Abstract
Objective
To improve understanding of SARS-CoV-2-transmission and prevention measures on cruise ships, we investigated a Norwegian cruise ship outbreak from July to August 2020 using a multidisciplinary approach after a rapid outbreak response launched by local and national health authorities.
Methods
We conducted a cross-sectional study among crew members using epidemiologic data and results from SARS-CoV-2 polymerase chain reaction (PCR) of nasopharynx-oropharynx samples, antibody analyses of blood samples, and whole-genome sequencing.
Results
We included 114 multinational crew members (71% participation), median age 36 years, and 69% male. The attack rate was 33%; 32 of 37 outbreak cases were seropositive 5-10 days after PCR. One PCR-negative participant was seropositive, suggesting a previous infection. Network-analysis showed clusters based on common exposures, including embarkation date, nationality, sharing a cabin with an infected cabin-mate (adjusted odds ratio [AOR] 3.27; 95% confidence interval [CI] 0.97-11.07, p = 0.057), and specific workplaces (mechanical operations: 9.17 [1.82-45.78], catering: 6.11 [1.83-20.38]). Breaches in testing, quarantine, and isolation practices before/during expeditions were reported. Whole-genome sequencing revealed lineage B.1.36, previously identified in Asia. Despite extensive sequencing, the continued transmission of B.1.36 in Norway was not detected.
Conclusions
Our findings confirm the high risk of SARS-CoV-2-transmission on cruise ships related to workplace and cabin type and show that continued community transmission after the outbreak could be stopped by implementing immediate infection control measures at the final destination.
Keywords: Outbreak investigation, Cruise ship, COVID-19, Epidemiology, Immunity, Whole-genome sequencing
Graphical abstract

SARS-CoV-2 was first reported from Wuhan, China, in 2019, leading to outbreaks of COVID-19 and rapid spread worldwide. Cruise ships are ideal incubators for respiratory viruses because of crowded places, close-contact settings, and confined and enclosed spaces (the 3 C's) (WHO, 2021). Consequently, a number of SARS-CoV-2 outbreaks have occurred on cruise ships causing considerable morbidity and mortality (Expert Taskforce for the COVID-19 Cruise Ship Outbreak, 2020; Kordsmeyer et al., 2021). Cruise operators need to ensure that cruises do not pose unacceptable health risks to passengers, crew, and the general public (ECDC, 2021). Early recognition and an effective outbreak management strategy are crucial to controlling SARS-CoV-2 transmission on board (Kordsmeyer et al., 2021, Walker et al., 2021). To our knowledge, this is the first study of a SARS-CoV-2 outbreak in northern European waters on a cruise ship registered in a Nordic country.
Outbreak Setting
In June 2020, the MS Roald Amundsen became the first cruise ship to return to service in Norway after a 3-month suspension because of the pandemic after being in storage in a small coastal municipality with a multinational crew on board. The ship had recently updated, but not fully implemented, outbreak prevention, control, and response plans (Wiersholm AS and DNV GL SA Law Firm, 2020). At the time, the national incidence of COVID-19 was low (3.7 cases/100,000 population/14 days), and no cases were reported from this municipality to the Norwegian Institute of Public Health (NIPH). The ship had a total capacity of 528 passengers. From July 6-17, 2020, approximately 90 crew members, recruited through a foreign staffing company, arrived in Norway, primarily from the Philippines. Before starting service, they had to provide COVID-19 certificates confirming negative results for tests taken 3-4 days before departure but were not tested in Norway before boarding the ship. They underwent 10 days of initial restrictions on board, including social distancing, respiratory and hand hygiene measures, and isolation in a single cabin if any symptoms developed. However, they were allowed to work during quarantine, except those employed in the kitchen.
Passengers and crew members had cabins on separate decks. Single or twin cabins were assigned to the staff based on job and rank; however, some were quarantined in twin cabins together with staff with an earlier embarkation date.
The MS Roald Amundsen completed two one-week voyages from Tromsø and around the Svalbard archipelago at 75° north and back; Expedition 1, July 17-24, 2020, included 210 passengers and 160 crew, and Expedition 2, July 24-31, 2020, included 181 passengers and 160 crew. Most passengers were Norwegian citizens, whereas 5% were from other European countries. Most crew members attended both expeditions.
Outbreak detection and public health response
On July 26, 2020, 2 days after returning home from Expedition 1, a passenger developed symptoms and had a SARS-CoV-2 positive test result 2 days later. The timeline was consistent with probable exposure on board (Lauer et al., 2020). On July 29, 2020, the local municipal medical officer notified the NIPH about the case, who then immediately contacted the ship and recommended alerting and testing of passengers, but no action was taken. During Expedition 2, several symptomatic crew members were reported unable to work but were not tested nor isolated.
After finishing Expedition 2, the ship arrived back in Tromsø on the morning of July 31, 2020. At midday, the NIPH was alerted by the Tromsø municipal chief medical officer that 4 sick crew members had been hospitalized and subsequently diagnosed with COVID-19. By then, all passengers had left the ship. An outbreak response was immediately launched by Tromsø municipality with support from the NIPH and Norwegian Directorate of Health. The objectives were to determine the origin and extent of the outbreak and implement infection control measures to prevent further spread in Norway.
On July 31, 2020, and during the following days, all passengers and crew on both expeditions had a nasopharynx-oropharynx swab taken, either in Tromsø or at their current location, which was tested by real-time polymerase chain reaction (PCR) for SARS-CoV-2. All close contacts of cases on both expeditions were traced and tested. After July 31, 2020, crew member cases were isolated in single cabins or at the hospital, whereas those who tested negative were quarantined onboard or in hotels in Tromsø.
The attack rate among crewmembers was 25.2% (42 of 167) compared with 7.2% (28 of 391) among passengers, with an increase in affected passengers from 1.4% on Expedition 1 to 13.8% on Expedition 2. All crew members who tested positive had attended both expeditions. Reported dates for symptom onset for crew members ranged from July 6 to August 11, and for passengers, from July 26 to August 8. Ten crew members were hospitalized because of severe illness (no fatalities).
After the outbreak response, we conducted a study to determine when and how the virus was introduced and to examine risk factors for transmission onboard. To do so, we examined samples and exposure data collected from the crew members while they were in quarantine or isolation in Tromsø, as they had a significantly higher attack rate than the passengers and were still in one geographic area. As only a quarter of the crew was infected, we also aimed to assess if the noninfected crewmembers were protected by immunity from earlier infections.
Methods
Study Design and Population
We conducted a cross-sectional study among the crew members present in Tromsø from August 9-11, 2020. The study coordinator gave a video-streamed informational talk to all crewmembers in quarantine or isolation on the ship on August 9, 2020, which they shared in real-time with their colleagues staying in hotels or the hospital. Afterward, the attendees received written study information and consent forms.
Data Collection
The coordinator and a laboratory team collected signed consent forms and drew venous blood samples from crew members agreeing to participate on August 10-11, 5-10 days after the nasopharynx-oropharynx sampling (NOPS) on July 31, 2020, to August 6, 2020. Participants staying in hotels or the hospital were sampled on-site. A case was defined as a participant attending Expedition 2 with a SARS-CoV-2-positive PCR result.
The NIPH received crew and passenger lists from the ship, including data on sex, age, country of origin, embarkation date, workplace, cabin number and type, symptom onset date, clinical symptoms, test date, and results that were included in the NIPH outbreak registry and used in our study. Type of work was categorized as administrative, passenger services, outdoor work, mechanical operations, and catering occupations (Table 1 ).
Table 1.
Demographics of study participants in a cruise ship outbreak of SARS-CoV-2, Tromsø, Norway from July 2020 to August 2020
| Characteristics | n | % |
|---|---|---|
| Total number of participants | 114 | 100.0 |
| Sex | ||
| Women | 35 | 30.7 |
| Men | 79 | 69.3 |
| Age group, years | ||
| 20-29 | 23 | 20.2 |
| 30-39 | 47 | 41.2 |
| 40-49 | 33 | 29.0 |
| ≥50 | 11 | 9.7 |
| Country of origin | ||
| Norway / Europe* | 33 | 29.0 |
| The Philippines | 81 | 71.0 |
| Type of work† | ||
| Administrative | 11 | 9.6 |
| Passenger services | 22 | 19.3 |
| Outdoor work | 21 | 18.8 |
| Mechanical operations | 10 | 8.8 |
| Catering | 50 | 43.9 |
| Cabin type | ||
| Single | 60 | 52.6 |
| Twin | 54 | 47.4 |
| Embarkation date | ||
| 5 July 2020 and earlier | 33 | 29.0 |
| 6-8 July 2020 | 45 | 39.4 |
| 9-24 July 2020 | 36 | 31.6 |
| SARS-CoV-2 PCR test | ||
| Negative | 77 | 67.5 |
| Positive | 37 | 32.5 |
A few European countries.
Administrative: officers and managers. Passenger services: reception, store and medical/spa. Outdoor work: expedition team and work on deck. Mechanical operations: engine work, electricity, and carpentry. Catering: kitchen, bar and restaurant
Laboratory Methods
NOPS were analyzed with a SARS-CoV-2 real-time reverse transcription PCR test with primers and probe in the E-gene (Corman et al., 2020), which was considered positive when the cycle threshold (Ct) value was ≤37. Plasma was tested for SARS-CoV-2 antibodies with the Abbott Architect SARS-CoV-2 immunoglobin G (IgG) and immunoglobin M (IgM) assays, detecting antinucleocapsid antibodies, and with the Liaison SARS-CoV-2 S1/S2 IgG and IgM assays, detecting antispike antibodies (Supplementary Table 1). Of note, IgG antibodies against spike is a correlate of neutralizing activity (Dispinseri et al., 2021). To avoid unspecific results, only samples positive in at least 2 different assays were considered seropositive. For selected samples, SARS-CoV-2 IgG was in addition analyzed by immunofluorescence staining of SARS-CoV-2 infected Vero cells (Henriksen et al., 2020).
We performed whole-genome sequencing (WGS) of SARS-CoV-2 using the Artic Network nCoV-2019 V3 protocol (Corona Methods Development Community, 2020) and an Illumina MiSeq sequencer, with some supplementary genome sequences obtained using the Swift Amplicon SARS-CoV-2 Panel (Swift Bioscience, Ann Arbor, United States) on Illumina NovoSeq at the Norwegian Sequencing Center. The resulting sequences were assembled into consensus genome sequences using pipelines available on GitHub (pipelines for the Artic data: https://github.com/folkehelseinstituttet/fhi-ncov-seq-pipelines). The most similar sequences in the global initiative on sharing all influenza data (GISAID) database EpiCoV were identified using Basic Local Alignment Search Tool (BLAST) and included in the phylogenetic analysis by neighbor-joining of maximum composite likelihood distances using the MEGA X software (Kumar et al., 2018).
Statistical Analyses
We linked outbreak registry data to laboratory data from the University Hospital of North Norway and WGS results from the NIPH. We used STATA Statistical Software Version 16.1 (StataCorp LLC, Texas, United States) for multivariable logistic regression analysis with outcome-variable SARS-CoV-2 test result and calculated adjusted odds ratios (AORs) with 95% confidence intervals (CIs). Variables with p-value >0.1 were included in the model. We considered 2-sided p-values <0.05 statistically significant. Collinearity was not a problem with variance inflation factor <2.0 for all variables. Interaction was assessed by including cross-product terms between all independent variables and gender. There was no statistically significant interaction in the full model. We used nonparametric tests to compare participants’ IgG titers in different embarkation periods and to compare Ct-values between IgG-positive and IgG-negative participants. The descriptive network-analysis was conducted using STATA, whereas the network was visualized and modularity determined using Gephi 0.9.2, an open-source, free software for network visualization and exploration (https://gephi.org). The network was weighted by the number of shared characteristics using the following four: grouped date of embarkation, sharing a cabin, workplace, and country of origin (weight 0 to 4).
Results
Crew member study population
In total, 71% (114 of 160) of eligible crew members with a median age of 36.4 years participated in the study, of whom 69% were male, 29% were Norwegian/European, and 71% Filipino (Table 1). The median number of days on board before PCR-testing was 24 (interquartile [IQR] 18-37). Most Norwegian/European participants (88%, n = 29) had single cabins compared with 38% (n = 31, p <0.001) of Filipino participants. Fifty (62%) Filipino participants shared a twin cabin with a fellow citizen, whereas only 4 (12%, p <0.001) Norwegian/European participants did so.
Outbreak study analyses
The attack rate among crew member participants was 32.5% (37 of 114), similar to that of the entire crew (25.8%, p = 0.182). The distribution of cases by date and symptom onset shows a slowly increasing outbreak with a sharp peak on July 31, and then decreasing incidence (Figure 1 ). Participants’ confirmed PCR-positive tests had mean a Ct value of 24.9 (range 15.4-36.8).
Figure 1.
Epidemiologic curve of 37 SARS-CoV-2 cases by date of symptom onset, July 2020 to August 2020.
Respiratory infection symptoms were reported by 25 (68%) cases, including 6, in addition, having lost their sense of smell and taste, and 2 reported gastroenteritis. Nine had “other” or unknown symptoms, and 3 were asymptomatic (Table 2 ).
Table 2.
Reported clinical symptoms among 37 cases in a cruise ship outbreak of SARS-CoV-2, Tromsø, Norway from July 2020 to August 2020
| Characteristics | n | % |
|---|---|---|
| Total no. SARS-CoV-2 cases | 37 | 100.0 |
| Clinical symptoms | ||
| Upper respiratory tract infection (URTI)* | 18 | 48.6 |
| Lower respiratory tract infection (LRTI)† | 7 | 18.9 |
| Unknown symptoms‡ | 7 | 18.9 |
| Other symptoms§ | 2 | 5.4 |
| Asymptomatic | 3 | 8.1 |
18 SARS-CoV-2 cases reported URTI, among which 4 had lost the sense of smell and taste and 2 others in addition reported diarrhea and abdominal pain.
7 cases reported LRTI, among which 2 had lost the sense of smell and taste.
7 cases reported date for symptom onset but no symptoms ("unknown symptoms").
One case reported fatigue only and another reported tachypnea only.
SARS-CoV-2 spread among the participants in most workplaces on board. The first cases retrospectively reporting symptom onset between July 20-22, 2020, worked in catering occupations and mechanical operations (Figure 1). Nine days later, symptoms among administrative and passenger service staff were reported. No cases were identified among participants working on deck, with electricity and carpentry, or with medical and spa services.
The attack rate was higher among Filipino participants (41%) than among those from Norway/Europe (12%, p = 0.006) (Table 3 ). In multivariable analyses, being defined as a case was associated with working with mechanical operations (AOR 8.26, 95% CI 1.54-44.16) and catering occupations (6.06, 1.78-20.67). Sharing a twin cabin with an infected cabin-mate was strongly associated with being a case in crude analysis (OR 7.20, 2.48-20.41) but was attenuated to 3.27 (0.97-11.07) in the full model. The mean serial interval between symptom onset in twin cabins with both persons infected was 4.3 days (range 1-8 days).
Table 3.
Characteristics associated with being a case of SARS-CoV-2 in a cruise ship outbreak, Tromsø, Norway from July 2020 to August 2020
| Characteristics | Cases n (%) | OR | 95% CI | p-value | AOR | 95% CI | p-value | N |
|---|---|---|---|---|---|---|---|---|
| Sex | 0.149 | 114 | ||||||
| Women | 8 (22.9%) | 1.00 | 35 | |||||
| Men | 29 (36.7%) | 1.96 | 0.79-4.87 | 79 | ||||
| Age group, years | 0.579 | 114 | ||||||
| 20-29 | 5 (21.7%) | 1.00 | 23 | |||||
| 30-39 | 15 (31.9%) | 1.69 | 0.53-5.41 | 47 | ||||
| 40-49 | 13 (39.4%) | 2.34 | 0.70-7.86 | 33 | ||||
| ≥ 50 | 4 (36.6%) | 2.06 | 0.42-9.97 | 11 | ||||
| Country of origin | 0.006 | 0.812 | 114 | |||||
| Norway / Europe* | 4 (12.1%) | 1.00 | 1.00 | 33 | ||||
| The Philippines | 33 (40.7%) | 4.98 | 1.60-15.51 | 1.20 | 0.26-5.47 | 81 | ||
| Type of work† | <0.001 | 0.005 | 114 | |||||
| Admin./passenger services/outdoor | 5 (9.3%) | 1.00 | 1.00 | 54 | ||||
| Mechanical operations | 5 (50.0%) | 9.80 | 2.09-45.89 | 9.17 | 1.82-45.78 | 10 | ||
| Catering | 27 (54.0%) | 11.50 | 3.93-33.71 | 6.11 | 1.83-20.38 | 50 | ||
| Cabin type | <0.001 | 0.057 | 114 | |||||
| Single, or twin with neg. cabin-mate | 23 (24.5%) | 1.00 | 1.00 | 94 | ||||
| Twin with pos. cabin-mate | 14 (70.0%) | 7.20 | 2.48-20.91 | 3.27 | 0.97-11.07 | 20 | ||
| Embarkation date | 0.006 | 0.525 | 114 | |||||
| 5 July 2020 and earlier | 10 (30.3%) | 1.00 | 1.00 | 33 | ||||
| 6-8 July 2020 | 22 (48.9%) | 2.20 | 0.86-5.66 | 1.52 | 0.51-4.55 | 45 | ||
| 9-24 July 2020 | 5 (13.4%) | 0.37 | 0.11-1.23 | 0.67 | 1.46-3.12 | 36 |
The multivariable model included country of origin, type of work, cabin type, and embarkation date.
OR = odds ratio; AOR = adjusted OR; N = denmoninator.
A few European countries
Administrative (officers, managers), passenger services (reception, store, medical/spa) and outdoor work (expedition team, work on deck) were combined as they had similar COVID-19 rates. Mechanical operations: engine work, electricity, and carpentry. Catering occupations: kitchen, restaurant and bar.
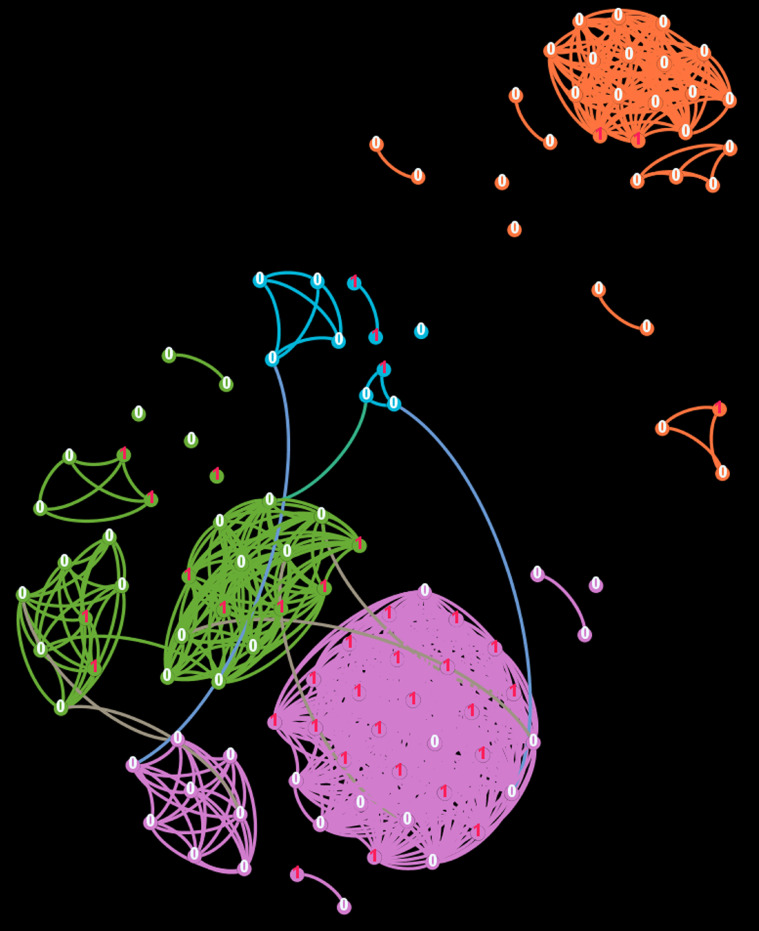
The network-analysis displayed four distinct clusters (Figure 2 ). The purple cluster (n = 42) had an attack rate of 50%, of which 71% had catering occupations, all had the embarkation date July 6-8, 2020, most were Filipino (98%) and stayed in twin cabins (76%) (Figure 2, Table 4 ). Two clusters (green, n = 31 and blue, n = 10) had an attack rate similar to that of the study population as a whole, and most participants were Filipino. The orange (n = 31) had an attack rate of 10%, well below the study population, and most participants were Norwegian or European (97%), had outdoor work (55%), single cabin (90%), and late embarkation (84%). Cases were neither more central in the network nor had more links than non-cases.
Figure 2.
Social network-analysis of 114 study participants, July 2020 to August 2020. The network was split into four different communities displayed in different colors: purple cluster (n = 42), green cluster (n = 31), orange cluster 2 (n = 31) and blue cluster (n = 10). Nodes with a red “1” indicates a case; white “0” indicates a noncase. Only links between participants with at least three different connections are shown. Four possible connections were included: sharing twin cabin, same country of origin, same work place, and same embarkation date.
Table 4.
Characteristics of 4 clusters in a social network-analysis of participants in a cruise ship outbreak of SARS-CoV-2, Tromsø, Norway from July 2020 to August 2020
| Modularity class (cluster)* | 0 | 1 | 2 | 3 | Total |
|---|---|---|---|---|---|
| Cluster color code | Purple | Green | Orange | Blue | |
| Number of participants | 42 | 31 | 31 | 10 | 114 |
| Characteristics | % | % | % | % | % |
| Sex | |||||
| Women | 31 | 23 | 39 | 30 | 31 |
| Men | 69 | 77 | 61 | 70 | 69 |
| Age groups, years | |||||
| 20-29 | 26 | 10 | 23 | 20 | 20 |
| 30-39 | 40 | 42 | 39 | 50 | 41 |
| 40-49 | 24 | 39 | 26 | 30 | 29 |
| ≥50 | 10 | 10 | 13 | 0 | 10 |
| Country of origin | |||||
| Norway / Europe† | 2 | 6 | 97 | 0 | 29 |
| The Philippines | 98 | 94 | 3 | 100 | 71 |
| Type of work‡ | |||||
| Administrative | 0 | 3 | 29 | 10 | 10 |
| Passenger services | 19 | 29 | 3 | 40 | 33 |
| Outdoor work | 5 | 6 | 55 | 0 | 13 |
| Mechanical operations | 5 | 13 | 6 | 20 | 9 |
| Catering | 71 | 48 | 6 | 30 | 44 |
| Type of cabin | |||||
| Single | 24 | 58 | 90 | 40 | 53 |
| Twin | 76 | 42 | 10 | 60 | 47 |
| Embarkation date | |||||
| July 5, 2020 and earlier | 0 | 100 | 6 | 0 | 29 |
| July 6-8, 2020 | 100 | 0 | 10 | 0 | 39 |
| July 9-24, 2020 | 0 | 0 | 84 | 100 | 32 |
| SARS-CoV-2 PCR test result | |||||
| Negative | 50 | 68 | 90 | 70 | 68 |
| Positive | 15 | 32 | 10 | 30 | 32 |
The color codes correspond to the 4 clusters in the network-analysis in Figure 2.
A few European countries.
Administrative work: officers and managers. Passenger services: reception, store and medical/spa. Outdoor work: expedition team and work on deck. Mechanical operations: engine work, electricity, and carpentry. Catering: restaurant, bar and kitchen.
Immunologic Responses
Antibody analysis of the 37 outbreak cases demonstrated SARS-CoV-2 seropositivity in 32 (87%), confirming their COVID-19 diagnosis (Figure 3 ). Except for one seropositive case with only IgG and 5 cases with only IgM, all seropositive cases had IgM and IgG antibodies. The quantitative antispike IgG assay revealed antibody titers from 18.5-166 AU/ml (mean value 70 AU/ml). There was a nonsignificant increase in antibody titers with increasing time after reported symptom onset (p = 0.092), and only samples taken more than 7 days after symptom start had titers above 100 AU/ml (Figure 4 ). The median Ct value of NOPS was 7.9 Ct-values lower in the 10 IgG-negative than the 27 IgG-positive cases (p = 0.130), indicating a 100-fold higher viral load among IgG-negative participants (Figure 5 ). Cases with a high viral load and a low or lacking IgG titer were presumably in an early phase of infection. In agreement with this, 10 of the 11 antispike IgG-negative cases reported symptom onset less than 14 days (2-12 days) before blood sampling (Figure 4).
Figure 3.
Serology results from 37 PCR-positive SARS-CoV-2 cases and 77 PCR-negative participants using 2 IgM and 2 IgG assays, July-August 2020.
Figure 4.
Antispike (Anti-S) IgG titer (AU/ml) from all 37 PCR-positive cases plotted based on the number of days between reported symptom onset and blood sampling. Samples with titers ≥ 15.0 AU/ml (dashed line) are defined as positive (red circles), and samples with titers < 12.0 AU/ml are negative (green circles). No equivocal samples with titers < 15.0 but ≥ 12.0 AU/ml were detected.
Figure 5.
SARS-CoV-2 RNA in a nasopharynx-oropharynx swab (NOPS) from all 37 PCR-positive cases grouped by the presence of SARS-CoV-2 IgG, July 2020 to August 2020. Individual PCR results are shown as Ct-values. Median Ct-values are indicated with a horizontal line within the box.
To detect participants that may have recovered from COVID-19, antibody analysis was performed for all 77 PCR-negative participants. Only one participant (from Asia) had both SARS-CoV-2 IgM and IgG, the latter detected by 2 assays (Figure 4). In 5 others, only IgM or IgG antibodies against the spike-protein were detected. These 6 plasma samples were further analyzed by immunofluorescence staining of SARS-CoV-2 infected Vero cells. Similar to positive control plasma (Figure 6 , panel A and B), the IgM and IgG-positive sample gave cytoplasmic staining of infected but not uninfected cells (Figure 6, panel C and D). For the other samples, only weak background staining was observed (Figure 6, panel E and F).
Figure 6.
Panel A to F. Microscope images showing immunofluorescence staining of SARS-CoV-2 infected Vero cells using serum as primary antibody, August 2020. A) Positive control 1: serum from the case with the highest antinucleocapsid IgG titer. B) Positive control 2: Serum from the case with highest antispike IgG titer. C) Serum from PCR-negative participant with antispike IgG and antinucleocapsid and antispike IgM. D) The same serum as in C) used on uninfected Vero cells. E) Serum from PCR-negative participant with only positive antispike IgG. F) The same serum as in E) used on uninfected Vero cells. Similar images were obtained from the 4 other PCR-negative participants. As secondary antibody goat antihuman IgG-Alexa Fluor 488 was used. DNA was stained with Draq 5. Cytoplasmic staining is observed in A-C.
We concluded that only one PCR-negative participant was seropositive for SARS-CoV-2, indicating a previous infection with COVID-19. Noteworthy, the two different IgM and IgG assays used gave identical qualitative results for 98% (112 of 114) and 94% (108 of 114) of samples (Figure 3).
Whole‐genome Sequencing
WGS yielded near-complete (>97% of the genome) sequences for 29 of the 37 PCR-positive samples, 90% coverage for 1 sample, low-coverage (17%-59%) for three, and no sequences for four samples because of low viral RNA (Ct >35). One common virus, lineage B.1.36, was identified (lineage attribution as of September 13, 2021) (GISAID EpiCoV, 2021). The sequences constituted a highly similar group within this lineage as only two mutations in the whole-genome, C14760T and G26828T, separated it from other B.1.36 viruses (Figure 7 ). The only other viruses belonging to this outbreak genetic group in Norway were viruses received in the National Reference Laboratory from non-participant crew members (n = 5), cases among passengers (n= 10), and their close contacts (n = 2). Apart from these, the closest sequence matches were viruses mostly detected in India and Saudi Arabia from April 2020 to February 2021. Within the outbreak cluster, further sub-grouping based on single nucleotide substitutions could be discerned.
Figure 7.
Phylogenetic analysis of SARS-CoV-2 whole genomes (coverage >98%) from cases in the outbreak on MS Roald Amundsen in Tromsø, Norway in July 2020 to August 2020. Outbreak associated viruses from cases among crew member participants (red dots, n = 29), crew member nonparticipants (blue, n = 5), passengers (green, n = 10) and their close contacts (gold, n = 2) were compared with the most similar sequences in the GISAID EpiCoV database, as well as to representatives of other lineage B.1.36 viruses in Norway during 2020. The scale bar corresponds to a distance of 0.00005 nucleotide substitutions per site (approx. 1.5 substitutions per viral genome).
Discussion
The findings from the epidemiologic, serologic, and WGS investigations are consistent with an evolving COVID-19 outbreak in this multinational, nonimmune study population, caused by a single virus introduction that was identified in all cases with a successful WGS result. Efficient infection control measures were quickly implemented just after the ship's arrival in Tromsø, and the outbreak was successfully contained.
We found that type of work, mechanical operations (i.e. engine work) and catering, were the main outbreak driver as also reported in other ship outbreaks (Expert Taskforce for the COVID-19 Cruise Ship Outbreak, 2020; Kakimoto et al., 2020; Kasper et al., 2020). The finding that sharing a cabin with an infected cabin-mate increased the odds of being a case agrees with a study among passengers aboard the Diamond Princess (Plucinski et al., 2021). We detected only a few cases among outdoor work, reinforcing the evidence that SARS-CoV-2 transmission is lower outdoors (Bulfone et al., 2021, Kasper et al., 2020).
The increased odds for COVID-19 among Filipino participants was strongly attenuated in the multivariable analysis suggesting that workplace and sharing a cabin were stronger risk factors than the country of origin. This may reflect the segregated community on cruise ships with a crew from low-income countries living in shared facilities and working in confined spaces, making them more vulnerable to infections.
The outbreak was probably initiated by an Asian crew member prior to the first expedition and subsequently spread among crew and passengers on two expeditions in Arctic waters, illustrating how increased international mobility results in the fast spread of infectious diseases. Although no crew member participants reported symptom onset between July 6-19, 2020, there was likely ongoing transmission during this period, either by unreported illness or asymptomatic SARS-CoV-2 transmission (Payne et al., 2020). A nonparticipant crew member reported symptom onset on July 6, 2020, and could have been the primary case. The increasing number of cases from Expedition 1 to 2 shows that there was more extensive transmission on the second voyage with likely spill-over from crew to passengers. Given a typical incubation period of 4-5 days (Lauer et al., 2020), this would suggest that exposure occurred simultaneously for many people onboard. It is likely that breaches in several of the intended control measures led to undetected transmission onboard in early to mid-July when preparing for departure.
With the exception of 5 cases, all cases had detectable levels of SARS-CoV-2 specific IgM and/or IgG antibodies, confirming their COVID-19 diagnosis. The measured antibody response seems to agree with other studies. SARS-CoV-2 IgM is usually detected by day 5-7 from symptom onset and IgG by day 7-10, with peak levels approximately by day 49 (Stephens and McElrath, 2020).
SARS-CoV-2 viral load in the upper respiratory tract peaks in the first week of illness and declines about 1000-fold within 10 days after diagnosis, contemporary with an increasing level of SARS-CoV-2 antibodies (Cevik et al., 2021, Leuzinger et al., 2021a, Leuzinger et al., 2021b). As our 10 IgG-negative cases (5 were also IgM negative), had median RNA load >100-fold higher than the 27 IgG-positive cases, they were most likely in an early phase of infection. In agreement with this, 10 of 11 cases that were lacking detectable IgG antibodies against the spike-protein reported symptom onset only 2-12 days before sampling, giving us confidence in the retrospectively reported symptom data.
SARS-CoV-2 IgM and IgG were only convincingly detected in one of 77 PCR-negative participants, increasing the number of seropositive participants to 33. This may have been a crew member with an infection before the outbreak or one of the first outbreak cases.
In 5 other PCR-negative participants, single-assay positive antispike IgM or IgG results were likely because of cross-reactivity of preexisting antibodies against seasonal coronavirus or other microbes, as previously described (Manthei et al., 2021). Since only one PCR-negative participant was seropositive, preexisting SARS-CoV-2 immunity did probably not contribute to limit the spread of the virus. Instead, cross-reactive T cells from previous infections with common human coronaviruses (Perez-Potti et al., 2021) or a strong innate immune response (Park and Iwasaki, 2020) may have offered some protection.
The first documented COVID-19 case outside China was reported in Thailand in mid-January 2020, only 6 months before Expedition 2. Using a combination of serology and PCR, we assume to have detected all previous and ongoing SARS-CoV-2 infections among the participating crew.
The rapid action from the municipality, NIPH, and the Norwegian Directorate of Health efficiently limited the extent of the outbreak. No onward transmission of viruses belonging to this outbreak genetic group has been detected in Norway after the outbreak, despite extensive nationwide sequencing of SARS-CoV-2. Although other B.1.36 lineage viruses subsequently appeared in Norway, only cases with a direct epidemiologic link to this outbreak had sequences that matched the outbreak strain. The most similar sequences are viruses from Asia, and the sequence data are consistent with the introduction from this region (GISAID EpiCoV, 2021).
Strengths and weaknesses of the study
The strengths include the multidisciplinary approach to generating detailed epidemiologic data and the inclusion of WGS data from most PCR-positive participants. Our access to the National Reference Laboratory database enabled comparison of the outbreak strain to the Norwegian WGS database to map any onward transmission in Norway, which, to our knowledge, has not been done in previous studies. The study was limited by the fact that only three-quarters of crew members consented to participation, no passengers were included, and data on social gatherings were not available, implying potentially significant missing links for studying virus transmission dynamics. Furthermore, symptom onset dates were obtained retrospectively for the early cases, which may have introduced selection and recall bias, and immune response was measured only once.
Implications of the study
We found that the combined use of epidemiology, serology, and WGS can describe a SARS-CoV-2 outbreak in detail. The study is important in understanding the dynamics of an outbreak to prevent and control further outbreaks on cruise ships. The outbreak occurred early in the pandemic as one of the first reported on a Norwegian vessel. Norwegian legislation related to the COVID-19 pandemic has since been amended, and national recommendations for coastal cruise vessels, including crew change, accordingly revised (Norwegian Maritime Authority, 2021). Current legislation and guidance explicitly state that cruise ship companies need to implement infection control measures as described in their safety management system that includes risk assessment, responsibility, resources needed, communication, education, and test strategies, and this is in accordance with the European recommendations (ECDC, 2021).
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. This work was not funded, and the authors’ employers had no role in writing the manuscript or the decision to submit it for publication.
Contributors
KG, GSS, OK, and PE designed the study. KG wrote the study protocol and applied for ethical approval. KG, SH, and GSS contributed to the collection of blood samples. KG, KS, and HS conducted the epidemiologic analyses. SH and CHR conducted and interpreted the serologic analyses. OH conducted the WGS sequence analysis. OH, KSJ, and KB interpreted the WGS. KG and CHR drafted the manuscript. All authors contributed to interpreting the analysis, developing the manuscript, and approving the final draft.
Ethical Approval
The study was approved by the Regional Committee for Medical and Health Research Ethics, North Norway (reference: 2020/170968) and the Data Protection Officer at UNN (2020/9635). The study complied with the Declaration of Helsinki. All participants signed an informed consent form before participation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors alone are responsible for the views expressed in this publication, and they do not necessarily represent the decisions or policies of their respective employers. The corresponding author had full access to all data in the study and had final responsibility for the decision to submit for publication.
The authors would also like to thank the following: All participants, technicians at the University Hospital of North Norway, and Norwegian Institute of Public Health who drew blood samples, performed SARS-CoV-2, PCR, serology, and WGS, Kathrine Kristoffersen and Trond Brattland at the Public Health Department in Tromsø municipality, and originating and submitting laboratories that have provided the SARS-CoV-2 sequences in the GISAID EpiFlu database used in this analysis.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2022.02.025.
Appendix. Supplementary materials
References
- Bulfone TC, Malekinejad M, Rutherford GW, Razani N. Outdoor Transmission of SARS- CoV-2 and Other Respiratory Viruses: A Systematic Review. J Infect Dis. 2021;223(4):550–561. doi: 10.1093/infdis/jiaa742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2(1):e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona Methods Development Community. nCoV-2019 sequencing protocol v3 (LoCost) V.3; 2020. Available from: https://www.protocols.io/view/ncov-2019-sequencing-protocol-v3-locost-bh42j8ye?version_warning=no&step=4 ). [Accessed July 30, 2021].
- Dispinseri S, Secchi M, Pirillo MF, Tolazzi M, Borghi M, Brigatti C, et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nature communications. 2021;12(1):2670. doi: 10.1038/s41467-021-22958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC. European Centre for Disease Prevention and Control. COVID-19: EU guidance for cruise ship operations; 2021. Available from: https://www.ecdc.europa.eu/en/publications-data/COVID-19-cruise-ship-guidance . [Accessed 7 December 2021].
- Expert Taskforce for the COVID-19 Cruise Ship Outbreak Epidemiology of COVID-19 Outbreak on Cruise Ship Quarantined at Yokohama, Japan, February 2020. Emerging infectious diseases. 2020;26(11):2591–2597. doi: 10.3201/eid2611.201165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GISAID EpiCoV. The Global Initiative on Sharing All Influenza Data database 2021. [Accessed 13 September 2021].
- Henriksen S, Trydal Ø, Sigurdsen SE, Tylden GD, Rinaldo CH. Antibody response in a family with COVID-19. Tidsskrift for den Norske laegeforening: tidsskrift for praktisk medicin. ny raekke. 2020;140(11) doi: 10.4045/tidsskr.20.0420. [DOI] [PubMed] [Google Scholar]
- Kakimoto K, Kamiya H, Yamagishi T, Matsui T, Suzuki M, Wakita T. Initial Investigation of Transmission of COVID-19 Among Crew Members During Quarantine of a Cruise Ship - Yokohama, Japan, February 2020. MMWR Morbidity and mortality weekly report. 2020;69(11):312–313. doi: 10.15585/mmwr.mm6911e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper MR, Geibe JR, Sears CL, Riegodedios AJ, Luse T, Von Thun AM, et al. An Outbreak of Covid-19 on an Aircraft Carrier. The New England journal of medicine. 2020;383(25):2417–2426. doi: 10.1056/NEJMoa2019375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordsmeyer AC, Mojtahedzadeh N, Heidrich J, Militzer K, von Münster T, Belz L, et al. Systematic Review on Outbreaks of SARS-CoV-2 on Cruise, Navy and Cargo Ships. International journal of environmental research and public health. 2021;18(10) doi: 10.3390/ijerph18105195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172(9):577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzinger K, Gosert R, Søgaard KK, Naegele K, Bielicki J, Roloff T, et al. Epidemiology and precision of SARS-CoV-2 detection following lockdown and relaxation measures. J Med Virol. 2021;93(4):2374–2384. doi: 10.1002/jmv.26731. [DOI] [PubMed] [Google Scholar]
- Leuzinger K, Osthoff M, Dräger S, Pargger H, Siegemund M, Bassetti S, et al. Comparing immunoassays for SARS-Coronavirus-2 antibody detection in patients with and without laboratory-confirmed SARS-Coronavirus-2 infection. J Clin Microbiol. 2021 doi: 10.1128/JCM.01381-21. Jcm0138121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthei DM, Whalen JF, Schroeder LF, Sinay AM, Li SH, Valdez R, et al. Differences in Performance Characteristics Among Four High-Throughput Assays for the Detection of Antibodies Against SARS-CoV-2 Using a Common Set of Patient Samples. American journal of clinical pathology. 2021;155(2):267–279. doi: 10.1093/ajcp/aqaa200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwegian Maritime Authority. Guidelines for crew changes in Norway during the COVID-19 pandemic; 2021. Available from: https://www.sdir.no/en/news/news-from-the-nma/guidelines-regarding-seafarers-signing-on-and-off-ships-in-norwegian-ports/. [Accessed 7 December 2021].
- Park A, Iwasaki A. Type I and Type III Interferons - Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell host & microbe. 2020;27(6):870–878. doi: 10.1016/j.chom.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne DC, Smith-Jeffcoat SE, Nowak G, Chukwuma U, Geibe JR, Hawkins RJ, et al. SARS-CoV-2 Infections and Serologic Responses from a Sample of U.S. Navy Service Members — USS Theodore Roosevelt, April 2020. MMWR Morb Mortal Wkly Rep. 2020;69:714–7212020. doi: 10.15585/mmwr.mm6923e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Potti A, Lange J, Buggert M. Deciphering the ins and outs of SARS-CoV-2-specific T cells. Nature immunology. 2021;22(1):8–9. doi: 10.1038/s41590-020-00838-5. [DOI] [PubMed] [Google Scholar]
- Plucinski MM, Wallace M, Uehara A, Kurbatova EV, Tobolowsky FA, Schneider ZD, et al. Coronavirus Disease 2019 (COVID-19) in Americans Aboard the Diamond Princess Cruise Ship. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2021;72(10):e448–ee57. doi: 10.1093/cid/ciaa1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens DS, McElrath MJ. COVID-19 and the Path to Immunity. Jama. 2020;324(13):1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LJ, Codreanu TA, Armstrong PK, Goodwin S, Trewin A, Spencer E, et al. SARS-CoV-2 infections among Australian passengers on the Diamond Princess cruise ship: A retrospective cohort study. PLoS One. 2021;16(9) doi: 10.1371/journal.pone.0255401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. COVID 19. Avoid the three C's; 2021 . [Accessed December 2021].
- Wiersholm AS and DNV GL SA Law Firm. Granskningsrapport. Utbrudd av covid-19 på Hurtigruten-skipet MS Roald Amundsen 17.- 31. juli 2020 (External Investigation Report); 2020. Available from: https://presse.hurtigruten.no/documents/granskningsrapport-100977.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.