Abstract
Investigating the metabolic effects of radiation is critical to understand the impact of radiotherapy, space travel, and exposure to environmental radiation. In patients undergoing hemopoietic stem cell transplantation, iron overload is a common risk factor for poor outcomes. However, no studies have interrogated the multi-organ effects of these treatments concurrently. Herein, we use a model that recapitulates transfusional iron overload, a condition often observed in chronically transfused patients. We applied an omics approach to investigate the impact of both iron load and irradiation on the host metabolome. Results revealed dose-dependent effects of irradiation in red blood cells, plasma, spleen, and liver energy and redox metabolism. Increases in polyamines and purine salvage metabolites were observed in organs with high oxygen consumption including the heart, kidney, and brain. Irradiation also impacted the metabolism of the duodenum, colon, and stool, suggesting a potential effect on the microbiome. Iron infusion affected the response to radiation in the organs and blood, especially in erythrocyte polyamines and spleen antioxidant metabolism, and affected glucose, methionine and glutathione systems and tryptophan metabolism in the liver, stool, and brain. Together, the results suggest that radiation impacts metabolism on a multi-organ level with a significant interaction of host iron status.
Keywords: Radiation, Iron, Metabolism, Organs, Mass Spectrometry, Polyamine, Methionine
Graphical Abstract
INTRODUCTION
Understanding the metabolic impact of irradiation holds several clinically relevant implications. For example, γ-emitting irradiators (137Cs and 60Co sources) have been used to deliver myeloablative doses of radiation prior to bone marrow transplantation in patients with both hematological disorders and malignancies.1 Over the past decades, radiotherapy has been a mainstay in the treatment of several cancers, including breast cancer, where treatment with 137Cs at doses of >10 Gy in multiple fractions per day (e.g., 4) have been reported both as external whole breast radiotherapy or interstitial irradiation to the tumor bed.2 Preclinical animal models of cell and gene therapy, as well as studies on hematopoietic stem cell function rely heavily on mouse models of radiation, prior to adoptive hematopoietic stem cell (HSC) transfer and/or bone marrow transplant. This is the case of studies on aging,3 (healthy and stress) hematopoiesis,4 and hematological malignancies.5 Similarly, common are the studies where genetic ablation of specific genes of interest results in a non-viable mouse in adult age, requiring bone marrow transplant of hematopoietic stem cells from knock out mice to irradiated wild type mice with the goal to investigate the role of a specific gene in adult/old age (e.g., protein L-isoaspartyl o-methyltransferase).6 Bone marrow irradiation techniques often require a lethal dose of irradiation to ensure adequate engraftment with donor cells. While mice are used extensively in transplantation studies involving bone marrow ablation, limited information is known about the impact of irradiation on systems metabolism.
Such information is relevant for example to determine the metabolic cost of chronic or acute irradiation exposure following space travel or terrorist attacks. Despite these security measures, limited information is available about the systemic impact of radiation from a diagnostic standpoint (e.g., for the rapid assessment of terrorist threats), which could be relevant to inform intervention strategies to mitigate the health toll on victims of such attacks.
In the near future, the National Aeronautics and Space Administration (NASA) and the recently established US Army Space Force are planning long duration and deep space travel for space exploration and research missions. Similar public and private efforts to explore and perhaps colonize proximal satellites and planets (e.g., missions to the Moon, the Lunar Gateway, or to Mars) will challenge astronauts with a prolonged exposure to microgravity7 and space radiation, such as high energy particles from galactic cosmic rays and potential particles from solar events.8 Manned missions to Mars are estimated to last between 650 and 920 days, exposing the astronauts to a cumulative dose equivalent of 870–1200 mSv (3–4.5 Gy).9 On the other hand, low Earth orbit missions – such as those in the International Space Station (ISS) – partially shield astronauts from radiations otherwise experienced in deep space, owing to the protective effect of Earth’s magnetosphere. Yet, ISS astronauts returning to Earth and animal experiments performed onboard have confirmed that spaceflight significantly impacts the cardiovascular and central nervous system, promotes muscle atrophy, cataract formation, and triggers immune system effects – in part also explained by the effects of microgravity. Results from the NASA twin study revealed that genetically identical twins – one on Earth and one on the ISS for 340 days – had differential metabolism, with differential levels of metabolic markers of genotoxic stress, inflammation, and altered amino acid metabolism, Krebs cycle metabolites and tryptophan metabolites (indoles), which all increased in twin inflight.10
Similar alterations to the microbiome (especially members of the bacterial taxa Lachnospiraceae and Enterococcaceae) and metabolites of potential microbial origin (including tryptophan-derived indoles) have been reported in plasma of humans surviving ionizing radiation exposure.11,12 Studies have also confirmed alterations in tryptophan/indole metabolism in low-dose uranium radiation exposure studies.13 Studies on serum and plasma have been reported at low dose of radiation (e.g., 4 Gy)14 or urine (with a total dose of radiation between 5–10 Gy over a 30 day period).15 Metabolomics studies examining the impact of bone marrow failure-inducing doses of radiation have also been published to document the effect on mice exposed to a higher radiation dose range (5–11 Gy). These identified a significant enrichment in purine metabolism, tricarboxylic acid cycle, fatty acids, acyl-carnitines, and amino acids in the spleen of animals upon early perturbations with γ irradiation.14 Despite several studies focusing on plasma, to the best of our knowledge, dose response studies focusing on plasma, blood and multiple organs have not been published yet, making it difficult to appreciate system-wide responses to radiation.
Furthermore, radiation can be used in HSC transplantation (HSCT). Although HSCT is the only cure for many malignant and nonmalignant conditions, its use is limited by significant morbidity and mortality.16–18 Transfusion-induced iron overload is common in patients undergoing HSCT.19 It is also a risk factor for poor HSCT outcomes.20,21 In mouse models that modify iron status (e.g., by dietary manipulation, chronic transfusion, or intravenous iron administration), we identified specific changes in the gut microbiota and in circulating microbial metabolites.22 In particular, Lactobacilli spp., which are notable for being less dependent on iron for growth,23 are decreased with increasing iron load. In tandem, fecal and plasma indole metabolites, which have anti-oxidative and anti-inflammatory properties that can modulate cytokine production and host immune responses,24–26 were reduced. Additionally, murine studies showed that irradiation exposure reduces the level of indoles which is associated with increased risk of graft versus host disease (GVHD), one of the most severe side effects of HSCT. 27 Thus, determining the impact and interaction of both gamma irradiation and iron levels on the host metabolome is the main focus of this study.
METHODS
Irradiation
Female wild-type C57Bl6 mice were purchased from Charles River Laboratories (Stone Ridge, NY) and used at 8–10 weeks of age. After one week of acclimatization in a pathogen-free facility, cohorts of mice were retro-orbitally infused with phosphate buffer saline (PBS) or 12.5 mg of iron dextran (Henry Shein Animal Health, Dublin, OH), twice a week for 2 weeks for a total of 50 mg of iron. After 2 days of rest, mice were then divided in groups and irradiated with 7, 8, 9, 10, 11 Gy of C-137 (n=5 per group) going from a non-lethal to a lethal dose for this mouse strain. Total dose was split in 2 doses 3 hours apart. Stools were freshly collected after the last dose of PBS/iron (pre-stool) and after irradiation (post-stool). At day +4 post irradiation, mice were euthanized and blood was collected by cardiac puncture in heparinized syringe. Plasma obtained after centrifugation at 4500 g for 10 minutes at +4°C. Tissues, including brain, colon, duodenum, heart, kidney, liver, spleen and stool (pre- or post-irradiation/iron infusion) were weighted and all the biological samples were stored at −80°C until further processing.
Iron-related measurements:
Spleen and liver were collected at necropsy and non-heme iron determined using a wet ashing procedure. In brief, the wet weight of samples was quantified and portions of liver and spleen were placed in 2 ml micro-tubes. Following desiccation at 65 °C for 24 h, 1 ml of acid digestion mixture (3 M hydrochloric acid, 10% trichloracetic acid) was added and samples were heated at 65 °C for an additional 24 h. The acidified sample (50 μl) was then incubated for 30 min with 200 μl of chromogen (1.6 M bathophenanthroline, 2 M sodium acetate, 11.5 M thioglycolic acid). Absorbance at 535 nm of samples and iron standards was measured in duplicate and mean values used for calculating total iron. Iron transferrin saturation in plasma was calculated using Iron/TIBC Reagent Set (Pointe Scientific, Canton, MI).
Metabolomics:
Metabolomics analyses were performed as extensively described in previous studies.28 A volume of 20μl of frozen plasma or RBC aliquots or 15 mg of ground tissue (GenoGrinder) was extracted in either 480μl or 1 ml, respectively of methanol:acetonitrile:water (5:3:2, v/v/v).29 After vortexing at 4°C for 30 min, extracts were separated from the protein pellet by centrifugation for 10 min at 10,000g at 4°C and stored at −80°C until analysis. Ultra-High-Pressure Liquid Chromatography-Mass Spectrometry analyses were performed using a Vanquish UHPLC coupled online to a Q Exactive mass spectrometer (Thermo Fisher, Bremen, Germany).30 Samples were analyzed using a 5 minute gradient as described.30,31 Solvents were supplemented with 0.1% formic acid for positive mode runs and 1 mM ammonium acetate for negative mode runs. MS acquisition, data analysis and elaboration was performed as described.
Statistical methods and data analysis.
Graphs and statistical analyses (unpaired T-test) were prepared with GraphPad Prism 8.0 (GraphPad Software, Inc, La Jolla, CA). Heat maps, hierarchical clustering analyses, partial least square-discriminant analyses, two-way ANOVA were calculated and plotted with MetaboAnalyst 5.0.32 Bar graphs were plotted with the free online tool Metabolite Autoplotter.33 Graphics that provide an overview of the experimental design were generated through the software Biorender.
RESULTS
Multi-organ characterization of the metabolic impact of irradiation
Cohorts of iron overloaded or control mice were irradiated with doses ranging from 7 to 11 Gy and metabolomics analyses were performed on RBCs, plasma, and the following organs: brain, colon, duodenum, heart, kidney, liver, spleen and stool (Supplementary Table 1). Results are reported in tabulated form in Supplementary Table 1, which includes metabolite names, KEGG IDs, retention times, accurate intact masses, along with all raw data. For each matrix tested in this study, results were graphed in Supplementary Figure 1, including unsupervised analyses, hierarchical clustering analysis of the significant metabolites by ANOVA and principal component analyses (PCAs), as well as bar plots for significant metabolites as a function of irradiation doses.
Irradiation impacts glycolysis, polyamine, sulfur metabolism and acyl-carnitines in RBCs, plasma, spleen, and liver
Metabolomics data suggest that irradiation causes metabolic damage to RBC energy and redox systems in a dose-dependent fashion (heat map in Figure 1.B), a phenomenon that was accompanied by changes in spleen and liver metabolomes. This is relevant in that spleen and liver are the organs where extravascular hemolysis, i.e., resident macrophage-dependent erythrophagocytosis is observed (Figure 1.C). In Figure 1.D we show a representative PCA of RBCs exposed to increasing doses of irradiation, with PC1 explaining 17% of the total variance across sample groups from mice irradiated with different doses vs untreated mice. Similar patterns were noted in the plasma, spleen and liver metabolomes, with the latter showing a subset of metabolites spiking at the lowest dose of radiation (7 Gy), or peaking at 10–11 Gy. Glycolysis (Figure 1.E), polyamines (especially putrescine), amino acids (especially arginine, glutamine, sulfur metabolites – methionine, taurine – Figure 1.F–G) ranked amongst the top metabolites impacted by irradiation in a dose-response fashion in the spleen. Similar effects were observed in RBCs with respect to glycolysis (decreases of fructose bisphosphate and pyruvate) and sulfur metabolites (taurine – Figure 1.H), purines (inosine and hypoxanthine – Figure 1.I) and polyamines (spermine, spermidine, putrescine – Figure 1.J). Notably, irradiation induced significant decreases in the levels of free carnitine, as well as several acyl-carnitines in RBCs from irradiated mice, suggestive of potential activation of the Lands cycle in red blood cells from irradiated mice even at low doses (Figure 2.A). Irradiation was also found to have an impact on the Krebs cycle in the liver – with peak levels of multiple carboxylic acids (citrate, fumarate and malate at Gy 8), polyamines (putrescine) and poly-unsaturated fatty acids (eicosapentaenoic acid) in the liver, the latter in an irradiation dose-dependent manner (Figure 2.B).
Figure 1 – Metabolomics analyses of red blood cells, plasma, liver and spleen following irradiation.
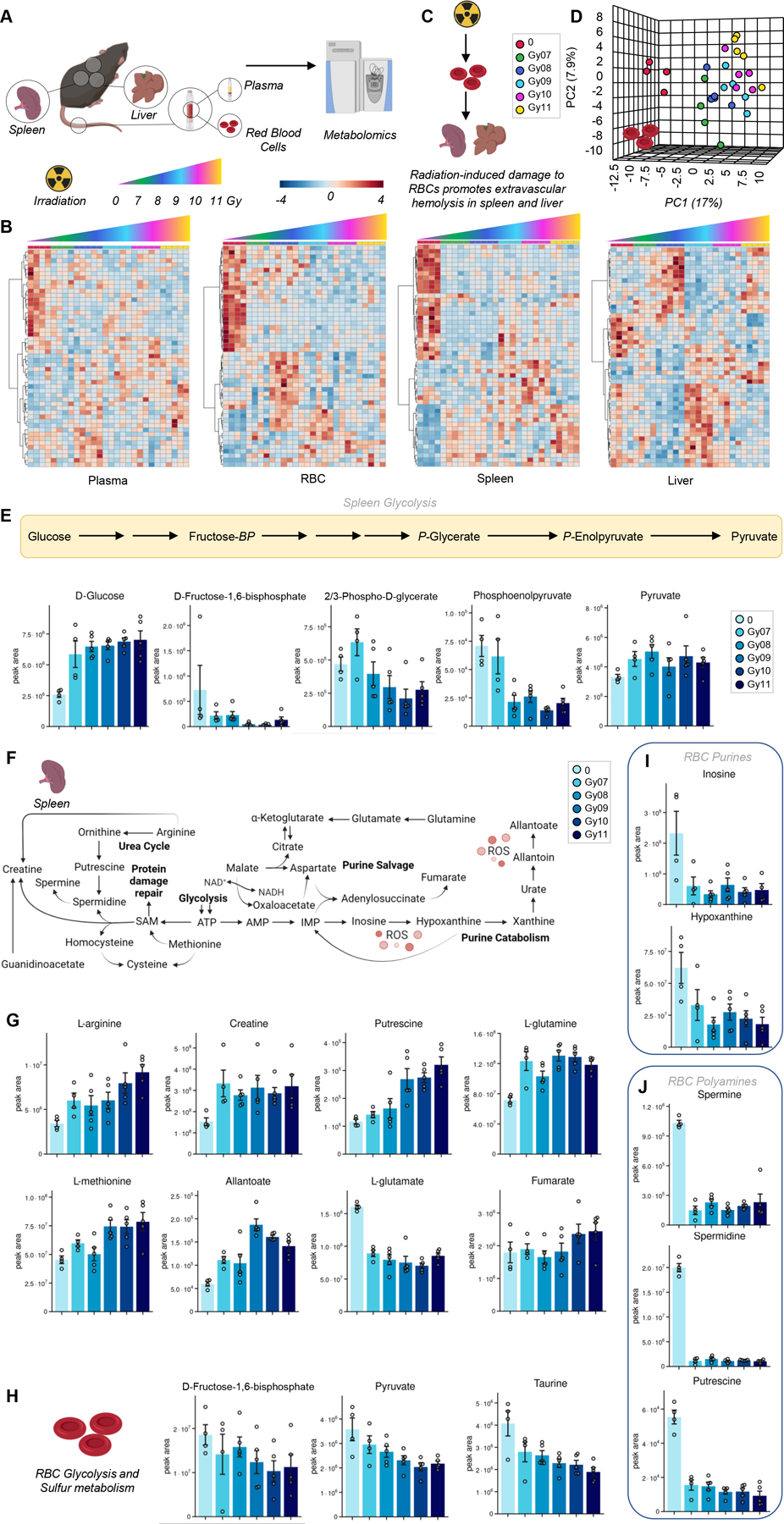
We performed metabolomics analyses of red blood cells (RBCs), plasma and organs where erythrophagocytosis occurs, spleen and liver as a function of irradiation (untreated: 0; or irradiated: from 7 to 11 Gy – A). Heat maps of metabolites significant by ANOVA are shown in B. Data suggest progressive damage to RBC energy and redox systems, which corresponds to increased metabolic changes at higher irradiation doses in spleen and liver (C). A representative principal component analysis of RBCs exposed to increasing doses of irradiation (D). Results point at a significant impact of irradiation on glycolytic (E), polyamine (overview of the pathway in F, bar plots in G) and sulfur/amino acid metabolism in the spleen. We report alteration of glycolysis, taurine (H) and purine (J) metabolism in RBCs of irradiated mice.
Figure 2 – Alterations of acyl-carnitine metabolism in RBCs from irradiated mice.
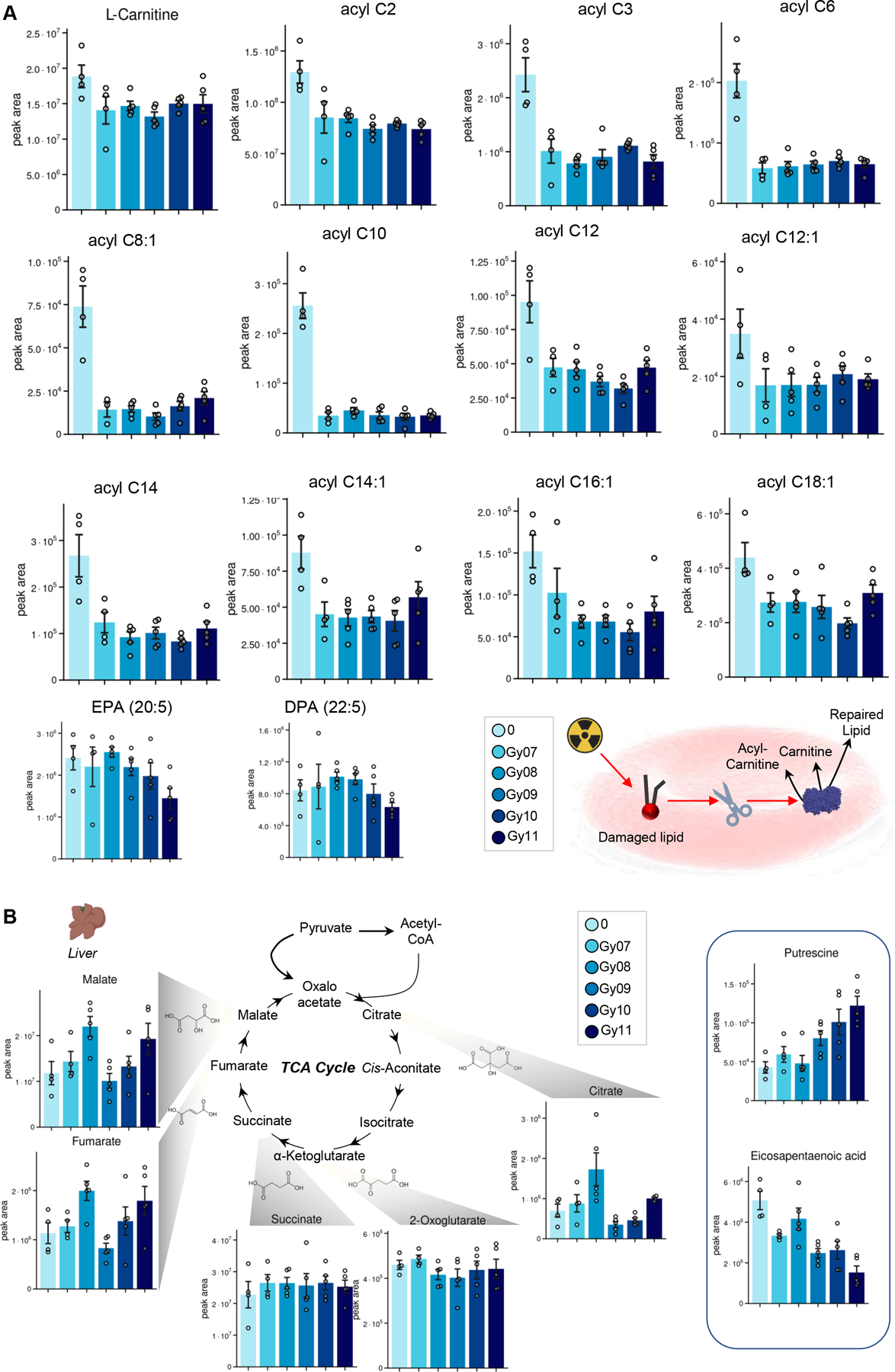
(A). Irradiation was also found to have an impact on the Krebs cycle, polyamines and poly-unsaturated fatty acids in the liver (B).
Metabolic impact of irradiation on mouse brain, liver and kidneys
Given the role of RBCs on oxygen transport and the damaging impact of radiation on RBC metabolism, we thus focused on metabolomics analyses of organs with high oxygen consumption, such as brain, heart and kidney (Figure 3.A). Hierarchical clustering analyses of the metabolites significant by ANOVA revealed radiation dose response patterns in the brain, heart and kidneys (Figure 3.B). This dependency on irradiation dose of metabolic phenotypes was mostly evident in the kidney, as shown by the PCA (Figure 3.C). Significant metabolic alterations were observed following irradiation in heart, kidney and brain (Figure 3.D–G). Across these organs, we observed increases in the levels of polyamines (putrescine in heart and kidney), purines and purine salvage metabolites (inosine, hypoxanthine, S-adenosylmethionine – decreasing in kidney and brain), amino acids (glutamine in kidney) and carboxylic acids (especially decreases in fumarate and malate). Interestingly, succinate was observed to increase in the brain and decrease in the heart following irradiation (Figure 3.D and F), suggesting organ-specific responses to irradiation.
Figure 3 – Metabolic impact of irradiation on mouse brain, liver and kidneys.
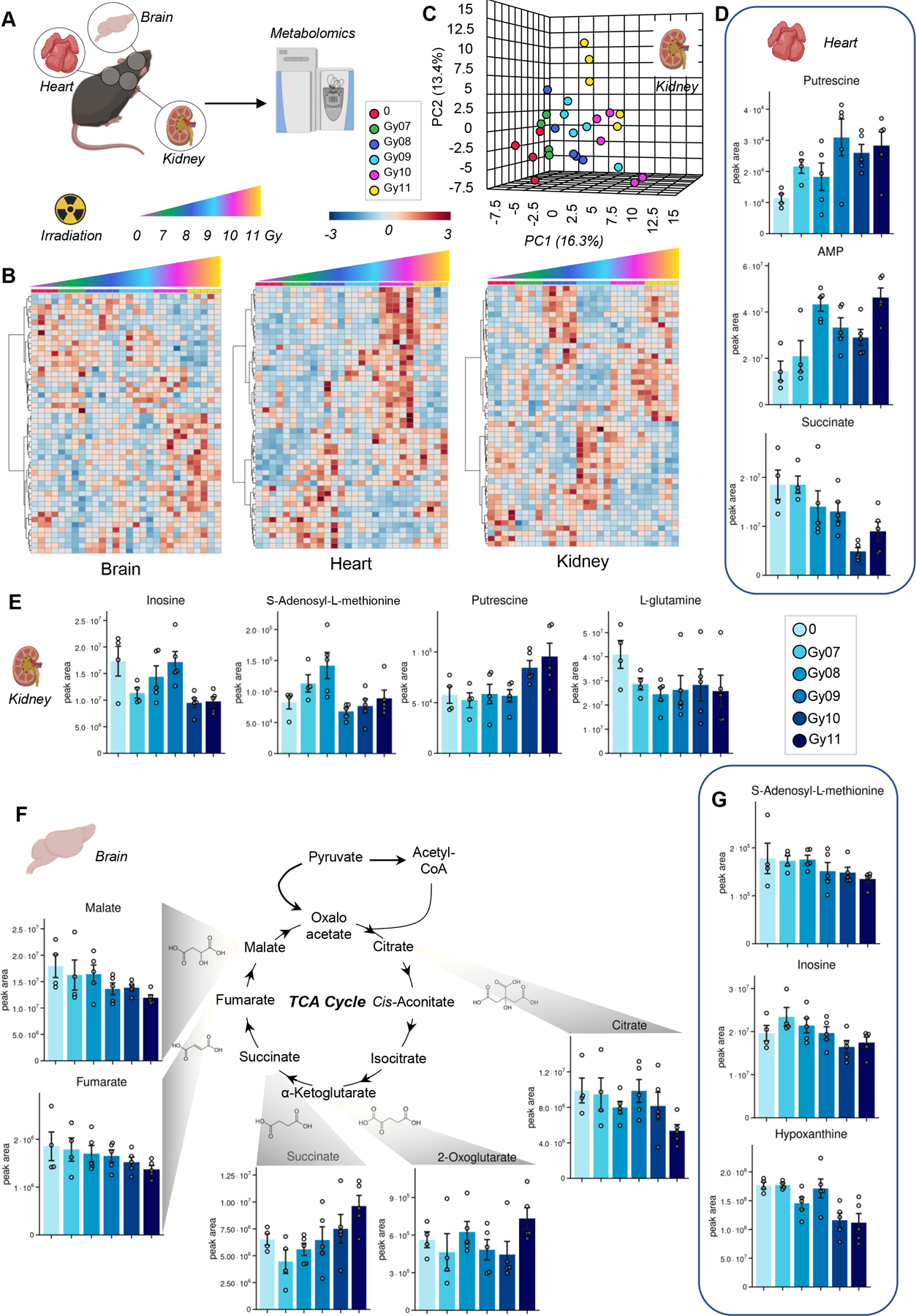
(A), as determined by hierarchical clustering of the metabolites significant by ANOVA (B) and principal component analysis (representative PCA for kidneys in C). In D-G, bar plots of significantly altered polyamines, purines, amino acids and carboxylic acids in heart, kidney and brain.
Metabolic impact of irradiation on mouse colon, duodenum and stool
Recent studies have suggested an impact of irradiation on the gut microbiome and associated metabolomes.11 To test whether these observations were recapitulated in our model, we performed metabolomics analysis was performed in colon, duodenum and stool in intravenous vehicle-treated mice, before and after irradiation i(pre- and post-stool – Figure 4.A). Statistical analyses via ANOVA informed the clustering of metabolites in the heat maps in Figure 4.B, which show a significant impact of irradiation on the metabolomes of these four tissues. Overall, results suggest that radiation induced alterations to metabolites in colon and stool compatible with a potential effect on the gut microbiome (Figure 4.C). Specifically, in the colon, irradiation promoted decreases in the levels of glycolytic metabolites (glucose, pyruvate, lactate), increases in polyamines (putrescine and spermidine) and altered levels of acyl-carnitines, with increases in C4- and C5-carnitine (intermediates of branched chain amino acid catabolism), and decreases in many other acyl-carnitines, especially hydroxylated forms (markers of fatty acid catabolism, with several odd chain fatty acyl-conjugated moieties of potential bacterial origin – Figure 4.D).
Figure 4 – Metabolic impact of irradiation on mouse colon, duodenum and stool.
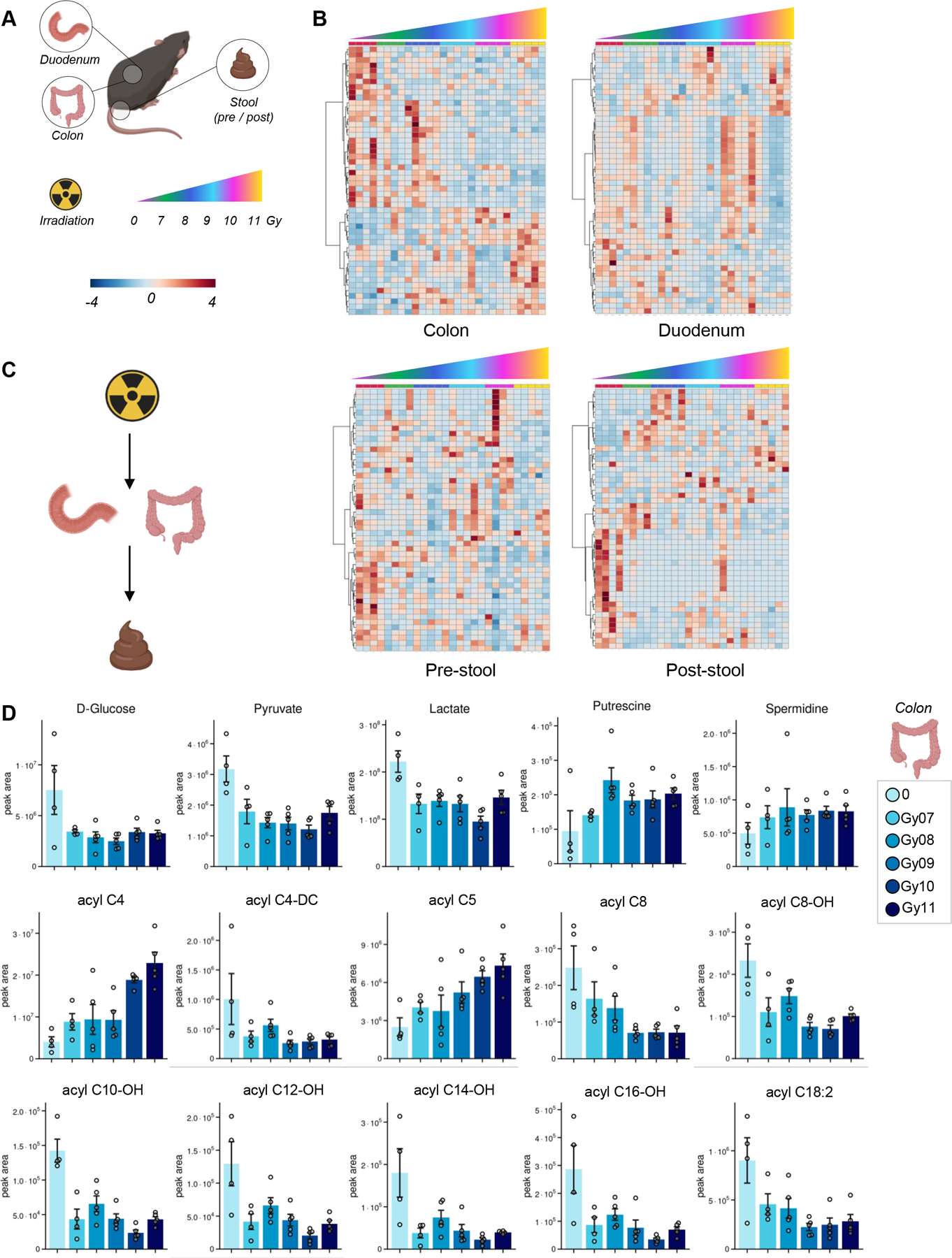
(A), as determined by ANOVA (heat maps in B show significant metabolic patterns). Results suggest that radiation induced alterations to metabolites in colon and stool compatible with a potential effect on the gut microbiome (C). In D, representative bar plots for top metabolites affected by irradiation in the colon, including glycolytic metabolites, polyamines and acyl-carnitines (especially hydroxylated forms).
Impact of iron infusion on the metabolic responses to irradiation in organs and blood
After appreciating the impact of irradiation on systems metabolism, we wondered whether these effects would be exacerbated by prior infusion of iron, a model that recapitulates transfusional iron overload, a condition often observed in chronically transfused patients with thalassemia or sickle cell disease, or myelodysplastic syndrome.34 Since some of these patients receiving transfusion may also undergo bone-marrow irradiation as part of HSCT procedures, we investigated whether iron overload would compound the metabolic impact of irradiation on systems metabolism (Figure 5.A).
Figure 5 – Impact of iron infusion on the metabolic responses to irradiation in organs and blood (cells –
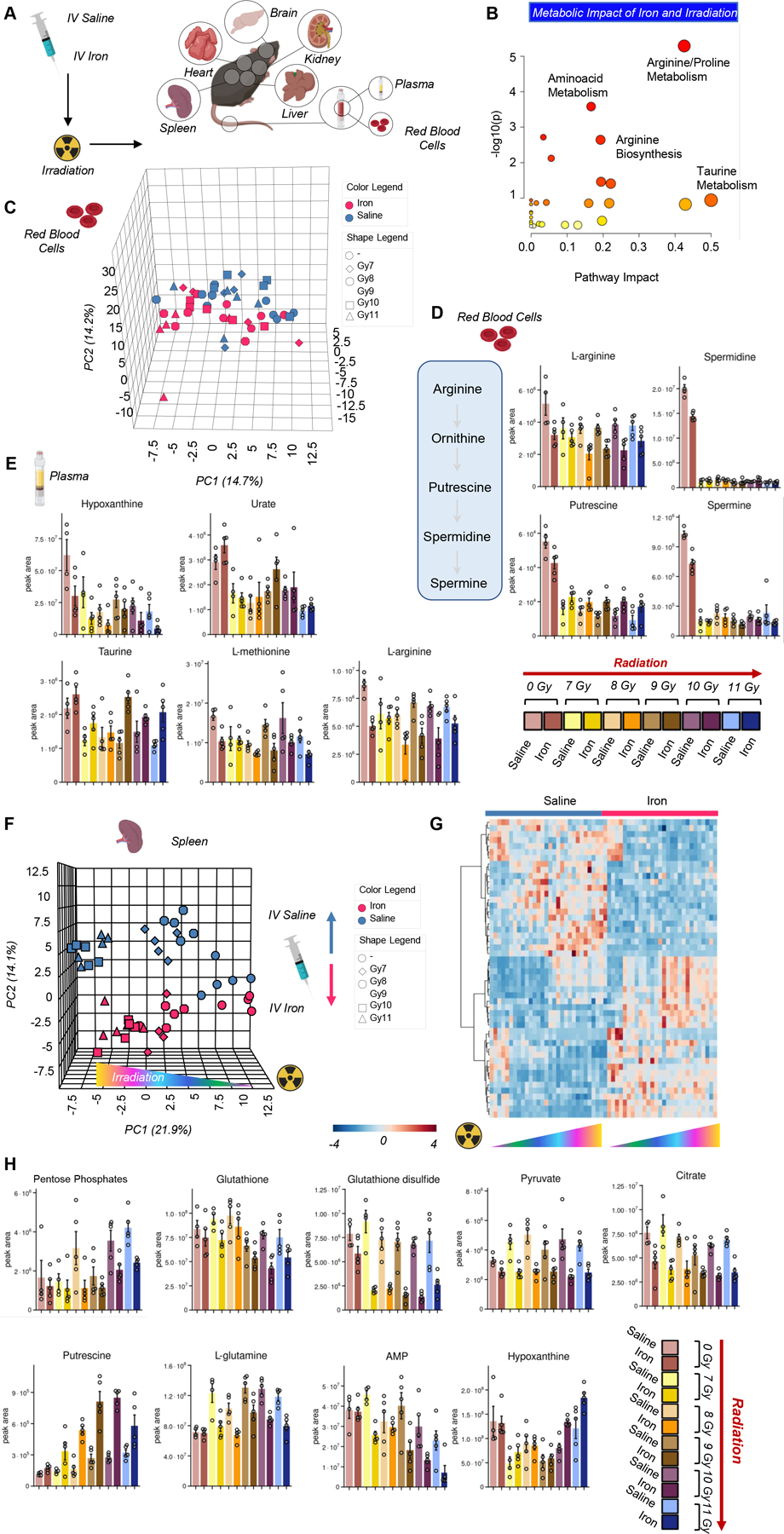
A). An overview of the top metabolic pathways affected by iron infusion and irradiation is provided in B. In C, principal component analysis shows a clear effect of iron (red vs blue for saline) and irradiation on the red blood cell metabolic phenotypes, with the highest effects noted on red blood cell arginine metabolism and polyamine synthesis (D), purine oxidation and sulfur metabolism in plasma (E). Spleen was the organ showing the highest metabolic changes after iron infusion and irradiation, as shown by the principal component analysis in F. In G, heat map of the spleen metabolites significant by two-way ANOVA. The top hits from this analysis are reported as bar plots in H.
Results are reported extensively in supplementary table 2 and all unsupervised and more targeted analyses are reported for each matrix in Supplementary Figure 2. Pathway analyses indicated that the top metabolic pathways affected by iron infusion and irradiation included arginine/proline metabolism (also including polyamines as downstream product of arginine metabolism), amino acid metabolism, especially taurine metabolism (Figure 5.B). Principal component analysis shows a clear effect of iron (red vs blue for saline) and irradiation on the metabolic phenotypes (RBCs are presented as an example in Figure 5.C). In RBCs, the highest effects of iron and irradiation were noted on arginine metabolism and polyamine synthesis While the levels of these metabolites decreased overall with radiation, iron infusion prior to radiation resulted in further decreases in arginine along with increased levels of putrescine and unchanged or decreased levels of the downstream polyamines spermidine and spermine, respectively. (Figure 5.D). Similar effects were noted in the levels of purines (hypoxanthine and urate), with iron inducing decreases in hypoxanthine and increases in urate, despite a trend towards decrease in irradiated mice for both metabolites (Figure 5.E). Iron infusion was accompanied by decreases in methionine and arginine, and increases in taurine levels, with a relatively negligible effect of irradiation dosage (Figure 5.E).
Spleen was the organ showing the highest metabolic change after iron infusion and irradiation, as shown by the PCA in (Figure 5.F), where sample grouping based on irradiation phenotypes was evident across PC1, explaining 22% of the total variance, and iron infusion on PC2, which explained ~14% of the total variance. The effect of intravenous iron infusion on the spleen metabolome was even more evident in the heat map of the metabolites significant by two-way ANOVA (Figure 5.G). The top hits from this analysis are reported as bar plots in Figure 5.H, and include several metabolites in the antioxidant responses (pentose phosphate isomers, reduced and oxidized glutathione), decreasing as a function of iron infusion. Similar trends were observed for pyruvate and citrate – both decreasing in the spleen following iron infusion. Increases in polyamines, especially putrescine, following irradiation in a dose-dependent fashion were further exacerbated by iron infusion (Figure 5.H), which also corresponded to increased glutamine and AMP consumption and hypoxanthine generation in the spleen.
Iron infusion and irradiation significantly impact glucose, sulfur and tryptophan metabolism in the liver, stool and brain
Iron infusion had a significant effect on liver metabolism (Figure 6.A), especially on glycolysis (glucose, fructose bisphosphate, glyceraldehyde 3-phosphate, phosphoglycerate), pentose phosphate pathway (pentose phosphate isomers) and reduced glutathione – all decreasing in iron infused mice (Figure 6.B). On the other hand, iron infusion boosted methionine and taurine metabolism, with several intermediates of this pathway increasing in the liver after iron infusion, including taurine and methionine, methionine sulfoxide, acetyl-choline, with significant decreases in hypotaurine (Figure 6.C).
Figure 6 – Iron infusion and irradiation significantly impact liver, stool and brain metabolism.
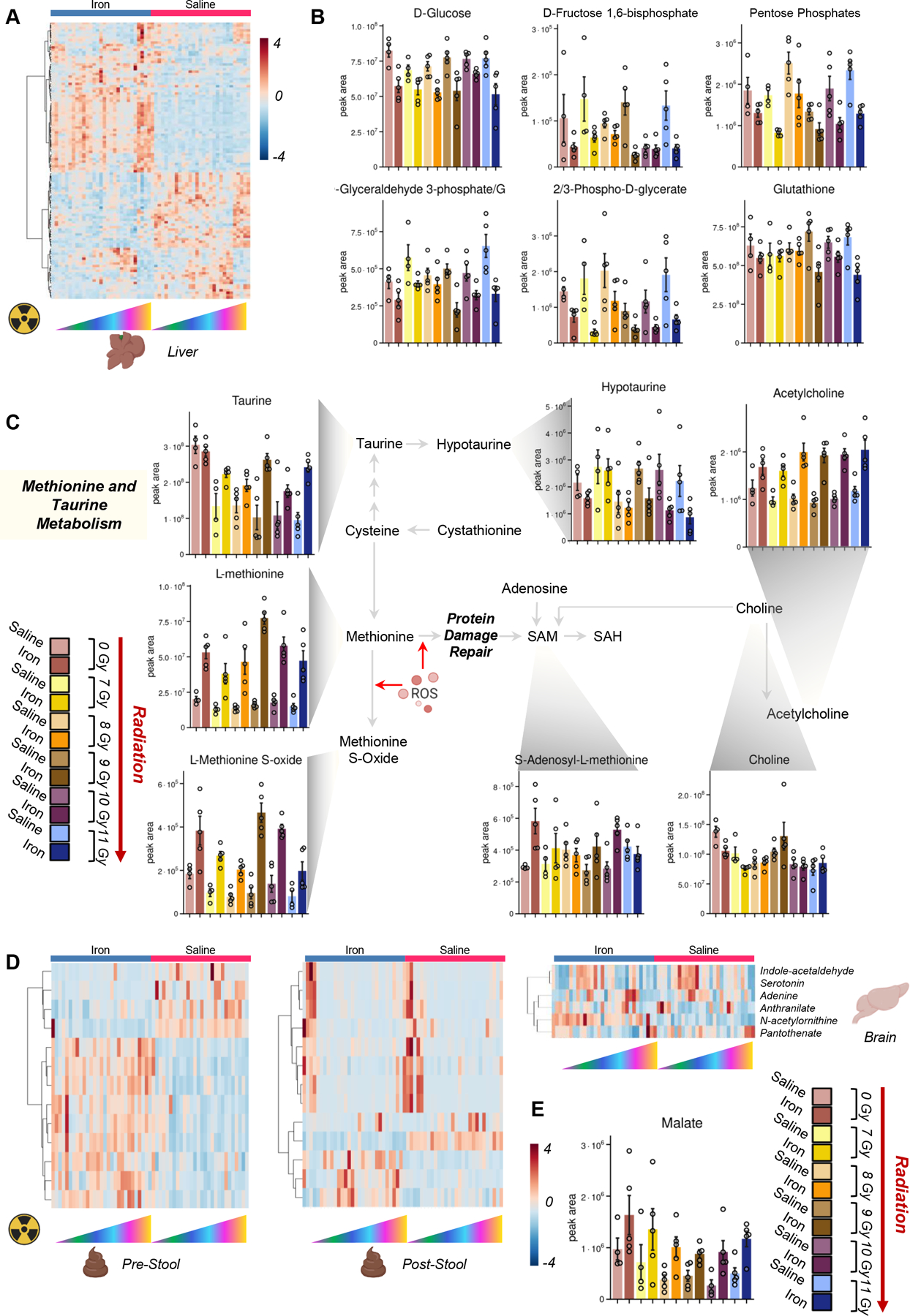
Heat map in A shows a significant impact of iron infusion and irradiation on the liver metabolome, with top significant metabolites with differential trends mapping in the glycolytic pathway, glutathione, taurine and sulfur homeostasis (B and C, respectively). Strong metabolic impacts of iron infusion and irradiation were observed in the stool (with iron infusion without irradiation – pre-stool; with iron infusion and after irradiation – post-stool – D) and in the brain, where several tryptophan metabolites (D) and carboxylic acids like malate (E) were altered by iron infusion.
Iron infusion also impacted stool (before and after irradiation – Figure 6.D) and the brain. In the brain, several tryptophan metabolites of potential bacterial origin and/or with a role in neurotransmission (indole-acetaldehyde, serotonin, anthranilate) were significantly impacted by iron infusion and irradiation (Figure 6.D), along with carboxylic acids like malate (Figure 6.E), which increased upon iron infusion.
Metabolic correlates to iron levels in the liver and the spleen
Finally, direct measurements of iron in the liver and the spleen highlighted an increase in organ levels of iron as a function of irradiation (~1.5 fold increase) and iron infusion (~15–20 fold increase – Figure 7.A–B). Correlation analyses of metabolomics data from all matrices tested in this study were performed against iron levels in the liver or spleen (Figure 7.C and D). Results confirm the pathway analyses described in the previous paragraphs, showing a significant impact of iron infusion on liver and spleen metabolites (top 25 correlates ranked by Spearman r; p<0.001 are reported in Figure 7.E), especially amino acids (asparagine, cysteine, tryptophan, phenylalanine, methionine, proline, threonine) indoles (e.g., indolepyruvate, hydroxyindoleacetate), but also heart carnosine levels– with some organ-specific trends (e.g., indolepyruvate increasing in liver and decreasing in the stool after iron infusion) (Figure 7.F–G).
Figure 7 – Direct measurements of iron in liver (A) and spleen (B) and metabolic correlates across organs (C-D), as color-coded in D.
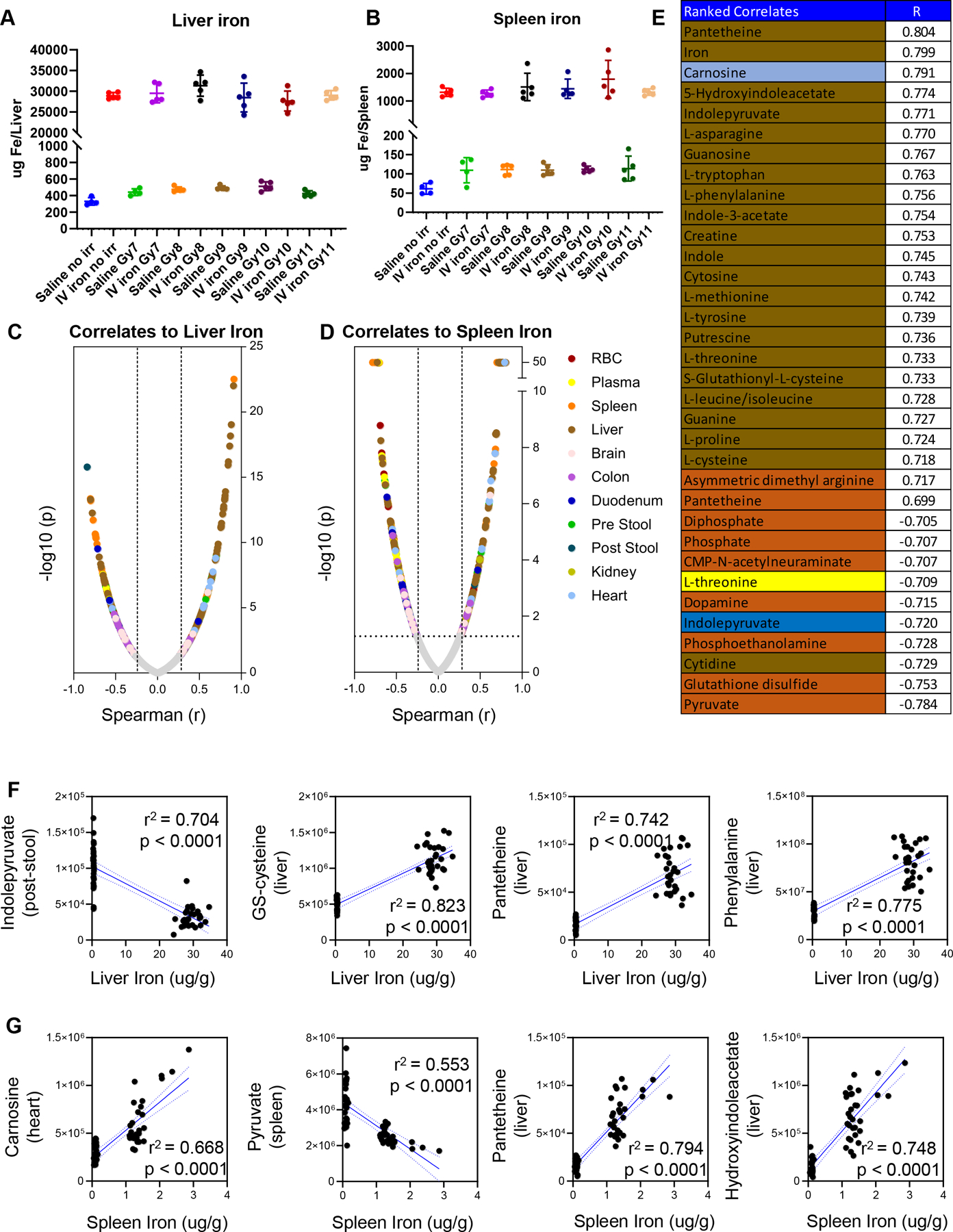
The same color code is applied to the top 25 significant correlates (Spearman) to spleen iron (strongly correlated to liver iron as well). In F and G, line plots show selected correlates to liver and spleen iron.
DISCUSSION
In the present study we report a comprehensive multi-organ characterization of systems responses to radiation doses from 7–11 Gy, in presence and absence of prior iron overload. As a result, we report a significant impact of radiation on arginine metabolism towards the increased synthesis of polyamines in all matrices/organs tested in this study, except for RBCs, where polyamine levels significantly decreased following radiation. These phenomena of polyamines – especially putrescine - increasing in all organs and decreasing in RBCs were further exacerbated by prior iron overload. Though the specific mechanisms underlying this observation are unclear, our findings recapitulate previous observations in cancer cells were c-Myc or p53-dependent signaling regulate some of the rate-limiting enzymes in the polyamine synthesis pathway, at the crossroads of methionine metabolism and purine salvage.35
Our results are also consistent with the radiation-induced activation of the rate-limiting enzyme of the polyamine synthesis pathway, ornithine decarboxylase, a phenomenon that had been described in the early 1990’s following exposure to ultraviolet radiation36 or in patients prone to developing skin cancer following prolonged sun exposure.37 Similar radiation-induced activation of ornithine decarboxylase was associated with poorer outcomes in rodent models of colon cancer, whereby activation of polyamine synthesis pathway abrogated the beneficial anti-radiation effects of the dietary sulfur compound diallyl sulfide.38 Of note, activation of the polyamine synthesis pathway was here associated with alterations in purine oxidation, decreasing in most organs, especially RBCs, upon radiation. This is an unexpected finding since we would have predicted that radiation induced decreases in energy metabolism (here observed in multiple organs at the glycolytic and mitochondrial level) would have corresponded to increased breakdown of high-energy phosphate purines (ATP, AMP – which we indeed observed) and increases in oxidant stress. Declines in glycolysis were noted upstream to pyruvate kinase, suggesting an effect of irradiation and iron levels (perhaps through Fenton chemistry) on redox sensitive reactive thiols in rate-limiting steps of glycolysis (especially glyceraldehyde 3-phosphate dehydrogenase39 and pyruvate kinase40). With respect to oxidant stress, the main signature identified here – as a function of radiation dose and exacerbated by iron infusion – was the activation of the pentose phosphate pathway, the decline in glutathione pools and oxidation of methionine and taurine in plasma and RBCs. Since methionine metabolism is linked to polyamine synthesis through purine salvage reaction and methylation processes, future studies should aim at understanding the impact of radiation and iron infusion on epigenetic regulation through methylation of proteins (e.g., to repair deamidation-induced isoaspartyl-damage, which could also target glycolytic enzymes mostly in the solvent accessible areas of their active sites41), RNA, histones and DNA.42 Of note, accumulation of taurine, methionine, methionine sulfoxide and acetyl-choline in the liver following iron infusion and irradiation is suggestive of a significant impact of these treatments on sulfur and one-carbon metabolism. These findings confirm decreases in circulating levels of taurine (e.g., in RBCs) reported in the past as a consequence of radiation,43 which is relevant in light of the protective role of this metabolite on boosting energy metabolism and antioxidant responses in RBCs.44 Of note, acute45 and prolonged46 stress on RBC elicit increased levels of acyl-carnitine levels accompanied by decreased deformability and increased vesiculation rate,45 as a function of alterations to the membranes and activation of the Lands cycle pathway to repair (oxidant) damage to (lyso)phospholipids.47 On the other hand, lower levels of acyl-carnitines are associated with increased susceptibility to osmotic and oxidant stress in healthy blood donors as a function of sex dimorphisms, especially RBCs from hypogonadic males undergoing testosterone-replacement therapy.48 Radiation in this study resulted in decreased levels of RBC acyl-carnitines. As such, it is possible that radiation itself may interfere with Lands cycle repair of membranes. Since the energy-less, oxidized and less deformable RBC rapidly undergoes extravascular hemolysis in the liver and spleen,49,50 it is also possible that some of the metabolic signatures observed in these organs following irradiation and iron infusion are not the result of a direct effect of these treatments and at least in part influenced by increased erythrophagocytosis selectively of the most damaged cells. In support, RBC exposed to the highest levels of radiation in this study (11 Gy) had higher levels of acyl-carnitines, suggesting more comprehensive damage to the RBC population as a whole.
Our data also expand on prior findings showing liver increases in the levels of taurine and methionine, suggestive that sulfur metabolism dysregulation follows organ-specific trends which could be at least in part explained by microbial dysbiosis. Indeed, deconjugation of taurine-conjugated bile acids is catalyzed by enzymes expressed by bacteria in the gut microbiome, whereby alterations of bile acid metabolism has been mechanistically implicated in inflammatory complications, such as macrophage activation, in the context of cholestasis,51 trauma-induced microbiome alterations following decreased enteric perfusion,52 bacterial infiltration in precancerous intraductal papillary mucinous neoplasms,53 iron overload54 and non-alcoholic fatty liver disease55. Similar considerations can be made for several short and odd chain (carnitine-conjugated) fatty acids, increasing in the colon and stool following irradiation, especially when preceded by intravenous iron infusion. These findings are consistent with prior reports on alterations of these metabolites as a function of the interactions between host genetics and the gut microbiome on the development of central nervous system autoimmune conditions.56 Interestingly, decline in highly-unsaturated fatty acids in the context of inflammation-induced neurodegenerative diseases such as Alzheimer’s57 or aging have been recently reported, suggesting that irradiation could phenocopy at least in part a model of accelerated (metabolic) aging4 and shortened healthspan in humans.58 In this view, it is worthwhile to note that alterations of one-carbon metabolism (especially methionine and choline) have been associated with impaired erythropoiesis in aging,59 neurocognitive deficit in infants60 and individuals with Down syndrome,61 which represents a model of accelerated aging,62 and overall shortened lifespan through regulation of the mechanistic Target of Rapamycin (mTOR).63
Finally, radiation and iron infusion-induced alterations of indole metabolites, which study recapitulate previous findings in the literature.8,11,12,64 Expanding on the literature, here we report that the brain metabolome was significantly impacted by iron and radiation, with tryptophan metabolites – including neuroactive serotonin – being the most impacted pathway in the brain. Impacts of iron infusion on brain metabolism were expected, in that iron is a cofactor in several neuron-specific functions, such as myelin and neurotransmitter (e.g., dopamine) synthesis.65 While no significant alterations of dopamine metabolism were observed in the brain, liver and other organs, phenylalanine levels positively correlated with hepatic iron levels upon intravenous infusion, an observation in keeping with the dependency of the phenylalanine to tyrosine conversion reaction to iron levels.66 Of note, alterations of indole/serotonin metabolism in the colon/stool and the brain provides further correlative evidence for the systemic impact of iron and radiation through alteration on the gut-brain axis67 and outcomes of HSCT.27,68,69
Limitations of this study are noted, in that only a single omics approach has been used to characterize the impact of iron and irradiation on systems metabolism. Future studies could involve other omics (e.g., transcriptomics, proteomics, lipidomics) to complement metabolomics data. Given the clear signatures in polyamine and kynurenine metabolism, steady state metabolomics data will need to be confirmed through metabolic tracing experiments, for example through the use of stable isotope-labeled arginine and tryptophan to further test the impact of irradiation and iron supplementation on the metabolism of these two amino acids in vivo.
In summary, this study provides for the first time a comprehensive, system-wide look at the impact of radiation and intravenous iron infusion on the metabolism of multiple organs in mice. The results revealed that irradiation impacted the metabolome of the spleen, RBC, plasma, brain, liver, heart, kidney, colon, duodenum and stool, and that these impacts were modulated by iron infusion. In the organs, radiation impacted arginine metabolism as reflected by increased levels of polyamines. In RBCs, the opposite trend was observed, consistent with the previously described radiation-induced activation of ornithine decarboxylase in nucleated cells. Decreases in levels of RBC antioxidants including taurine and glutathione, and polyamines, the apparent activation of the pentose phosphate pathway, as well as decreases in acyl-carnitines, may indicate increased oxidant damage and membrane lipid remodeling (activation of the Lands cycle) in RBCs after irradiation. Our results also suggest that radiation and iron infusion modulate tryptophan metabolism. Moreover, the brain metabolome was significantly impacted by radiation and iron, which has not been described until now. Finally, sulfur metabolism appeared to be dysregulated in an organ-specific manner, potentially owing to microbial dysbiosis. Increases in colon and stool fatty acids and indole/tryptophan metabolites after radiation, especially in the iron treated groups, may also reflect interactions between the gut microbiome and host genome. These findings further detail the impacts of irradiation across multiple organs and shed light on the effect of iron availability on radiosensitivity.
Supplementary Material
Supplementary Figure 1 – Multivariate analyses and vectorial versions of figures 1–4, divided by organ.
Supplementary Figure 2 – Multivariate analyses and vectorial versions of figures 5–7, divided by organ.
Supplementary Table 1.xlsx - All raw data and elaborations
ACKNOWLEDGMENTS
This research was supported by funds from the Boettcher Webb-Waring Investigator Award (ADA), RM1GM131968 (ADA) from the National Institute of General and Medical Sciences, and R01HL146442 (ADA), R01HL149714 (ADA), R01HL148151 (ADA), R21HL150032 (ADA), R21HL145319 (EAH) from the National Heart, Lung, and Blood Institute.
Footnotes
CONFLICT OF INTEREST
Though unrelated to the contents of this manuscripts, the authors declare that AD is a founder of Omix Technologies Inc.. AD is also a consultant for Altis Biosciences LLC., Rubius Inc. and Forma Inc. AD is a consultant for Hemanext Inc. All the other authors disclose no conflicts of interest relevant to this study.
SUPPORTING INFORMATION
Supporting Information is available free of charge at the Journal Website.
DATA AVAILABILITY
Raw mass spectrometry data for this study are available for free download at the Metabolomics Workbench website, with the following IDs: ST002031-ST002041.
References
- 1.Gibson BW; Boles NC; Souroullas GP; Herron AJ; Fraley JK; Schwiebert RS; Sharp JJ; Goodell MA, Comparison of Cesium-137 and X-ray Irradiators by Using Bone Marrow Transplant Reconstitution in C57BL/6J Mice. Comp Med 2015, 65, (3), 165–72. [PMC free article] [PubMed] [Google Scholar]
- 2.Fentiman IS; Deshmane V; Tong D; Winter J; Mayles H; Chaudary MA, Caesium(137) implant as sole radiation therapy for operable breast cancer: a phase II trial. Radiother Oncol 2004, 71, (3), 281–5. [DOI] [PubMed] [Google Scholar]
- 3.Henry CJ; Casás-Selves M; Kim J; Zaberezhnyy V; Aghili L; Daniel AE; Jimenez L; Azam T; McNamee EN; Clambey ET; Klawitter J; Serkova NJ; Tan AC; Dinarello CA; DeGregori J, Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J Clin Invest 2015, 125, (12), 4666–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dong S; Wang Q; Kao YR; Diaz A; Tasset I; Kaushik S; Thiruthuvanathan V; Zintiridou A; Nieves E; Dzieciatkowska M; Reisz JA; Gavathiotis E; D’Alessandro A; Will B; Cuervo AM, Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones CL; Stevens BM; D’Alessandro A; Reisz JA; Culp-Hill R; Nemkov T; Pei S; Khan N; Adane B; Ye H; Krug A; Reinhold D; Smith C; DeGregori J; Pollyea DA; Jordan CT, Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 2018, 34, (5), 724–740.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Alessandro A; Hay A; Dzieciatkowska M; Brown BC; Morrison EJ; Hansen KC; Zimring JC, Protein-L-isoaspartate O-methyltransferase is required for in vivo control of oxidative damage in red blood cells. Haematologica 2021, 106, (10), 2726–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michaletti A; Gioia M; Tarantino U; Zolla L, Effects of microgravity on osteoblast mitochondria: a proteomic and metabolomics profile. Sci Rep 2017, 7, (1), 15376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laiakis EC; Shuryak I; Deziel A; Wang YW; Barnette BL; Yu Y; Ullrich RL; Fornace AJ Jr.; Emmett MR, Effects of Low Dose Space Radiation Exposures on the Splenic Metabolome. Int J Mol Sci 2021, 22, (6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simonsen LC; Slaba TC; Guida P; Rusek A, NASA’s first ground-based Galactic Cosmic Ray Simulator: Enabling a new era in space radiobiology research. PLoS Biol 2020, 18, (5), e3000669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garrett-Bakelman FE; Darshi M; Green SJ; Gur RC; Lin L; Macias BR; McKenna MJ; Meydan C; Mishra T; Nasrini J; Piening BD; Rizzardi LF; Sharma K; Siamwala JH; Taylor L; Vitaterna MH; Afkarian M; Afshinnekoo E; Ahadi S; Ambati A; Arya M; Bezdan D; Callahan CM; Chen S; Choi AMK; Chlipala GE; Contrepois K; Covington M; Crucian BE; De Vivo I; Dinges DF; Ebert DJ; Feinberg JI; Gandara JA; George KA; Goutsias J; Grills GS; Hargens AR; Heer M; Hillary RP; Hoofnagle AN; Hook VYH; Jenkinson G; Jiang P; Keshavarzian A; Laurie SS; Lee-McMullen B; Lumpkins SB; MacKay M; Maienschein-Cline MG; Melnick AM; Moore TM; Nakahira K; Patel HH; Pietrzyk R; Rao V; Saito R; Salins DN; Schilling JM; Sears DD; Sheridan CK; Stenger MB; Tryggvadottir R; Urban AE; Vaisar T; Van Espen B; Zhang J; Ziegler MG; Zwart SR; Charles JB; Kundrot CE; Scott GBI; Bailey SM; Basner M; Feinberg AP; Lee SMC; Mason CE; Mignot E; Rana BK; Smith SM; Snyder MP; Turek FW, The NASA Twins Study: A multidimensional analysis of a year-long human spaceflight. Science 2019, 364, (6436). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo H; Chou WC; Lai Y; Liang K; Tam JW; Brickey WJ; Chen L; Montgomery ND; Li X; Bohannon LM; Sung AD; Chao NJ; Peled JU; Gomes ALC; van den Brink MRM; French MJ; Macintyre AN; Sempowski GD; Tan X; Sartor RB; Lu K; Ting JPY, Multi-omics analyses of radiation survivors identify radioprotective microbes and metabolites. Science 2020, 370, (6516). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P ÓB; Vaitheesvaran B; Saha S; Hartil K; Chen EI; Goldman D; Fleming WH; Kurland IJ; Guha C; Golden A, Intestinal microbiota-derived metabolomic blood plasma markers for prior radiation injury. Int J Radiat Oncol Biol Phys 2015, 91, (2), 360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grison S; Martin JC; Grandcolas L; Banzet N; Blanchardon E; Tourlonias E; Defoort C; Favé G; Bott R; Dublineau I; Gourmelon P; Souidi M, The metabolomic approach identifies a biological signature of low-dose chronic exposure to cesium 137. J Radiat Res 2012, 53, (1), 33–43. [DOI] [PubMed] [Google Scholar]
- 14.Li HH; Lin YT; Laiakis EC; Goudarzi M; Weber W; Fornace AJ Jr., Serum Metabolomic Alterations Associated with Cesium-137 Internal Emitter Delivered in Various Dose Rates. Metabolites 2020, 10, (7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goudarzi M; Weber W; Mak TD; Chung J; Doyle-Eisele M; Melo D; Brenner DJ; Guilmette RA; Fornace AJ, Development of urinary biomarkers for internal exposure by cesium-137 using a metabolomics approach in mice. Radiat Res 2014, 181, (1), 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saber W; Cutler CS; Nakamura R; Zhang MJ; Atallah E; Rizzo JD; Maziarz RT; Cortes J; Kalaycio ME; Horowitz MM, Impact of donor source on hematopoietic cell transplantation outcomes for patients with myelodysplastic syndromes (MDS). Blood 2013, 122, (11), 1974–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anasetti C; Logan BR; Lee SJ; Waller EK; Weisdorf DJ; Wingard JR; Cutler CS; Westervelt P; Woolfrey A; Couban S; Ehninger G; Johnston L; Maziarz RT; Pulsipher MA; Porter DL; Mineishi S; McCarty JM; Khan SP; Anderlini P; Bensinger WI; Leitman SF; Rowley SD; Bredeson C; Carter SL; Horowitz MM; Confer DL; Blood; Marrow Transplant Clinical Trials, N., Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med 2012, 367, (16), 1487–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majhail NS; Chitphakdithai P; Logan B; King R; Devine S; Rossmann SN; Hale G; Hartzman RJ; Karanes C; Laport GG; Nemecek E; Snyder EL; Switzer GE; Miller J; Navarro W; Confer DL; Levine JE, Significant improvement in survival after unrelated donor hematopoietic cell transplantation in the recent era. Biol Blood Marrow Transplant 2015, 21, (1), 142–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sirvent A; Auquier P; Oudin C; Bertrand Y; Bohrer S; Chastagner P; Poiree M; Kanold J; Thouvenin S; Perel Y; Plantaz D; Tabone MD; Yakouben K; Gandemer V; Lutz P; Sirvent N; Vercasson C; Berbis J; Chambost H; Leverger G; Baruchel A; Michel G, Prevalence and risk factors of iron overload after hematopoietic stem cell transplantation for childhood acute leukemia: a LEA study. Bone Marrow Transplant 2017, 52, (1), 80–87. [DOI] [PubMed] [Google Scholar]
- 20.Pullarkat V; Blanchard S; Tegtmeier B; Dagis A; Patane K; Ito J; Forman SJ, Iron overload adversely affects outcome of allogeneic hematopoietic cell transplantation. Bone Marrow Transplant 2008, 42, (12), 799–805. [DOI] [PubMed] [Google Scholar]
- 21.Meyer SC; O’Meara A; Buser AS; Tichelli A; Passweg JR; Stern M, Prognostic impact of posttransplantation iron overload after allogeneic stem cell transplantation. Biol Blood Marrow Transplant 2013, 19, (3), 440–4. [DOI] [PubMed] [Google Scholar]
- 22.La Carpia F; Wojczyk BS; Annavajhala MK; Rebbaa A; Culp-Hill R; D’Alessandro A; Freedberg DE; Uhlemann AC; Hod EA, Transfusional iron overload and intravenous iron infusions modify the mouse gut microbiota similarly to dietary iron NPJ: Biofilms and Microbiomes 2019, 5, (1), 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imbert M; Blondeau R, On the iron requirement of lactobacilli grown in chemically defined medium. Curr Microbiol 1998, 37, (1), 64–6. [DOI] [PubMed] [Google Scholar]
- 24.Zelante T; Iannitti RG; Cunha C; De Luca A; Giovannini G; Pieraccini G; Zecchi R; D’Angelo C; Massi-Benedetti C; Fallarino F; Carvalho A; Puccetti P; Romani L, Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 2013, 39, (2), 372–85. [DOI] [PubMed] [Google Scholar]
- 25.Roager HM; Licht TR, Microbial tryptophan catabolites in health and disease. Nat Commun 2018, 9, (1), 3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamas B; Richard ML; Leducq V; Pham HP; Michel ML; Da Costa G; Bridonneau C; Jegou S; Hoffmann TW; Natividad JM; Brot L; Taleb S; Couturier-Maillard A; Nion-Larmurier I; Merabtene F; Seksik P; Bourrier A; Cosnes J; Ryffel B; Beaugerie L; Launay JM; Langella P; Xavier RJ; Sokol H, CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016, 22, (6), 598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swimm A; Giver CR; DeFilipp Z; Rangaraju S; Sharma A; Ulezko Antonova A; Sonowal R; Capaldo C; Powell D; Qayed M; Kalman D; Waller EK, Indoles derived from intestinal microbiota act via type I interferon signaling to limit graft-versus-host disease. Blood 2018, 132, (23), 2506–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issaian A; Hay A; Dzieciatkowska M; Roberti D; Perrotta S; Darula Z; Redzic J; Busch MP; Page GP; Rogers SC; Doctor A; Hansen KC; Eisenmesser EZ; Zimring JC; D’Alessandro A, The interactome of the N-terminus of band 3 regulates red blood cell metabolism and storage quality. Haematologica 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.D’Alessandro A; Howie HL; Hay AM; Dziewulska KH; Brown BC; Wither MJ; Karafin M; Stone EF; Spitalnik SL; Hod EA; Francis RO; Fu X; Thomas T; Zimring JC, Hematologic and systemic metabolic alterations due to Mediterranean class II G6PD deficiency in mice. JCI Insight 2021, 6, (14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nemkov T; Reisz JA; Gehrke S; Hansen KC; D’Alessandro A, High-Throughput Metabolomics: Isocratic and Gradient Mass Spectrometry-Based Methods. Methods Mol Biol 2019, 1978, 13–26. [DOI] [PubMed] [Google Scholar]
- 31.Nemkov T; Stefanoni D; Bordbar A; Issaian A; Palsson BO; Dumont LJ; Hay AM; Song A; Xia Y; Redzic JS; Eisenmesser EZ; Zimring JC; Kleinman S; Hansen KC; Busch M; D’Alessandro A, Blood donor exposome and impact of common drugs on red blood cell metabolism. JCI Insight 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang Z; Chong J; Zhou G; de Lima Morais DA; Chang L; Barrette M; Gauthier C; Jacques P; Li S; Xia J, MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 2021, 49, (W1), W388–w396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietzke M; Vazquez A, Metabolite AutoPlotter - an application to process and visualise metabolite data in the web browser. Cancer Metab 2020, 8, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rapido F; Brittenham GM; Bandyopadhyay S; La Carpia F; L’Acqua C; McMahon DJ; Rebbaa A; Wojczyk BS; Netterwald J; Wang H; Schwartz J; Eisenberger A; Soffing M; Yeh R; Divgi C; Ginzburg YZ; Shaz BH; Sheth S; Francis RO; Spitalnik SL; Hod EA, Prolonged red cell storage before transfusion increases extravascular hemolysis. J Clin Invest 2017, 127, (1), 375–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lane DJR; Bae DH; Siafakas AR; Suryo Rahmanto Y; Al-Akra L; Jansson PJ; Casero RA Jr.; Richardson DR, Coupling of the polyamine and iron metabolism pathways in the regulation of proliferation: Mechanistic links to alterations in key polyamine biosynthetic and catabolic enzymes. Biochim Biophys Acta Mol Basis Dis 2018, 1864, (9 Pt B), 2793–2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosen CF; Gajic D; Drucker DJ, Ultraviolet radiation induction of ornithine decarboxylase in rat keratinocytes. Cancer Res 1990, 50, (9), 2631–5. [PubMed] [Google Scholar]
- 37.Terleth C; van Laar T; Schouten R; van Steeg H; Hodemaekers H; Wormhoudt T; Cornelissen-Steijger PD; Abrahams PJ; van der Eb AJ, A lack of radiation-induced ornithine decarboxylase activity prevents enhanced reactivation of herpes simplex virus and is linked to non-cancer proneness in xeroderma pigmentosum patients. Cancer Res 1997, 57, (19), 4384–92. [PubMed] [Google Scholar]
- 38.Baer AR; Wargovich MJ, Role of ornithine decarboxylase in diallyl sulfide inhibition of colonic radiation injury in the mouse. Cancer Res 1989, 49, (18), 5073–6. [PubMed] [Google Scholar]
- 39.Reisz JA; Wither MJ; Dzieciatkowska M; Nemkov T; Issaian A; Yoshida T; Dunham AJ; Hill RC; Hansen KC; D’Alessandro A, Oxidative modifications of glyceraldehyde 3-phosphate dehydrogenase regulate metabolic reprogramming of stored red blood cells. Blood 2016, 128, (12), e32–42. [DOI] [PubMed] [Google Scholar]
- 40.van de Wetering C; Manuel AM; Sharafi M; Aboushousha R; Qian X; Erickson C; MacPherson M; Chan G; Adcock IM; ZounematKermani N; Schleich F; Louis R; Bohrnsen E; D’Alessandro A; Wouters EF; Reynaert NL; Li J; Wolf CR; Henderson CJ; Lundblad LKA; Poynter ME; Dixon AE; Irvin CG; van der Vliet A; van der Velden JL; Janssen-Heininger YM, Glutathione-S-transferase P promotes glycolysis in asthma in association with oxidation of pyruvate kinase M2. Redox Biol 2021, 47, 102160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reisz JA; Nemkov T; Dzieciatkowska M; Culp-Hill R; Stefanoni D; Hill RC; Yoshida T; Dunham A; Kanias T; Dumont LJ; Busch M; Eisenmesser EZ; Zimring JC; Hansen KC; D’Alessandro A, Methylation of protein aspartates and deamidated asparagines as a function of blood bank storage and oxidative stress in human red blood cells. Transfusion 2018, 58, (12), 2978–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng Q; Weng K; Li S; Xu R; Wang Y; Wu Y, A Perspective of Epigenetic Regulation in Radiotherapy. Front Cell Dev Biol 2021, 9, 624312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Desai TK; Maliakkal J; Kinzie JL; Ehrinpreis MN; Luk GD; Cejka J, Taurine deficiency after intensive chemotherapy and/or radiation. Am J Clin Nutr 1992, 55, (3), 708–11. [DOI] [PubMed] [Google Scholar]
- 44.Bertolone L; Roy MK; Hay AM; Morrison EJ; Stefanoni D; Fu X; Kanias T; Kleinman S; Dumont LJ; Stone M; Nemkov T; Busch MP; Zimring JC; D’Alessandro A, Impact of taurine on red blood cell metabolism and implications for blood storage. Transfusion 2020, 60, (6), 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nemkov T; Skinner SC; Nader E; Stefanoni D; Robert M; Cendali F; Stauffer E; Cibiel A; Boisson C; Connes P; D’Alessandro A, Acute Cycling Exercise Induces Changes in Red Blood Cell Deformability and Membrane Lipid Remodeling. Int J Mol Sci 2021, 22, (2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bissinger R; Nemkov T; D’Alessandro A; Grau M; Dietz T; Bohnert BN; Essigke D; Wörn M; Schaefer L; Xiao M; Beirne JM; Kalo MZ; Schork A; Bakchoul T; Omage K; Kong L; Gonzalez-Menendez I; Quintanilla-Martinez L; Fehrenbacher B; Schaller M; Dhariwal A; Birkenfeld AL; Grahammer F; Qadri SM; Artunc F, Proteinuric chronic kidney disease is associated with altered red blood cell lifespan, deformability and metabolism. Kidney Int 2021, 100, (6), 1227–1239. [DOI] [PubMed] [Google Scholar]
- 47.Wu H; Bogdanov M; Zhang Y; Sun K; Zhao S; Song A; Luo R; Parchim NF; Liu H; Huang A; Adebiyi MG; Jin J; Alexander DC; Milburn MV; Idowu M; Juneja HS; Kellems RE; Dowhan W; Xia Y, Hypoxia-mediated impaired erythrocyte Lands’ Cycle is pathogenic for sickle cell disease. Sci Rep 2016, 6, 29637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander K; Hazegh K; Fang F; Sinchar D; Kiss JE; Page GP; DʼAlessandro A; Kanias T, Testosterone replacement therapy in blood donors modulates erythrocyte metabolism and susceptibility to hemolysis in cold storage. Transfusion 2021, 61, (1), 108–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roussel C; Morel A; Dussiot M; Marin M; Colard M; Fricot-Monsinjon A; Martinez A; Chambrion C; Henry B; Casimir M; Volle G; Depond M; Dokmak S; Paye F; Sauvanet A; Le Van Kim C; Colin Y; Georgeault S; Roingeard P; Spitalnik SL; Ndour PA; Hermine O; Hod EA; Buffet PA; Amireault P, Rapid clearance of storage-induced microerythrocytes alters transfusion recovery. Blood 2021, 137, (17), 2285–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.D’Alessandro A, In vivo clearance of stored red blood cells. Blood 2021, 137, (17), 2275–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Kasmi KC; Vue PM; Anderson AL; Devereaux MW; Ghosh S; Balasubramaniyan N; Fillon SA; Dahrenmoeller C; Allawzi A; Woods C; McKenna S; Wright CJ; Johnson L; D’Alessandro A; Reisz JA; Nozik-Grayck E; Suchy FJ; Sokol RJ, Macrophage-derived IL-1beta/NF-kappaB signaling mediates parenteral nutrition-associated cholestasis. Nat Commun 2018, 9, (1), 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiener G; Moore HB; Moore EE; Gonzalez E; Diamond S; Zhu S; D’Alessandro A; Banerjee A, Shock releases bile acid inducing platelet inhibition and fibrinolysis. J Surg Res 2015, 195, (2), 390–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morgell A; Reisz JA; Ateeb Z; Davanian H; Reinsbach SE; Halimi A; Gaiser R; Valente R; Arnelo U; Del Chiaro M; Chen MS; D’Alessandro A, Metabolic Characterization of Plasma and Cyst Fluid from Cystic Precursors to Pancreatic Cancer Patients Reveal Metabolic Signatures of Bacterial Infection. J Proteome Res 2021, 20, (5), 2725–2738. [DOI] [PubMed] [Google Scholar]
- 54.Ding H; Zhang Q; Yu X; Chen L; Wang Z; Feng J, Lipidomics reveals perturbations in the liver lipid profile of iron-overloaded mice. Metallomics 2021, 13, (10). [DOI] [PubMed] [Google Scholar]
- 55.Soderborg TK; Clark SE; Mulligan CE; Janssen RC; Babcock L; Ir D; Young B; Krebs N; Lemas DJ; Johnson LK; Weir T; Lenz LL; Frank DN; Hernandez TL; Kuhn KA; D’Alessandro A; Barbour LA; El Kasmi KC; Friedman JE, The gut microbiota in infants of obese mothers increases inflammation and susceptibility to NAFLD. Nat Commun 2018, 9, (1), 4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montgomery TL; Künstner A; Kennedy JJ; Fang Q; Asarian L; Culp-Hill R; D’Alessandro A; Teuscher C; Busch H; Krementsov DN, Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc Natl Acad Sci U S A 2020, 117, (44), 27516–27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lonnemann N; Hosseini S; Marchetti C; Skouras DB; Stefanoni D; D’Alessandro A; Dinarello CA; Korte M, The NLRP3 inflammasome inhibitor OLT1177 rescues cognitive impairment in a mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A 2020, 117, (50), 32145–32154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson LC; Parker K; Aguirre BF; Nemkov TG; D’Alessandro A; Johnson SA; Seals DR; Martens CR, The plasma metabolome as a predictor of biological aging in humans. Geroscience 2019, 41, (6), 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutchak PA; Laxman S; Estill SJ; Wang C; Wang Y; Wang Y; Bulut GB; Gao J; Huang LJ; Tu BP, Regulation of Hematopoiesis and Methionine Homeostasis by mTORC1 Inhibitor NPRL2. Cell Rep 2015, 12, (3), 371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Freedman R; Hunter SK; Law AJ; D’Alessandro A; Noonan K; Wyrwa A; Camille Hoffman M, Maternal choline and respiratory coronavirus effects on fetal brain development. J Psychiatr Res 2020, 128, 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Galletti P; De Bonis ML; Sorrentino A; Raimo M; D’Angelo S; Scala I; Andria G; D’Aniello A; Ingrosso D; Zappia V, Accumulation of altered aspartyl residues in erythrocyte proteins from patients with Down’s syndrome. FEBS J 2007, 274, (20), 5263–77. [DOI] [PubMed] [Google Scholar]
- 62.Culp-Hill R; Zheng C; Reisz JA; Smith K; Rachubinski A; Nemkov T; Butcher E; Granrath R; Hansen KC; Espinosa JM; D’Alessandro A, Red blood cell metabolism in Down syndrome: hints on metabolic derangements in aging. Blood Adv 2017, 1, (27), 2776–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Plummer JD; Johnson JE, Extension of Cellular Lifespan by Methionine Restriction Involves Alterations in Central Carbon Metabolism and Is Mitophagy-Dependent. Front Cell Dev Biol 2019, 7, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.La Carpia F; Wojczyk BS; Annavajhala MK; Rebbaa A; Culp-Hill R; D’Alessandro A; Freedberg DE; Uhlemann AC; Hod EA, Transfusional iron overload and intravenous iron infusions modify the mouse gut microbiota similarly to dietary iron. NPJ Biofilms Microbiomes 2019, 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hare D; Ayton S; Bush A; Lei P, A delicate balance: Iron metabolism and diseases of the brain. Front Aging Neurosci 2013, 5, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lehmann WD; Heinrich HC, Impaired phenylalanine-tyrosine conversion in patients with iron-deficiency anemia studied by a L-(2H5)phenylalanine-loading test. Am J Clin Nutr 1986, 44, (4), 468–74. [DOI] [PubMed] [Google Scholar]
- 67.Jones CB; Davis CM; Sfanos KS, The Potential Effects of Radiation on the Gut-Brain Axis. Radiat Res 2020, 193, (3), 209–222. [DOI] [PubMed] [Google Scholar]
- 68.Michonneau D; Latis E; Curis E; Dubouchet L; Ramamoorthy S; Ingram B; de Latour RP; Robin M; de Fontbrune FS; Chevret S; Rogge L; Socié G, Metabolomics analysis of human acute graft-versus-host disease reveals changes in host and microbiota-derived metabolites. Nat Commun 2019, 10, (1), 5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galloway-Peña JR; Peterson CB; Malik F; Sahasrabhojane PV; Shah DP; Brumlow CE; Carlin LG; Chemaly RF; Im JS; Rondon G; Felix E; Veillon L; Lorenzi PL; Alousi AM; Jenq RR; Kontoyiannis DP; Shpall EJ; Shelburne SA; Okhuysen PC, Fecal Microbiome, Metabolites, and Stem Cell Transplant Outcomes: A Single-Center Pilot Study. Open Forum Infect Dis 2019, 6, (5), ofz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 – Multivariate analyses and vectorial versions of figures 1–4, divided by organ.
Supplementary Figure 2 – Multivariate analyses and vectorial versions of figures 5–7, divided by organ.
Supplementary Table 1.xlsx - All raw data and elaborations
Data Availability Statement
Raw mass spectrometry data for this study are available for free download at the Metabolomics Workbench website, with the following IDs: ST002031-ST002041.


