| TB | EBC [c] | Saliva | ||||||
| ZY7 | DDDTSAVLQ↓SGFRCGRGC [d] | 1884.8 | 0 | 3.2 − 55.3 | 27.7 | 33.4 | 68.4 | Arg and Cys bridge, zwitterion |
| MS7 | DDDTSAVLE×SGFRCGRGC | 1855.8 | −1 | - - | - - | - - | - - | scramble sequence |
| NR10 | DDDTSAVLQ↓SGFACGAGC | 1714.6 | −2 | > 100 | - - | - - | - - | Cys bridge |
| NC11 | DDDTSAVLQ↓SGFAGGAGG | 1622.7 | −2 | > 100 | - - | - - | - - | no Arg/Cys bridge |
| DS12 | DDDTSAVLQ↓SGFrcGrGc [e] | 1884.3 | 0 | 2.7 − 73.2 | 114.4 | 94.2 | 142.4 | D-amino acid substitution |
| TE3 | RGRTSAVLQ↓SGFRC | 1535.8 | +4 | 2.5 − 3.1 | - - | - - | - - | no shielding site |
| OR8 | DDDTSAVLQ↓SGFR | 1408.6 | −1 | > 100 | - - | - - | - - | single Arg |
| YR9 | DDDGDDDTSAVLQ↓SGFRRGRR | 2336.1 | −1 | 0.3 − 5.1 | 3.4 | 9.5 | - - | quadruple Arg |
| YF15 | SGFRRGRR | 989.6 | +5 | < 0.3 | - - | - - | - - | fragment |
The electrophoretic property at pH 8.0; note that all the peptides contain a free N-terminus and an amidated C-terminus.
The operation window was determined in Tris-buffer (TB) media at 10 min readout time.
EBC: exhaled breath condensate.
↓ designates the Mpro cleavage site; × designates unrecognized site for Mpro.
The single-letter in uppercase is L-amino acid (AA), while the single-letter in lowercase is D-amino acid.
The functional peptide is used under label-free conditions with AuNPs as a readout to detect the presence of Mpro. This is advantageous because surface labelling techniques often require precise control over the bio-nano functionalization and may involve frequently unanticipated particle aggregation.[20] Here, we chose bis(p-sulfonatophenyl)phenylphosphine (BSPP) to modify the 13-nm citrate-AuNPs made by Turkevich method, which fulfill with two guiding parameters.[21] First, the BSPP ligand is rich in terminal sulfonate groups and thus gives the nanoparticle strong negative charges, i.e., the zeta potentials (ζ) are −26.5 ± 1.5 mV. Second, the nanoparticles are colloidally stable in various media due to the effective phosphorus-to-gold coordination, large steric hindrance from the bulky aromatic rings, and strong electrostatic double layer repulsions.[22] BSPP-capped AuNPs have a compact ligand layer and thus favor tight interparticle interactions, e.g., the hydrodynamic diameter (DH) is 19.1 nm with a PDI of 0.10 (Figure S1).[23] More characterizations of the ligand exchange and the resulting particles such as TEM, DLS, UV-Vis, and 1H NMR are given in Figure S1.
We here selected Mpro peptidase as a model example to test our label-free peptide and BSPP-AuNPs. Mpro regulates viral replication and transcription and is therefore an attractive therapeutic and diagnostic target for SARS-CoV-2.[3, 4] We first validated our hypothesis by incubating BSPP-AuNPs with either ZY7 parent peptide or its pre-digested fragments. The full cleavage was confirmed by HPLC and ESI-MS (Figures 2b and S2). Figure 2c shows the color change as a function of the peptide concentration and time. The addition of ZY7 fragments (6−50 μM) to the BSPP gold dispersion caused an instant color shift from ruby red to purple. Such a color evolution continuously built with increasing fragments concentration and reaction time. In comparison, BSPP-AuNPs coexist with the ZY7 parent peptide with no change of the color (Figure S3b).
Characterizations of the gold aggregates and several peptide derivatives of ZY7 were subsequently employed for a mechanistic study. TEM images in Figure 1c and hydrodynamic diameter (DH) by dynamic light scattering (DLS, Figure 2d) clearly indicate the formation of AuNP aggregates in the presence of ZY7 fragments. This size increases commensurate with alternations in surface charge. For example, the zeta potentials of BSPP-AuNPs mixed with increasing amount of ZY7 fragments showed a sizable increase from original value to nearly neutral, e.g., −26.5 vs. −2.5 mV; see Figure 2e. We attribute this increasing charge to the positive guanidine groups in arginine-rich fragment adsorbed onto the surface sulfonate via electrostatic attractions.[15] This neutralized the surface charge and compromised the interparticle repulsions thus resulting in the collapse of colloidal system.
To confirm this, we synthesized an analogue peptide NR10, where the arginine residues were substituted with neutral alanine; see Table 1. In this case, the absence of arginine determinants caused no aggregation of the AuNPs irrespective of proteolytic cleavage (Figure S3). This manifested in no change in DH (i.e., approx. 20 nm) and zeta potentials, albeit there was a slight decrease in the surface potential due to weak association of NR10 of −2 charge on the particle surfaces (i.e., −30 vs. −40 mV). We therefore conclude that the aggregation of BSPP-AuNPs was dominated by the positively charged arginine residues rather than disulfide bridging. To this end, we quantitatively assessed the color changes through analyses of the progression of UV-Vis spectra. In Figure 2f–g, the time-lapsed absorbance profiles of AuNPs with ZY7 parent peptide remain essentially the same indicating a preserved dispersity. The ZY7 fragment led to a sizable decrease in the SPR at 520 nm commensurate with a noticeably raising band at 600 nm. This is a clear indication of AuNP aggregation and color change.[12] We thus define a ratiometric signal, Abs600/Abs520, to quantify the aggregation and color change.
The above mechanism favors electrostatic-induced flocculation of AuNPs and was further corroborated by a reversible colorimetric experiment, employing several different surfactants (Figure S4). For example, the addition of ionic surfactants such as cetyltrimethylammonium bromide (CTAB) and sodium dodecyl sulfate (SDS) recovered colloidal AuNPs from the aggregates. Excess anionic SDS scavenges the positively charged peptide fragments, thus restoring the electrostatic double layer repulsions between particles.[24]
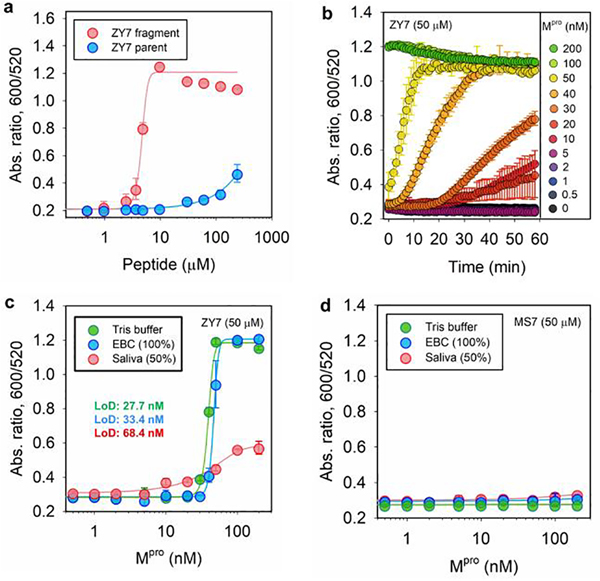
To study the limit of detection (LoD) for Mpro based on our sensing system, we first optimized the concentration of ZY7 peptide in an assay. Experimentally, BSPP-AuNPs (3.8 nM, 100 μL) were incubated with peptides and the optical measurements (Abs600/Abs520) were recorded at 10 min after AuNP addition. The minimum amount of fragment needed to induce a noticeable color change has to exceed 3.2 μM, but parent ZY7 should not cause particle aggregation and only slightly affects the particle dispersity at concentrations higher than 55.3 μM (Figure 3a). Excess ZY7 peptide likely increases the ionic strength and screens the repulsive forces between particles thus making the system colloidally unstable. Accordingly, we defined a working window for the assays based on ZY7 as 3.2−55.3 μM; see Table 1. In practice, a concentration of 50 μM has been observed to yield the best result and was defined as the optimal peptide concentration for the rest of this study.
Figure 3.
Operation window of the sensing system and limit of detection (LoD) for Mpro. (a) Ratiometric signal (Abs600/Abs520) collected from BSPP-AuNPs (3.8 nM, 100 μL) incubated with various amount of ZY7 parent (blue) and fragments (red). The working window for ZY7 substrate is 3.2−55.3 μM. (b) Time progression of absorbance ratio in the enzyme assay, where a fixed amount of ZY7 substrate (50 μM) is incubated with increasing concentrations Mpro (0−200 nM). The test media is exhaled breath condensate (EBC). The data points were read every 1 min for 1 h. (c) The absorbance ratio as a function of Mpro concentration, where ZY7 substrate (50 μM) is used. The determined LoD for Mpro is: 27.7 nM in Tris buffer (TB), 33.4 nM in EBC matrix, and 68.4 nM in saliva. Note that the readout time is 20 min for saliva. The linear regions used to calculate LoDs can be found in Figure S5d. Error bar = standard deviation. (d) The absorbance ratio as a function of Mpro concentration in three biofluids, where MS7 (control) substrate (50 μM) is used. Readout time is set to be 10 min.
To understand the clinical value of the system, we tested Mpro detection in three different matrices including Tris buffer (TB), exhaled breath condensate (EBC), and diluted pooled saliva (50%). The enzyme assay was performed by incubating a defined concentration of ZY7 substrate (i.e., 50 μM) with various concentrations of spiked Mpro (i.e., 0−200 nM) in a 20 μL volume for 3 h. Subsequently, 100 μL of BSPP gold dispersion (3.2 nM) was added as a readout mechanism. All concentrations are with respect to a 120 μL volume; see more details in Supporting Information. Representative aggregation kinetics in EBC within 1 hour are shown in Figure 3b. Similar data collected in other biological media were supplemented in Figure S5. The trend clearly indicates that higher amount of Mpro led to more particle aggregations or rapid color change and vice versa. Importantly, BSPP-AuNPs were stable in the presence of Mpro alone and in all tested buffers (Figure S5).
Next, we set 10 min as the readout time for pursuing a rapid protease detection. Figure 3c shows the ratio of Abs600/Abs520 as a function of Mpro concentration. Thus, the LoD for Mpro was determined to be: 27.7 nM in Tris buffer (TB), 33.4 nM in EBC matrix, and 68.4 nM in 50% saliva. The method for LoD calculation is reported in previous literature.[25] This suggests that our sensor is best performing in TB and EBC media, while other complex matrices such as saliva would yield sensitivity in about 3-fold lower or more. No aggregation occurred in human plasma presumably negative proteins that scavenge the positively charged peptide fragments.[26] The clinical-relevant level of Mpro in novel SARS-CoV-2 infected cells still lacks rigor reports.[27] Nevertheless, the protease LoD of our sensor is similar to other nanoparticle-based protease assays (i.e., in low nanomolar ranges), but is much less time-consuming (i.e., t =10 min).[5, 10, 26, 28, 29] On the contrary, control experiment using a scrambled peptide sequence, MS7, showed no AuNP aggregation at any concentrations of Mpro (Figure 3d). For MS7 peptide, we substituted the indispensable glutamine (Q) at P1 site with its analogue, glutamic acid (E), and therefore no specific recognition by Mpro, circumventing the formation of aggregating fragments; see Table 1. Importantly, the colorimetric detection can be carried out in a one-pot assay. The enzymatic cleavage and particle aggregation took place simultaneously in-situ, albeit a longer readout time was needed (~1 h). The LoD of the one-pot assay for Mpro is about 40.9 nM (Figure S6).
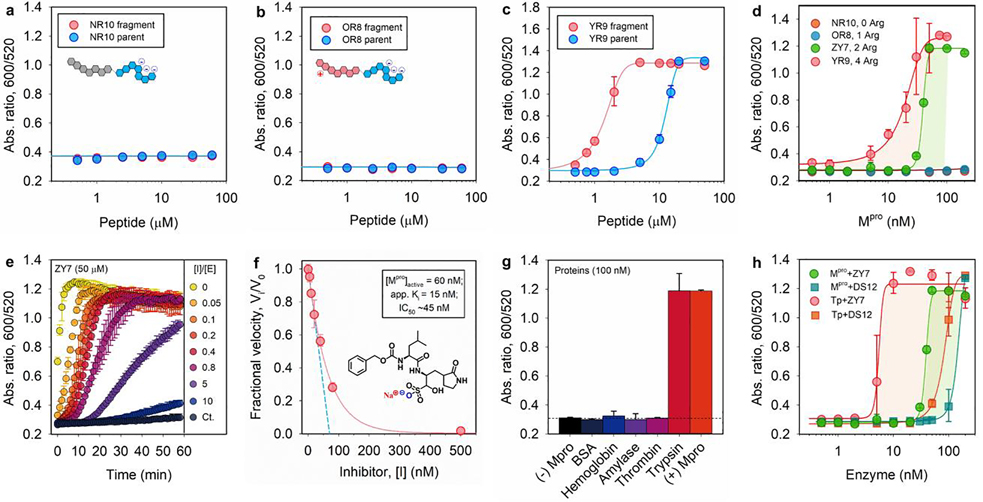
We briefly describe the perspective to improve our protease detection limit. The above modulation of particle dispersity is based on the electrophoretic property of substrates, and thus we further explored the effect of tuning arginine number from 0, 1, 2, and 4, which corresponds to NR10, OR8, ZY7, and YR9 peptide, respectively (Table 1). According to Figure 4a,b, the aggregating sequences containing zero or single guanidine side chain produce no optical signal change. These non-aggregating systems have low ionic valence and attenuated electrostatic interactions according to the Schulze-Hardy rule.[15, 29] Compared to the ZY7 fragment of two arginine residues, the inclusion of four arginine residues in YR9 strongly interferes with the particle surface potential and requires substantially lower concentrations to induce aggregations (e.g., 0.3 vs. 3.2 μM, Figure 4c). This also resulted in an eight-fold improvement in sensitivity to Mpro (Figure 4d). That is, the LoD is improved from 27.7 to 3.4 nM in TB by using YR9 peptide of high ionic valence (Table 1).
Figure 4.
Tunning sensitivity and specificity of the sensor. (a-c) The operation window of Mpro sensors based on NR10, OR8, and YR9 peptide, which contains 0, 1, and 4 arginine residues in its aggregating sequence, respectively; see Table 1. (d) The Mpro LoD of sensors based on four peptides of varying number of arginine: no LoD observed for NR10 and OR8 substrate; 27.7 and 3.4 nM is determined for YR9 and ZY7 substrate, respectively. (e) Time progression of ratiometric signal (Abs600/Abs520) in inhibitor assays. Increasing molar ratio of [inhibitor]/[Mpro] from 0−10 was employed. The control curve (Ct.) designates inhibitor only without Mpro additive. (f) A typical inhibition titration curve fitted with the Morrison equation (Eqn. S2) is shown for the competitive inhibitor, GC376.[30] Inset shows the chemical structure of GC376 inhibitor. A Henderson equation was applied to resolve the apparent inhibitor dissociation constant, Ki (app) =15 nM, and active enzyme concentration, [E]0 =60 nM (out of 100 nM). The IC50 is 45 nM. (g) Sensor activation by other mammalian proteins (100 nM), including bovine serum albumin (BSA), hemoglobin, α-amylase (100 U/mL), thrombin, and trypsin. Assay with and without Mpro is included as positive and negative control. (h) The response of sensors based on ZY7 and DS12 substrate to Mpro or trypsin in Tris buffer. LoD for Mpro is 27.7 and 114.4 nM for ZY7 and DS12, respectively; LoD for trypsin is 9.7 and 30.3 nM for ZY7 and DS12, respectively.
Next, we used a known competitive inhibitor (GC376) for Mpro to study the inhibition assays.[30] Figure 4e shows the results of assaying an increasing molarity of GC376 (i.e., 0−1 μM) in the presence of constant amount of Mpro (100 nM) and ZY7 substrate (50 μM). Note that the inhibitor itself does not affect the particle dispersity; see the control line in Figure 4e. As anticipated, the aggregation kinetics were largely retarded due to low activities of pre-mixed Mpro with the inhibitor. Examination of the absorbance ratio at 15 min yields a typical inhibitor titration curve (Figure 4f). A linear form of the Morrison equation derived by Henderson (Eqn. S2, Figure S6) was applied to evaluate the titrated Mpro concentration ([E]0=60 nM) and the potency of GC376 inhibitor [Ki (app) =15 nM, IC50 =45 nM].[31] This half maximal inhibitory concentration (IC50) is lower than the majority of reported values[3, 32] most likely due to the different assay conditions, such as incubation time and buffer pH.[5, 29] Overall, this inhibitor assay demonstrated that our sensing system can be employed to rapidly screen for anti-Mpro therapeutic agents.
We further cross-tested several mammalian proteins against our sensing system, including bovine serum albumin (BSA), hemoglobin, α-amylase (i.e., digests α−1,4-glucosidic bond in starch), thrombin (i.e., cleaves Arg-Gly bond in fibrinogen), and trypsin (i.e., cleaves at C-terminus of Arg or Lys). As shown in Figure 4g, no particle aggregation was detected in the presence of BSA and hemoglobin, indicating good compatibility of the sensor with common proteins. Similarly, no optical signal was addressed by addition of other enzymes such as α-amylase or thrombin. Nonetheless, arginine-specific proteases such as trypsin caused false positive results. ESI-MS data confirmed that ZY7 substrate was recognized by trypsin at the C-terminal of arginine at P4’ site, exposing a short CGRGC fragment that causes the AuNP aggregation (Figure S2i). Indeed, this short fragment was also found when the assay was performed in saliva (Figure S2h) leading to background signal (the pooled saliva could contain Arg-gingipains implicated in periodontal disease).[33] As a viable strategy to suppress such a side effect and use our sensor in other clinical samples containing trypsin-like enzymes, we mutated the aggregating amino acids with their D-enantiomers to create a new DS12 peptide; see Table 1. The results assaying from the DS12 substrate showed improved proteolytic resistance to trypsin (Figure 4h). Remarkably, no background activation was observed for the sensor based on DS12 in saliva media (see Figure S5j). Despite these expectations, the LoD for Mpro using DS12 substrate showed an approximately 3-fold reduction comparing to that of ZY7 substrate (Table 1). The loss in sensitivity is in alignment with previous reports of a 4-fold reduction in Mpro efficiency on alteration of the P4’ arginine.[19]
The relevance of peptide/AuNP-based protease detection resides in its simplicity where no complex bioconjugation techniques or readout instrumentation are required. This allows for the large-scale implementation of qualitive diagnostics for protease markers. Tests like these can be easily affixed to routinely used face coverings turning them into easy-to-use diagnostic kits. We have previously reported the design and integration of a sticker-based testing platform that can be attached to face coverings for bio-detection.[34] Herein, we adapted the test onto a flow-cell inspired Mpro sensing strip (Figure 5a,b). The strip consisted of two pad lanes: (i) A test lane decorated with 50 μM of ZY7 peptide, and (ii) a positive control lane loaded with 50 μM of peptide fragment (YF15). This peptide fragment is the most potent aggregating domain from all the peptides listed in Table 1. 60 μL of BSPP-AuNPs was housed in the blister pack and used for readout or color agent (Figure 5b). In principle, one pink and one purple lane would signify a COVID-negative test, while two purple lanes indicate a positive test for the presence of Mpro.
Figure 5.
Portable device for protease detection. (a) Structural illustration of the flow-cell device on a face covering for colorimetric sensing. The vents allow the transmitted respiratory droplets to be concentrated on the absorbent pad. The reagent yielding color change is packed in the reservoir. More details about the flow strip can be found in Table S1. (b) A white-light image of the assembled sensing trip, equipped with a BSPP-AuNP dispersion in the blister pack, ZY7 peptide on the test lane (left, dash box), and YF15 peptide on the positive-control lane (right, dash box). Scale bar of 1 cm is shown. (c) The sensing strip’s LoD for Mpro is estimated to be 30–40 nM. Monodispersed BSPP-AuNPs appear as pink-red, while the clustered gold aggregates by peptide fragments appear as violet-blue. (d) Strip testing of Mpro marker on COVID-negative participants (NP#, n=10). All test lanes show pink-red in the absence of SARS-CoV-2 protease. SEM images of non-aggregating AuNPs on the red lane (e) and clustered AuNPs (f) on the purple lane. (g) Sensor testing on aqueous EBC matrices collected from COVID-negative subjects (n=10). No absorbance ratio change/false positive was noticed.
We determined the LoD for Mpro on the sensing strip. As illustrated in Figure 5c, various concentrations of Mpro in EBC (0−100 nM, with respect to 30 μL volume) were tested at 10 min. The test lanes incubated with Mpro more than 40 nM started to appear as purple color, indicating positive test results. We thus conclude that the sensor LoD for Mpro is similar in both solution and semi-solid (i.e., polyester pad) phase, 30−40 nM.
In a real scenario for a COVID-infected patient, the strip/pad affixed on a face covering would collect the respiratory droplets that harbor Mpro biomarker (for 8 h or a normal workday). Meanwhile, on the absorbent pad the concentrated Mpro proteases start to cleave the pre-loaded ZY7 peptide. At the end of the day, the BSPP-AuNPs in the blister pack are released and moved to the absorbent pad via capillary action to interact with the peptides for 10 min readout. The present sensing device has been demonstrated to be suitable support for detecting α-amylase in our previous work.[34] As a proof-of-concept experiment, we show here ten EBC samples collected from the COVID-negative volunteers (i.e., verified by PCR test) and showed no false positives (Figure 5d). This result was also collaborated with solution-phase assays where no alterations in ratiometric signals were detected (Figure 5g). Ongoing work focuses on evaluating these with EBC and saliva from COVID-positive subjects.
Conclusion
A universal formula of zwitterionic peptide, (Asp)n−(AA)x−(Arg)m, is reported to modulate the dispersity of AuNPs and applied for a colorimetrical detection of Mpro, a protease implicated in SARS-CoV-2 viral replication. We built a label-free peptide (ZY7) that carries net neutral charge causing no aggregation of negatively charged BSPP-AuNPs. While enzymatic cleave of ZY7 by Mpro releases a positively charged fragment that triggers the collapse of gold suspension via electrostatic interactions. By quantifying the color change with measurable absorbance ratio, we have determined that our sensor performs best in EBC with an LoD of 33.4 nM using the ZY7 peptide. This rapid and reproducible color change provides a simple platform for inhibitor-screening targeting at Mpro. We assayed ten COVID-negative human subjects and found no false-positive result. Overall, the present AuNP-based colorimetric assay does not require tedious bioconjugations or specific instrumentations, and hence can be easily integrated into qualitative diagnostic kits for daily use.
Supplementary Material
Acknowledgements
The authors thank internal funding from the UC Office of the President (R00RG2515) and the National Institutes of Health (R01 DE031114; R21 AG065776; R21 AI157957) for financial support. This work was supported in part by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1650112. C.M. acknowledges support from the Achievement Reward for College Scientists (ARCS) Foundation. The electron microscopy work was performed in part at the San Diego Nanotechnology Infrastructure (SDNI) of University of California San Diego, a member of the National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant ECCS-1542148). We thank Dr. Ke Li and Lei Fu for the fruitful discussions on the molecular dynamic simulation of the peptide.
Footnotes
Supporting information for this article is given via a link at the end of the document.
References
- [1].de Graaf M, van Beek J, Koopmans MP, Nat. Rev. Microbiol. 2016, 14, 421. [DOI] [PubMed] [Google Scholar]
- [2].V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V, Nat. Rev. Microbiol. 2021, 19, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, Zhang B, Li X, Zhang L, Yang H, Nature 2020, 582, 289. [DOI] [PubMed] [Google Scholar]
- [4].Steuten K, Kim H, Widen JC, Babin BM, Onguka O, Lovell S, Bolgi O, Cerikan B, Neufeldt CJ, Bogyo M, ACS Infect. Dis. 2021, 7, 1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Medintz IL, Clapp AR, Brunel FM, Tiefenbrunn T, Tetsuo Uyeda H, Chang EL, Deschamps JR, Dawson PE, Mattoussi H, Nat. Mater. 2006, 5, 581. [DOI] [PubMed] [Google Scholar]
- [6].Sanman LE, Bogyo M, Annu. Rev. Biochem. 2014, 83, 249; I. L. H. Ong, K.-L. Yang, Analyst 2017, 142, 1867; W. Rut, K. Groborz, L. Zhang, X. Sun, M. Zmudzinski, B. Pawlik, X. Wang, D. Jochmans, J. Neyts, W. Młynarski, R. Hilgenfeld, M. Drag, Nat. Chem. Biol. 2021, 17, 222. [DOI] [PubMed] [Google Scholar]
- [7].Zhou W, Gao X, Liu D, Chen X, Chem. Rev. 2015, 115, 10575; A. Heuer-Jungemann, N. Feliu, I. Bakaimi, M. Hamaly, A. Alkilany, I. Chakraborty, A. Masood, M. F. Casula, A. Kostopoulou, E. Oh, K. Susumu, M. H. Stewart, I. L. Medintz, E. Stratakis, W. J. Parak, A. G. Kanaras, Chem. Rev. 2019, 119, 4819; Y. Mantri, J. V. Jokerst, ACS Nano 2020, 14, 9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ, Nature 1996, 382, 607; C. J. Murphy, T. K. Sau, A. M. Gole, C. J. Orendorff, J. Gao, L. Gou, S. E. Hunyadi, T. Li, J. Phys. Chem. B 2005, 109, 13857; P. K. Jain, X. Huang, I. H. El-Sayed, M. A. El-Sayed, Acc. Chem. Res. 2008, 41, 1578; M. Retout, H. Valkenier, E. Triffaux, T. Doneux, K. Bartik, G. Bruylants, ACS Sens. 2016, 1, 929; M. Grzelczak, L. M. Liz-Marzán, R. Klajn, Chem. Soc. Rev. 2019, 48, 1342; J. Li, K. Pu, Chem. Soc. Rev. 2019, 48, 38. [DOI] [PubMed] [Google Scholar]
- [9].Chang C-C, Chen C-P, Wu T-H, Yang C-H, Lin C-W, Chen C-Y, Nanomaterials 2019, 9, 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Guarise C, Pasquato L, De Filippis V, Scrimin P, Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Häkkinen H, Nat. Chem 2012, 4, 443; J. Zhou, M. N. Creyer, A. Chen, W. Yim, R. P. M. Lafleur, T. He, Z. Lin, M. Xu, P. Abbasi, J. Wu, T. A. Pascal, F. Caruso, J. V. Jokerst, J. Am. Chem. Soc. 2021, 143, 12138. [DOI] [PubMed] [Google Scholar]
- [12].Jin Z, Sugiyama Y, Zhang C, Palui G, Xin Y, Du L, Wang S, Dridi N, Mattoussi H, Chem. Mater. 2020, 32, 7469. [Google Scholar]
- [13].Chen J, Ma Y, Du W, Dai T, Wang Y, Jiang W, Wan Y, Wang Y, Liang G, Wang G, Adv. Funct. Mater 2020, 30, 2001566; M. N. Creyer, Z. Jin, C. Moore, W. Yim, J. Zhou, J. V. Jokerst, ACS Appl. Mater. Interfaces 2021, 13, 45236. [Google Scholar]
- [14].Li K, Galle Kankanamge SR, Weldeghiorghis TK, Jorn R, Kuroda DG, Kumar R, J. Phys. Chem. C 2018, 122, 4747; S. Wang, L. Du, Z. Jin, Y. Xin, H. Mattoussi, J. Am. Chem. Soc. 2020, 142, 12669; M. Xu, W. Yim, J. Zhou, J. Zhou, Z. Jin, C. Moore, R. Borum, A. Jorns, J. V. Jokerst, IEEE Nanotechnol. Mag. 2021, 15, 8. [Google Scholar]
- [15].Bian T, Gardin A, Gemen J, Houben L, Perego C, Lee B, Elad N, Chu Z, Pavan GM, Klajn R, Nat. Chem. 2021, 13, 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim Y, Johnson RC, Hupp JT, Nano Lett 2001, 1, 165; G. Sener, L. Uzun, A. Denizli, ACS Appl. Mater. Interfaces 2014, 6, 18395. [Google Scholar]
- [17].Liu X, Zhang Q, Knoll W, Liedberg B, Wang Y, Adv. Mater. 2020, 32, 2000866. [DOI] [PubMed] [Google Scholar]
- [18].Wang Y, Fedin I, Zhang H, Talapin DV, Science 2017, 357, 385; Z. Jin, L. Du, C. Zhang, Y. Sugiyama, W. Wang, G. Palui, S. Wang, H. Mattoussi, Bioconjugate Chem. 2019, 30, 871. [DOI] [PubMed] [Google Scholar]
- [19].Shan YF, Xu GJ, Acta. Biochim. Biophys. Sin. 2005, 37, 807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bartczak D, Kanaras AG, Langmuir 2011, 27, 10119; D. Wu, J. Zhou, M. N. Creyer, W. Yim, Z. Chen, P. B. Messersmith, J. V. Jokerst, Chem. Soc. Rev. 2021, 50, 4432. [DOI] [PubMed] [Google Scholar]
- [21].Turkevich J, Stevenson PC, Hillier J, Discuss. Faraday Soc 1951, 11, 55; C. J. Loweth, W. B. Caldwell, X. Peng, A. P. Alivisatos, P. G. Schultz, Angew. Chem. Int. Ed. 1999, 38, 1808; X. Ji, X. Song, J. Li, Y. Bai, W. Yang, X. Peng, J. Am. Chem. Soc. 2007, 129, 13939. [Google Scholar]
- [22].Yon M, Pibourret C, Marty J-D, Ciuculescu-Pradines D, Nanoscale Adv. 2020, 2, 4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jokerst JV, Lobovkina T, Zare RN, Gambhir SS, Nanomedicine 2011, 6, 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jung J-M, Savin G, Pouzot M, Schmitt C, Mezzenga R, Biomacromolecules 2008, 9, 2477; J. Zhou, M. Xu, Z. Jin, R. M. Borum, N. Avakyan, Y. Cheng, W. Yim, T. He, J. Zhou, Z. Wu, Y. Mantri, J. V. Jokerst, Angew. Chem. Int. Ed. 2021, 60, 26357. [DOI] [PubMed] [Google Scholar]
- [25].Armbruster DA, Pry T, Clin. Biochem. Rev. 2008, 29 Suppl 1, S49. [PMC free article] [PubMed] [Google Scholar]
- [26].Ding X, Ge D, Yang K-L, Sens. Actuators B Chem. 2014, 201, 234. [Google Scholar]
- [27].Martínez-Fleta P, Alfranca A, González-Álvaro I, Casasnovas JM, Fernández-Soto D, Esteso G, Cáceres-Martell Y, Gardeta S, López-Sanz C, Valés-Gómez M, J. Immunol. Res. 2020, 205, 3130; J. Wenzel, J. Lampe, H. Müller-Fielitz, R. Schuster, M. Zille, K. Müller, M. Krohn, J. Körbelin, L. Zhang, M. Schwaninger, Nat. Neurosci. 2021, 24, 1522. [DOI] [PubMed] [Google Scholar]
- [28].Zhao W, Chiuman W, Lam JCF, Brook MA, Li Y, Chem. Commun 2007, 3729; Y. Cheng, R. M. Borum, A. E. Clark, Z. Jin, C. Moore, P. Fajtová, A. J. O’Donoghue, A. F. Carlin, J. V. Jokerst, Angew. Chem. Int. Ed. 2021, 10.1002/anie.202113617 [Google Scholar]
- [29].Xue W, Zhang G, Zhang D, Analyst 2011, 136, 3136. [DOI] [PubMed] [Google Scholar]
- [30].Hung H-C, Ke Y-Y, Huang SY, Huang P-N, Kung Y-A, Chang T-Y, Yen K-J, Peng T-T, Chang S-E, Hsu JT-A, Antimicrob. Agents Chemother. 2020, 64, e00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Henderson PJ, Biochem. J. 1972, 127, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ma C, Wang J, Proc. Natl. Acad. Sci. 2021, 118, e2024420118; Z. Li, Y. Lin, Y.-Y. Huang, R. Liu, C.-G. Zhan, X. Wang, H.-B. Luo, Proc. Natl. Acad. Sci. 2021, 118, e2024937118.33568498 [Google Scholar]
- [33].Kaman WE, Galassi F, de Soet JJ, Bizzarro S, Loos BG, Veerman EC, van Belkum A, Hays JP, Bikker FJ, J. Clin. Microbiol. 2012, 50, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jin Z, Jorns A, Yim W, Wing R, Mantri Y, Zhou J, Zhou J, Wu Z, Moore C, Penny WF, Jokerst JV, Anal. Chem. 2021, 93, 11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.