ABSTRACT
Bladder cancer (BC) is one of the most common cancers world-wide with a poor prognosis. Non-SMC (Structural Maintenance of Chromosomes)-condensin I complex subunit H (NCAPH) is a regulatory subunit of the condensin I complex and plays an important role in tumorigenesis and progression in several types of cancers. However, the role of NCAPH in BC remains unknown. In this study, we tried to reveal the biological functions of NCAPH in BC. We detected the expressions of NCAPH in BC and adjacent tissues, and BC cells lines. Subsequently, the gain- and loss-of-function experiments were performed to determine the effects of NCAPH on BC cell proliferation, apoptosis, and activation of the MEK/ERK signaling pathway in vitro. Moreover, we used BALB/c nude mice and established a xenograft model to investigate whether silence NCAPH using shRNA targeting NCAPH (shNCAPH) can inhibit BC tumor growth in vivo. The results showed NCAPH was overexpressed in BC tissues compared to adjacent tissues and highly expressed in BC cell lines. Additionally, overexpression of NCAPH promoted cell proliferation and inhibited apoptosis in SW780 cells. Conversely, knockdown of NCAPH reduced cell proliferation and enhanced apoptosis in UMUC3 cells. Furthermore, we found that the NCAPH activated the MEK/ERK signaling pathway in BC cells. MEK1/2 inhibitor U0126 blocked the increase of cell proliferation regulated by NCAPH overexpression. Knockdown of NCAPH significantly inhibited tumor growth in mice. Our results suggest that NCAPH might play an important role in BC progression and provide the potential marker in the diagnosis of BC.
KEYWORDS: NCAPH, bladder cancer, MEK/ERK
Introduction
Bladder cancer (BC) is the 10th most common cancer world-wide, with a yearly incidence of approximately 549,000 cases in 2018 [1]. Bladder cancer ranks higher among men and it is the sixth most common cancer world-wide in men and ninth leading cause of cancer death [1]. Many efforts were made to study the cancer risk factors and develop the prevention and treatment [2,3]. However, the mechanisms for bladder cancer occurrence and development still remain unclear and it is worthwhile to expend the effort to study them and provide the potential marker in the diagnosis and gene treatment for bladder cancer.
Condensin plays a key role in mitotic chromosome assembly and segregation during mitosis and there are two subtype condensins including condensin I and Condensin II. Condensin I and condensin II both contain a set of subunits [4]. Condensin I protein complex is composed of two structural maintenances of chromosomes 2 (SMC2) and SMC4 and three non-SMC-condensin I complex subunits NCAPD2, NCAPG, and NCAPH [5]. In eukaryotes, there are at least six SMC family proteins (SMC1-SMC6) and they form three heterodimers. SMC1-SMC3 heterodimer form the cohesin complex with sister-chromatid cohesion proteins Scc1 and Scc3 to mediate sister-chromatid cohesion. Another complex containing SMC5-SMC6 heterodimer takes part in DNA repair and checkpoint responses. SMC2-SMC4 heterodimer functions as the core both in condensin I protein complex and condensin II protein complex to mediate chromosome condensation and segregation during mitosis [6–9]. The functions of condensin I complex subunits on the cancer progression have been reported. It has been proven that the SMC2 and SMC4 could promote cancer cell proliferation [10–13]. Additionally, condensin I complex subunit NCAPD2 and NCAPG also function as oncogenic genes in cancer development [14–17].
In a recent study, it has also been found that NCAPH serves as an oncogene in various types of cancers. For instance, NCAPH is highly expressed in endometrial cancer, breast cancer, and prostate cancer tissues compared with normal tissues and it is associated with poor prognosis in these cancers [18–20]. Moreover, NCAPH promotes cell proliferation and migration in many cancers [21–23]. Knockdown of NCAPH induces cell-cycle arrest at the G2/M phase and increases apoptosis in non-small-cell lung cancer and colon cancer cells [24,25]. However, the functions of NCAPH in BC are still unknown. Therefore, in this present study, we tried to uncover the expression of NCAPH in BC tissues and the effects of NCAPH on cell proliferation and apoptosis in BC cells. In addition, it is well known that activation of the MEK/ERK signaling pathway promotes oncogenesis through regulating cell proliferation and apoptosis in bladder cancer [26–28]. Shrestha et al. found that the expressions of condensin complex members including NCAPH were increased in MEK-inhibitor (MEKi)–resistant cell lines in low-grade serous ovarian carcinoma [29]. Moreover, inhibition of MEK/ERK signaling pathway decreases the expressions of cyclin-dependent kinase 1 (CDK1) and its activator cyclin B1 [30,31]. Suppression of CDK1 and cyclin B1 could induce cell-cycle arrest at the G2/M phase [32,33]. Therefore, we determined that whether NCAPH affects the cell proliferation and aoptosis and activation of the MEK/ERK signaling pathway in BC cells in vitro. Moreover, we used a previously established xenograft model using BALB/c nude mice with small modification [34] and injected with shRNA targeting NCAPH (shNCAPH) stable transfected cell line to investigate whether silence NCAPH can inhibit BC tumor growth in vivo. Hopefully, our findings could provide a potential marker in the diagnosis of BC.
Materials and methods
GEPIA database analysis
GEPIA, an online database, is used to analyze RNA-seq data of tumor tissue and normal tissue samples from the TCGA and GTEx database (http://gepia.cancer-pku.cn/index.html). Therefore, we used GEPIA to obtain the expressions of NCAPH in BC tissues and normal tissues. Cancer type was bladder urothelial carcinoma (BLCA) and the log scale log2 (TPM+1) was used to transform the expression data for plotting. Then relative the expressions of NCAPH in BC tissues (n = 404) and normal tissues (n = 28) were obtained. * represents p value (P) < 0.05.
Tissue specimens and cell lines
In the current study, 30 pairs of human BC tumor tissues and adjacent tissues and a total of 60 BC tumor tissues from Biobank, Shengjing Hospital of China Medical University were used for detecting mRNA expression of NCAPH (NM_015341.5) by quantitative real-time polymerase chain reaction (qRT-PCR). Fourteen pairs of paraffin-embedded BC tumor tissues and adjacent tissues were used for detecting protein expression of NCAPH using immunohistochemistry (IHC). Prior patient consents were obtained and the study was approved by the ethics committee of the Shengjing Hospital of China Medical University, and all experiments were conducted in accordance with the provisions of the Declaration of Helsinki. The human ureteral epithelial SV-HUC-1 cells and BC cell lines UMUC3, 5637 and SW780 were purchased from the Procell (Wuhan, China). The BC cell line J82 was purchased from the iCell Bioscience Inc (Shanghai, China).
Cell culture and transfection
SV-HUC-1 cells were grown in Ham’s F-12 K medium (PM150910, Procell) and UMUC3 and J82 cells were cultured in MEM medium (PM150410, Procell). A total of 5637 cells were maintained in RPMI-1640 medium (31,800–014, Gibco, Grand Island, NY, USA) and SW780 cells were cultured in Leibovitz’s L-15 medium (PM151010, Procell). All medium was supplemented with 10% fetal bovine serum (FBS, 11,011, TIANHANG, Hangzhou, China) and the cell lines were maintained in an incubator with a humidified atmosphere of 5% CO2 at 37°C. The NCAPH cDNA sequence was inserted into a plasmid vector pcDNA3.1 (GenScript, Nanjing, China) to obtain a NCAPH-overexpressing plasmid and three siRNAs (siNCAPH-1, siNCAPH-2 and siNCAPH-3) targeting NCAPH (JTS, Wuhan, China) were used for silencing NCAPH. The sequences of three NCAPH siRNAs were:
siNCAPH-1:
Sense, 5ʹ-CUGCCAGCCACAAUGAAUATT-3ʹ;
antisense, 5ʹ-UAUUCAUUGUGGCUGGCAGTT-3ʹ.
siNCAPH-2:
Sense, 5ʹ-CCAGCAGGAGUAUUGACAUTT-3ʹ;
antisense, 5ʹ-AUGUCAAUACUCCUGCUGGTT-3ʹ.
siNCAPH-3:
Sense, 5ʹ-GUGCUACUGAAAUGGGAACTT-3ʹ;
antisense, 5ʹ-GUUCCCAUUUCAGUAGCACTT-3ʹ.
Then NCAPH overexpressing plasmid was transfected into SW780 cells and NCAPH siRNAs were transfected into UMUC3 cells with Lipofectamin 2000 reagent (11,668–019, Invitrogen, Carlsbad, CA, USA) for further experiments.
BC tumor xenograft model
Male BALB/c nude mice (6-week old, 16 ± 1 g) were purchased from HFK Bioscience (Beijing, China) were received freely sterilized food and acidified water in a 12 h light–dark cycle and pathogen-free environment under the temperature of 22 ± 1°C. Twelve healthy mice were randomly divided into (n = 6 per group, 3 mice per cage) shNC and shNCAPH group. The vector containing shRNA targeting NCAPH (shNCAPH) and negative control (shNC) were constructed. Then shNCAPH or shNC was transfected into UMUC3 cells and the stable transfected cell line were selected with G418 (500 μg/ml, 11,811,023, Invitrogen). The 5 × 106 cells were injected subcutaneously into the right armpit of each mouse to develop a xenograft model. The mice were euthanized and xenograft tumor tissues were immediately harvested at 25 days post implantation. Tumor volumes were determined using the following formula: length × width2 × 0.5. The samples from six mice from each group were subjected to immunohistochemistry (IHC), quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and Western blot analysis. The experiment complied with the Guide for the Care and Use of laboratory animals and all experiments using animals had received prior approval by the ethics committee of the Shengjing Hospital of China Medical University.
IHC
Paraffin-embedded human BC tumor tissues and adjacent tissues and xenograft tumor tissues from nude mice were processed for the protein analysis. Briefly, sections were prepared and underwent deparaffinized and antigen retrieval. Sections were subsequently incubated with 3% H2O2 (10,011,218, Sinopharm, Shanghai, China) and blocked with goat serum (SL038, Solarbio, Beijing, China). Then the sections were subsequently incubated with primary antibody anti-NCAPH (1:100, 11,515-1-AP, Proteintech, Wuhan, China) or anti-Ki67 (1:100, A2094, ABclonal, Wuhan, China) overnight at 4°C and incubated with HRP-conjugated secondary antibody (1:500, #31,460, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C for 1 h. The signals were detected with DAB (DA1010, Solarbio). Sections were subsequently counterstained with hematoxylin and scanned using a microscope (BX53, Olympus, Tokyo, Japan).
qRT-PCR
Total RNA was isolated from cells (1 × 106) and tumor tissues (≈50 mg) with total RNA Extraction Kit (DP419, Tiangen, Beijing, China) and then concentrations of total RNA were determined. Subsequently, total RNA (1.5 μg) from each group was used to synthesize into cDNA using oligo (dT)15 and random primer and then 1 μl of cDNA was subjected to qRT-PCR analysis. The PCR primers of NCAPH and GAPDH were as follows: NCAPH forward: 5’-GCCAGCCACAATGAATAA-3ʹ, reverse: 5’-TGCAGATCAAAGACCCTC-3ʹ, GAPDH forward: 5’-GACCTGACCTGCCGTCTAG-3ʹ, reverse: 5’-AGGAGTGGGTGTCGCTGT-3ʹ. The qRT-PCR was performed using SYBR Green (SY1020, Solarbio) with an Exicycler 96 real-time PCR system (BIONEER, Daejeon, Korea) and the following reaction conditions: 94°C for 5 min, followed by 40 cycles at 94°C for 10 s, 60°C for 20s, and 72°C for 30s. The relative mRNA levels were normalized to GAPDH and analyzed using the 2−ΔΔ CT method [35].
Western blotting
Cells and tumor tissues were lyzed in RIPA buffer (R0010, Solarbio) containing PMSF (P0100, Solarbio). Protein concentrations were measured by BCA Protein Assay Kit (PC0020, Solarbio) and equal amount of protein (10–20 μg) was separated on SDS-polyacrylamide gel electrophoresis. After being transferred onto polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore, Billerica, MA, USA), the membranes were blocked with 5% skim milk for 1 h and then incubated overnight at 4°C with the primary antibodies anti-NCAPH (1:1000, DF3967, Affinity, Cincinnati, OH, USA), anti-CDK1 (1:1000, A11420, ABclonal), anti-Cyclin B1 (1:1000, A19037, ABclonal), anti-p-MEK1/2 (1:500, AP0209, ABclonal), anti-MEK1/2 (1:500, AF6385, Affinity), anti-p-ERK1/2 (1:1000, AF8208, Affinity), anti-ERK1/2 (1:1000, AF6240, Affinity) and anti-GAPDH (1:10,000, 60,004–1, Proteintech). The proteins subsequently were incubated with HRP-conjugated secondary antibodies (1:3000) at 37°C for 1 h and visualized using the enhanced chemiluminescence (ECL, PE0010, Solarbio) detection system.
Cell proliferation
Cell proliferation was determined by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay and bromodeoxyuridine (BrdU) staining. For MTT assay, cells were seeded into 96-well plates (5 × 103 cells/well). After transfection, the cells were incubated for 0, 24, 48, and 72 h and added with MTT (KGA311, KeyGen Biotech, Nanjing, China). Four hour later, the supernatant was moved and the optical density of each well was measured by a microplate reader (800Ts, BioTek, Winooski, Vermont, USA) at 490 nm after 150 μl DMSO (ST038, Beyotime, Shanghai, China) treatment. For BrdU staining, cell proliferation was determined by BrdU incorporation to assess DNA synthesis. The transfected cells were cultured for 48 h and treated with 30 μg/ml BrdU (BD Pharmingen, San Diego, CA, USA) for 45 min. Then cells were fixed and incubated with anti-BrdU antibody (1:100, A20304, ABclonal) overnight at 4°C. The cells were incubated with secondary antibody (1:200) for 1 h and the cells were observed and recorded under a microscope.
Cell cycle and apoptosis analysis
Cell cycle and apoptosis analysis were performed by flow cytometry. For cell cycle analysis, the cells were fixed with 70% ethanol at 4°C for 2 h and then cells were incubated with propidium iodide (PI) and RNase A (C1052, Beyotime) for 30 min. For apoptosis analysis, the cells were incubated with Annexin V-FITC and PI (C1062, Beyotime) at room temperature for 10–20 min. After incubation, cell cycle and apoptosis were analyzed using a flow cytometer (NovoCyte, San Diego, CA, USA).
TdTmediated dUTP nick end labeling (TUNEL) assay
The effect of NCAPH on cell apoptosis was also assessed using the TUNEL In Situ Cell Death Detection Kit (12,156,792,910, Roche, Basel, Switzerland). The fixed cell slides were permeabilized in 0.1% Triton X-100 (ST795, Beyotime) and then incubated with TUNEL solution at 37°C for 1 h. Finally, the fluorescence of cells was detected under a microscope.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8.0 (GraphPad Software Inc., San Diego, CA, USA) and all data were presented as the mean ± standard deviation (SD). Differences in the results among groups were compared using Student’s t-test or multi-way analysis of variance (ANOVA) test. The chi-square test was used to analyze the relationship between NCAPH mRNA expression and clinical pathological characteristics. Statistical significance was defined as p < 0.05.
Results
NCAPH expression is overexpressed in BC tissues and cell lines
We used GEPIA database to analyze the expression of NCAPH in BC tissues (n = 404) and normal tissues (n = 28). The results showed that NCAPH is overexpressed in BC tissues compared with normal non-tumor tissues (Figure 1(a)). To investigate the role of NCAPH in bladder cancer, we collected 30 pairs of BC carcinoma tissues and adjacent non‐tumor tissues and the mRNA expression of NCAPH was detected. The qRT-PCR analysis showed that NCAPH mRNA levels in carcinoma tissues were remarkably higher than that in paired adjacent tissues (Figure 1(b)). Immunohistochemical analysis determined the protein levels of NCAPH were increased in BC carcinoma tissues compared with the adjacent non‐tumorous samples (Figure 1(c)). Clinicopathological association analyses of the 60 BC carcinoma tissues revealed that NCAPH mRNA expression was positively associated with tumor stage, lymph node metastasis, TNM stage, histopathological grade and tumor size (Table 1). Next, the mRNA expression of NCAPH in human ureteral epithelial SV-HUC-1 cells and BC cell lines was detected. The results showed that the BC cell lines, including UMUC-3, 5637, J82 and SW780, displayed significantly higher mRNA levels than SV-HUC-1 cells (Figure 1(d)). These results indicate that NCAPH expression is overexpressed in BC tissues and cell lines.
Figure 1.
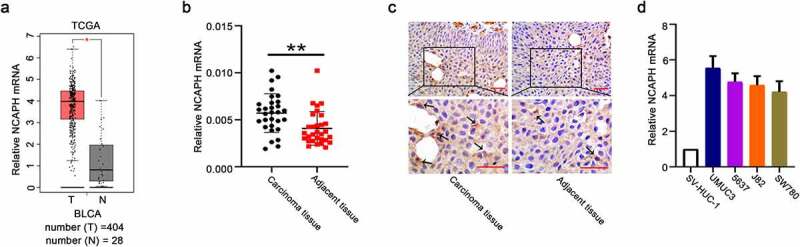
NCAPH is highly expressed in BC tissues and cell lines. (a) The relative mRNA levels of NCAPH in BC tissues and normal tissues were analyzed using the GEPIA database (T, tumor, n = 404; N, normal, n = 28, *P < 0.05). (b) The mRNA levels of NCAPH in BC tissues and adjacent tissues were determined by qRT‐PCR. (c) The expressions of NCAPH in BC tissues and adjacent tissues were detected using immunohistochemistry staining. Scale bar, 50 μm. The black arrow indicated the NCAPH protein expression (brown staining). (d) The mRNA levels of NCAPH in human ureteral epithelial SV-HUC-1 cells and BC cell lines (UMUC3, 5637, J82 and SW780) were evaluated using qRT‐PCR. The data were presented as the mean ± SD. **P < 0.01, *P < 0.05.
Table 1.
Analysis of the association between expression of NCAPH and clinicopathological characteristics of bladder cancer
| Clinical parameters | NCAPH expression level |
P value | |
|---|---|---|---|
| High | Low | ||
| Gender | |||
| Male | 23 | 28 | 0.14812 |
| Female | 7 | 2 | |
| Age | |||
| ≤60 | 23 | 23 | 0.76019 |
| >60 | 7 | 7 | |
| Tumor stage | |||
| pTa-T1 | 13 | 29 | 2.4E-05 |
| T2-T4 | 17 | 1 | |
| Lymph node metastasis | |||
| Positive | 6 | 0 | 0.03142 |
| Negative | 24 | 30 | |
| Metastasis | |||
| M0 | 29 | 30 | 1 |
| M1 | 1 | 0 | |
| TNM stage | |||
| I–II | 21 | 30 | 0.00382 |
| III–IV | 9 | 0 | |
| Histopathological grade | |||
| G1-G2 | 5 | 15 | 0.01371 |
| G3 | 25 | 15 | |
| Tumor size | |||
| ≤3 cm | 8 | 26 | 9.5E-06 |
| >3 cm | 22 | 4 | |
| Tumor number | |||
| Unifocal | 18 | 15 | 0.60376 |
| Multifocal | 12 | 15 | |
NCAPH promotes the cell proliferation of BC cells
The results in Figure 1(d) showed that the mRNA level of NCAPH is higher in UMUC3 cells and lower in SW780 cells than other cell lines; therefore, we chose these two cell lines for further study. To investigate the potential role of NCAPH in BC cells, we transfected SW780 cells with NCAPH overexpression vector to achieve ectopic expression of NCAPH and we transfected UMUC3 cells with siRNAs targeting NCAPH to silence NCAPH. The results in Figure 2(a,b) showed that NCAPH overexpression vector significantly increased NCAPH mRNA and protein levels in SW780 cells. The NCAPH siRNAs could significantly reduce NCAPH mRNA and protein levels in UMUC3 cells (Figure 2(c,d)). We chose NCAPH siRNA-2 and siRNA-3 for further study because of their better transfection efficiencies. Next, the BrdU assay and MTT assay were performed to determine the effect of NCAPH on cell proliferation of BC cells. The results showed that overexpression of NCAPH remarkably increased the BrdU-positive cell number and cell growth rates in SW780 cells and knockdown of NCAPH significantly inhibited the BrdU-positive cell number and cell growth rates in UMUC3 cells (Figure 2(e-h)).
Figure 2.
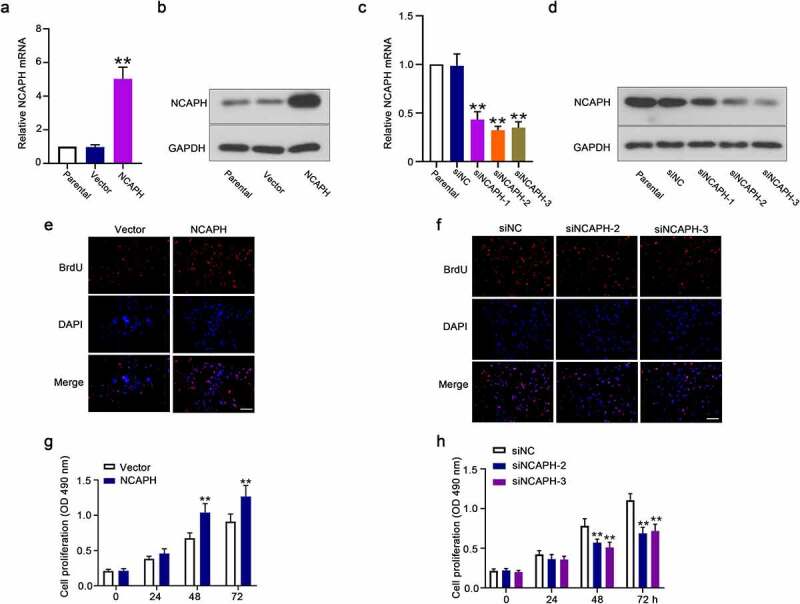
NCAPH promotes the cell proliferation of BC cells. (a,b) The mRNA and protein levels of NCAPH in SW780 cells transfected with NCAPH expression vector and control vector were determined by qRT‐PCR and Western blot. (c,d) The mRNA and protein levels of NCAPH in UMUC3 cells transfected with NCAPH siRNA1-3 and negative control (siNC) were determined by qRT‐PCR and Western blot. (e-h) The effect of NCAPH on cell proliferation of BC cells was determined by BrdU assay (Scale bar, 100 μm) and MTT assay. The data were presented as the mean ± SD. **P < 0.01. In g and h, no significant difference between Vector and NCAPH, siNC and siNCAPH-2/3 group at 0 and 24 h.
Cell cycle distribution analysis confirmed the effect of NCAPH on BC cell proliferation. We found that overexpression of NCAPH increased the ratio of S cell population (Figure 3(a,c)). And knockdown of NCAPH increased the ratio of cell population in G2/M phases instead of G1 and S phases (Figure 3(b,e)). The protein levels of cyclin B1 and CDK1 were dramatically increased in NCAPH overexpressed SW780 cells and decreased in NCAPH silenced UMUC3 cells compared with the control cells (Figure 3(d,f)). The results indicate that NCAPH promotes cell proliferation and plays an important role in cycle distribution in BC cells.
Figure 3.
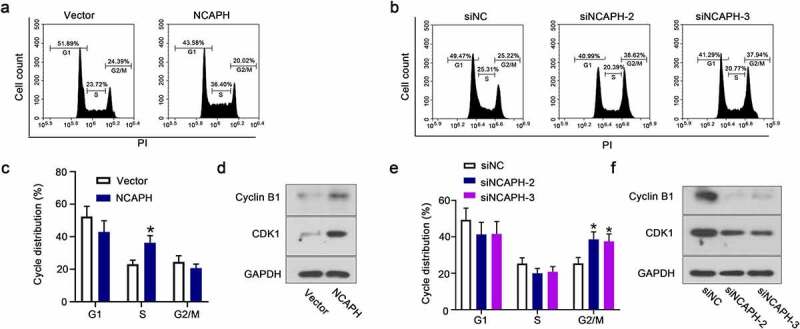
NCAPH regulates the cell cycle transition of BC cells. (a,c) SW780 cells were transfected NCAPH expression vector and control vector and the proportion of cells in each cell-cycle phase was analyzed by flow cytometry. (b,e) UMUC3 cells were transfected NCAPH siRNAs and negative control (siNC) and the proportion of cells in each cell-cycle phase was analyzed by flow cytometry. (d,f) The protein levels of Cyclin B1 and CDK1 were detected by Western blot. The data were presented as the mean ± SD. *P < 0.05.
NCAPH inhibits the cell apoptosis of BC cells
To investigate the role of NCAPH in cell apoptosis of BC cells, the transfected cells were collected and stained with annexin V and PI and then accessed by flow cytometry. The ratio of apoptotic cells was significantly decreased in NCAPH expression vector transfected SW780 cells and increased in NCAPH siRNAs transfected UMUC3 cells (Figure 4(a,b)). The results were confirmed by TUNEL staining assay. Overexpression of NCAPH reduced the TUNEL-positive cell number in SW780 cells and knockdown of NCAPH increased the TUNEL-positive cell number in UMUC3 cells (Figure 4(c,d)). Our data indicate that NCAPH inhibits the cell apoptosis of BC cells.
Figure 4.
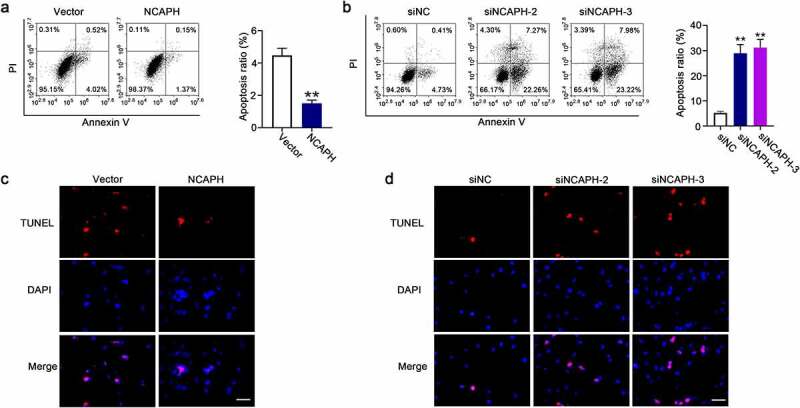
NCAPH inhibits the cell apoptosis of BC cells. (a,b) SW780 cells were transfected NCAPH expression vector and control vector and UMUC3 cells were transfected NCAPH siRNAs and negative control (siNC), the cells were stained with Annexin V and PI, and cell apoptosis was analyzed flow cytometry. (c,d) TUNEL assay was used to determine the effect of NCAPH on cell apoptosis of BC cells. Scale bar, 50 μm. The data were presented as the mean ± SD. **P < 0.01.
NCAPH promotes progress of BC through MEK/ERK signaling pathway
It is well known that activation of MEK/ERK signaling pathway is important for progression of bladder cancer including cell proliferation and apoptosis. Therefore, we wondered whether the expression of NCAPH is correlated with the activation of MEK/ERK signaling pathway in BC cells. In this study, we evaluated the activation of MEK/ERK signaling pathway in NCAPH overexpressing and knockdown BC cells. The results in Figure 5(a,b) displayed that the phosphorylation levels of MEK and ERK were dramatically increased in NCAPH overexpressing SW780 cells and decreased in NCAPH silenced UMUC3 cells compared with the their controls. In order to verify our hypothesis, MEK1/2 inhibitor U0126 was used to block the activation of MEK/ERK signaling pathway in NCAPH overexpressing SW780 cells. The activation of MEK/ERK signaling pathway was determined by Western blot and the effect of U0126 on cell proliferation in NCAPH overexpressing SW780 cells were evaluated by BrdU and MTT assay. The results indicated that U0126 inhibited NCAPH-promoted the activation of MEK/ERK signaling pathway and reversed the increase of cell proliferation regulated by NCAPH (Figure 5(c-e)). The results suggested that NCAPH could promote progress of BC through MEK/ERK signaling pathway.
Figure 5.
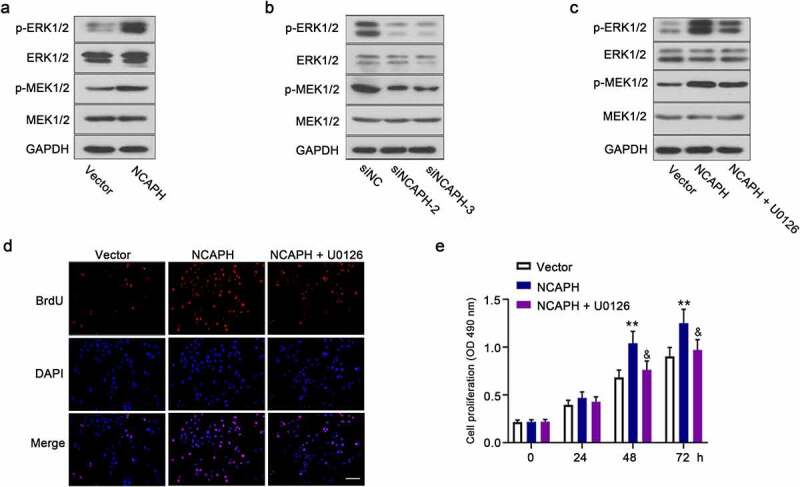
NCAPH promotes the activation of MEK/ERK signaling pathway in BC cells. (a,b) The protein levels of p-MEK1/2, MEK1/2, p-ERK1/2 and ERK1/2 in NCAPH overexpressing SW780 cells and NCAPH silenced UMUC3 cells were detected by Western blot. (c) SW780 cells were transfected NCAPH expression vector and treated with MEK1/2 inhibitor 10 μM U0126, the protein levels of p-MEK1/2, MEK1/2, p-ERK1/2 and ERK1/2 were detected by Western blot. (d,e) The cell proliferation of BC cells was determined by BrdU assay (Scale bar, 100 μm) and MTT assay. The data were presented as the mean ± SD. Vector versus NCAPH, **P < 0.01; NCAPH versus NCAPH + U0126, &P < 0.05. No significant difference between Vector and NCAPH, NCAPH and NCAPH + U01260 group at 0 and 24 h.
Knockdown of NCAPH suppresses the BC tumor growth in vivo
To further validate the role of NCAPH on progress of BC, we performed a xenograft model in vivo. The results showed that NCAPH shRNA significantly inhibited the mRNA and protein levels of NCAPH in tumor tissues (Figure 6(a,b)). Tumor volume was significantly decreased in NCAPH silenced mice (Figure 6(c,d)). In addition, the expression of Ki‐67, a cell proliferation marker, was significantly decreased in shNCAPH injected tumors (Figure 6(e)). These results indicate that the knockdown of NCAPH suppresses tumor growth of BC in vivo.
Figure 6.
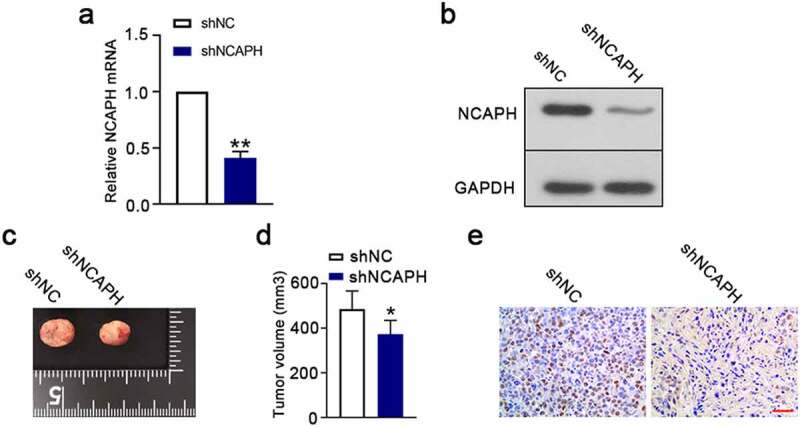
Knockdown of NCAPH suppresses tumor growth in vivo. (a,b) A total of 5 × 106 shRNA-mediated NCAPH silenced UMUC3 cells were subcutaneously injected into nude mice. The tumor tissues were immediately harvested at 25 days post implantation. The mRNA and protein levels of NCAPH in tumor tissues were determined by qRT‐PCR and Western blot. (c,d) Tumor volume was evaluated at day 25. (e) The expression of Ki67 (brown staining) in tumor tissues was detected using immunohistochemistry staining. Scale bar, 50 μm. The data were presented as the mean ± SD. **P < 0.01, *P < 0.05.
Discussion
BC is one of most common cancers world-wide with a poor prognosis and high mortality. And the mechanisms of progression of BC still need to be studied extensively and it will provide more potential markers in the diagnosis and treatment for BC. In this study, we focused on the effects of NCAPH on BC progression.
Human NCAPH, a regulatory subunit of the condensin I complex, plays an important role in maintaining the function of condensin to regulate the sister-chromatid cohesion and separation during mitosis in proliferative cells [4,36]. NCAPH, especially in cancer cells, is important for regulating cell proliferation and cell death. A study found that overexpression of NCAPH facilitates proliferation, migration, and invasion of hepatocellular carcinoma cells in vitro and knockdown of NCAPH inhibits tumor growth in mice xenograft models in vivo [22]. Wang et al. reported that the transcription of NCAPH could be activated by transcription factor E2F1 and depletion of NCAPH significantly inactivated AKT/SGK signaling pathway in cervical carcinoma [23]. Moreover, in colon cancer and non-small-cell lung cancer, it has been found that depletion of NCAPH could induce cell cycle arrest at the G2/M phase and cell apoptosis [24,25]. These studies determined the oncogenic role of NCAPH in these cancers. In BC, the data from both GEPIA database and our results in this study showed that NCAPH is comparatively overexpressed in BC tissues than normal or adjacent non-tumor tissues. We analyzed the associations between mRNA expression of NCAPH and the clinicopathological characteristics and our results showed that NCAPH expression was positively related to tumor stage, lymph node metastasis, TNM stage, histopathological grade and tumor size. Moreover, higher expression levels of NCAPH were also observed in BC cell lines than that in human ureteral epithelial SV-HUC-1 cells. We determined that overexpression of NCAPH promoted cell proliferation and inhibited cell apoptosis of BC cells, whereas knockdown of NCAPH exhibited the opposite effects on BC cells. Moreover, we validated that knockdown of NCAPH suppressed tumor growth of BC in vivo using a xenograft model. Our results indicated that NCAPH plays an important role in promoting BC progression.
MEK1 and MEK2 are dual-specificity kinases that phosphorylate ERK1 and ERK2 to activate ERK1/2 signaling pathway and thence promote inappropriate cell proliferation and survival [37]. Activation of the MEK/ERK signaling pathway triggers in the development of many kinds of cancers and MEK/ERK signaling pathway is one of the main activating pathways in cancers [38]. In BC, NK2 homeobox 8 (Nkx2.8) functions as a tumor-suppressor to inhibit cell proliferation by inactivating MEK/ERK signaling pathway [39]. Zhao et al. found that knockdown of kinesin family member 15 (KiF15) inhibits phosphorylation levels c-Raf, MEK and ERK and cell proliferation of BC cells [40]. Moreover, treatment of MEK/ERK inhibitor U0126 enhances the inhibitory effect of GP130 inhibitor SC144 on the expression of BCL-XL and BC cell proliferation [27]. Additionally, Sun et al. demonstrated that activation of ERK1/2 could promote pro-apoptotic protein BIM degradation, which might lead to inhibit cell apoptosis [41]. Inhibition of MEK/ERK signal transduction could induce G2/M arrest and suppress the expressions of CDK1 and cyclin B1 [30,31,42]. Moreover, downregulation of CDK1 and cyclin B1 inhibits cell proliferation by inducing G2/M arrest in many types of cancer cells [32,33,43]. Our results showed that knockdown of NCAPH-induced cell arrest at G2/M, inhibited CDK1 and cyclin B1 expressions and inactivated MEK/ERK signaling pathway. Moreover, in low-grade serous ovarian carcinoma, the expressions of condensin complex members including NCAPH were increased in MEK-inhibitor (MEKi)–resistant cell lines [29]. It indicates that there might be a regulatory mechanism between MEK/ERK signaling pathway and condensin, and NCAPH might affect cell proliferation through regulating expressions of CDK1 and cyclin B1 in MEK/ERK-dependent pathway. In this study, our results showed that phosphorylation levels of MEK and ERK were enhanced in NCAPH overexpressing cells and reduced in NCAPH silenced cells. Additionally, MEK1/2 inhibitor U0126 blocked the activation of MEK/ERK signaling pathway in NCAPH overexpressing SW780 cells and reversed the increase of cell proliferation regulated by NCAPH overexpression. The results indicate that it is very likely that NCAPH promotes malignant biological properties through MEK/ERK signaling pathway in BC. However, we didn’t investigate the regulatory mechanism of NCAPH on MEK/ERK activation in this present study. A deeper study of the effects of NCAPH on the activation of MEK/ERK signaling pathway would be considered in the future.
In conclusion, our results showed that NCAPH is highly expressed in BC tissues and NCAPH could promote cell proliferation and inhibit cell apoptosis of BC cells. Additionally, NCAPH increased the activation of the ERK/MAPK signaling pathway. Our results indicated that NCAPH plays an important role in BC progression through regulating MEK/ERK signaling pathway and our results might provide a novel diagnostic and therapeutic target in BC.
Funding Statement
This study was supported by grants from the 345 Talent Project of Shengjing Hospital to Bo Li (M0317) and the Surface Project of Natural Science Foundation of Liaoning Province (2021-MS-174).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data available on request from the authors.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Cumberbatch MGK, Jubber I, Black PC, et al. Epidemiology of bladder cancer: a systematic review and contemporary update of risk factors in 2018. Eur Urol. 2018;74(6):784–795. [DOI] [PubMed] [Google Scholar]
- [3].Bladder cancer: diagnosis and management of bladder cancer: © NICE (2015) . Bladder cancer: diagnosis and management of bladder cancer. BJU Int. 2017;120(6):755–765. [DOI] [PubMed] [Google Scholar]
- [4].Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26(15):1659–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ono T, Losada A, Hirano M, et al. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115(1):109–121. [DOI] [PubMed] [Google Scholar]
- [6].Ball AR Jr., Yokomori K. The structural maintenance of chromosomes (SMC) family of proteins in mammals. Chromosome Res. 2001;9(2):85–96. [DOI] [PubMed] [Google Scholar]
- [7].Losada A, Hirano T. Dynamic molecular linkers of the genome: the first decade of SMC proteins. Genes Dev. 2005;19(11):1269–1287. [DOI] [PubMed] [Google Scholar]
- [8].Harvey SH, Krien MJ, O’Connell MJ. Structural maintenance of chromosomes (SMC) proteins, a family of conserved ATPases. Genome Biol. 2002;3(2):Reviews3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Yatskevich S, Rhodes J, Nasmyth K. Organization of Chromosomal DNA by SMC Complexes. Annu Rev Genet. 2019;53(1):445–482. [DOI] [PubMed] [Google Scholar]
- [10].Yan Y, Liu C, Zhang J, et al. SMC4 knockdown inhibits malignant biological behaviors of endometrial cancer cells by regulation of FoxO1 activity. Arch Biochem Biophys. 2021;712:109026. [DOI] [PubMed] [Google Scholar]
- [11].Zhang C, Kuang M, Li M, et al. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Sci Rep. 2016;6(1):34508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Murakami-Tonami Y, Kishida S, Takeuchi I, et al. Inactivation of SMC2 shows a synergistic lethal response in MYCN -amplified neuroblastoma cells. Cell Cycle. 2014;13(7):1115–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Han YH, Wan Y, Xiong H, et al. Structural maintenance of chromosomes 2 is identified as an oncogene in bladder cancer in vitro and in vivo. Neoplasma. 2020;67(2):364–370. [DOI] [PubMed] [Google Scholar]
- [14].Zhang Y, Liu F, Zhang C, et al. Non-SMC condensin i complex subunit D2 is a prognostic factor in triple-negative breast cancer for the ability to promote cell cycle and enhance invasion. Am J Pathol. 2020;190:37–47. [DOI] [PubMed] [Google Scholar]
- [15].Jing Z, He X, Jia Z, et al. NCAPD2 inhibits autophagy by regulating Ca(2+)/CAMKK2/AMPK/mTORC1 pathway and PARP-1/SIRT1 axis to promote colorectal cancer. Cancer Lett. 2021;520:26–37. [DOI] [PubMed] [Google Scholar]
- [16].Jiang L, Ren L, Chen H, et al. NCAPG confers trastuzumab resistance via activating SRC/STAT3 signaling pathway in HER2-positive breast cancer. Cell Death Dis. 2020;11(7):547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gong C, Ai J, Fan Y, et al. NCAPG promotes the proliferation of hepatocellular carcinoma through PI3K/AKT signaling. Onco Targets Ther. 2019;12:8537–8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Qiu X, Gao Z, Shao J, et al. NCAPH is upregulated in endometrial cancer and associated with poor clinicopathologic characteristics. Ann Hum Genet. 2020;84(6):437–446. [DOI] [PubMed] [Google Scholar]
- [19].Lu H, Shi C, Wang S, et al. Identification of NCAPH as a biomarker for prognosis of breast cancer. Mol Biol Rep. 2020;47(10):7831–7842. [DOI] [PubMed] [Google Scholar]
- [20].Cui F, Hu J, Xu Z, et al. Overexpression of NCAPH is upregulated and predicts a poor prognosis in prostate cancer. Oncol Lett. 2019;17(6):5768–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shimomura H, Sasahira T, Nakashima C, et al. Non-SMC condensin I complex subunit H (NCAPH) is associated with lymphangiogenesis and drug resistance in oral squamous cell Carcinoma. J Clin Med. 2019;9(1):72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun C, Huang S, Wang H, et al. Non-SMC condensin I complex subunit H enhances proliferation, migration, and invasion of hepatocellular carcinoma. Mol Carcinog. 2019;58(12):2266–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang M, Qiao X, Cooper T, et al. HPV E7-mediated NCAPH ectopic expression regulates the carcinogenesis of cervical carcinoma via PI3K/AKT/SGK pathway. Cell Death Dis. 2020;11(12):1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kim B, Kim SW, Lim JY, et al. NCAPH is required for proliferation, migration and invasion of non-small-cell Lung Cancer cells. Anticancer Res. 2020;40(6):3239–3246. [DOI] [PubMed] [Google Scholar]
- [25].Yin L, Jiang LP, Shen QS, et al. NCAPH plays important roles in human colon cancer. Cell Death Dis. 2017;8(3):e2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jiang T, Lu X, Yang F, et al. LMTK3 promotes tumorigenesis in bladder cancer via the ERK/MAPK pathway. FEBS Open Bio. 2020;10(10):2107–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li X, He S, Tian Y, et al. Synergistic inhibition of GP130 and ERK signaling blocks chemoresistant bladder cancer cell growth. Cell Signal. 2019;63:109381. [DOI] [PubMed] [Google Scholar]
- [28].Cao R, Meng Z, Liu T, et al. Decreased TRPM7 inhibits activities and induces apoptosis of bladder cancer cells via ERK1/2 pathway. Oncotarget. 2016;7(45):72941–72960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Shrestha R, Llaurado Fernandez M, Dawson A, et al. Multiomics characterization of low-grade serous ovarian carcinoma identifies potential biomarkers of MEK inhibitor sensitivity and therapeutic vulnerability. Cancer Res. 2021;81(7):1681–1694. [DOI] [PubMed] [Google Scholar]
- [30].Chiu CY, Kuo KK, Kuo TL, et al. The activation of MEK/ERK signaling pathway by bone morphogenetic protein 4 to increase hepatocellular carcinoma cell proliferation and migration. Mol Cancer Res. 2012;10(3):415–427. [DOI] [PubMed] [Google Scholar]
- [31].Liu L, Zhu Y, Xu Y, et al. Prevention of ERK activation involves melatonin-induced G(1) and G(2) /M phase arrest in the human osteoblastic cell line hFOB 1.19. J Pineal Res. 2012;53(1):60–66. [DOI] [PubMed] [Google Scholar]
- [32].Lee MH, Cho Y, Kim DH, et al. Menadione induces G2/M arrest in gastric cancer cells by down-regulation of CDC25C and proteasome mediated degradation of CDK1 and cyclin B1. Am J Transl Res. 2016;8(12):5246–5255. [PMC free article] [PubMed] [Google Scholar]
- [33].Paruthiyil S, Cvoro A, Tagliaferri M, et al. Estrogen receptor β causes a G2 cell cycle arrest by inhibiting CDK1 activity through the regulation of cyclin B1, GADD45A, and BTG2. Breast Cancer Res Treat. 2011;129(3):777–784. [DOI] [PubMed] [Google Scholar]
- [34].Zhao J, Shi L, Zeng S, et al. Importin-11 overexpression promotes the migration, invasion, and progression of bladder cancer associated with the deregulation of CDKN1A and THBS1. Urol Oncol. 2018;36(6):311.e1-.e13. [DOI] [PubMed] [Google Scholar]
- [35].Winer J, Jung CK, Shackel I, et al. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270(1):41–49. [DOI] [PubMed] [Google Scholar]
- [36].Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nat Rev Genet. 2010;11(6):391–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Caunt CJ, Sale MJ, Smith PD, et al. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15(10):577–592. [DOI] [PubMed] [Google Scholar]
- [38].Maik-Rachline G, Hacohen-Lev-Ran A, Seger RNERK. Mechanism of translocation, substrates, and role in cancer. Int J Mol Sci. 2019;20(5):1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yu C, Zhang Z, Liao W, et al. The tumor-suppressor gene Nkx2.8 suppresses bladder cancer proliferation through upregulation of FOXO3a and inhibition of the MEK/ERK signaling pathway. Carcinogenesis. 2012;33(3):678–686. [DOI] [PubMed] [Google Scholar]
- [40].Zhao H, Bo Q, Wu Z, et al. KIF15 promotes bladder cancer proliferation via the MEK-ERK signaling pathway. Cancer Manag Res. 2019;11:1857–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Luciano F, Jacquel A, Colosetti P, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. [DOI] [PubMed] [Google Scholar]
- [42].Yin X, Zhang R, Feng C, et al. Diallyl disulfide induces G2/M arrest and promotes apoptosis through the p53/p21 and MEK-ERK pathways in human esophageal squamous cell carcinoma. Oncol Rep. 2014;32(4):1748–1756. [DOI] [PubMed] [Google Scholar]
- [43].Sang Eun H, Seong Min K, Ho Jeong L, et al. Scutellarein induces fas-mediated extrinsic apoptosis and G2/M cell cycle arrest in Hep3B Hepatocellular Carcinoma Cells. Nutrients. 2019;11(2):263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data available on request from the authors.


