Abstract
Coronaviruses are important human pathogens, among which the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent for the COVID-19 pandemic. To combat the SARS-CoV-2 pandemic, there is a pressing need for antivirals, especially broad-spectrum antivirals that are active against all seven human coronaviruses (HCoVs). For this reason, we are interested in developing antiviral assays to expedite the drug discovery process. Here, we provide the detailed protocol for the cytopathic effect (CPE) assay and the plaque assay for human coronaviruses 229E (HCoV-229E), HCoV-OC43, and HCoV-NL63, to identify novel antivirals against HCoVs. Neutral red was used in the CPE assay, as it is relatively inexpensive and more sensitive than other reagents. Multiple parameters including multiplicity of infection, incubation time and temperature, and staining conditions have been optimized for CPE and plaque assays for HCoV-229E in MRC-5, Huh-7, and RD cell lines; HCoV-OC43 in RD, MRC-5, and BSC-1 cell lines, and HCoV-NL63 in Vero E6, Huh-7, MRC-5, and RD cell lines. Both CPE and plaque assays have been calibrated with the positive control compounds remdesivir and GC-376. Both CPE and plaque assays have high sensitivity, excellent reproducibility, and are cost-effective. The protocols described herein can be used as surrogate assays in the biosafety level 2 facility to identify entry inhibitors and protease inhibitors for SARS-CoV-2, as HCoV-NL63 also uses ACE2 as the receptor for cell entry, and the main proteases of HCoV-OC43 and SARS-CoV-2 are highly conserved. In addition, these assays can also be used as secondary assays to profile the broad-spectrum antiviral activity of existing SARS-CoV-2 drug candidates.
Keywords: Human Coronavirus, 229E, OC43, NL63, Antiviral
Background
Humans have been battling viruses throughout history. Viral epidemics and pandemics have caused devastating economic, social, and political unrest, in addition to significant morbidity and mortality ( Shang et al., 2021 ). There are seven human coronaviruses: SARS-CoV ( Gagneur et al., 2002 ), MERS-CoV ( Zumla et al., 2015 ), and SARS-CoV-2 ( Hui et al., 2020 ), which cause severe acute respiratory syndrome; and four common human coronaviruses HCoV-229E, HCoV-NL63, HCoV-OC43, and HCoV-HKU1, which cause a significant portion of upper and lower respiratory tract infections in humans worldwide (Mesel- Lemoine et al., 2012 ). Although vaccines are the mainstay for viral prophylaxis, antiviral drugs are essential complements and play a vital role in viral disease containment, especially during the lag period between the initial viral outbreak and the delivery of effective vaccines. Furthermore, vaccines may lose potency and become ineffective when mutations emerge among circulating viruses ( Collier et al., 2021 ; Lopez Bernal et al., 2021 ; Williams and Burgers, 2021). As the third coronavirus outbreak in human history, the COVID-19 pandemic is a timely reminder of the urgent need for broad-spectrum antiviral drugs that can be rapidly deployed for the prevention and treatment of emerging and re-emerging viral diseases.
We have routinely used the CPE and plaque assays to test antivirals against many different types of viruses, including influenza, enterovirus, and coronavirus ( Hu et al., 2017a , 2017b, 2017c, 2017d, 2018, 2021a, 2021b; Li, F. et al., 2016, 2017; Ma et al., 2019 , 2020a, 2020b; Musharrafieh et al., 2019a , 2019b, 2020; Smail et al., 2021 ; Wang et al., 2018 ; Xia et al., 2021 ; Zhang et al., 2018 , 2019, 2020). The CPE and plaque assays have several key advantages over alternative methods. First, these assays measure viral infectivity instead of a given viral protein or gene, as both assays directly report the potency of testing compounds in inhibiting the replication of infectious viruses. Second, CPE and plaque assays are more reproducible than the TCID50 assay ( Nadgir et al., 2013 ). Third, both CPE and plaque assays are relatively inexpensive compared to RT-qPCR ( Corman et al., 2020 ), immunoassays ( Liu et al., 2005 ; Payne et al., 2006 ), viral flow cytometry ( Brussaard et al., 2000 ), or Transmission Electron Microscope (TEM) (Roingeard, 2008), and do not require expensive reagents or specialized instruments. In this study, we have established two cell-based antiviral assays, the CPE assay and the plaque assay for HCoV-229E, HCoV-NL63, and HCoV-OC43. These assays can be used as alternatives for the primary screening of SARS-CoV-2 antivirals in BSL-2 facilities, and also as secondary assays to characterize the broad-spectrum antiviral activity of potential drug candidates. The CPE assay is appropriate for high-throughput screenings in a 96-well plate format, while the plaque assay is comparatively labor-intense and time-consuming, and is more suitable as a secondary assay to confirm the antiviral activity of initial hits identified from the primary CPE assay ( Ratnam et al., 1995 ). Overall, these assays are expected to expedite the discovery of new antivirals against coronaviruses in BSL-2 facilities. In this report, we describe the detailed protocols for utilizing the CPE assay (flowchart illustrated in Figure 1) and the plaque assay (flowchart illustrated in Figure 2) to test antiviral activity of drug candidates against HCoV-229E, HCoV-NL63, and HCoV-OC43. Multiple parameters for the assays have been optimized, including the cell lines, multiplicity of infection, incubation time and temperature, and staining conditions. The optimized CPE and plaque assays conditions for HCoV-229E, HCoV-OC43 and HCoV-NL63 are listed in Table 1. Both assays have been calibrated with the positive control compounds remdesivir ( Wang et al., 2020 ) and GC-376 ( Ma et al., 2020c ). Alternatively, CPE and plaque assays protocols for HCoV-229E, HCoV-OC43, and HCoV-NL63 can be found elsewhere ( Schmidt et al., 1979 ; Gerna et al., 1980 ; Herzog et al., 2008 ; Bracci et al., 2020 ; Hirose et al., 2021 ; Schirtzinger et al., 2021 ).
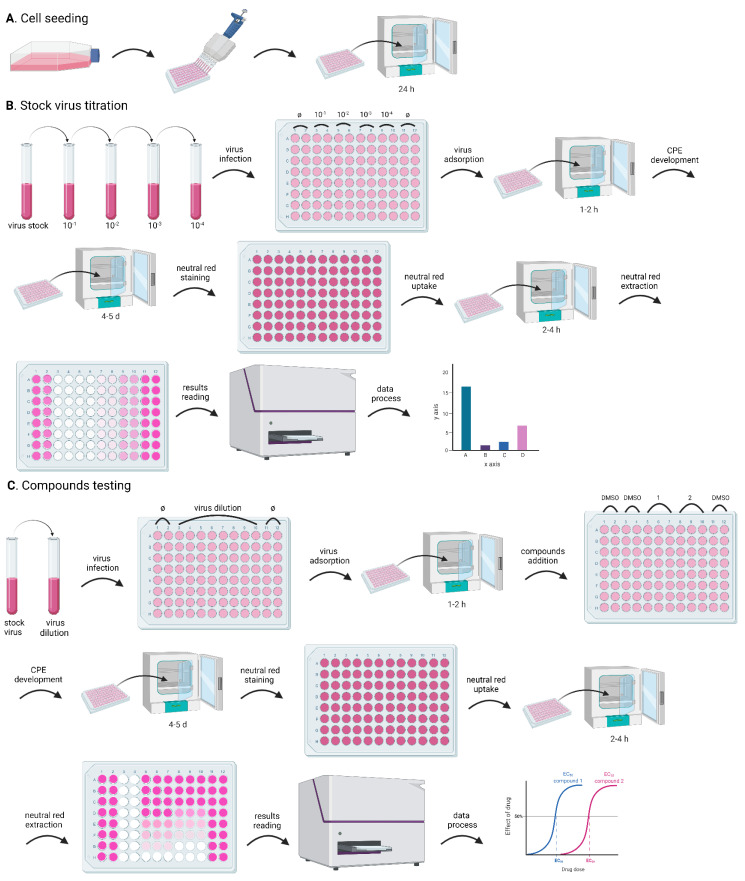
Figure 1. Flowchart for the CPE assay.
The whole process includes: cell seeding, to prepare cells in a 96-well plate for the experiments; stock virus titration, to optimize the assay conditions; and compound testing, for the evaluation of the antiviral activity of test compounds. Both stock virus titration and compound testing comprise the following steps: dilute the stock virus to obtain the desired MOI; infect the cells in the 96-well plate with a small volume (100 µL) of the diluted virus; incubate the 96-well plate in the incubator for 1-2 h, to facilitate viral attachment (virus adsorption); add the test compounds (only for the compound testing experiment); incubate the 96-well plate in the incubator to develop CPE; stain the cells with neutral red; extract neutral red from the cells; quantify neutral red by measuring absorbance; analyze data. A. Preparation of cells in the 96-well plate for the CPE assay; B. Major steps of stock virus titration in the CPE assay; C. Assessment of antiviral activity of test compounds in the CPE assay.
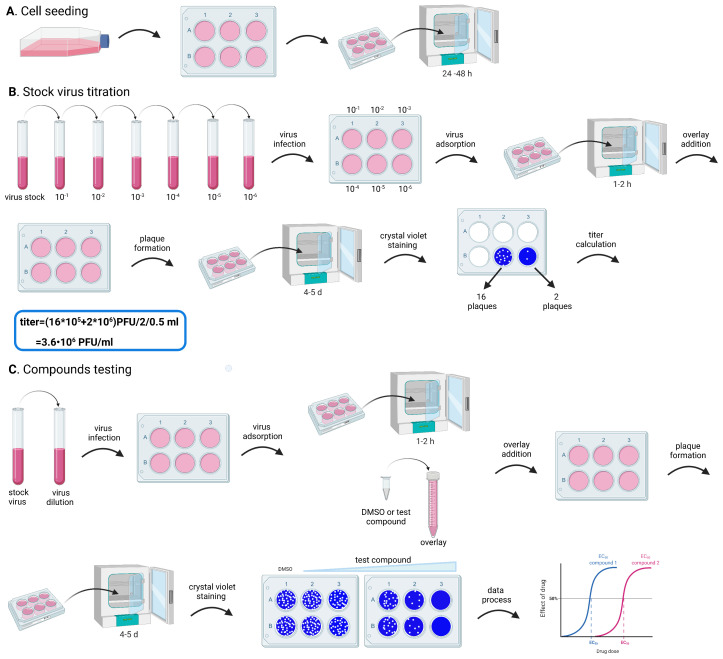
Figure 2. Flowchart for the plaque assay.
The whole process includes: cell seeding, to prepare cells in a 6-well plate for the experiments; stock virus titration, to optimize the assay conditions; and compound testing, for the evaluation of the antiviral activity of test compounds. Both stock virus titration and compound testing comprise the following steps: dilute the stock virus, to obtain the desired MOI; infect the cells in the 6-well plate with a small volume (500 µL) of the diluted virus; incubate the 6-well plate in the incubator for 1-2 h, to facilitate viral attachment (virus adsorption); add an avicel overlay containing the test compounds (only for the compound testing experiment); incubate the 6-well plate in the incubator, to allow plaque formation; stain the cells with crystal violet; analyze data. A. Preparation of cells in the 6-well plate for the plaque assay; B. Major steps of stock virus titration in the plaque assay; C. Assessment of antiviral activity of test compounds in the plaque assay.
Table 1. Cell lines and incubation temperature and time for HCoV-229E, HCoV-OC43, and HCoV-NL63 in the CPE and plaque assays in this protocol.
| Virus | Assay | Plate | Cell line | Seeding density; volume per well | Total wells needed for x compounds | Incubation Temperature (°C) | Incubation Time (day) |
| HCoV-OC43 | CPE | 96-well | RD | 2.5×104 cells/mL; 100 µL | 6(x+1)*3 | 33 | 4.5 |
| plaque assay | 6-well | RD | 2.5×104 cells/mL; 3 mL | (6x+1)*2 | 33 | 4.5 | |
| HCoV-229E | CPE | 96-well | MRC-5 | 5×104 cells/mL; 100 µL | 6(x+1)*3 | 33 | 5.5 |
| plaque assay | 6-well | RD | 2.5×104 cells/ml; 3 mL | (6x+1)*2 | 33 | 5.5 | |
| HCoV-NL63 | CPE | 96-well | Vero E6 | 2×104 cells/mL; 100 µL | 6(x+1)*3 | 37 | 4 |
| plaque assay | 6-well | Vero E6 | 2×104 cells/mL; 3 mL | (6x+1)*2 | 37 | 4 |
Materials and Reagents
Flat-bottomed 96-well tissue culture plates (Genesee, catalog number: 25-109)
96-well storage plate (Thermo Scientific, catalog number: AB-1058)
96-well deep well storage plates (ThermoFisher Scientific, catalog number: 260251)
50 mL conical centrifuge tubes, sterile, polypropylene (Genesee, catalog number: 25-108)
15 mL conical centrifuge tubes, sterile, polypropylene (Genesee, catalog number: 25-106)
10 mL serological pipets, sterile (Genesee, catalog number: 25-104)
25 mL serological pipets, sterile (Genesee, catalog number: 25-106)
1.7 mL DNase/RNase-free tubes (Genesee, catalog number: 25-282)
1,000 µL pipette tips, low binding (Genesee, catalog number: 24-160R)
200 µL pipette tips, low binding (Genesee, catalog number: 24-150R)
10 µL pipette tips, low binding (Genesee, catalog number: 24-121R)
Tissue culture treated flasks, 600 mL, vent (Genesee, catalog number: 25-211)
Tissue culture treated flasks, 250 mL, vent (Genesee, catalog number: 25-209)
Sterile syringe filter, 0.2 µm (Fisher Scientific, catalog number: 09-740-100)
6-well cell culture plates (Genesee, catalog number: 25-105)
HCoV-OC43 virus (BEI Resources, catalog number: NR-52725)
HCoV-229E virus (BEI Resources, catalog number: NR-52726)
HCoV-NL63 virus (BEI Resources, catalog number: NR-470)
Human rhabdomyosarcoma cell line, RD (ATCC® CCL-136TM)
Human fibroblast cell line, MRC-5 (ATCC® CCL-171TM)
HEK-293T-hACE2 cell line (BEI Resources, catalog number: NR-52511)
A549-hACE2 cell line (BEI Resources, catalog number: NR-53821)
Vero E6 cell line (ATCC® CRL-1586TM)
HCT-8 cell line (ATCC® CCL-244TM)
Huh-7 cell line (Millipore Sigma, catalog number: 01042712-1VL, a kind gift from Dr. Tianyi Wang at the University of Pittsburgh)
BHK-21 cell line (ATCC® CCL-10TM)
BSC-1 cell line (ATCC® CCL-26TM, a kind gift from Dr. Kui Li at the University of Tennessee Health Science Center)
Calu-3 cell line (ATCC® HTB-55TM)
Caco-2 cell line (ATCC® HTB-37TM)
Trypsin-EDTA, 0.25% 1×, phenol red (Genesee, catalog number: 25-510)
Dulbecco's modified Eagle medium (DMEM) (Genesee, catalog number: 25-501)
Eagle's minimum essential medium (EMEM) (ATCC® 30-2003TM)
FBS (Gibco, catalog number: 26140-095)
Penicillin-Streptomycin (P/S) 100× solution (Genesee, catalog number: 25-512)
Glacial acetic acid (CAMEO chemicals, catalog number: UN2789)
Na2HPO4·2H2O (Sigma, catalog number: 04272-1KG)
KH2PO4 (Sigma, catalog number: P9791-1KG)
NaCl (Fisher Scientific, catalog number: BP358-10)
KCl (Sigma, catalog number: P3911-1KG)
Ethanol (ThermoFisher, catalog number: A405-20)
MgCl2·6H2O (Sigma, catalog number: M2670-1KG)
CaCl2 (Sigma, catalog number: C1016-2.5KG)
DMEM powder (Gibco, catalog number: 12-800-017)
HEPES, sodium salt (Gold Biotechnology, catalog number: H-401-2.5)
NaHCO3 (Millipore Sigma, catalog number: S5761-5KG)
Condensed HCl, 36.5 to 38% (Fisher Scientific, catalog number: A144-212)
Avicel microcrystalline cellulose (FMC BioPolymer, Philadelphia, PA)
Crystal violet (Fisher Scientific, catalog number: AAB2193236)
Nano pure, or distilled and deionized water
Phosphate Buffered Saline (PBS) (see Recipes)
Neutral red (3-amino-7-dimethylamino-2-methyl-phenazine hydrochloride) (Sigma, catalog number: N4638)
10× PBS stock (see Recipes)
100× MgCl2 stock (see Recipes)
100× CaCl2 stock (see Recipes)
DPBS, calcium and magnesium free (see Recipes)
PBS, with calcium and magnesium (see Recipes)
Neutral red stock solution (see Recipes)
Complete culture medium (see Recipes)
Neutral red working solution (see Recipes)
Neutral red de-staining solution (see Recipes)
2× DMEM (see Recipes)
1.2% avicel (see Recipes)
0.2% crystal violet (see Recipes)
Equipment
Two cell culture incubators (Eppendorf, Galaxy® 170 R), humidified, 5% CO2/95% air, with temperatures set up at 33°C and 37°C, respectively
Inverted microscope Olympus CKX53 (ThermoFisher Scientific, catalog number: NC1991101)
MultiskanTM FC Microplate Photometer (ThermoFisher Scientific, catalog number: 51119000)
CountessTM 3 Automated Cell counter (ThermoFisher Scientific, catalog number: AMQAX2000)
Fume hood (for hazardous chemicals)
Microtiter plate shaker (Fisher Scientific, catalog number: 88-861-023)
Hot plate stirrer (Fisher Scientific, catalog number: 11-520-49SH)
PIPETMAN Classic P10 (Genesee, catalog number: 37-100P10)
PIPETMAN Classic P100 (Genesee, catalog number: 37-100P100)
PIPETMAN Classic P1000 (Genesee, catalog number: 37-100P1K)
PIPETMAN Classic P20 (Genesee, catalog number: 37-100P20)
PIPETMAN Classic P200 (Genesee, catalog number: 37-100P200)
Reagent reservoir, sterile (VWR, catalog number: 89094-662)
Vortex Mixer (Fisher Scientific, catalog number: 02-215-414)
Centrifuge (Beckman, catalog number: A99465)
Bottle Top Vacuum Filters (Genesee, catalog number: 25-235)
SartoriusTM BiohitTM PicusTM NxT Electronic Pipettes, 50-1,200 µL, 12 Channels (SartoriusTM, catalog number: LH745491)
SartoriusTM BiohitTM PicusTM NxT Electronic Pipettes, 0.2-10 µL, 12 Channels (SartoriusTM, catalog number: LH745421)
HandE-Vac Handheld Aspirating System (Argos TechnologiesTM, catalog number: 10-987-042)
Software
ImageJ (National Institutes of Health, version 1.50 c, https://imagej.nih.gov/ij/download.html)
Prism GraphPad (https://www.graphpad.com)
Procedure
-
Virus propagation
HCoV-OC43 was propagated in HCT-8 or RD cell lines. HCoV-229E was propagated in MRC-5 or Huh-7 cell lines. HCoV-NL63 was initially propagated in the HEK-293T-hACE2 cell line, followed by additional 3-4 passages in Vero E6 or Huh-7 cell lines.
Note: For all cell lines we have tried, only the HEK-293T-hACE2 cell line produces a high titer of HCoV-NL63, and the propagated HCoV-NL63 virus was accommodated in the Vero E6 or Huh-7 cell lines to allow CPE development and plaque formation.
-
Cell seeding (Day 0)
Note: All procedures should be carried out under aseptic conditions and in the sterile environment of a laminar flow cabinet.
For a T75 flask of cell culture at 90-95% confluency, aspirate the 20 mL of cell culture medium and rinse the cells with DPBS buffer (10 mL of buffer is used for a 75-cm2 flask) by gentle agitation to remove any remaining serum, which might inhibit the action of trypsin. Repeat this step once.
Add trypsin-EDTA solution (3 mL is used for a 75-cm2 flask) to the cell monolayer in the flask, agitate gently, and incubate the flask at 37°C for 5-10 min.
Lightly tap the flask to detach the cells and add prewarmed (to room temperature) complete culture medium (10 mL for a 75-cm2 flask). Gently disperse the medium by pipetting with a serological pipette over the cell monolayer surface several times to ensure >95% recovery of cells. Pipet up and down for a few times to make sure that a homogeneous suspension is obtained.
Transfer the cell suspension to a 15 mL sterile centrifuge tube and centrifuge at 500 × g for 10 min at room temperature (20-30°C) to spin down the cells.
Aspirate the supernatant and add 10 mL of complete culture medium to resuspend the cells.
Count the cell density of the suspension in step e using a cell counter.
Transfer approximately 1 × 106 cells into a 175-cm2 flask and bring the total volume of the cell suspension to 40 mL with complete culture medium. This will allow cells to achieve 70-80% confluency 18-24 h after seeding.
Incubate the flask in a cell culture incubator (humidified, 5% CO2/95% air, 37°C).
-
Virus infection (Day 1)
Check the cell confluency in the 175-cm2 flask using a phase-contrast inverted microscope; cells should be at 70-80% confluency.
Warm DMEM medium in a 37°C metallic beads bath (a replacement for water bath to avoid potential contamination due to microbial growth in the water) and thaw the virus stocks (the initial virus was obtained from BEI Resources and stored at -80°C).
Prepare virus infection medium by adding FBS into DMEM to a final concentration of 2%. For 50 mL of DMEM, add 1 mL of FBS.
Dilute 100 µL of the virus stock (from step b) into 10 mL of virus infection medium and mix well by pipetting up and down a few times.
Aspirate the medium in the 175-cm2 flask and rinse the cells with PBS, calcium and magnesium buffer (50 mL of buffer is used for a 175-cm2 flask).
Aspirate the PBS, calcium, and magnesium buffer.
Infect the cells in the 175-cm2 flask by adding 10 mL of virus dilution prepared in step d. Gently swirl the flask to allow the virus dilution to evenly cover the cell monolayer.
Incubate the infected cells in the 175-cm2 flask in a cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E; 37°C for HCoV-NL63) for 1-2 h. Swirl the flask every 15 min to allow the virus dilution to evenly cover the cell monolayer to facilitate virus attachment to the infected cells.
Aspirate the virus dilution in the 175-cm2 flask and add 40 mL of virus infection medium into the flask.
Incubate the 175-cm2 flask in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E; 37°C for HCoV-NL63) for 3 days.
-
Virus collection (Day 4)
-
Collect the supernatant in the 175-cm2 flask in a 50 mL sterile centrifuge tube and centrifuge at 4,000 × g at room temperature (20-30°C) for 10 min. This step spins down the cells or cell debris in the supernatant.
Note: HCoVs achieve the highest viral titer 2-3 days post infection, which is before significant CPE will be observed, and the titer starts to decrease afterwards. Thus, the virus should be collected at no more than 3 days post infection.
Transfer supernatant to another 50 mL sterile centrifuge tube using a 10 mL serological pipette. Filter the supernatant through a sterile 0.2 µm filter to remove any remaining cell debris. Aliquot the virus into sterile 1.5 mL DNase/RNase-free tubes at 1 mL per tube. Label each tube with the virus name and the date collected. Flash freeze the tubes (recommended but not necessary) and store all tubes in a -80°C freezer for future use.
-
-
-
CPE assay - Titrate the virus stock
We have examined different cell lines for HCoV-OC43, HCoV-229E, and HCoV-NL63 in both the CPE and plaque assays, and the results are listed in Table 2. HCoV-OC43 can develop complete CPE in the RD, MRC-5, and BSC-1 cell lines. HCoV-229E can develop complete CPE in MRC-5, Huh-7, and RD cell lines. HCoV-NL63 can develop complete CPE in Vero E6, Huh-7, MRC-5, and RD cell lines. In this study, the following conditions were shown as representative examples: HCoV-OC43 CPE assay in the RD cell line, HCoV-229E CPE assay in the MRC-5 cell line, and HCoV-NL63 in the Vero E6 cell line.
-
Cell seeding (Day 0)
Steps a-f is the same as in A. Virus propagation.
Dilute the cells with complete culture medium. At least 12 mL of media with a density of approximately 2.5 × 104 cells/mL is needed for one 96-well plate. This will allow the cells to achieve 80-90% confluency at 18-24 h after seeding and be ready for virus infection.
-
Mix the cell suspension by pipetting up and down a few times and transfer the suspension to a sterile reservoir. Dispense 100 µL of the cell suspension into each well of the 96-well plate using a multichannel pipette.
Note: Mixing the cell suspension several times to ensure it is homogeneous. Repeat periodically for even distribution.
Cover the plate with a matched lid and incubate the cells in the cell culture incubator (humidified, 5% CO2/95% air, 37°C) for 18-24 h.
-
Virus infection (Day 1)
Note: Removing and replacing reagents in the wells needs to be performed gently and carefully to avoid disturbing the cell monolayer. A brief centrifugation step (200 × g for 5 min) can be included if necessary.
Check the cell confluency in the 96-well plate, using a phase-contrast inverted microscope; cells should be 80-90% confluency.
Warm DMEM medium at 37°C in a beads basket and thaw one tube of HCoV virus stock from the -80°C freezer.
Prepare virus infection medium by adding FBS into DMEM to a final concentration of 2%. For 30 mL of DMEM, add 0.6 mL of FBS.
Serially dilute the propagated HCoV virus from 101 to 104 folds: Label four 15 mL sterile centrifuge tubes with 10-1, 10-2, 10-3, and 10-4, and add 2.7 mL of virus infection medium into each tube. In the 10-1 tube, add 300 µL of propagated HCoV virus stock, and mix well by vortexing at a moderate speed for 10 s; in the 10-2 tube, add 300 µL of the virus dilution from the 10-1 tube, and mix well by vortexing at moderate speed for 10 s; in the 10-3 tube, add 300 µL of the virus dilution from the 10-2 tube, and mix well by vortexing at moderate speed for 10 s; in the 10-4 tube, add 300 µL of the virus dilution from the 10-3 tube, and mix well by vortexing at moderate speed for 10 s.
Aspirate the medium in the 96-well cell culture plate using the HandE-Vac Handheld Aspirating System and rinse the cells by adding 200 µL of PBS, calcium, and magnesium buffer into each well using a multichannel pipette.
On the lid of the 96-well plate, label columns 1-2 and 11-12 (No virus), columns 3-4 (10-1), columns 5-6 (10-2), columns 7-8 (10-3), and columns 9-10 (10-4) as illustrated in Figures 3A, 3F, 3G, 4A, 4F, 4G, 4H, 5A, 5F, and 5G.
Aspirate the PBS, calcium and magnesium buffer using the HandE-Vac Handheld Aspirating System.
-
To the 96-well plate prepared in step e, add 100 µL of virus infection medium into columns 1-2 and 11-12 (No virus), 100 µL of virus dilution from the 10-1 tube into columns 3-4 (10-1), 100 µL of virus dilution from the 10-2 tube into columns 5-6 (10-2), 100 µL of virus dilution from the 10-3 tube into columns 7-8 (10-3), and 100 µL of virus dilution from the 10-4 tube into columns 9-10 (10-4).
Note: a small volume of 100 µL of virus dilution was used for infecting to facilitate virus attachment to the cells.
Incubate the plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E; 37°C for HCoV-NL63) for 1-2 h to ensure viral adsorption.
Add 100 µL of virus infection medium into each well in the 96-well plate to bring the total volume in each well to 200 µL, which is suitable for cell maintenance longer than 3 days.
Incubate the plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E; humidified, 5% CO2/95% air, 37°C for HCoV-NL63) for 4-5 days.
-
Microscopic evaluation (Days 2, 3, 4, and 5)
Examine the cells using a phase-contrast inverted microscope daily post-infection, and record changes in cell morphology and CPE development.
-
Neutral red uptake assay (Day 5)
Note: Steps a-k do not require sterile conditions or sterilized materials.
Centrifuge the neutral red working solution (prepared one day prior to the neutral red uptake assay and incubated in the 37°C cell culture incubator overnight) at 4,000 × g for 5 min at room temperature (20-30°C) to remove any precipitations.
Aspirate the virus infection medium in the cells and wash the cells by adding 200 µL of PBS, calcium and magnesium buffer into each well.
Gently transfer the neutral red working solution (step a) into a reagent reservoir.
Add 100 µL of neutral red working solution into each well using the 12-channel Electronic Pipette.
-
Incubate the plate in the 37°C incubator (5% CO2) for 2-4 h.
Note: For the RD and MRC-5 cell lines, the optimal incubation time is 2 h, and longer incubation times will result in significant loss of sensitivity. For the Vero E6 and A549-hACE2 cell lines, the incubation time can be up to 4 h without affecting assay sensitivity.
Aspirate the neutral red working solution.
Wash the cells by adding 200 µL of PBS, calcium and magnesium buffer into each well.
-
Aspirate the washing buffer and remove any residual buffer by gently tapping the plate upside down on paper towels or let it air dry.
Note: the wells need to be completely dry before addition of neutral red de-staining solution.
Add 100 µL of neutral red de-staining solution into each well.
Shake the plate rapidly on a microtiter plate shaker at 200 rpm for 15 min to allow the de-staining solution to completely extract the neutral red from the cells and to form a homogeneous solution.
Measure the absorbance of neutral red extract at 540 nm in a spectrophotometer, save the data in .text format for analysis. The data in the .text format file will be transferred into Excel for data processing.
Discard the de-staining solution into a proper liquid waste container, and discard the plate.
-
-
CPE assay - Testing inhibitors
-
Cell seeding (Day 0)
The whole procedure in this part (steps a-i) is exactly the same as in B. CPE assay - Titrate the virus.
-
Virus infection (Day 1)
Check the confluency of the cells in the 96-well plate using a phase-contrast inverted microscope; cells should be 80-90% confluency.
Warm the DMEM medium at 37°C in a beads basket and thaw one tube of propagated HCoV virus stored in -80°C freezer.
Prepare dilutions of test compounds (GC-376 and remdesivir) in DMSO by creating eight three-fold dilutions (0.5 log unit), starting from the highest concentration of 1 mM for each compound. The concentrations of the resulting stock solutions are 1, 0.3, 0.1, 0.03, 0.01, 0.003, 0.001, and 0.0003 mM.
Make virus infection medium by adding FBS into DMEM to a final concentration of 2%. For 30 mL of DMEM, add 0.6 mL of FBS.
Dilute HCoV stock virus for infecting. To obtain a S/B ratio greater than five, the optimal dilution for HCoV-229E is the 102 -fold dilution (equal to 2 × 103 PFU/mL) of the stock virus, that is 200 µL of HCoV-229E stock virus in 19.8 mL of virus infection medium. The optimal dilution for HCoV-NL63 is the 103 -fold dilution (equal to 7.2 × 103 PFU/mL) of the stock virus, that is 20 µL of HCoV-NL63 stock virus in 19.98 mL of virus infection medium. The optimal dilution for HCoV-OC43 is 102 -fold dilution (equal to 1.38 × 104 PFU/mL) of the stock virus, that is 200 µL HCoV-OC43 stock virus in 19.8 mL virus infection medium.
Aspirate the medium in the 96-well plate using the HandE-Vac Handheld Aspirating System and rinse the cells by adding 200 µL of PBS, calcium, and magnesium buffer into each well using a multichannel pipette. On the plate lid, label columns 1-2 and 11-12 as ‘No virus’; columns 3-4 as ‘virus’; columns 5-7 as ‘remdesivir’; and columns 8-10 as ‘GC-376’, as illustrated in Figures 3B, 4B, and 5B.
Aspirate the PBS, calcium and magnesium buffer using the HandE-Vac Handheld Aspirating System.
Add 100 µL of virus infection medium into columns 1-2 and 11-12 (No virus) in the 96-well cell culture plate, and 100 µL of virus dilution (step e) into columns 3-10.
Incubate the 96-well plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E, 37°C for HCoV-NL63) for 1-2 h.
Add 4 µL of DMSO or each concentration of the test compounds GC-376 and remdesivir (step c) into 400 µL of virus infection medium in a 96-well Deep well storage plate, pipette up and down a few times to mix well.
Transfer 100 µL of virus infection medium containing DMSO, remdesivir or GC-376 serial concentrations (in step j) into columns 3-4 (Virus), 5-7 (remdesivir), or 8-10 (GC-376), respectively, in the 96-well plate.
Incubate the 96-well plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E, 37°C for HCoV-NL63) for 3-5 days.
-
Microscopic evaluation (Days 2, 3, 4, and 5)
Examine the cells using a phase-contrast inverted microscope daily post infection., Complete CPE should develop in columns 3-4 (Virus) by 4 to 5 days post infection.
Note: Finding the proper endpoint of the CPE assay is critical when testing compounds. It is ideal to perform neutral red uptake assay when the virus group gives ~95% CPE
-
Neutral red uptake assay (Day 5)
Steps a-k are exactly the same as steps a-k in B. CPE assay - Titrate the virus.
-
-
Plaque assay - Titrate the virus
-
Cell seeding (Day 0)
Steps a-f is the same as in A. Virus propagation.
Dilute the cells with complete culture medium. At least 20 mL of approximately 2.5 × 104 cells/mL cell stock is needed for one 6-well plate.
Mix the cell suspension to ensure homogeneity by pipetting up and down several times and dispense 3 mL of the cell suspension into each well of the 6-well plate.
Cover the plate with a lid and incubate the cells in the cell culture incubator (humidified, 5% CO2/95% air, 37°C) for 24 to 48 h.
-
Virus infection (Day 1)
Check confluency of the cells in the 6-well plate using a phase-contrast inverted microscope; cells should be >95% confluency at ~48 h after seeding.
Warm DMEM medium at a 37°C in a beads basket, and thaw one tube of propagated HCoV virus stored in -80°C freezer.
Prepare virus infection medium by adding FBS into DMEM to a final concentration of 2%. For 10 mL of DMEM, add 200 µL of FBS.
Prepare the serial dilution of HCoV virus from 101 to 106-fold: Label six 15 mL sterile centrifuge tubes with 10-1, 10-2, 10-3, 10-4, 10-5, and 10-6. Add 1.8 mL of virus infection medium into each tube. In the 10-1 tube, add 200 µL of HCoV virus stock, and mix well by vortexing at moderate speed for 10 s; in the 10-2 tube, add 200 µL of virus dilution from the 10-1 tube, and mix well by vortexing at moderate speed for 10 s; in the 10-3 tube, add 200 µL of virus dilution from the 10-2 tube, and mix well by vortexing at moderate speed for 10 s; in the 10-4 tube, add 200 µL of virus dilution from the 10-3 tube, and mix well by vortexing at moderate speed for 10 s; in the 10-5 tube, add 200 µL of virus dilution from the 10-4 tube, mix well by vortexing at moderate speed for 10 s; in the 10-6 tube, add 200 µL ofvirus dilution from the 10-5 tube, and mix well by vortexing at moderate speed for 10 s.
Aspirate the growth medium in the 6-well plate and rinse the cells with 3 mL of PBS, calcium, and magnesium buffer in each well. Gently swirl the plate to wash the cells thoroughly. On the plate cover, label each well with ’10-1, 10-2, 10-3, 10-4, 10-5 and 10-6’, as illustrated in Figures 6A, 6F, 6G, 7A, 7F, 7G, 7H, 8A, 8F, and 8G.
Aspirate the PBS, calcium, and magnesium buffer.
-
Add 500 µL of 10-1, 10-2, 10-3, 10-4, 10-5, and 10-6 virus dilutions from step d into the corresponding wells. Gently swirl the plate to allow the virus inoculum to evenly cover the cell monolayer.
Note: The desired volume of virus inoculum for infecting the cells in a 6-well plate will be 250 µL to 1 mL per well, to allow complete coverage of the cell monolayer and efficient adsorption of viruses.
Incubate the infected cells in the 6-well plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E, 37°C for HCoV-NL63) for 1-2 h.
During the incubation, prepare overlay containing 0.6% Avicel in DMEM plus 2% FBS by mixing equal volumes of 2× DMEM and 1.2% Avicel. For one 6-well plate, mix 12.5 mL of 2× DMEM and 12.5 mL of 1.2% Avicel with 500 µL of FBS.
Aspirate viral inoculum from step h and wash the unbound virus in each well with 3 mL of PBS, calcium and magnesium buffer. Gently swirl the plate to wash the cells thoroughly.
Aspirate the PBS, calcium and magnesium buffer.
Add 4 mL of overlay from step i into each well in the 6-well plate.
-
Incubate the 6-well plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E, 37°C for HCoV-NL63) for 4-5 days.
Note: The optimal incubation time for HCoV-OC43 is about 4 days and 15 h. The optimal incubation time for HCoV-229E is about 5 days and 15 h. The optimal incubation time for HCoV-NL63 is about 3 days and 15 h. However, an incubation time that is 6 h shorter or longer than the optimal time will not significantly affect the results.
-
Plate staining (Day 5)
Aspirate the Avicel overlay.
Remove the residual Avicel by washing the cells with 3 mL of PBS, calcium and magnesium buffer each well.
Aspirate the PBS, calcium and magnesium buffer.
Add 1.5 mL of 0.2% crystal violet into each well, incubate at room temperature for 30 min.
Discard the crystal violet into a liquid waste container for biohazard materials, immerse the plate into tap water to remove the extra crystal violet dye.
Leave the plate on a bench to allow it to air dry and take an image of the plate.
-
-
Plaque assay - Test antiviral activity of compounds
-
Cell seeding (Day 0)
Steps a-i are exactly the same as in D. Plaque Assay - Titrate the virus, except that seeding cells in five 6-well plates for testing two compounds to determine the EC50s.
-
Virus infection (Day 1)
Steps a-b, follow the same procedure as in D. Plaque Assay - Titrate the virus.
Prepare virus dilution for infecting. The optimal condition for testing compounds in the plaque assay is to have 80-150 plaques in each well of a 6-well plate. Based on the titration results in Figure 6A, the optimal dilution for HCoV-229E is 800-fold dilution (equal to 250 PFU/mL) of stock virus. That is, for five 6-well plates, add 25 µL of stock virus into 20 mL of virus infection medium, and mix well for infecting. Based on the titration results in Figure 7A, the optimal dilution for HCoV-NL63 is 30,000-fold dilution (equal to 240 PFU/mL) of stock virus. That is, for five 6-well plates, add 6.7 × 10-4 µL of stock virus into 20 mL of virus infection medium, and mix well for infecting. However, it is preferred to serially dilute the HCoV-NL63 stock virus 10-5-fold, and add 67 µL of the 10-5-fold dilution into 20 mL of virus infection medium, and mix well for infecting. Based on the titration results in Figure 8A, the optimal dilution for HCoV-OC43 is 5,000-fold (equal to 276 PFU/mL) dilution of stock virus. That is, for five 6-well plates, add 4 µL of stock virus into 20 mL of virus infection medium. However, it is preferred to first dilute the HCoV-OC43 stock virus 10-1-fold, and add 40 µL of the 10-1-fold dilution into 20 mL of virus infection medium, to minimize variations.
Aspirate the growth medium in the 6-well plate and rinse the cells with 3 mL of PBS, calcium, and magnesium buffer in each well. Gently swirl the plate to wash the cells thoroughly.
Aspirate the PBS, calcium, and magnesium buffer.
Add 500 µL of diluted virus from step c into each well of the five 6-well plates. Gently swirl the plate to allow the virus inoculum to evenly cover the cell monolayer.
Incubate the infected cells in the 6-well plates in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E, 37°C for HCoV-NL63) for 1-2 h.
During the incubation, prepare the overlay solution which consists of 0.6% Avicel in DMEM, 2% FBS, and different concentrations of the test compounds or DMSO (control). For five 6-well plates, mix 65 mL of 2× DMEM and 65 mL of 1.2% Avicel with 2.6 mL of FBS. Aliquot 8.5 mL of the overlay to each tube (for two wells in duplicates), label the name and concentration of the test compound on each tube, and add the corresponding concentration of the compound into the overlay in each tube, vortex the tube to mix the compound, and overlay thoroughly.
Aspirate the viral inoculum and wash the unbound virus with 3 mL of PBS, calcium and magnesium buffer. Gently swirl the plate to wash the cells thoroughly. On the plate lid, label the name and concentration of the test compounds for each well.
Aspirate the PBS, calcium, and magnesium buffer.
Transfer 4 mL of overlay containing the test compound from step g into the corresponding wells in the 6-well plate.
Incubate the 6-well plate in the cell culture incubator (humidified, 5% CO2/95% air, 33°C for HCoV-OC43 and HCoV-229E, 37°C for HCoV-NL63) for 4-5 days.
-
Plate staining (Day 5)
Steps a-f, follow the same procedure as in D. Plaque Assay - Titrate the virus.
Note: The optimal incubation time for the three CoVs in the plaque assay was determined in Plaque assay - Titrate the virus. It’s recommended to record the time that the incubation of the plates starts, to determine when it is ready for staining.
-
Table 2. Cell lines tested in the HCoV-OC43, HCoV-229E, and HCoV-NL63 CPE and plaque assays and results.
| Virus | Cellular receptor | Cell lines tested in CPE and/or plaque assay | Results (work-Y, not work-N) |
|---|---|---|---|
| HCoV-OC43 | 9-Oacetylated sialic acid (9-O-Ac-Sia) ( Hulswit et al., 2019 ) | RD | CPE assay-Y; plaque assay-Y |
| MRC-5 | CPE assay-Y; plaque assay-Y | ||
| Huh-7 | CPE assay-N; plaque assay-N | ||
| Vero | CPE assay-N; plaque assay-N | ||
| BHK-21 | CPE assay-partial CPE observed; Plaque assay-N | ||
| BSC-1 | CPE assay-Y; plaque assay-Y | ||
| Calu-3 | CPE assay-N; plaque assay-N | ||
| HCoV-229E | Human aminopeptidase N (hAPN) (Li, Z. et al., 2019; Yeager et al., 1992 ) | RD | CPE assay-Y; plaque assay-Y |
| Huh-7 | CPE assay-Y; plaque assay-Y | ||
| MRC-5 | CPE assay-Y; plaque assay-Y | ||
| Vero | CPE assay-N; plaque assay-N | ||
| BHK-21 | CPE assay-N; plaque assay-N | ||
| Calu-3 | CPE assay-N; plaque assay-N | ||
| HCoV-NL63 | Angiotensin-converting enzyme2 (ACE2) ( Hofmann et al., 2005 ; Wu et al., 2009 ) | MRC-5 | CPE assay-Y; plaque assay-Y |
| Huh-7 | CPE assay-Y; plaque assay-Y | ||
| Vero | CPE assay-Y; plaque assay-Y | ||
| BHK-21 | CPE assay-N; plaque assay-N | ||
| Caco-2 | CPE assay-N; plaque assay-N | ||
| RD | CPE assay-Y; plaque assay-Y | ||
| Calu-3 | virus can replicate, but no CPE observed, no plaque formation |
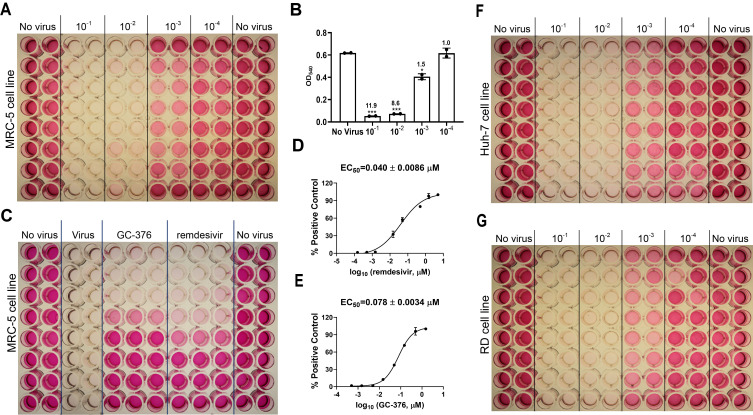
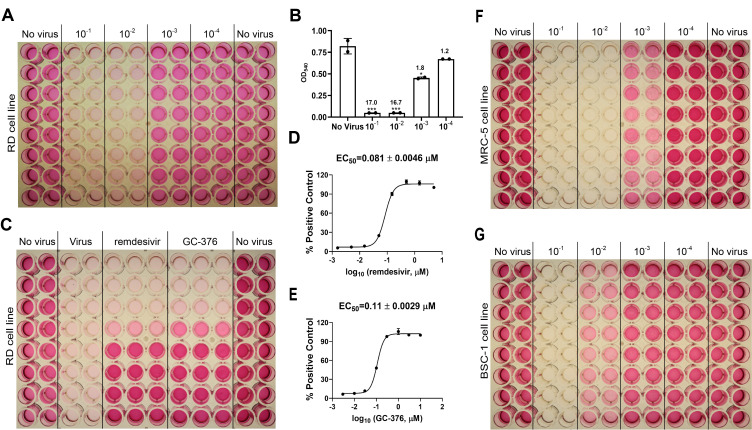
Figure 3. CPE assay for HCoV-229E.
A. Titration of HCoV-229E in the MRC-5 cell line. B. Results of HCoV-229E titration in MRC-5 cell line in CPE assay. The signal to background (S/B) ratios are labeled for each virus dilution. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test). C. Determination of EC50 values for GC-376 and remdesivir against HCoV-229E in the CPE assay with the MRC-5 cell line. D. EC50 curve fitting for remdesivir obtained in GraphPad Prism 8. E. EC50 curve fitting for GC-376 obtained in GraphPad Prism 8. F. Titration of HCoV-229E in Huh-7 cell line. G. Titration of HCoV-229E in RD cell line. The images are representatives of three independent repeats.
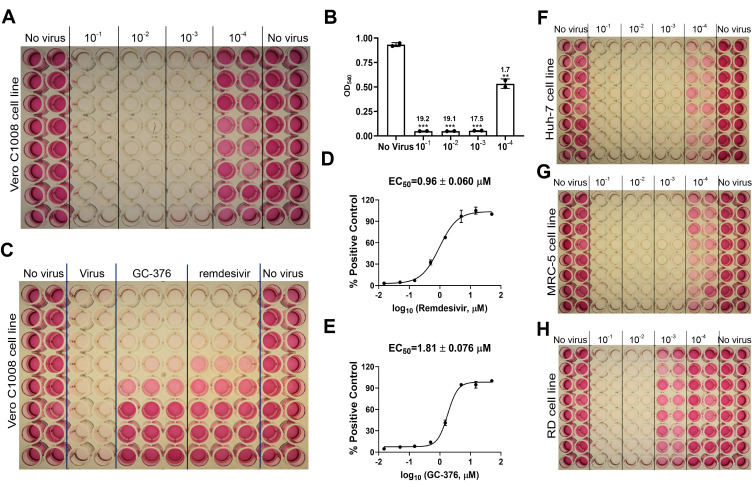
Figure 4. CPE assay for HCoV-NL63.
A. Titration of HCoV-NL63 in the Vero E6 cell line. B. Results of HCoV-NL63 titration in Vero E6 cell line in CPE assay. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test). The S/B ratios are labeled for each virus dilution. C. Determination of EC50 values for GC-376 and remdesivir against HCoV-NL63 in the CPE assay. D. EC50 curve fitting for remdesivir obtained in GraphPad Prism 8. E. EC50 curve fitting for GC-376 obtained in GraphPad Prism 8. F. Titration of the HCoV-NL63 in Huh-7 cell line; G. Titration of HCoV-NL63 in MRC-5 cell line. H. Titration of HCoV-NL63 in RD cell line. The images are representatives of three independent repeats.
Figure 5. CPE assay for HCoV-OC43.
A. Titration of HCoV-OC43 in the RD cell line. B. Results of HCoV-OC43 titration in RD cell line in CPE assay. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test). The S/B ratios are labeled for each virus dilution. C. Determination of EC50 values for GC-376 and remdesivir against HCoV-OC43 in CPE assay; D. EC50 curve fitting for remdesivir obtained in Prism 8. E. EC50 curve fitting for GC-376 obtained in Prism 8. F. Titration of HCoV-OC43 in the MRC-5 cell line. G. Titration of HCoV-OC43 in the BSC-1 cell line. The images are representatives of three independent repeats.
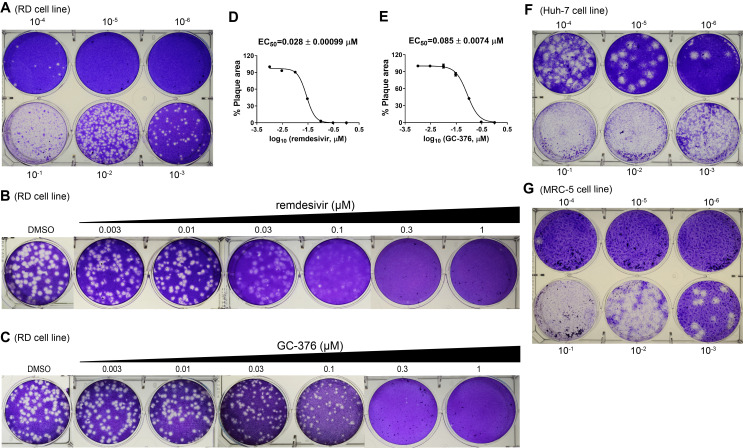
Figure 6. Plaque reduction assay for HCoV-229E.
A. Titration of HCoV-229E in the RD cell line. B. Determination of EC50 value for remdesivir against HCoV-229E in plaque assay. C. Determination of EC50 value for GC-376 against HcoV-229E in plaque assay. D. EC50 curve fitting for remdesivir obtained in GraphPad Prism 8. E. EC50 curve fitting for GC-376 obtained in GraphPad Prism 8. F. Titration of HcoV-229E in the Huh-7 cell line. G. Titration of HCoV-229E in the MRC-5 cell line. The images are representatives of three independent repeats.
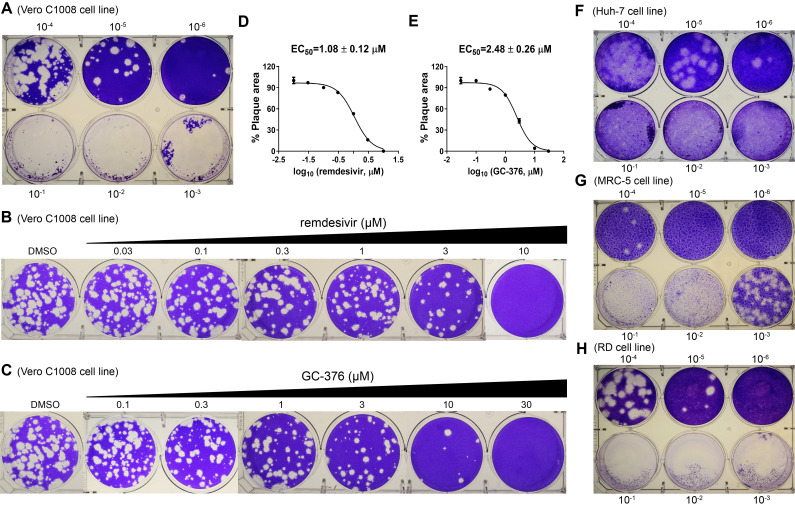
Figure 7. Plaque reduction assay for HCoV-NL63.
A. Titration of HCoV-NL63 in the RD cell line. B. Determination of EC50 value for remdesivir against HCoV-NL63 in plaque assay. C. Determination of EC50 value for GC-376 against HCoV-NL63 in plaque assay. D. EC50 curve fitting for remdesivir obtained in GraphPad Prism 8. E. EC50 curve fitting for GC-376 obtained in GraphPad Prism 8. F. Titration of HCoV-NL63 in the Huh-7 cell line. G. Titration of HCoV-NL63 in the MRC-5 cell line. H. Titration of HCoV-NL63 in the RD cell line. The images are representatives of three independent repeats.
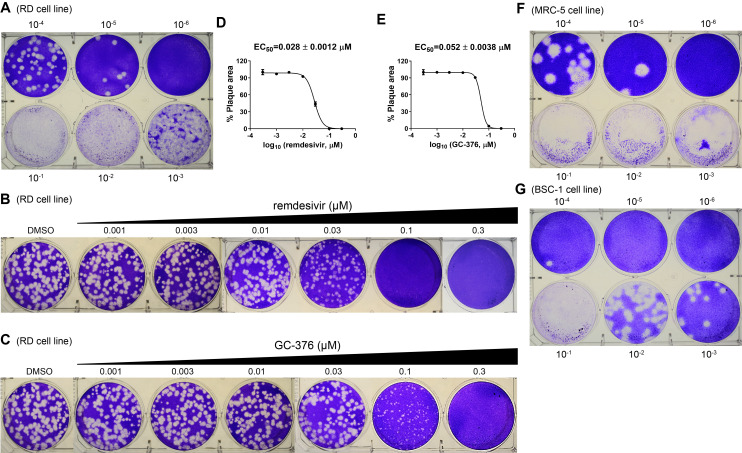
Figure 8. Plaque reduction assay for HCoV-OC43.
A. Titration of HCoV-OC43 in the RD cell line. B. Determination of EC50 value for remdesivir against HCoV-OC43 in plaque assay. C. Determination of EC50 value for GC-376 against HCoV-OC43 in plaque assay. D. EC50 curve fitting for remdesivir obtained in GraphPad Prism 8. E. EC50 curve fitting for GC-376 obtained in GraphPad Prism 8. F. Titration of HCoV-OC43 in the MRC-5 cell line. G. Titration of HCoV-OC43 in the BSC-1 cell line. The images are representatives of three independent repeats.
Data analysis
-
For virus titration in the CPE assay
Take the average reading of each group: No virus, 10-1, 10-2, 10-3, and 10-4 dilutions. Plot the data in GraphPad Prism 8 and calculate the signal to background ratio (S/B) for each virus dilution group by dividing the average reading of the No virus group by the average reading of each virus dilution group (Figures 3C, 4C, 5C). Choose the virus dilution with the lowest virus amount that gives S/B > 5 for compound testing.
Note: Omit the reading of the outer wells of the plate (wells in row A and row H, columns 1 and 12) and only use data in the inner wells for data processing. The data in the outer wells are not reliable due to the higher rate of evaporation causing inconsistent cell growth compared to the inner wells. An alternative way is to fill the outer wells with PBS or media only when setting up the plates during cell seeding.
-
For testing inhibitors in the CPE assay
In GraphPad Prim 8, create ‘New table & graph’, select ‘XY’, and on ‘options’, enter ‘3’ replicate values for Y. Input log (concentration of testing compound) in column X, and the three replicate values of absorbance at 540 nm corresponding to each concentration of the test compound in column Y. Click on ‘Analyze’ button, select ‘Normalize’, enter the average value of absorbance at 540 nm of the ‘Virus’ group as 0% and the average value of absorbance at 540 nm of the ‘No virus’ group as 100%. The EC50s of test compounds were determined by fitting the curves with nonlinear regression using log (inhibitor) vs response with variable slopes (Figures 3D, 3E; 4D, 4E; 5D, 5E).
Note: Only use cells in the inner wells for data processing.
-
For virus titration in the plaque assay
Calculation of virus titer: The virus titer will be calculated by taking the average of the last two wells which show countable plaques. For the titration result of HCoV-229E (Figure 6A), the 10-4 well had 12 plaques, so the titer is 12 × 104 PFU/0.5 mL, which corresponds to 2.4 × 105 PFU/mL; the 10-3 well had 80 plaques, so the titer is 80 × 103 PFU/0.5 mL, which corresponds to 1.6 × 105 PFU/mL. The final titer will be (2.4 × 105 PFU/mL + 1.6 × 105 PFU/mL)/2 = 2 × 105 PFU/mL. For the titration result of HCoV-NL63 (Figure 7A), the 10-6 well had 4 plaques, so the titer is 4 × 106 PFU/0.5 mL, which corresponds to 8 × 106 PFU/mL; the 10-5 well had 32 plaques, so the titer is 32 × 105 PFU/0.5 mL, which corresponds to 6.4 × 106 PFU/mL. The final titer will be (8 × 106 PFU/mL + 6.4 × 106 PFU/mL)/2 = 7.2 × 106 PFU/mL. For the titration result of HCoV-OC43 (Figure 8A), the 10-5 well had 8 plaques, so the titer is 8 × 105 PFU/0.5 mL, which corresponds to 1.6 × 106 PFU/mL; the 10-4 well had 58 plaques, so the titer is 58 × 104 PFU/0.5 mL, which corresponds to 1.16 × 106 PFU/mL. The final titer will be (1.6 × 106 PFU/mL + 1.16 × 106 PFU/mL)/2 = 1.38 × 106 PFU/mL. The results of HCoV-229E, HCoV-NL63, and HCoV-OC43 titer calculation are summarized in Table 3.
-
For testing inhibitors in the plaque assay
Quantify the plaque area in each well of the 6-well plate in ImageJ (a step by step detailed instruction for how to measure area percentage using ImageJ can be found at this website: https://cs.appstate.edu/ret/imageJ/PClabs/imlab/measure/pctarea.html). In GraphPad Prim 8, create ‘New table & graph’, select ‘XY’, and on ‘options’, enter ‘2’ replicate values for Y. Input log (concentration of testing compound) in column X, and the two replicate values of plaque area quantified in ImageJ corresponding to each concentration of the test compound in column Y. Click on ‘Analyze’ button, select ‘Normalize’, enter 0 as 0% and the average value of plaque area of the ‘DMSO’ wells as 100%. The EC50s of test compounds were determined by fitting the curves with nonlinear regression using log (inhibitor) vs response with variable slopes (Figures 3D, 3E; 4D, 4E; 5D, and 5E).
Note: Plaque area was used for quantification because it provides more accurate results, as plaques formed from the same virus infection are not uniform and vary in size. Also, in the plaque assay, some compounds show antiviral activity by reducing the sizes of the plaques at certain lower concentrations and completely inhibiting plaque formation at higher concentrations.
Table 3. Titer calculation for HCoV-229E, HCoV-NL63, and HCoV-OC43.
| Virus | Plaque assay titration results | Plaque counting | Viral titer (PFU/mL) |
| HCoV-229E | Figure 6A | 10-4 well: 12 | (12×104/0.5+80×103/0.5)/2=2×105 |
| 10-3 well: 80 | |||
| HCoV-NL63 | Figure 7A | 10-6 well: 4 | (4×106/0.5+32×105/0.5)/2=7.2×106 |
| 10-5 well: 32 | |||
| HCoV-OC43 | Figure 8A | 10-5 well: 8 | (8×105/0.5+58×104/0.5)/2=1.38×106 |
| 10-4 well: 58 |
Notes
Troubleshooting
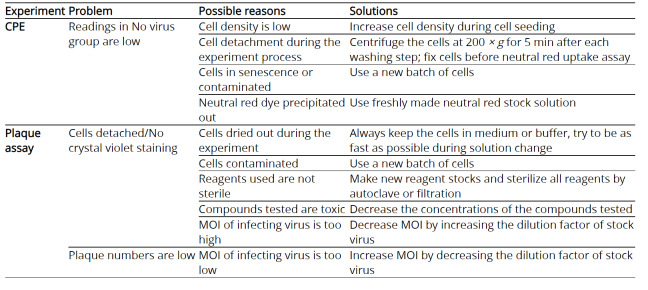
The trouble shooting advice can be found in Table 4.
Table 4. Troubleshooting information for CPE and plaque assay.
|
Recipes
-
10× PBS stock
Dissolve 17.8 g Na2HPO4·2H2O, 2.4 g KH2PO4, 80 g NaCl and 2 g KCl in 1 L of nano pure H2O. Autoclave and store at 4°C for up to 2 months.
-
100× MgCl2 stock
Dissolve 10.165 g MgCl2·6H2O in 1 L of nano pure H2O. Filter with 0.2 µm vacuum-driven filter and store at 4°C for up to 2 months.
-
100× CaCl2 stock
Dissolve 9.989 g CaCl2 in 1 L of nano pure H2O. Filter with 0.2 µm vacuum-driven filter and store at 4°C for up to 2 months.
-
DPBS, calcium and magnesium-free
Add 100 mL of the 10× PBS stock into 900 mL of nano pure water, autoclave and store at room temperature (20-30°C) for up to 2 months.
-
PBS, with calcium and magnesium
For 1 L, add 100 mL of the 10× PBS stock, 10 mL of 100× MgCl2 stock, 10 mL of 100× CaCl2 stock into 880 mL of nano pure water. Filter with 0.2 µm vacuum-driven filter and store at room temperature (20-30°C) for up to 2 months.
-
Neutral red stock solution (4 mg/mL)
Weigh 400 mg neutral red powder, dissolve in 100 mL of DPBS by stirring for 2 h, and filter the solution with 0.2 µm vacuum-driven filter. Protect the solution from light with foil and store at room temperature (20-30°C), for up to 2 months.
-
Complete culture medium (culture medium plus 10% FBS and 1%P/S, vol/vol)
For 500 mL of DMEM, add 50 mL of FBS and 5 mL of P/S. Mix well and store at 4°C for up to 2 weeks.
-
Neutral red working solution (50 µg/mL)
Dilute 1:80 of the neutral red stock solution with DMEM in centrifuge tubes under sterile conditions. For example, mix 150 µL of neutral red stock solution with 11.85 mL of DMEM for one 96-well plate. Incubate overnight in the incubator where cells are cultured.
Note: Neutral red working solution should be prepared one day before use and incubated in the CO2 cell culture incubator overnight.
-
Neutral red de-staining solution (50% ethanol, 49% deionized water, 1% glacial acetic acid, vol/vol)
Add 500 mL of ethanol (96%), 490 mL of nano pure water, 10 mL of glacial acetic acid, mix well by stirring for 15 min, and store at room temperature (20-30°C), for up to 2 months.
-
2× DMEM
Dissolve 2 packets of DMEM powder and 7.4 g NaHCO3 into 1 L of nano pure H2O, add 10 mL 1 M of HEPES buffer (pH7.2), and 20 mL of 100× P/S. Adjust pH to ~7.2 with concentrated HCl. Filter with 0.2 µm vacuum-driven filter and store at 4°C for up to 2 weeks.
-
1.2% avicel (wt/vol)
Weigh 12 g Avicel powder and add into 1 L of nano pure H2O, mix by stirring for 1-2 h at room temperature. Autoclave and store at room temperature (20-30°C), for up to 3 months.
-
0.2% crystal violet (wt/vol)
Weigh 2 g crystal violet, add into 200 mL of methanol and mix to completely dissolve the crystal violet. Add 800 mL of nano pure water, mix well and store at room temperature (20-30°C), for up to 6 months.
Acknowledgments
JW was supported by the National Institute of Allergy and Infectious Diseases of Health (NIH) (grants AI147325, AI157046, and AI158775) and the Arizona Biomedical Research Commission Centre Young Investigator grant (ADHS18-198859). YH was supported by the NIH training grant T32 GM008804.
Competing interests
The authors declare no competing interests.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
Q&A
Post your question about this protocol in Q&A and get help from the authors of the protocol and some of its users.
References
- 1. Bracci N., Pan H. C., Lehman C., Kehn-Hall K. and Lin S. C.(2020). Improved plaque assay for human coronaviruses 229E and OC43. PeerJ 8: e10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brussaard C. P., Marie D. and Bratbak G.(2000). Flow cytometric detection of viruses. J Virol Methods 85(1-2): 175-182. [DOI] [PubMed] [Google Scholar]
- 3. Collier D. A., De Marco A., Ferreira I., Meng B., Datir R., Walls A. C., Kemp S. S., Bassi J., Pinto D., Fregni C. S., et al.(2021). Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 593(7857): 136-141. [DOI] [PubMed] [Google Scholar]
- 4. Corman V. M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D. K., Bleicker T., Brunink S., Schneider J., Schmidt M. L., et al.(2020). Detection of 2019 novel coronavirus(2019-nCoV) by real-time RT-PCR. Euro Surveill 25(3): 2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gagneur A., Sizun J., Vallet S., Legr M. C., Picard B. and Talbot P. J.(2002). Coronavirus-related nosocomial viral respiratory infections in a neonatal and paediatric intensive care unit: a prospective study. J Hosp Infect 51(1): 59-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gerna G., Cattaneo E., Cereda P. M., Revelo M. G. and Achilli G.(1980). Human coronavirus OC43 serum inhibitor and neutralizing antibody by a new plaque-reduction assay. Proc Soc Exp Biol Med 163(3): 360-366. [DOI] [PubMed] [Google Scholar]
- 7. Herzog P., Drosten C. and Muller M. A.(2008). Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol J 5: 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hirose R., Watanabe N., Bandou R., Yoshida T., Daidoji T., Naito Y., Itoh Y. and Nakaya T.(2021). A Cytopathic Effect-Based Tissue Culture Method for HCoV-OC43 Titration Using TMPRSS2-Expressing VeroE6 Cells. mSphere 6(3): e00159-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B. and Pohlmann S.(2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A 102(22): 7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu Y., Hau R. K., Wang Y., Tuohy P., Zhang Y., Xu S., Ma C. and Wang J.(2018). Structure-Property Relationship Studies of Influenza A Virus AM2-S31N Proton Channel Blockers. ACS Med Chem Lett 9(11): 1111-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu Y., Kitamura N., Musharrafieh R. and Wang J.(2021). Discovery of Potent and Broad-Spectrum Pyrazolopyridine-Containing Antivirals against Enteroviruses D68, A71, and Coxsackievirus B3 by Targeting the Viral 2C Protein. J Med Chem 64(12): 8755-8774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y., Ma C., Szeto T., Hurst B., Tarbet B. and Wang J.(2021). Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses. ACS Infect Dis 7(3): 586-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hu Y., Musharrafieh R., Ma C., Zhang J., Smee D. F., DeGrado W. F. and Wang J.(2017). An M2-V27A channel blocker demonstrates potent in vitro and in vivo antiviral activities against amantadine-sensitive and-resistant influenza A viruses . Antiviral Res 140: 45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu Y., Wang Y., Li F., Ma C. and Wang J.(2017). Design and expeditious synthesis of organosilanes as potent antivirals targeting multidrug-resistant influenza A viruses. Eur J Med Chem 135: 70-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu Y., Zhang J., Musharrafieh R., Hau R., Ma C. and Wang J.(2017). Chemical Genomics Approach Leads to the Identification of Hesperadin, an Aurora B Kinase Inhibitor, as a Broad-Spectrum Influenza Antiviral. Int J Mol Sci 18(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hu Y., Zhang J., Musharrafieh R. G., Ma C., Hau R. and Wang J.(2017). Discovery of dapivirine, a nonnucleoside HIV-1 reverse transcriptase inhibitor, as a broad-spectrum antiviral against both influenza A and B viruses. Antiviral Res 145: 103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hui D. S., E I. A., Madani T. A., Ntoumi F., Kock R., Dar O., Ippolito G., McHugh T. D., Memish Z. A., Drosten C., et al.(2020). The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health- The latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis 91: 264-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hulswit R. J. G., Lang Y., Bakkers M. J. G., Li W., Li Z., Schouten A., Ophorst B., van Kuppeveld F. J. M., Boons G. J., Bosch B. J., et al.(2019). Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A. Proc Natl Acad Sci U S A 116(7): 2681-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li F., Hu Y., Wang Y., Ma C. and Wang J.(2017). Expeditious Lead Optimization of Isoxazole-Containing Influenza A Virus M2-S31N Inhibitors Using the Suzuki-Miyaura Cross-Coupling Reaction. J Med Chem 60(4): 1580-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li F., Ma C., Hu Y., Wang Y. and Wang J.(2016). Discovery of Potent Antivirals against Amantadine-Resistant Influenza A Viruses by Targeting the M2-S31N Proton Channel. ACS Infect Dis 2(10): 726-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z., Tomlinson A. C., Wong A. H., Zhou D., Desforges M., Talbot P. J., Benlekbir S., Rubinstein J. L. and Rini J. M.(2019). The human coronavirus HCoV-229E S-protein structure and receptor binding. Elife 8: e51230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu I. J., Chen P. J., Yeh S. H., Chiang Y. P., Huang L. M., Chang M. F., Chen S. Y., Yang P. C., Chang S. C., Wang W. K. et al.(2005). Immunofluorescence assay for detection of the nucleocapsid antigen of the severe acute respiratory syndrome(SARS)-associated coronavirus in cells derived from throat wash samples of patients with SARS. J Clin Microbiol 43(5): 2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., et al.(2021). Effectiveness of Covid-19 Vaccines against the B.1.617.2(Delta) Variant. N Engl J Med 385(7): 585-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma C., Hu Y., Townsend J. A., Lagarias P. I., Marty M. T., Kolocouris A. and Wang J.(2020). Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors. ACS Pharmacol Transl Sci 3(6): 1265-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ma C., Hu Y., Zhang J., Musharrafieh R. and Wang J.(2019). A Novel Capsid Binding Inhibitor Displays Potent Antiviral Activity against Enterovirus D68. ACS Infect Dis 5(11): 1952-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ma C., Hu Y., Zhang J. and Wang J.(2020). Pharmacological Characterization of the Mechanism of Action of R523062, a Promising Antiviral for Enterovirus D68. ACS Infect Dis 6(8): 2260-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma C., Sacco M. D., Hurst B., Townsend J. A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M. T., Chen Y., et al.(2020). Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res 30(8): 678-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mesel-Lemoine M., Millet J., Vidalain P. O., Law H., Vabret A., Lorin V., Escriou N., Albert M. L., Nal B. and Tangy F.(2012). A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J Virol 86(14): 7577-7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Musharrafieh R., Kitamura N., Hu Y. and Wang J.(2020). Development of broad-spectrum enterovirus antivirals based on quinoline scaffold. Bioorg Chem 101: 103981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Musharrafieh R., Ma C., Zhang J., Hu Y., Diesing J. M., Marty M. T. and Wang J.(2019). Validating Enterovirus D68-2Apro as an Antiviral Drug Target and the Discovery of Telaprevir as a Potent D68-2Apro Inhibitor . J Virol 93(7): e02221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Musharrafieh R., Zhang J., Tuohy P., Kitamura N., Bellampalli S. S., Hu Y., Khanna R. and Wang J.(2019). Discovery of Quinoline Analogues as Potent Antivirals against Enterovirus D68(EV-D68). J Med Chem 62(8): 4074-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nadgir S. V., Hensler H. R., Knowlton E. R., Rinaldo C. R., Rappocciolo G. and Jenkins F. J.(2013). Fifty percent tissue culture infective dose assay for determining the titer of infectious human herpesvirus 8. J Clin Microbiol 51(6): 1931-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Payne A. F., Binduga-Gajewska I., Kauffman E. B. and Kramer L. D.(2006). Quantitation of flaviviruses by fluorescent focus assay. J Virol Methods 134(1-2): 183-189. [DOI] [PubMed] [Google Scholar]
- 34. Ratnam S., Gadag V., West R., Burris J., Oates E., Stead F. and Bouilianne N.(1995). Comparison of commercial enzyme immunoassay kits with plaque reduction neutralization test for detection of measles virus antibody. J Clin Microbiol 33(4): 811-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roingeard P.(2008). Viral detection by electron microscopy: past, present and future. Biol Cell 100(8): 491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schirtzinger E. E., Kim Y. and Davis A. S.(2021). Improving human coronavirus OC43(HCoV-OC43) research comparability in studies using HCoV-OC43 as a surrogate for SARS-CoV-2. J Virol Methods 299: 114317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmidt O. W., Cooney M. K. and Kenny G. E.(1979). Plaque assay and improved yield of human coronaviruses in a human rhabdomyosarcoma cell line. J Clin Microbiol 9(6): 722-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shang Y., Li H. and Zhang R.(2021). Effects of Pandemic Outbreak on Economies: Evidence From Business History Context. Front Public Health 9: 632043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Smail S. W., Saeed M., Twana A., Khudhur Z. O., Younus D. A., Rajab M. F., Abdulahad W. H., Hussain H. I., Niaz K. and Safdar M.(2021). Inflammation, immunity and potential target therapy of SARS-COV-2: A total scale analysis review. Food Chem Toxicol 150: 112087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang Y., Hu Y., Xu S., Zhang Y., Musharrafieh R., Hau R. K., Ma C. and Wang J.(2018). In Vitro Pharmacokinetic Optimizations of AM2-S31N Channel Blockers Led to the Discovery of Slow-Binding Inhibitors with Potent Antiviral Activity against Drug-Resistant Influenza A Viruses . J Med Chem 61(3): 1074-1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., et al.(2020). Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 395(10236): 1569-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Williams T. C. and Burgers W. A.(2021). SARS-CoV-2 evolution and vaccines: cause for concern? Lancet Respir Med 9(4): 333-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu K., Li W., Peng G. and Li F.(2009). Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci U S A 106(47): 19970-19974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xia Z., Sacco M., Hu Y., Ma C., Meng X., Zhang F., Szeto T., Xiang Y., Chen Y. and Wang J.(2021). Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir. ACS Pharmacol Transl Sci 4(4): 1408-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yeager C. L., Ashmun R. A., Williams R. K., Cardellichio C. B., Shapiro L. H., Look A. T. and Holmes K. V.(1992). Human aminopeptidase N is a receptor for human coronavirus 229E. Nature 357(6377): 420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang J., Hu Y., Foley C., Wang Y., Musharrafieh R., Xu S., Zhang Y., Ma C., Hulme C. and Wang J.(2018). Exploring Ugi-Azide Four-Component Reaction Products for Broad-Spectrum Influenza Antivirals with a High Genetic Barrier to Drug Resistance. Sci Rep 8(1): 4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang J., Hu Y., Hau R., Musharrafieh R., Ma C., Zhou X., Chen Y. and Wang J.(2019). Identification of NMS-873, an allosteric and specific p97 inhibitor, as a broad antiviral against both influenza A and B viruses. Eur J Pharm Sci 133: 86-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang J., Hu Y., Wu N. and Wang J.(2020). Discovery of Influenza Polymerase PA-PB1 Interaction Inhibitors Using an In Vitro Split-Luciferase Complementation-Based Assay . ACS Chem Biol 15(1): 74-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zumla A., Hui D. S. and Perlman S.(2015). Middle East respiratory syndrome. Lancet 386(9997): 995-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]