Extended Data Fig. 4 |. Functions and localization of RAD51AP1 in the DSB response.
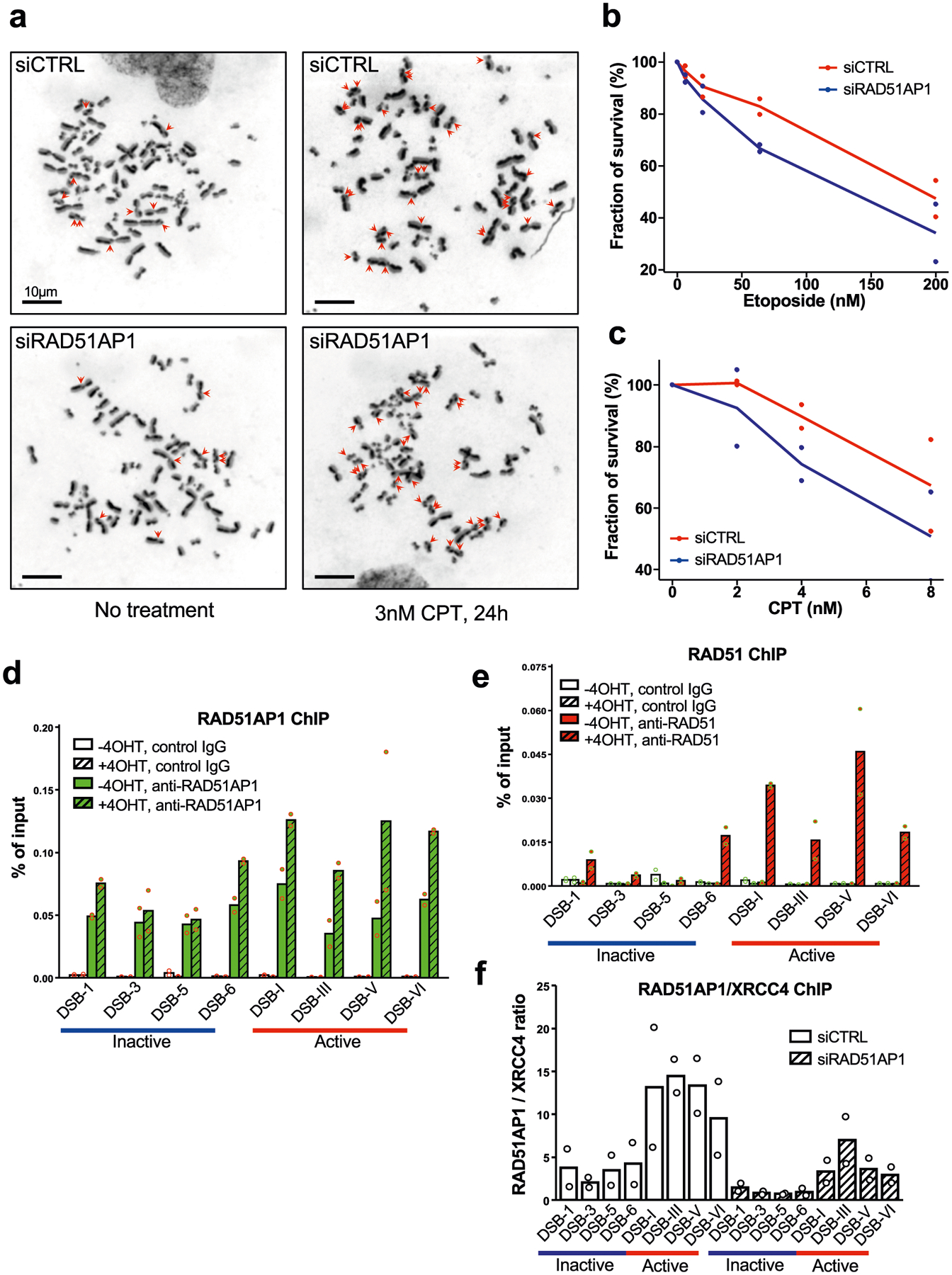
a, Representative mitotic spreads showing SCEs. Samples were prepared from U2OS cells transfected with control or RAD51AP1 siRNA, and treated with 3 nM CPT or vehicle for 24 h. Scale bars, 10 μm. b, c, Survival analyses of cells treated with etoposide (b) or CPT (c). Data are mean (n = 2 independent experiments). d, e, ChIP–qPCR analysis of RAD51AP1 (d) or RAD51 (e) at sites of AsiSI-induced DSBs in transcriptionally active and inactive regions. AsiSI-ER was activated by 4-OHT. f, RAD51AP1 ChIP signals at several transcriptionally active and inactive AsiSI sites were normalized to XRCC4 ChIP signals. XRCC4, which binds to DSBs independently of transcription, serves as a reference for RAD51AP1. RAD51AP1 knockdown substantially reduced RAD51AP1 ChIP signals, confirming the specificity of RAD51AP1 ChIP. Data in a–c are mean (n = 2 independent experiments).
