Abstract
Neural crest (NC) cells are a dynamic population of embryonic stem cells that create various adult tissues in vertebrate species including craniofacial bone and cartilage and the peripheral and enteric nervous systems. NC development is thought to be a conserved and complex process that is controlled by a tightly-regulated gene regulatory network (GRN) of morphogens, transcription factors, and cell adhesion proteins. While multiple studies have characterized the expression of several GRN factors in single species, a comprehensive protein analysis that directly compares expression across development is lacking. To address this lack in information, we used three closely related avian models, Gallus gallus (chicken), Coturnix japonica (Japanese quail), and Pavo cristatus (Indian peafowl), to compare the localization and timing of four GRN transcription factors, PAX7, SNAI2, SOX9, and SOX10, from the onset of neurulation to migration. While the spatial expression of these factors is largely conserved, we find that quail NC cells express SNAI2, SOX9, and SOX10 proteins at the equivalent of earlier developmental stages than chick and peafowl. In addition, quail NC cells migrate farther and more rapidly than the larger organisms. These data suggest that despite a conservation of NC GRN players, differences in the timing of NC development between species remain a significant frontier to be explored with functional studies.
Keywords: Neural crest, Specification, EMT, PAX7, SNAI2, SOX9, SOX10, Chicken, Quail, Peafowl
1. Introduction
Neural crest (NC) cells are a multipotent population of cells that arise from the epithelial ectodermal germ layer, undergo an epithelial to mesenchymal transition (EMT), migrate, and differentiate into many different cell and tissue derivatives (Martik and Bronner, 2017). During early neurulation, the lateral edges of the elevated neural plate border begin to express NC progenitor proteins, including PAX7 (Basch et al., 2006). Then, as the neural plate border fuses together to form the neural tube, NC cells are specified in the most dorsal region and begin to upregulate definitive NC markers, such as SNAI2 and SOX9 (Cheung and Briscoe, 2003; Spokony et al., 2002; Taneyhill et al., 2007). As development progresses, avian NC cells activate SOX10 prior to delaminating from the neural tube and initiate an EMT driven by changes in the expression of transcriptional regulators of cell adhesion molecules, cell adhesion, and in cell polarity (Cheng et al., 2000; Honore et al., 2003). During EMT and early migration, premigratory NC cells express E-cadherin (Dady et al., 2012; Dady and Duband, 2017; Rogers et al., 2018) and upregulate the expression of migratory cadherins (Cadherin-11 and Cadherin-7) (Kawano et al., 2002; Manohar et al., 2020; Vallin et al., 1998). Following EMT, migratory NC cells reduce expression of E-cadherin, modulating collective migration downstream of SIP1/ZEB2 (Rogers et al., 2013) and acquire a mesenchymal and migratory state. Next, they travel through distinct pathways to reach a variety of destinations, where they undergo differentiation (Bronner and LeDouarin, 2012). Any abnormalities that arise during the development of NC cells can lead to a variety of craniofacial birth defects, as well as a number of rare neurocristopathies (Pilon, 2021).
NC cell development is regulated by a complex gene regulatory network (GRN) consisting of morphogens that drive rapid changes in the expression of transcription factors, which then regulate the expression of cadherins (Simoes-Costa and Bronner, 2015). While there are hundreds of genes with reported functions in the GRN, there is still a lack of information about the endogenous spatiotemporal localization of their encoded proteins across species. Here, we quantify the onset, localization, and timing of NC cell development via comparative analysis of GRN proteins in three avian species, Gallus gallus (chicken), Coturnix japonica (quail), and Pavo cristatus (peafowl), by focusing on the expression of PAX7, SOX9, SNAI2, and SOX10. PAX7, a paired-box transcription factor, is one of the earliest markers of NC development, as it functions to specify the neural plate border and define the cells that will be competent to form the NC cell population (Basch et al., 2006; Khudyakov and Bronner-Fraser, 2009). SNAI2 is a zinc-finger transcription factor that is highly conserved among vertebrate species and known to play an important role in EMT, functioning in a regulatory loop with Cadherin-6B during delamination (Schiffmacher et al., 2016; Taneyhill et al., 2007). SOX9, a high-mobility-group (HMG) domain-containing transcription factor, is a marker of prospective and early migratory NC cells (Basch et al., 2006). SOX10 is an SRY-related HMG-box family transcription factor that is necessary and sufficient to drive NC migration (Cheng et al., 2000; Honore et al., 2003; McKeown et al., 2005). Multiple studies have utilized analyses of gene expression via in situ hybridization and RNA-sequencing to characterize the timing of NC cell development in multiple species (Khudyakov and Bronner-Fraser, 2009; Soldatov et al., 2019; Williams et al., 2019). However, a comprehensive analysis of protein expression between species and across NC development is necessary to provide a framework for detailed functional analysis in the future.
Work from multiple organisms identified that NC GRN factors undergo post-transcriptional (Hutchins and Bronner, 2018) and post-translational (Hausser et al., 2019; Lander et al., 2011; Lee et al., 2012) modifications. Therefore, it is necessary to combine transcriptional analyses with protein expression studies to form a more complete picture of NC development. Using three closely related avian species, we characterize the spatiotemporal expression of PAX7, SOX9, SNAI2, and SOX10. Some key differences appear in quail, including earlier expression of the NC specifiers, SOX9, SNAI2, and SOX10, earlier NC cell migration out of the neural tube marked by HNK1, and differences in cell migration modes. Further, NC EMT timing and mechanisms differ between the three avians. These results suggest that there are several key differences in the timing of NC cell development between species, emphasizing the need for functional in vivo protein studies in the future.
2. Material and methods
2.1. Avian embryos
Fertilized chicken and quail eggs were obtained from UC Davis Hopkins Avian Facility and incubated at 37 °C to the desired stages according to the criteria of Hamburger and Hamilton (HH) (Hamburger and Hamilton, 1951). Fertilized peafowl eggs were gifted from Dr. Pauline Perez. Fertilized eggs were incubated for different amounts of time to reach specific Hamburger Hamilton (HH)-equivalent stages, with quail being the fastest developing, followed by chick, then peafowl. To reach HH8, quail embryos were incubated 26–29 h, chick 28–32 h, and peafowl 36–42 h. Embryos were prepared for cryosectioning by equilibrating in 5% sucrose for 30 min to 1 h at room temperature, then were transferred to 15% sucrose overnight in 4 °C. Embryos were incubated in 10% gelatin overnight at 37 °C, flash frozen in liquid nitrogen and stored at −80 °C prior to sectioning. Embryos were then cyrosectioned in 14–16 μm thick sections at −27 °C on a Microm NX70 cryostat.
2.2. Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described (Manohar et al., 2020; Rogers et al., 2013). Embryos were fixed on filter paper in 4% paraformaldehyde (PFA) in phosphate buffer for 15 min at room temperature. After fixation, embryos were washed in 1X TBS (500 mM Tris-HCl pH 7.4, 1.5 M NaCl, and 10 mM CaCl2) containing 0.1% Triton X-100 (TBST + Ca2+). Embryos were incubated in blocking buffer (TBST + Ca2+ with 10% donkey serum) for 1 h at room temperature. Primary antibodies were diluted in blocking buffer and incubated with embryos for 24–48 h at 4 °C (Table 1). After incubation with primary antibodies, embryos were washed with TBST + Ca2+ and incubated with Alexa Fluor secondary antibodies diluted (1:500) in blocking buffer for 12–24 h at 4 °C. Embryos were then washed with TBST + Ca2+ and post-fixed in 4% PFA for 1 h at room temperature or 12–24 h at 4 °C. All embryos were imaged in whole mount and section using Zeiss Imager M2 with Apotome capability and Zen optical processing software.
Table 1.
Antibodies used in study.
| Antibody target | Dilution | Species | Isotype | Immunogen | Source |
|---|---|---|---|---|---|
| SOX10 | 1:500 | Mouse | Mouse IgG2a | SOX10 fusion protein Ag23191 | Protein tech (66786-1-Ig) |
| SOX9 | 1:500 | Human | Rabbit IgG | KLH-conjugated linear peptide corresponding to the C-terminal sequence of human SOX9 | Millipore (AB5535) |
| PAX7 | 1:5–1:10 | Chicken | Mouse IgG1 | Amino acids 418-427 | DSHB (PAX7) |
| SNAI2 | 1:250 | Human | Rabbit IgG | Whole protein | Cell signaling (9585S) |
| HNK1 | 1:2 | Chicken | Mouse IgM | Chicken optic nerve | DSHB (3H5) |
2.3. In situ hybridization chain reaction (HCR)
HCR was performed using the protocol suggested by Molecular Technologies (Choi et al., 2018) with minor modifications. Chicken and quail embryos were fixed in 4% paraformaldehyde in phosphate buffer for 1 h at room temperature, washed in 1X PBS with 0.1% Tween-20, and were dehydrated in a series of 25%, 50%, 75%, and 100% methanol. Embryos were stored at −20 °C for up to 2 weeks prior to beginning HCR protocol. Embryos were rehydrated, but were not incubated with proteinase-K as suggested by the protocol. Embryos were incubated with 10 pmol of probes dissolved in hybridization buffer overnight at 37 °C. After washes on the second day, embryos were incubated with 15–30 pmol of hairpins H1 and H2 diluted in amplification buffer at room temperature overnight. Embryos were subsequently washed in 2X SSC+ 0.1% Tween-20 and imaged using a Zeiss Apotome.2 as described above.
2.4. Imaging
Fluorescence images were taken using Zeiss Imager M2 with Apotome.2 and Zen software (Carl Zeiss). Whole mount embryos were imaged using a EC Plan-Neofluar 10x/0.30 WD = 5.2 M27. Embryo sections were imaged using a Plan-Apochromat 20x/0.8 WD = 0.55 M27.
2.5. Neural tube size analysis
Neural tube size was measured using Adobe Illustrator. Vectors were drawn from the dorsal to the ventral sides of the neural tube and lateral to lateral to measure width of the neural tube. The distance measured was then converted into microns using the in-image scale bar. Graphs were generated using GraphPad 9. An ordinary one-way ANOVA statistical test was used to compare data sets.
2.6. Cell migration and cell count analyses
Cell migration was measured using Adobe Illustrator. A line was drawn from the dorsal midline of the neural tube to the end of the last migrated cell for SOX9, SNAI2, and SOX10+ cells at stages HH9 and HH11. The distance measured was then converted into microns using the in-image scale bar. Graphs were generated using GraphPad 9. An unpaired t-test was used for statistical analysis. Cells were counted in FIJI using the Cell Counter plug-in and plotted using GraphPad 9. The percentage of co-positive cells was calculated by dividing the number of cells expressing one protein over the number of the same cells expressing another protein. An ordinary one-way ANOVA statistical test was used to compare data sets.
2.7. Protein localization analysis
Overlay of protein expression in whole mount and sections was performed in FIJI by subtracting background and superimposing all embryos from that stage to demonstrate that protein expression is consistent between embryos within the same stage as in (Simoes-Costa and Bronner, 2015).
2.8. Fluorescence intensity analysis
The medial-lateral fluorescence intensity was calculated in FIJI by drawing a segmented line either from ventrolateral to dorsal or dorsal to lateral neural tube at a line width of 200 and a distance of 200 μm. A fluorescence intensity graph was generated through the plot profile plugin, which was subtracted from background. The fluorescence intensity was then normalized by dividing each point by the highest fluorescence intensity value.
2.9. Protein sequence and domain analysis
Chick and quail DNA and amino acid sequences were obtained from the National Center for Biotechnology Information (NCBI) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Peafowl DNA sequence contigs were obtained from the NCBI Pavo Cristatus genome sequencing and assembly (Accession PRJNA413288) (Dhar et al., 2019). Both quail and chick sequences were used as queries to identify peafowl orthologous NC GRN genes. Reassembled DNA contigs were translated using Expasy Translate software (Ison et al., 2013). Full length amino acid sequences were then input into the Simple Modular Architecture Research Tool (SMART) Program (Letunic and Bork, 2018; Letunic et al., 2021) to create domain architecture maps that were exported as *.svg files and annotated in Adobe Illustrator. Multiple sequence alignments and comparisons were performed using Jalview (Procter et al., 2021; Waterhouse et al., 2009), Clustal Omega, and MView (Madeira et al., 2019).
3. Results
Neural crest cells have a large degree of plasticity, and prior work indicated that even between the closely related chick and quail, NC cells exhibit differences from gene expression, timing, and contributions to derivatives (Le Douarin et al., 2004; Rothstein et al., 2018). To define the similarities and differences in the spatial and temporal expression and localization of multiple NC-specific GRN genes proteins, we performed HCR and IHC on embryos from multiple avian species. We collected embryos at multiple HH-equivalent developmental stages, performed HCR (quail, chick) and IHC (quail, chick, peafowl), we characterized the temporal similarities and differences in expression, quantified the percentage of cells expressing each protein, and identified the differences in cranial NC cell migration between quail, chick, and peafowl. Here we present a detailed analysis of NC cell protein localization from neurulation (HH7) to late migration (HH15). The novelty in this study lies in our focus on spatiotemporal protein expression as a complement to previous RNA-sequencing (Williams et al., 2019) and in situ hybridization studies focusing on gene expression (Khudyakov and Bronner-Fraser, 2009).
3.1. Onset of NC gene expression differs in quail and chick
Previous NC gene expression studies performed separately in quail (Sakai et al., 2006) and chick (Khudyakov and Bronner-Fraser, 2009) identified differential onset of genes such as Sox9. To characterize expression of these genes side-by-side in embryos, we performed in situ Hybridization Chain Reaction (HCR) across multiple stages of quail and chick embryos using probes for Snai2, Sox9, Sox10, and Pax7. HCR identified that quail embryos upregulate Snai2 gene expression by 2 somite stage (SS) (data not shown) in the neural plate border, then robustly express Snai2, and weakly express Sox9 in the dorsal neural folds by 3 SS (Fig. 1A–1C, 1A1–1C1). In contrast, chick embryos do not express Snai2 or Sox9 at 3 SS stage, but do express the NC progenitor marker, Pax7 (Fig. 1M–1O). Sox9 and Sox10 are expressed at 4 SS in quail embryos (Fig. 1D–1F, 1D1–1F1), but in chick embryos, only Sox9 is expressed, and Sox10 is absent, at 4 SS (Fig. 1P–1R). By 4 SS-6 SS stages, Snai2, Sox9, and Sox10 are strongly expressed in quail premigratory NC cells (Fig. 1G–1L, 1G1–1 L1) and by 4 SS and 5 SS, respectively, Snai2 is strongly expressed, and Sox9 and Sox10 are weakly expressed, in chick (Fig. 1P–1X).
Fig. 1. Onset of quail NC gene expression is earlier than chick.
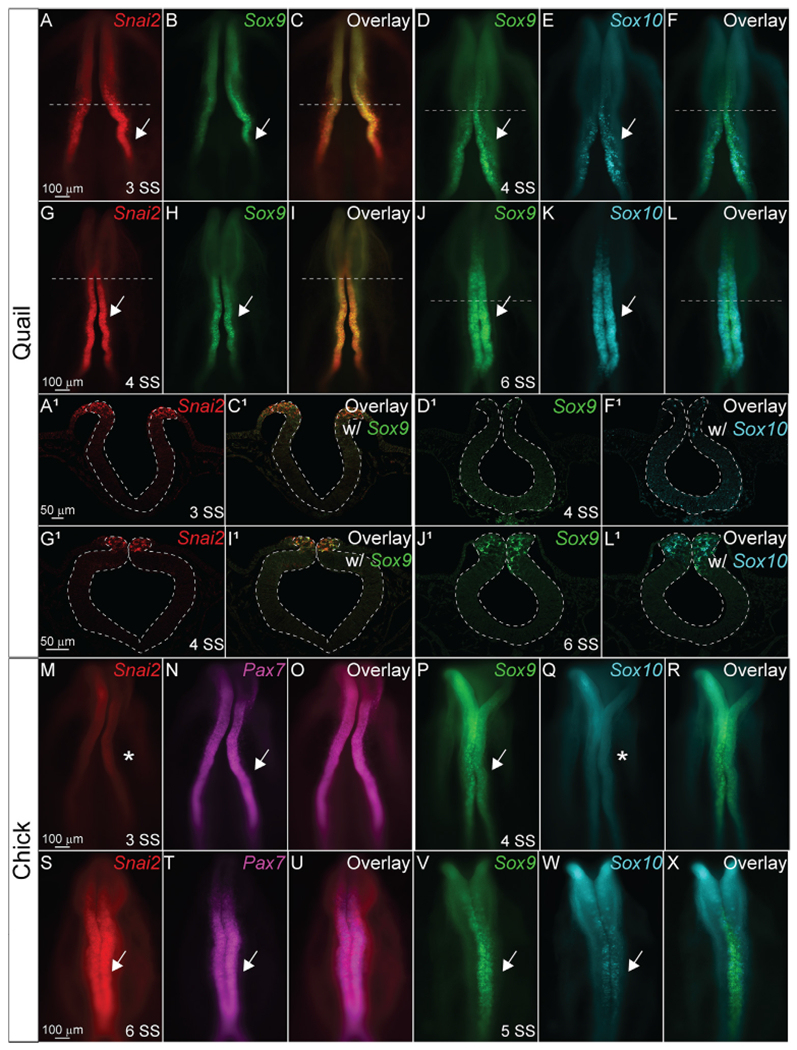
Hybridization chain reaction (HCR) using probes for Snai2, Sox9, Sox10, and Pax7 in (A-L1) quail and (M–X) chick embryos. (A, G, A1, G1, M, and S) are Snai2 (red). (B, H, D, J, D1, J1, P, V) are Sox9 (green). (E, K, Q, W) are Sox10 (cyan). (N, T) are Pax7 (magenta). (C, C1, F, F1, I, I1, L, L1, O, U, R, and X) show overlays of two genes. Arrows indicate positive signal, while asterisks indicate no signal. Scale bars as as marked in rows.
3.2. PAX7 timing and localization is similar between chick, quail, and peafowl
To define the similarities and differences between NC protein expression in chick, quail, and peafowl we performed IHC for PAX7, a NC progenitor marker, from neurulation (HH7, 1–3 SS) to migration (HH11, 13–15 SS) stages. Further, we created composite expression overlays by aligning multiple embryos to identify if the spatial patterning is consistent between individuals. In confirmation with previous gene and protein expression studies in chick (Basch et al., 2006; Khudyakov and Bronner-Fraser, 2009), PAX7 protein is expressed in neural plate border cells at HH7 (Fig. 2A–2C′, 1SS) in quail, chick, and peafowl embryos, and continues to be expressed as the neural tube fuses at HH8 (Fig. 2D–2F′, 4–5 SS). There exists a moderate amount of mediolateral variability in the location of the neural plate border in quail embryos (Fig. 2A’), but the spatial localization of PAX7 appears consistent between the three organisms at HH8 (Fig. 2D–2F’, 4–5 SS). By HH9, PAX7+ NC cells have begun to migrate laterally out of the now fused dorsal midbrain in all three organisms (Fig. 2G–2I’, 7 SS).
Fig. 2. PAX7 expression timing in quail, chick, and peafowl whole embryos.
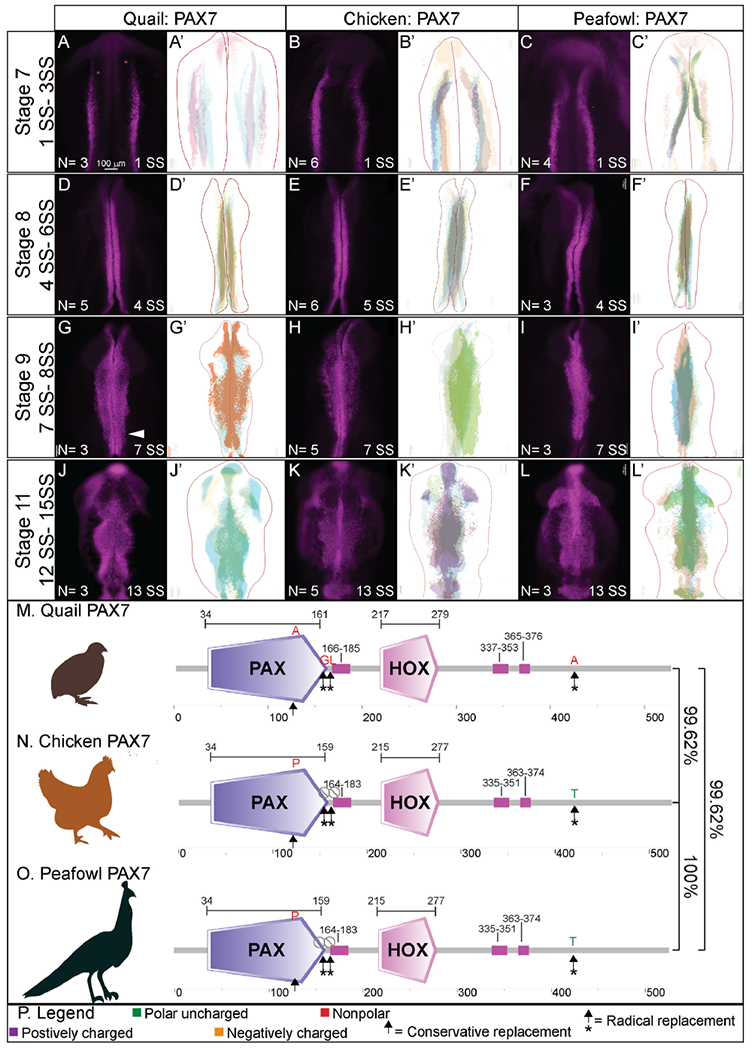
IHC for PAX7 expression in (A-C′) HH7 (1 SS) neural plate border, (D-F′) HH8 (5 SS) dorsal neural tube, (G-I′) HH9 (7 SS) EMT stage NC cells, and (J-L′) HH11 (13 SS), neural tube and migratory NC cells. (A, D, G, J) in quail, (B, E, H, K) in chick, and (C, F, I, L) in peafowl embryos. (A′, B′, C′) schematic overlays of multiple embryos. Quail exhibits more posterior midline PAX7+ expression at HH9 (G, arrowhead). Number of embryos analyzed at each stage and used for schematic overlays indicated in IHC panels. Scale bar is 100 μm and all images were taken at the same magnification. (M–O) Amino acid sequences were aligned and compared, then analyzed in SMART to obtain domain diagrams. PAX (purple) and HOX (pink) domains are indicated on images and small pink boxes are low complexity domains. (N, O) Chick and peafowl PAX7 proteins are identical and (M) quail PAX7 has two amino acid replacements, one conservative (same type of amino acid, black arrow), one radical replacement (black arrow with asterisk), and two additional amino acids (black arrows with asterisks). (P) Legend for (M–O). Purple is positively charged amino acid, green is polar uncharged, yellow is negatively charged, red is nonpolar. Scale bar for all whole mount images is 100 μm and is marked in the first image.
Some key differences appear at HH9 between species. In quail, PAX7+ NC cells appear to migrate in denser collectives compared to the chick (compare Fig. 2G to 2H). In peafowl, PAX7+ cells appear to have delayed migration out of the midline (compare Fig. 2I–2G and 2H). Analysis of fluorescence intensity indicates similar PAX7 levels in the dorsal neural tube of chick and quail at HH8, and lower fluorescence intensity in peafowl (Supp. Fig. 1A–1D), but higher levels in the quail and peafowl dorsal neural tube at HH9 (Supp. Fig. 1E–1H) and HH11 (Supp. Fig. 1I–1L) compared to chick. Further, the laterally migrating cells extend more posteriorly in quail (Fig. 2H, white arrow). At HH11, PAX7 expression remains strong in the midline of chick embryos, suggesting continued migration out of the neural tube (Fig. 2J, 2J′, 13 SS). In peafowl, PAX7+ cells are largely still close to the midline, suggesting a later migration out of the neural tube (Fig. 2L and 2L’).
To begin to understand differences in PAX7 protein sequences between species, we performed multiple sequence alignment of the full-length chick, quail, and peafowl amino acid sequences. Chick and quail genomes are sequenced, annotated, and available on NCBI and KEGG, while the peafowl genome was reassembled using the available genome in NCBI (Dhar et al., 2019). Multiple sequence alignment identified that the PAX7 protein sequence is 100% conserved between chick and peafowl (Fig. 2M, 2O, Supp. Fig. 2M), while both sequences share 99.62% sequence identity with quail (Fig. 2N). The two sequences have 2 amino acid substitutions, one conservative (proline to alanine) and one radical (threonine to alanine) from chick and peafowl to quail (Fig. 2M–2P, arrows) and quail PAX7 has two amino acids inserted in the N-terminal region glycine and leucine) that chick and peafowl are lacking (Fig. 2N).
To resolve PAX7 timing and localization, embryos were sectioned in the transverse plane and imaged at the midbrain axial level. At HH7 (3 SS), PAX7 marks the neural plate border cells that will be competent to form NC prior to neural tube closure (Fig. 3A–3C). Quail and peafowl neurulation appears to occur at a slightly faster rate than in chick. As the neural tube fuses at HH8 (5 SS), chick and peafowl NC remain premigratory within the dorsal neural tube (Fig. 3E and F), while quail cells appear to migrate laterally out of the closing neural tube (Fig. 3D). At HH9 (7 SS), PAX7+ premigratory NC progenitors remain in the dorsal neural tube in all organisms, and NC cells migrate dorsolaterally in chick (Fig. 3H), and dorsally in quail (Fig. 3G) and peafowl (Fig. 3I). At this stage, peafowl NC cells are less densely packed at 7 SS compared to quail and chick. As migration continues at HH11 (13 SS), PAX7 expression is reduced in the leading cells at ventrolateral edge of the NC population in all organisms, but it remains expressed in the dorsal neural tube (Fig. 3J–3L).
Fig. 3. PAX7 expression timing in quail, chick, and peafowl sections.
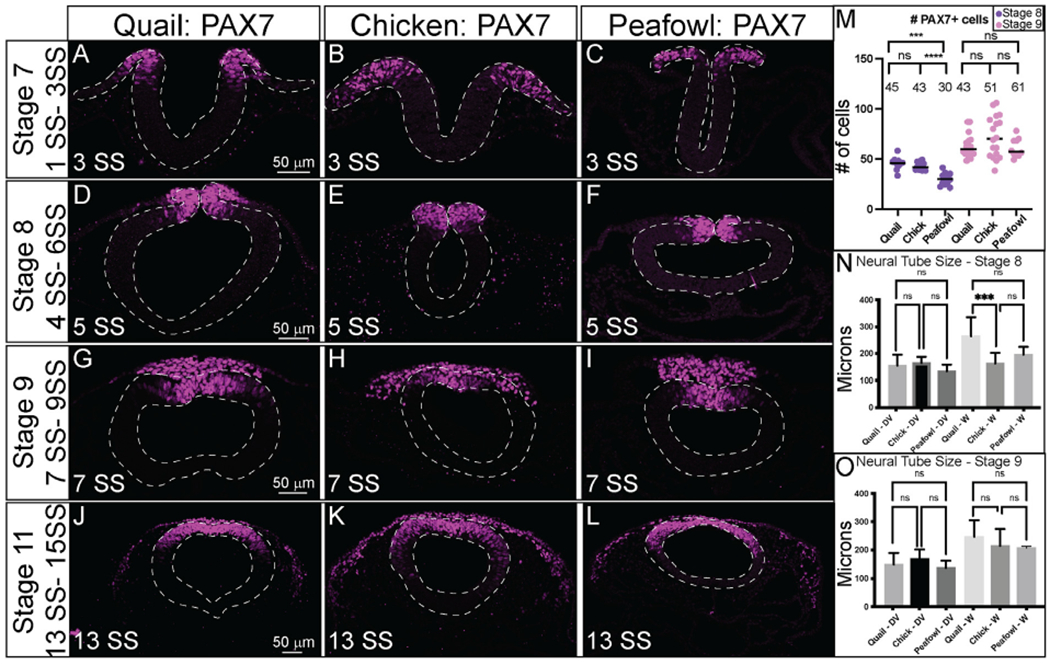
Transverse sections showing IHC for PAX7 in multiple stages of quail, chick, and peafowl embryos in (A–C) HH7, (D–F) HH8, (G–I) HH9, and (J–L) HH11. Dorsal is to the top and ventral is to the bottom. PAX7 expressed in all organisms by HH7 in the neural plate border (A–C), remains in the dorsal neural tube during neurulation (D–F), is expressed in premigratory and migratory NC cells (G–I), and is maintained in the dorsal neural tube and migratory NC cells (J–K). Scale bars are 50 μm and are as marked in first panel of each row. (M) The number of PAX7+ cells were quantified for HH8 and HH9, p = 0.001 between quail and peafowl, and p = 0.0002 between chick and peafowl, but no statistical difference was observed between chick and quail at HH8 (p = 0.990) or between any species at HH9 (p = 0.654 for chick and quail, p = 0.675 for chick and peafowl, and p = 0.999 for quail and peafowl). Ordinary one-way ANOVA statistical test used. (N) Neural tube size comparison at HH8. Chick dorsal-ventral (DV) compared to quail or peafowl DV is not significant, but width of the quail neural tube is larger than chick. Ordinary one-way ANOVA statistical test used (DV, p = 0.9756 and W, p = 0.002). (O) Neural tube size comparison at HH9. The DV and width of the neural tubes are no longer statistically significant. Ordinary one-way ANOVA statistical test used (DV, p = 0.3783 and W, p = 0.7398). Scale bar is marked in the first image of each row.
Given the differences in neural tube closure, we sought to determine if there were any differences in the number of PAX7+ cells in chick, quail, and peafowl at pre- and post-migratory stages (HH8 and HH9). However, at HH8, there is no statistical difference between the number of PAX7+ cells between chick and quail; however, there are significantly fewer cells in the peafowl compared to both the chick (Fig. 3M, p = 0.0002) and quail (Fig. 3M, p = 0.001). By HH9, there is no statistical difference in the number of PAX7+ NC cells in any of the organisms (Fig. 3M). In addition, we measured the differences in neural tube dorsal-ventral length and width between the three organisms, noting that at HH8, the width of quail neural tube is significantly larger than in chick (Fig. 3N, p = 0.0002), but by HH9, the differences in width are no longer significant (Fig. 3K, p = 0.4). For both HH8 and HH9, we did not observe statistically significant differences in dorsal-ventral lengths. We noted that in quail embryos, the larger neural tubes at HH8 were followed by more collective NC cell migration at HH9, raising interesting questions regarding differences in NC migration mechanisms in quail. To better understand these cell migration variances, we analyzed the expression of EMT inducers, SOX9 and SNAI2, and measured the migratory distances of cells positive for these proteins in the three species.
3.3. SNAI2 is expressed early in the neural plate border in quail
SNAI2 gene expression has been identified as early as HH8 and HH6.5 in chick and quail, respectively, with expression observed in the dorsal neural folds and neural plate border at the putative mid- and hindbrain levels (Basch et al., 2006; Khudyakov and Bronner-Fraser, 2009; Sakai et al., 2006). To determine if the protein localization in chick, quail, and peafowl is similar to its reported expression in chick at late HH7/early HH8 (3–4 SS) (Taneyhill et al., 2007), we performed IHC for SNAI2 in various stages. We observed SNAI2 protein strongly in the dorsal neural folds as early as stage HH7 (3 SS) in quail and peafowl (Fig. 4A and 4C), however, SNAI2 was only weakly expressed in chick at 3 SS and was more robust at 5 SS (Fig. 4B–4B’). At HH8 (4–6SS), SNAI2 is expressed in the fusing midbrain neural tube in chick, quail, and peafowl, but quail and peafowl SNAI2 expression extends more anteriorly into the diencephalon (Fig. 4A, 4B, 4D, 4E’, white arrows). At HH9 (7SS), SNAI2+ cells are undergoing EMT in all three organisms (Fig. 4G–4I′). Whereas quail NC cells strongly express SNAI2 in the midline, chick and peafowl SNAI2+ cells appear more uniformly distributed (compare Fig. 4G and 4I to 4H). At HH11, SNAI2+ migratory cells have not extended as far ventrolaterally or anteriorly in peafowl compared to chick and quail (Fig. 4L, 4L’, asterisk). The SNAI2 amino acid sequences are highly conserved in all three organisms (Fig. 4M–4P, Supp. Fig. 2M). Specifically, the amino acid sequences of chick and peafowl are 100% similar, and those sequences are 99.63% similar to the quail sequence, with a single conservative amino acid change on the N-terminus from glutamic acid to aspartic acid from chick and peafowl to quail (compare Fig. 4M, O to 4N).
Fig. 4. SNAI2 expression timing in quail, chick, and peafowl whole embryos.
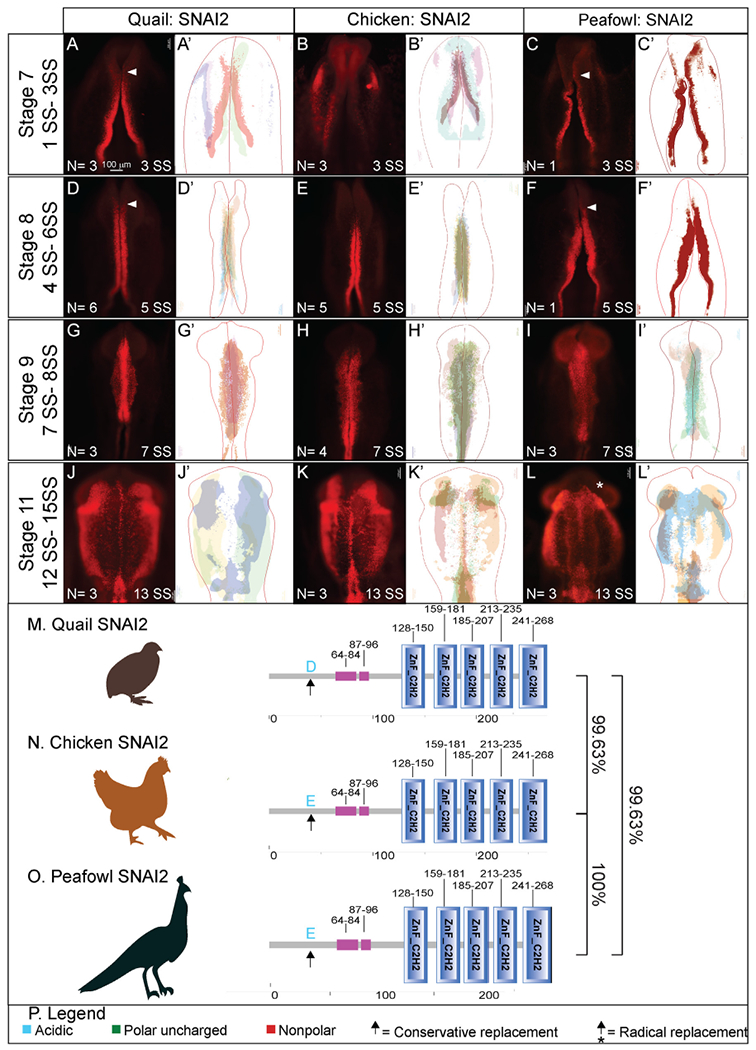
IHC for SNAI2 expression in (A-C′) HH7 (1 SS) neural plate border, (D-F′) HH8 (5 SS) dorsal neural tube, (G-I′) HH9 (7 SS) EMT stage NC cells, and (J-L′) HH11 (13 SS), neural tube and migratory NC cells. (A, D, G, J) in quail, (B, E, H, K) in chick, and (C, F, I, L) in peafowl embryos. (A′, B′, C′) schematic overlays of multiple embryos. Quail and peafowl exhibit more anterior SNAI2+ expression than chick between HH7 to HH8 (compare A, D to B, E and C, F, arrowhead). Number of embryos analyzed at each stage and used for schematic overlays indicated in IHC panels. Scale bar is 100 μm and all images were taken at the same magnification. (M–O) Amino acid sequences were aligned and compared, then analyzed in SMART to obtain domain diagrams. Zinc finger domains (blue) and small pink boxes are low complexity domains are shown on schematics. (N, O) Chick and peafowl SNAI2 proteins are identical and (M) quail SNAI2 has one conservative amino acid replacement (same type of amino acid, black arrow). (P) Legend for (M–O). Blue is acidic amino acid, green is polar uncharged, red is nonpolar. Scale bar for all whole mount images is 100 μm and is marked in the first image.
In transverse section, it is clear that SNAI2 expression in quail (Fig. 5A, HH7, 1 SS) precedes that of chick (Fig. 5B, HH7, 2 SS) and peafowl (Fig. 5C, HH7, 1 SS) in the neural plate border. At HH8 (5 SS), quail SNAI2+ cells are starting to undergo EMT or migrate out of the neural tube while chick and peafowl cells remain within the neuroepithelium (Fig. 5D–5F, 5 SS). As with previous markers, at HH9 (7 SS), quail SNAI2+ cells appear to migrate more collectively during EMT compared to chick, while the chick embryos maintain stronger SNAI2 expression in the premigratory population than either quail or peafowl (Fig. 5G–5I). By HH11 (13–14 SS) SNAI2 is expressed in migrating NC cells in chick, quail, and peafowl with very few SNAI2+ premigratory NC cells (Fig. 5J–5L, asterisk). As SNAI2 drives NC EMT (Taneyhill et al., 2007) and is expressed earlier in quail, we analyzed the number of total SNAI2+ cells and the distance that SNAI2+ cells migrated from the midline. There were significantly more quail SNAI2+ NC cells at HH8 in quail embryos compared to chick, but by HH9, this difference was lost (Fig. 5M). We compared the distance that SNAI2+ cells migrated from the midline to the leading edge in all three organisms, and at HH9, both chick and quail cells migrated farther ventrolaterally than peafowl, but by HH11, the distance between chick or quail with peafowl was no longer significantly different (Fig. 5N, p = 0.004 for chick and peafowl and p = 0.008 for quail and peafowl, p = 0.052 for chick and quail at HH9). However, at HH11 quail SNAI2+ NC cells migrated further than chick SNAI2+ cells (Fig. 5N, p = 0.043). Fluorescence intensity analyses demonstrated higher intensity for SNAI2+ cells in quail premigratory NC cells than chick at the NC leading edge (Supp. Fig. 2A–2C). Early migratory NC cells had similar intensity between species for migrating NC cells (Supp. Fig. 2H–2K).
Fig. 5. SNAI2 expression timing in quail, chick, and peafowl sections.
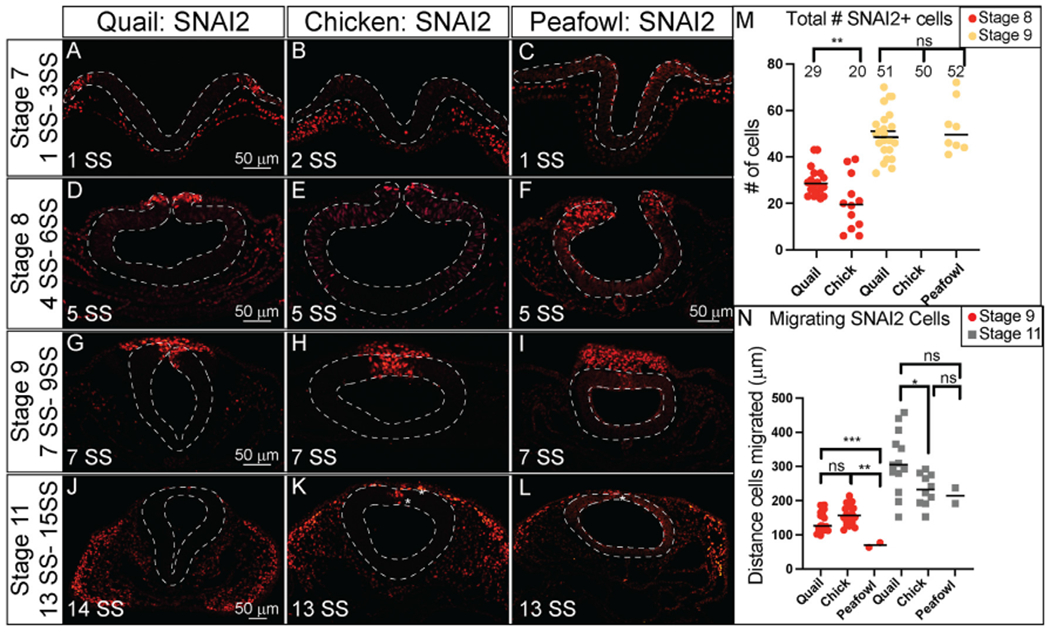
Transverse sections showing IHC for SNAI2 in multiple stages of quail, chick, and peafowl embryos in (A–C) HH7, (D–F) HH8, (G–I) HH9, and (J–L) HH11. Dorsal is to the top and ventral is to the bottom. In quail, SNAI2 is expressed at HH7 in the neural plate border (compare A to B and C), is expressed in the dorsal neural tube during neurulation/NC specification at HH8 in all three organisms (D–F), is expressed in premigratory and migratory NC cells at HH9 during EMT (G–I), and is maintained in the dorsal neural tube in a small number of premigratory NC cells but is highly expressed in migratory NC cells at HH11 (J–K). Scale bars are 50 μm and are as marked in first panel of each row. (M) The number of SNAI2+ cells were quantified for HH8 and HH9, at HH8 quail had statistically more cells than chicken, but by HH9 that statistical significance was no longer observed. Ordinary one-way ANOVA statistical test was used (HH8, p = 0.0571 and HH9, p = 0.9975). (N) Cell migration was measured from the neural tube midline to the furthest migrated cell for HH9 and HH11. At HH9, chicken and quail cells migrated further than peafowl (p = 0.004 chick and peafowl and p = 0.008 for quail and peafowl, p = 0.052 for chick and quail). At HH11, quail cells have migrated further than chick (p = 0.043 for chick and quail, p = 0.946 for chick and peafowl, and p = 0.209 for quail and peafowl). Scale bar is marked in the first image of each row or in panel F.
3.4. SOX9 is more robustly expressed in quail and peafowl during specification
The Sox9 gene is expressed at HH8 in chick in the closing neural folds (Fig. 1) (Basch et al., 2006; Betancur et al., 2009; Yardley and Garcia-Castro, 2012) and as early as HH6 in the anterior neural folds of quail embryos (Sakai et al., 2006). To identify whether the protein expression mirrors the spatiotemporal gene expression, we performed IHC for SOX9 in multiple stages of chick, quail, and peafowl embryos. In contrast to PAX7, SOX9 protein was not expressed prior to late HH7 in any of the three species. At HH7, quail and peafowl SOX9 expression is visible in several NC cells within the closing neural tube (Fig. 6A, 6C, 3 SS). In contrast, chick SOX9 is barely apparent in the dorsal neural tube at late HH7 in whole mount (Fig. 6A, 6A′, 3 SS). At HH8, SOX9 is expressed in the dorsal neural tube at the midbrain level in all three organisms, but is more robust in the quail and peafowl (Fig. 6D–6F′, 5 SS). At HH9 during EMT, SOX9+ cells are expressed in migrating NC cells (Fig. 6G–6I′, 7 SS). Similar to PAX7 expression, chick SOX9+ cells appear more dispersed during migration than quail or peafowl SOX9+ cells, which appear more condensed in quail and retained to the midline in peafowl (compare Fig. 6H–6G and 6I). Additionally, peafowl SOX9+ cells have not migrated as far as those of chick and quail at this stage. By HH11, SOX9 is still expressed in migrating NC cells, as well as the otic placode in all species (Fig. 6J–6L′, 13 SS). Few SOX9+ cells remain in the neural tube at this stage (Fig. 6J, 6K, 6L white arrows). Also, peafowl SOX9+ cells have not migrated as far anteriorly in peafowl at this stage (Fig. 6L, 6L’). As with the PAX7 and SNAI2 sequence alignments, chick and peafowl SOX9 amino acid sequences are 100% conserved and are 99.39% similar to the quail SOX9 sequence (Fig. 6M–6O, Supp. Fig. 3M). There are both conservative (serine to asparagine) and radical replacements (lysine to glutamic acid and alanine to threonine) in the sequences as well as a lost amino acid (glutamine) in chick and peafowl compared to quail (Fig. 6M–6P).
Fig. 6. SOX9 expression timing in chick, quail, and peafowl whole embryos.
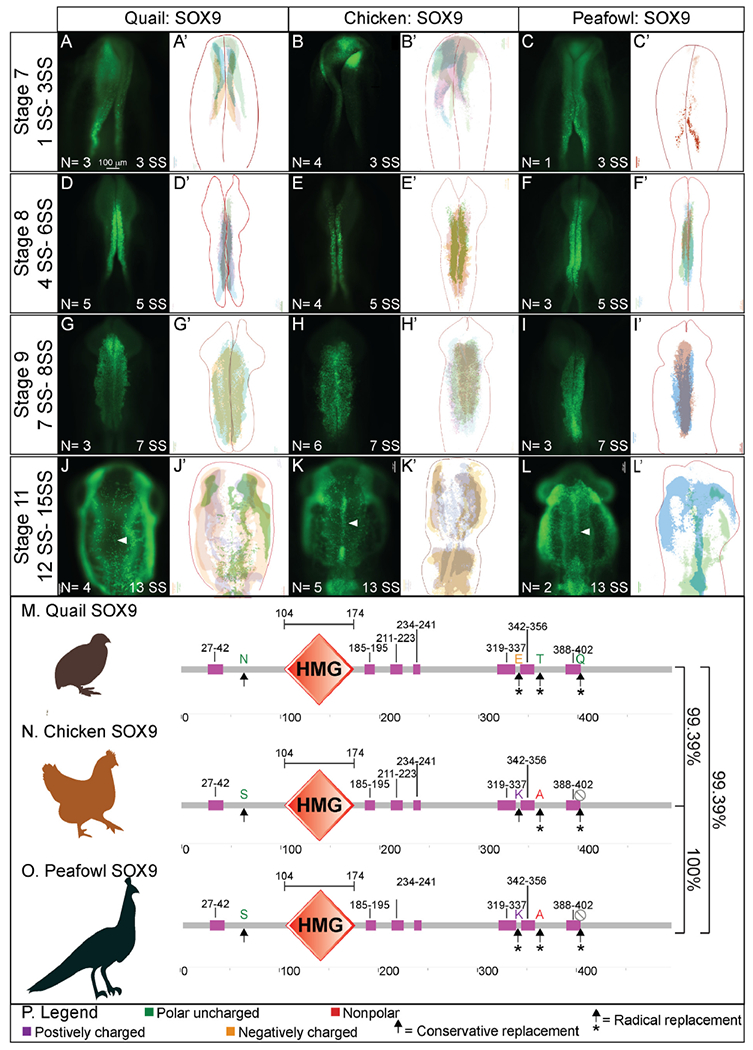
IHC for SOX9 expression in (A-C′) HH7 (1 SS) neural plate border, (D-F′) HH8 (5 SS) dorsal neural tube, (G-I′) HH9 (7 SS) EMT stage NC cells, and (J-L′) HH11 (13 SS), neural tube and migratory NC cells. (A, D, G, J) in quail, (B, E, H, K) in chick, and (C, F, I, L) in peafowl embryos. (A′, B′, C′) schematic overlays of multiple embryos. Chick and peafowl maintain more SOX9+ cells in premigratory cells at HH11 than quail (compare K and L to J, arrowhead). Number of embryos analyzed at each stage and used for schematic overlays indicated in IHC panels. Scale bar is 100 μm and all images were taken at the same magnification. (M–O) Amino acid sequences were aligned and compared, then analyzed in SMART to obtain domain diagrams. HMG domains (orange) and low complexity domains (pink) are shown in diagram. (N, O) Chick and peafowl SOX9 proteins are identical and (M) quail SOX9 has four amino acid replacements, one conservative (same type of amino acid, black arrow), two radical replacements (black arrow with asterisk), and one additional amino acid (black arrows with asterisks). (P) Legend for (M–O). Purple is positively charged amino acid, green is polar uncharged, yellow is negatively charged, red is nonpolar. Scale bar for all whole mount images is 100 μm and is marked in the first image.
In sections, quail SOX9 is upregulated in premigratory NC cells by late HH7, while chick SOX9 is expressed at very low levels in the dorsal neural folds (Fig. 7A and 7B, 3 SS). Due to limited embryo availability, we were unable to obtain a section in peafowl at this stage. SOX9 is also expressed in the notochord at all stages tested in all three species (Fig. 7A–7K). Similar to PAX7+ cells, quail SOX9+ NC cells begin migrating at the onset of neural tube closure at HH8 (5 SS, arrowhead) while chick and peafowl cells remain in the neural tube (Fig. 7C–7E). We confirmed that quail cells migrate more collectively at HH9 as they are more densely packed compared to chick and peafowl (Fig. 7F–7H, 7 SS). At HH11, most chick and quail SOX9+ cells have migrated out of the neural tube while peafowl still has some premigratory cells remaining (Fig. 7I–7K, 14–15 SS).
Fig. 7. SOX9 expression timing in quail, chick, and peafowl sections.
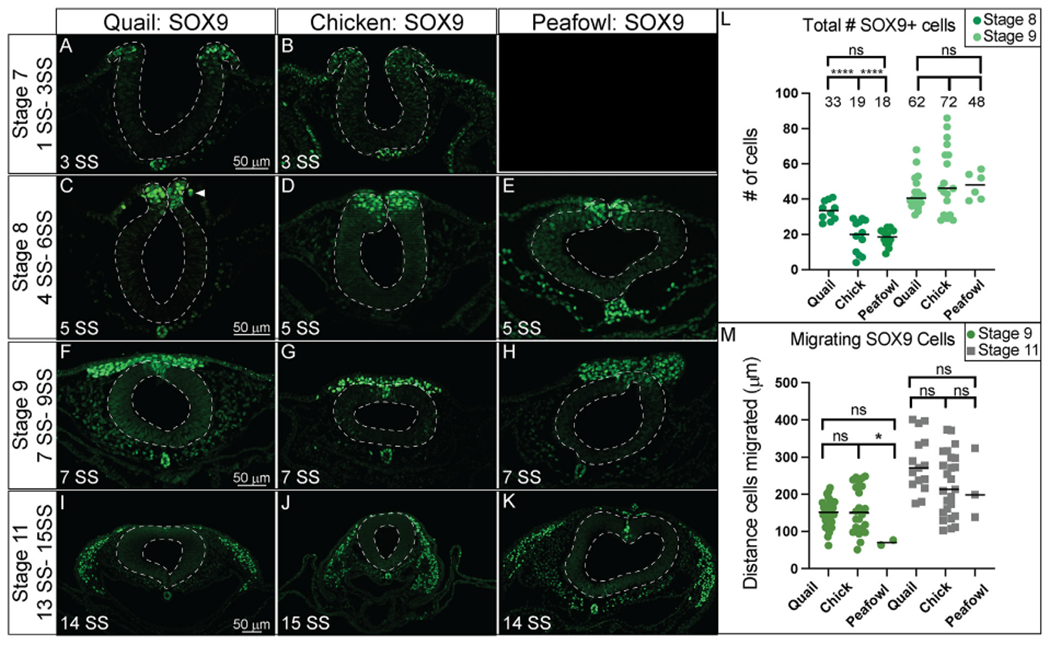
Transverse sections showing IHC for SOX9 in multiple stages of quail, chick, and peafowl embryos in (A, B) HH7 (not obtained for peafowl), (C–E) HH8, (F–H) HH9, and (I–K) HH11. Dorsal is to the top and ventral is to the bottom. SOX9 is expressed in quail at HH7 in the neural plate border/dorsal neural tube (compare B to A), but we were unable to obtain peafowl sections at this stage to compare. SOX9 is expressed in the dorsal neural tube during neurulation/NC specification at HH8 in all three organisms (C–E), is expressed in premigratory and migratory NC cells at HH9 during EMT (F–H), and is maintained in the dorsal neural tube in a small number of premigratory NC cells (most prevalent in peafowl), but is highly expressed in migratory NC cells at HH11 (I–K). Scale bars are 50 μm and are as marked in first panel of each row. (L) The number of SOX9+ cells were quantified for HH8 and HH9. At HH8, quail expressed more SOX9+ cells than chick (p = 0.0001) and peafowl (p = 0.0001), but by HH9 this difference was no longer statistically significant (p = 0.997). Ordinary one-way ANOVA statistical test used. (M) Cell migration was measured from the neural tube midline to the furthest migrated cell for HH9 and HH11. No statistical significance was observed, except between quail and peafowl at HH9 (p = 0.0373). Scale bar is marked in the first image of each row.
To determine if the timing of NC specification by SOX9 expression was equivalent between species, we counted the total number of NC cells positive for SOX9 at stages HH8 and HH9 (Fig. 7L). We identified that there are an average of approximately 1.7X more SOX9+ cells in quail embryos compared to chick and peafowl embryos at HH8 (4–6 SS) (Fig. 7L). At HH9, the average number of SOX9+ NC cells is no longer statistically different between organisms although there is variability in the total number of cells expressing SOX9 (Fig. 7L). To identify if the differences in timing of quail NC specification lead to increased migratory distance, we measured the distance between the leading SOX9+ NC cells from the midline of the embryos. While there are no statistical differences in migration distance between chick and quail or chick and peafowl at HH9, quail SOX9+ cells migrated further compared to peafowl (Fig. 7J). By HH11 there was no statistical difference in migration distance from the midline in any of the species suggesting that the early onset of specification does not necessarily lead to faster NC migration (Fig. 7J). These results suggest that there may be differences in NC timing between avian species. Additional analysis of SOX9 fluorescence intensity in the dorsal neural tube at HH8 confirms that SOX9 levels in the dorsal neural tube of chick, quail, and peafowl are similar at HH8 7 (Supp. Fig. 3A–3D). However, at HH11, chick and peafowl SOX9 levels in the dorsal neural tube exhibited stronger intensity than quail, while quail cells had stronger intensity in the migratory regions (Supp. Fig. 3I–3L).
3.5. SOX10 expression occurs just prior to EMT in all species
Previous analysis of Sox10 gene expression in chick identified expression in the cranial neural folds at HH8+, in migrating NC cells at HH9, and in otic placodes at HH10+ (Basch et al., 2006; Betancur et al., 2010; McKeown et al., 2005). Quail Sox10 gene expression has been reported as barely detectable at HH8 but is strongly expressed in the cranial neural folds and migrating NC cells by HH9 (Sakai et al., 2006). Our analysis detected Sox10 transcripts as early as 4 SS in quail and 5 SS in chick (Fig. 1). IHC for SOX10 protein shows similar localization to Sox10 gene expression. Chick SOX10 protein is barely detectable at HH8, but quail and peafowl SOX10 is expressed more strongly in the dorsal neural folds (Fig. 8A–8C′, 5 SS). By HH9, SOX10 is expressed in early migrating NC cells in all three organisms (Fig. 8D–F′). Chick SOX10+ cells are more dispersed, while quail and peafowl SOX10+ cells appear more closely associated (Fig. 8D–F′, 7 SS). By HH11, migrating NC cells continue to express SOX10 sparsely in the midline and more intensely in the ventrolateral NC populations (Fig. 8G–I′, 13 SS). SOX10 is also expressed in the otic placodes in all organisms at HH11 (Fig. 8G–I′). Of the protein sequences compared, the SOX10 amino acid sequence exhibited the most differences between species (Supp. Fig. 4M). In the case of SOX10, amino acid sequence alignment showed that chick and peafowl SOX10 are 98.92% similar (Fig. 8N, 8O), chick and quail have 98.48% similar protein sequences (Fig. 8M, 8N), and quail and peafowl have 99.13% sequence similarity (Fig. 8M, 8O). The quail SOX10 amino acid sequence has 104 amino acids on the terminal end that do not appear to exist in either chick or peafowl sequences, and therefore, similarity was calculated by aligning only conserved portions of the proteins (Fig. 8M, Supp. Fig. 4M). There are two conservative amino acid replacements in the N-terminus (aspartic acid to glutamic acid and asparagine to serine) in chick and peafowl compared to quail (compare Fig. 8M–O). The remaining amino acid changes are derived from radical replacements. In the case of SOX10, the peafowl sequence mirrors the chick sequence and differs from quail at positions 108 (aspartic acid to glutamic acid), 137 (asparagine to serine), and 417 (threonine to alanine). The peafowl sequence mirrors the quail sequence and differs from chick at positions 139 (proline to serine), 143 (glycine to serine), 299 (alanine to serine), and 307 (threonine to alanine). At position 142, chick and quail mirror each other and differ from peafowl (alanine to serine) (Fig. 8M, 8N, Supp. Fig. 4M). Although the protein expression timing differs and the localization is similar between organisms, future work will focus on defining if the protein functions are conserved between species.
Fig. 8. SOX10 expression timing in quail, chick, and peafowl whole embryos.
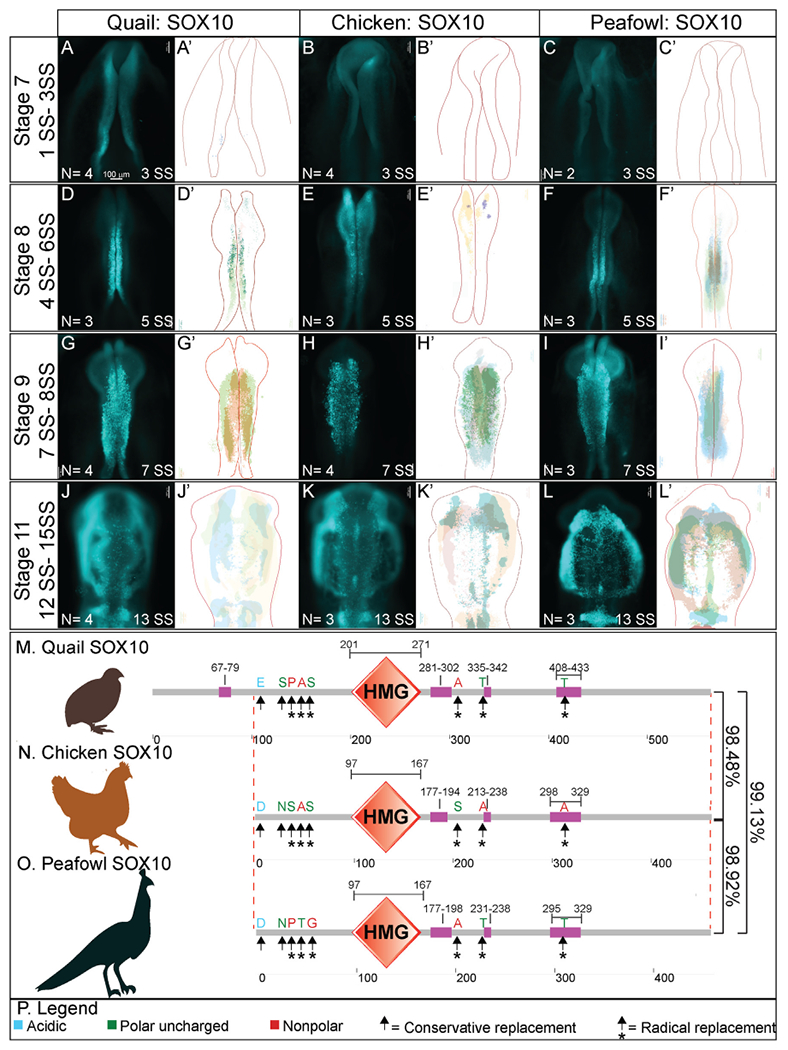
IHC for SOX10 expression in (A-C′) HH8 (5 SS) dorsal neural tube, (D-F′) HH9 (7 SS) EMT stage NC cells, and (G-I′) HH11 (13 SS), neural tube and migratory NC cells. (A, D, G) in quail, (B, E, H) in chick, and (C, F, I) in peafowl embryos. (A′- C′, D′- F′, G′- 1′) schematic overlays of multiple embryos. Peafowl embryos have delayed migration of SOX10+ cells compared to chick and quail as the cells remain visible in the dorsal focal plane (compare I to G and H). Number of embryos analyzed at each stage and used for schematic overlays indicated in IHC panels. Scale bar is 100 μm and all images were taken at the same magnification. (J–L) Amino acid sequences were aligned and compared, then analyzed in SMART to obtain domain diagrams. HMG domains (orange) and low complexity domains (pink) are shown on schematics. (J–L) Peafowl SOX10 differs from the other proteins profiled here. It shares amino acid sequences with both chick and quail (same type of amino acid, black arrow, different type of amino acid black arrow plus asterisk), and has one amino acid that is unique from both chick and quail, which resemble each other. (M) Legend for (J–L). Blue is acidic amino acid, green is polar uncharged, red is nonpolar. Scale bar for all whole mount images is 100 μm and is marked in the first image.
In transverse section, SOX10 protein is expressed in a limited number of premigratory NC cells, co-localizing with a small proportion of the PAX7, SNAI2, and SOX9+ populations in all species after HH8 (Fig. 9A–9F). At late HH8, SOX10 appears localized to premigratory NC cells in the neural tube, but is expressed as early as 4 SS in quail (Figs. 8A, Fig. 9D–F, 5 SS). At HH9, very few cells in the neural tube are SOX10+ in chick, quail, or peafowl, while most migrating cells express SOX10 (Fig. 9G–I, 7 SS). As with previous markers, SOX10+ migratory NC cells are more densely packed in quail compared to chick (Fig. 9G–I). By HH11, SOX10 is expressed in migratory cells, and SOX10+ quail NC cells continue to migrate collectively into the ventrolateral regions of the head in contrast to the more mesenchymal chick and peafowl cells (Fig. 9J–L, 13 SS).
Fig. 9. SOX10 and HNK1 expression timing in quail, chick, and peafowl sections.
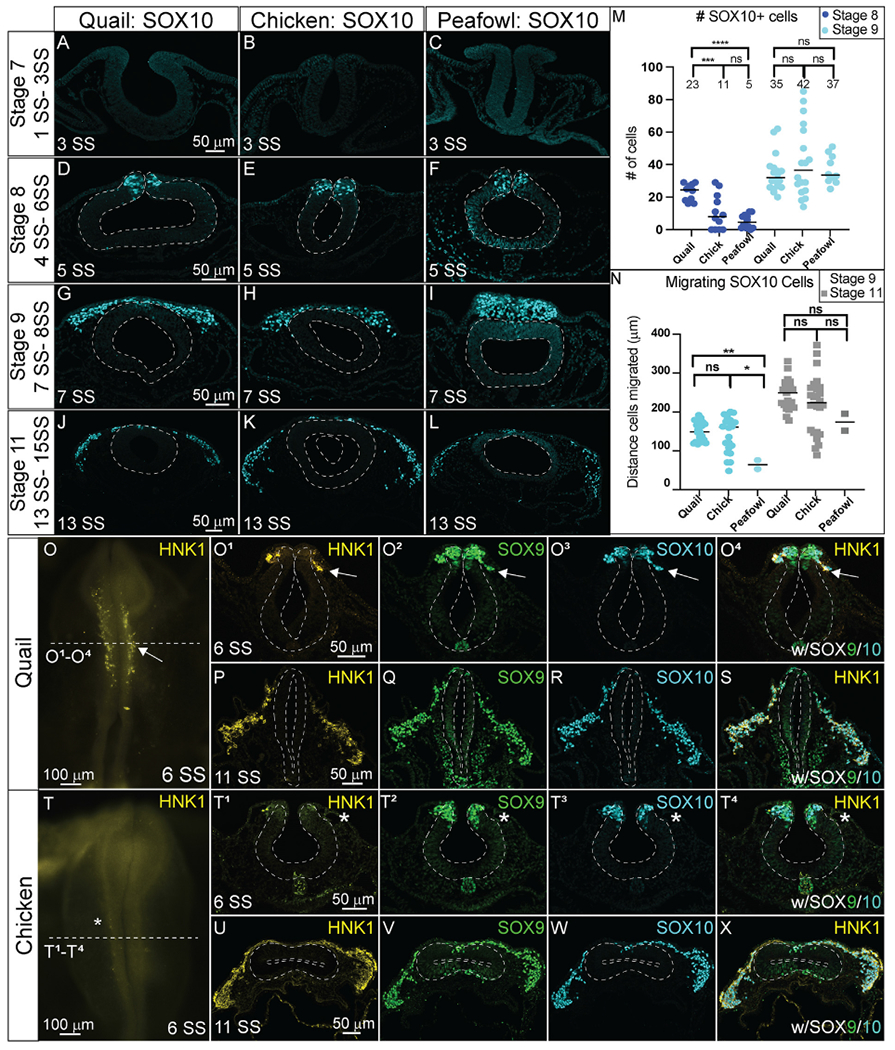
Transverse sections showing IHC for (A–N) SOX10 and (O–X) HNK1 in multiple stages of chick, quail, and peafowl embryos in (A-C, O– O4) HH8, (D–F) HH9, (G–I) HH11, and (P– S, U- X) HH10. Dorsal is to the top and ventral is to the bottom in section images while anterior is to the top and posterior to the bottom in whole mount images. SOX10 is not expressed at HH7 in any of the embryos (A–C) but is expressed in the dorsal neural tube during neurulation/NC specification at HH8 in all three organisms (D–F), is expressed in premigratory and migratory NC cells at HH9 during EMT (G–I), and is maintained in the dorsal neural tube in a small number of premigratory NC cells but is highly expressed in migratory NC cells at HH11 (J–L). (M) The number of SOX10+ cells were quantified for HH8 and HH9. At HH8, quail expressed more SOX10+ cells than chick (p = 0.001) and peafowl (p = 0.001), but by HH9 this difference was no longer statistically significant (p = 0.990). An ordinary one-way ANOVA statistical test was used. (N) Cell migration was measured from the neural tube midline to the furthest migrated cell for HH9 and HH11. At HH9, chick (p = 0.019) and quail (p = 0.009) cells migrated further than peafowl, but no statistically significant difference in distance was observed between quail and chick (p = 0.760). At HH11, no statistical difference was observed between species (p = 0.288 for chick and quail, p = 0.556 for chick and peafowl, and p = 0.244 for quail and peafowl). An Ordinary one-way ANOVA statistical test was used. Scale bars are as indicated in each whole mount panel or the first panel of each section row.
To confirm developmental differences in NC timing at premigratory stages, we counted the number of SOX10+ cells at HH8 and 9. At HH8 (3–5 SS), there were 2-fold more SOX10+ cells in quail embryos compared to chick and 4-fold more SOX10+ cells in quail embryos compared to peafowl (Fig. 9M), but this difference was lost by HH9. At HH9, peafowl cells migrated a significantly shorter distance compared to either chick or quail. In contrast to SNAI2+ cells, there were no differences in the distances migrated by SOX10+ NC cells in any species at HH 11 (Fig. 9N). Fluorescence intensity measurements of premigratory SOX10 positive cells at HH8 in sectioned embryos showed higher intensity for quail and peafowl compared to chick, consistent with whole mount and section data (Supp. Fig. 4A–4D). However, by HH9, fluorescence intensity is similar between chick and quail, and by HH11, the majority of SOX10+ cells have migrated out of the neural tube and are migrating laterally (Supp. Fig. 4E–4L).
Noting that quail SOX10 had an earlier onset and that quail NC cells appeared to migrate at an earlier developmental stage than either chick or peafowl, we characterized the expression of a migratory cell marker, HNK1 in quail and chick embryos (Fig. 9O–9X). HNK1 expression was upregulated in quail as early as 6 SS, and the HNK1+ cells co-localized with both SOX9 and SOX10 at HH stages 8–11 (Fig. 9O1–9O4, 9P–9S, 6 SS- 11 SS). In contrast, HNK1 was not expressed at 6 SS in chick embryos (Fig. 9T–9T4), but was expressed in early migrating NC cells by 7 SS (Supp. Fig. 5). HNK1 expression in chick remained co-localized with SOX9 and SOX10 in migratory NC cells similar to quail embryos (Fig. 9U–9X, Supp. Fig. 5).
3.6. Heterogeneous protein expression identifies subsets of NC populations in all organisms
Prior work established that the premigratory NC population is multipotent (Bhattacharya et al., 2018; Buitrago-Delgado et al., 2015; Kerosuo et al., 2015) and that at the transcript level, multiple NC-related genes coding for transcription factors are expressed throughout the dorsal neural tube (Lignell et al., 2017; Williams et al., 2019). However, information defining the timing and localization of the NC proteins during NC specification and EMT is still lacking. To identify the extent of overlapping and discrete expression of the NC progenitor protein PAX7 with specifier/EMT driving proteins (SNAI2, SOX9, SOX10), we performed multiplexed IHC during specification (HH8, 4–6 SS) and EMT (HH9, 7–9 SS) stages. The number of NC cells expressing each protein was quantified in transverse sections from multiple embryos at equivalent HH stages (Figs. 10 and 11). Comparing the total number of PAX7, SOX9, and SOX10+ cells in chick, quail, and peafowl at HH8 identified both similarities and differences in NC developmental timing between species. There are no significant differences in the number of PAX7+ cells in chick and quail at HH8, but there are significantly fewer PAX7+ cells in peafowl at HH8 compared to both chick and quail (Fig. 10A, 10C, 10G, 10K, chick to peafowl). Further, as shown above, there were more SOX9 and SOX10+ NC cells in quail than either chick or peafowl at HH8 (Fig. 10D, 10E, 10H, 10I, 10L, 10M). During specification in quail, 77% and 51% of PAX7+ cells co-express SOX9 and SOX10, respectively at HH8 (4–6 SS) (Figs. 10B and 10C–10F). In chick (HH8, 4–6 SS), 48% and 24% of PAX7+ cells are also positive for SOX9 or SOX10, respectively (Figs. 10B and 10G–10J). In peafowl embryos, 61% and 17% of PAX7+ cells co-express SOX9 and SOX10, respectively (Fig, 10B, 10K–10N). These data suggest that there is a more rapid onset of NC specifiers in quail than chick or peafowl embryos, with the NC program beginning at earlier HH stages. In contrast to the low to moderate co-expression of SOX9 and SOX10 proteins with PAX7, SNAI2 appeared to be expressed in more NC cells at HH8 than the other specifiers in all three organisms (Fig. 10O–10Z).
Fig. 10. Comparative expression of PAX7, SOX9, SOX10, and SNAI2 during specification in quail, chick, and peafowl.
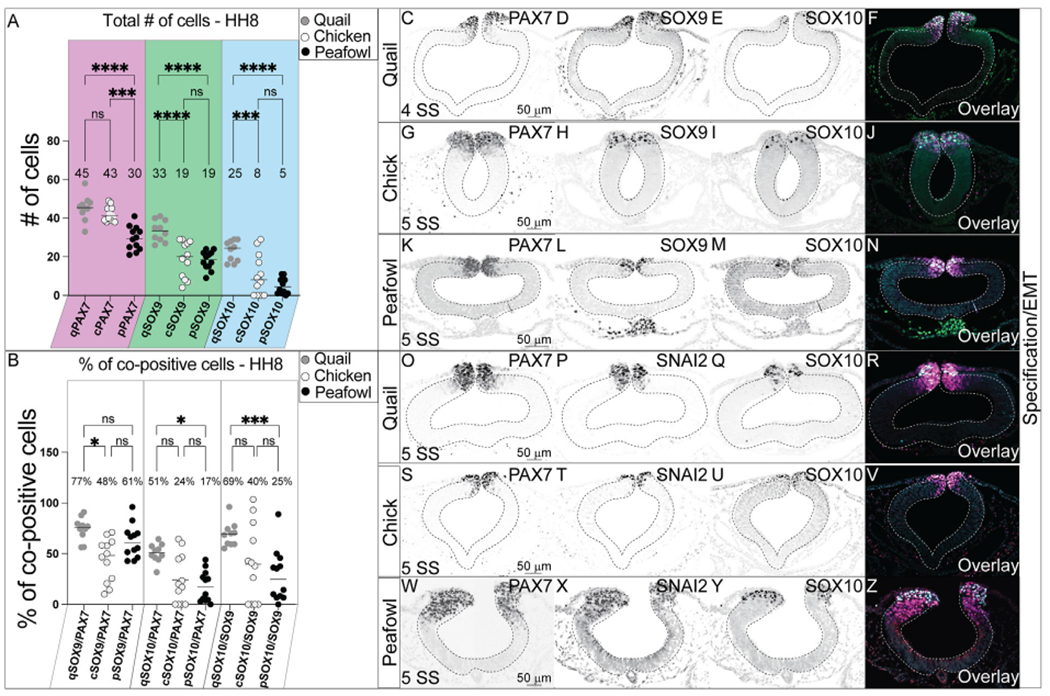
(A) The number of PAX7+ cells is not significantly different between chicken and quail at HH8, but both chick and quail have a higher number of PAX7+ cells than peafowl (p = 0.001 between quail and peafowl, p = 0.0002 between chick and peafowl, but no statistical difference was observed between chick and quail at HH8 (p = 0.990). For SOX9+ cells, quail has more than both chick and peafowl (p = 0.001 for both). For SOX10+ cells, quail has more cells than both chick and peafowl (p = 0.001 for both). An ordinary one-way ANOVA statistical test was used. (B) Quail has a higher percentage of co-positive cells at HH8 for SOX9/PAX7 compared to chick (p = 0.034) and a higher percentage of co-positive cells for SOX10/PAX7 (p = 0.015) and SOX10/SOX9 (p = 0.0009) compared to peafowl. An ordinary one-way ANOVA statistical test was used. IHC for PAX7, SOX9, and SOX10 in (C– F) quail, (G–J) chick, and (K– N) peafowl at HH8 (5 SS). IHC for PAX7, SNAI2, and SOX10 in (O–R) quail, (S– V) chick, and (W–Z) peafowl at HH8 (5 SS). Black and white images are single protein expression at indicated stage and color images are overlay of all three markers. Scale bars are as indicated in first panel of each row.
Fig. 11. Comparative expression of PAX7, SOX9, SOX10, and SNAI2 during EMT in quail, chick, and peafowl.
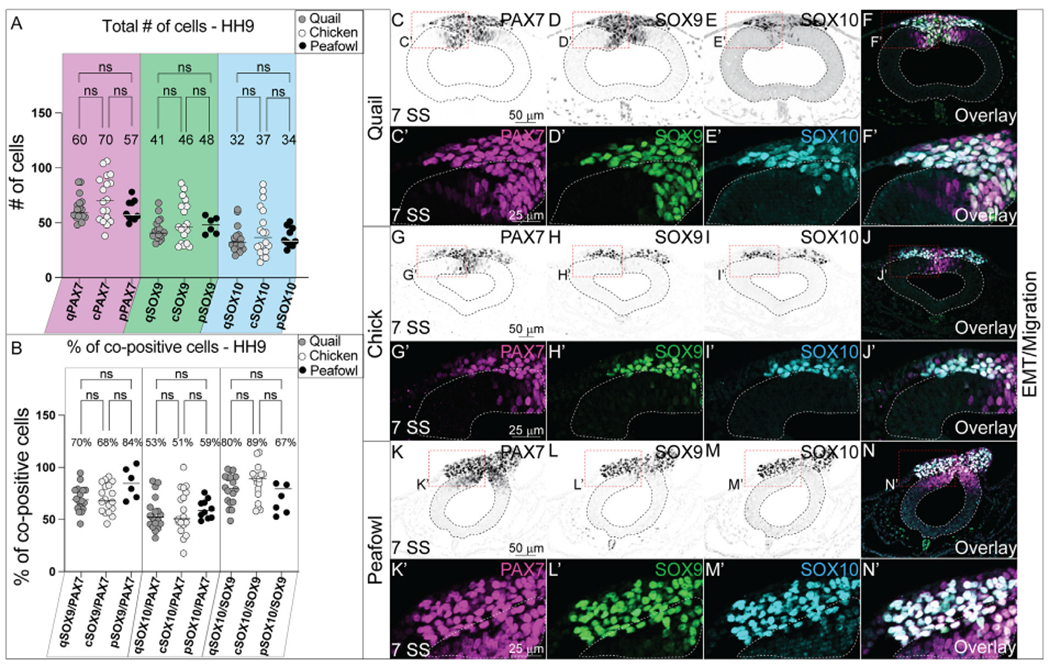
(A) The total number of cells for PAX7, SOX9, and SOX10 are not statistically significant between quail, chick, and peafowl. An ordinary one-way ANOVA statistical test was used (PAX7, p = 0.675–0.999, SOX9, p = 0.878–0.999, SOX10, p = 0.895–0.999). (B) Percentage of co-positive cells at HH9 between quail, chick, and peafowl shows no statistical difference. An ordinary one-way ANOVA statistical test was used (SOX9/PAX7, p = 0.9999, SOX10/PAX7, p = 0.9998, SOX10/SOX9, p = 0.8684). IHC for PAX7, SOX9, and SOX10 in (C– F′) quail, (G- J′) chick, and (K– N′) peafowl at HH9 (7 SS). Black and white images are single protein expression at indicated stage and color images are overlay of all three markers. Red dashed boxes indicate zoomed images in (C′- F′, G′- J′, and K′- N′). Neural tubes are indicated by dashed lines. Scale bars are as indicated in first panel of each row.
During EMT (HH9, 7–9 SS), the total number of NC cells expressing PAX7, SOX9, and SOX10 individually or together are no longer significantly different between chick, quail, or peafowl (Figs. 11A and 11C–11N′). At this stage, the early migrating NC cell populations look very similar (Fig. 11F, 11J, 11N) in all organisms, and the percent of cells co-positive for PAX7 cells co-expressing SOX9 is 68% in chick, 70% in quail, and 84% in peafowl, but is not statistically significant (Fig. 11B, 11C′, 11D′, 11G′, 11K′, 11L′). At the same stage, the percent of PAX7+ NC cells co-expressing SOX10 is 51% in chick, 53% in quail, and 59% in peafowl and is not statistically different (Fig. 11B, 11C′, 11E′, 11I′, 11K′, 11M′). Further, at HH9, the percent of SOX9 cells that co-express SOX10 is 89% in chick, 80% in quail, and 67% in peafowl, but the ratios are not statistically different from each other (Figs. 11B and 11D–11N). Our results support the proposed hierarchical NC gene regulatory network (GRN) that begins with PAX7 expression in a large population of NC progenitors, followed by the succession of the NC specifiers SOX9, SNAI2, and SOX10 (Meulemans and Bronner-Fraser, 2004; Nikitina et al., 2008; Steventon et al., 2005); however, they suggest that the timing of these proteins differs between species.
3.7. Differential timing and expression during differentiation
Based on their differential rates of specification and migration as identified by early expression of NC specifiers and faster migration in quail, we hypothesized that there may be differences in protein localization during NC differentiation. Quail, chick, and peafowl embryos were cultured to HH15 (26 SS) and IHC for SOX9, SOX10, and PAX7 was performed (Fig. 12). We noted some anatomical differences in the developing embryos (rostral shape, heart size, branchial arch (BA) patterning, and anterior prominence) (Fig. 12A–12C, 12D–12F). We also witnessed apparent differences in protein expression at these stages. Unlike chick and peafowl, quail embryos appear to express SOX10 in the developing eye (Fig. 12B), and only weakly SOX9 in the trigeminal ganglia (TG) (Fig. 12A), while both proteins are strongly expressed in the chick and peafowl TG (Fig. 12B and C). Also, peafowl embryos strongly express SOX9 in their otic vesicles (OV), while the others only express SOX10 (compare Fig. 12A and 12D to 12G). These analyses suggest that at the same HH stage during differentiation phases, there are differences in avian developmental program markers at the molecular level. Transverse sections of the developing embryos maintain similar expression profiles and patterning with some differences. Namely, PAX7 expression extends more ventrally in the quail neural tube compared to chick, and PAX7 expression appears to be maintained for longer distances in migrating NC in chick compared to quail embryos (Fig. 12C1–12C3, 12 F1–12F3).
Fig. 12. SOX9/SOX10 localization during differentiation differs between avian species.
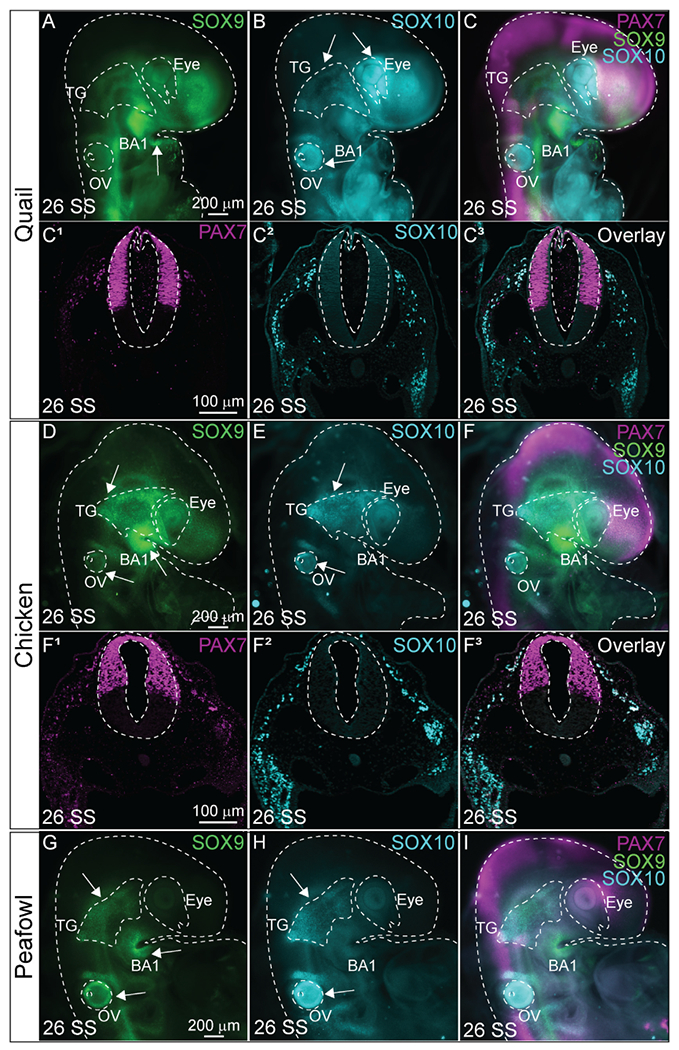
IHC in (A- C3) quail, (D- F3) chick, and (G–I) peafowl embryos for (A, D, G) SOX9, (B, C2, E, F2, H) SOX10, and (C, C3, F, F3, I) overlay with PAX7 in 26 SS embryos. Differences: (B) SOX10 is strongly expressed in the quail eye, but not chick and peafowl. (G) SOX9 is prominently expressed in the peafowl otic vesicle (OV), but not quail or chick. (A, B, D, E, G, H) The trigeminal ganglia (TG) strongly expresses SOX9 and SOX10 in chick and peafowl, but expression is weak in quail. Embryos are outlined with dashed lines. Whole mount embryos are anterior to the top facing right and sections are dorsal to the top. Scale bars are as marked in first panel of each row.
4. Discussion
We compared the spatiotemporal localization and number of PAX7, SNAI2, SOX9, and SOX10+ cells in chick, quail, and peafowl embryos from early neurula stages (HH7) to late migration and differentiation stages (HH15). We have identified that the spatial location of specified NC cells is conserved in chick, quail, and peafowl embryos. Specifically, all three organisms develop NC crest cells within the dorsal neuroepithelium, and these cells undergo a true EMT: delaminating from the neural tube, collectively migrating, and undergoing mesenchymalization during migration (Figs. 3, 5, 7 and 9). In addition, although PAX7 protein expression and timing appears conserved in all species at late HH7 in the dorsal neural folds, the early onset of quail SNAI2 protein differs from expression in chick and peafowl (Figs. 4 and 5) (Taneyhill et al., 2007). Further, in quail, SOX9, SNAI2, and SOX10 are expressed 1–2 SS stages earlier than in chick and peafowl, NC cells appear to undergo EMT at an earlier stage, and the quail cells migrate further and faster. For the most part, the amino acid sequences of these proteins are highly conserved between all three species, with peafowl and chick sequences more similar to each other than those in quail (Figs. 2, 4 and 6). However, the SOX10 amino acid sequence exhibits intriguing differences (Fig. 8) that we intend to study in the future. There appears to be a similar program at play that regulates specification and EMT of NC cells, but how these factors are related hierarchically in each organism remains unclear. Thus, this work provides a framework to define similarities and differences in the molecular mechanisms regulating NC cell development between closely related species with different external phenotypes and developmental timelines.
4.1. Differences in developmental timing
While analyzing the expression of each protein, we found that quail not only expressed SNAI2, SOX9, and SOX10 at earlier equivalent developmental time points than chick and peafowl, but quail NC cells also appeared to migrate out of the neural tube and upregulate HNK1 expression earlier than chick and peafowl (HH8 versus HH9) (Figs. 3, 5, 7 and 9). This expedited NC development was particularly interesting as we identified that at HH8, quail neural tubes were significantly larger in diameter when compared to chick, but by HH9, this difference was no longer observed. In addition, we also found that at HH8, quail embryos had more SOX9 and SOX10+ cells. Taken together, we hypothesize that due to a larger neural tube and shorter developmental timeline in quail, NC GRN proteins are likely activated earlier to initiate earlier cell migration out of the neural tube. These results are consistent with the scaling pattern differences seen in the ventral neural tube between chick and zebra finch (Uygur et al., 2016), but they do not explain the differences in the heterochronous onset of protein expression when comparing all three organisms.
When comparing ventral neural tube patterning between the zebra finch and the chick, it was apparent that ventral neural tube patterning in the zebra finch occurs on a smaller scale and over a shorter time than in chick and that these differences in scaling are in response to a gradient of the morphogen Sonic hedgehog (Shh) (Uygur et al., 2016). Although early chick neural tube patterning (Ericson et al., 1997; Luo et al., 2006) and NC specification and EMT (Martik and Bronner, 2017; Rogers and Nie, 2018) are well characterized, less is known about the factors that drive NC induction and EMT in quail (Schneider, 2018), and virtually nothing has been previously shown about embryogenesis in general or NC-specific development in peafowl embryos.
Links between signaling molecules like Wnt, Shh, and BMP, and dorsoventral patterning, including NC formation, are well established in frog and chick (Bhattacharya et al., 2018; Chalpe et al., 2010; Garnett et al., 2012; Ossipova and Sokol, 2011; Piloto and Schilling, 2010; Shi et al., 2011). Further, cooperation between SOX9, SNAI2, and BMP signaling drives NC specification and EMT in chick and quail (Sakai et al., 2006; Zhang and Klymkowsky, 2009), however, there is a lack of information about the expression, timing, or signaling of these morphogens in other avian species during NC specification and EMT. Based on the rapid onset of quail NC specification and EMT, it is likely that the signals driving these processes also have earlier onset. As SNAI2 has been linked to the BMP4 and Wnt signaling pathways previously in Xenopus and mouse embryos, it is possible that those morphogen pathways begin signaling at earlier stages in quail (Sakai et al., 2005; Tribulo et al., 2004).
Together, these cross-species studies suggest mechanisms that explain differences in developmental timing and species-specific scaling to better understand developmental growth between closely related avian species. Further studies are necessary to understand the mechanisms, which drive earlier quail NC cell development compared to chick. Compared to late stage limb development, NC specification (HH8) and EMT (HH9) occur significantly earlier in development. We identified that quail NC cells were specified approximately 4–6 h faster than chick and 8–10 h faster than peafowl based on time of incubation. Quail embryos incubate for 17 days, whereas chick embryos incubate for 20 and peafowl for 30 days prior to hatching, so it is logical that quail undergo faster development. However, we did not identify a specific linear scale of development based on normal developmental timelines or hatched size of embryo (quail < chick < peafowl); rather, quail NC cells appeared to develop more quickly than the other avians despite their somite-based staging. Further, at the early stages analyzed, the three types of embryos were not significantly different in overall size (see scale bars in Figs. 1–12). Our results support the idea of scaled developmental processes, but also suggest that quail NC cell development is more rapid regardless of embryonic size, and that some proteins may play different roles in quail compared to chick embryos based on their stage of onset.
4.2. Heterogeneous protein expression
To further quantify the similarities and differences in NC cell development between quail and chick, we counted the total number of cells, and the proportion of cells co-expressing different factors. We identified that NC cells have spatially and temporally heterogeneous NC protein expression, and that the population is not a homogenous cell population, complementing previous studies using single cell RNA-sequencing at NC specification stages (HH8) and spatial genomic analysis (SGA) at NC EMT stages (HH9) in chick embryos (Lignell et al., 2017; Williams et al., 2019). Single cell RNA-sequencing (scRNA-seq) and HCR analyses performed at HH8-9 and HH10 identified high levels of Pax7 and Snai2 transcripts that decreased from HH8 to HH10, and that chromatin accessibility changes for NC specifiers between HH8 and HH10, suggesting that cis-regulatory landscapes are dynamic during different stages of NC development (Williams et al., 2019). SGA, a quantitative single cell profiling technique, confirmed that markers of premigratory NC cells inhabit multiple stem cell niches within the dorsal neural tube, but that these groups are spatially distinct (Lignell et al., 2017).
Our multiplexed spatiotemporal protein expression analyses support prior work demonstrating hierarchical temporal NC GRN gene expression with PAX7 protein expressed earliest in the neural plate border followed by SNAI2, then SOX9, and finally, SOX10 (Khudyakov and Bronner-Fraser, 2009). However, not all PAX7+ cells express NC specifier proteins. Rather, it is clear that only small subsets of premigratory PAX7+ cells actively express SOX9 and SOX10 proteins during specification (HH8) for example (Fig. 10), and in quail, the co-expressing subset is significantly larger at HH8 (74% versus 44%). The early expression of bonafide NC markers, SOX9 and SNAI2, in quail versus chick and peafowl suggests that quail NC cells are specified earlier, which is supported by their early migration, but also may dictate novel protein functions and protein-protein interactions due to the different embryonic environments. Future work will focus on defining species-specific roles of these factors. These results not only highlight initial differences in the timing of protein expression of NC specifiers between avian species, but also demonstrate that gene expression studies should be complemented by protein localization analyses. Transcript expression implies a developmental state, but ultimately, the protein is the factor that drives cell state changes. Work in multiple systems has identified that mRNA and protein expression levels are not always correlated (Gry et al., 2009; Guo et al., 2008; Koussounadis et al., 2015). However, mRNA expression analyses remain strong indicators of changing developmental states for some cell types and would be bolstered by paired protein analyses.
4.3. Sequence similarities and differences
Genome sequence analyses of multiple avian species place peafowl closest to chick in a molecular phylogeny created using homologous proteins (Dhar et al., 2019). Our work focused on identifying similarities and differences in well-studied NC transcription factors and supports these relationships. Specifically, we identified that the amino acid sequences of three out of four NC transcription factors (PAX7, SNAI2, SOX9) show that the peafowl and chick sequences are 100% similar. However, the SOX10 amino acid sequence differs in all three organisms. Strikingly, quail SOX10 putatively has 104 additional amino acids on its N-terminus compared to chick and peafowl and 6/8 amino acid substitutions are radical replacements, where the amino acid is exchanged into another with different properties. There are multiple examples of biologically relevant single amino acid point mutations that dictate changes in protein sequence, structure, and function during disease and development (Airoldi et al., 2010). The classic example of a single amino acid change resulting in disease is that of Sickle Cell Anemia (Coleman and Inusa, 2007). Specific to neural crest cells, work in zebrafish identified that a single amino acid in the receptor tyrosine kinase protein, ERBB3, is necessary for its role in the formation of normal Sox10+ NC cells (Buac et al., 2008). In addition, a nonsynonymous mutation in a single amino acid in the LBH transcription factor in Cichlids is associated with alternative NC development and craniofacial adaptation (Powder et al., 2014). Therefore, although the spatial localization of PAX7, SNAI2, SOX9, and SOX10 proteins is similar between species, the differences in the timing of expression and their amino acid sequences provide a possibility of dissimilar folding, interactions, or functions in quail versus chick and peafowl, which result in differential development and dissimilar NC-related phenotypes.
4.4. Implications for developmental studies
Using multiplexed HCR and IHC, we have created a resource for the cell and developmental biology communities that can be used to compare specific NC gene and protein expression, and identifies differences in NC developmental programs, between closely related species. The heterochronous expression in the onset of SNAI2, SOX9, and SOX10 between the three species suggests that there may not be a single NC GRN program, but rather, that different animals may express these conserved genes and proteins with distinctive timing and possibly unique functions, that may account, at least in part, for later phenotypic differences. For example, even at equivalent developmental stages based on somite number, the embryos consistently exhibit phenotypic differences including neural tube size (Fig. 3) and rostral/craniofacial development (Fig. 12). Differences in NC development may be an indication of an exchange in the development of one cell type versus another (neural versus NC). However, these differences do not necessarily scale with animal size or incubation time. The rapid onset of quail NC formation and EMT is later offset by either slowed quail cell migration or increased speed of chick and peafowl NC cells. However, of note, chick and peafowl cells may migrate faster at later stages as their embryos will grow significantly larger than quail embryos as development proceeds, while quail cell migration may slow as the cells approach their regions of differentiation. The smaller number of peafowl NC cells expressing these proteins may be due to the animals having proportionately smaller head to body ratios, actively using different developmental programs, or could simply be due to artifacts limited by sample size. Further work is necessary to confirm that the proteins that are expressed at earlier stages in quail actually play a role (different or similar) at earlier stages of development. However, we find the new information confirming temporal differences gene and protein expression and localization in even closely related species intriguing, and hope that it provides discussion points and support for research in emerging and non-traditional model organisms.
Supplementary Material
Acknowledgements
We thank our colleagues from the Rogers Lab at UC Davis in the Department of Anatomy, Physiology, and Cell Biology who provided insight and expertise. Huge thank you to Dr. Pauline Perez for providing fertilized peafowl eggs for our study. The authors declare no competing financial interests.
Funding
This work was supported by UC Davis startup funds and UC Davis CAMPOS [CDR], National Institutes of Health (NIH) [R15-HD092170-01 to CDR, which funded ACA], the UC Davis Environmental Health Sciences Center (EHSC) under Award Number P30-ES023513 of the NIEHS, NIH [pilot grant from parent grant to CDR, which funded CE]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the UC Davis EHSC, nor the NIH.
Glossary
- EMT
Epithelial to mesenchymal transition
- NC
Neural crest
- GRN
Gene regulatory network
- HCR
In situ hybridization chain reaction
- IHC
Immunohistochemistry
- HH
Hamburger Hamilton
- SS
Somite stage
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ydbio.2021.12.018.
References
- Airoldi CA, Bergonzi S, Davies B, 2010. Single amino acid change alters the ability to specify male or female organ identity. Proc. Natl. Acad. Sci. U. S. A 107, 18898–18902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI, 2006. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441, 218–222. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T, 2010. Genomic code for Sox10 activation reveals a key regulatory enhancer for cranial neural crest. Proc. Natl. Acad. Sci. U.S.A 107, 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancur PA, Sauka-Spengler T, Bronner-Fraser M, 2009. c-Myb, Ets-1 and Sox9 directly activate a Sox10 core enhancer in delaminating cranial neural crest. Dev. Biol 331, 438–438. [Google Scholar]
- Bhattacharya D, Rothstein M, Azambuja AP, Simoes-Costa M, 2018. Control of neural crest multipotency by Wnt signaling and the Lin28/let-7 axis. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner ME, LeDouarin NM, 2012. Development and evolution of the neural crest: an overview. Dev. Biol 366, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buac K, Watkins-Chow DE, Loftus SK, Larson DM, Incao A, Gibney G, Pavan WJ, 2008. A Sox10 expression screen identifies an amino acid essential for Erbb3 function. PLoS Genet 4, e1000177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buitrago-Delgado E, Nordin K, Rao A, Geary L, LaBonne C, 2015. NEURODEVELOPMENT. Shared regulatory programs suggest retention of blastula-stage potential in neural crest cells. Science 348, 1332–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalpe AJ, Prasad M, Henke AJ, Paulson AF, 2010. Regulation of cadherin expression in the chicken neural crest by the Wnt/beta-catenin signaling pathway. Cell Adhes. Migrat 4, 431–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Cheung M, Abu-Elmagd MM, Orme A, Scotting PJ, 2000. Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain. Res. Dev. Brain. Res 121, 233–241. [DOI] [PubMed] [Google Scholar]
- Cheung M, Briscoe J, 2003. Neural crest development is regulated by the transcription factor Sox9. Development 130, 5681–5693. [DOI] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA, 2018. Third-generation in Situ Hybridization Chain Reaction: Multiplexed, Quantitative, Sensitive, Versatile, Robust, vol. 145. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman E, Inusa B, 2007. Sickle cell anemia: targeting the role of fetal hemoglobin in therapy. Clin. Pediatr 46, 386–391. [DOI] [PubMed] [Google Scholar]
- Dady A, Blavet C, Duband JL, 2012. Timing and kinetics of E-to N-cadherin switch during neurulation in the avian embryo. Dev. Dynam 241, 1333–1349. [DOI] [PubMed] [Google Scholar]
- Dady A, Duband JL, 2017. Cadherin interplay during neural crest segregation from the non-neural ectoderm and neural tube in the early chick embryo. Dev. Dynam 246, 550–565. [DOI] [PubMed] [Google Scholar]
- Dhar R, Seethy A, Pethusamy K, Singh S, Rohil V, Purkayastha K, Mukherjee I, Goswami S, Singh R, Raj A, Srivastava T, Acharya S, Rajashekhar B, Karmakar S, 2019. De novo assembly of the Indian blue peacock (Pavo cristatus) genome using Oxford Nanopore technology and Illumina sequencing. GigaScience 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Rashbass P, Schedl A, Brenner-Morton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J, 1997. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell 90, 169–180. [DOI] [PubMed] [Google Scholar]
- Garnett AT, Square TA, Medeiros DM, 2012. BMP, Wnt and FGF signals are integrated through evolutionarily conserved enhancers to achieve robust expression of Pax3 and Zic genes at the zebrafish neural plate border. Development 139, 4220–4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gry M, Rimini R, Stromberg S, Asplund A, Ponten F, Uhlen M, Nilsson P, 2009. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom 10, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiao P, Lei S, Deng F, Xiao GG, Liu Y, Chen X, Li L, Wu S, Chen Y, Jiang H, Tan L, Xie J, Zhu X, Liang S, Deng H, 2008. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin 40, 426–436. [DOI] [PubMed] [Google Scholar]
- Hamburger V, Hamilton HL, 1951. A series of normal stages in the development of the chick embryo. J. Morphol 88, 49–92. [PubMed] [Google Scholar]
- Hausser J, Mayo A, Keren L, Alon U, 2019. Central dogma rates and the trade-off between precision and economy in gene expression. Nat. Commun 10, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honore SM, Aybar MJ, Mayor R, 2003. Sox10 is required for the early development of the prospective neural crest in Xenopus embryos. Dev. Biol 260, 79–96. [DOI] [PubMed] [Google Scholar]
- Hutchins EJ, Bronner ME, 2018. Draxin acts as a molecular rheostat of canonical Wnt signaling to control cranial neural crest EMT. J. Cell Biol 217, 3683–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ison J, Kalas M, Jonassen I, Bolser D, Uludag M, McWilliam H, Malone J, Lopez R, Pettifer S, Rice P, 2013. EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics 29, 1325–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano R, Matsuo N, Tanaka H, Nasu M, Yoshioka H, Shirabe K, 2002. Identification and characterization of a soluble cadherin-7 isoform produced by alternative splicing. J. Biol. Chem 277, 47679–47685. [DOI] [PubMed] [Google Scholar]
- Kerosuo L, Nie S, Bajpai R, Bronner ME, 2015. Crestospheres: long-term maintenance of multipotent, premigratory neural crest stem cells. Stem Cell Rep 5, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khudyakov J, Bronner-Fraser M, 2009. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev. Dynam 238, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koussounadis A, Langdon SP, Um IH, Harrison DJ, Smith VA, 2015. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep 5, 10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander R, Nordin K, LaBonne C,2011. The F-box protein Ppa is a common regulator of core EMT factors Twist, Snail, Slug, and Sip1. J. Cell Biol 194, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin NM, Creuzet S, Couly G, Dupin E, 2004. Neural crest cell plasticity and its limits. Development 131, 4637–4650. [DOI] [PubMed] [Google Scholar]
- Lee PC, Taylor-Jaffe KM, Nordin KM, Prasad MS, Lander RM, LaBonne C, 2012. SUMOylated SoxE factors recruit Grg4 and function as transcriptional repressors in the neural crest. J. Cell Biol 198, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P, 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46, D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Khedkar S, Bork P, 2021. SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49, D458–D460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lignell A, Kerosuo L, Streichan SJ, Cai L, Bronner ME, 2017. Identification of a neural crest stem cell niche by Spatial Genomic Analysis. Nat. Commun 8, 1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Ju MJ, Redies C, 2006. Regionalized cadherin-7 expression by radial glia is regulated by Shh and Pax7 during chicken spinal cord development. Neuroscience 142, 1133–1143. [DOI] [PubMed] [Google Scholar]
- Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R, 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47, W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manohar S, Camacho-Magallanes A, Echeverria C, Rogers CD, 2020. Cadherin-11 is required for neural crest specification and survival. Front. Physiol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martik ML, Bronner ME, 2017. Regulatory logic underlying diversification of the neural crest. Trends Genet 33, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown SJ, Lee VM, Bronner-Fraser M, Newgreen DF, Farlie PG, 2005. Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev. Dynam 233, 430–444. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M, 2004. Gene-regulatory interactions in neural crest evolution and development. Dev. Cell 7, 291–299. [DOI] [PubMed] [Google Scholar]
- Nikitina N, Sauka-Spengler T, Bronner-Fraser M, 2008. Dissecting early regulatory relationships in the lamprey neural crest gene network. Proc. Natl. Acad. Sci. U. S. A 105, 20083–20088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossipova O, Sokol SY, 2011. Neural crest specification by noncanonical Wnt signaling and PAR-1. Development 138, 5441–5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon N, 2021. Treatment and prevention of neurocristopathies. Trends Mol. Med 27, 451–468. [DOI] [PubMed] [Google Scholar]
- Piloto S, Schilling TF, 2010. Ovo1 links Wnt signaling with N-cadherin localization during neural crest migration. Development 137, 1981–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powder KE, Cousin H, McLinden GP, Craig Albertson R, 2014. A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol. Biol. Evol 31, 3113–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procter JB, Carstairs GM, Soares B, Mourao K, Ofoegbu TC, Barton D, Lui L, Menard A, Sherstnev N, Roldan-Martinez D, Duce S, Martin DMA, Barton GJ, 2021. Alignment of biological sequences with Jalview. Methods Mol. Biol 2231, 203–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Nie S, 2018. Specifying Neural Crest Cells: from Chromatin to Morphogens and Factors in between. Wiley Interdiscip Rev Dev Biol, p. e322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Saxena A, Bronner ME, 2013. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. J. Cell Biol 203, 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Sorrells LK, Bronner ME, 2018. A catenin-dependent balance between N-cadherin and E-cadherin controls neuroectodermal cell fate choices. Mech. Dev 152, 44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein M, Bhattacharya D, Simoes-Costa M, 2018. The molecular basis of neural crest axial identity. Dev. Biol 444 (Suppl. 1), S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D, Suzuki T, Osumi N, Wakamatsu Y, 2006. Cooperative action of Sox9, Snail2 and PKA signaling in early neural crest development. Development 133, 1323–1333. [DOI] [PubMed] [Google Scholar]
- Sakai D, Tanaka Y, Endo Y, Osumi N, Okamoto H, Wakamatsu Y, 2005. Regulation of Slug transcription in embryonic ectoderm by beta-catenin-Lef/Tcf and BMP-Smad signaling. Dev. Growth Differ 47, 471–482. [DOI] [PubMed] [Google Scholar]
- Schiffmacher AT, Xie V, Taneyhill LA, 2016. Cadherin-6B proteolysis promotes the neural crest cell epithelial-to-mesenchymal transition through transcriptional regulation. J. Cell Biol 215, 735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider RA, 2018. Neural crest and the origin of species-specific pattern. Genesis 56, e23219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Severson C, Yang J, Wedlich D, Klymkowsky MW, 2011. Snail2 controls mesodermal BMP/Wnt induction of neural crest. Development 138, 3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M, Bronner ME, 2015. Establishing neural crest identity: a gene regulatory recipe. Development 142, 242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, Bock C, Farlik M, Piacentino ML, Boismoreau F, Hilscher MM, Yokota C, Qian X, Nilsson M, Bronner ME, Croci L, Hsiao WY, Guertin DA, Brunet JF, Consalez GG, Ernfors P, Fried K, Kharchenko PV, Adameyko I, 2019. Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364. [DOI] [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet JP, 2002. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development 129, 421–432. [DOI] [PubMed] [Google Scholar]
- Steventon B, Carmona-Fontaine C, Mayor R, 2005. Genetic network during neural crest induction: from cell specification to cell survival. Semin. Cell Dev. Biol 16, 647–654. [DOI] [PubMed] [Google Scholar]
- Taneyhill LA, Coles EG, Bronner-Fraser M, 2007. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development 134, 1481–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribulo C, Aybar MJ, Sanchez SS, Mayor R, 2004. A balance between the antiapoptotic activity of Slug and the apoptotic activity of msx1 is required for the proper development of the neural crest. Dev. Biol 275, 325–342. [DOI] [PubMed] [Google Scholar]
- Uygur A, Young J, Huycke TR, Koska M, Briscoe J, Tabin CJ, 2016. Scaling pattern to variations in size during development of the vertebrate neural tube. Dev. Cell 37, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallin J, Girault JM, Thiery JP, Broders F, 1998. Xenopus cadherin-11 is expressed in different populations of migrating neural crest cells. Mech. Dev 75, 171–174. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ, 2009. Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RM, Candido-Ferreira I, Repapi E, Gavriouchkina D, Senanayake U, Ling ITC, Telenius J, Taylor S, Hughes J, Sauka-Spengler T, 2019. Reconstruction of the global neural crest gene regulatory network in vivo. Dev. Cell 51, 255–276 e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yardley N, Garcia-Castro MI, 2012. FGF signaling transforms non-neural ectoderm into neural crest. Dev. Biol 372, 166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Klymkowsky MW, 2009. Unexpected functional redundancy between twist and slug (Snail2) and their feedback regulation of NF-kappaB via nodal and cerberus. Dev. Biol 331, 340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


