Abstract
The role of Artificial Intelligence and Machine Learning in cancer research offers several advantages, primarily scaling up the information processing and increasing the accuracy of the clinical decision-making. The key enabling tools currently in use in Precision, Digital and Translational Medicine, here named as ‘Intelligent Systems’ (IS), leverage unprecedented data volumes and aim to model their underlying heterogeneous influences and variables correlated with patients’ outcomes. As functionality and performance of IS are associated with complex diagnosis and therapy decisions, a rich spectrum of patterns and features detected in high-dimensional data may be critical for inference purposes. Many challenges are also present in such discovery task. First, the generation of interpretable model results from a mix of structured and unstructured input information. Second, the design, and implementation of automated clinical decision processes for drawing disease trajectories and patient profiles. Ultimately, the clinical impacts depend on the data effectively subjected to steps such as harmonisation, integration, validation, etc. The aim of this work is to discuss the transformative value of IS applied to multimodal data acquired through various interrelated cancer domains (high-throughput genomics, experimental biology, medical image processing, radiomics, patient electronic records, etc.).
Subject terms: Translational research, Scientific community
Background: the role of intelligent systems in cancer research
Artificial Intelligence (AI) refers to functions and processes by which machines learn from data how to intelligently use the available information and establish associations between variables. In oncology, complex datasets reflect the presence of many quantifiable dimensions needing Machine Learning (ML) solutions to recommend, decide and especially predict over time while new data are accrued. Although AI and ML are often interchangeably used terms, they are complementary and can be reconciled under the term ‘Intelligent Systems’ (IS) (‘learning health systems’ has also appeared with clinical data in precision medicine [1]). A corresponding glossary appears at the end (see Table 1). In general, cancer researchers leverage IS for their ability to conduct inference by algorithmic rules which eventually lead to informed clinical decisions. These algorithms can analyse and integrate multiple data modalities facilitating the confluence of information from heterogeneous sources. Then, by combining computing power and mathematical thinking with domain knowledge and context-specific information, IS deals with scalability and generalisability. Therefore, IS solutions assist the cancer expert in key tasks such as identifying triggers for diagnosis, or intervention/treatment, and support the decisions taken under conditions of uncertainty. As this uncertainty can have many causes, and only some are controllable, efficiency is desired together with the applicability of principles such as stability and predictability, both necessary to ensure reproducibility metrics and establish high-quality predictions [2].
Table 1.
Glossary.
| AI = Artificial Intelligence |
| Digital tools able to perform tasks commonly associated with intelligent beings. |
| ML = Machine Learning |
| Methodological solutions that work algorithmically to find consistent patterns and reliable features in large amounts of data aimed at deciding and formulating predictions based on new data. |
| IS = Intelligent Systems |
| Software tools that support expert decision-makers in acquiring information from data, learning patterns from the processing of information and using the knowledge to facilitate the decisions. |
| EHR = Electronic Health Records |
| Digital version of the patient’s paper chart and medical history. |
| EM = Ensemble Modelling |
| A computational process by which multiple diverse base models are used to predict an outcome. |
| DL = Deep Learning |
| Deep learning is a class of algorithms employing multiple layers to extract informative features from the raw input data and use them to learn and generalise. |
| TL = Transfer Learning |
| Family of techniques enabling researchers to infer about a problem by using the knowledge gained from a model applied to a similar problem. |
| RL = Reinforcement Learning |
| Algorithmic process of learning by doing to make a sequence of decisions based on rewarding desired behaviours and/or penalising undesired ones. |
| IoMT = Internet of Medical Things |
| The collection of medical devices and applications that connect to healthcare IT systems through online computer networks. |
| VoI = Value of Information |
| An analytic method that quantifies the potential benefits of additional information before making decisions under uncertain conditions. |
To better delineate what cancer patient outcomes can be inferred from all types of acquired information, the IS role is to design and implement strategies that assemble and engineer a wealth of data features. These features are used to define signatures and patterns carrying information at a molecular level and combining evidence obtained at cell biology, pathology and radiology levels. Then, to exploit the interdependence between these domains, the definition of suitable standards is required [3]. This is a major harmonisation effort, with three major impacts involved: (i) the identification of diagnostic markers, (ii) The monitoring of patients’ response to treatment and (iii) the delivery of prognostication paths. In such regards, IS contribute to lower both infrastructural barriers (data management related costs, computational requirements for fast data processing, expertise needed to ensure quality processes, etc.) and ensure further developments toward:
-
A.
Easier access to Big Data database resources and repositories (genes, ncRNAs, proteins, peptides, small molecules, monoclonal antibodies, bio-images, registries, etc.);
-
B.
Fast deployment of scientific findings related to molecular profiles (including RNA), genetic information, electronic health records (EHR), medical imaging archives, etc.
-
C.
Digitally annotated biobanks with biological samples associated to clinical information and analyses performed on experimental data;
-
D.
Drug banks with sophisticated semantic frameworks and efficient algorithms to facilitate repurposing and repositioning.
These resources suggest a rich research agenda. A few major directions are: (i) repurposing existing drugs for treatment decisions supported by multi-sourced protein–protein interaction networks [4], (ii) developing ML algorithms predictive of anticancer drug efficacy [5], (iii) advancing semantics to deploy public cancer genomic datasets as linked data and offer automated analytics and visualisation [6], in addition to other emerging applications [7]. Accessibility, especially concerning cloud-sourced data, and heterogeneity of independently created ontologies and vocabularies represent real bottlenecks. There is still limited and inconsistent data reuse, which calls for creating shared models and performance measures for efficient data warehousing approaches. It is worth mentioning that focusing mainly on the algorithmic performance of methods designed for diagnostic or therapeutic scopes may be a limiting factor. The ability to infer patient’s outcomes depends on multidimensional decisions embedded within the IS-supported clinical workflow, for instance calibrating the measurement of under/overtreatment effects or identifying early detection or relapse to guide specialistic care (management of treatment intensity effects, reassessment of timing in monitoring toxicity, etc.) during follow-up or surveillance.
At the methodological level, a first critical issue is the training of algorithms over multiple datasets to build first descriptive and then predictive models aimed at generalisation. For example, Deep Learning (DL) may be blind to the underlying cancer biology but still offer (a) the power of digesting huge information from patient sample tissues via large volumes of digitised biopsy slides across many cancer types, (b) the alignment of such detailed maps with therapy factors, (c) the combination of tissue characteristics with treatment response and the correlation with identified patterns to tell what cancer drugs work best at the individual level or find clues in tissue microenvironment for establishing vulnerabilities to immunotherapy drugs. In general, cancer dynamics have inherent complexity that cannot be just algorithmically trained. The mechanisms governing such dynamics involve dysregulated cell signalling networks with crosstalk between many variables that operate at different time scales, which complicates data collection. Some variables may induce response due to reprogramming, for instance at the metabolic level. Mutations affecting treatment may require updates or corrections of earlier predictions, especially for personalising the cure or improve/accelerate trial practices with priority assigned to establishing synthetic control groups or modulating drug doses for prevention of resistance.
Precision medicine is gaining clear benefits from IS [8]. Immune profiling (IP) offers a good example. A recent study [9] on the PD-L1 biomarker to predict a patient’s response to treatment was performed via tumour-immune cell interactions analysed spatially and computationally by AI-assisted multiplexing approaches. About the same marker, another study [10] identified factors explaining the success of target immunotherapy treatment in urothelial cancer by using information from tumour and immune cells together with patient clinical and outcome data [11]. An ML algorithm performed over 36 features from multimodal data to find the 20 most associated with a specific response to PD-L1 inhibitor and deliver predictive marks of potential tumour adverse immune cells in patients after treatment. The key factor was the integration of data types: without just one of these types, the predictive performance vanished.
Significance for oncology
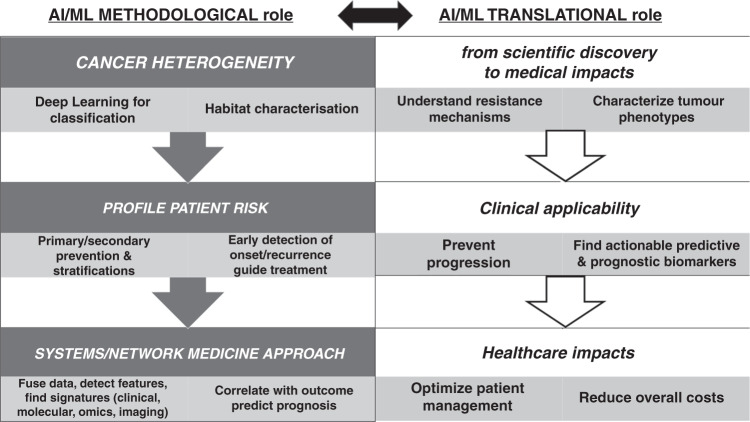
The recent advances in radiology had strong impacts on early diagnosis and timely treatment, reducing the chances of misdiagnosis and therapy-related complications. Benefits involve less overtreatment in the short term and less mortality in the long term. IS contributed in various ways, measurable at prevention (primary and secondary), diagnostics, therapy and prognostication (see Fig. 1) levels.
Fig. 1. Impacts Overview.
AI and ML methodological and translational roles in Precision Oncology.
Prevention
The stratification of patients according to their risk profile is often associated with the design of screening methods able to detect cancer at the early stage [12]. One reason for using IS approaches is to ensure the scalability of results to large-scale population screening. Most improvements in screening outcomes have been obtained from medical imaging, and this presents cost barriers. Although advanced learning with large complex datasets has become possible with various strategies and tools, often the limitation is in terms of the clinical applicability of the proposed solutions. These tend to leverage only some of the patients’ characteristics and thus justify even more the need to profile large patient cohorts across many stratifying factors. Typically, an increased detection power of disease marks captured from multimodal data at an early stage leads to a much more accurate patients’ classification. In the end, a full assessment of the actual capability of IS algorithms requires a demonstration of comparable performance with clinicians’ interpretations, although an excess of focus on this aspect may diminish some potential gains. The reference goes to accuracy and efficiency achievable in triaging practices, for instance, for which more studies are needed [13].
Diagnosis
Medical imaging has gradually become the reference field for IS contributions [14–20]. Here, the spectrum of investigations in cancer covers primary versus metastatic lesion detection, image segmentation and classification, discrimination between benign and malignant nodules, histological subtyping, and many others. DL has established new standards through the learning of features directly computed from data. Complex solutions, typically Convolutional Neural Networks (CNN) or Recurrent Neural Networks (RNN), can execute training over large sets of labelled Regions of Interest (ROI) from where to learn the parameters useful to the generalisation of results. CNN applications and demonstrations have for instance covered skin cancer from a dataset of 12,940 clinical images [21] and metastatic breast cancer (BC) [22], and in BC it was also shown the feasibility of fusing CNN with other feature extractors [23].
DL is very effective in identifying variation by automatically inspecting complex patterns [24]. An application is provided by ExPecto [25], an experimentally validated tool that links genome-wide sequence data genetic mutations with disease prediction. In general, the ability to decrease variability depends on the heterogeneity of data and requires algorithmic training followed by testing and validation. This normally happens via accurate separation of tasks into independent datasets to optimise parameter tuning and performance assessment. With regards to imaging variation, the field of radiomics [26–30] currently inspires most of the algorithmic thinking toward predictive modelling for cancer detection, characterisation, and monitoring. This is because the imaging features extracted by ML algorithms are subject to sequential analysis steps aimed at prioritising their discrimination power and ensuring control of data redundancies, selection operated according to relevance and model tractability.
Therapy
Optimal therapeutic strategies are considered those able to overcome identified gaps and develop successful, sustainable and scalable interventions that improve the outcomes at the individual as well as population levels. IS present a wealth of methods that accurately measure response prediction and allow to develop and refine treatment options for cancer patients [31]. Correspondingly, they have revealed useful to advance drug discovery. IS may especially improve predictability, accuracy (with effects on effectiveness and safety), and speed/timeline of drug discovery [32]. It is possible to analyse large-scale AI-empowered and ML-driven drug discovery platforms centred on large warehoused data and predictive algorithms tailored to specific proteins with their structures as a target. An example is the scalable cloud computing ML-driven SpliceCore platform (https://www.envisagenics.com/) with a > 5 ml RNA splicing errors database supporting identification, testing and validation of hypothetical drug targets that specifically target RNA splice variants causing cancer (such as triple-negative breast cancer and other genetic diseases). To stress the importance of therapeutic targeting of splicing in cancer [33], another tool worth mentioning is SpliceAI (https://github.com/Illumina/SpliceAI), a 32-layer deep CNN operating on pre-mRNA sequences [34]. Then, the cloud-based platform Ligand Express (https://www.cyclicarx.com/) that screens small-molecule drugs against proteomes and determines polypharmacological profiles. Here, an AI-assisted identification of protein targets is operated by a structure-based drug-centric technology (MatchMaker, a DL engine) determining drug’s effects and visualising predicted drug–protein interactomes with coverage of both on- and off-target interactions (expected and unanticipated, respectively).
Computational drug development predictions face necessarily the validation stage to minimise the uncertainty about the efficacy with which any single drug or combination of drugs may eventually lead to better outcomes, something risky and demanding for new compounds or large-scale clinical trials. IS augment the possibility of diversifying drug pipelines [35, 36]. In addition, used drugs are increasingly investigated for repositioning/repurposing scopes, implying high computational demand for evaluating the possible application in new disease domains of libraries of approved drugs (about 1500 FDA-approved drugs can be potentially matched with about 10000 potential targets, numbers destined to change quickly anyhow). Some interesting in silico approaches refer to pharmacophore modelling and docking-based virtual screening for repurposing small-molecule drugs against multidrug-resistant cancers [37]. The role of resistance in cancer has stimulated research from prevention to control directions and effectively accelerated experimental and validation phases. The results have led to developing more targeted therapies to (i) identify neoantigens and immunotherapies [38, 39], (ii) understand toxicity from chemotherapy and radiotherapy and more automated planning of treatment programs in radiology [40–44], (iii) prioritise surgery [45].
Prognostication
Survival prediction determines how patient profiles match the available treatment options. A key characteristic in cancer is the inherent risk of metastasis that generates recurrence. Metastatic tumours, especially recurring ones with acquired resistance to therapy, influence cancer management because the probabilistic quantification of this type of risk is the most uncertain cancer management phase [46]. Usually, biomarkers determine this phase by looking at the disease at a molecular level. IS have increasingly leveraged the algorithmic processing of patient information inducing response to therapy under many influencing factors, from genetics and immune status to environment and lifestyle. In association with factors objectively measurable (histopathological characteristics or effects of adjuvant therapies) and crucial for determining disease markers, instruments such as risk scores and nomograms [47] may guide the therapy phases by quantifying chances of recurrence and prognostic assessment via various genetic and/or molecular markers and signatures. Recourse to DL (and other unsupervised algorithms) may offer black-box solutions with little insight into the processes determining their final output. Graph-based integrations may help integrate multi-omics data and improve the prediction accuracy of clinical outcomes [48]. In addition, beyond the problems of inaccurate, incomplete or biased data towards specific populations, a further complication is due to algorithms that may misrepresent the context without the necessary specificity. More importantly, the nonlinear complexity of biological processes indicates the necessity of matching quantitative modelling with the context-related complexities [49].
Leveraging data features in modelling cancer
Among the tools traditionally used in quantitative cancer research, it is common to find those modelling mechanistically and predicting tumour progression/growth. Growth, and resilience to survive, suggest that tumours are complex adaptive systems facing heterogeneous environments and dealing with variables measurable at various length and time scales (oncogenes, tumour suppressor genes, cell–cell communication, etc.). Models of the dynamics of growth in tumours have mostly followed principles of statistical physics and dynamical systems accounting for both inter-tumour processes and host–tumour interactions. This way, inference on cancer heterogeneity (and evolution) has been mainly explorative of the likelihood that different advantageous and deleterious mutations survive in the tumour cell population. Outstanding open questions remain and refer to multiscale nonlinear model parameterisations, the role of resistance, subclone detection among various other factors that might limit predictability [50]. At a methodological level, it is interesting to address the IS developments that occurred in two major areas of clinical impacts in cancer: (1) data science analytics to manage issues such as aggregation, augmentation, imbalance issues, etc. (2) algorithmic treatment of the complexity that affects the dimensions of data, with reinforcement learning deserving special attention (see the next section) when a dynamic rather than a static treatment of the data is necessary.
Ensemble models
IS have emphasised the potential significance of a more integrative use of multitype omics evidence. For instance, more accuracy is gained in assessing the cancer metastatic potential, in prioritising the patient-specific variables that once quantified may help the clinicians individualise the treatment options, in increasing the depth of risk profiles and the precision of outcome estimates, in planning cost-effective follow-up actions. Data integration is a critical structural component that calls for adaptive strategies at the algorithm level. Ensemble modelling (EM) is for example an approach that engages multiple models to predict outcomes by the means of different algorithms and/or different training sets. Then, EM aggregates the results in a prediction with reduced generalisation error.
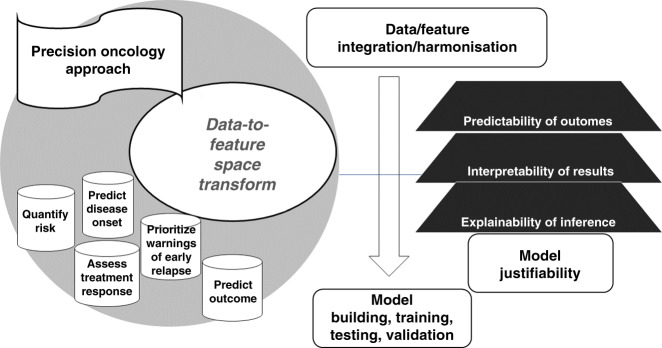
Substantial criticism points to the ability of learning strategies to explain the results they achieve, beyond converging to optimality. As a rule, highly complex models can lead to low interpretability and poor explanation of what mechanisms underlie predictions. In oncology, data modalities may reveal the insufficient size, and model results may present limited interpretability [51]. This means that data integration must be contextualised at the level of input raw data, features (extraction, selection and combination) and finally model outcome predictions. These levels are more or less convoluted, therefore characterising the strategies for data-to-feature-to-prediction transforms that enable IS performance assessment through measures and scores associated with the risk of disease onset, relapse, treatment response (Fig. 2).
Fig. 2. Methodological Approach.
Role of features in IS modelling.
Transfer learning
The bottleneck of data significance is often insufficiency of information. This problem calls for alternative learning approaches aimed to augment the space of features using, for instance, surrogate datasets. Such practice is foundational in Transfer Learning (TL) approaches [52]. TL algorithms reuse a model pre-trained over a new problem to improve its predictive performance by transferring knowledge about a related problem learned from auxiliary data. Examples are provided by drug sensitivity prediction [53], cancer classification [54], automatic lung CT image segmentation [55], etc. Domain-specific models can use importance/attribution methods to estimate the feature’s contribution to the predictions (using prior or context knowledge).
Differentiating features in cancer domains
The processing of NGS datasets has become increasingly time effective and routinely assisted by ML tools for executing accurate read alignment and robust variant calling tasks discriminating coding and non-coding bioentities (mRNA and ncRNA) by sophisticated multiplexed omics strategies. In terms of characterisation, a typical investigation target is subtyping, as this might affect treatment. A recent study [56] discovered patterns characterised by feature similarity and used to computationally classify five new subtypes within primary luminal A-type breast tumours. The impacts involve further development of targeted drugs and potential improvement of therapies/immunotherapies at an individualised patient level. Another pan-cancer study [57] used DL with multimodal data to find molecular subtypes, together with genetic mutations, gene expression signatures, and pathology biomarkers, by exploiting histology from tissue slides of >5000 patients across multiple solid tumours. In addition, a key aspect is timely monitoring to track progression or regression or target the emergence of resistance. This goes beyond traditional metrics (RECIST) to assign a central role to the interplay between different tumour cells and the immune response in the tumour microenvironment (TME). Emerging DL applications in digital pathology treat this domain to better stratify patients and identify relevant biomarkers predictive of clinical outcomes and treatment response [58].
Cancer immunograms [59] describing the interactions between cancer and the immune system, and in general, the integrated picture of immune oncological profiling [60], are present among the IS research developments. Gene expression signatures are ideally either prognostic (telling about patient prognosis with therapy or not) or predictive (telling about treatment benefit, typically within randomised controlled clinical trials). However, the fact of being non-specifically immune-related can bring to the loss of important information associated with patient outcomes. Other data augmentation strategies can be supported by supplementary patient data points provided from new data sources (patient-generated outcomes, sensors, wearables, implantable electronic devices, etc.). Here, the risk is learning features from data with some types of imbalance (skewed distributions force classifiers to be trained with bias). The problem is what model should be used, considering that both under- and over-sampling approaches can, respectively, downsize or upsize the classes depending on their large or small size. Clustering methods can mitigate under-sampling effects in data while common solutions to over-sampling are considered SMOTE [61] and ADASYN [62]. Selection and collider biases are also problems. Associations between risk factors and outcomes can be induced at the individual versus sample level, thus affecting the likelihood of an individual being sampled and distorting the associations between the variables in the sample. Of relevance here are a double negative effect, lack of generalisation (beyond the sample), and inaccurate inference (within the sample), especially with collider bias (various tools are nevertheless available [63]).
Predictive analytics
Predictive analytics holds great promises for clinical decisions and genomic-driven risk stratifications but crucially needs prospective validation and bias control to become an automatically embedded feature of any IS learning approach. Data heterogeneity, although highly informative, plays against a comprehensive coverage of endpoints. Support may come from EHR capturing multifaceted data, identifying patients that meet inclusion-exclusion criteria and delineating reachable endpoints. Within omics, data integration strategies try to process data both horizontally (same omics across different studies, thus automatically reducing the patient stratification depth), and vertically (different omics applied to the same samples to directly deal with multidimensionality, genotype-phenotype correlation, and biomarker discovery). However, the applicability of these strategies is ultimately cancer type-dependent and particularly challenging when inference is conducted at a pan-cancer level. Here, some form of control of complexity should be reflected onto models extracting features to verify significant associations across cancers and/or estimating the effects of perturbed conditions. For example, to uncover latent molecular signatures, integrative omics approaches should aim at profiling specific omics-related data with harmonised criteria. As cancer heterogeneity remains in large part undeciphered, it is worth considering approaches able to balance the role of influential variables that are either partially observable/measurable or latent. These variables potentially trigger early detection in cancer and are usable in latent variable models to provide accurate quantifications of risk of relapse and warning signals of emerging resistance [64].
Reinforcement learning
Harnessing cancer data for clinical translation has guided IS toward emerging directions [65]. IS development represents an inherently elastic process that adapts rapidly to contextualised data structures and targets the salient characteristics helping the models to be efficient and accurate. A trade-off between structural data and algorithmic complexity guides model selection depending on the relevance of the needed information and experiments. In such regards, the value of information (VoI) inherent to the context is an important factor defined in principle by a ratio of benefits over costs, i.e., survival or quality of life gains versus costs of discovering biomarkers, administering therapy, avoid side effects and similar criteria. VoI tends to correlate value also with data volumes (genomics, proteomics, histopathology, radiology, etc.) due to the analyses needed by them (say, tissue diagnostic analysis [66], informed decision-making, optimal trial design, etc.) [67, 68].
Among their scopes, IS allow to better prioritise modifiable factors and accelerate the identification of patients at high risk of adverse events. This is the area in which reinforcement learning (RL) approaches may support the development of personalised treatments. RL aims at optimising the prediction performance by processing a series of trial and error moves while considering the feedback received from the environment (or context) and the rewards assigned to the trials. As the goal is to learn adaptively (policy) how to maximise a reward function (health), the target with treatment becomes a dynamic process (drug administration, how it evolves and how personalised is the policy). Examples come from adaptive radiotherapy in synergy with immune modulators [69], with AI assisting with the adaptive planning process, or dynamic treatment regimes [70–72]. In both cases, a nonlinear feedback control problem is manifested and the support to decisions comes from selected actions for inferring the outcomes given the uncertainty of their interactions with the environment and possible delayed effects (feedback). RL has been evaluated in cancer screening [73] and to enable clinical decision systems at the point of care aligned with evidence-based data. In general, RL responds to the need of achieving superior reproducibility, quality control, interpretability, and generalisability standards (toward different endpoints within the same patient cohort, different patient cohorts within same cancer, different cancers). Hence, comparing datasets through benchmarks for assessing robust validation, even with deficient data, is a crucial issue [74].
Emerging applications
Integrative radiomics
Radiomics represents a natural complement to radiology in the assessment of early prediction of response to treatment. By characterising non-invasively intratumor heterogeneity by a myriad of features, radiomics adds value integrable with clinical information when assessing treatment outcomes. A main goal of radiomics is correlating the radiomic phenotypes with the genomic profile. Radio-genomics [75, 76] goes in this direction to better predict genotypic traits based on raw images and transformed features. Similar principles hold for radio-metabolics [77] and theranostic [78] and other hybrid approaches like pathoradiomics (a recent study [79] differentiates NSCLC subtypes and novel tumour-immune response pathomics-based DL classifications in TME [80]). There are challenges associated with the clinical translation of radiomic results (as per the recommendations related to applications that come from the National Clinical Trials Network [81]), although the potential discoveries have been only weakly validated so far and need to be consolidated. Other major problems are summarised in a few needed actions: (i) standardisation of the image acquisition parameters, (ii) harmonisation of results, (iii) share of pre-/post-processing steps in image analyses. In such regards, fused radiomics [82] (especially multimodality [83, 84]) and integrated radiomics (joint profiling with genomics, metabolomics, pathomics, holomics, etc. [85]) are the domains that promise to progress in the next future.
Of general interest is the variation problem [86]. Problems of a technical nature, together with the inherent aspect of patient variability, call for statistics and signal processing techniques that can deliver stable feature indexes and robust models. The features with explainable variability are considered the most informative for inference and predictive for generalisability. In such regards, habitat delineation [87–89] is a strong direction of study aimed to identify subregions with distinct characteristics, such as radio resistance. After the initial focus on image mining [90] and the significance established for delta radiomics [91–93], the delineation of habitats has gained popularity because of the specificity assigned to the tumour partitioning targeted to TME studies and impacting future immunotherapy. Standard validation strategies and multiple approaches are needed to prove performance quality and reproducibility aspects at the level of previously adopted standards, such as the radiomic quality score (checklist to ensure the quality) and the phantoms used to investigate reproducibility and stability of CT, PET or MRI radiomic features at single or multiple sites.
Internet of medical things
Internet of Medical Things (IoMT) [94, 95] is strongly connected with ML and in particular DL concerning fine-tuning segmentation and sub-type identification problems for which joint radio-pathomic signatures are sought [96]. Here, imaging findings have been useful to predict response to treatment via DL algorithms offering increased efficiency (speed and cost reduction) and accuracy (improved quality of images and their interpretation). What is still lacking is the generalisability of the approaches and the ability to translate the high-tech content results of these studies into clinically valid applications. Both discriminative and generative DL models are employed and network refinements are implemented by a mix of feature engineered methods (mathematically transformed to reduce error in modelling the target) and non-engineered methods, both types finding utilisation also in distributed radiomics (e.g., multicentre studies [97–99]).
Outlook
Building systems-level value of information
Methods’ specificity can concretise into data overfitting when the performance is measured for new data over which the results should be generalised. Being the use of DL pervasive, until this suffers from a certain lack of full interpretability due to the black-box learning paradigm, a good part of data-intensive cancer research can have only a reduced impact. The data integration strategies aimed to gain statistical power do not follow standard solutions yet. In principle, the VoI of integrated datasets may be summed linearly or combined in more sophisticated ways depending on the systems’ uncertainty level, thus driving decisional processes to take advantage of decision trees and similar model structures.
Discovery is usually leveraging hypothesis generation that iterates through an automated cycle (measuring, validating, and automating) ending with predictive modelling for decision support. The IS added value in this cycle is ultimately defined in terms of better outcomes obtained at a lower cost, which implies the fusion of streams of information over time (Fig. 3). In radiomics, for instance, focusing on signatures and profiles requires detection of ‘hard data’ vulnerabilities [100], which might be amplified when searching for robust gene expression programs through multiple normalisations [101], elucidation of phenotype transitions, insights into therapy resistance, characterisation of TME, etc. These factors motivate an increased use of systems and network approaches for future applications. On the other hand, ‘soft data’ such as EHR, personalised and dynamic treatment data, disease trajectories, risk profiles, and scores for patient stratification are products of dynamical processes centred on patients. As such, the data distributions are subject to drift over time, causing the ML model to lose predictive power.
Fig. 3. Integrated View.
The patient trajectory within the cancer data workflow.
The sketch in Fig. 3 represents the complexity underlying the process of assembling various data types for stratification purposes at both population and individualised scales. The data distributions depend on factors that are endogenous and exogenous to the patient. Therefore, full reliability of data through the process requires continuous testing. Explaining the possible ways uncertainty can affect models remains quite difficult as well as it is problematic the interpretability of results through the usual metrics. In perspective, support to clinical decision systems in oncology will require dynamic IS calibration strategies for uncertainty control.
Impacts in clinical practice
Two main factors limit the impact of big data and IS in clinical practice. First, in view of the multiple data dimensions associated with novel drug discovery strategies and therapies, changes may be expected in the clinical trials [102–104]. Second, an increased input heterogeneity tends to make inference more convoluted and requiring specialised methodologies assigning specific roles to various types of intervention solutions (coming from software engineering, analytics, decision processes, etc.) in support of clinical evidence [105, 106]. With few exceptions [107–109], most translational studies are retrospective and not prospective [110]. This limits the ability to measure the IS generalisation performance and calls for high-quality reporting to mitigate potential problems related to data and algorithmic biases, confounders, etc. [111]. In turn, this further stimulates the adoption of model confidence measures to facilitate cross-domain applicability and reproducibility of findings [112].
Important initiatives are TRIPOD-ML [112, 113], SPIRIT-AI and CONSORT-AI [114, 115]. Algorithm-related aspects like quality control, model recalibration and retraining performances should be checked under varying conditions but also account for features reassessment with evolving clinical and operational practices. FDA guidelines were recently introduced with a regulatory framework for software-guided medical devices that must include predetermined change control plans. The sphere of opacity or ambiguity in IS models is also involved with the DL processing of unstructured data that represents an additional complex step toward inference (like multiple hidden layers or special optimisation functions). Some promising models have mitigated the overfitting effects: for instance, Deep Forest (or gcForest), applied to lung cancer staging with a decision-fusion strategy conservative in the parameters used over multimodal genetic data [116].
The clinical feasibility of large data IS studies depends on the generalisability of results across different populations (due to age, gender, ethnicity factors), despite the effects of biases (from training over historical data with size gaps and disparities) and confounders on the model outcomes. Consequently, validation concerns (choice of the dataset, selected criteria for significance assigned to variables, model discrimination, etc.) emerge. The usefulness or expected performance of sound algorithms is not sufficient to justify adoption and integration into clinical workflows. Accepting less accuracy from a 'black-box' learning machine may lead to gains in terms of transparency and interpretability, but assuming this balance is achieved, the risk is to rely on oversimplified models guiding the complex decision. Ultimately, the key question remains whether model results are beneficial to patients. Therefore, any adopted metric should prioritise and quantitatively measure the net benefit of using a certain model to guide actions and decisions with an appropriate outcome-driven model design. The continuous expansion of phenotypic knowledge bases of reference clinical datasets across populations and open software analytics platforms allowing reproducibility of results support this direction offering opportunities for testing and validating models by cross-referencing the findings over independent datasets while fine-tuning the methods (i.e., by ranking performance factors and applying standards for algorithm fairness and best practice recommendations to train-retrain and calibrate). Such developments will be likely inducing a revision of the model prediction paradigm [117, 118] and exerting effects on effective translation to clinical practice together with further propelling prospective studies.
Acknowledgements
The author acknowledges NSF support from grant NSF 19-500. DMS 1918925/1922843 (years: 08/01/2019 – 08/01/2022).
Author contributions
All contributions were from the single author.
Funding
None.
Ethics approval and consent to participate
Not applicable.
Consent to publish
Not applicable.
Competing interests
The author declares no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McNutt TR, Benedict SH, Low DA, Moore K, Shpitser I, Jiang W, et al. Using big data analytics to advance precision radiation oncology. Int J Radiat Oncol Biol Phys. 2018;101:285–91. doi: 10.1016/j.ijrobp.2018.02.028. [DOI] [PubMed] [Google Scholar]
- 2.Yu B. Three principles of data science: predictability, computability, and stability. KDD ‘17: In: Proceedings of the 23rd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. ACM Digital Library; 2017. p. 5.
- 3.Hulsen T, Jamuar SS, Moody AR, Karnes JH, Varga O, Hedested S, et al. From big data to precision medicine. Front Med. 2019;6:34. doi: 10.3389/fmed.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabasi A-L, et al. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat Commun. 2018;9:2691. doi: 10.1038/s41467-018-05116-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kong J, Lee H, Kim D, Han SK, Ha D, Shin K, et al. Network-based machine learning in colorectal and bladder organoid models predicts anti-cancer drug efficacy in patients. Nat Commun. 2020;11:5485. doi: 10.1038/s41467-020-19313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamdar MR, Fernández JD, Polleres A, Tudorache T, Musen MA. Enabling Web-scale data integration in biomedicine through linked open data. NPJ Digit Med. 2019;2:90. doi: 10.1038/s41746-019-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan HCS, Shan H, Dahoun T, Vogel H, Yuan S. Advancing drug discovery via artificial intelligence. Trends Pharmacol Sci. 2019;40:592–604. doi: 10.1016/j.tips.2019.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Seyhan AA, Carini C. Are innovation and new technologies in precision medicine paving a new era in patients centric care? J Transl Med. 2019;17:114. doi: 10.1186/s12967-019-1864-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koelzer VH, Sirinukunwattana K, Rittscher J, Mertz KD. Precision immunoprofiling by image analysis and artificial intelligence. Virchows Arch. 2019;474:511–22. doi: 10.1007/s00428-018-2485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leiserson MDM, Syrgkanis V, Gilson A, Dudik M, Gillett S, Chayes J, et al. A multifactorial model of T cell expansion and durable clinical benefit in response to a PD-L1 inhibitor. PLoS ONE. 2018;13:e0208422. doi: 10.1371/journal.pone.0208422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snyder A, Nathanson T, Funt SA, Ahuja A, Buros Novik J, Hellmann MD, et al. Contribution of systemic and somatic factors to clinical response and resistance to PD-L1 blockade in urothelial cancer: An exploratory multi-omic analysis. PLoS Med. 2017;14:e1002309. doi: 10.1371/journal.pmed.1002309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parikh RB, Gdowski A, PAtt DA, Hertler A, Mermel C, Bekelman JE. Using big data and predictive analytics to determine patient risk in oncology. Am Soc Clin Oncol Educ Book. 2019;39:e53–e58. doi: 10.1200/EDBK_238891. [DOI] [PubMed] [Google Scholar]
- 13.Sechopoulos I, Mann RM. Stand-alone artificial intelligence—the future of breast cancer screening? Breast. 2020;49:254–60. doi: 10.1016/j.breast.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kann BH, Thompson R, Thomas CR, Dicker A, Aneja S. Artificial intelligence in oncology: current applications and future directions. Oncology. 2019;33:45–63. [PubMed] [Google Scholar]
- 15.Patel SK, George B, Rai V. Artificial Intelligence to decode cancer mechanism: beyond patient stratification for precision oncology. Front Phys. 2020;11:1177. [DOI] [PMC free article] [PubMed]
- 16.Rattan R, Kataria T, Banerjee S, Goyal S, Gupta D, Pandita A, et al. Artificial intelligence in oncology, its scope and future prospects with specific reference to radiation oncology. Br J Radiol. 2019;1:1. doi: 10.1259/bjro.20180031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weikert T, Cyriac J, Yang S, Nesic I, Parmar V, Stieltjes B. A practical guide to artificial intelligence based analysis in radiology. Invest Radiol. 2020;55:1–7. doi: 10.1097/RLI.0000000000000600. [DOI] [PubMed] [Google Scholar]
- 18.Nagy M, Radakovich N, Nazha A. Machine learning in oncology: what should clinicians know? JCO Clin Cancer Inform. 2020;4:799–810. doi: 10.1200/CCI.20.00049. [DOI] [PubMed] [Google Scholar]
- 19.Tseng H-H, Wei L, Luo Y, Ten Haken RK, El Naqa I. Machine learning and imaging informatics in oncology. Oncology. 2020;98:344–62. doi: 10.1159/000493575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaffray DA, Das S, Jacobs PM, Jeraj R, Lambin P. How advances in imaging will affect precision radiation oncology. Int J Radiat Oncol Biol Phys. 2018;101:292–8. doi: 10.1016/j.ijrobp.2018.01.047. [DOI] [PubMed] [Google Scholar]
- 21.Esteva A, Kuprel B, Novoa R, Ko J, Swetteret SM, Blau HM. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. J Am Med Assoc. 2017;318:2199–210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys. 2017;44:5162–71. doi: 10.1002/mp.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levine AB, Schlosser C, Grewal J, Coope R, Jones SJM, Yip S. Rise of the machines: advances in deep learning for cancer diagnosis. Trends Canc. 2019;5:157–69. doi: 10.1016/j.trecan.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Theesfeld CL, Yao K, Chen KM, Wong AK, Troyanskaya OG. Deep learning sequence-based ab initio prediction of variant effects on expression and disease risk. Nat Genet. 2918;50:1171–9. doi: 10.1038/s41588-018-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambin P, Leijenaar RTH, Deist TM, Perlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev. 2017;14:749–62. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 27.Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18:500–10. doi: 10.1038/s41568-018-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, et al. Artificial intelligence and machine learning in radiology: opportunities, challeneges, pitfalls, and criteria for success. J Am Coll Radiol. 2018;15:504–8. doi: 10.1016/j.jacr.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 29.Parekh VS, Jacobs MA. Deep learning and radiomics in precision medicine. Exp Rev Precis Med Drug Dev. 2019;4:59–72. doi: 10.1080/23808993.2019.1585805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Avanzo M, Wei L, Stancanello J, Vallières M, Rao A, Morin O, et al. Machine and deep learning methods for radiomics. Med Phys. 2020;47:e185–202. doi: 10.1002/mp.13678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakellaropoulos T, Vougas K, Narang S, Koinis F, Kotsinas A, et al. A deep learning framework for predicting response to therapy in cancer. Cell Rep. 2019;29:3367–73. doi: 10.1016/j.celrep.2019.11.017. [DOI] [PubMed] [Google Scholar]
- 32.Liang G, Fan W, Luo H, Zhu X. The emerging roles of artificial intelligence in cancer drug development and precision therapy. Biomed Pharmacother. 2020;128:110255. doi: 10.1016/j.biopha.2020.110255. [DOI] [PubMed] [Google Scholar]
- 33.Lee SC, Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22:976–86. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaganathan K, Kyriazopoulou Panagiotopoulou S, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–48.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Segler MHS, Preuss M, Waller MP. Planning chemical syntheses with deep neural networks and symbolic AI. Nature. 2018;555:604. doi: 10.1038/nature25978. [DOI] [PubMed] [Google Scholar]
- 36.Vamathevan J, Clark D, Czodrowski P, Dunham I, Ferran E, Lee G, et al. Applications of machine learning in drug discovery and development. Nat Rev Drug Discov. 2019;18:463–77. doi: 10.1038/s41573-019-0024-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinić J, Efferth T, García-Sosa AT, Grahovac J, Padrón JM, Pajeva I, et al. Repurposing old drugs to fight multidrug resistant cancers. Drug Resist Updat. 2020;52:100713. doi: 10.1016/j.drup.2020.100713. [DOI] [PubMed] [Google Scholar]
- 38.Bulik-Sullivan B, Busby J, Palmer CD, Davis MJ, Murphy T, Clark A, et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat Biotechnol. 2019;37:55–63. doi: 10.1038/nbt.4313. [DOI] [PubMed] [Google Scholar]
- 39.Nazha A, Sekeres MA, Bejar R, Rauh MJ, Othus M, Komrokji RS, et al. Genomic biomarkers to predict resistance to hypomethylating agents in patients with myelodysplastic syndromes using artificial intelligence. JCO Prec Oncol. 2019;3:1–11. doi: 10.1200/PO.19.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nasief H, Zheng C, Schott D, Hall W, Tsai S, Erickson B, et al. A machine learning based delta-radiomics process for early prediction of treatment response of pancreatic cancer. npj Precis Oncol. 2019;3:25. doi: 10.1038/s41698-019-0096-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lou B, Doken S, Zhuang T, Wingerter D, Gidwani M, Mistry N, et al. An image-based deep learning framework for individualizing radiotherapy dose. Lancet Digit Health. 2019;1:e136–147. doi: 10.1016/S2589-7500(19)30058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Z, Ren W, Li S, Liu J, Sun Y, Yan J, et al. Radiomic analysis in contrast-enhanced CT: predict treatment response to chemoradiotherapy in esophageal carcinoma. Oncotarget. 2017;8:104444–54. doi: 10.18632/oncotarget.22304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen D, Long T, Jia X, Lu W, Gu X, Iqbal Z, et al. A feasibility study for predicting optimal radiation therapy dose distributions of prostate cancer patients from patient anatomy using deep learning. Sci Rep. 2019;9:1076. doi: 10.1038/s41598-018-37741-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen D, Jia X, Sher D, Lin MH, Iqbal Z, Liu H, et al. D radiotherapy dose prediction on head and neck cancer patients with a hierarchically densely connected U-net deep learning architecture. Phys Med Biol. 2019;64:065020. doi: 10.1088/1361-6560/ab039b. [DOI] [PubMed] [Google Scholar]
- 45.Hollon TC, Pandian B, Adapa AR, Urias E, Save AV, Khalsa SSS, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020;26:52–58. doi: 10.1038/s41591-019-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halabi S, Li C, Luo S. Developing and validating risk assessment models of clinical outcomes in modern oncology. JCO Prec Oncol. 2019;3:PO.19.000068. doi: 10.1200/PO.19.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blyuss O, Zaikin A, Cherepanova V, Munblit D, Kiseleva EM, Prytomanova OG, et al. Development of PancRISK, a urine biomarker-based risk score for stratified screening of pancreatic cancer patients. Br J Cancer. 2020;122:692–6. doi: 10.1038/s41416-019-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, Joung J-G, Sohn K-A, Shin H, Park YR, Ritchie MD, et al. Knowledge boosting: a graph-based integration approach with multi-omics data and genomic knowledge for cancer clinical outcome prediction. J Am Med Inform Assoc. 2015;22:109–20. doi: 10.1136/amiajnl-2013-002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cook DP, Vanderhyden BC. Context specificity of the EMT transcriptional response. Nat Commun. 2020;11:2142. doi: 10.1038/s41467-020-16066-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lipinski KA, Barber LJ, Davies MN, Ashenden M, Sottoriva A, Gerlinger M. Cancer evolution and the limits of predictability in precision cancer medicine. Trends Cancer. 2016;2:49–63. doi: 10.1016/j.trecan.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Azuaje F. Artificial Intelligence for precision oncology: beyond patient stratification. Npj Prec Oncol. 2019;3:6. doi: 10.1038/s41698-019-0078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pan SJ, Yang Q. A survey on transfer learning. IEEE Tr Knowl Data Eng. 2010;22:1345–59. [Google Scholar]
- 53.Turki T, Wei Z, Wang, TL J. A transfer learning approach via procrustes analysis and mean shift for cancer drug sensitivity prediction. J Bioinform Comput Biol. 2018;16:1840014. doi: 10.1142/S0219720018400140. [DOI] [PubMed] [Google Scholar]
- 54.Sevakula RK, Singh V, Verma NK, Kumar C, Cui Y. Transfer learning for molecular cancer classification using deep neural networks. IEEE/ACM Trans Comput Biol Bioinform. 2019;16:2089–2100. doi: 10.1109/TCBB.2018.2822803. [DOI] [PubMed] [Google Scholar]
- 55.Vu CC, Siddiqui ZA, Zamdborg L, Thompson AB, Quinn TJ, Castillo E, et al. Deep convolutional neural networks for automatic segmentation of thoracic organs-at-risk in radiation oncology - use of non-domain transfer learning. J Appl Clin Med Phys. 2020;21:108–13. doi: 10.1002/acm2.12871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poudel P, Nyamundanda G, Patil Y, Cheang MCU, Sadanandam A. Heterocellular gene signatures reveal luminal-A breast cancer heterogeneity and differential therapeutic responses. npj Breast Cancer. 2019;5:21. doi: 10.1038/s41523-019-0116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer. 2020;1:789–99. doi: 10.1038/s43018-020-0087-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bi WL, Hosny A, Schabath MB, Giger ML, Birkbak NJ, Mehrtash A, et al. Artificial intelligence in cancer imaging: clinical challenges and applications. CA Canc J Clin. 2019;69:127–57. doi: 10.3322/caac.21552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blank CU, Haanen JB, Ribas A, Schumacher TN. Cancer Immunology. The “cancer immunogram”. Science. 2016;352:658–60. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 60.Lyons YA, Wu SY, Overwijk WW, Baggerly KA, Sood AK. Immune cell profiling in cancer: molecular approaches to cell-specific identification. npj Prec Oncol. 2017;1:26. doi: 10.1038/s41698-017-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority oversampling technique. J Art Intell Res. 2002;16:321–257. [Google Scholar]
- 62.He H, Bai Y, Garcia EA, Li S. ADASYN: adaptive synthetic sampling approach for imbalanced learning. In: 2008 IEEE international joint conference on neural networks. (IEEE Xplore ed.), IEEE; 2008. p. 1322–8.
- 63.Griffith GJ, Morris TT, Tudball MJ, Herbert A, Mancano G, Pike L, et al. Collider bias undermines our understanding of COVID-19 disease risk and severity. Nat Commun. 2020;11:5749. doi: 10.1038/s41467-020-19478-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bueno MJ, Mouron S, Quintela-Fandino M. Personalising and targeting antiangiogenic resistance: a complex and multifactorial approach. Br J Cancer. 2017;116:1119–25. doi: 10.1038/bjc.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dlamini Z, Francies FZ, Hull R, Marima R. Artificial intelligence (AI) and big data in cancer and precision oncology. Computat Str Biotech J. 2020;18:2300–11. doi: 10.1016/j.csbj.2020.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Halama N. Machine learning for tissue diagnostics in oncology: brave new world. Br J Cancer. 2019;121:431–3. doi: 10.1038/s41416-019-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tuffaha HW, Gordon LG, Scuffham PA. Value of information analysis in oncology: the value of evidence and evidence of value. J Oncol Pract. 2014;10:e55–62. doi: 10.1200/JOP.2013.001108. [DOI] [PubMed] [Google Scholar]
- 68.Kunst NR, Alarid-Escudero F, Paltiel AD, Wang S-Y. A value of information analysis of research on the 21-gene assay for breast cancer management. Value Health. 2019;22:1102–10. doi: 10.1016/j.jval.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beaton L, Bandula S, Gaze MN, Sharma RA. How rapid advances in imaging are defining the future of precision radiation oncology. Br J Cancer. 2019;120:779–90. doi: 10.1038/s41416-019-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Linn KA, Laber EB, Stefanski LA. iqLearn: interactive Q-Learning in R. J Stat Softw. 2015;64:i01. doi: 10.18637/jss.v064.i01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tseng HH, Luo Y, Cui S, Chien JT, Ten Haken RK, El Naqa I. Deep reinforcement learning for automated radiation adaptation in lung cancer. Med Phys. 2017;44:6690–705. doi: 10.1002/mp.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Petersen BK, Yang J, Grathwohl WS, Cockrell C, Santiago C, An G, et al. Deep reinforcement learning and simulation as a path toward precision medicine. J Comput Biol. 2019;26:597–604. doi: 10.1089/cmb.2018.0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ali I, Hart GR, Gunabushanam G, Liang Y, Muhammad W, Nartowt B, et al. Lung nodule detection via deep reinforcement learning. Front Oncol. 2018;8:108. doi: 10.3389/fonc.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu S, See KC, Ngiam KY, Celi LA, Feng M. Reinforcement learning for clinical decision support in critical care: comprehensive review. J Med Intern Res. 2020;2287:e18477. doi: 10.2196/18477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mazurowski MA. Radiogenomics: what it is and why it is important. J Am Coll Radiol. 2015;12:862–6. doi: 10.1016/j.jacr.2015.04.019. [DOI] [PubMed] [Google Scholar]
- 76.Wu J, Tha KK, Xing L, Li R. Radiomics and radiogenomics for precision radiotherapy. J Radiat Res. 2018;59:i25–i31. doi: 10.1093/jrr/rrx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Papanikolaou N, Matos C, Koh DM. How to develop a meaningful radiomic signature for clinical use in oncologic patients. Cancer Imag. 2020;20:33. doi: 10.1186/s40644-020-00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Keek SA, Leijenaar RTH, Jochems A, Woodruff HC. A review on radiomics and the future of theranostics for patient selection in precision medicine. Br J Radiol. 2018;91:20170926. doi: 10.1259/bjr.20170926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alvarez-Jimenez C, Sandino AA, Prasanna P, Gupta A, Viswanath SE, Romero E. Identifying cross-scale associations between radiomic and pathomic signatures of non-small cell lung cancer subtypes: preliminary results. Cancers. 2020;12:3663. doi: 10.3390/cancers12123663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Saltz JH, Gupta R. Artificial intelligence and the interplay between tumor and immunity, Ch. 10. In: Artificial Intelligence and Deep Learning in Pathology. (Stanley C ed.), Elsevier; 2021. p. 211–35.
- 81.Nie K, Al-Hallaq H, Li A, Benedict SH, Sohn JW, Moran JM, et al. NCTN assessment of current applications of radiomics in oncology. Int J Rad Oncol. 2019;104:302–15. doi: 10.1016/j.ijrobp.2019.01.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lv W, Ashrafinia S, Ma J, Lu L, Rahmim A. Multi-level multi-modality fusion radiomics: application to PET and CT imaging for prognostication of head and neck. Cancer IEEE J Biomed Health Inform. 2020;24:2268–77. doi: 10.1109/JBHI.2019.2956354. [DOI] [PubMed] [Google Scholar]
- 83.Wei L, Osman S, Hatt M, El Naqa I. Machine learning for radiomics-based multimodality and multiparametric modeling. Q J Nucl Med Mol Imag. 2019;63:323–38. doi: 10.23736/S1824-4785.19.03213-8. [DOI] [PubMed] [Google Scholar]
- 84.Papp L, Spielvogel CP, Rausch I, Hacker M, Beyer T. Personalized medicine through hybrid imaging and medical big data analysis. Front Phys. 2018;6:51. [Google Scholar]
- 85.Hagiwara A, Fujita S, Ohno M, Aoki S. Variability and standardization of quantitative imaging. Integr Radiol. 2020;55:601–16. doi: 10.1097/RLI.0000000000000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mühlberg A, Katzmann A, Heinemann V, Kärgel R, Wels M, Taubmann O, et al. The Technome—a predictive internal calibration approach for quantitative imaging biomarker research. Sci Rep. 2020;10:1103. doi: 10.1038/s41598-019-57325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sala E, Merna E, Himoto Y, Veeraraghavan H, Brenton JD, Snyder A, et al. Unravelling tumour heterogeneity using next-generation imaging: radiomics, radiogenomics, and habitat imaging. Clin Radiol. 2017;72:3–10. doi: 10.1016/j.crad.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are. Data Radiol. 2016;278:563–77. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gillies RJ, Balagurunathan Y. Perfusion MR imaging of breast cancer: insights using ‘habitat imaging’. Radiology. 2018;288:36–37. doi: 10.1148/radiol.2018180271. [DOI] [PubMed] [Google Scholar]
- 90.Sollini M, Antunovic L, Chiti A, Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imag. 2019;46:2656–72. doi: 10.1007/s00259-019-04372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fave X, Zhang L, Yang J, Mackin D, Balter P, Gomez D, et al. Delta-radiomics features for the prediction of patient outcomes in non-small cell lung cancer. Sci Rep. 2017;7:588. doi: 10.1038/s41598-017-00665-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jeon SH, Song C, Chie EK, Kim B, Kim YH, Chang W, et al. Delta-radiomics signature predicts treatment outcomes after preoperative chemoradiotherapy and surgery in rectal cancer. Radiat Oncol. 2019;14:43. doi: 10.1186/s13014-019-1246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin P, Yang PF, Chen S, Shao Y-Y, Xu L, Wu Y, et al. A delta-radiomics model for preoperative evaluation of neoadjuvant chemotherapy response in high-grade osteosarcoma. Cancer Imag. 2020;20:7. doi: 10.1186/s40644-019-0283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gatouillat A, Badr Y, Massot B, Sejdić E. Internet of medical things: a review of recent contributions dealing with cyber-physical systems in medicine. IEEE Internet Things J. 2018;5:3810–22. [Google Scholar]
- 95.Han T, Nunes VX, Souza LFDF, Marques AG, Silva ICL, Marcos Aurelio AF, et al. Internet of medical things—based on deep learning techniques for segmentation of lung and stroke regions in CT scans. IEEE Access. 2020;8:71117–35. [Google Scholar]
- 96.Souza LFF, Silva ICL, Marques AG, Silva FHDS, Nunes VX, Hassan MM, et al. Internet of medical things: an effective and fully automatic iot approach using deep learning and fine-tuning to lung CT segmentation. Sensors. 2020;20:E6711. doi: 10.3390/s20236711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sun C, Tian X, Liu Z, Li W, Li P, Chen J, et al. Radiomic analysis for pretreatment prediction of response to neoadjuvant chemotherapy in locally advanced cervical cancer: a multicentre study. eBioMedicine. 2019;46:160–9. doi: 10.1016/j.ebiom.2019.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dissaux G, Visvikis D, Da-Ano R, Pradier O, Chajon E, Barillot I, et al. Pretreatment 18F-FDG PET/CT radiomics predict local recurrence in patients treated with stereotactic body radiotherapy for early-stage non-small cell lung cancer: a multicentric study. J Nucl Med. 2020;61:814–20. doi: 10.2967/jnumed.119.228106. [DOI] [PubMed] [Google Scholar]
- 99.Li ZC, Bai H, Sun Q, Li Q, Liu L, Zou Y, et al. Multiregional radiomics features from multiparametric MRI for prediction of MGMT methylation status in glioblastoma multiforme: a multicentre study. Eur Radiol. 2018;28:3640–50. doi: 10.1007/s00330-017-5302-1. [DOI] [PubMed] [Google Scholar]
- 100.Welch ML, McIntosh C, Haibe-Kains B, Milosevic MF, Wee L, Dekker A, et al. Vulnerabilities of radiomic signature development: the need for safeguards. Radioth Oncol. 2019;130:2–9. doi: 10.1016/j.radonc.2018.10.027. [DOI] [PubMed] [Google Scholar]
- 101.Capobianco E, Valdes C, Sarti S, Jiang Z, Poliseno L, Tsinoremas NF. Ensemble modeling approach targeting heterogeneous RNA-Seq data: application to melanoma pseudogenes. Sci Rep. 2017;7:17344. doi: 10.1038/s41598-017-17337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ho D. Artificial intelligence in cancer therapy. Science. 2020;367:982–3. doi: 10.1126/science.aaz3023. [DOI] [PubMed] [Google Scholar]
- 103.Shah P, Kendall F, Khozin S, Goosen R, Hu J, Laramie J, et al. Artificial intelligence and machine learning in clinical development: a translational perspective. npj Digit Med. 2019;2:69. doi: 10.1038/s41746-019-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Toh TS, Dondelinger F, Wang D. Looking beyond the hype: applied AI and machine learning in translational medicine. EBioMedicine. 2019;47:607–15. doi: 10.1016/j.ebiom.2019.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu R, Rizzo S, Whipple S, Pal N, Pineda AL, Lu M, et al. Evaluating eligibility criteria of oncology trials using real-world data and AI. Nature 2021. [DOI] [PMC free article] [PubMed]
- 106.Capobianco E. Imprecise data and their impact on translational research in medicine. Front Med. 2020;7:82. doi: 10.3389/fmed.2020.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bezemer T, de Groot MC, Blasse E, Ten Berg MJ, Kappen TH, Bredenoord AL, et al. Factor in clinical decision support systems. J Med Intern Res. 2019;21:e11732. doi: 10.2196/11732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, et al. Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med. 2020;12:eaax7533. doi: 10.1126/scitranslmed.aax7533. [DOI] [PubMed] [Google Scholar]
- 109.Liu Y, Kohlberger T, Norouzi M, Dahl GE, Smith JL, Mohtashamian A, et al. Artificial intelligence–based breast cancer nodal metastasis detection: insights into the black box for pathologists. Arch Pathol Lab Med. 2019;143:859–68. doi: 10.5858/arpa.2018-0147-OA. [DOI] [PubMed] [Google Scholar]
- 110.Steiner DF, MacDonald R, Liu Y, Truszkowski P, Hipp JD, Gammage C, et al. Impact of deep learning assistance on the histopathologic review of lymph nodes for metastatic breast cancer. Am J Surg Path. 2018;42:1636–46. doi: 10.1097/PAS.0000000000001151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kelly CJ, Karthikesalingam A, Suleyman M, Corrado G, King D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17:195. doi: 10.1186/s12916-019-1426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet. 2019;393:1577–9. doi: 10.1016/S0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]
- 113.Faes L, Liu X, Wagner SK, Fu DJ, Balaskas KA. Clinician’s guide to artificial intelligence: how to critically appraise machine learning studies. Transl Vis Sci Technol. 2020;9:33. Erratum in: Transl Vis Sci Technol. 2020;9:7. [DOI] [PMC free article] [PubMed]
- 114.CONSORT-AI and SPIRIT-AI Steering Group. Reporting guidelines for clinical trials evaluating artificial intelligence interventions are needed. Nat Med. 2019;25:1467–8. doi: 10.1038/s41591-019-0603-3. [DOI] [PubMed] [Google Scholar]
- 115.Liu X, Faes L, Calvert MJ, Denniston AK. CONSORT/SPIRIT-AI Extension Group. Extension of the CONSORT and SPIRIT statements. Lancet. 2019;394:1225. doi: 10.1016/S0140-6736(19)31819-7. [DOI] [PubMed] [Google Scholar]
- 116.Dong Y, Yang W, Wang J, Zhao J, Qiang Y. MLW-gcForest: a multi-weighted gcForest model towards the staging of lung adenocarcinoma based on multi-modal genetic data. BMC Bioinform. 2019;20:578. doi: 10.1186/s12859-019-3172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nestor B, McDermott MBA, Chauhan G, Naumann T, Hughes MC, Goldenberg A, et al. Rethinking clinical prediction: why machine learning must consider year of care and feature aggregation. In: Machine Learning for Health (ML4H): Workshop at NeurIPS. 2018. arXiv:1811.07216 [cs.LG].
- 118.Davis SE, Greevy RA, Fonnesbeck C, Lasko TA, Walsh CG, Matheny ME. A nonparametric updating method to correct clinical prediction model drift. J Am Med Inform Assoc. 2019;26:1448–57. doi: 10.1093/jamia/ocz127. [DOI] [PMC free article] [PubMed] [Google Scholar]