Abstract
The prevalence of infections by nontuberculous mycobacteria is increasing, having surpassed tuberculosis in the United States and much of the developed world. Nontuberculous mycobacteria occur naturally in the environment and are a significant problem for patients with underlying lung diseases such as bronchiectasis, chronic obstructive pulmonary disease, and cystic fibrosis. Current treatment regimens are lengthy, complicated, toxic and they are often unsuccessful as seen by disease recurrence. Mycobacterium abscessus is one of the most commonly encountered organisms in nontuberculous mycobacteria disease and it is the most difficult to eradicate. There is currently no systematically proven regimen that is effective for treating M. abscessus infections. Our approach to drug discovery integrates machine learning, medicinal chemistry and in vitro testing and has been previously applied to Mycobacterium tuberculosis. We have now identified several novel 1-(phenylsulfonyl)-1H-benzimidazol-2-amines that have weak activity on M. abscessus in vitro but may represent a starting point for future further medicinal chemistry optimization. We also address limitations still to be overcome with the machine learning approach for M. abscessus.
Keywords: Drug discovery, machine learning, Mycobacterium abscessus, Mycobacterium tuberculosis, nontuberculous mycobacteria
1. Introduction
Tuberculosis is a well-known disease affecting the global population caused by Mycobacterium tuberculosis; however, nontuberculous mycobacteria (NTM) are an under-researched area and there is currently no standard drug regimen [1]. Nontuberculous mycobacterial infections are of particular concern to vulnerable patients with pulmonary diseases like cystic fibrosis [1], where treatments can conflict with ongoing chemotherapies. Unlike tuberculous mycobacteria, nontuberculous mycobacteria do not need a living host to transmit the organism and are present in water and soil [2]. They are able to survive in harsh environments lacking nutrients that inhibit the growth of other bacteria [2], and one such organism, Mycobacterium abscessus, is therefore very difficult to treat due to its resistance to common antibiotics, as well as the lack of correlation between in vitro and in vivo efficacy [2, 3]. Thus, drug discovery for M. abscessus infections is perhaps more difficult than for other bacteria and is certainly in need of novel tools to assist it.
Drug discovery studies for M. abscessus frequently start with screening of a large library of molecules at a single concentration, either of already approved drugs [4, 5], well-known collections like the Pathogen Box [6, 7], or general libraries of diverse molecules [8]. However, most often, these high-throughput screening (HTS) methods only produce a very small number of ‘hit’ compounds (the definition of which varies from study to study) from the primary screen, and only a few of these molecules are usually confirmed to inhibit growth in a dose-dependent manner. These screening methods have been discussed as an inefficient method of drug discovery for M. abscessus in particular [8]. Confirmed lead compounds are then tested individually [9-11] or further optimized by a synthetic series [12] in vitro and in vivo to evaluate the potential for clinical treatment. Parallel studies may be undertaken to identify the mechanism of action and the biological target. Other studies have utilized a small, targeted set of compounds for specific goals, like compounds that had not been previously identified as antibiotics [13]. In summary, most researchers involved in M. abscessus drug discovery therefore tend to screen molecule libraries as part of a proof-of-concept followed by further experimental optimization [14-16].
Another route utilized to discover compounds for M. abscessus has been to leverage activities of compounds against a similar organism, such as M. tuberculosis where data is more plentiful, and then try to repurpose these molecules for M. abscessus treatment. Such efforts have tended to be more successful in identifying highly potent compounds with known targets [17]. Moreira et al. found that filtering a fragment library by first testing compounds against M. tuberculosis yielded a higher hit rate against NTM, including M. abscessus [18]; similar reasoning has led other studies to use the previously mentioned Pathogen Box, which includes >100 known M. tuberculosis active compounds [7]. The various approaches that have been used in M. abscessus drug discovery have been reviewed recently illustrating that while there has been a heavy focus on HTS and drug repurposing, there has been little interest in synthesis of novel molecules [1].
An alternative approach showing some promise in drug discovery is the development and increased application of HTS alongside computational methods like machine learning that has been applied to drug discovery for M. tuberculosis [19, 20]. These machine learning methods have the advantage of leveraging public data to prioritize a subset of compounds and potentially obtain a higher hit rate than random library screening [21], limiting the cost and time spent on failed leads. In the current study, we have utilized our Assay Central software, to apply a Bayesian machine learning approach to building models for public and our own data for M. abscessus drug discovery, building upon previous efforts that culminated in promising in vitro results for M. tuberculosis [22], Neisseria gonorrhoeae [23] and Staphylococcus aureus [24]. This current study demonstrates how we used a machine learning model to assist in the selection of novel compounds for testing in vitro.
2. Methods
2.1. Chemicals and reagents
All reagents and solvents were purchased from commercial suppliers and used without further purification. 1H spectra were measured on Bruker AC-300 (300 MHz). Chemical shifts were measured in DMSO-d6, using tetramethylsilane as an internal standard, and reported as units (ppm) values. Mass spectra were recorded on Finnigan MAT INCO 50 mass spectrometer (EI, 70 eV) with direct injection. The purity of the final compounds was analyzed on an Agilent 1290 Infinity II HPLC system coupled to Agilent 6460 triple-quadrupole mass spectrometer equipped with an electrospray ionization source. Elemental analysis (% C, H, N) was carried out by an elemental analyzer EURO EA. Melting points were determined on Electrothermal 9001 (10 °C per min) and are uncorrected. Merck KGaA silica gel 60 F254 plates were used for analytical thin-layer chromatography. Yields refer to purified products and are not optimized. All final compounds are > 95 % pure.
Compounds were resuspended in DMSO (MilliporeSigma). Resazurin sodium salt, tyloxapol, phorbol 12-myristate 13-acetate (PMA), Triton X 100, and kanamycin sulfate salt were purchased from MilliporeSigma.
2.2. Resazurin Microtitre Assay (REMA)
Selected test compounds were prepared in DMSO and kanamycin, a positive control, was prepared in deionized water and sterile filtered. Using non-treated polystyrene 96-well plates (Corning), drugs were serially two-fold diluted in triplicate (unless specified otherwise) in 7H9 broth (Difco) supplemented with albumin dextrose saline (ADS; 10 g/L bovine serum albumin fraction V, 4 g/L dextrose, 1.6 g/L NaCl), 0.5% glycerol, and 0.1% Tyloxapol (7AGT). Mycobacterium abscessus ATCC 19977 (smooth) was grown in 7AGT until mid-logarithmic growth was reached. The M. smegmatis strain used was mc2155 (ATCC 700084) and the M. tuberculosis strain was H37Rv. Cells were passed through a 40 μM cell strainer and allowed to settle. Culture cell density was measured by optical density (OD600) and diluted to reach a final density of 1 X 105 cells/well. All wells, including test compounds and kanamycin controls, contained a final concentration of 1% DMSO and 200 μL total volume. The kanamycin control was diluted in 2-fold steps ranging from 0.2-82.5 μM. Plates were incubated for 48hrs at 37°C, 100 rpm before adding 20 μL resazurin solution (125 μg/mL in phosphate buffered saline). Following the addition of resazurin, plates were incubated in the dark for an additional 24 hrs. Fluorescence was measured with an excitation at 544nm and emission at 590nm with a Molecular Devices, SpectraMax M2 microplate reader (California, USA).
2.3. Cell cytotoxicity assay
Compound cytotoxicity was measured by using human monocytes (THP1, ATCC TIB-202) and the CellTiter-Glo 2.0 assay (Promega) following the manufacturers protocol. Briefly, test compounds were dissolved in DMSO and kanamycin was dissolved in deionized water at a final concentration of 100 μM. THP1 cells were cultured in RPMI 1640 [+] L-Glutamine with 10% fetal bovine serum (FBS) (Gibco) in a humidified 37°C, 5% CO2 incubator. Phorbol 12-myristate 13-acetate (PMA) was added for a final concentration of 50 ng/mL and THP1 cells were plated at 1x105 cell/well in a 96 well plate (Corning 3610). After 48hrs of incubation, cells were incubated with PMA free media for 24hrs before exposure to compounds. Cells were exposed to compounds (100 μM), 1% Triton X 100, 1% DMSO, or kanamycin (100 μM) for 48hrs. Test compounds and kanamycin had a final concentration of 1% DMSO. The plate was then equilibrated to room temperature for 30 minutes before the addition of the CellTiter-Glo 2.0 reagent. The plate was mixed for 2 minutes on an orbital shaker then incubated for 10 minutes at room temperature before luminescence signal (RLU = relative light unit) was measured using a Tecan Infinite 200 Pro plate reader (Zurich, Switzerland).
2.4. Machine learning with Assay Central®
Assay Central® is a proprietary software used to build machine learning models from high-quality datasets and generate predictions. It applies extended-connectivity fingerprint descriptors from the Chemistry Development Kit library [25] and a Bayesian algorithm previously described for other drug discovery projects [24, 26-34]. Structure-activity datasets were collated in Molecular Notebook (Molecular Materials Informatics, Inc. in Montreal, Canada) and were curated through a series of scripts to detect and correct any problematic data (i.e. multiple components, salt removal, potentially inaccurate structure depiction). Performance metrics generated from internal five-fold cross-validation are included with each model. These include a receiver operator characteristic curve, recall, precision, specificity, Cohen’s kappa, Matthews Correlation Coefficient, and balanced accuracy.
Predictions are generated from resulting Bayesian model by first enumerating all training data fingerprints and calculating a given fingerprint’s “contribution” to an active classification from the ratio of its presence in active and inactive molecules; the summation of contributions of the fingerprints in a prospective molecule produces the probability-like prediction score [35]. Scores greater than 0.5 are considered an active prediction.
Data was curated from the literature [5-7, 9, 14, 15, 18, 36-44] the NIAID ChemDB HIV, Opportunistic Infection and Tuberculosis Therapeutics Database [45] (downloaded in 2017), and 19 other compounds tested as a proof-of-concept. The data consisted of MIC/MIC50/MIC90 values, set to a threshold of 500 μM. Models were also generated solely from the specific chemical series tested herein and from ChEMBLv 27 MIC data. Batches of compounds were sent and periodically predicted with all Assay Central models to prioritize in vitro testing and the model was updated over time.
2.5. Statistical analysis
GraphPad Prism version 8.2 was used for data analysis. The log(inhibitor) vs. response, variable slope (four parameters) equation was used to determine the best fit curve. For the cell cytotoxicity experiment one-way ANOVA and Dunnet's multiple comparison test were used to determine statistical significance.Percent inhibition was calculated as previously described with the following modifications [15].
3. Results
3.1. Machine learning
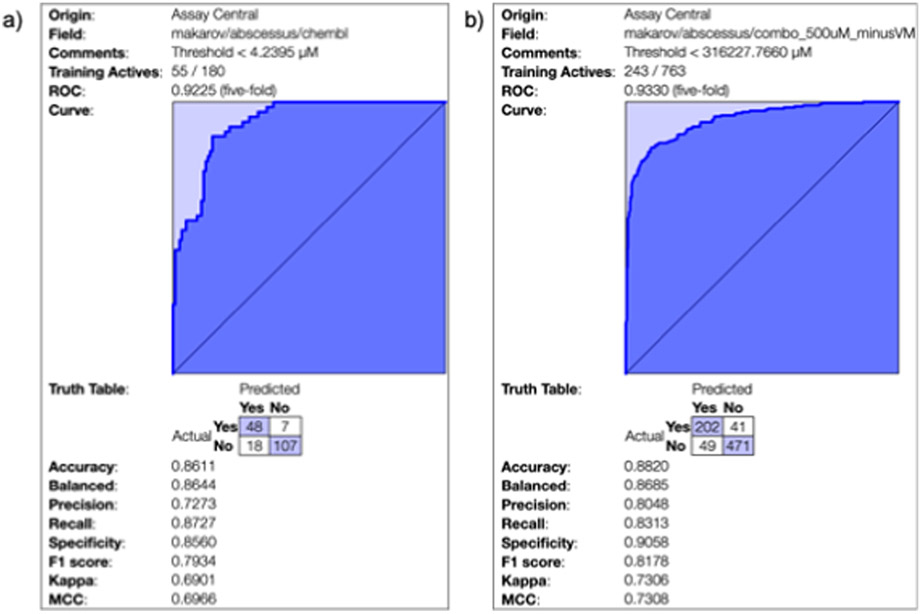
Two machine learning models for M. abscessus inhibitors were generated with Assay Central in this study and five-fold cross-validation was performed to generate metrics in order to assess the model performance. Each binary model was generated from different curated datasets. A model was generated with public data available in ChEMBL [46] however this dataset provides an example of the issues in M. abscessus drug discovery. Out of the over 800 MIC values present in the database, 313 have mixture of activities that cannot be properly merged (i.e. combinations of “>” and “=” measurements). The remaining values were consolidated into 180 compounds total (Figure 1a) and this model has a five-fold cross validation Receiver Operator Characteristic (ROC) of 0.92 and a balanced accuracy of 0.86 (other statistics were also excellent, Figure 1). The second M. abscessus Bayesian model used literature data and other compounds from our initial in vitro screens, in which we set the threshold as 500 μM (Figure 1b) which resulted in a model with a five-fold cross validation ROC of 0.93 and balanced accuracy of 0.86. A model that was generated just from our data generated in this study had poor metrics (data not shown) and was very specific to the chemical series being explored herein. We did not find any improvement when combining models (data not shown). Also, as both models have different optimal cut-offs we could not use one dataset as a test set for the other. After an initial screen of several compounds from our library of over 4000 molecules (synthesis described for several compounds relevant to this study in section 3.1), these Bayesian models were then used to assist in selecting additional compounds by scoring them to prioritize for in vitro testing over several rounds. We therefore used the models to prioritize compounds throughout the project as our in vitro testing capabilities were limited.
Figure 1: M. abscessus Bayesian models 5-fold cross validation.
(a), ChEMBLv27 MIC values (b) and the literature model used for predictions at a threshold of 500 μM with preliminary in vitro tested compounds.
3.2. Synthetic route for 1-(phenylsulfonyl)-1H-benzimidazol-2-amine and 1-benzoyl-1H-benzimidazol-2-amine derivatives
Most of all the studied 2-aminobenzimidazole derivatives were obtained by the reaction of 2-aminobenzimidazole with substituted sulfonyl chlorides or acid chloranhydrides in pyridine with formation aim phenylsulfonyl- and benzoylbenzimidazoles (Scheme 1, step c, d). 2-aminobenzimidazoles with substituents in the benzene ring were obtained by successive transformation including the reduction of a 2-nitroaniline derivative by catalytic hydrogenation of Pd/C (10 mol%) in EtOH (Scheme 1, step a), and in the case of 5-chloro-3-nitropyridine-2- amine, iron was used as a reducing agent in the presence of hydrochloric acid (Scheme 3, step a). For annulation of the imidazole ring, a condensation reaction of o-phenylenediamine derivatives with cyanogen bromide was carried out (Scheme 1, step b). 2-aminobenzimidazole substituted at the amino group was obtained by condensation of 2-aminobenzimidazole with benzaldehyde to form a Schiff base and subsequent reduction at the double bond with sodium borohydride (Scheme 2). All synthesized and tested compounds were > 97% pure and stable in working solutions, their analytical and spectra data presented in Supplementary information.
Scheme 1.
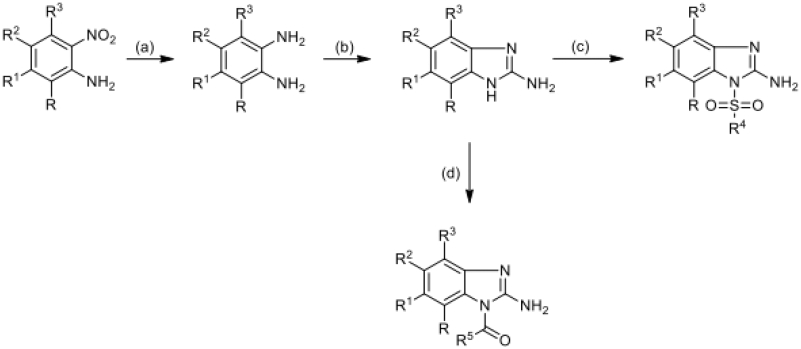
Reagents: (a) Pd/C, EtOH; (b) CNBr, MeOH, H2O; (c) ClSO2R4, Py; (d) ClCOR5, Py
Scheme 3.
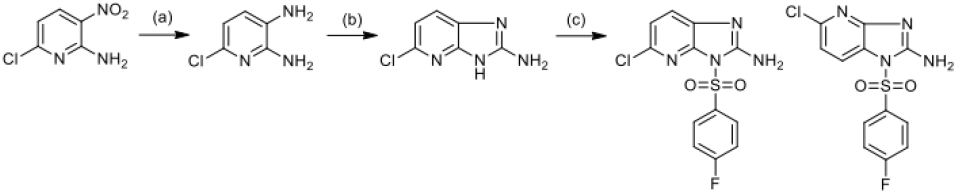
Reagents: (a) Fe,HCl, EtOH, H2O; (b) CNBr, MeOH, H2O; (c) ClSO2(C6H4)F-p, Py
Scheme 2.
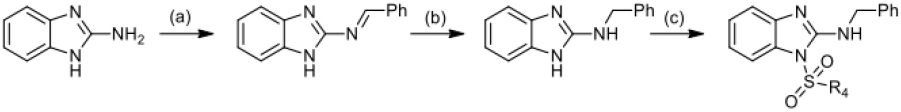
Reagents: (a) PhCOH, p-TSA, toluene; (b) NaBH4, EtOH; (c) ClSO2R4, Py
Scheme 1.
Step a).
Solution of 2-nitro-4-trifluoromethylaniline (4.85 mmol) in EtOH (100 ml) was treated by 0.1g Pd/C (10% mol), hydrogen is passed for 2 hours until the absorption stops. The reaction mass was filtered off from coal through a layer of silica gel, washed with alcohol (3 * 20 ml) and evaporated, the oily residue is cooled, triturated with hexane and filtered off. Diamino compound was obtained with yield 0.77 g (90%), which was used for the next step without further purification.
Step b).
Solution of 3,4-diaminobenzotrifluoride (2 mmol) in MeOH (8 ml) and H2O (8 ml) was treated by BrCN (6 mmol) and heated at 50 °C for 1 hour. Then the reaction mass was evaporated by half, the evaporation residue was cooled and neutralized with aqueous ammonia to pH=8, extracted with EtOAc, the organic layers were combined and washed with water, dried with Na2SO4, evaporated and the oily residue is triturated with hexane. Aim product was isolated with yeald 0.33 g (82%) and used for the next step without further purification.
Solution of 4,5-dimethyl-o-phenylenediamine (2.6 mmol) in MeOH (14 ml) and H2O (14 ml) was treated by BrCN (7.8 mmol) and reaction mixture was heated at 50 °C for 1 hour. Then the reaction mass is evaporated by half, the residue after evaporation is cooled and made alkaline with aqueous ammonia to pH=8, almost immediately a thick beige precipitate is formed. It is cooled, the precipitate is filtered off, washed with water and recrystallized from H2O (yield 69%).
Step c).
2-Aminobenzimidazole (3.8 mmol) is dissolved in a minimum amount of pyridine and 4-fluorobenzenesulfonyl chloride (3.8 mmol) is added dropwise, the reaction mixture is stirred at room temperature for 3 hours. It is diluted with cold water, cooled, the precipitate formed is filtered off and washed with water. Compound 11426122 was recrystallized from EtOH (yield 70%).
Step d).
2-Aminobenzimidazole (1.13 mmol) is dissolved in a minimum amount of pyridine and 2,4-difluorobenzoyl chloride (1.13 mmol) is added dropwise, the reaction mixture is stirred at room temperature for 3 hours. It is diluted with cold water, cooled, the precipitate formed is filtered off and washed with water. Compound 11926223 was purified by recrystallization from MeOH (23%).
Scheme 2
Step a).
Suspension of 2-aminobenzimidazole (1.5 mmol) in toluene (6 ml) was treated by benzaldehyde (2.25 mmol) dropwise and a catalytic amount of p-toluenesulfonic acid is added. The reaction mass is boiled with stirring for 1 hour, cooled, the precipitate formed is filtered off. Recrystallized from CH3CN (57%).
Step b).
Sodium borohydride (6.8 mmol) was added portionwise to a suspension of the compound from step a) (1.7 mmol) in EtOH (5 ml), then was heated at 70 °C for 1 hour. The reaction mixture brightens noticeably and flakes of precipitate appear, then it is cooled to room temperature and diluted with cold water. The formed precipitate is cooled and filtered off, washed with water. The final amino compound was recrystallized from mixture hexane/acetone with yield 78%.
Then synthesis follow according to (Step c) described for the scheme of synthesis Scheme 1.
Scheme 3
Step a).
Suspension of 2-amino-6-chloro-3-nitropyridine (11.5 mmol) in 8 ml of EtOH and 2 ml of H2O was treated by 36% hydrochloric acid (0.092 ml) and iron (120 mmol). The reaction mixture was boiled for 1 hour, then the iron was filtered off and washed several times with hot EtOH. The filtrate was evaporated, the evaporation residue was dissolved in ethyl acetate and washed with water (4 * 50 ml). The organic layer was separated, dried with Na2SO4, activated carbon is added and the mixture is left stirring overnight at room temperature. The charcoal was filtered off through a layer of silica gel, the filtrate was evaporated. Diaminopyridine was obtained with yield 1.05 g (64%) and used for the next step without further purification. Then synthesis follow according to (Step b, c) described for the scheme of synthesis Scheme 1.
3.3. REMA
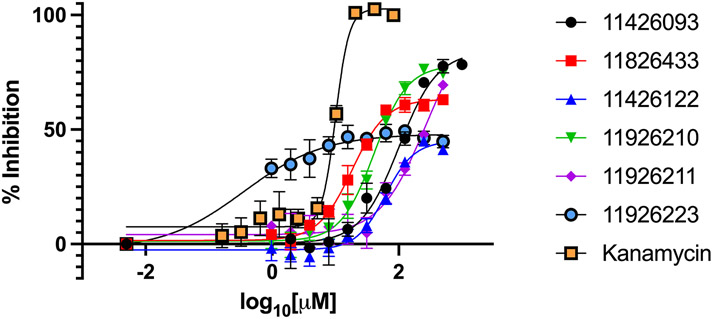
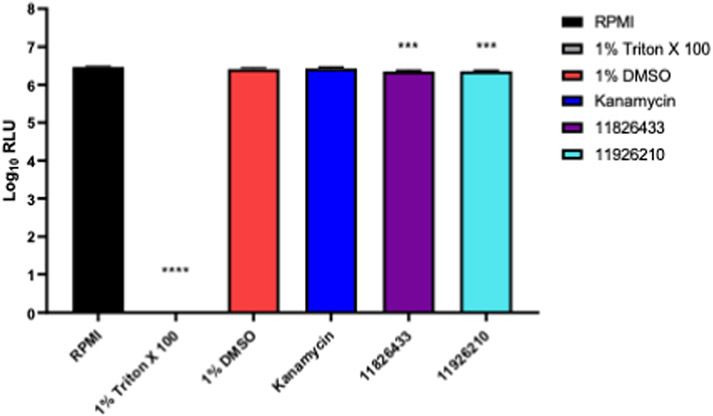
A resazurin microtiter assay (REMA) was used to determine the in vitro efficacy of compounds tested against M. abscessus, M. tuberculosis, and M. smegmatis. M. smegmatis is a nonpathogenic NTM that is often used as a model-mycobacteria in the laboratory. Firstly, we screened a small selection of compounds from an in-house chemical library with many biological active heterocycles and found the benzimidazole derivative 11426093 which showed activity on M. abscessus. Guided by our machine learning models we then synthesized and tested 110 new derivatives (described previously in section 3.1) and six novel 1-(phenylsulfonyl)-1H-benzimidazol-2-amine compounds were identified with 50% inhibition of M. abscessus growth at concentrations <500 μM (Table 1, Figure 2). The best performing compounds identified were 11826433 and 11926210, which inhibited 50% of M. abscessus growth at 267 μM and 262 μM, respectively. Both these compounds have several methyl groups and the phenylsulfonyl moiety what increases their lipophilicity. However, the six hit compounds required a much higher concentration than kanamycin to prevent growth, and in all cases 100% inhibition was not achieved even with the highest concentrations tested at 1 mM. In future to be of further utility, these compounds will likely require hit-to-lead modification to increase in vitro efficacy. Additionally, the two best performing compounds 11826433, 11926210 were shown to only have slight cytotoxicity against human monocytes (THP-1 cell) at 100 μM (Figure 3). Triton X-100 was used as a cytotoxicity positive control.
Table 1.
Concentration (μM) required to inhibit Mycobacteria 50% compared to a kanamycin control.
| IC50 (μM) | ||||
|---|---|---|---|---|
| Compounda | M. abs | M. tb | M. smeg | |
| Kanamycin (control) | 29 ± 6 | 14 ± 2 | 5 ± 2 | |
| 11426093 |
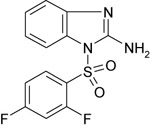
|
342 ± 116 | NT | NT |
| 11826433 |
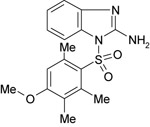
|
267 ± 79 | NT | NT |
| 11426122 |
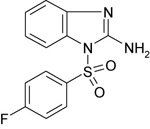
|
452 ± 2 | NT | NT |
| 11926210 |
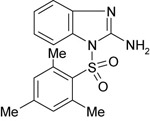
|
262 ± 42 | NT | NT |
| 11926211 |
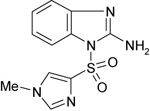
|
315b | NT | NT |
| 11926223 |
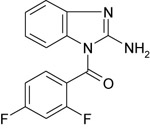
|
478b | NT | NT |
| 10726016 |
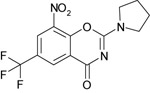
|
NI | 216b | 14b |
| 10726028 |
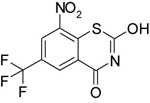
|
NI | 315b | 109b |
Results shown are the means ± SDs from 1 to 11 independent experiments with each independent assay always being performed with technical triplicate wells.
Experiments were only performed once. NI, no inhibitory activity; NT, not tested.
Figure 2: Dose dependent inhibition of M. abscessus as measured by REMA.
Results shown are means ± SDs from a representative plot with assays performed in triplicate. Compounds tested were compared to a kanamycin control to determine percent inhibition.
Figure 3: Cytotoxicity of compounds against human monocytes (THP-1) as measured by CellTiter-Glo 2.0.
Results shown are means ± SDs from a single independent experiment with assays performed in triplicate. One-way ANOVA and Dunnet's multiple comparison test were used to determine statistical significance. *** = P <0.001, **** = P <0.0001 compared to a RPMI only condition. RLU = Relative light unit.
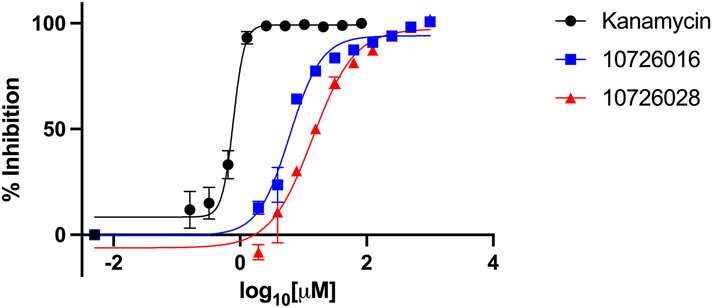
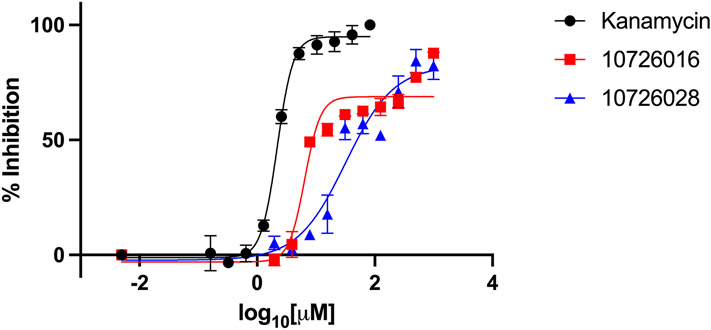
For M. smegmatis and M. tuberculosis, compounds 10726016 and 10726028 were selected by our Bayesian models and showed some activity against these bacteria, inhibiting 50% of M. smegmatis growth at 14 μM and 109 μM, respectively (Table 1, Figure 4). 10726016 and 10726028 also inhibited 50% of M. tuberculosis growth at 216 μM and 315 μM, respectively (Table 1, Figure 5). The species-specific nature of these two compounds was evident as neither exhibited 50% inhibition on the highly resistant M. abscessus.
Figure 4: Dose dependent inhibition of M. smegmatis as measured by REMA.
Results shown are means ± SDs from a single independent experiment performed in triplicate. Compounds tested were compared to a kanamycin control to determine percent inhibition.
Figure 5: Dose dependent inhibition of M. tuberculosis as measured by REMA.
Results shown are means ± SDs from a single independent experiment performed in triplicate. Compounds tested were compared to a kanamycin control to determine percent inhibition.
4. Discussion
In the current study we have used a combination of machine learning and medicinal chemistry approaches (as we have done previously for M. tuberculosis) to select compounds for in vitro testing against M. abscessus. In the process of this project, we identified a new class of compounds 1-(phenylsulfonyl)-1H-benzimidazol-2-amines that showed relatively weak in vitro activity compared with kanamycin as our positive control. Compared with work by others assessing compounds against this bacterium and reviewed recently [1], we can conclude that drug discovery for this bacterium is indeed very difficult. However, previous studies have also predominantly focused on testing existing drugs rather than identifying novel chemical series as we have done herein. This compound class does however provide an accessible starting point for future optimization as the chemistry involved is straightforward and the molecule does not have any obvious liabilities. Target identification would be important in order to further develop this series.
We previously applied machine learning approaches to identify new leads for M. tuberculosis using the naïve Bayesian approach [47, 48], but we are not aware of any similar efforts to apply machine learning methods to M. abscessus drug discovery outside of this work. In this study we used two models generated with both literature data alone or a combination of literature and our own data from the current study to assist in selection of compounds for testing. These models were used at various stages to prioritize due to the limited in vitro testing resources and could also be further applied to score other commercial libraries for selection of compounds for testing in future. Our previous applications of Bayesian models to M. tuberculosis was more successful in selecting more active molecules. For example, machine learning models that included bioactivity and cytotoxicity data ranked compounds for testing [49] and identified actives [22]. Several examples of M. tuberculosis machine learning models that were used to score vendor libraries to find actives in vitro [50], or screened available libraries of compounds with promising hit rates of 15-71% which exceeded the 0.6 – 1.5% usually seen with HTS screening [20, 50, 51]. We have also combined different M. tuberculosis models [52] and tested with 1,924 molecules leading to enrichments in finding actives of 11.8 fold [53]. We also previously generated massive M. tuberculosis models with 345,011 molecules in them but found we did not see improved predictivity over smaller data sets consisting of thousands of molecules [54]. Our application of Bayesian and other machine learning approaches was also extended to model data from treatment studies of M. tuberculosis infected mice with promising external validation with additional compounds not in the model [55, 56]. Our most recent efforts combined in vivo and in vitro M. tuberculosis data and evaluated different machine learning methods with external test sets, where we concluded that the Bayesian algorithm was comparable to deep learning methods [30]. In this current study we have used published literature data alone or in combination with our own data in order to generate models which possess good 5-fold cross validation statistics (5-fold ROC > 0.92). These models did not however perform particularly well in predicting very active compounds for M. abscessus as the datasets were much smaller, representing hundreds of compounds (and likely less diverse) and consisted of fewer likely high-quality actives when compared with those models used previously for M. tuberculosis which consisted of thousands of molecules and many actives. These M. abscessus models would certainly gain from the addition of further in vitro data and in particular additional actives from larger screens. Such machine learning models may likely assist with future hit-lead optimization although as we have shown there are certainly considerable challenges still to overcome with developing antibacterials against M. abscessus.
Supplementary Material
Acknowledgments
We kindly acknowledge NIH NIGMS funding to develop the software from R44GM122196-02A1 and Dr. Alex M. Clark (Molecular Materials Informatics, Inc.) for Assay Central® support. EK and VM were supported by the Russian Science Foundation under grant 21-15-00042.
Dr. Mohamed Nasr is thanked for assistance with obtaining the NIAID ChemDB HIV, Opportunistic Infection and Tuberculosis Therapeutics Database. Interested parties can contact NIH NIAID and it has restrictions on reuse.
Footnotes
Conflicts of interest
S.E. is owner, and K.M.Z. are employees of Collaborations Pharmaceuticals, Inc. All others have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Egorova A, Jackson M, Gavrilyuk V, Makarov V. Pipeline of anti-Mycobacterium abscessus small molecules: Repurposable drugs and promising novel chemical entities. Med Res Rev 2021. doi: 10.1002/med.21798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lopeman RC, Harrison J, Desai M, Cox JAG. Mycobacterium abscessus: Environmental Bacterium Turned Clinical Nightmare. Microorganisms 2019;7 doi: 10.3390/microorganisms7030090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Ganapathy US, Dartois V, Dick T. Repositioning rifamycins for Mycobacterium abscessus lung disease. Expert Opin Drug Discov 2019;14:867–878. doi: 10.1080/17460441.2019.1629414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chopra S, Matsuyama K, Hutson C, Madrid P. Identification of antimicrobial activity among FDA-approved drugs for combating Mycobacterium abscessus and Mycobacterium chelonae. J Antimicrob Chemother 2011;66:1533–1536. doi: 10.1093/jac/dkr154 [DOI] [PubMed] [Google Scholar]
- [5].Aziz DB, Low JL, Wu ML, Gengenbacher M, Teo JWP, Dartois V, Dick T. Rifabutin Is Active against Mycobacterium abscessus Complex. Antimicrob Agents Chemother 2017;61 doi: 10.1128/AAC.00155-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jeong J, Kim G, Moon C, Kim HJ, Kim TH, Jang J. Pathogen Box screening for hit identification against Mycobacterium abscessus. PLoS One 2018;13:e0195595. doi: 10.1371/journal.pone.0195595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Low JL, Wu ML, Aziz DB, Laleu B, Dick T. Screening of TB Actives for Activity against Nontuberculous Mycobacteria Delivers High Hit Rates. Front Microbiol 2017;8:1539. doi: 10.3389/fmicb.2017.01539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Malin JJ, Winter S, van Gumpel E, Plum G, Rybniker J. Extremely Low Hit Rate in a Diverse Chemical Drug Screen Targeting Mycobacterium abscessus. Antimicrob Agents Chemother 2019;63 doi: 10.1128/AAC.01008-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim TS, Choe JH, Kim YJ, Yang CS, Kwon HJ, Jeong J, Kim G, Park DE, Jo EK, Cho YL, Jang J. Activity of LCB01-0371, a Novel Oxazolidinone, against Mycobacterium abscessus. Antimicrob Agents Chemother 2017;61 doi: 10.1128/AAC.02752-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hanh BTB, Kim TH, Park JW, Lee DG, Kim JS, Du YE, Yang CS, Oh DC, Jang J. Etamycin as a Novel Mycobacterium abscessus Inhibitor. Int J Mol Sci 2020;21 doi: 10.3390/ijms21186908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sarathy JP, Ganapathy US, Zimmerman MD, Dartois V, Gengenbacher M, Dick T. TBAJ-876, a 3,5-Dialkoxypyridine Analogue of Bedaquiline, Is Active against Mycobacterium abscessus. Antimicrob Agents Chemother 2020;64 doi: 10.1128/AAC.02404-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Graham J, Wong CE, Day J, McFaddin E, Ochsner U, Hoang T, Young CL, Ribble W, DeGroote MA, Jarvis T, Sun X. Discovery of benzothiazole amides as potent antimycobacterial agents. Bioorg Med Chem Lett 2018;28:3177–3181. doi: 10.1016/j.bmcl.2018.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kirkwood ZI, Millar BC, Downey DG, Moore JE. Antimycobacterial activity of nonantibiotics associated with the polypharmacy of cystic fibrosis (CF) against mycobacterium abscessus. Int J Mycobacteriol 2018;7:358–360. doi: 10.4103/ijmy.ijmy_142_18 [DOI] [PubMed] [Google Scholar]
- [14].Gupta R, Netherton M, Byrd TF, Rohde KH. Reporter-Based Assays for High-Throughput Drug Screening against Mycobacterium abscessus. Front Microbiol 2017;8:2204. doi: 10.3389/fmicb.2017.02204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Richter A, Strauch A, Chao J, Ko M, Av-Gay Y. Screening of Preselected Libraries Targeting Mycobacterium abscessus for Drug Discovery. Antimicrob Agents Chemother 2018;62 doi: 10.1128/AAC.00828-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Berube BJ, Castro L, Russell D, Ovechkina Y, Parish T. Novel Screen to Assess Bactericidal Activity of Compounds Against Non-replicating Mycobacterium abscessus. Front Microbiol 2018;9:2417. doi: 10.3389/fmicb.2018.02417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dupont C, Viljoen A, Dubar F, Blaise M, Bernut A, Pawlik A, Bouchier C, Brosch R, Guerardel Y, Lelievre J, Ballell L, Herrmann JL, Biot C, Kremer L. A new piperidinol derivative targeting mycolic acid transport in Mycobacterium abscessus. Mol Microbiol 2016;101:515–529. doi: 10.1111/mmi.13406 [DOI] [PubMed] [Google Scholar]
- [18].Moreira W, Lim JJ, Yeo SY, Ramanujulu PM, Dymock BW, Dick T. Fragment-Based Whole Cell Screen Delivers Hits against M. tuberculosis and Non-tuberculous Mycobacteria. Front Microbiol 2016;7:1392. doi: 10.3389/fmicb.2016.01392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing hit identification in Mycobacterium tuberculosis drug discovery using validated dual-event Bayesian models. PLoS One 2013;8:e63240. doi: 10.1371/journal.pone.0063240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ekins S, Reynolds RC, Kim H, Koo MS, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian models leveraging bioactivity and cytotoxicity information for drug discovery. Chem Biol 2013;20:370–378. doi: 10.1016/j.chembiol.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ekins S, Casey AC, Roberts D, Parish T, Bunin BA. Bayesian models for screening and TB Mobile for target inference with Mycobacterium tuberculosis. Tuberculosis (Edinb) 2014;94:162–169. doi: 10.1016/j.tube.2013.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ekins S, Reynolds R, Kim H, Koo M-S, Ekonomidis M, Talaue M, Paget SD, Woolhiser LK, Lenaerts AJ, Bunin BA, Connell N, Freundlich JS. Bayesian Models Leveraging Bioactivity and Cytotoxicity Information for Drug Discovery. Chemistry & biology 2013;20:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pereira JC, Daher SS, Zorn KM, Sherwood M, Russo R, Perryman AL, Wang X, Freundlich MJ, Ekins S, Freundlich JS. Machine Learning Platform to Discover Novel Growth Inhibitors of Neisseria gonorrhoeae. Pharm Res 2020;37:141. doi: 10.1007/s11095-020-02876-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Dalecki AG, Zorn KM, Clark AM, Ekins S, Narmore WT, Tower N, Rasmussen L, Bostwick R, Kutsch O, Wolschendorf F. High-throughput screening and Bayesian machine learning for copper-dependent inhibitors of Staphylococcus aureus. Metallomics 2019;11:696–706. doi: 10.1039/c8mt00342d [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Willighagen EL, Mayfield JW, Alvarsson J, Berg A, Carlsson L, Jeliazkova N, Kuhn S, Pluskal T, Rojas-Cherto M, Spjuth O, Torrance G, Evelo CT, Guha R, Steinbeck C. The Chemistry Development Kit (CDK) v2.0: atom typing, depiction, molecular formulas, and substructure searching. J Cheminform 2017;9:33. doi: 10.1186/s13321-017-0220-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Anantpadma M, Lane T, Zorn KM, Lingerfelt MA, Clark AM, Freundlich JS, Davey RA, Madrid PB, Ekins S. Ebola Virus Bayesian Machine Learning Models Enable New in Vitro Leads. ACS Omega 2019;4:2353–2361. doi: 10.1021/acsomega.8b02948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ekins S, Gerlach J, Zorn KM, Antonio BM, Lin Z, Gerlach A. Repurposing Approved Drugs as Inhibitors of Kv7.1 and Nav1.8 to Treat Pitt Hopkins Syndrome. Pharm Res 2019;36:137. doi: 10.1007/s11095-019-2671-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ekins S, Puhl AC, Zorn KM, Lane TR, Russo DP, Klein JJ, Hickey AJ, Clark AM. Exploiting machine learning for end-to-end drug discovery and development. Nat Mater 2019;18:435–441. doi: 10.1038/s41563-019-0338-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hernandez HW, Soeung M, Zorn KM, Ashoura N, Mottin M, Andrade CH, Caffrey CR, de Siqueira-Neto JL, Ekins S. High Throughput and Computational Repurposing for Neglected Diseases. Pharm Res 2018;36:27. doi: 10.1007/s11095-018-2558-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lane T, Russo DP, Zorn KM, Clark AM, Korotcov A, Tkachenko V, Reynolds RC, Perryman AL, Freundlich JS, Ekins S. Comparing and Validating Machine Learning Models for Mycobacterium tuberculosis Drug Discovery. Mol Pharm 2018;15:4346–4360. doi: 10.1021/acs.molpharmaceut.8b00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Russo DP, Zorn KM, Clark AM, Zhu H, Ekins S. Comparing Multiple Machine Learning Algorithms and Metrics for Estrogen Receptor Binding Prediction. Mol Pharm 2018;15:4361–4370. doi: 10.1021/acs.molpharmaceut.8b00546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sandoval PJ, Zorn KM, Clark AM, Ekins S, Wright SH. Assessment of Substrate-Dependent Ligand Interactions at the Organic Cation Transporter OCT2 Using Six Model Substrates. Mol Pharmacol 2018;94:1057–1068. doi: 10.1124/mol.117.111443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang PF, Neiner A, Lane TR, Zorn KM, Ekins S, Kharasch ED. Halogen Substitution Influences Ketamine Metabolism by Cytochrome P450 2B6: In Vitro and Computational Approaches. Mol Pharm 2019;16:898–906. doi: 10.1021/acs.molpharmaceut.8b01214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zorn KM, Lane TR, Russo DP, Clark AM, Makarov V, Ekins S. Multiple Machine Learning Comparisons of HIV Cell-based and Reverse Transcriptase Data Sets. Mol Pharm 2019;16:1620–1632. doi: 10.1021/acs.molpharmaceut.8b01297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Clark AM, Dole K, Coulon-Spektor A, McNutt A, Grass G, Freundlich JS, Reynolds RC, Ekins S. Open Source Bayesian Models. 1. Application to ADME/Tox and Drug Discovery Datasets. J Chem Inf Model 2015;55:1231–1245. doi: 10.1021/acs.jcim.5b00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shen GH, Wu BD, Wu KM, Chen JH. In Vitro activities of isepamicin, other aminoglycosides, and capreomycin against clinical isolates of rapidly growing mycobacteria in Taiwan. Antimicrob Agents Chemother 2007;51:1849–1851. doi: 10.1128/AAC.01551-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kozikowski AP, Onajole OK, Stec J, Dupont C, Viljoen A, Richard M, Chaira T, Lun S, Bishai W, Raj VS, Ordway D, Kremer L. Targeting Mycolic Acid Transport by Indole-2-carboxamides for the Treatment of Mycobacterium abscessus Infections. J Med Chem 2017;60:5876–5888. doi: 10.1021/acs.jmedchem.7b00582 [DOI] [PubMed] [Google Scholar]
- [38].Franz ND, Belardinelli JM, Kaminski MA, Dunn LC, Calado Nogueira de Moura V, Blaha MA, Truong DD, Li W, Jackson M, North EJ. Design, synthesis and evaluation of indole-2-carboxamides with pan anti-mycobacterial activity. Bioorg Med Chem 2017;25:3746–3755. doi: 10.1016/j.bmc.2017.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Fernandez-Roblas R, Martin-de-Hijas NZ, Fernandez-Martinez AI, Garcia-Almeida D, Gadea I, Esteban J. In vitro activities of tigecycline and 10 other antimicrobials against nonpigmented rapidly growing mycobacteria. Antimicrob Agents Chemother 2008;52:4184–4186. doi: 10.1128/AAC.00695-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Falkinham JO 3rd, Macri RV, Maisuria BB, Actis ML, Sugandhi EW, Williams AA, Snyder AV, Jackson FR, Poppe MA, Chen L, Ganesh K, Gandour RD. Antibacterial activities of dendritic amphiphiles against nontuberculous mycobacteria. Tuberculosis (Edinb) 2012;92:173–181. doi: 10.1016/j.tube.2011.12.002 [DOI] [PubMed] [Google Scholar]
- [41].Disratthakit A, Doi N. In vitro activities of DC-159a, a novel fluoroquinolone, against Mycobacterium species. Antimicrob Agents Chemother 2010;54:2684–2686. doi: 10.1128/AAC.01545-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Baranyai Z, Kratky M, Vinsova J, Szabo N, Senoner Z, Horvati K, Stolarikova J, David S, Bosze S. Combating highly resistant emerging pathogen Mycobacterium abscessus and Mycobacterium tuberculosis with novel salicylanilide esters and carbamates. Eur J Med Chem 2015;101:692–704. doi: 10.1016/j.ejmech.2015.07.001 [DOI] [PubMed] [Google Scholar]
- [43].Pang H, Li G, Wan L, Jiang Y, Liu H, Zhao X, Zhao Z, Wan K. In vitro drug susceptibility of 40 international reference rapidly growing mycobacteria to 20 antimicrobial agents. Int J Clin Exp Med 2015;8:15423–15431. [PMC free article] [PubMed] [Google Scholar]
- [44].Cieslik W, Spaczynska E, Malarz K, Tabak D, Nevin E, O'Mahony J, Coffey A, Mrozek-Wilczkiewicz A, Jampilek J, Musiol R. Investigation of the antimycobacterial activity of 8-hydroxyquinolines. Med Chem 2015;11:771–779. doi: 10.2174/1573406410666150807111703 [DOI] [PubMed] [Google Scholar]
- [45].Anon. NIAID ChemDB HIV, Opportunistic Infection and Tuberculosis Therapeutics Database. 2018. [Google Scholar]
- [46].Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, Davies M, Dedman N, Karlsson A, Magarinos MP, Overington JP, Papadatos G, Smit I, Leach AR. The ChEMBL database in 2017. Nucleic Acids Res 2017;45:D945–D954. doi: 10.1093/nar/gkw1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ekins S, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Hohman M, Bunin B. A Collaborative Database And Computational Models For Tuberculosis Drug Discovery. Mol BioSystems 2010;6:840–851. [DOI] [PubMed] [Google Scholar]
- [48].Ekins S, Kaneko T, Lipinksi CA, Bradford J, Dole K, Spektor A, Gregory K, Blondeau D, Ernst S, Yang J, Goncharoff N, Hohman M, Bunin B. Analysis and hit filtering of a very large library of compounds screened against Mycobacterium tuberculosis Molecular bioSystems 2010;6:2316–2324. [DOI] [PubMed] [Google Scholar]
- [49].Gamo F-J, Sanz LM, Vidal J, de Cozar C, Alvarez E, Lavandera J-L, Vanderwall DE, Green DVS, Kumar V, Hasan S, Brown JR, Peishoff CE, Cardon LR, Garcia-Bustos JF. Thousands of chemical starting points for antimalarial lead identification. Nature 2010;465:305–310. [DOI] [PubMed] [Google Scholar]
- [50].Ekins S, Reynolds RC, Franzblau SG, Wan B, Freundlich JS, Bunin BA. Enhancing Hit Identification in Mycobacterium tuberculosis Drug Discovery Using Validated Dual-Event Bayesian Models PLOSONE 2013;8:e63240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ekins S, Casey AC, Roberts D, Parish T, Bunin BA. Bayesian Models for Screening and TB Mobile for Target Inference with Mycobacterium tuberculosis Tuberculosis (Edinburgh, Scotland) 2014;94:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ekins S, Freundlich JS, Reynolds RC. Fusing dual-event datasets for Mycobacterium Tuberculosis machine learning models and their evaluation. Journal of chemical information and modeling 2013;53:3054–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ekins S, Freundlich JS, Hobrath JV, Lucile White E, Reynolds RC. Combining computational methods for hit to lead optimization in Mycobacterium tuberculosis drug discovery. Pharm Res 2014;31:414–435. doi: 10.1007/s11095-013-1172-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Ekins S, Freundlich JS, Reynolds RC. Are Bigger Data Sets Better for Machine Learning? Fusing Single-Point and Dual-Event Dose Response Data for Mycobacterium tuberculosis. Journal of chemical information and modeling 2014;54:2157–2165. doi: 10.1021/ci500264r [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Ekins S, Perryman AL, Clark AM, Reynolds RC, Freundlich JS. Machine Learning Model Analysis and Data Visualization with Small Molecules Tested in a Mouse Model of Mycobacterium tuberculosis Infection (2014-2015). J Chem Inf Model 2016;56:1332–1343. doi: 10.1021/acs.jcim.6b00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ekins S, Pottorf R, Reynolds RC, Williams AJ, Clark AM, Freundlich JS. Looking back to the future: predicting in vivo efficacy of small molecules versus Mycobacterium tuberculosis. J Chem Inf Model 2014;54:1070–1082. doi: 10.1021/ci500077v [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.