ABSTRACT
The functions of exosomes in allergic diseases including asthma have aroused increasing concerns. This paper focuses on the effects of exosomes derived from human bone marrow-mesenchymal stem cells (hBM-MSCs) on the proliferation of bronchial smooth muscle cells in asthma and the mechanism involved. Exosomes were extracted from hBM-MSCs and identified. Human BSMCs were induced with transforming growth factor (TGF)-β1 to mimic an asthma-like condition in vitro and then treated with exosomes. A mouse model with asthma was induced by ovalbumin (OVA) and treated with exosomes for in vivo study. The hBM-MSC-derived exosomes significantly reduced the abnormal proliferation and migration of TGF-β1-treated BSMCs. microRNA (miR)-188 was the most enriched miRNA in exosomes according the microarray analysis, and JARID2 was identified as a mRNA target of miR-188. Either downregulation of miR-188 or upregulation of JARID2 blocked the protective effects of exosomes on BSMCs. JARID2 activated the Wnt/β-catenin signaling pathway. In the asthmatic mice, hBM-MSC-derived exosomes reduced inflammatory cell infiltration, mucus production, and collagen deposition in mouse lung tissues. In conclusion, this study suggestes that hBM-MSC-derived exosomes suppress proliferation of BSMCs and lung injury in asthmatic mice through the miR-188/JARID2/Wnt/β-catenin axis. This study may provide novel insights into asthma management.
KEYWORDS: Human bone marrow-mesenchymal stem cells, exosomes, microRNA-188, JARID2, wnt/β-catenin, asthma, bronchial smooth muscle cells
1. Introduction
Asthma, which burdens over 300 million population globally, is a frequent respiratory disease featured with chronic airway inflammation, respiratory symptoms, and variable expiratory airflow obstruction, followed by airway wall thickening, airway narrowing, and increased mucus [1,2]. Persistent airflow obstruction is caused by the increasing mucus production in the airway lumen and the airway remodeling that involves pathologies including excessive subepithelial collagen deposition (fibrosis), goblet cell hyperplasia, airway smooth muscle hyperplasia, reduced epithelial and cartilage integrity, and increased vascularity [3,4]. These lead to the common clinical symptoms of asthma including persistent coughing, chest tightness, shortness of breath, and wheezing [5]. Conventional drugs including inhaled corticosteroids are effective for patients with mild-to-moderate asthma, but patients with severe asthma remain refractory despite the recommended maximal therapeutic regimens and have limited options [6–8]. Understanding molecular mechanisms responsible for asthma development and exploring new strategies for asthma management are of great significance.
Exosomes are nano-sized extracellular vesicles (Evs) released from the endosome that play key roles in regulating pathological and physiological conditions including allergic airway diseases by transferring cargos such as nucleic acids (microRNA [miRNA], mRNA, DNA), proteins, enzymes, and metabolites [9]. Mesenchymal stem cells (MSCs) are multipotent stromal precursor cells in neonatal and adult tissues that exert key functions in allergic diseases due to the tissue repair ability, immunosuppressive characteristics, and the release of biological factors [10]. MSCs secret exosomes with the capacity to promote wound healing and accelerate lung tissue regeneration, which may also function in suppressing airway remodeling in asthma [11]. The most frequently used sources of MSCs are bone marrow (BM-MSCs), umbilical cord (UCB-MSCs) and adipose tissue (AD-MSCs) [12]. Transforming growth factor-β1 (TGF-β1) is a key mediator upregulated in asthma that mediates fibrotic responses and recruitment of inflammatory cells, and it can also regulate the production of extracellular matrix proteins and vascular endothelial cell growth factors, contributing to increased proliferation of airway smooth muscle cells (ASMCs) and vascular remodeling in the asthmatic airway [13–15]. TGF-β1 treatment has been frequently used to induce airway smooth muscle cell proliferation in asthma-related researches [16,17]. In this study, we aimed to explore the potential functions of human BM-MSCs (hBM-MSCs)-derived exosomes in asthma using transforming growth factor (TGF)-β1-treated bronchial smooth muscle cell (BSMC) models and ovalbumin (OVA)-challenged asthmatic mouse models [18].
miRNAs are a major class of cargos carried by exosomes, and the exosomal miRNAs frequently exert crucial functions in the pathogenesis of asthma [19]. They may regulate inflammatory responses and signaling pathways by governing expression of target genes through direct binding [19]. Here, our microarray analysis confirmed a significant increase in miR-188 expression in exosome-treated BSMCs. miR-188 has been recognized as a suppressor of the lung cancer cells and the lung cancer stem cells [20,21]. Downregulation of miR-188 was found in ASMCs after inflammatory stimulation [22]. In addition, our subsequent bioinformatics analyses suggested jumonji and AT-rich interaction domain containing 2 (JARID2) as a target of miR-188. JARID2 has been reported to be involved in TGF-β-induced epithelial–mesenchymal transition (EMT) of lung cancer cells [23]. However, the functions of miR-188 and JARID2 in airway remodeling and the pathogenesis of asthma remain unknown. Here, we hypothesized that hBM-MSC-derived exosomes possibly mediate asthma progression through a miR-188/JARID2 axis.
2. Materials and methods
2.1. Ethical approval
This study was approved by the Animal Ethics Committee of Shengjing Hospital of China Medical University. All animal procedures were performed in accordance with the Guide for the Care and Use of Laboratory Animals issued by the National Institutes of Health (NIH, Bethesda, Maryland, USA). Great attempts were made to minimize the number and pain of animals.
2.2. Cell culture and treatment
Human BSMCs (CAT No. CP-H102) were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, Hubei, China) and identified by immunofluorescence staining of alpha-smooth-muscle-actin. All cells were free of HIV-1, HBV, HCV, mycoplasma, bacterial, yeast, or fungal infections. BSMCs were cultured in Dulbecco’s modified Eagle’s medium (Gibco Company, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) at 37°C in humidified air enriched with 5% CO2. Human recombinant TGF-β1 (#240-B, R&D Systems Inc., Minneapolis, MN, USA) was administrated into BSMCs at 10 ng/mL for continuous 24 h to mimic an asthma-like condition in vitro. Cells treated with an equal dose of phosphate-buffered saline (PBS) were set as control. XAV-939 (Cat. No. 15147, MedChemExpress, Monmouth Junction, NJ, USA), a Wnt/β-catenin-specific inhibitor, was used to treat the BSMCs at 5 nM to inactivate the Wnt/β-catenin pathway in cells [24]. Dimethyl Sulfoxide (DMSO) was used as control.
hBM-MSCs (ATCC® PCS-500-012™) were acquired from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in MSC growth medium (Cyagen Biosciences Inc., Suzhou, Jiangsu, China) at 37°C with 5% CO2. Cells at passage 3 were incubated with antibodies against CD105 (ab11414), CD73 (ab81720), CD90 (ab23894), CD34 (ab8536), and CD14 (ab182032), CD19 (ab31947), and with the goat anti-mouse IgG H&L (FITC) (ab6785) or the isotype control IgG (all purchased from Abcam Inc., Cambridge, MA, USA). After that, the samples were analyzed using a FACSVerse instrument (BD Biosciences, San Jose, CA, USA) and FlowJo software (TreeStar Inc., Ashland, OR, USA). MSCs were further cultured in osteogenic or adipogenic differentiation medium (Cyagen Biosciences), and the osteogenic and adipogenic differentiation abilities of cells were examined using alizarin red staining and oil red O staining kits (LMAIBio Co., Ltd., Shanghai, China), respectively.
The miR-188 mimic (CAUCCCUUGCAUGGUGGAGGG)/inhibitor (GUAGGGAACGUACCACCUCCC) were procured from GenePharma Co., Ltd. (Shanghai, China). The CDS sequence of JARID2 (NM_001267040.1) was cloned and inserted into the pcDNA 3.1 vectors to construct JARID2-overexpression vector (pcDNA-JARID2) by GenePharma. The empty vector was used as control. All vectors were transfected into cells using a Lipofectamine 3000 kit (Invitrogen, Thermo Fisher Scientific Inc., Waltham, MA, USA) according to the kit’s instructions. In short, cells were cultured in 96-well plates at 2 × 104 cells/well until reaching a 70–90% confluence. Thereafter, 0.15 μL and 0.3 μL Lipofectamine 3000 Reagent, respectively, was diluted in 5 μL Opti-MEM Medium (Thermo Fisher Scientific). Next, 0.2 μg pcDNA vector was mixed and diluted in 0.4 μL P3000™ reagent and 10 μL Opti-MEM medium. The miRNA mimic/inhibitor were diluted in a similar manner except for the addition of P3000™ Reagent. The diluted miRNA mimic/inhibitor or pcDNA for transfection were added into an equal volume of Lipofectamine 3000 Reagent for 15 min of incubation at room temperature. The mixture was loaded into the cells at a dose of 10 μL per well. After 48 h of incubation at 37°C, the transfection efficiency was determined.
2.3. Isolation of hBM-MSC-derived exosomes
An Optima L-100XP Ultracentrifuge (Beckman Coulter Inc., Chaska, MN, USA) was used to extract and purify exosomes from the hBM-MSC medium using differential ultracentrifugation method. Exosomes in culture medium were first exhausted after ultracentrifugation at 100,000 g. Thereafter, the hBM-MSCs were cultured in exosome-free medium for 48 h. After that, the supernatant was successively centrifuged at 300 g for 10 min, 2,000 g for 10 min, and at 10,000 g for 30 min to discard the dead cells and cell debris. Next, the supernatant was further collected and ultra-centrifuged at 100,000 g for 70 min. Next, the exosomes were collected and observed under a transmission electron microscope (TEM, FEI Tecnai G2 F30, Oregon, USA). The particle size distribution was examined by Zetasizer Nano (Malvern Panalytical Co., Ltd., Malvern, Worcestershire, UK). The expression of exosome-specific biomarkers CD81 (ab109201, Abcam) and tumor susceptibility 101 (TSG101, ab125011, Abcam) was determined by Western blot analysis. The protein concentration of the extracted exosomes was determined using a bicinchoninic acid (BCA) kit, and 20 μg exosomes was used each time to treat the BSMCs.
2.4. Absorption of exosomes by BSMCs
The extracted exosomes were labeled by the red fluorescence PKH26 (Sigma-Aldrich Chemical Company, St Louis, MO, USA) according to the kit’s instructions. The exosomes were washed in PBS, and 20 μg/mL exosome suspension in PBS was cultured with BSMCs with serum-free medium at 37°C for 8 h, 16 h or 24 h, respectively. After that, the cells were fixed in 4% paraformaldehyde, and the nuclei were stained by Hoechst 33,342 (MedChemExpress, Monmouth Junction, NJ, USA). Absorption of exosomes by BSMCs was observed under a confocal laser scanning microscope (Leica Biosystems, Solms, Germany). Relative luciferase activity in cells was evaluated using the Image Pro Plus 6.0 Software (NIH).
2.5. Cell counting kit-8 (CCK-8) method
A CCK-8 kit (Beyotime Biotechnology Co., Ltd, Shanghai, China) was used to examine the proliferation ability of cells. In brief, 2,000 cells were seeded in 96-well plates. Each well was loaded with 10 μL CCK-8 solution at 0 h, 12 h, 24 h and 48 h solution, followed by another 2 h of incubation. The optical density (OD) at 450 nm was determined using a spectrophotometer (Shanghai Metash Instruments Co., Ltd., Shanghai, China).
2.6. 5-ethynyl-2ʹ-deoxyuridine (EdU) labeling assay
BSMCs were cultured in 24-well plates (1 × 105 cells/well) and added with EdU solution (C10310-1, Guangzhou RiboBio Co., Ltd., Guangzhou, Guangdong, China) till a final concentration of 10 μM. After a 2-h culture, cells were fixed and then permeabilized by 0.5% Triton-100 in PBS. Next, suspended cells were stained with Apollo®567 solution (RiboBio Co., Ltd) for 30 min and further with 1× Hoechst 33,342 for 30 min. The number of EdU-positive cells was observed under a fluorescence microscope (Olympus Optical Co., Ltd, Tokyo, Japan).
2.7. Flow cytometry
An Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) cell apoptosis kit (Thermo Fisher Scientific) was used and flow cytometry was applied to measure apoptosis of cells. In short, the BSMCs were sorted on 6-well plates (1 × 105 cells/well) till an 80% cell confluence. After transfection, the cells were centrifuged and resuspended in 500 μL binding buffer. Next, the cells were incubated with 5 μL Annexin V-FITC and PI avoiding light exposure at room temperature for 15 min. After dual staining, the cells were analyzed using the FACSVerse flow cytometer and the FlowJo Software to measure cell apoptosis.
2.8. Transwell assay
Migration of BSMCs was determined using 24-well polycarbonate membrane inserts (8.0 µm pore, Corning Glass Works, Corning, NY, USA) without Matrigel. Cells (2 × 104 cells/well) were suspended in serum-free medium (Gibco Company) and then loaded in the apical chambers, while 10% FBS-supplemented medium was filled into the basolateral chambers. The Transwell chambers were placed in a 37°C incubator for 48 h. Thereafter, the cells on the upper surface of the membrane were discarded using cotton swabs, and cells migrated to the lower surface of the membrane were fixed and stained with 0.5% crystal violet. The number of migratory cells in five randomly selected fields were observed under the microscope.
2.9. miRNA microarray analysis
Differentially expressed miRNAs after exosome treatment were screened using the SurePrint Human miRNA Microarrays (Agilent Technologies, Palo Alto, CA, USA) according to the protocol of the Exosome miRNA Profiling with the Agilent miRNA Microarray Platform. In brief, total RNA was processed using a miRNA complete labeling and hybridization kit and a miRNA Spike-in kit (Agilent). The RNA samples were hybridized with the miRNA microarrays, which were further scanned by a microarray scanner (Agilent). Data were obtained by the Agilent feature extraction v.12.0.0.7. The success of labeling and hybridization was evaluated using the QC metrics. The number of determinable miRNAs was estimated by the IsGeneDetected parameter.
2.10. Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNA from tissues or cells was isolated utilizing a TRIzol kit (Thermo Fisher Science). For mRNA quantification, the isolated RNA was reverse-transcribed to cDNA using an RT kit (Thermo Fisher Scientific). After that, real-time qPCR was performed using LightCycler®96 on a FastStart™SYBR Green system (Roche Ltd., Basel, Switzerland). GAPDH was used as the internal reference. Expression of miRNAs was determined using a miRNeasy kit (Qiagen GmbH, Hilden, Germany) and miRCURY LNA miRNA PCR Assays (Qiagen), and U6 was used as the internal reference. Relative gene expression was measured by the 2−ΔΔCT method. The primers are listed in Table 1.
Table 1.
Primer sequences for RT-qPCR
| Gene | Primer sequence (5ʹ-3ʹ) |
|---|---|
| hsa-miR-188 | F: TCCCTTGCATGGTGGAG |
| R: GAACATGTCTGCGTATCTC | |
| hsa-JARID2 | F: GGACAAAGGCGTCCTCAATGAC |
| R: GCAGGCTCCTTGCTGAAACACA | |
| mmu-miR-188 | F: ATCCCTTGCATGGTGGAG |
| R: GAACATGTCTGCGTATCTC | |
| mmu-JARID2 | F: GCCTGCATAAAGAAGCACCTCAG |
| R: GAGGTCAGTCACTTGCTGCATG | |
| mmu-β-catenin | F: AGCCCAGTCCCATTAGTTGC |
| R: GATGCCTACTGACTCGCTCC | |
| mmu-miR-124 | F: GTGTTCACAGCGGACCTT |
| R: GAACATGTCTGCGTATCTC | |
| mmu-miR-130a | F: AGTGCAATGTTAAAAGGGCA |
| R: GAACATGTCTGCGTATCTC | |
| mmu-miR-410 | F: ATATAACACAGATGGCCTGT |
| R: GAACATGTCTGCGTATCTC | |
| mmu-miR-223 | F: GTCAGTTTGTCAAATACCCC |
| R: GAACATGTCTGCGTATCTC | |
| hsa-UBR7 | F: GAACAGGGAAAGGATGATGTCCG |
| R: AGCTCCTGAAGTTTGCAGCCAG | |
| hsa-FOXN2 | F: GGTTCCTTATGGTGTGTTGATCC |
| R: GTTTCGCACCTGGTTCTGCTTG | |
| hsa-CD2AP | F: CCAAAGCCTGAACTGATAGCTGC |
| R: GGACTTGTGGAGCTGCTGGTTT | |
| hsa-miR-124 | F: TTCACAGCGGACCTTGA |
| R: GAACATGTCTGCGTATCTC | |
| hsa-miR-130a | F: CACATTGTGCTACTGTCT |
| R: GAACATGTCTGCGTATCTC | |
| hsa-miR-410 | F: TATAACACAGATGGCCTG |
| R: GAACATGTCTGCGTATCTC | |
| hsa-miR-223 | F: CGTGTATTTGACAAGCTG |
| R: GAACATGTCTGCGTATCTC | |
| GAPDH | F: GTCTCCTCTGACTTCAACAGCG |
| R: ACCACCCTGTTGCTGTAGCCAA | |
| U6 | F: CTCGCTTCGGCAGCACAT |
| R: TTTGCGTGTCATCCTTGCG |
RT-qPCR, reverse transcription quantitative polymerase chain reaction; hsa, homo sapiens; mmu, mus musculus; miR-188, microRNA-188; JARID2, jumonji and AT-rich interaction domain containing 2; UBR7, ubiquitin protein ligase E3 component n-recognin 7; FOXN2, forkhead box N2; CD2AP, CD2 associated protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.; F: forward; R: reverse.
2.11. Western blot analysis
Cells were lyzed in cold radio-immunoprecipitation assay lysis buffer (Beyotime). An equal amount of protein sample was run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride membranes. After being blocked by 5% bovine serum albumin, the membranes were co-cultured with the primary antibodies against JARID2 (1:1,000, ab192252, Abcam), β-catenin (1:5,000, ab32572, Abcam), and GAPDH (1:2,000, ab128915, Abcam) at 4°C for 12 h, and then with the secondary antibody goat anti-rabbit immunoglobulin G (IgG) H&L (HRP) (1:10,000, ab205718, Abcam) at 37°C for 2 h. The signal of the target protein bands was examined using the Pierce ECL Western blotting substrate (Thermo Fisher Scientific), and the protein level was evaluated using the Image Pro Plus 6.0 software.
2.12. Dual-luciferase reporter gene assay
The putative binding sequence between miR-188 and the 3ʹUTR of JARID2 was first predicted in StarBase (http://starbase.sysu.edu.cn/). Next, the wild-type (WT) JARID2 3ʹUTR sequence was amplified and inserted into the psiCHECK™-2-vector (Promega, Corp., Madison, WI, USA) to establish JARID2-WT vector, and the mutant-type (MT) vector containing mutant sequence of JARID2 was constructed as well. These vectors were co-transfected with miR-188 mimic or negative control (NC) mimic into BSMCs using a Lipofectamine 3000 kit (Invitrogen) according to the protocol. After 48 h, the cells were harvested and lysed, and the luciferase activity in cells was examined using a Dual-Luciferase® Reporter 1000 Assay System (Promega).
2.13. RNA immunoprecipitation (RIP) assay
A RIP assay was performed using an EZ-magna RIP kit (Millipore Corp., Billerica, MA, USA) in accordance with the kit’s instructions. In brief, the BSMCs, at an 80–90% confluence, were lyzed and then incubated in complete RIP lysis buffer that contained magnet beads and anti-Argonaute-2 (Ago-2, Abcam) or anti-IgG. After that, the samples were incubated with proteinase K to isolate the immunoprecipitated RNA. Expression of miR-188 and JARID2 in the precipitate was determined by RT-qPCR.
2.14. A mouse model with asthma induced by OVA
Thirty BALB/c mice (6–8 weeks old, 20 ± 2 g) were acquired from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). Animals were housed in a temperature-controlled room in a 12-h dark/light cycle with ad libitum access to drinking water and standard feed. All animals were acclimated for 1 week before any experimental procedures. The mice were allocated into control group, OVA group and OVA + exo group, 10 in each group. For the OVA (Grade V, Sigma-Aldrich) treatment, mice were sensitized through an intraperitoneal injection of 20 μg OVA in 2 mg Alum Adjuvant (Thermo Fisher Scientific) in 200 μL sterile normal saline once a week for 2 weeks. After that, mice were challenged with 5% aerosolized OVA in saline through the respiratory tract three times a week from week 3 to week 10. Mice in the control group were treated with an equal volume of normal saline. Mice in the OVA + exo group were additionally injected with 20 μg/mL exosomes through caudal vein from week 7 to week 10 after OVA exposure. At 24 h after the last injection, the mice were euthanized through an intraperitoneal injection of 150 mg/kg pentobarbital sodium. The mouse lung tissues were collected. The right lungs were used for gene detection, and the left ones were used for pathological examination.
2.15. Pathological examination
The left lung tissues were fixed in 4% paraformaldehyde for 24 h, and then dehydrated, embedded in paraffin, cut into 4-μm sections, and dewaxed routinely. The sections were loaded on glass slides. After that, the inflammatory response in tissues was examined by hematoxylin and eosin (HE) staining (Sigma-Aldrich), and the mucus production in tissues was examined by periodic acid schiff (PAS) staining (Solarbio Science & Technology Co., Ltd., Beijing, China). The staining results were observed under the microscope.
2.16. Fluorescence in situ hybridization (FISH) assay
A miRNA FISH probe kit (Genepharma) was applied according to the manufacturer’s instructions to examine miR-188 expression. For cellular experiments, the cells were fixed in 4% paraformaldehyde and then incubated with 0.5% Triton X-100 on ice for 10 min. Next, the cells were pre-hybridized at 37°C for 30 min, and then with 1 µM miR-188 probe at 37°C overnight. After that, the cells were counter-stained with 4ʹ, 6-diamidino-2-phenylindole (1 µg/mL) for 10 min. The staining was visualized under a fluorescence microscope (Carl Zeiss, Oberkochen, Germany) and quantified using the ImageJ. The FISH assay on lung tissue sections (4 μm) was performed in a similar manner.
2.17. Data analysis
All statistical analyses were performed on Prism Version 8.0 (GraphPad, GraphPad, La Jolla, CA, USA). Data were collected from at least three independent experiments and presented as mean ± standard deviation (SD). Differences were analyzed by the t test (two groups) and one-way or two-way analysis of variance (ANOVA, over two groups). p less than 0.05 (p < 0.05) was considered to show a significant difference.
3. Results
3.1. Identification of the hBM-MSCs and their derived exosomes
The acquired hBM-MSCs were first identified using flow cytometry, which confirmed positive expression of CD105, CD73, and CD90 while negative expression of CD34, CD14, or CD19 in cells (Figure 1)). To further confirm the differentiation capacities of the hBM-MSCs, the cells were cultured and subjected to 3 weeks of induction of osteogenic differentiation and adipogenic differentiation. The alizarin red staining observed the formation of calcareous nodules, while the oil red O staining observed the formation of red oil droplets after adipogenic differentiation (Figure 1)), which indicated the differentiation potentials of the hBM-MSCs. The hBM-MSC-derived exosomes were collected after differential centrifugation. The particles were in typical oval shape under the TEM (Figure 1)). In addition, the subsequent nanoparticle tracking analysis confirmed the diameters of the particles were approximately 30–120 nm (Figure 1)). Furthermore, the presence of the exosome-specific proteins was determined by Western blot analysis, which suggested that the levels of CD81 and TSG101 were significantly higher in the exosomes than those in hBM-MSCs (Figure 1)).
Figure 1.
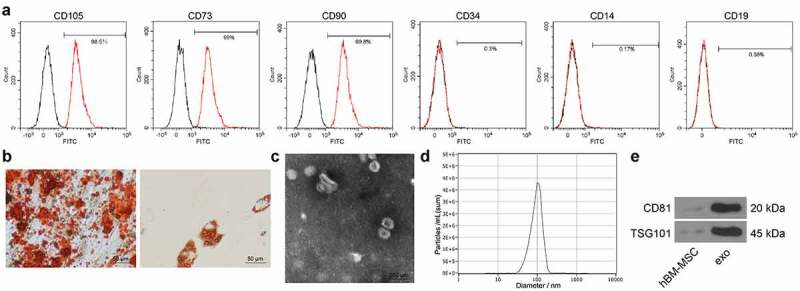
Identification of the hBM-MSCs and the derived exosomes. (a), expression of the surface marker proteins of hBM-MSCs determined by flow cytometry; (b), osteogenic and adipogenic differentiation abilities of hBM-MSCs examined by alizarin red staining (left) and oil red O staining (right), respectively; (c), shapes of the extracted particles observed under a TEM; (d), distribution of the particle size determined by nanoparticle tracking analysis; (e), protein levels of exosome-specific marker proteins CD81 and TSG101 determined by Western blot analysis. Three independent experiments were performed.
3.2. Exosomes inhibit proliferation and migration of TGF-β1-induced BSMCs
To explore whether exosomes can be absorbed by BSMCs, the exosomes were labeled by PKH26 and then co-cultured with the BSMCs (Figure 2)). It was found that the exosomes were absorbed by the BSMCs with the increase of time. After that, the BSMCs were treated with TGF-β1 to mimic an asthma-like condition in vitro, and this group was defined as the model group. The TGF-β1-induced BSMCs were further administrated with 20 μg exosomes. The CCK-8 assay suggested that TGF-β1 significantly increased proliferation of BSMCs, but this increase was blocked by the exosomes (Figure 2)). Likewise, the EdU labeling assay suggested that the DNA replication activity of the BSMCs was increased by TGF-β1 but then inhibited by exosomes (Figure 2)). The apoptosis of cells was further determined by flow cytometry. It was found that TGF-β1 reduced the number of apoptotic BSMCs, while further administration of exosomes increased the apoptosis of cells (Figure 2)). In addition, the Tranwell assay suggested that the migration of BSMCs was induced by TGF-β1 but blocked by exosomes (Figure 2)).
Figure 2.
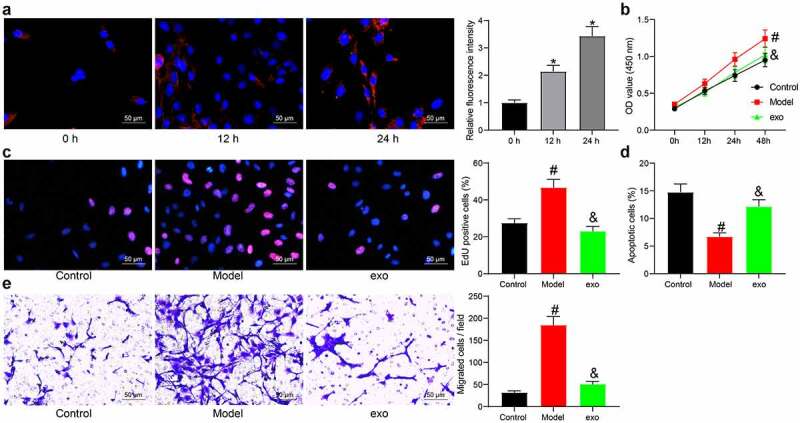
Exosomes inhibit proliferation and migration of TGF-β1-induced BSMCs. (a), absorption of exosomes by BMSCs examined by PKH26 labeling (one-way ANOVA, *p < 0.05 vs 0 h); (b), proliferation of cells determined by the CCK-8 method (two-way ANOVA, #p < 0.05 vs control; &p < 0.05 vs model); (c), DNA replication activity of cells determined by EdU labeling assay (one-way ANOVA, #p < 0.05 vs control; &p < 0.05 vs model); (d), apoptosis of cells determined by flow cytometry (one-way ANOVA, #p < 0.05 vs control; &p < 0.05 vs model); (e), migration ability of cells measured by Transwell assay (one-way ANOVA, #p < 0.05 vs control; &p < 0.05 vs model). Data were presented as mean ± SD from three independent experiments.
3.3. Exosomes convey miR-188 into BSMCs
To confirm the potential cargo delivered by exosomes that was involved in the above events, we performed a miRNA microarray analysis. First, we found that the content of recognizable miRNA in exosomes was increased with the increase of the exosome concentration, and a dose of 20 μg was sufficient enough for miRNA detection (Figure 3)). Next, we used this dose of exosomes to treat the BSMCs (Model group vs exo group), and the RNA sample from cells was collected and hybridized to screen the differentially expressed miRNAs. The top five differentially expressed miRNAs with good recognition are presented in Figure 3). miR-188, which was suggested as the most differentially expressed in exosome-treated BSMCs, was selected for the subsequent investigation. Next, the expression of miR-188 in the exosome resuspension and in the supernatant collected after the final centrifugation was determined using RT-qPCR, and an enrichment of miR-188 was found in the exosome resuspension (Figure 3)). Moreover, the FISH assay results indicated that the exosome treatment significantly elevated the miR-188 expression in BSMCs in a time-dependent manner (Figure 3)).
Figure 3.
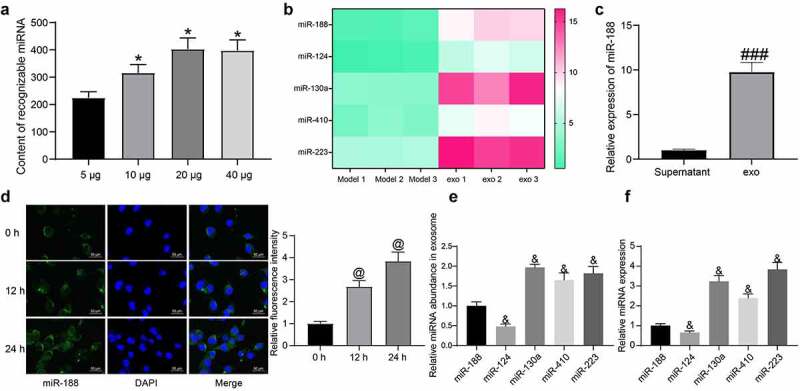
Exosomes convey miR-188 into BSMCs. (a), impact of different concentration of exosomes on the hybridization effect of miRNA microarrays (one-way ANOVA, *p < 0.05 vs 5 μg); (b), top five differentially expressed miRNAs in BSMCs after exosome treatment; (c), miR-188 expression in exosome resuspension and in the supernatant collected after the final centrifugation determined by RT-qPCR (unpaired t test, ###p < 0.001 vs supernatant); (d), in-situ miR-188 expression in BSMCs after exosome treatment determined by the FISH assay (one-way ANOVA, @p < 0.05 vs 0 h); (e), relative abundance of the five miRNAs in exosomes examined by RT-qPCR (one-way ANOVA, &p < 0.05 vs miR-188); (f), basic expression of the five miRNAs in TGF-β1-treated BMSCs (&p < 0.05 vs miR-188). Data were presented as mean ± SD from three independent experiments.
The abundance of the five miRNAs in exosomes was then analyzed. However, it was observed that miR-188 was not the miRNA showing the highest abundance in exosomes (Figure 3)). RT-qPCR further showed that the basic expression of miR-188 in BMSCs without exosome treatment was quite low among the five miRNAs (Figure 3)). These results showed that in addition to the abundance of miRNA in exosomes, the effect of exosome treatment on miRNA differential expression is also related to the basic expression of miRNA in recipient cells.
3.4. Downregulation of miR-188 blocks the effects of exosomes on BSMCs
To confirm the role of miR-188 in exosomes, miR-188 inhibitor and the NC inhibitor were transfected into hBM-MSCs, and the successful transfection was confirmed by RT-qPCR (Figure 4)). Then, the exosomes from these hBM-MSCs were extracted and named exo-inhibitor and exo-NC, respectively. Thereafter, the RT-qPCR results identified decreased expression of miR-188 in exo-inhibitor (Figure 4)). BSMCs in the model group (treated with TGF-β1) were treated with exo-inhibitor and exo-NC, respectively. Thereafter, the FISA assay results suggested that the expression of miR-188 in exo-inhibitor-treated BSMCs was significantly reduced compared to the exo-NC treated cells (Figure 4)), The CCK-8 method showed that the proliferation ability of BSMCs was increased after exo-inhibitor treatment (Figure 4)). Likewise, the DNA replication ability of cells was enhanced as well when miR-188 was inhibited (Figure 4)). The flow cytometry results further showed that the pro-apoptotic function of exosomes on TGF-β1-treated BSMCs was blocked when miR-188 was suppressed (Figure 4)). In addition, the Transwell assay suggested that the migration potential of the model BSMCs was enhanced after exo-inhibitor treatment (Figure 4)).
Figure 4.
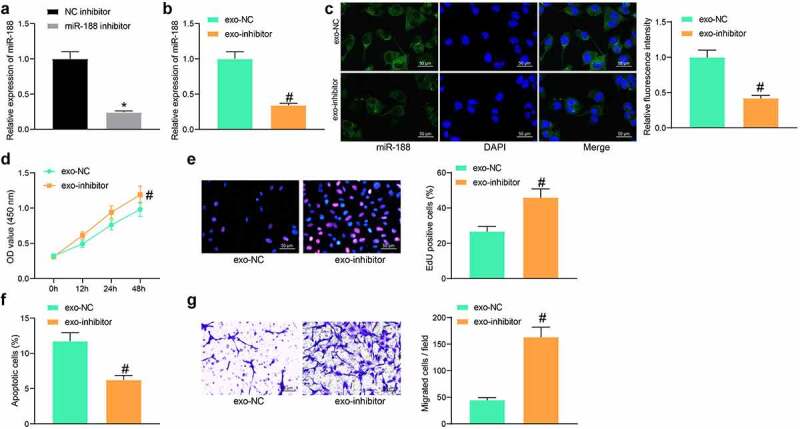
Downregulation of miR-188 blocks the effects of exosomes on BSMCs. (a), expression of miR-188 in hBM-MSCs after miR-188 inhibitor transfection determined by RT-qPCR (unpaired t test, *p < 0.05); (b), miR-188 expression in exo-inhibitor and in exo-NC determined by RT-qPCR (unpaired t test, #p < 0.05); (c), in-situ miR-188 expression in BSMCs after exosome treatment determined by the FISH assay (unpaired t test, #p < 0.05); (d), proliferation of cells determined by the CCK-8 method (unpaired t test, #p < 0.05); (e), DNA replication activity of cells determined by EdU labeling assay (unpaired t test, #p < 0.05); (f), apoptosis of cells determined by flow cytometry (unpaired t test, #p < 0.05); (g), migration ability of cells measured by Transwell assay (unpaired t test, #p < 0.05). Data were presented as mean ± SD from three independent experiments.
3.5. miR-188 directly targets JARID2
The focus of the study was then shifted to identifying potential downstream molecules. We then predicted the candidate mRNA targets of miR-188 using five bioinformatics systems including miRDB (http://mirdb.org/), TargetScan (http://www.targetscan.org/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), StarBase, and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php). A total of four genes were found to be intersected, including UBR7, JARID2, FOXN2 and CD2AP (Figure 5)). After that, miR-188 mimic and NC mimic were transfected into BSMCs, and the successful transfection was confirmed by RT-qPCR again (Figure 5)). Thereafter, we examined the mRNA expression of UBR7, JARID2, FOXN2 and CD2AP in cells using RT-qPCR. Among the four candidate genes, only JARID showed a decline in cells after miR-188 mimic transfection (Figure 5)). The Western blot analysis additionally confirmed a decline in the protein level of JARID2 in cells after miR-188 upregulation (Figure 5)). To further verify the binding relationship between miR-188 and JARID2, we obtained the putative binding sequence between miR-188 and JARID2 from StarBase (Figure 5)), based on which JARID2-WT or JARID-MT luciferase reporter vectors were constructed. The vectors were co-transfected with miR-188 mimic or NC mimic into BSMCs. Forty-eight hours later, it was found that the luciferase activity in cells co-transfected with miR-188 mimic and JARID-WT was notably declined (Figure 5)), while other transfections led to no changes in luciferase activity in cells. In addition, an RIP assay was performed, which suggested that anti-Ago2 could combine and enrich miR-188 and JARID2 compared to anti-IgG, and miR-188 mimic further enhanced the enrichments of miR-188 and JARID2 by anti-Ago2 (Figure 5)).
Figure 5.
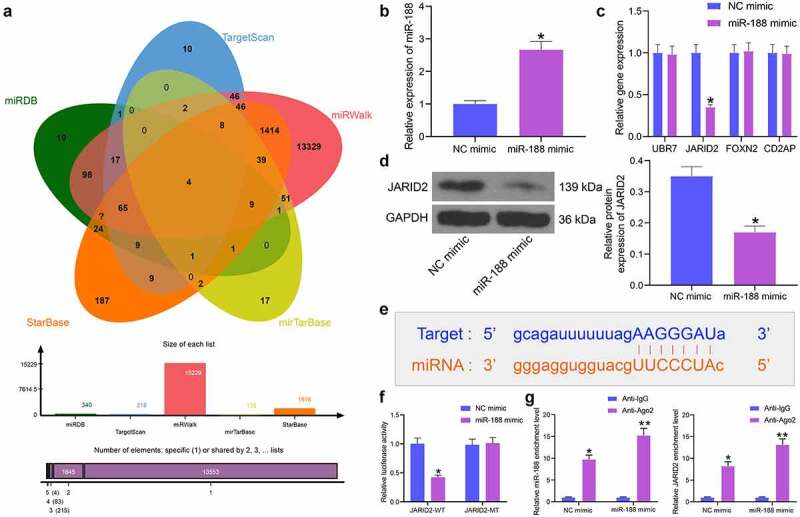
miR-188 directly targets JARID2. (a), a Venn diagram for the intersected target mRNAs of miR-188 predicted using five bioinformatic systems; (b), transfection efficiency of miR-188 in BSMCs determined by RT-qPCR (unpaired t test, *p < 0.05); (c), mRNA expression of UBR7, JARID2, FOXN2 and CD2AP in cells detected by RT-qPCR (two-way ANOVA,*p < 0.05); (d), protein expression of JARID2 in cells measured by Western blot analysis (unpaired t test, *p < 0.05); (e), putative binding sequence between miR-188 and JARID2 obtained from StarBase; (f), binding relationship between miR-188 and JARID2 validated through a dual luciferase reporter gene assay (two-way ANOVA,*p < 0.05); (g), binding relationship between miR-188 and JARID2 validated through an RIP assay (two-way ANOVA, *p < 0.05, **p < 0.01). Data were presented as mean ± SD from three independent experiments.
3.6. Overexpression of JARID2 activates the Wnt/β-catenin pathway and blocks the effects of exosomes on BSMCs
As previously reported, JARID2 is a potential activator of the Wnt/β-catenin pathway [25], whose aberrant activation might promote abnormal proliferation of ASMCs and airway remodeling [26,27]. To explore whether JARID2 exerts a similar function here, overexpression of JARID2 was further introduced in BMSCs in different groups through administration of pcDNA-JARID2, followed by the administration of the Wnt/β-catenin inhibitor XAV-939. The transfection efficiency of pcDNA-JARID2 was identified by RT-qPCR again. The pcDNA-JARID2 significantly elevated the JARID2 expression in exosome-treated BSMCs, but XAV-939 did not affect the JARID2 expression in cells (Figure 6)). Then, the protein level of β-catenin in cells was determined by Western blot analysis. In model cells treated with TGF-β1, the β-catenin expression was significantly increased, whereas exosomes blocked the abnormal activity of β-catenin. This signaling pathway was reactivated upon the additional upregulation of JARID2, but the XAV-939 treatment suppressed the β-catenin level elevated by JARID2 (Figure 6)).
Figure 6.
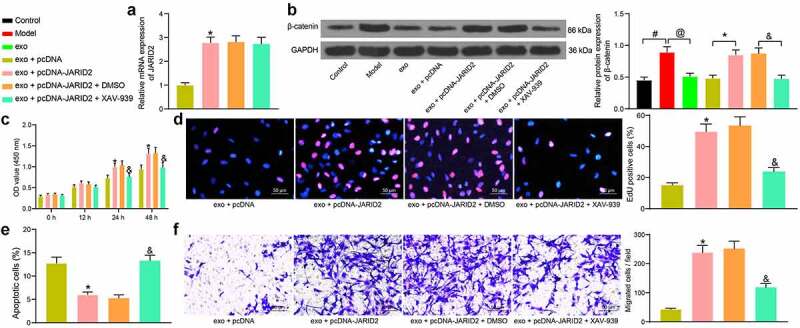
Overexpression of JARID2 activates the Wnt/β-catenin pathway and blocks the effects of exosomes on BSMCs. (a), mRNA expression of JARID2 in hBM-MSCs after pcDNA-JARID2 transfection and XAV-939 treatment determined by RT-qPCR (one-way ANOVA, *p < 0.05 vs exo + pcDNA); (b), protein level of β-catenin in cells evaluated by Western blot analysis (one-way ANOVA, #p < 0.05 vs control, @p < 0.05 vs model, *p < 0.05 vs exo + pcDNA, &p < 0.05 vs exo + pcDNA-JARID2 + DMSO); (c), proliferation of cells determined by the CCK-8 method (two-way ANOVA, *p < 0.05 vs exo + pcDNA; &p < 0.05 vs exo + pcDNA-JARID2 + DMSO); (d), DNA replication activity of cells determined by EdU labeling assay (one-way ANOVA, *p < 0.05 vs exo + pcDNA; &p < 0.05 vs exo + pcDNA-JARID2 + DMSO); (e), apoptosis of cells determined by flow cytometry (one-way ANOVA, *p < 0.05 vs exo + pcDNA group; &p < 0.05 vs exo + pcDNA-JARID2 + DMSO); (f), migration ability of cells measured by Transwell assay (one-way ANOVA, *p < 0.05 vs exo + pcDNA; &p < 0.05 vs exo + pcDNA-JARID2 + DMSO). Data were presented as mean ± SD from three independent experiments.
Rescue experiments concerning the functions of JARID2 and the Wnt/β-catenin pathway were performed. The exosome-treated BSMCs were further transfected with pcDNA-JARID2, followed by XAV-939 or DMSO treatment. The CCK-8 and EdU labeling assays suggested that the proliferation and DNA replication activity of cells were recovered after JARID2 overexpression but suppressed again by XAV-939 (Figure 6). The flow cytometry results showed that the apoptosis of BSMCs induced by exosomes was reduced by JARID2, but the cell apoptosis was re-activated after XAV-939 treatment (Figure 6)). According to the Transwell assay, overexpression of JARID2 increased the migratory potential of the BSMCs, but the cell migration was reduced following XAV-939-mediated Wnt/β-catenin inhibition (Figure 6)). These results validated that the inhibitory function of exosomal miR-188 in cells was achieved through the inhibition of the JARID2/Wnt/β-catenin axis.
3.7. Exosomes relieve lung injury in mice induced with OVA
Animal experiments were performed to verify the protective function of exosomes in vivo. A mouse model was induced by OVA treatment and followed by exosome treatment (Figure 7)). The expression of miR-188, JARID2 and β-catenin in the homogenate of mouse lung tissues was determined by RT-qPCR. It was found that miR-188 was downregulated, while JARID2 and β-catenin were upregulated in mouse lung tissues after OVA exposure. These trends were reversed by the additional treatment of exosomes (Figure 7)). The FISH assay also suggested that the in-situ expression of miR-188 in the airway tissues of OVA-treated mice was significantly reduced, but exosome treatment elevated miR-188 expression in the tissues (Figure 7)). The pathological presentations in the lung airway tissues were examined (Figure 7)). Compared to the normal ones, the OVA-challenged mice showed significant inflammatory cell infiltration (HE staining) and increased mucus production (PAS staining). All these pathological changes were relieved by the exosome treatment. These results indicated that the hBM-MSC-derived exosomes showed a similar protective function against lung injury in asthmatic mice.
Figure 7.
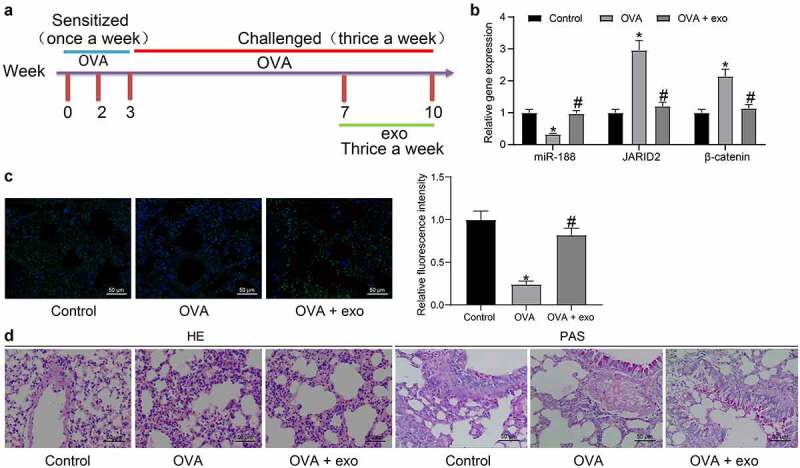
Exosomes relieve lung injury in mice induced with OVA. (a), a diagram for the establishment of the mouse model with OVA-induced asthma and the time points for different treatments; (b), expression of miR-188 and mRNA expression of JARID2 and β-catenin in the homogenate of mouse lung tissues determined by RT-qPCR (two-way ANOVA, *p < 0.05 vs control, #p < 0.05 vs OVA); (c), expression of miR-188 in the lung airway tissues of mice examined by the FISH assay (one-way ANOVA, *p < 0.05 vs control, #p < 0.05 vs OVA); (d), pathological changes in mouse lung tissues determined by HE staining and PAS staining. N = 10 in each group. Data were presented as mean ± SD from three independent experiments.
4. Discussion
The lung is a complex organ encompassing an extend array of structural and immune cells in the parenchyma and airway. Intercellular communication is critical for lung function, and exosomes are hopeful tools in lung biology and in asthma development [28]. Airway remodeling is a well-recognized characteristic of asthma induced by proliferation and hypertrophy of smooth muscle, and increased smooth muscle cell diameter is frequently found in airways of patients with severe asthma [29,30]. This study demonstrates that hBM-MSC-derived exosomes carry miR-188 to reduce aberrant proliferation and migration of BSMCs through suppressing the JARID2/Wnt/β-signaling pathway. The exosomes also have protective functions against OVA-induced lung injury in mice.
Studies have revealed the anti-inflammatory and anti-tissue remodeling functions of MSCs in allergic asthma [31,32]. The research by Bonfield et al. suggested that administration of BM-MSCs alleviated airway inflammation in an OVA-challenged asthmatic mouse model [33]. However, safety concerns over MSC injection such as the following uncontrolled cell replication, vascular occlusion, and potential mal-differentiation of MSCs might not be overlooked, leaving the exosomes that contain biologically active molecules with less disadvantages as alternative tools [34]. Importantly, the MSC secretomes including Evs, have shown therapeutic potentials in several animal models recently [35,36]. The anti-inflammatory and anti-apoptotic properties of the MSC-derived exosomes in cardiovascular diseases also indicated their potential effects on other chronic inflammatory conditions [37]. The BM-MSC-derived exosome has recently been reported as a promising therapeutic candidate for severe COVID-19 [38]. In addition, clinical trials are being conducted concerning the clinical potential of exosome-based treatment for acute respiratory distress syndrome [39]. In the present study, after identification of the extracted exosomes using the specific surface biomarkers CD81 and TSG101. The hBM-MSC-derived exosomes were used to treated the BSMCs, after which the abnormal proliferation and migration of BSMCs were decreased while the apoptosis of cells was encouraged. In the OVA-challenged mice, the exosomes relieved inflammatory cell infiltration, mucus production and collagen deposition in murine lung tissues. These results indicated that the hBM-MSC-derived exosomes potentially reduce airway remodeling and inflammatory responses and alleviate the pathogenesis of asthma.
miRNAs are a major type of exosome cargoes and delivered into target cells to exert diverse functions, and they are frequently involved in the physiological and pathological process in severe asthma [19,40,41]. For instance, exosomal miR-1470 from MSCs was found to enhance the proportion of CD4+CD25+FOXP3+ regulatory T cells in asthmatic patients [42]. Therefore, we performed a miRNA microarray analysis to analyze the enriched miRNAs. It was found that miR-188 was upregulated by approximately 10 times in BSMCs after the exosome treatment. A study by Andrew R et al. confirmed decreased expression of miR-188 in ASMCs after a proinflammatory stimulus [22]. To confirm the biological function of miR-188 in asthma, downregulation of miR-188 was further introduced in the TGF-β1-treated BSMCs second to exosome treatment. Then, the anti-proliferative and pro-apoptotic functions of exosomes were blocked. Expression of miR-188 was also confirmed to be reduced the murine lung tissues after OVA challenge but recovered after exosome treatment. These results indicated that miR-188 is at least partially responsible for the protective events of hBM-MSC-derived exosomes in asthma.
Our subsequent bioinformatic analyses using five prediction systems suggested UBR7, JARID2, FOXN2 and CD2AP as four common target genes of miR-188, while only JARID2 showed a significant decline in BSMCs after miR-188 upregulation. The binding between miR-188 and JARID2 was further confirmed using luciferase reporter and RIP assays. JARID has been demonstrated to participate in the EMT process of lung cancer cells [23,43]. Here, to explore whether JARID2 affects asthma progression, similar rescue experiments were performed through upregulating JARID2 in the TGF-β1-treated BSMCs after exosome treatment. Consequently, high JARID2 expression blocked the effects of exosomes again. The JARID2 has been reported as a positive regulator of the Wnt/β-catenin pathway [25]. This signaling is a fundamental mechanism participating in multiple biological activities including cell proliferation, development and morphogenesis, while its regulation in the development of airway remodeling has been also reported in asthmatic patients [44]. Downregulation of this pathway alleviated airway remodeling in asthmatic rats [26]. In this paper, we observed that the β-catenin expression in BSMCs was positively regulated by JARID2 as well. Also, high activity of β-catenin in the OVA-challenged mice was suppressed by exosomes, indicating that the exosomes potentially suppresses the JARID2/Wnt/β-catenin axis through carrying miR-188.
In summary, the present study demonstrates that exosomes derived from hBM-MSCs might suppress airway remodeling and lung injury, thereby suppressing the pathological development of asthma through conveying miR-188 and the subsequent suppression of the JARID2/Wnt/β-catenin axis (Figure 8). These findings offered novel evidence that exosomes showed potent protective functions against asthma development.
Figure 8.
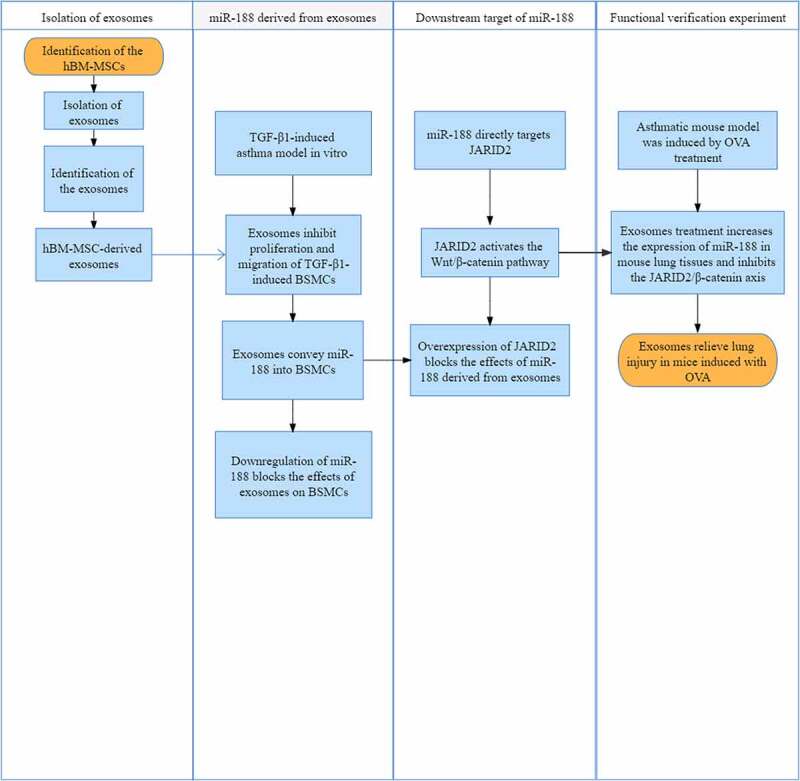
The study roadmap.
Acknowledgments
The authors are thankful to Guiding plan of Liaoning Natural Science Foundation (2019-ZD-0778).
Funding Statement
This work was supported by Guiding plan of Liaoning Natural Science Foundation [2019-ZD-0778].
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
References
- [1].Nanda A, Wasan AN.. Asthma in adults. Med Clin North Am. 2020;104:95–108. [DOI] [PubMed] [Google Scholar]
- [2].Schatz M, Rosenwasser L. The allergic asthma phenotype. J Allergy Clin Immunol Pract. 2014;2:645–648. quiz 9. [DOI] [PubMed] [Google Scholar]
- [3].Boonpiyathad T, Sozener ZC, Satitsuksanoa P, et al. Immunologic mechanisms in asthma. Semin Immunol. 2019;46:101333. [DOI] [PubMed] [Google Scholar]
- [4].Wang YC, Jaakkola MS, Lajunen TK, et al. Asthma-COPD overlap syndrome among subjects with newly diagnosed adult-onset asthma. Allergy. 2018;73:1554–1557. [DOI] [PubMed] [Google Scholar]
- [5].Peters MC, Wenzel SE. Intersection of biology and therapeutics: type 2 targeted therapeutics for adult asthma. Lancet. 2020;395:371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kepil Ozdemir S, Bavbek S. Prospects for new and emerging therapeutics in severe asthma: the role of biologics. Expert Rev Respir Med. 2017;11:505–512. [DOI] [PubMed] [Google Scholar]
- [7].Li X. Hot topic: precision medicine for asthma-has the time come? Curr Allergy Asthma Rep. 2019;19:45. [DOI] [PubMed] [Google Scholar]
- [8].Opina MT, Moore WC. Phenotype-driven therapeutics in severe asthma. Curr Allergy Asthma Rep. 2017;17:10. [DOI] [PubMed] [Google Scholar]
- [9].Hough KP, Deshane JS. Exosomes in allergic airway diseases. Curr Allergy Asthma Rep. 2019;19:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Karabey B, Daglioglu ST, Azbar N, et al. Bacterial and archeal dynamics of a labscale HYBRID gas fermentation bioreactor fed with CO2 and H2. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2019;54:1348–1355. [DOI] [PubMed] [Google Scholar]
- [11].Porro C, Lepore S, Trotta T, et al. Isolation and characterization of microparticles in sputum from cystic fibrosis patients. Respir Res. 2010;11:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hass R, Kasper C, Bohm S, et al. Different populations and sources of human mesenchymal stem cells (MSC): a comparison of adult and neonatal tissue-derived MSC. Cell Commun Signal. 2011;9:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Makinde T, Murphy RF, Agrawal DK. The regulatory role of TGF-beta in airway remodeling in asthma. Immunol Cell Biol. 2007;85:348–356. [DOI] [PubMed] [Google Scholar]
- [14].Shi S, Jin L, Zhang S, et al. MicroRNA-590-5p represses proliferation of human fetal airway smooth muscle cells by targeting signal transducer and activator of transcription 3. Arch Med Sci. 2018;14:1093–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Woodman L, Siddiqui S, Cruse G, et al. Mast cells promote airway smooth muscle cell differentiation via autocrine up-regulation of TGF-beta 1. J Immunol. 2008;181:5001–5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Fang P, Xue Y, Zhang Y, et al. SIRT7 regulates the TGF-beta1-induced proliferation and migration of mouse airway smooth muscle cells by modulating the expression of TGF-beta receptor I. Biomed Pharmacother. 2018;104:781–787. [DOI] [PubMed] [Google Scholar]
- [17].Yang Z, Qu Z, Yi M, et al. MiR-204-5p inhibits transforming growth factor-beta1-induced proliferation and extracellular matrix production of airway smooth muscle cells by regulating six1 in asthma. Int Arch Allergy Immunol. 2020;181:239–248. [DOI] [PubMed] [Google Scholar]
- [18].Kim DI, Song MK, Lee K. Comparison of asthma phenotypes in OVA-induced mice challenged via inhaled and intranasal routes. BMC Pulm Med. 2019;19:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].van Den Berge M, Tasena H. Role of microRNAs and exosomes in asthma. Curr Opin Pulm Med. 2019;25:87–93. [DOI] [PubMed] [Google Scholar]
- [20].Lv DQ, Li HY, Wu XM, et al. MiR-188 inhibits proliferation and promotes apoptosis of lung adenocarcinoma cells by targeting SIX1 to negatively regulate ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:721–727. [DOI] [PubMed] [Google Scholar]
- [21].Yang X, Wang B, Chen W, et al. MicroRNA-188 inhibits biological activity of lung cancer stem cells through targeting MDK and mediating the Hippo pathway. Exp Physiol. 2020;105:1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kuhn AR, Schlauch K, Lao R, et al. MicroRNA expression in human airway smooth muscle cells: role of miR-25 in regulation of airway smooth muscle phenotype. Am J Respir Cell Mol Biol. 2010;42:506–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tange S, Oktyabri D, Terashima M, et al. JARID2 is involved in transforming growth factor-beta-induced epithelial-mesenchymal transition of lung and colon cancer cell lines. PLoS One. 2014;9:e115684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huang SM, Mishina YM, Liu S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. [DOI] [PubMed] [Google Scholar]
- [25].Zhu F, Li H, Liu Y, et al. miR-155 antagomir protect against DSS-induced colitis in mice through regulating Th17/Treg cell balance by Jarid2/Wnt/beta-catenin. Biomed Pharmacother. 2020;126:109909. [DOI] [PubMed] [Google Scholar]
- [26].Huang Y, Wang L, Jia XX, et al. Vitamin D alleviates airway remodeling in asthma by down-regulating the activity of Wnt/beta-catenin signaling pathway. Int Immunopharmacol. 2019;68:88–94. [DOI] [PubMed] [Google Scholar]
- [27].Liu Z, Mei L, He Z. Long non-coding RNA00882 contributes to platelet-derived growth factor-induced proliferation of human fetal airway smooth muscle cells by enhancing Wnt/beta-catenin signaling via sponging miR-3619-5p. Biochem Biophys Res Commun. 2019;514:9–15. [DOI] [PubMed] [Google Scholar]
- [28].Canas JA, Sastre B, Rodrigo-Munoz JM, et al. Exosomes: a new approach to asthma pathology. Clin Chim Acta. 2019;495:139–147. [DOI] [PubMed] [Google Scholar]
- [29].Ojiaku CA, Cao G, Zhu W, et al. TGF-beta1 evokes human airway smooth muscle cell shortening and hyperresponsiveness via Smad3. Am J Respir Cell Mol Biol. 2018;58:575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang M, Du Y, Xu Z, et al. Functional effects of WNT1-Inducible signaling pathway protein-1 on bronchial smooth muscle cell migration and proliferation in OVA-induced airway remodeling. Inflammation. 2016;39:16–29. [DOI] [PubMed] [Google Scholar]
- [31].Hong GH, Kwon HS, Lee KY, et al. hMSCs suppress neutrophil-dominant airway inflammation in a murine model of asthma. Exp Mol Med. 2017;49:e288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun YQ, Deng MX, He J, et al. Human pluripotent stem cell-derived mesenchymal stem cells prevent allergic airway inflammation in mice. Stem Cells. 2012;30:2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bonfield TL, Koloze M, Lennon DP, et al. Human mesenchymal stem cells suppress chronic airway inflammation in the murine ovalbumin asthma model. Am J Physiol Lung Cell Mol Physiol. 2010;299:L760–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Du YM, Zhuansun YX, Chen R, et al. Mesenchymal stem cell exosomes promote immunosuppression of regulatory T cells in asthma. Exp Cell Res. 2018;363:114–120. [DOI] [PubMed] [Google Scholar]
- [35].Cosenza S, Toupet K, Maumus M, et al. Mesenchymal stem cells-derived exosomes are more immunosuppressive than microparticles in inflammatory arthritis. Theranostics. 2018;8:1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shigemoto-Kuroda T, Oh JY, Kim DK, et al. MSC-derived extracellular vesicles attenuate immune responses in two autoimmune murine models: type 1 diabetes and uveoretinitis. Stem Cell Reports. 2017;8:1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Huang L, Ma W, Ma Y, et al. Exosomes in mesenchymal stem cells, a new therapeutic strategy for cardiovascular diseases? Int J Biol Sci. 2015;11:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sengupta V, Sengupta S, Lazo A, et al. Exosomes derived from bone marrow mesenchymal stem cells as treatment for severe COVID-19. Stem Cells Dev. 2020;29:747–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wilson JG, Liu KD, Zhuo H, et al. Mesenchymal stem (stromal) cells for treatment of ARDS: a phase 1 clinical trial. Lancet Respir Med. 2015;3:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mortaz E, Alipoor SD, Varahram M, et al. Exosomes in severe asthma: update in their roles and potential in therapy. Biomed Res Int. 2018;2018:2862187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Mousavi SR, Ahmadi A, Jamalkandi SA, et al. Involvement of microRNAs in physiological and pathological processes in asthma. J Cell Physiol. 2019;234:21547–21559. [DOI] [PubMed] [Google Scholar]
- [42].Zhuansun Y, Du Y, Huang F, et al. MSCs exosomal miR-1470 promotes the differentiation of CD4(+)CD25(+)FOXP3(+) Tregs in asthmatic patients by inducing the expression of P27KIP1. Int Immunopharmacol. 2019;77:105981. [DOI] [PubMed] [Google Scholar]
- [43].Terashima M, Tange S, Ishimura A, et al. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292:82–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kwak HJ, Park DW, Seo JY, et al. The Wnt/beta-catenin signaling pathway regulates the development of airway remodeling in patients with asthma. Exp Mol Med. 2015;47:e198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


