ABSTRACT
High levels of transcription and alternative splicing are recognized hallmarks of gene expression in the testis and largely driven by cells in meiosis. Because of this, the male meiosis stage of the cell cycle is often viewed as having a relatively permissive environment for gene expression. In this review, we highlight recent findings that identify the RNA binding protein RBMXL2 as essential for male meiosis. RBMXL2 functions as a “guardian of the transcriptome” that protects against the use of aberrant (or “cryptic”) splice sites that would disrupt gene expression. This newly discovered protective role during meiosis links with a wider field investigating mechanisms of cryptic splicing control that protect neurons from amyotrophic lateral sclerosis and Alzheimer’s disease. We discuss how the mechanism repressing cryptic splicing patterns during meiosis evolved, and why it may be essential for sperm production and male fertility.
KEYWORDS: Cryptic splicing, meiosis, neuronal disease
Pre-mRNA RNA splicing is a crucial mechanism in eukaryotes and is required to enable expression of protein-coding RNAs (mRNAs) from most mammalian genes. Splicing joins together exons within nascent RNA transcripts, thus creating open reading frames from split genes. Splicing is carried out by a molecular machine called the spliceosome [1]. For accurate pre-mRNA splicing the spliceosome needs to precisely identify short consensus sequences called splice sites at exon-intron junctions and join these together. Because of their short length, sequences similar to splice sites (but not selected by the spliceosome) can occur somewhat frequently within genes. Such infrequently used splice sites have the potential to be selected by the spliceosome but are generally not used, so are referred to as “cryptic” in this review (Figure 1). Cryptic splice site sequences are only weakly recognized by the spliceosome and may be located within repetitive sequences and repressed by nuclear RNA binding proteins [2–4]. However, cryptic splice sites can become activated under certain conditions, including some neurological diseases, and their selection can disrupt production of full-length proteins [5]. Some nuclear RNA binding proteins play key roles in repressing the selection of cryptic splicing patterns within the nervous system. These include TDP43 protein that represses cryptic splicing patterns in neurons but becomes disrupted in amyotrophic lateral sclerosis (ALS) leading to the death of motor neurons [6–9]. Cryptic exons are also included in the hippocampus of patients with Alzheimer’s disease [10]. Through its role in cryptic splicing repression TDP43 has been identified as a “guardian of the transcriptome” that is essential for neuron survival [7]. Whether repression of cryptic splicing is important outside of the nervous system has been less well understood. Here, we highlight recent research that reveal a male germ cell-specific nuclear RNA binding protein that operates as a newly discovered guardian of the transcriptome during meiosis.
Figure 1.
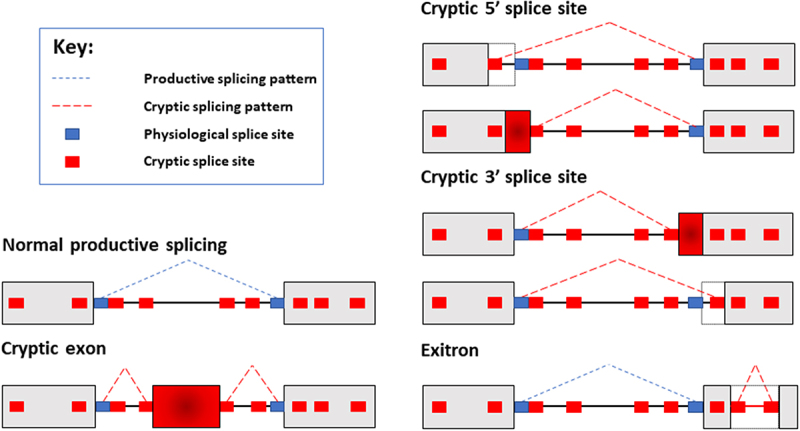
Schematic diagram of cryptic splicing patterns. Most genes are split between exons (shown as gray boxes here) and introns (shown as connecting lines between the boxes). Normal patterns of splice site selection will involve the spliceosome recognizing bona fide splice sites, and joining exons together to create mRNAs. In this example, normal productive splicing is indicated with dashed blue lines. Cryptic splice sites (smaller red boxes) resemble physiological splice sites (smaller blue boxes), and are found within both introns and exons. While normally these cryptic splice sites are ignored by the spliceosome, potentially they could act as decoy sites splice sites for spliceosome selection. Use of cryptic splice sites would produce different mRNAs from genes. Here the normal splicing patterns is shown as a broken blue line joining the physiological splice sites. Examples of cryptic splicing are indicated with dashed red lines. These cryptic splicing events are inclusion of a cryptic exon embedded deep within an intron; selection of cryptic 5ʹ and 3ʹ splice sites; and aberrant recognition of cryptic splice sites within an exon, leading to the interior of this exon being aberrantly recognized as an intron (in a cryptic splicing event known as an exitron).
The testis is considered a relatively permissive site for gene expression patterns. Most human genes produce multiple different mRNAs by using alternative splice sites or by using different combinations of exons. Such alternative splicing permits single genes to produce multiple mRNA isoforms to help amplify the information embedded in the genome. Particularly high levels of alternative splicing have been detected in the testis and in the brain compared to other tissues [11–13]. Alternative splicing patterns can evolve rapidly between species and early analyses detected higher levels of evolutionary divergent splicing in the testis compared to other tissues. This includes the brain, where alternative mRNA isoforms were more likely to be frequently conserved between species than alternative splice isoforms in the testis [11]. More recent comparative transcriptomic analyses confirm some newly evolved exons are exclusively expressed within the testis but suggest there may also be broadly similar levels of conserved mRNA splice isoforms in the testes compared to other tissues [14–16]. The more recently evolved splicing events within the testis are less likely to play a fundamental biological role than more evolutionarily ancient alternative splicing events that have been maintained under selective pressure. However, some recently evolved exons within the testis might later evolve into more generally useful mRNA isoforms via an evolutionary model, which is called the “testis-first” hypothesis. This hypothesis suggests that splicing permissiveness in the testis enables genes to “try out” new exon combinations before they can later be placed under selective pressure [17]. As well as high levels of alternative splicing, there are also particularly high levels of transcription within the testis compared with most other tissues – both in amounts of RNA produced and numbers of genes transcribed [12,18,19].
The human testis produces between 45 and 207 million sperm a day, making it one of the most active developmental pathways still operating in adults [20,21]. The testis contains populations of germ cells (in the developmental pathway leading to sperm) and somatic cells (including Sertoli cells that support germ cell development and Leydig cells that produce testosterone). A population of mitotically active cells called spermatogonia that are early in the germ cell developmental pathway differentiate into cells called spermatocytes. Spermatocytes undergo meiosis, a special form of cell division that produces haploid daughter cells via two sequential divisions. The first meiotic division is preceded by a long prophase that lasts around 2 weeks in mice, referred to as meiotic prophase I. This is divided up into five sequential sub-stages called leptotene, zygotene, pachytene, diplotene and diakinesis – all characterized by distinct chromosomal behaviors. During meiotic prophase I chromosomes condense, and non-sister chromatids form crossovers and undergo genetic recombination. Subsequently, cells separate sister chromatids through a second cell division called meiosis II. This produces haploid spermatids that after meiosis differentiate into spermatozoa (Figure 2).
Figure 2.
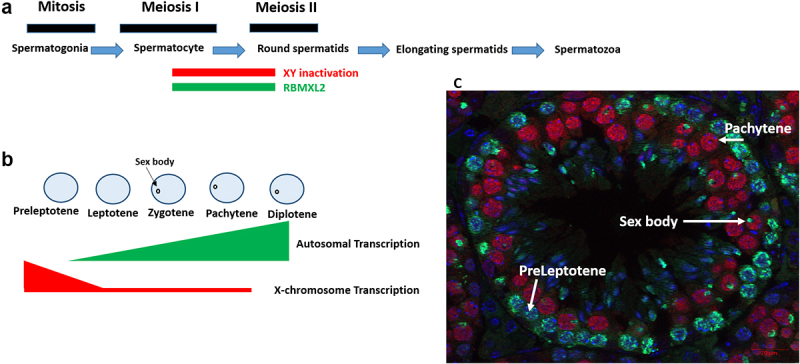
RBMXL2 is expressed during diplotene and pachytene of male meiotic prophase. a. Mouse germ cell development, showing the expression window of RBMXL2 and time period of XY inactivation. b. Transcription patterns of the X chromosome and autosomes during meiotic prophase. c. Seminiferous tubule counterstained with antibodies specific to RBMXL2 protein (pseudocoloured red, detected within nuclei of cells in pachytene in this tubule) and γH2AX (green color, detected within nuclei of pre-leptotene cells and within the sex bodies of pachytene cells).
The cell types that are responsible for the high levels of splicing and gene transcription in the testis have been identified as spermatocytes [12,16,22,23]. Recent transcriptomic analyses of purified testicular cell types reveal that alternative splicing and gene expression levels peak during mid to late pachytene and diplotene stages of meiosis (Figure 2) [24]. In contrast, leptotene, zygotene and early pachytene are transcriptionally quiescent [24–27]. Further RNA sequencing analyses of purified mouse germ cell types detected extensive transcription of both genes and intergenic regions during pachytene and diplotene and in round spermatids, pinpointing these particular cell types as being major contributors to the high levels of testis gene expression and transcriptome complexity [28]. The more “permissive” gene expression environment in the testis may perhaps occur as a result of relaxed chromatin folding. High levels of autosomal transcription during pachytene and diplotene are driven by patterns of open chromatin, including increased levels of the epigenetic mark H3K4me2 (a marker of active promoters) and decreased CpG methylation (a modification normally associated with patterns of gene repression) [12,29]. Furthermore, bursts of meiotic gene expression are driven by activation of super enhancers bound by the MYBL1 and SCML2 transcription factors [30].
A permissive gene environment during meiosis would be consistent with some relaxation of splicing fidelity being tolerated. Despite this, recent data suggest that the interesting parallels between gene expression programs in the brain and testis [13] also extend to a requirement to repress cryptic splicing patterns that would cause cell death. Humans and mice (and likely all placental mammals) express a testis-specific RNA binding protein called RBMXL2 (also known as heterogeneous nuclear ribonucleoproteins G-testis or hnRNP-GT) [31,32]. Mutations have been detected within infertile men for the human RBMXL2 gene [33]. Mouse RBMXL2 protein is expressed in pachytene and diplotene spermatocytes, the stages of meiosis that have the highest levels of transcription and alternative splicing (Figure 2) [34]. Genetic deletion of the mouse Rbmxl2 gene causes cell death during meiotic diplotene, thereby reducing testis size and preventing sperm production [34]. Detailed molecular analysis of this mouse model show that RBMXL2 protein prevents the spliceosome selecting cryptic splice sites during the pachytene and diplotene stages of male meiosis [34] – thus performing a similar molecular role to TDP43 in neurons. Both RBMXL2 and TDP43 are members of a group of proteins called hnRNPs (heterogeneous nuclear ribonucleoproteins) that bind to nuclear RNAs as they are transcribed to control their splicing patterns. Although they both operate as “guardians of transcriptomes”, RBMXL2 protein is only expressed in spermatocytes and spermatids. In contrast, TDP43 protein is expressed more ubiquitously. Despite this, point mutations affecting TDP43 specifically cause neuronal cell death [35]. Interestingly, while complete genetic knockout of TDP43 causes embryonic death in mice [36,37], conditional genetic knockout of TDP43 in the testis causes male infertility [38].
Genes encoding mRNAs that are incorrectly spliced in the absence of RBMXL2 are enriched in functions associated with meiosis, chromosome segregation and spermatogenesis (Sara Luzzi unpublished). The inappropriate selection of cryptic splice sites in spermatocytes in the absence of RBMXL2 protein might therefore cause spermatocyte cell death by preventing proper expression of key genes needed for meiosis. RBMXL2 regulates splicing patterns of over a hundred genes during meiotic prophase. Important genes that contain cryptic splice sites that are repressed by RBMXL2 protein include Brca2 (encoding a DNA repair protein involved in genetic recombination) and Meioc (which encodes a cytoplasmic protein that is critical during meiotic prophase) [39–41]. Exactly which RBMXL2 target genes are most important for spermatocyte survival is unknown. Complicating this prediction, the phenotypes caused by splicing errors within a narrow window of meiosis might be different from traditional genetic knockouts that assess when a gene is first needed in a developmental pathway. For example, Meioc genetic knockout spermatocytes have an unusually short meiotic prophase and do not reach pachytene or diplotene – the developmental window in which RBMXL2 protein is expressed (Figure 2). Similarly, genetic knockout of the Brca2 gene prevents germ cell development at an early stage before germ cells enter meiosis [41]. The effects of changing Meioc and Brca2 RNA processing pathways during the narrow window of meiotic prophase when RBMXL2 is normally expressed are not well understood and difficult to predict. An alternative cause of spermatocyte cell death in the absence of RBMXL2 may be through genotoxic damage via formation of RNA:DNA hybrids (R-loops). The loss of splicing factors can cause normally intronic regions to be included within incorrectly spliced mRNAs (rather than being removed by splicing). R-loops form as a result of transcription involving local melting of DNA close to the elongating RNA polymerase, and intronic regions remaining within the pre-mRNA being able to base pair with the melted DNA duplex (forming R loops), leading to DNA damage [42]. In principle it should be possible to potentially correct aberrant splicing patterns caused by loss of RBMXL2 during meiosis. However, while individual cryptic splice sites can be therapeutically targeted using antisense oligonucleotides [43], it would be difficult to use this approach to correct the hundreds of targets that are normally controlled by RBMXL2 during meiosis.
Understanding RBMXL2 function fits into a bigger picture involving the evolution of new genes and regulation of gene expression patterns in the body. The RBMXL2 gene originated via retro-transposition of an mRNA encoded by the X-linked gene RBMX approximately 65 million years ago. As a result of this, the RBMXL2 gene does not contain introns [32]. RBMX is an RNA binding protein important for controlling splicing, transcription and genome stability [44,45]. Unpaired regions of the X and Y chromosomes (including also RBMX gene) become transcriptionally silent during pachytene, within a heterochromatic structure called sex body (or XY body) (Figure 2). This process is termed meiotic sex chromosome inactivation (MSCI) [46], and leads to either reduced or complete loss of sex-linked gene expression for the reminder of meiosis (over ~9 days in mice and even longer in humans [47]). Most retrogenes decay rapidly after they are formed. However, many essential X-linked genes that are transcriptionally silenced by MSCI are functionally replaced by retrogenes that are only expressed in the testis [48].
RBMXL2 is only expressed during and immediately after meiosis, so how are the genes controlled by RBMXL2 in the testis normally spliced in other tissues that do not express RBMXL2? A possible answer is that the splice events that are controlled by RBMXL2 during meiosis might be controlled instead by RBMX in other cell types within the body. In this scenario RBMXL2 may functionally replace RBMX function during meiotic prophase, either as a direct “like for like” replacement or as a more specialized replacement that has evolved to control specific gene expression pathways needed for meiosis [49–51]. However, whether RBMX and RBMXL2 proteins have similar functional activity is not yet fully answered. RBMXL2 mainly operates as a splicing repressor during meiotic prophase in mice. In contrast, global data from human cells has characterized the properties of RBMX protein mainly as a splicing activator that binds to specifically methylated pre-mRNA to slow progression of RNA polymerase II transcription thus facilitating spliceosome function and activating exon inclusion [52]. RBMX is also mutated in the X-linked intellectual disability syndrome Shashi syndrome, where it leads to increased p53 activity and neuronal defects via splicing activation of MDM4 exon 6 [53].
Why is it important to repress cryptic splice sites during meiosis? Conventional splice sites have evolved to enable exons to be precisely joined together and maintain protein-coding open reading frames. Since cryptic splice sites are usually not used they are not under the same selective pressure as bona fide splice sites. Cryptic splice site inclusion into mRNAs often disrupts protein-coding reading frames by introducing premature termination codons (PTCs). In cells that are not dividing by meiosis, PTC-containing mRNAs are degraded by an RNA stability pathway called Nonsense Mediated Decay (NMD) that prevents translation into truncated proteins that could be harmful to the cell. However, uniquely in the testes PTC-containing transcripts can become stabilized. This stabilization occurs during meiosis, thus increasing the likelihood of mRNAs originating from cryptic splicing being translated into potentially toxic proteins. The reason for this stabilization is because of meiotic-associated changes in the NMD pathway. One of the core protein components of the NMD pathway is a protein called UPF3B that is encoded by a gene on the X chromosome that is turned off during meiosis by MCSI. As a consequence its autosomal paralogue gene UPF3A becomes active when germ cells enter pachytene [54]. Genetic deletion of UPF3A induces meiotic defects in a mouse model showing UPF3A expression is critical for meiosis [54]. However, UPF3A protein has only weak activity in the NMD pathway because of an amino acid substitution compared to UPF3B. In fact, while UPF3B promotes mRNA degradation via NMD, UPF3A may operate as an NMD repressor [54]. Hence meiotic expression of UPF3A may lead to translation of some PTC-containing mRNAs and represent a possible reason why it is particularly important to repress cryptic splicing events during meiosis.
Other RNA binding proteins are also essential for splicing control during meiosis and have been recently reviewed [55–57]. However, the molecular defects that appear during mouse meiotic prophase in the absence of RBMXL2 protein, involving a high frequency of aberrantly selected cryptic splicing events, are largely distinct from those that have been identified for other splicing regulators during meiosis. One example is the splicing regulator PTBP2 that is expressed at high levels in spermatocytes [58]. Although PTBP2 protein represses cryptic splice sites in other cell types, its role during germ cell development seems more consistent as a master regulator of developmentally regulated splicing [58,59]. Genetic knockout of Ptbp2 causes male germ cells to be prematurely sloughed off into the lumen of seminiferous tubules and defects to accumulate in the Sertoli cells cytoskeleton, suggesting impaired interactions between somatic and germ cells without PTBP2 protein [58]. This testicular phenotype correlates with disrupted splicing patterns detected for ~200 genes normally controlled by PTBP2, mainly with roles in Sertoli-germ cell communication. In contrast, more than 60% of the splicing events controlled by RBMXL2 during meiosis involve repression of cryptic splice sites rather than regulation of already known alternative splice events [34].
How does cryptic splicing repression by RBMXL2 integrate with other recently discovered aspects of splicing control during meiosis? High-throughput RNA sequencing analysis of purified meiotic spermatocytes and spermatids show that 10% of the alternative splicing events during meiosis occur via intron retention [23,24,60]. These intron-retained mRNAs play a key role in developmental gene expression. Stable mRNAs containing retained introns are transcribed in spermatocytes and remain nuclear for a few days before being spliced and translated in the post-meiotic stages of spermatogenesis to encode crucial proteins in sperm development [23]. Mechanistically, intron retention involves the repression of splice sites, leading to whole introns being retained within mRNAs. The retained introns detected in spermatocytes and round spermatids have weak splice sites, suggesting a model where the extremely high levels of transcription during meiosis may overload the splicing machinery thus leading to intron retention. Despite RBMXL2’s established role in repressing the selection of cryptic splice sites during meiosis, global changes in intron retention were not detected in the Rbmxl2 knockout mouse model [34]. However, slower patterns of intron removal during meiosis may make some pre-mRNAs more vulnerable to cryptic exons being mistakenly selected by the spliceosome in mice that do not express RBMXL2 protein.
A key question for the future is why do spermatocytes die without RBMXL2 protein, and to what extent does this resemble neuronal cell death in cells depleted for TDP43 activity? Is cell death caused by the loss of specific important proteins as a result of cryptic splicing events in protein coding mRNAs or does cell death result from genotoxic damage caused by accumulation of R-loops from incorrectly spliced mRNAs (Figure 3)? What is the mechanism by which RBMXL2 represses cryptic splicing patterns? Does RBMXL2 bind to sequences in pre-mRNAs near to cryptic splice sites to sterically occlude the spliceosome? Or does RBMXL2 protein bind and antagonize the function of splicing activator proteins [61] preventing them from activating selection of cryptic splice sites that are otherwise poised for selection by the spliceosome? Could RBMXL2 repress cryptic exons by stabilizing formation of stalled spliceosomes on nascent RNA [62,63] (Figure 3)? Does RBMXL2 only repress cryptic splicing patterns or is it also involved in other aspects of meiosis (Figure 3)? Finally, do RBMX and RBMXL2 perform similar functions in cryptic splicing repression? This last question is of wide importance: RBMX protein has been implicated in controlling chromosome biology and DNA repair as well as splicing and transcription and is also mutated in an X-linked intellectual disability syndrome [44,53,64]. Supporting a wider biological role, RBMX also controls transcription of the CBX5 gene within leukemia cells [65,66]. Further mechanistic investigations of RBMXL2 and RBMX functions will help to address these issues and should reveal further gene expression pathways that operate during human development and disease.
Figure 3.

Mechanistic models to explain the impact of cryptic splicing on meiosis, and possible additional roles of RBMXL2. a. RBMXL2 could repress cryptic splicing by either counteracting the function of splicing activators; sterically blocking access of the spliceosome to cryptic splice sites; or promoting stalling of spliceosome assembly at cryptic splice sites. In the absence of RBMXL2 protein cryptic splicing could hinder production of proteins important for meiosis (Model 1) or alternatively impair transcription elongation and promote formation of R-loops (Model 2). Both scenarios would lead to meiotic arrest. b. Both RBMX and RBMXL2 are known to regulate splicing in somatic and germ cells respectively. Could RBMXL2 also have a function in other pathways known to be regulated by RBMX such as RNA polymerase II transcription and DNA repair/cell division?.
Funding Statement
This work was funded by the BBSRC (grants Biotechnology and Biological Sciences Research Council BB/S008039/1 and BB/P006612/1) and the King Fahad Medical City, Ministry of Health, Kingdom of Saudi Arabia. No potential competing interest was reported by the authors.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Papasaikas P, Valcárcel J, Eu JV.. Valcárcel (J. Special Issue: 40 Years of TiBS the spliceosome: the Ultimate RNA Chaperone and Sculptor. Trends Biochem Sci. 2016;41(1):33–45. [DOI] [PubMed] [Google Scholar]
- [2].Zarnack K, König J, Tajnik M, et al. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].McClory SP, Lynch KW, Ling JP.. HnRNP L represses cryptic exons. RNA. 2018;24(6):761–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Attig J, Agostini F, Gooding C, et al. Heteromeric RNP Assembly at LINEs Controls Lineage-Specific RNA processing. Cell. 2018;174(5):1067–1081.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sibley CR, Blazquez L, Ule J. Lessons from non-canonical splicing. Nat Rev Genet. 2016;17(7):407–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jp L, Pletnikova O, Jc T, et al. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science. 2015;349(6248):978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Donde A, Sun M, Ling JP, et al. Splicing repression is a major function of TDP-43 in motor neurons. Acta Neuropathol Internet]. 2019; 138:813–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tan Q, Yalamanchili HK, Park J, et al. Extensive cryptic splicing upon loss of RBM17 and TDP43 in neurodegeneration models. Hum Mol Genet. 2016;25(23):70–81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fratta P, Sivakumar P, Humphrey J, et al. Mice with endogenous TDP-43 mutations exhibit gain of splicing function and characteristics of amyotrophic lateral sclerosis. EMBO J. 2018;37(11);e98684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sun M, Bell W, Kd L, et al. Cryptic exon incorporation occurs in Alzheimer’s brain lacking TDP-43 inclusion but exhibiting nuclear clearance of TDP-43. Acta Neuropathol. 2017;133(6):923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kan Z, Garrett-Engele PW, Johnson JM, et al. Evolutionarily conserved and diverged alternative splicing events show different expression and functional profiles. Nucleic Acids Res. 2005;33(17):5659–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Soumillon M, Necsulea A, Weier M, et al. Cellular Source and Mechanisms of High Transcriptome Complexity in the Mammalian Testis. Cell Rep. 2013;3(6):2179–2190. [DOI] [PubMed] [Google Scholar]
- [13].Naro C, Cesari E, Sette C. Splicing regulation in brain and testis: common themes for highly specialized organs [Internet]. Cell Cycle [cited 2021. Jun 17]; 20(5–6):480–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Merkin J, Russell C, Chen P, et al. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science. 2012;338(6114):1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mazin PV, Khaitovich P, Cardoso-Moreira M, et al. Alternative splicing during mammalian organ development. Nat Genet. 2021;133(6):923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Schmid R, Grellscheid SN, Ehrmann I, et al. The splicing landscape is globally reprogrammed during male meiosis. Nucleic Acids Res. 2013;41(22):10170–10184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Carelli FN, Hayakawa T, Go Y, et al. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016;26(3):301–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramsköld D, Wang ET, Burge CB, et al. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol. 2009;5(12):e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cardoso-Moreira M, Halbert J, Valloton D, et al. Gene expression across mammalian organ development. Nature. 2019;571(7766):505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Amann RP, Howards SS. Daily spermatozoal production and epididymal spermatozoal reserves of the human male. J Urol. 1980;124(2):211–215. [DOI] [PubMed] [Google Scholar]
- [21].Griswold MD. Spermatogenesis: the Commitment to Meiosis. Physiol Rev. 2016;96(1):1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gamble J, Chick J, Seltzer K, et al. An expanded mouse testis transcriptome and mass spectrometry defines novel proteins. Reproduction. 2020;159(1):15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Naro C, Jolly A, Di Persio S, et al. An Orchestrated Intron Retention Program in Meiosis Controls Timely Usage of Transcripts during Germ Cell Differentiation. Dev Cell. 2017;41(1):82–93.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Chen Y, Zheng Y, Gao Y, et al. Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res. 2018;28(9):879–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J Cell Biol. 1964;22(3):521–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Turner JMA, Mahadevaiah SK, Fernandez-Capetillo O, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37(1):41–47. [DOI] [PubMed] [Google Scholar]
- [27].Paronetto MP, Messina V, Barchi M, et al. Sam68 marks the transcriptionally active stages of spermatogenesis and modulates alternative splicing in male germ cells. Nucleic Acids Res. 2011;39(12):4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ernst C, Eling N, Martinez-Jimenez CP, et al. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat Commun. 2019;10(1):1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Maezawa S, Yukawa M, Alavattam KG, et al. Dynamic reorganization of open chromatin underlies diverse transcriptomes during spermatogenesis. Nucleic Acids Res. 2018;46(2):593–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Maezawa S, Sakashita A, Yukawa M, et al. Super-enhancer switching drives a burst in gene expression at the mitosis-to-meiosis transition. Nat Struct Mol Biol. 2020;27(10):978–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Venables JP. RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum Mol Genet. 2000;9(5):685–94 . [DOI] [PubMed] [Google Scholar]
- [32].Elliott DJ, Venables JP, Newton CS, et al. An evolutionarily conserved germ cell-specific hnRNP is encoded by a retrotransposed gene. Hum Mol Genet. 2000;9(14):2117–2124. [DOI] [PubMed] [Google Scholar]
- [33].Gh W, Gianotten J, Nj L, et al. Heterogeneous nuclear ribonucleoprotein G‐T (HNRNP G‐T) mutations in men with impaired spermatogenesis. Mol Hum Reprod. 2004;10(4):265–269. [DOI] [PubMed] [Google Scholar]
- [34].Ehrmann I, Crichton JH, Gazzara MR, et al. An ancient germ cell-specific RNA-binding protein protects the germline from cryptic splice site poisoning. Elife Internet]. [cited 2019. 8 Aug 27]; e39304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Buratti E, Fe B. TDP-43: gumming up neurons through protein-protein and protein-RNA interactions. Trends Biochem Sci. 2012;37(6):237–247. [DOI] [PubMed] [Google Scholar]
- [36].Sephton CF, Good SK, Atkin S, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285(9):6826–6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wu L-S, Cheng W-C, Hou S-C, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48(1):56–62. [DOI] [PubMed] [Google Scholar]
- [38].Reddi PP. Transcription and Splicing Factor TDP-43: role in regulation of gene expression in testis. Semin. Reprod. Med. 2017;35(2):167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Soh YQS, Mikedis MM, Kojima M, et al. Meioc maintains an extended meiotic prophase I in mice. PLoS Genet. 2017;13(4):e1006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Abby E, Tourpin S, Ribeiro J, et al. Implementation of meiosis prophase i programme requires a conserved retinoid-independent stabilizer of meiotic transcripts. Nat Commun. 2016;7:10324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Connor F, Bertwistle D, Mee PJ, et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17(4):423–430. [DOI] [PubMed] [Google Scholar]
- [42].Tam AS, Stirling PC. Splicing, genome stability and disease: splice like your genome depends on it! Curr. Genet. 2019;65:905–912. [DOI] [PubMed] [Google Scholar]
- [43].Erdos MR, Cabral WA, Tavarez UL, et al. A targeted antisense therapeutic approach for Hutchinson–Gilford progeria syndrome. Nat Med. 2021;27(3):536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Elliott DJ, Dalgliesh C, Hysenaj G, et al. RBMX family proteins connect the fields of nuclear RNA processing, disease and sex chromosome biology. Int J Biochem Cell Biol. 2019;108:1–6. [DOI] [PubMed] [Google Scholar]
- [45].Adamson B, Smogorzewska A, Fd S, et al. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat Cell Biol. 2012;14(3):318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Turner JMA. Meiotic sex chromosome inactivation. Development. 2007;134(10):1823–1831. [DOI] [PubMed] [Google Scholar]
- [47].MacDonald CC. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond (2018 update). Wiley Interdiscip. Rev. RNA. 2019;:10(4):e1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Emerson JJ, Kaessmann H, Betrán E, et al. Extensive gene traffic on the mammalian X Chromosome. Science. 2004;303(5657):537–540. [DOI] [PubMed] [Google Scholar]
- [49].Jiang L, Li T, Zhang X, et al. RPL10L Is Required for male meiotic division by compensating for rpl10 during meiotic sex chromosome inactivation in mice. Curr Biol. 2017;27(10):1498–1505.e6. [DOI] [PubMed] [Google Scholar]
- [50].Wang PJ. X chromosomes, retrogenes and their role in male reproduction. Trends Endocrinol Metab. 2004;15(2):79–83. [DOI] [PubMed] [Google Scholar]
- [51].Long M, Emerson JJ. Meiotic Sex Chromosome Inactivation: compensation by Gene Traffic. Curr Biol. 2017;27(13):R659–61. [DOI] [PubMed] [Google Scholar]
- [52].Zhou KI, Shi H, Lyu R, et al. Regulation of Co-transcriptional Pre-mRNA Splicing by m6A through the Low-Complexity Protein hnRNPG. Mol Cell. 2019;76(1):70–81.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cai T, Cinkornpumin JK, Yu Z, et al. Deletion of RBMX RGG/RG motif in Shashi-XLID syndrome leads to aberrant p53 activation and neuronal differentiation defects. Cell Rep. 2021;36(2):109337. [DOI] [PubMed] [Google Scholar]
- [54].Shum EY, Jones SH, Shao A, et al. The Antagonistic Gene Paralogs Upf3a and Upf3b Govern Nonsense-Mediated RNA Decay. Cell. 2016;165(2):382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Licatalosi DD Roles of RNA-binding proteins and post-transcriptional regulation in driving male germ cell development in the mouse. In: Advances in Experimental Medicine and Biology . ; 2016;907:123–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Legrand JMD, Hobbs RM. RNA processing in the male germline: mechanisms and implications for fertility. Semin Cell Dev Biol. 2018;79:80–91. [DOI] [PubMed] [Google Scholar]
- [57].Song H, Wang L, Chen D, et al. The function of pre-mRNA alternative splicing in mammal spermatogenesis. Int J Biol Sci. 2020;16(1):38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hannigan MM, Zagore LL, Licatalosi DD. Ptbp2 Controls an Alternative Splicing Network Required for Cell Communication during Spermatogenesis. Cell Rep. 2017;19(12):2598–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Jp L, Chhabra R, Jd M, et al. PTBP1 and PTBP2 Repress Nonconserved Cryptic Exons. Cell Rep. 2016;17(1):104–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Naro C, Sette C. Timely-regulated intron retention as device to fine-tune protein expression. Cell Cycle. 2017;16(14):1321–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nasim MT, Chernova TK, Chowdhury HM, et al. HnRNP G and Tra2β: opposite effects on splicing matched by antagonism in RNA binding. Hum Mol Genet. 2003;12(11):1337–1348. [DOI] [PubMed] [Google Scholar]
- [62].Vaquero-Garcia J, Barrera A, Gazzara MR, et al. A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife. 2016;5:e11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Chiou N-T, Shankarling G, Lynch KW. hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol Cell. 2013;49(5):972–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Shashi V, Xie P, Schoch K, et al. The RBMX gene as a candidate for the Shashi X-linked intellectual disability syndrome. Clin Genet. 2015;88(4):1067–1081.e17. [DOI] [PubMed] [Google Scholar]
- [65].Prieto C, Nguyen DTT, Liu Z, et al. Transcriptional control of CBX5 by the RNA-binding proteins RBMX and RBMXL1 maintains chromatin state in myeloid leukemia. Nat Cancer. 2021;2(7):318–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Prieto C, Nguyen D, Vu LP, et al. RNA binding protein rbmx is required in acute myeloid leukemia by regulating the transcriptional activity of the Heterochromatin Protein HP1α. Blood. 2018;132(Supplement 1):883.29945954 [Google Scholar]


