Abstract
Background:
Plasminogen activator inhibitor-1 (PAI-1), a key inhibitor of plasminogen activators (PAs) tissue-type PA (tPA) and urokinase-type PA (uPA) plays a crucial role in many (patho)physiological processes (e.g., cardiovascular disease, tissue fibrosis) as well as in many age-related pathologies. Therefore, much effort has been put into the development of small molecule or antibody-based PAI-1 inhibitors.
Objective:
To elucidate the molecular mechanism of nanobody-induced PAI-1 inhibition.
Methods and Results:
Here we present the first crystal structures of PAI-1 in complex with two neutralizing nanobodies (Nbs). These structures, together with biochemical and biophysical characterization, reveal that Nb VHH-2g-42 (Nb42) interferes with the initial PAI-1/PA complex formation, whereas VHH-2w-64 (Nb64) redirects the PAI-1/PA interaction to PAI-1 deactivation and regeneration of active PA. Furthermore, whereas vitronectin does not have an impact on the inhibitory effect of Nb42, it strongly potentiates the inhibitory effect of Nb64, which may contribute to a strong inhibitory potential of Nb64 in vivo.
Conclusions:
These findings illuminate the molecular mechanisms of PAI-1 inhibition. Nb42 and Nb64 can be used as starting points to engineer further improved antibody-based PAI-1 inhibitors or guide the rational design of small molecule inhibitors to treat a wide range of PAI-1-related pathophysiological conditions.
Keywords: cardiovascular diseases, crystallography, X-ray, fibrinolysis, plasminogen activator inhibitor 1, single-domain antibodies
1 ∣. INTRODUCTION
Plasminogen activator inhibitor-1 (PAI-1), a 45-kDa glycoprotein, is the main physiological inhibitor of tissue-type (tPA) and urokinase-type (uPA) plasminogen activators (PAs) that represent important components of the fibrinolytic system.1 Numerous studies have demonstrated that elevated levels of PAI-1 are a risk factor for various thrombotic diseases.2-4 Experimental animal studies provided evidence that inhibition of PAI-1 activity results in a profibrinolytic effect.5-9 Furthermore, studies in animal models as well as epidemiologic and clinical studies in humans have shown that PAI-1, apart from its role in cardiovascular disease, is also involved in various other pathophysiological processes by acting through multiple pathways.1,10-12
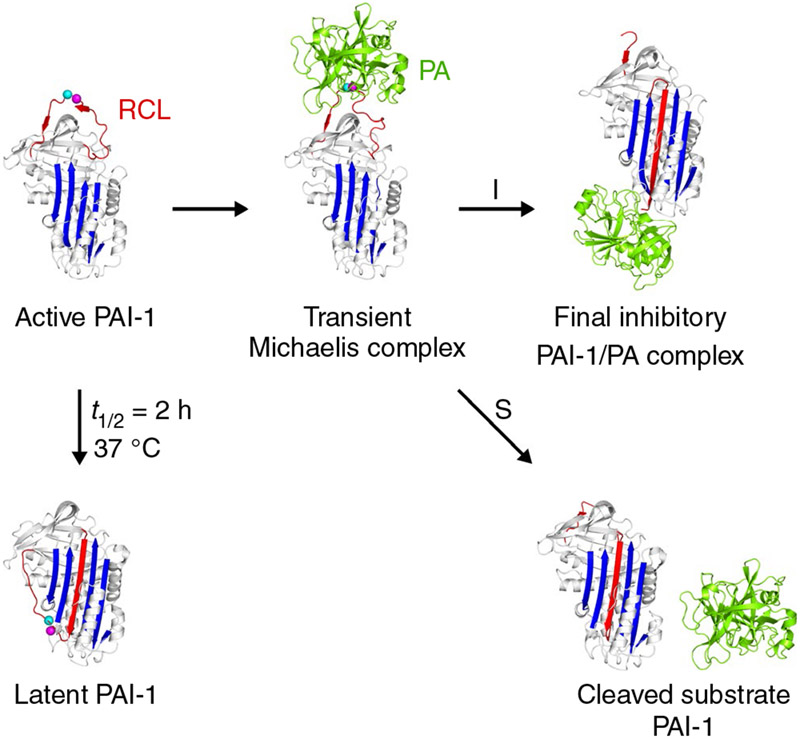
Structural studies have distinguished three conformations of PAI-1: active, cleaved, and latent (Figure 1).13,14 In the active conformation, PAI-1 inserts its flexible surface-exposed reactive center loop (RCL) that presents a substrate-mimicking peptide sequence (P1-P1′ corresponding to Arg346-Met347) into the active site of the PA to form a transient Michaelis complex. Following this initial docking step, an acyl-enzyme intermediate is formed by cleavage of the P1-P1′ bond. This triggers a major conformational change in which the cleaved RCL is rapidly inserted into the central β-sheet A of PAI-1 to form an extra antiparallel strand (s4A). Simultaneously, the bound PA is translocated 70 Å to the opposite side of the PAI-1 molecule, where the PA remains stably attached because of distortion of its catalytic triad.15,16 The substrate form of PAI-1 varies from the active conformation in that the acyl-enzyme intermediate is supposedly more prone to hydrolysis resulting in the release of the PA and yielding cleaved PAI-1.17 Finally, active PAI-1 spontaneously converts into a nonreactive stable latent form by insertion of the RCL without prior cleavage, thereby making P1-P1′ inaccessible for its target PA.18 In plasma and the extracellular matrix, vitronectin (Vn) binds to active PAI-1 with high affinity, thereby slowing down this transition and stabilizing the active form.19,20
FIGURE 1.
Schematic overview of the plasminogen activator inhibitor-1 (PAI-1) conformations as well as the inhibitory (I) and substrate (S) branch upon interaction with the plasminogen activators (PAs) tissue-type PA (tPA) and urokinase-type PA (uPA). PAI-1 is light gray; the central β-sheet A of the PAI-1 molecule is blue; the flexible reactive center loop (RCL) is red; Arg346 and Met347 (P1-P1′) of the reactive center are indicated by magenta and cyan spheres, respectively. The PA is green. PDB structures 1DVN, 1DB2, 5BRR, 3EOX, 1EZX, and 1H4W were used to generate this figure
The involvement of PAI-1 in various pathophysiological conditions makes it an attractive target for the development of specific inhibitors. PAI-1 inhibition can be achieved through three mechanisms: (a) by a direct interference with the formation of the PAI-1/PA Michaelis complex, (b) by accelerating the conversion of PAI-1 to its latent form, or (c) by inducing the PAI-1 substrate behavior. Over the past 2 decades, several PAI-1 inhibitors including small molecules, peptides, antibodies, and antibody derivatives have been discovered and characterized. However, to the best of our knowledge, to date none of those are suitable for therapeutic use in humans, presumably because of toxicity and selectivity issues.21 Better understanding of the molecular mechanisms of PAI-1 inhibition is therefore necessary to guide the rational design of improved PAI-1 modulators.
Our laboratory has generated a variety of monoclonal antibodies (mAbs), antibody fragments, and nanobodies (Nbs) that impair the functional activity of PAI-1.6,22,23 Nbs (~15 kDa) are recombinant antigen-binding variable domains derived from heavy-chain-only antibodies found in sera of Camelidae.24 Within a panel of Nbs, VHH-2g-42 (Nb42) and VHH-2w-64 (Nb64) exhibit distinct inhibitory properties toward PAI-1.23 According to biochemical characterization, Nb64 switched the PAI-1/PA interaction to the substrate pathway, whereas Nb42 appeared to interfere with the initial steps of the PAI-1/PA complex formation.23
Beyond the very few structures of PAI-1 complexes with small molecule or peptide inhibitors25-27 and only one recent structure of PAI-1 in complex with an antibody Fab fragment,28 no structures of PAI-1/Nb complexes have been described. Here, we report the first atomic structures of two stabilized active PAI-1 mutants, PAI-1-W175F and PAI-1-N150H-K154T-Q301P-Q319L-M354I (PAI-1-stab), each in a triple complex with Nb42 and Nb64 as well as PAI-1-stab in complex with Nb42. These structures, complemented with biochemical and biophysical characterization, reveal the mechanisms of PAI-1 inhibition. This deeper understanding of PAI-1 inhibition at the molecular level is essential to improve the inhibitory properties of these antibody-based profibrinolytics and to guide rational design of small molecule PAI-1 inhibitors.
2 ∣. METHODS
2.1 ∣. Cloning, expression, and purification of recombinant PAI-1 and Nbs
A Nb library was previously constructed, and a panel of Nbs with distinct inhibitory properties toward PAI-1 activity were characterized and reported.23 The selected Nbs, wild-type PAI-1 (PAI-1-wt), PAI-1-W175F, and PAI-1-stab were expressed in Escherichia coli as His-tagged small ubiquitin-like modifier (SUMO) fusion proteins29 using auto-induction ZYP-5052 media.30 Native proteins were prepared by treating the fusion proteins with SUMO hydrolase and purified by subtractive immobilized metal affinity chromatography, ion exchange chromatography, and gel filtration at 4°C.
2.2 ∣. Crystallization and data collection
To prepare the PAI-1-stab/Nb42 complex, PAI-1-stab was incubated with Nb42 (1:1.1 molar ratio, 22°C, 30 minutes) and concentrated to 5.6 mg mL−1 at 4°C. The complex was crystallized using the sitting-drop vapor diffusion method, mixing an equal volume of protein solution with precipitant (1 mol L−1 succinic acid, 0.1 mol L−1 HEPES, 1% w/v PEG2000, pH 7.0) at 20°C. For cryoprotection, the drop was overlaid with paraffin oil and the crystals were pulled through the oil before flash-cooling in liquid nitrogen. The triple PAI-1-stab/Nb42/Nb64 complex (1:1.1:1.1 molar ratio) was prepared in a similar way as the PAI-1-stab/Nb42 complex and concentrated to 6.8 mg mL−1. Optimized crystals were grown by mixing the protein solution with precipitant (0.1 mol L−1 Bis-Tris pH 6.5, 17% w/v PEG3350, 3% v/v methanol) at a 2:1 volume ratio in a sitting-drop vapor diffusion format at 20°C and cryoprotected by transferring the crystals through paraffin oil. The PAI-1-W175F/Nb42/Nb64 complex was prepared in a similar way as PAI-1-stab/Nb42/Nb64 and concentrated to 6.8 mg mL−1. Optimized crystals were grown by mixing the protein solution with precipitant (0.1 mol L−1 Bis-Tris pH 6.5, 10% w/v PEG3350) at a volume ratio of 2:1 using hanging-drop vapor diffusion at 4°C. Crystals were cryoprotected by briefly transferring the crystal to crystallization solution supplemented with 20% v/v ethylene glycol.
X-ray diffraction data were collected at 100 K using the PROXIMA2 beamline of SOLEIL Synchrotron and the ID-30A and ID-30B beamlines at the European Synchrotron Radiation Facility (ESRF). Data collection and refinement statistics are listed in Table 1.
TABLE 1.
Data collection and refinement statistics
| PAI-1- W175F/ Nb42/Nb64 |
PAI-1-stab/ Nb42/Nb64 |
PAI-1-stab/ Nb42 |
|
|---|---|---|---|
| PDB ID | 6GWN | 6GWP | 6GWQ |
| Data collection | |||
| Space group | P 21 | P 21 | C 2 |
| Cell dimensions | |||
| a, b, c (Å) | 45.48, 71.42, 96.60 | 43.68, 70.80, 98.51 | 89.87, 123.65, 64.98 |
| α,β,γ (°) | 90, 100.54, 90 | 90, 97.48, 90 | 90, 129.94, 90 |
| Resolution (Å) | 94.97-2.03 (2.05-2.03) | 48.84-2.28 (2.34-2.28) | 49.82-2.32 (2.35-2.32) |
| Rmerge | 0.056 (0.721) | 0.093 (1.06) | 0.076 (0.533) |
| I/σ(I) | 12.22 (2.00) | 10.29 (2.01) | 9.97 (2.00) |
| CC1/2 | 0.997 (0.784) | 0.994 (0.75) | 0.995 (0.746) |
| Completeness (%) | 98 (96) | 97(99) | 100 (100) |
| Redundancy | 3.7 (3.5) | 3.8 (4.0) | 3.9 (3.7) |
| Refinement | |||
| No. of complexes/ASU | 1 | 1 | 1 |
| Reflections used in refinement | 38 685 (1158) | 26 555 (1889) | 23 423 (751) |
| Reflections used for Rfree | 1957 (66) | 1362 (101) | 1153 (36) |
| Rwork | 0.209 | 0.203 | 0.206 |
| Rfree | 0.249 | 0.224 | 0.231 |
| No. of atoms | 4676 | 4750 | 3596 |
| Protein | 4567 | 4686 | 3577 |
| Water | 109 | 64 | 19 |
| B factors (Å2) | |||
| Protein | 55.89 | 61.43 | 55.09 |
| Water | 51.87 | 49.24 | 51.91 |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.009 | 0.009 |
| Bond angles (°) | 1.06 | 1.22 | 1.20 |
Note: Diffraction data were collected from a single crystal. The values in parentheses are for the highest resolution shell.
2.3 ∣. Structure determination, refinement, and analysis
The obtained diffraction data were processed using autoPROC in default settings, with a high-resolution cutoff on CC1/2 0.60.31 The data were initially phased by molecular replacement using the structure of PAI-1-W175F (Protein Data Bank [PDB]: 3Q0232) or PAI-1-stab (PDB: 1DB233) in active conformation as a search model using PHASER.34 Nb42 and Nb64 were initially modelled based on homologous Nb structures that were selected using a protein basic local alignment search tool (BLAST) (PDB: 5JA8 and 5JA9, respectively35). The structures of the PAI-1/Nb complexes were improved by iterative rounds of manual rebuilding in Coot and refinement in phenix.refine.36 Final models have been deposited to the PDB under the accession codes 6GWN (PAI-1-W175F/Nb42/Nb64), 6GWP (PAI-1-stab/Nb42/Nb64), and 6GWQ (PAI-1-stab/Nb42). PyMOL (The PyMOL Molecular Graphics System, version 2.0.7, Schrödinger, LLC) was used to visualize and superimpose models and to compute root-mean-squared deviations for all Cα atoms (Cα RMSD). The interfaces present in the complexes were analyzed using the PISA software.37
2.4 ∣. Size exclusion chromatography and inline small-angle X-ray scattering (SAXS)
SAXS experiments were performed at 288 K on the PAI-1/Nb complexes in a buffer containing 20 mmol L−1 Bis-Tris pH 6, 300 mmol L−1 NaCl and 5% v/v glycerol at the SWING beamline (SOLEIL Synchrotron). Protein samples (40-50 μL, ~10 mg mL−1) were loaded on an Agilent Bio SEC-3, 300Å, 4.6 × 300-mm high-performance liquid chromatography column and were eluted into the SAXS capillary cell at a flow rate of 0.3 mL min−1. SAXS data for both buffer (180 frames, 750 ms exposure time, and 250 ms dead time) and PAI-1/Nb complex (254 frames, same exposure and dead times) were collected. Buffer frames were processed, averaged, and subtracted from each protein frame (excluding outliers) using Scatter.38 For each sample, five scattering curves collected around the peak maximum were selected using CORMAP39 and averaged and scaled using Scatter.38 Solution-scattering profiles from the atomic structures of the complexes were calculated and fitted to the experimental averaged scattering curves using CRYSOL.40 Models for fitting the SAXS data for the triple complex were based on the crystal structure of PAI-1-W175F/Nb42/Nb64 (PDB: 6GWN). Models for fitting the SAXS data for binary complexes were based on the same crystal structure by taking chains A and B (PAI-1-W175F/Nb42) or A and C (PAI-1-W175F/Nb64).
2.5 ∣. Effect of Nbs on the stoichiometry of inhibition (SI) of uPA and tPA by PAI-1
The SI, defined as the number of moles of PAI-1 required for the inactivation of one mole of PA, for the reaction between free or monomeric Vn-bound PAI-1-wt and two-chain tPA (tctPa) or uPA (tcuPA) (all from Molecular Innovations Inc) in the presence of Nbs was estimated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as described previously.41,42 Briefly, PAI-1-wt (3.0-4.0 μmol L−1) (in the absence or presence of Vn at 1.2- to 1.5-fold molar excess) was preincubated with Nb (6.0-8.0 μmol L−1) for 10 minutes on ice and 2 minutes at room temperature, then mixed with an equal volume of 3.6-8.0 μmol L−1 tcuPA or tctPA (1.5- to 2.0-fold molar excess) and incubated 10 minutes at room temperature. The gels were stained with GelCode Blue Safe Protein Stain, and scanned and analyzed using a Molecular Imager equipped with Quantity One (version 4.2.3) software (Bio-Rad Laboratories). The amount of PAI-1 (unreactive plus latent, cleaved, and complexed with PA) was estimated from the intensity of the corresponding bands on gel scans.
2.6 ∣. Effect of Nbs on the rate of RCL insertion during the reaction of NBD P9 PAI-1 with tPA and uPA
Time-dependent RCL insertion during the reaction of PAI-1 with tctPA and tcuPA was measured by incubating various amounts of the PA (0-5.0 μmol L−1) with 18-24 nmol L−1 NBD (N-((2-(iodoacetoxy)ethyl)-N-methyl)amino-7-nitrobenz-2-oxa-3-diazole) P9 PAI-1 (Molecular Innovations Inc) (in the absence or presence of Vn at 1.2- to 1.5-fold molar excess) alone or in in the presence of 180-24 nmol L−1 Nb42, Nb64 or both Nbs in a manner similar to that used for studies of mAbs.17,43,44 A microvolume stopped-flow reaction analyzer (SX-20; Applied Photophysics Ltd), equipped with a fluorescence detector and a thermostated cell (25°C) was used to record the time change in NBD fluorescence emission through a 515-nm cutoff filter (excitation at 490 nm). The first-order observed rate constant (kobs) was calculated by fitting the recorded change in NBD fluorescence with a single exponential equation Ft = F∞+Ae−(kobs)t, where Ft is the fluorescence emission at time t, F∞ is the final fluorescence, and A is the amplitude. The fitting was done using Pro-Data Viewer software (Applied Photophysics Ltd). Dependences of kobs on the concentration of PA ([PA]) were plotted using SigmaPlot 12.0. The values of klim and Km were estimated by fitting a hyperbolic equation kobs = klim*[PA]/(Km+[PA]). (klim is kobs at infinite [PA] and Km is the [PA] at kobs = klim/2. The values of the second-order rate constant klim/Km were estimated by fitting a linear equation kobs = klim/Km *[PA]+A0 to the data. All experiments were carried out in 0.1 mol L−1 Hepes/NaOH pH 7.5 buffer.
2.7 ∣. PAI-1 neutralization assay
The functional properties of Nb42 and Nb64 were determined by assessing their ability to inhibit active PAI-1. PAI-1 activity was determined by a chromogenic plasminogen-coupled assay described by Verheijen et al.45 and performed as previously described23 with minor modifications. PAI-1-wt (168 ng mL−1 with an activity of ~60% of the theoretical maximum value) was incubated with buffer or a 10-fold molar excess of Vn, followed by incubation with serial two-fold dilutions of Nb, resulting in a molar ratio between 0.5- and 256-fold molar excess of Nb over PAI-1. To evaluate the ability of the Nbs to inhibit PAI-1 activity towards uPA, a similar assay was conducted with minor modifications. Following incubation of PAI-1-wt (120 ng mL−1 with an activity of ~60% of the theoretical maximum value) with buffer or Vn, and subsequent incubation with Nb, 50 μL of uPA (5 IU mL−1) was added. After incubation, a substrate solution containing plasminogen (2 μmol L−1) and S-2403 (0.6 mmol L−1) was added. A standard curve spanning 5 IU mL−1 to 1 IU mL−1 uPA was used to quantify the amount of residual uPA activity. To quantify the additive effects of Nb42 and Nb64 on PAI-1 neutralization, dose-response curves for Nb42 were evaluated in the presence of Nb64 at fixed concentrations (1- to 16-fold molar excess over PAI-1). The percentage of PAI-1 inhibition (ie neutralization of PAI-1 activity) was calculated from the residual PAI-1 activity measured in the presence of Nbs.
2.8 ∣. Statistical analysis
Inhibition curves were fitted using the non-linear regression [Inhibitor] vs. response – Variable slope (four parameters) model on GraphPad Prism 7.03 (GraphPad Software). Maximum inhibition and half maximal inhibitory concentration (IC50) values were calculated for each experimental repeat and reported as mean ± SD (n ≥ 3). Statistical analyses were performed with the two-tailed unpaired t-test. P < 0.05 was considered statistically significant.
Stopped-flow fluorescence traces were analyzed using a Pro-Data Viewer (Applied Photophysics Ltd). Assuming pseudo-first-order kinetics with [PA] at least 5-fold higher than that of NBD P9 PAI-1 or its complex with Nbs, a single exponential equation was fit to the stopped flow traces to calculate kobs. The quality of the fit was estimated by visual analysis of the plots of the residuals (deviation of the fitted function from the actual data). The values of kobs (n ≥ 3; standard error < 10%) were plotted against [PA]. Plots were fitted by hyperbolic or linear equation to calculate parameters (klim, Km, and klim/Km), using nonlinear least squares fitting with the Levenberg-Marquardt algorithm (SigmaPlot 12.0 for Windows; SPSS Inc). Correlation coefficients (r) calculated from curve fittings were used as a parameter of the goodness of fit; r2 of the fit was greater than 0.90 for all the kinetic data.
3 ∣. RESULTS
3.1 ∣. Overview of the PAI-1/Nb complex structures
The crystal structures of the triple complexes PAI-1-W175F/Nb42/Nb64 and PAI-1-stab/Nb42/Nb64 as well as of the binary complex PAI-1-stab/Nb42 were determined at 2.0, 2.3, and 2.3 Å resolution, respectively (Table 1). Superimposition of the structures of the two triple complexes yielded a Cα RMSD of 1.093 A, excluding the flexible RCL, showing that they are essentially the same. In addition, the crystal structure of the PAI-1-stab/Nb42 binary complex is very similar to the corresponding substructure of the triple PAI-1-W175F complex (Cα RMSD of 1.709 Å). Therefore, only the PAI-1-W175F/Nb42/Nb64 structure will be used to discuss the interactions between PAI-1 and both Nbs.
The structure of PAI-1 in the complex (Figure 2A) represents the well-studied active conformation seen for various serpins.46 As expected, a major part of the RCL, residues 335 to 348, is disordered in the electron density maps and is therefore not included in the final model of the complex. Comparison of the PAI-1-W175F structure in the complex and the isolated PAI-1-W175F structure (PDB: 3Q02,32 chain A) shows that no major conformational changes are induced upon interaction of PAI-1 with the inhibitory Nbs (Cα RMSD of 0.678 Å).
FIGURE 2.
Cartoon representation of the crystal structure of PAI-1 complexed with Nb42 and Nb64. A, Overall structure, consisting of PAI-1 (orange), Nb42 (cyan) and Nb64 (green). The flexible reactive center loop (RCL) is colored red. Residues 335 to 348 of the RCL were not observed in the crystal structure and are presented by a dashed line. Framework regions of Nb42 and Nb64 are cyan and green, respectively; the complementarity-determining regions (CDR) 1, CDR2, and CDR3 are yellow, brown, and blue, respectively. The residues closer than 4 Å to Nbs are magenta. B, Close-up view of the PAI-1/Nb42 interface. C, Close-up view of the PAI-1/Nb64 interface
SAXS data, collected to verify whether the obtained crystal structures of PAI-1/Nb complexes represent the true structure in solution, convincingly support the crystallographic conformations in each case (Figure S1).
3.2 ∣. Interactions between PAI-1 and Nb42
The structures reveal that Nb42 binds to a conformational epitope on PAI-1 which comprises the loop connecting strands 1 and 2 of β-sheet B (s1B and s2B) as well as the proximal portions of each strand (Figure 2A). The 608 Å2 interface is stabilized mainly by polar interactions made by residues on all Nb CDRs (Figure 2B). CDR1 Thr31 and Gln35, CDR2 Asn53, and CDR3 Arg100 and Leu101 interact with PAI-1 through the formation of 5 hydrogen bonds and a salt bridge (Figure S2B, C and Table S1).
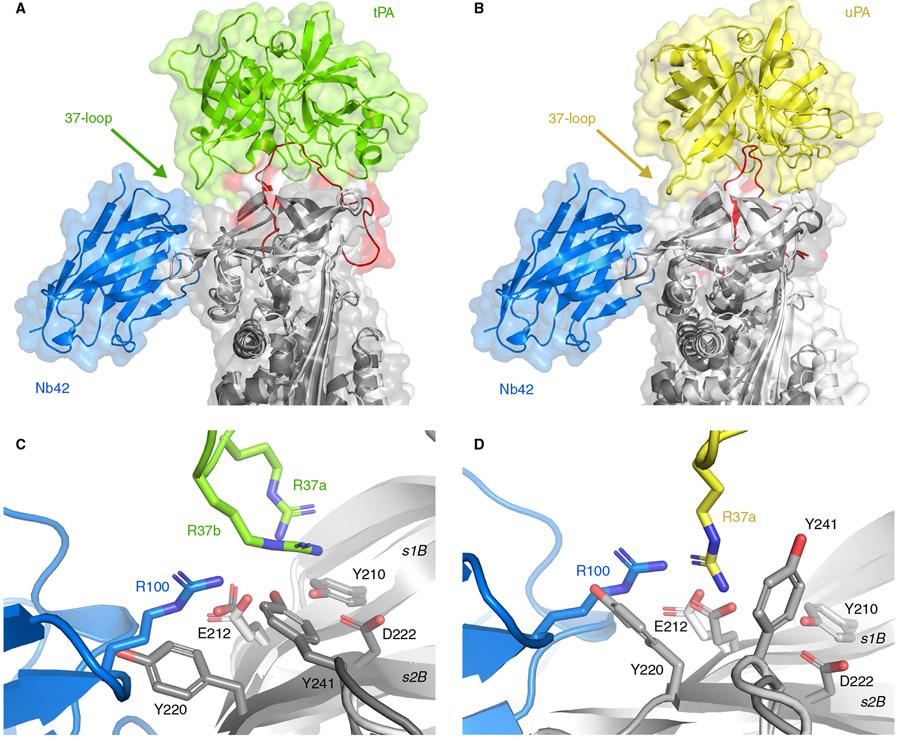
Next, we have superimposed the PAI-1-W175F/Nb42/Nb64 structure with the previously established structures of Michaelis complexes of PAI-1 with S195A mutants of tPA (Figure 3A) and uPA (Figure 3B), respectively (PDB: 5BRR47 and 3PB148). Such superimposition reveals that the binding of Nb42 occurs at a distance from the RCL and thus should not interfere with the interaction of the RCL and the active site of the PA, which was confirmed by fluorescence measurements using NBD P1′ PAI-1 (data not shown). Simultaneously, there is a small overlap in the binding region in PAI-1 for Nb42 and an additional exosite binding region in PAI-1 for both PAs (Figure 3C, D). Indeed, a negatively charged patch formed by the side chains of Tyr210, Glu212, Tyr241, and Asp222 on β-sheet B of PAI-1 makes electrostatic interactions with the so-called 37-loop of tPA and uPA.47,48 This loop includes two consecutive positive charges of Arg37a and Arg37b. In the uPA complex, there is likewise a salt bridge between PAI-1 Glu212 and the sole charged residue Arg37a. Additionally, the aromatic side chain of Tyr220 interacts with uPA Arg37a through a cation-π interaction. In this respect, it is notable that the PAI-1/Nb42 interface includes a salt bridge formed between PAI-1 Glu212 and CDR3 Arg100. We thus conclude that the binding of Nb42 interferes with the interaction of PAs with this exosite on PAI-1 and thereby could affect the initial Michaelis complex formation.
FIGURE 3.
Superimposition of the current crystal structure of PAI-1 (light gray) complexed with Nb42 (blue) and the previously determined structures of the Michaelis complexes between PAI-1 (dark gray) and tPA (green) (A, PDB: 5BRR), and PAI-1 (dark gray) and uPA (yellow) (B, PDB: 3PB1). The RCL of PAI-1 is red. C, Detail of the interaction interfaces of the PAI-1/Nb42 complex and the PAI-1/tPA complex. D, Detail of the interaction interfaces of the PAI-1/Nb42 complex and the PAI-1/uPA complex
3.3 ∣. Interactions between PAI-1 and Nb64
Nb64 binds to an epitope located at the opposite side of the PAI-1 molecule with respect to the RCL (Figure 2A). The larger part of the epitope is formed by the exposed portion of the hI-s5A loop (residues 300 to 314) (Figure 2C). Nb64 has its CDR3 folded over the FR2 region and binds PAI-1 in a sideways orientation, resulting in a slightly larger interaction interface of 802 Å2 compared with Nb42. This interface is stabilized by 10 hydrogen bonds and 3 salt bridges, mainly involving CDR2 (Ser54, Asp55, and Tyr60) and CDR3 (Arg102, Arg105, Asn106, and Tyr108) (Figure S2D, E and Table S1). Notably, interactions between PAI-1 and the FR2 region (Glu44 and Arg45) as well as a stacking interaction between PAI-1 Phe302 and the disulfide bond formed between Cys50 (CDR2) and Cys104 (CDR3) contribute to the sideways binding of Nb64 to PAI-1 (Figure S2E).
Interestingly, this interface in PAI-1, comprising hC and the hI-s5A loop, was previously identified as a major determinant of the epitope of the substrate inducing mAb MA-8H9D4.49 Previous studies showed that MA-8H9D4 neither affects binding of PAs to PAI-1, nor induces significant deacylation of the PAs from preformed PAI-1/PA complexes.17 This indicates that, as reported for MA-8H9D4, Nb64 acts after initial complex formation and prior to the formation of the final inhibitory complex, possibly by preventing disordering of the active site of the PA.17
3.4 ∣. The reaction of PAI-1 with tPA and uPA is redirected toward the substrate branch by Nb64, but not by Nb42, irrespective of the presence of Vn
To evaluate the effects of both Nbs on PAI-1 functionality, we analyzed the outcome of the reaction between PAI-1-wt and tctPA and tcuPA by SDS-PAGE. Nb42 and Nb64 affected the PAI-1/PA interaction differently, as evident from the analysis of the reaction products (PAI-1/PA-complex, unreactive or latent PAI-1, and cleaved substrate PAI-1) (Figure 4). The amounts of latent PAI-1 did not change in the presence of one or both Nbs. Thus, neither Nb significantly accelerates the transition from active to latent PAI-1 (Figure 4). Furthermore, Nb42 did not significantly change the distribution between complexed and cleaved PAI-1 upon the interaction with both PAs (Figure 4 and Table 2), indicating that Nb42 affects the initial complex formation. In contrast, Nb64 almost completely redirected the PAI-1 reaction toward the substrate branch for both PAs. As a result, a dramatic decrease in the inhibitory complex coincided with a simultaneous increase in cleaved PAI-1 and free PA (Figure 4). The extent (SI value) of this redirection was more pronounced for uPA (SI = 27 ± 5) than tPA (SI = 15.6 ± 3.0). Nb42 did not significantly alter the effects of Nb64 on the PAI-1/tPA interaction. However, the simultaneous presence of both Nbs (Figure 4) affects the SI for uPA, decreasing it to the values similar to that for tPA (16.9 and 12.4, respectively, Table 2). Vn did not have an impact on the effect of Nb42 on the outcome of the PAI-1/PA reactions. In contrast, for Nb64, the presence of Vn increased the SI for the PAI-1/tPA reaction (15.6 ± 3.0 to > 30) and decreased the SI for the PAI-1/uPA reaction (27 ± 5 to 10.0 ± 2.0). This decrease, however, only reflects a minor effect on the overall reaction outcome (96% to 90% substrate reaction).
FIGURE 4.
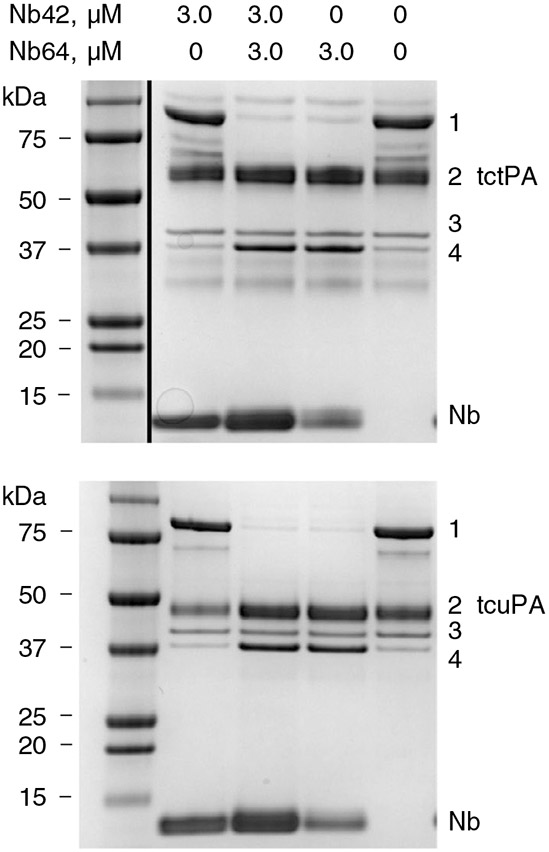
Outcome of the reactions of two-chain tPA (tctPA) (top) and tcuPA (bottom) with PAI-1-wt in the presence of Nb42, Nb64 or both Nbs. SDS-PAGE analysis of products of the reaction between 1.8 μmol L−1 human recombinant tctPA (top) or 1.8 μmol L−1 of human recombinant tcuPA with human recombinant PAI-1-wt (1.5 μmol L−1) alone and in the presence of 3.0 μmol L−1 of Nb42, Nb64, or both Nbs. Positions of PAI-1-wt/PA inhibitory complex (1), PA (2), unreactive/latent PAI-1-wt (3), and cleaved (4) PAI-1-wt, are indicated to the right of the gels. The ladder in the top image was rearranged to the left-hand side
TABLE 2.
The kinetic parametersa and stoichiometry of inhibition (SI)b for the reaction of two-chain tPA (tctPA) and tcuPA with complexes of free or vitronectin-bound NBD P9 PAI-1 or PAI-1-wt with Nb42, Nb64 and both Nbs
| None |
Nb42 |
Nb64 |
Nb42/Nb64 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
klim s−1 |
Km μmol L−1 |
klim/Km (μmol/ L)−1 s−1 |
SI |
klim s−1 |
Km μmol L−1 |
klim/Km (μmol/ L)−1 s−1 |
SI |
klim s−1 |
Km μmol L−1 |
klim/Km (μmol/ L)−1 s−1 |
SI |
klim s−1 |
Km μmol L−1 |
klim/Km (μmol/ L)−1 s−1 |
SI | ||
| −Vn | |||||||||||||||||
| tctPA | 1.5 ± 0.1 | 0.22 ± 0.04 | 6.8 | 1.1 ± 0.2 | 5.0 ± 0.4 | 1.2 ± 0.3 | 4.2 | 1.1 ± 0.2 | 1.4 ± 0.1 | 0.17 ± 0.03 | 8.2 | 15.6 ± 3.0 | 5.2 ± 0.6 | 1.4 ± 0.3 | 3.7 | 16.9 ± 0.3 | |
| tcuPA | 8.7 ± 0.4 | 0.77 ± 0.09 | 11.3 | 1.1 ± 0.2 | ND | ND | 0.70 ± 0.04 | 1.2 ± 0.2 | 4.4 ± 0.4 | 0.30 ± 0.07 | 14.7 | 27 ± 5 | ND | ND | 0.89 ± 0.04 | 12.4 ± 2.0 | |
| +Vnc | |||||||||||||||||
| tctPA | 2.2 ± 0.1 | 0.21 ± 0.02 | 10.5 | 1.0 ± 0.1 | ND | ND | 1.70 ± 0.06 | 1.0 ± 0.1 | 1.3 ± 0.1 | 0.18 ± 0.02 | 7.2 | >30 | ND | ND | 1.10 ± 0.03 | >30 | |
| tcuPA | 7.6 ± 0.5 | 0.44 ± 0.08 | 17.3 | 1.0 ± 0.1 | ND | ND | 0.85 ± 0.03 | 1.1 ± 0.1 | 2.5 ± 0.2 | 0.26 ± 0.05 | 9.6 | 10.0 ± 2.0 | ND | ND | 0.87 ± 0.02 | 14.0 ± 3.0 | |
klim (first-order rate constant of RCL insertion at infinite concentration of the PA), Km (concentration of the PA at kobs = klim/2), and klim/Km (specificity of the reaction).
The number of moles of PAI-1 or PAI-1/Nb complex required to inhibit 1 mole of the PA.
NBD P9 PAI-1 or PAI-1-wt was incubated for 10 min with human recombinant vitronectin (Vn) (1.2- to 1.5-fold molar excess) before adding Nbs.
ND = not determined (no plateau for kobs was reached within the tested PA concentrations up to 5.0 μimol L−1).
3.5 ∣. The rate of RCL insertion during the interaction of PAI-1 with both PAs is dramatically affected by Nb42, but not by Nb64, irrespective of the presence of Vn
NBD P9 PAI-1,50 an active PAI-1 mutant in which the fluorescent NBD probe is introduced at position P9 (N-terminal to the P1-P1′ bond) in the RCL, was used to determine the effects of the Nbs on the rate of RCL insertion. The values of the first-order limiting rate constant of RCL insertion (klim), the [PA] at half-saturation (Km) and klim/Km for the reaction of free NBD P9 PAI-1 and its Nb complexes with PAs are shown in Table 2. Binding of Nb42 to NBD P9 PAI-1 resulted in an increase in the klim (1.5 to 5.0 s−1) and Km (0.22 to 1.2 μmol L−1), decreasing the specificity of the PAI-1/tPA reaction (klim/Km) from 6.8 to 4.2 (μmol/L)−1 s−1 (Table 2). A more pronounced effect was observed for the reaction of NBD P9 PAI-1 with uPA, resulting in a 16-fold decrease in the klim/Km (from 11.3 to 0.7 (μmol/L)−1 s−1; P < 0.05) (Table 2). In the presence of Vn, the kinetic parameters were affected similarly (Table 2). While binding of Nb64 did not significantly change klim or Km for the reaction of NBD P9 PAI-1 with tPA, it resulted in a more than two-fold decrease in both the klim (from 8.7 to 4.4 s−1; P < 0.05) and the Km (from 0.77 to 0.3 μmol L−1; P < 0.05) with only a slight increase in the klim/Km (from 11.3 to 14.7 μmol L−1 s−1; P < 0.05) for the reaction with uPA. The presence of Vn did not affect the changes in the klim and Km induced by Nb64 for the PAI-1/tPA reaction; however, it resulted in a slight decrease in the Km for the reaction with uPA (from 4.4 to 2.5 s−1). Furthermore, when Vn was present, the specificity for both PAI-1/PA reactions decreased (Table 2). The simultaneous presence of Nb64 and Nb42 did not significantly change klim or Km for the reaction of NBD P9 PAI-1 with tPA compared with Nb42 alone (5.2 vs 5.0 s−1 and 1.4 vs 1.2 μmol L−1, respectively; P > 0.05). This observation indicates that the effects on the kinetics of RCL insertion for tPA and uPA in the presence of both Nbs are primarily due to Nb42. Interestingly, binding of Nb42 to the exosite for both PAs on PAI-1 affected the PAI-1/tPA and PAI-1/uPA reactions differently, which illuminates the differences in exosite interactions between PAI-1 and both PAs.
3.6 ∣. Both Nb42 and Nb64 prevent PAI-1 from inactivating PAs independently of Vn, whereas Vn strongly potentiates the inhibitory effect of Nb64
Of note, the crystal structures of the PAI-1/Nb complexes provide a rational explanation of the previously obtained affinity data for both Nbs, including the effect of glycosylation and mouse and rat PAI-1 cross-reactivity (Supporting Information). Next, to understand the ability of both Nbs to interfere with the PAI-1/PA interaction in semi-physiological conditions, we carried out further functional analysis. These showed that both Nbs inhibit PAI-1-wt activity in a dose-dependent manner (Figure S4A) when followed by incubation with tPA, reaching a maximum inhibition of 30% ± 6% and 55% ± 5% for Nb42 and Nb64, respectively (Figure 5A), with corresponding IC50 values of 87 ± 46 and 16 ± 5 nmol L−1 (Figure 5B). For Nb42, the values observed for the PAI-1/uPA interaction are similar to those for the PAI-1/tPA interaction. Interestingly, when PAI-1 is incubated with uPA, the maximum inhibition by Nb64 is increased to 88% ± 4% (P < 0.0001) with a corresponding lower IC50 value of 4.6 ± 0.6 nmol L−1 (P < 0.01) (Figure 5A, B). Furthermore, whereas binding of Vn to PAI-1 did not change the inhibitory effects of Nb42, it strongly potentiated the inhibitory effects of Nb64, resulting in an increased maximum PAI-1 inhibition of 93% ±6% (P < 0.0001) with a lower IC50 value (2.8 ± 0.8 nmol L−1, P < 0.0001) when incubated with tPA, and a lower IC50 value (2.0 ± 0.5 nmol L−1, P < 0.0001) when incubated with uPA (Figure 5A, B). Nb42 and Nb64 were also evaluated as a mixture for their capacity to inhibit PAI-1-wt activity. Incubation of PAI-1 with Nb42 in the presence of a fixed concentration of Nb64 clearly increased the maximal observed effect of Nb42, reaching a maximum inhibition of 67% ± 10% with an IC50 value of 4 ± 2 nmol L−1 (Figure S4B, C). This observed effect did not significantly differ from the theoretically calculated values for an additive effect (P = 0.5185, Figure S4D).
FIGURE 5.

Inhibition of PAI-1 activity by Nb42 and Nb64. A, Maximum inhibition of the activity of free or vitronectin-bound (+Vn) PAI-1-wt, by Nb42 and Nb64 in the presence of tPA (black bars) or uPA (white bars). B, IC50 values for the inhibition of free or Vn-bound (+Vn) PAI-1-wt by Nb42 and Nb64 in the presence of tPA (black bars) or uPA (white bars). All data are represented as mean ± SD; n ≥ 3. ** P < 0.01, **** P < 0.0001
4 ∣. DISCUSSION
In the past, numerous attempts have been made to discover and develop PAI-1 inhibitors and to optimize their properties. Beyond the several crystal structures of PAI-1 in active, latent or cleaved conformations,32,51,52 only a handful of structures containing PAI-1 in complex with inhibitory peptides or small molecules have been reported.25-27 Compared with these ligand-bound structures, the epitopes of PAI-1 neutralizing mAbs and Nbs have been mapped to different regions of the PAI-1 molecule.53 Although several molecular mechanisms of PAI-1 neutralization by mAbs were discovered,1,22,42,54 only one crystal structure of PAI-1 complexed with an inhibitory antibody fragment was reported.28 The structure containing the Fab fragment of the neutralizing antibody MEDI-57928 reveals that the fragment binds the RCL from P2 to P3′, including the reactive center P1-P1′ residues, and the adjacent exosite for binding the so-called 37-loop of PAs.
In this study, we have determined the crystal structures of complexes of two stabilized active PAI-1 mutants, PAI-1-W175F and PAI-1-stab, with two inhibitory Nbs, Nb42 and Nb64. Nb42 exclusively binds to strands s1B and s2B in PAI-1, previously identified as a region involved in making exosite interactions with the 37-loop of tPA and uPA.47,48 The importance of this 37-loop/exosite interaction with respect to PA specificity and the rate of Michaelis complex formation was demonstrated by studies using 37-loop mutants9,55 or antibodies specifically binding to this region of the PA.56 Notable differences between exosite interactions of tPA and uPA exist, resulting in a stronger and more extensive PAI-1/tPA exosite interaction.47,48,57 The current structures suggest that Nb42 sterically interferes with PA binding or affects the PAI-1/PA Michaelis complex by interacting with key residues in the exosite for the PA 37-loop. SDS-PAGE analysis of the effect of Nb42 on the PAI-1/PA reaction did not reveal any role for Nb42 in redirecting the reaction toward the substrate branch or in accelerating latency transition, indicating that Nb42 acts before the formation of the Michaelis complex. Furthermore, the observed effect of Nb42 on the kinetics of the interaction between NBD P9 PAI-1 and PAs illuminates the differences in exosite interactions between PAI-1 and both PAs. As mentioned, exosite interactions stabilize the PAI-1/tPA Michaelis complex and slow down the dissociation of the acylated proteinase from the initial binding site on the PAI-1 molecule,58 which becomes a limiting step for the rate of RCL insertion. Binding of Nb42 results in a simultaneous increase in the klim and Km for the reaction of RCL insertion and causes a similar, but much more pronounced, effect on the rate of RCL insertion with uPA, dramatically decreasing klim/Km. This could reflect faster dissociation of the acylated proteinase or even a change in the rate-limiting step of the reaction (ie RCL insertion instead of dissociation). Thus, Nb42 possesses a novel mechanism of PAI-1 neutralization, that is, destabilizing both the Michaelis complex and acyl-enzyme bound to the initial binding site on PAI-1, via interference with exosite interactions that do not block the inhibitory reaction between PAI-1 and PAs or change the SI. This mechanism is considerably different from previously described antibodies MEDI-579,28 MA-56A7C10,54 and MA-42A2F6,54 which also bind directly to residues in the RCL, thereby blocking the accessibility of the P1-P1′ bond for the PA active site.
It was previously reported that Nb64 redirects the inhibitory pathway of PAI-1 to the substrate pathway upon the interaction with its target PA.23 The epitope determined by alanine scanning identified residues from hB, hC, and the hI-s5A loop as major determinants of the epitope.23 By structural analysis of the PAI-1/Nb64 complex we could confirm the crucial involvement of the latter loop. Importantly, Nb64 redirects the PAI-1 mechanism toward substrate reaction, exceeding 90%, which correlates with previously reported biochemical data.23 Our structural data suggest that the key residues of the Nb64 epitope should be localized very close to the position of the PA after translocation to the opposite side of PAI-1. This strongly indicates that Nb64 prevents the final locking step required for the irreversible PAI-1/PA complex formation, possibly by preventing distortion of the active site of the PA, as previously reported for MA-8H9D4.17
Notably, when both Nbs are bound to PAI-1, additivity in the effects was observed. Nb42 does not significantly contribute to the effects of Nb64 in redirecting the PAI-1 mechanism toward the substrate pathway. At the same time, Nb64 does not affect the changes in the rate of RCL insertion induced by Nb42. These findings confirm that both Nbs act independently while interfering with different stages of the PAI-1/PA interaction.
As expected, based on the structures, the effect of Nb42 was not affected by the presence of vitronectin. Strikingly, Vn strongly potentiated the inhibitory effects of Nb64. Previous studies on the effects of Vn-binding to PAI-1 revealed a considerable stabilization of regions in PAI-1 remote from the Vn binding site, including the hI-s5A loop.59 Conformational stabilization of this region, including the epitope of Nb64, increases the potency of Nb64, as reflected by the substantial improvement in maximum inhibitory activity reaching approximately 90% in the presence of Vn, and the low nanomolar IC50 values. Of note, previous in vivo evaluations of PAI-1 inhibitory mAbs, displaying 58% to 80% inhibition, indicate that complete inhibition of PAI-1 activity is not a prerequisite to achieve a desirable profibrinolytic effect.6
Taken together, these crystal structures of PAI-1 in complex with two neutralizing Nbs, together with biochemical and biophysical characterization provide detailed and novel insights into the molecular mechanisms of PAI-1 inhibition and highlight the versatility of Nbs in modulating various stages of the PAI-1/PA interaction. Remarkably, while the inhibitory capacity of Nb42 is retained in the presence of Vn, Vn strongly potentiates the inhibitory effects of Nb64, and may contribute to a strong inhibitory potential of Nb64 in vivo.
Supplementary Material
Essentials.
High plasminogen activator inhibitor-1 (PAI-1) levels are a risk factor for cardiovascular diseases.
PAI-1 inhibitory nanobodies were characterized by biochemical and biophysical methods.
Three crystal structures of PAI-1 in complex with nanobodies that modulate PAI-1 were solved.
These nanobodies modulate distinct stages of the PAI-1/plasminogen activator interaction.
ACKNOWLEDGMENTS
X-ray data collection was made possible through the PROXIMA2 and SWING beamlines, Synchrotron SOLEIL, France, as well as the ID-30A and ID-30B beamlines, European Synchrotron Radiation Facility, France. The authors are grateful to the staff of these facilities for generous assistance and to Piotr Kabelis and Sara Verhaegen, who assisted with protein expression, purification, and crystallization. This study was partially supported by research funding from the Research Foundation - Flanders (FWO) (grant G072915N to Paul J. Declerck and Sergei V. Strelkov) and the National Institutes of Health (1-R01-HL130402-01A1 to Andrey A. Komissarov, Galina Florova, and Steven Idell). Machteld Sillen is a recipient of a PhD fellowship of the Research Foundation – Flanders (FWO).
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICT OF INTEREST
The authors declare no competing interests.
REFERENCES
- 1.Declerck PJ, Gils A. Three decades of research on plasminogen activator inhibitor-1: a multifaceted serpin. Seminars in Thrombosis and Hemostasis. New York: Thieme Medical Publishers, 2013; 356–364. [DOI] [PubMed] [Google Scholar]
- 2.Meltzer ME, Lisman T, de Groot PG, et al. Venous thrombosis risk associated with plasma hypofibrinolysis is explained by elevated plasma levels of TAFI and PAI-1. Blood. 2010;116:113–121. [DOI] [PubMed] [Google Scholar]
- 3.Thögersen AM, Jansson JH, Boman K, et al. High plasminogen activator inhibitor and tissue plasminogen activator levels in plasma precede a first acute myocardial infarction in both men and women: evidence for the fibrinolytic system as an independent primary risk factor. Circulation. 1998;98:2241–2247. [DOI] [PubMed] [Google Scholar]
- 4.Margaglione M, Di Minno G, Grandone E, et al. Abnormally high circulation levels of tissue plasminogen activator and plasminogen activator inhibitor-1 in patients with a history of ischemic stroke. Arterioscler Thromb. 1994;14:1741–1745. [DOI] [PubMed] [Google Scholar]
- 5.Izuhara Y, Yamaoka N, Kodama H, et al. A novel inhibitor of plasminogen activator inhibitor-1 provides antithrombotic benefits devoid of bleeding effect in nonhuman primates. J Cereb Blood Flow Metab. 2010;30:904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van De Craen B, Scroyen I, Abdelnabi R, et al. Characterization of a panel of monoclonal antibodies toward mouse PAI-1 that exert a significant profibrinolytic effect in vivo. Thromb Res. 2011;128:68–76. [DOI] [PubMed] [Google Scholar]
- 7.Wyseure T, Rubio M, Denorme F, et al. Innovative thrombolytic strategy using a heterodimer diabody against TAFI and PAI-1 in mouse models of thrombosis and stroke. Blood. 2015;125:1325–1332. [DOI] [PubMed] [Google Scholar]
- 8.Florova G, Azghani A, Karandashova S, et al. Targeting of plasminogen activator inhibitor 1 improves fibrinolytic therapy for tetracycline-induced pleural injury in rabbits. Am J Respir Cell Mol Biol. 2015;52:429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Florova G, Azghani AO, Karandashova S, et al. Targeting plasminogen activator inhibitor-1 in tetracycline-induced pleural injury in rabbits. Am J Physiol Lung Cell Mol Physiol. 2018;314:L54–L68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Placencio VR, DeClerck YA. Plasminogen activator inhibitor-1 in cancer: rationale and insight for future therapeutic testing. Cancer Res. 2015;75:2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rabieian R, Boshtam M, Zareei M, Kouhpayeh S, Masoudifar A, Mirzaei H. Plasminogen activator inhibitor type-1 as a regulator of fibrosis. J Cell Biochem. 2018;119:17–27. [DOI] [PubMed] [Google Scholar]
- 12.Vaughan DE, Rai R, Khan SS, Eren M, Ghosh AK. Plasminogen activator inhibitor-1 Is a marker and a mediator of senescence. Arterioscler Thromb Vasc Biol. 2017;37:1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sancho E, Declerck PJ, Price NC, Kelly SM, Booth NA. Conformational studies on plasminogen activator inhibitor (PAI-1) in active, latent, substrate, and cleaved forms. Biochemistry. 1995;34:1064–1069. [DOI] [PubMed] [Google Scholar]
- 14.Declerck PJ, De Mol M, Vaughan DE, Collen D. Identification of a conformationally distinct form of plasminogen activator inhibitor-1, acting as a noninhibitory substrate for tissue-type plasminogen activator. J Biol Chem. 1992;267:11693–11696. [PubMed] [Google Scholar]
- 15.Huntington JA, Read RJ, Carrell RW. Structure of a serpin-protease complex shows inhibition by deformation. Nature. 2000;407:923–926. [DOI] [PubMed] [Google Scholar]
- 16.Perron MJ, Blouse GE, Shore JD. Distortion of the catalytic domain of tissue-type plasminogen activator by plasminogen activator inhibitor-1 coincides with the formation of stable serpin-proteinase complexes. J Biol Chem. 2003;278:48197–48203. [DOI] [PubMed] [Google Scholar]
- 17.Komissarov AA, Declerck PJ, Shore JD. Mechanisms of conversion of plasminogen activator inhibitor 1 from a suicide inhibitor to a substrate by monoclonal antibodies. J Biol Chem. 2002;277:43858–43865. [DOI] [PubMed] [Google Scholar]
- 18.Mottonen J, Strand A, Symersky J, et al. Structural basis of latency in plasminogen activator inhibitor-1. Nature. 1992;355:270–273. [DOI] [PubMed] [Google Scholar]
- 19.Declerck PJ, De Mol M, Alessi MC, et al. Purification and characterization of a plasminogen activator inhibitor 1 binding protein from human plasma. Identification as a multimeric form of S protein (vitronectin). J Biol Chem. 1988;263:15454–15461. [PubMed] [Google Scholar]
- 20.Lawrence DA, Palaniappan S, Stefansson S, et al. Characterization of the binding of different conformational forms of plasminogen activator inhibitor-1 to vitronectin. Implications for the regulation of pericellular proteolysis. J Biol Chem. 1997;272:7676–7680. [DOI] [PubMed] [Google Scholar]
- 21.Fortenberry YM. Plasminogen activator inhibitor-1 inhibitors: a patent review (2006-present). Expert Opin Ther Pat. 2013;23:801–815. [DOI] [PubMed] [Google Scholar]
- 22.Debrock S, Declerck PJ. Neutralization of plasminogen activator inhibitor-1 inhibitory properties: identification of two different mechanisms. Biochim Biophys Acta. 1997;1337:257–266. [DOI] [PubMed] [Google Scholar]
- 23.Zhou X, Hendrickx ML, Hassanzadeh-Ghassabeh G, Muyldermans S, Declerck PJ. Generation and in vitro characterisation of inhibitory nanobodies towards plasminogen activator inhibitor 1. Thromb Haemost. 2016;116:1032–1040. [DOI] [PubMed] [Google Scholar]
- 24.Muyldermans S Nanobodies: natural single-domain antibodies. Annu Rev Biochem. 2013;82:775–797. [DOI] [PubMed] [Google Scholar]
- 25.Xue Y, Björquist P, Inghardt T, et al. Interfering with the inhibitory mechanism of serpins: crystal structure of a complex formed between cleaved plasminogen activator inhibitor type 1 and a reactivecentre loop peptide. Structure. 1998;6:627–636. [DOI] [PubMed] [Google Scholar]
- 26.Li SH, Reinke AA, Sanders KL, et al. Mechanistic characterization and crystal structure of a small molecule inactivator bound to plasminogen activator inhibitor-1. Proc Natl Acad Sci USA. 2013;110:E4941–E4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fjellström O, Deinum J, Sjögren T, et al. Characterization of a small molecule inhibitor of plasminogen activator inhibitor type 1 that accelerates the transition into the latent conformation. J Biol Chem. 2013;288:873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vousden KA, Lundqvist T, Popovic B, et al. Discovery and characterisation of an antibody that selectively modulates the inhibitory activity of plasminogen activator inhibitor-1. Sci Rep. 2019;9:1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weeks SD, Drinker M, Loll PJ. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr Purif. 2007;53:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. [DOI] [PubMed] [Google Scholar]
- 31.Vonrhein C, Flensburg C, Keller P, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67:293–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen JK, Thompson LC, Bucci JC, et al. Crystal structure of plasminogen activator inhibitor-1 in an active conformation with normal thermodynamic stability. J Biol Chem. 2011;286:29709–29717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nar H, Bauer M, Stassen JM, Lang D, Gils A, Declerck PJ. Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation. J Mol Biol. 2000;297:683–695. [DOI] [PubMed] [Google Scholar]
- 34.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadži S, Garcia-Pino A, Haesaerts S, et al. Ribosome-dependent Vibrio cholerae mRNAse HigB2 is regulated by a β-strand sliding mechanism. Nucleic Acids Res. 2017;45:4972–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV, Grosse-Kunstleve RW, Echols N, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 38.Förster S, Apostol L. Bras WJJoAC. Scatter: software for the analysis of nano-and mesoscale small-angle scattering. J Appl Crystallograp. 2010;43:639–646. [Google Scholar]
- 39.Franke D, Jeffries CM, Svergun DI. Correlation Map, a goodness-of-fit test for one-dimensional X-ray scattering spectra. Nat Methods. 2015;12:419–422. [DOI] [PubMed] [Google Scholar]
- 40.Svergun D, Barberato C, Koch MHJJoac. CRYSOL–a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallograp. 1995;28:768–773. [Google Scholar]
- 41.Komissarov AA, Florova G, Idell S. Effects of extracellular DNA on plasminogen activation and fibrinolysis. J Biol Chem. 2011;286:41949–41962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Florova G, Karandashova S, Declerck PJ, Idell S, Komissarov AA. Remarkable stabilization of plasminogen activator inhibitor 1 in a "molecular sandwich" complex. Biochemistry. 2013;52:4697–4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Komissarov AA, Andreasen PA, Bødker JS, Declerck PJ, Anagli JY, Shore JD. Additivity in effects of vitronectin and monoclonal antibodies against alpha-helix F of plasminogen activator inhibitor-1 on its reactions with target proteinases. J Biol Chem. 2005;280:1482–1489. [DOI] [PubMed] [Google Scholar]
- 44.Komissarov AA, Zhou A, Declerck PJ. Modulation of serpin reaction through stabilization of transient intermediate by ligands bound to alpha-helix F. J Biol Chem. 2007;282:26306–26315. [DOI] [PubMed] [Google Scholar]
- 45.Verheijen JH, Chang GT, Kluft C. Evidence for the occurrence of a fast-acting inhibitor for tissue-type plasminogen activator in human plasma. Thromb Haemost. 1984;51:392–395. [PubMed] [Google Scholar]
- 46.Gettins PG. Serpin structure, mechanism, and function. Chem Rev. 2002;102:4751–4804. [DOI] [PubMed] [Google Scholar]
- 47.Gong L, Liu M, Zeng T, et al. Crystal structure of the Michaelis complex between tissue-type plasminogen activator and plasminogen activators inhibitor-1. J Biol Chem. 2015;290:25795–25804.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin Z, Jiang L, Yuan C, et al. Structural basis for recognition of urokinase-type plasminogen activator by plasminogen activator inhibitor-1. J Biol Chem. 2011;286:7027–7032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naessens D, Gils A, Compernolle G, Declerck PJ. Elucidation of a novel epitope of a substrate-inducing monoclonal antibody against the serpin PAI-1. J Thromb Haemost. 2003;1:1028–1033. [DOI] [PubMed] [Google Scholar]
- 50.Shore JD, Day DE, Francis-Chmura AM, et al. A fluorescent probe study of plasminogen activator inhibitor-1. Evidence for reactive center loop insertion and its role in the inhibitory mechanism. J Biol Chem. 1995;270:5395–5398. [DOI] [PubMed] [Google Scholar]
- 51.Stout TJ, Graham H, Buckley DI, Matthews DJ. Structures of active and latent PAI-1: a possible stabilizing role for chloride ions. Biochemistry. 2000;39:8460–8469. [DOI] [PubMed] [Google Scholar]
- 52.Dewilde M, Strelkov SV, Rabijns A, Declerck PJ. High quality structure of cleaved PAI-1-stab. J Struct Biol. 2009;165:126–132. [DOI] [PubMed] [Google Scholar]
- 53.Bijnens AP, Ngo TH, Gils A, et al. Elucidation of the binding regions of PAI-1 neutralizing antibodies using chimeric variants of human and rat PAI-1. Thromb Haemost. 2001;85:866–874. [PubMed] [Google Scholar]
- 54.Bijnens AP, Gils A, Stassen JM, et al. The distal hinge of the reactive site loop and its proximity: a target to modulate plasminogen activator inhibitor-1 activity. J Biol Chem. 2001;276:44912–44918. [DOI] [PubMed] [Google Scholar]
- 55.Tachias K, Madison EL. Variants of tissue-type plasminogen activator that display extraordinary resistance to inhibition by the serpin plasminogen activator inhibitor type 1. J Biol Chem. 1997;272:14580–14585. [DOI] [PubMed] [Google Scholar]
- 56.Petersen HH, Hansen M, Schousboe SL, Andreasen PA. Localization of epitopes for monoclonal antibodies to urokinase-type plasminogen activator: relationship between epitope localization and effects of antibodies on molecular interactions of the enzyme. Eur J Biochem. 2001;268:4430–4439. [DOI] [PubMed] [Google Scholar]
- 57.Qureshi T, Goswami S, McClintock CS, Ramsey MT, Peterson CB. Distinct encounter complexes of PAI-1 with plasminogen activators and vitronectin revealed by changes in the conformation and dynamics of the reactive center loop. Protein Sci. 2016;25:499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Olson ST, Swanson R, Day D, Verhamme I, Kvassman J, Shore JD. Resolution of Michaelis complex, acylation, and conformational change steps in the reactions of the serpin, plasminogen activator inhibitor-1, with tissue plasminogen activator and trypsin. Biochemistry. 2001;40:11742–11756. [DOI] [PubMed] [Google Scholar]
- 59.Trelle MB, Hirschberg D, Jansson A, et al. Hydrogen/deuterium exchange mass spectrometry reveals specific changes in the local flexibility of plasminogen activator inhibitor 1 upon binding to the somatomedin B domain of vitronectin. Biochemistry. 2012;51:8256–8266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.