Abstract
Colorectal cancer (CRC) is the most common malignancy of the gastrointestinal tract. The stroma and the tumoral microenvironment (TME) represent ecosystem-like biological networks and are new frontiers in CRC. The present study demonstrates the use of a novel machine learning-based superpixel approach for whole slide images to unravel this biology. Findings of significance include the association of low proportionated stromal area, high immature stromal percentage, and high myxoid stromal ratio (MSR) with worse prognostic outcomes in CRC. Overall, stromal computational markers outperformed all others at predicting clinical outcomes. MSR may be able to prognosticate patients independent of pathological stage, representing an optimal way to effectively prognosticate CRC patients which circumvents the need for more extensive molecular and/or computational profiling. The superpixel approaches to the TME demonstrated here can be performed by a trained pathologist and recorded during synoptic cancer reporting with appropriate quality assurance. Future clinical trials will have the ultimate say in determining whether we can better tailor the need for adjuvant therapy in patients with CRC.
Keywords: Superpixel Segmentation, Tumoral Microenvironment, Colorectal Cancer, Frontiers Beyond the Microscope, Stromal Differentiation
Introduction
Colorectal cancer (CRC) is diverse and notable for being genetically inherited, although mostly it occurs sporadically.1 Today, paradigms in CRC are transitioning from the tumor to the tumor microenvironment (TME). Approaches that encompass the TME could be incorporated with other clinical, molecular, and computational findings in order to carve out a more precision-based patient care. The TME represents an ecosystem-like biological network that regulates both the extracellular matrix (ECM) and immune cell functioning.2
Challenges exist in the prognostication and management of patients with CRC, often secondary to difficulties in applying existing guidelines and adopting newer concepts.3 Previous work has been undertaken to understand the role of stromal differentiation (SD)4 and tumor budding (TB)5 in CRC. SD has been most extensively studied in CRC, although also demonstrated in cancers of the breast,6 cervix,7 and stomach.8 While the significance and reproducibility of TB in a true clinical environment is also a matter of concern to some.9 Overall, immature SD and high TB are seen as poor signatures, demonstrated to be associated with higher pathological stage and poor prognostic outcomes.10 Myxoid and immature are synonymous descriptors for this type of stromata, as immature stroma contains by definition at least a 40× field of myxoid change.4
The tumor–stroma-ratio (TSR) has also been examined extensively in CRC.11 It represents the ratio of tumor to stroma and is measured manually by a pathologist via conventional light microscopy (CLM). Questions regarding manual assessment and reproducibility are concerning, and traditionally, the TSR is performed only in the invasive front of the tumor and does not measure tumor-associated stroma in its entirety. Despite this, the TSR is seen as a promising prognostic tool, and tumors that harbor significant amounts of stroma have been shown to have worse prognostic outcomes, suggesting a role for tumor-node-metastasis classification.11
Current trends are pushing for more computation-based approaches as we enter the digital era of pathology. Most recently, Guedj et al.12 evaluated the prognosticating potential of digital image analysis (DIA) for quantifying the stromal compartments of intrahepatic cholangiocarcinoma. More specifically, DIA was used to calculate a proportionated stromal area (PSA), defined by the stroma to tumor area ratio, and low values were found to be associated with worse outcomes.
In breast cancer, DIA has found triple-negative biomarker status and high stroma to be associated with a poor prognosis, whereas in luminal tumors, high stroma was associated with favorable prognostic outcomes.13 The foregoing suggests that the significance of stroma may be tumoral subtype dependent in cancer, justifying the importance of validating DIA in CRC.
Previous publications did not differentiate tumor-associated stromata based on myxoid differentiation. It may be valuable to not only look at the stroma quantitatively, but to also look at the stroma qualitatively by assessing the degree of immature, myxoid differentiation.
Building on PSA,12 here we propose the significance of immature stromal percentage (ISP), defined as the absolute percentage of immature, myxoid stroma; and myxoid stromal ratio (MSR), defined as the ratio of myxoid to the collagenous stroma. Tumors with significant immature (myxoid) differentiation may have worse survival outcomes, and if so, then both the quality and quantity of the stroma in CRC are important.
Current paradigms are shifting away from manual qualitative assessment and towards quantitative scoring.14 Approaches demonstrated here can calculate tumor–stroma ratios, while also quantifying the proportions of different subtypes of tumor stromata. With so many open-access digital pathology solutions, more opportunities are becoming available for transitioning to computational methodologies. The tumoral microenvironment harbors significant biological diversity,15 and the present study will use superpixel methods for whole slide images (WSIs) to unravel this biology.
Materials and Methods
Institutional Review Board
This present study received approval from the Institutional Review Board (IRB) of the Human Research Protection Program licensing committee at Northwell Health (IRB number: 18-0128). Research methods followed all relevant guidelines and regulations, and no patient consent was required by the review board for this study.
Study Design
This was a retrospective study of colorectal carcinoma patients diagnosed in our health system (2014–2017). Case selection and retrieval took place from a larger cohort of 226 CRC patients which were published in a previous study,16 60 of which were selected at random. Cases were only included if they contained completed synoptic summaries and appropriately documented AJCC pathological staging information. Patients diagnosed as stage Tis stage were also excluded due to a lack of representative desmoplastic stroma. Cases with insufficient clinical information, follow-up, and tissue specimen availability were also excluded. We did not deploy any other methods of stratification or matching by age or anatomical extent of disease. For DIA, only one block was selected by a pathologist (SH) for each case, relating to the slide containing the largest portion of tumor. Additional quality assurance steps included slide reviewal for any folded, blurred, or obstructed morphologies. Slides were also evaluated for tumor budding, tumor-infiltrating lymphocytes (TIL), and stromal differentiation. Histological scoring required consensus between two observers (SH and MN). Additional clinicopathological data were retrieved from the electronic medical records for each case and cancer-free survival (CFS) data were provided by the cancer registry of Northwell Health. We included the following variables for exploratory analyses: CFS, age, gender, tumor budding score, pathological stage, TIL, lymph node stage, tumor grade, stromal differentiation, mismatch repair (MMR) status, Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (KRAS) and B-Raf Proto-Oncogene (BRAF).
Slide Digitalization
Histological features were assessed using virtual slides scanned at 20× on the Leica Aperio AT2 (Leica Biosystems, Buffalo Grove, IL) whole slide scanner. WSIs were viewed at up to × 400 magnification with ImageScope, ver. 12.3.2.8013, Leica Microsystems. All digital slides were reviewed for the quality following scanning and any out-of-focus areas required rescanning. All digital slides were downloaded in TIFF image format with variable degrees of JPEG image compression. WSI’s were deidentified and labeled with a sticker as 1–60. All WSI data transfer and storage were still undertaken in a Health Insurance Portability and Accountability Act of 1996 compliant manner.
Superpixel Image Segmentation
We incorporated QuPath,17 an open-source software for WSI analysis in order to evaluate the tumoral microenvironment. This was performed on QuPath version 0.2.1. Qupath comes built with adaptable and trainable machine learning (ML) algorithms. We chose to use superpixel image segmentation (SIS) in our study to quantify both stromal (myxoid and collagenous) and tumoral components. All WSI files and were imported and orientated appropriately. An artificial neural (DL) network was developed and trained from annotated patches selected from the 60 patients. No definitive training, testing, and validation cohorts were used in this study. Patches were selected to represent the tissue heterogeneity present in the data set and efforts were made to account for other staining and technical imperfections. Tumor was defined as cancer epithelial cells well to poorly differentiated and excluded benign colonic crypts. Myxoid stroma was defined as basophilic to gray extracellular matrix material. Collagen was defined as eosinophilic mature collagen fibers, stratified into multiple fibrous bands at tumoral front. Ignore patches covered a wide spectrum of tissue from the WSIs. Area of fat, red blood cells, and necrosis were included into ignore patches as well as areas of the WSI not containing tissue. Quality control (QC) comparisons of SIS methods to the original WSI were compared manually by two observers (SH and DW).
Following training, superpixels were automatically grouped based on pixel similarities between different labeled cellular populations. In our study, this was demonstrated as red (tumor epithelium), yellow (collagenous stroma), and blue (myxoid stroma). The entire area of the tumor and associated stroma was selected for annotation from an entire WSI as shown in Fig. 1. We classified the PSA as performed by Guedj et al.,12 defined by stromal to tumor area ratio. Secondly, we developed the theoretical concepts of ISP and MSR for the purpose of this study. ISP is defined as the absolute percentage of immature, myxoid stroma, whereas MSR is defined as the ratio of myxoid to collagenous stroma.
Fig. 1.
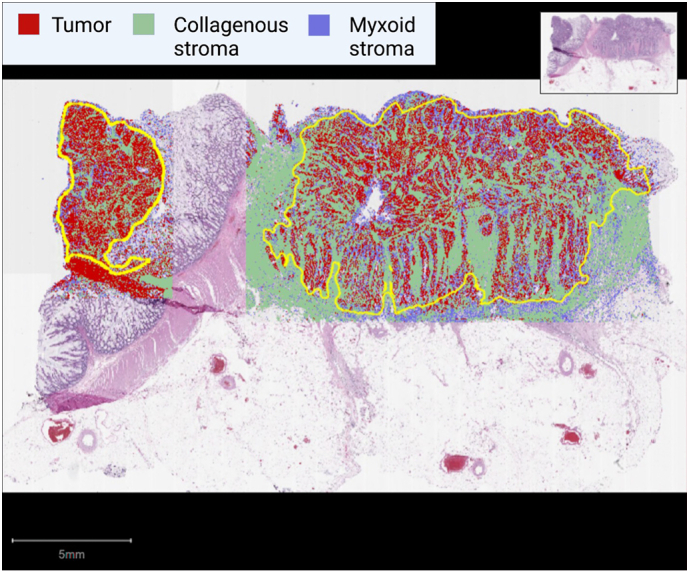
Representative virtual slide analyzed by QuPath. Tumor and associated stromal portions are delineated (yellow) and super-pixelated for tumor (red), collagenous stroma (green), and myxoid stroma (blue). Only areas selected for annotations area included into component percentages.
Tumor Budding
For tumor budding assessment, we assessed “hotspot” regions of tumor with the highest grade of tumor budding. To find hotspot regions, we scanned at least 10 individual fields at medium power (10× objective) at the invasive front for each case. The counting of tumor buds was performed in the hotspot region (20× objective lens) according to the 3-tier grading system proposed by the International Tumor Budding Consensus Conference.18 The grading was as follows: Bd1: 0–4 buds, Bd2: 5–9 buds, and Bd3: >10 buds.
Tumor-Infiltrating Lymphocytes
TILs were defined as small blue mononuclear cells which infiltrating between tumor cells. Tumors were assessed with a 4-tier scale at the deepest point of the invasive tumor. This was previously validated for the quantification of inflammatory in CRC by Klintrup et al.19 A score of 0 with no inflammatory cells, 1 with mild patchy increase in mononuclear cells, 2 with a moderate (bandlike), and 3 with florid (cuplike) inflammation. Scores 2 and 3 often are seen in conjunction with the destruction of cancer cell islands. Scoring was classified as low grade (0–1) and high grade (2–3).
Stromal Differentiation
Scoring of SD was primarily based on 3-tier system proposed by Ueno et al.20 The ECM was analyzed at the extramural desmoplastic front initially at low magnification (4×). Myxoid stroma was defined as an amorphous stromal substance made of amphophilic material with a basophilic to gray extracellular matrix. This was usually intermingled with randomly oriented hyalinized collagen. Stroma was regarded as immature when at least a 40× field of myxoid change was observed. Intermediate stroma was defined as keloid-like collagen, thick, and hypocellular collagen bundles with bright eosinophilic hyalinization and a minimum width of 20-μM.21 We categorized stroma as mature when the fibrotic stroma did not contain myxoid or keloid type stroma and was predominantly comprised of fine and mature, stratified collagen fibers.
Mismatch Repair Status
Immunohistochemistry was performed in order to determine MMR status on formalin-fixed and paraffin-embedded tumor sections cut at 4-μM thickness and stained on a Ventana Bench Mark Autostainer (Ventana Medical System, Tucson, AZ). The following rabbit monoclonal pre-diluted primary antibodies were used: MLH-1(M1, Ventana), PMS2 (EPR 3947, Ventana), MSH2(G219-1129, Ventana), and MSH6 (44, Ventana). Technical methodologies and quality assurance were undertaken by certified histotechnologists at the immunohistochemical laboratory of Long Island Jewish Medical Center (Northwell Health System, New Hyde Park, NY). Cases showing loss of immunohistochemical staining (<1%) for any of the following stains: MLH1, PMS2, MSH2, and MSH6 were considered MMR-deficient.
Next Generation Genomic Sequencing
Genomic alterations were tested by next generation sequencing from formalin-fixed, paraffin-embedded tissue at Genpath laboratories (Elwood Park, NJ). Nucleic acid was extracted with a non-degraded concentration greater than 1 ng/μL, which was subjected to PCR-based amplification. Both coding and non-coding regions of the selected genes were enriched and sequenced on the Illumina MiSeq instrument (San Diego, CA) with paired ends and 175 base pair reads. The mapping of the read data was based on reference build GRCh37/hg19. Single nucleotide variants, insertions, and deletions were identified on a customizable bioinformatics platform with an allele frequency greater than 5%.
Statistical Analysis
Comparative analysis was performed using the non-paired t-test to examine differences in the clinicopathological profile. The Kaplan–Meier method was used to evaluate CFS rate as a function of time. The log-rank method was used to compare differences between groups. These statistical analysis was performed using Prism Graphpad version 8.4.2. Heatmaps, non-linear regression, and univariate and multivariate analyses of CFS using cox proportional-hazard regression were performed on SPSS statistics version 23.0.0.3. A p-value <0.05 was considered statistically significant.
Results
Clinicopathological and Patient Characteristics
This study comprised data from 60 patients with colorectal adenocarcinoma who underwent surgical resection at our health system. Surgeries include block resection (1), right hemicolectomy (20), left hemicolectomy (27), transverse colectomy (5), and recto-sigmoidectomy (7). The mean age for our patient cohort was 67.4. There was a slight male predominance: 60% male (36), 40% female (24) to 51%. Tumor budding grades were as follows: absent or low (24), intermediate or high (36). Tumoral stage was as follows: pT1–2 (13), pT3–pT4 (47). For TIL, scoring was as follows: low (40) and high (20). Lymph node status was as follows: N0 (32), N1–2 (28). Tumor differentiation was as follows: well-differentiated (44), moderately to poorly differentiated (16). Stromal differentiation was as follows: 27 (45%) mature stroma, and 33 (55%) patients with immature/intermediate. Out of 60 patients, 46 (77%) had intact MMR, whereas 14 (23%) patients were MMR-deficient. Molecular testing was performed on a subset of patients: 9 (15%) had BRAF testing and 12 (20%) had KRAS mutation status.
Superpixel Image Segmentation
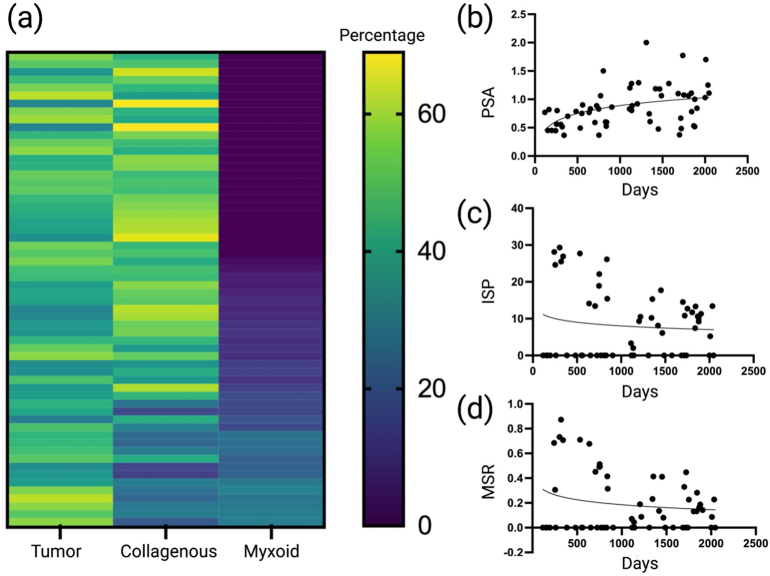
We found a spectrum of tumoral, non-myxoid, and myxoid stromal components by SIS. Heatmaps for percentages can be viewed in Fig. 2a, whereas non-linear regression curves can be seen in Fig. 2 for PSA (b), ISP (c), and MSR (d), respectively. Based on manual analysis, non-linear regression curves for CFS, cut-off vales were designated as 0.9 (PSA), 10% (ISP), and 0.19 (MSR).
Fig. 2.
Heatmaps and non-linear regression analyses. (a) Heatmap demonstrated the spectrum of tumoral, non-myxoid, and myoid stromal percentages. (b–d) Non-linear regression for cancer-free survival data and PSA (b), ISP (c), and MSR (d). PSA, Proportionated Stromal Area; ISP, Immature Stromal Percentage; MSR, Myxoid Stromal Ratio.
Means were calculated for PSA, ISP, and MSR in order to perform t-test analysis for the clinicopathological profile. Firstly, high tumor budding was found to be associated with a lower PSA (0.008). Pathological stage was found the be associated with a lower PSA (0.010). Stromal differentiation was found to be associated with high ISP (<0.001) and high MSR (<0.001). MMR deficiency was found to correlate with PSA (0.013), ISP (0.005), and MMR (0.022). The remaining variables were not significant. The results for t-test analyses for stromal computational signatures can be viewed in Table 1.
Table 1.
t-Test analysis for stromal computational signatures. PSA, Proportionated Stromal Area; ISP, Immature Stromal Percentage; Myxoid Stromal Ratio; TIL, Tumor Infiltrating Lymphocytes; MMR, Mismatch repair; KRAS, Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; BRAF, B-Raf Proto-Oncogene. Significant features (p≤0.05) are shown in bold.
| Ratio/Percentage |
PSA |
p value | ISP |
p value | MSR |
p value |
|---|---|---|---|---|---|---|
| Variable | Mean | Mean | Mean | |||
| Age | 0.081 | 0.070 | 0.077 | |||
| <=65 | 1.009 | 11.99 | 0.265 | |||
| >65 | 0.831 | 7.103 | 0.151 | |||
| Gender | 0.887 | 0.377 | 0.648 | |||
| Male | 0.905 | 7.947 | 0.201 | |||
| Female | 0.890 | 10.32 | 0.197 | |||
| Tumor budding | 0.008 | 0.086 | 0.055 | |||
| TBD1 | 1.052 | 6.160 | 0.120 | |||
| TBD2 or TBD3 | 0.793 | 10.72 | 0.242 | |||
| Pathological stage | 0.010 | 0.129 | 0.092 | |||
| ≤ PT2 | 1.121 | 5.236 | 0.097 | |||
| PT3–PT4 | 0.828 | 10.01 | 0.222 | |||
| TIL | 0.829 | 0.262 | 0.227 | |||
| Low | 0.887 | 7.668 | 0.189 | |||
| High | 0.912 | 10.83 | 0.201 | |||
| Lymph node sStage | 0.397 | 0.363 | 0.398 | |||
| N0 | 0.935 | 10.01 | 0.218 | |||
| N1–2 | 0.852 | 7.61 | 0.165 | |||
| Tumor grade | 0.635 | 0.246 | 0.192 | |||
| G1–2 | 0.911 | 7.977 | 0.168 | |||
| G3 | 0.858 | 11.42 | 0.261 | |||
| Stromal differentiation | 0.576 | <0.001 | <0.001 | |||
| Immature/intermediate | 0.871 | 15.91 | 0.347 | |||
| Mature | 0.927 | 0.322 | 0.004 | |||
| MMR | 0.013 | 0.005 | 0.022 | |||
| Intact | 0.962 | 7.183 | 0.154 | |||
| Deficient | 0.679 | 17.01 | 0.321 | |||
| KRAS | 0.712 | 0.951 | 0.983 | |||
| Wild type | 0.858 | 7.738 | 0.173 | |||
| Mutation | 0.779 | 7.350 | 0.170 | |||
| BRAF | 0.662 | 0.238 | 0.297 | |||
| Wild type | 0.958 | 9.075 | 0.187 | |||
| Mutation | 0.750 | 0.000 | 0.000 |
Cancer-Free Survival
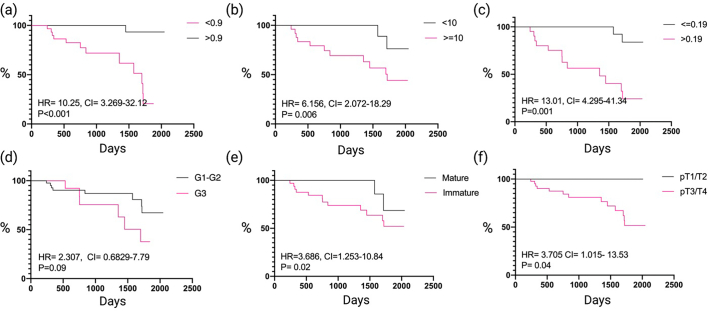
CFS data were collected for all the 60 patients with a mean follow-up time of 1415 days. After setting the positivity cutoff to 0.9 for PSA, 10% for ISP and 0.19 for MSR, we found that low PSA (HR: 10.25 (CI: 3.26–32.12), p= < 0.0001), ISP (HR: 6.156 (CI: 2.07–18.29), p=0.006) and MSR (HR: 13.01 (CI: 4.29–41.43), p=0.001), SD (HR: 3.66 (CI: 1.25–10.84), p=0.02) and tumoral stage (HR: 3.70 (CI: 1.01–13.53), p=0.04) to be associated with poor CFS on Kaplan–Meier survival analysis, tumor grade was not significant (HR: 2.30 (CI: 0.68–7.79), p=0.09) (Fig. 3).
Fig. 3.
Kaplan–Meier Survival Analysis. (a) Cancer-free survival between PSA low and high groups. (b) Cancer-free survival between ISP low and high groups. (c) Cancer-free survival between MSR low and high groups. (d) Cancer-free survival between mature and immature stroma groups. (e) Cancer-free survival between low (G1–2) and high grade (G3) groups. (f) Cancer-free survival between low (T1/2) and high (T3/4) tumoral stage. HR, Hazzard Ratio; CI, Confidence Interval; G, Grade; T, Stage.
Based upon cox-regression of CFS, pathological stage was found to be a poor prognostic factor on univariate analysis (HR: 3.705 (CI: 1.015–13.53), p=0.03); but not on multivariate analysis (HR: 0.405 (CI: 0.140–1.160), p=0.095). High TILs were found to have prognostic significant and be protective only on multivariate analysis (HR: 0.120 (CI: 0.018–0.809), p=0.029). Low PSA was found to be associated with worse prognostic outcomes on univariate (HR: 21.970 (CI: 2.829–170.7), p=0.003) and multivariate analyses (HR: 7.69 (CI: 2.217–100.00), p=0.018). High ISP was found to be associated with poor prognostic outcomes on univariate (HR: 1.189 (CI: 1.106–1.278), p= < 0.001) and multivariate analysis (HR: 1.253 (CI: 1.078–1.458), p < 0.003). High MSR was found to be associated with poor outcomes on univariate (HR: 5.613 (CI: 1.947–16.19), p=0.028) and multivariate (HR: 7.194 (CI: 1.197–43.70), p=0.030) analysis. Stromal differentiation was found to have prognostic significance on univariate (HR: 3.715 (CI: 1.829–16.64), p=0.048) and multivariate analysis (HR: 1.230 (CI: 1.03–-1.462), p=0.019). The remaining clinicopathological variables were not significant (p>0.05) on cox proportional hazard regression analysis (Table 2).
Table 2.
Univariate and multivariate analyses of cancer-free survival using the cox proportional-hazard regression. CI, Confidence Interval; HR, Hazard Ratio; TIL, Tumor Infiltrating Lymphocytes; PSA, Proportionated Stromal Area; ISP, Immature Stromal Percentage; Myxoid Stromal Ratio; MMR, Mis-match repair; KRAS, Ki-ras2 Kirsten rat sarcoma viral oncogene homolog; BRAF, B-Raf Proto-Oncogene. Significant features (p≤0.05) are shown in bold.
| Variable | Frequency | Univariate HR | CI | p value | Multivariate HR | CI | p value |
|---|---|---|---|---|---|---|---|
| Age | 0.987 | 0.943–1.032 | 0.560 | 1.051 | 0.982–1.125 | 0.151 | |
| <=65 | 22 (36.67%) | ||||||
| >65 | 38 (63.33%) | ||||||
| Gender | 1.247 | 0.429–3.624 | 0.685 | 1.489 | 0.476–4.652 | 0.494 | |
| Male | 36 (60%) | ||||||
| Female | 24 (40%) | ||||||
| Tumor budding | 2.815 | 0.784–10.10 | 0.112 | 1.011 | 0.01–7.83 | 0.173 | |
| TBD1 | 24 (40%) | ||||||
| TBD2 or TBD3 | 36 (60%) | ||||||
| Pathological stage | 3.705 | 1.015–13.53 | 0.047 | 0.405 | 0.140–1.170 | 0.095 | |
| ≤PT2 | 13 (21.67%) | ||||||
| PT3–PT4 | 47 (78.33%) | ||||||
| TIL | 0.404 | 0.111–1.467 | 0.168 | 0.120 | 0.018–0.809 | 0.029 | |
| Low | 40 (66.67%) | ||||||
| High | 20 (33.33%) | ||||||
| Lymph node stage | 0.577 | 0.180–1.848 | 0.355 | 0.909 | 0.205–4.033 | 0.900 | |
| N0 | 32 (53.33%) | ||||||
| N1–N2 | 28 (46.67%) | ||||||
| PSA | 21.739 | 2.829–170.7 | 0.003 | 7.69 | 2.217–100.0 | 0.018 | |
| >=0.9 | 25 (41.67%) | ||||||
| <0.9 | 35 (58.33%) | ||||||
| ISP | 1.189 | 1.106–1.278 | <0.001 | 1.253 | 1.078–1.458 | 0.003 | |
| <10 | 37 (61.67%) | ||||||
| >=10 | 23 (38.33%) | ||||||
| MSR | 5.613 | 1.947–16.19 | 0.028 | 7.194 | 1.197–43.70 | 0.030 | |
| <0.19 | 39 (65%) | ||||||
| >0.19 | 21 (35%) | ||||||
| Tumor grade | 2.815 | 0.784–10.10 | 0.112 | 1.454 | 0.300–7.043 | 0.642 | |
| G1 | 44 (73.33%) | ||||||
| G2–G3 | 16 (26.67%) | ||||||
| Stromal differentiation | 3.715 | 1.829–16.64 | 0.048 | 1.230 | 1.035–1.462 | 0.019 | |
| Mature | 27 (45%) | ||||||
| Immature/intermediate | 33 (55%) | ||||||
| MMR | 2.607 | 0.903–7.526 | 0.076 | 0.456 | 0.0533–3.958 | 0.477 | |
| Intact | 46 (76.67%) | ||||||
| Loss | 14 (23.33%) | ||||||
| KRAS | 2.828 | 0.17–47.146 | 0.469 | ||||
| Wild type | 8 (66.67%) | ||||||
| Mutation | 4 (33.33%) | ||||||
| BRAF | 1.31 | 0.11–3.52 | 0.97 | ||||
| Wild type | 8 (88.89%) | ||||||
| Mutation | 1 (11.11%) |
Discussion
The advent of open-access image analysis software has been paramount to the success of digital pathology. Qupath17 works to meet the need for a friendly, understandable digital pathology solution. It offers a comprehensive panel for image analysis and here, we demonstrate the utility of superpixels for quantifying different stromal parameters computationally: PSA, ISP, and MSR.
Importantly, PSA is not the same as the TSR.11 The PSA technique includes the entire tumor area and is computational. Traditionally, the TSR is calculated only at the invasive tumoral border and is performed manually by a pathologist on glass slides, allowing for observer bias. In colon cancer, high stroma ratios resulting in a low TSR at the invasive tumor front are associated with poor outcomes.11
However, we found the opposite effect for PSA, with low stroma ratios having worse prognostic outcomes. This may be secondary to the fact that PSA encompasses the entire tumor, not just the invasive front, and evaluating the invasive front in isolation may overestimate the stroma to tumor ratio. Low PSA could correlate with more tumor, which could also mean more tumor budding. Whereas for TSR, which is performed at the tumor invasive front, more stroma and less tumor could mean more desmoplastic, myxoid stroma, possibly accounting for the prognostic outcomes.22
Regarding DIA, our findings are also consistent with what was found in luminal tumors of the breast13 and in intrahepatic cholangiocarcinoma,12 where higher proportions of tumor (low PSA) were found to be associated with worse overall survival outcomes. A key point to consider is the variation in DIA methods between the two mentioned studies and this publication. In the breast publication,13 a single region of interest was identified from a WSI and tissue microarrays were created from tumor hotpots. In this publication, DIA was performed on the entire tumor from a complete WSI, also seen in study on intrahepatic cholangiocarcinoma.12 Using the entire WSI may be the better solution, especially considering the significant tumor heterogeneity present in CRC. We felt the most pragmatic approach was to use the slide with the largest section of tumor that most accurately represented the tumoral microenvironment.
Superpixel methods23 are gaining traction in the field of medicine and recent attempts have been made to combine superpixels with DL. In dermatology, superpixel models based on DL have been found to outperform other comparable methodologies.24 The advantages for SIS is that it can represent the structure of an image in adaptive sizes and shapes, with the ability to improve classification performance, especially for noisy classification and boundary misclassification.25 While DL is a methodology which applies non-linear transformations and high-level model abstractions in large databases.26 Future studies could continue to combine applications in DL with superpixels to further our understanding of the tumoral microenvironment, such is demonstrated here.
Recent advances for DL in CRC were made by Skrede et al.,27 who applied convolutional neural networks to WSIs, and developed an assay (DoMore-v.1) which differentiated prognostic groups independent of tumor stage in large patient cohorts. The authors suggest the assays superiority compared to other genomic and pathological prognostic markers, as the assay predicted cancer-specific survival in stage II (HR: 2.71, 95% CI: 1.25–5.86, p=0.011) and stage III patients (HR: 4.09, 95% CI: 2.77–6.03, p < 0.0001).27
More practically, the differentiation of the ECM leads to characteristic immature, myxoid stroma seen on routine histological evaluation. Preclinical studies of tumors growing in mice have suggested collagen to govern chemotherapeutic delivery,28 whereas ECM degradation has also been found to improve drug uptake and response.2 Deciphering the ECM in CRC could facilitate novel therapeutic approaches.2 Most likely, myxoid degeneration decreases the physical barrier, leading to improved therapeutic delivery.
In our study, we were able to more accurately characterize SD through DIA by calculating the ISP and MSR, which predicted patient outcomes and clinical profiles better than the manual analysis of SD. Manual analysis is more prone to interobserver variability, whereas the advent of DIA can allow for improved reproducibility.29 DIA techniques could be used to tailor the need for adjuvant chemotherapy, although clinical studies will be needed to determine this.
One significant pitfall in our study was that we were unable to validate our findings in separate validation and testing cohorts. Although the present work can serve as a pilot study, it will be important for future studies to validate model performance in different patient cohorts. Overfitting is a fundamental issue in applied machine inteligence,30 it will be important to test model fitting in diverse patient populations before implementing and adopting computational approaches for patient care.
Interestingly, MSR was not found to be associated with pathological stage, suggesting that it may be able to predict clinical outcomes independent of anatomical extent of disease. This may be the key to unraveling the clinical heterogeneity present in CRC. Today, the use of adjuvant treatment in stage II colon cancer has garnered much controversy,31 and treatment recommendations range from observation, to single agent chemotherapy or combination regimens. There is a need to better tailor the role of adjuvant therapy in CRC and an assessment of tumoral differentiation and anatomical extent of disease may be insufficient in truly addressing the questions which need answers.
High-risk stromal computational features (PSA <0.9, ISP >10%, MSR >0.19) could represent the CRC mesenchymal phenotype, associated with an increased migratory capacity, tumor cell invasion, and enhanced modulation of a diverse ecosystem of ECM components. We also found this computational mesenchymal phenotype to corelate with MMR deficiency. Regarding the molecular classification of CRC major intrinsic subtypes, C-type has been found to be the smallest and most distinct molecular subtype, upregulated in markers related to the EMT and MMR deficiency.32 Patients with C-type tumors are commonly referred to as the mesenchymal subtype, have a poor baseline prognosis, and show no benefit from adjuvant 5FU chemotherapy.32 Regarding the most recent consensus molecular subtypes of CRC,33 13% of tumors have been found to have a mixed molecular phenotype between CMS1 (microsatellite instability) and CMS4 (mesenchymal). More work examining the relationship between stromal features on WSIs and its relationship to the molecular profile in CRC is needed.
In summary, we developed a novel superpixel method to assess the TME in CRC which outperformed traditional metrics for prognostication currently used in clinical practice. This study supports the tumoral microenvironment as a prime candidate for future applications in digital pathology, and the techniques described in this body of work can be performed by a surgical pathologist with appropriate quality assurance.
Data Availability
Pathology data and the statistical analyses for the current study are available from the corresponding author upon reasonable request.
Funding
No funding was provided to produce this manuscript.
Author Contributions
MN, DW, and SH developed the theoretical formalism. SH, DW, and CA contributed to the acquisition of data and whole slide images. DW performed the analytic calculations and performed the numerical simulations. SH, DW, CA, and MN contributed to the final version of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
Acknowledgments
We thank Alexander Perry and Kathy Quinn for their role as research coordinators. Figures were constructed at Biorender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpi.2022.100009.
Appendix A. Supplementary data
Supplementary material
References
- 1.Jasperson K.W., Tuohy T.M., Neklason D.W., Burt R.W. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–2058. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henke E., Nandigama R., Ergün S. Extracellular matrix in the tumor microenvironment and its impact on cancer therapy. Front Mol Biosci. 2020;6 doi: 10.3389/fmolb.2019.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson H., Kirsch R., Messenger D., Driman D. A review of current challenges in colorectal cancer reporting. Arch Pathol Lab Med. 2019;143:869–882. doi: 10.5858/arpa.2017-0475-RA. [DOI] [PubMed] [Google Scholar]
- 4.Hacking S.M., Chakraborty B., Nasim R., Vitkovski T., Thomas R. A holistic appraisal of stromal differentiation in colorectal cancer: biology, histopathology, computation, and genomics. Pathol Res Pract. 2021;220 doi: 10.1016/j.prp.2021.153378. [DOI] [PubMed] [Google Scholar]
- 5.Koelzer V.H., Zlobec I., Lugli A. Tumor budding in colorectal cancer--ready for diagnostic practice? Hum Pathol. 2016;47:4–19. doi: 10.1016/j.humpath.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Zhai Q., Fan J., Lin Q., et al. Tumor stromal type is associated with stromal PD-L1 expression and predicts outcomes in breast cancer. PLoS One. 2019;14 doi: 10.1371/journal.pone.0223325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao L., Sun P.-L., He Y., Yao M., Gao H. Desmoplastic reaction and tumor budding in cervical squamous cell carcinoma are prognostic factors for distant metastasis: a retrospective study. Cancer Manag Res. 2020;12:137–144. doi: 10.2147/CMAR.S231356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kemi N.A., Eskuri M., Pohjanen V.-M., Karttunen T.J., Kauppila J.H. Histological assessment of stromal maturity as a prognostic factor in surgically treated gastric adenocarcinoma. Histopathology. 2019;75:882–889. doi: 10.1111/his.13934. [DOI] [PubMed] [Google Scholar]
- 9.Hacking S.M. Tumor budding or tumor baloney? Virchows Arch. 2021;479:435–436. doi: 10.1007/s00428-021-03112-7. [DOI] [PubMed] [Google Scholar]
- 10.Ueno H., Ishiguro M., Nakatani E., et al. Prognostic value of desmoplastic reaction characterisation in stage II colon cancer: prospective validation in a Phase 3 study (SACURA Trial) Br J Cancer. 2021;124(6):1088–1097. doi: 10.1038/s41416-020-01222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Pelt G.W., Sandberg T.P., Morreau H., et al. The tumour-stroma ratio in colon cancer: the biological role and its prognostic impact. Histopathology. 2018;73:197–206. doi: 10.1111/his.13489. [DOI] [PubMed] [Google Scholar]
- 12.Guedj N., Blaise L., Cauchy F., Albuquerque M., Soubrane O., Paradis V. Prognostic value of desmoplastic stroma in intrahepatic cholangiocarcinoma. Modern Pathol. 2021;34:408–416. doi: 10.1038/s41379-020-00656-y. [DOI] [PubMed] [Google Scholar]
- 13.Millar E.K., Browne L.H., Beretov J., et al. Tumour stroma ratio assessment using digital image analysis predicts survival in triple negative and luminal breast cancer. Cancers (Basel) 2020;12 doi: 10.3390/cancers12123749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lara H., Li Z., Abels E., et al. Quantitative image analysis for tissue biomarker use: a white paper from the digital pathology association. Appl Immunohistochem Mol Morphol. 2021;29(7):479–493. doi: 10.1097/PAI.0000000000000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quail D.F., Joyce J.A. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu D., Hacking S., Vitkovski T., Nasim M. Superpixel image segmentation of VISTA expression in colorectal cancer and its relationship to the tumoral microenvironment. Scient Rep. 2021;11:17426. doi: 10.1038/s41598-021-96417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankhead P., Loughrey M.B., Fernández J.A., et al. QuPath: open source software for digital pathology image analysis. Scient Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lugli A., Kirsch R., Ajioka Y., et al. Recommendations for reporting tumor budding in colorectal cancer based on the International Tumor Budding Consensus Conference (ITBCC) 2016. Mod Pathol. 2017;30:1299–1311. doi: 10.1038/modpathol.2017.46. [DOI] [PubMed] [Google Scholar]
- 19.Klintrup K., Mäkinen J.M., Kauppila S., et al. Inflammation and prognosis in colorectal cancer. Eur J Cancer. 2005;41:2645–2654. doi: 10.1016/j.ejca.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Ueno H., Jones A.M., Wilkinson K.H., Jass J.R., Talbot I.C. Histological categorisation of fibrotic cancer stroma in advanced rectal cancer. Gut. 2004;53:581–586. doi: 10.1136/gut.2003.028365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ueno H., Kajiwara Y., Ajioka Y., et al. Histopathological atlas of desmoplastic reaction characterization in colorectal cancer. Jpn J Clin Oncol. 2021;51:1004–1012. doi: 10.1093/jjco/hyab040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J., Shen Z., Deng Z., Mei L. Impact of tumor–stroma ratio on the prognosis of colorectal cancer: a systematic review. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.738080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang M., Liu X., Gao Y., Ma X., Soomro N.Q. Superpixel segmentation: a benchmark. Signal Process Image Commun. 2017;56:28–39. [Google Scholar]
- 24.Blanco G., Traina A.J.M., Traina C., Jr., et al. A superpixel-driven deep learning approach for the analysis of dermatological wounds. Comput Methods Programs Biomed. 2020;183 doi: 10.1016/j.cmpb.2019.105079. [DOI] [PubMed] [Google Scholar]
- 25.Shi C., Pun C.-M. Superpixel-based 3D deep neural networks for hyperspectral image classification. Pattern Recog. 2018;74:600–616. [Google Scholar]
- 26.Vargas R., Mosavi A., Ruiz R. Deep learning: a review. Adv Intel Syst Comput. 2017;5 [Google Scholar]
- 27.Skrede O.J., De Raedt S., Kleppe A., et al. Deep learning for prediction of colorectal cancer outcome: a discovery and validation study. Lancet. 2020;395:350–360. doi: 10.1016/S0140-6736(19)32998-8. [DOI] [PubMed] [Google Scholar]
- 28.Brown E., McKee T., diTomaso E., et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9:796–800. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- 29.Riber-Hansen R., Vainer B., Steiniche T. Digital image analysis: a review of reproducibility, stability and basic requirements for optimal results. Apmis. 2012;120:276–289. doi: 10.1111/j.1600-0463.2011.02854.x. [DOI] [PubMed] [Google Scholar]
- 30.Ying X. An overview of overfitting and its solutions. J Phys Conf Ser. 2019;1168 [Google Scholar]
- 31.Varghese A. Chemotherapy for stage II colon cancer. Clin Colon Rectal Surg. 2015;28:256–261. doi: 10.1055/s-0035-1564430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roepman P., Schlicker A., Tabernero J., et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134:552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guinney J., Dienstmann R., Wang X., et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Pathology data and the statistical analyses for the current study are available from the corresponding author upon reasonable request.