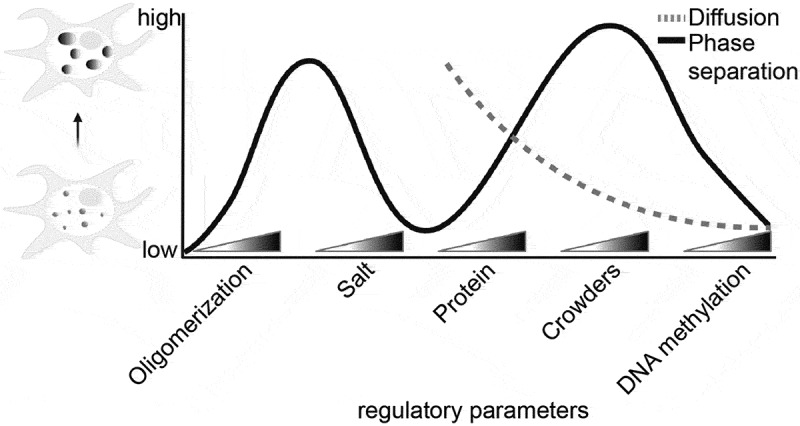
ABSTRACT
Heterochromatin is the highly compacted form of chromatin with various condensation levels hallmarked by high DNA methylation. MeCP2 is mostly known as a DNA methylation reader but has also been reported as a heterochromatin organizer. Here, we combine liquid–liquid phase separation (LLPS) analysis and single-molecule tracking with quantification of local MeCP2 concentrations in vitro and in vivo to explore the mechanism of MeCP2-driven heterochromatin organization and dynamics. We show that MeCP2 alone forms liquid-like spherical droplets via multivalent electrostatic interactions and with isotropic mobility. Crowded environments and DNA promote MeCP2 LLPS and slow down MeCP2 mobility. DNA methylation, however, restricts the growth of heterochromatin compartments correlating with immobilization of MeCP2. Furthermore, MeCP2 self-interaction is required for LLPS and is disrupted by Rett syndrome mutations. In summary, we are able to model the heterochromatin compartmentalization as well as MeCP2 concentration and heterogeneous motion in the minimal in vitro system.
KEYWORDS: DNA methylation, heterochromatin, liquid-liquid phase separation, MeCP2, protein–protein interaction, Rett syndrome, single molecule tracking
Graphical abstract
Introduction
The eukaryotic nucleus is compartmentalized into various dynamic subdomains with specific physico-chemical properties. This compartmentalization takes place in the absence of separating membranes inside the nucleus. As a result, the subnuclear concentration and/or exclusion of nuclear factors changes locally and different nuclear metabolic processes are segregated apart. This de facto reduces entropy and enhances the overall efficiency and specificity of nuclear metabolic processes [1]. Yet the underlying mechanisms regulating the formation, maintenance, and dynamics of such membrane-less functional organelles is under debate with recent studies suggesting a liquid–liquid phase separation (LLPS) model for nuclear and genome sub-compartmentalization [2–6]. The study of nucleosome arrays (NAs) in vitro by Gibson and colleagues revealed that NAs form liquid-like droplets under physiological conditions via LLPS, which could be modulated by DNA and histone post-translational modifications [5]. The latter were also shown to contribute to the overall NA mobility using single molecule kinetic analysis [7,8]. Furthermore, the finding that acetylated NAs formed distinct droplets from the ones formed by unmodified NAs led to the hypothesis of LLPS mediating compartmentalization of the genome into active and inactive domains.
Inactive chromatin, also termed heterochromatin, is generally highly compacted and transcriptionally silenced. In mammalian genomes, it comprises the majority of the genome and contains a large proportion of repeat DNA elements including interspersed and tandem repeats [9]. These have been shown to play a major role in organizing the genome within the nucleus and to have additional roles in regulating gene expression, and genome stability [10–17]. The most compacted form of heterochromatin present in all cell types (constitutive heterochromatin) is located at and around the centromeres (centromeric and peri-centromeric) and in mouse cells forms aggregates called chromocenters. These consist mainly of satellite DNA tandem repeats and make up 10% of the mouse genome [18]. In these heterochromatic regions, the mobility of both nucleosomes and other factors is reduced compared to other areas in the nucleus [19,20].
Molecularly, heterochromatin is distinguished by specific histone posttranslational modifications, in particular H3K9me2/3, and its complete absence was shown to dissolve heterochromatin compartments [21,22]. The H3K9me2/3 is recognized by ‘reader’ heterochromatin protein 1 (HP1) via the chromo domain [21]. HP1 was shown to drive chromocenter formation via LLPS, depending on dimerization and mediated by phosphorylation and interactive partners like H3K9 trimethylated histones [2–4,15]. Yet all three HP1 homologs (HP1α, HP1β, and HP1γ) showed no predominant enrichment (2 ~ 3 fold) in chromocenters compared to chromatin compaction levels (~2 fold) in mouse fibroblasts [23]. HP1α was also shown to be less expressed compared to other heterochromatin associated proteins such as MeCP2 and SUV39H1 [23]. Moreover, HP1α was shown not to significantly affect heterochromatin clustering and compaction [23,24]. A recent study proposed rather that HP1α LLPS ability was weak and not required for heterochromatin compartmentalization [25]. Altogether, these studies indicate that heterochromatin adopts a liquid-like membraneless compartment state, but it remains unclear which proteins or protein complexes are responsible [26].
Another major distinguishing molecular feature of heterochromatin is DNA methylation. In mammals, cytosine-based methylation takes place predominantly but not exclusively at the dinucleotide mCpG and was shown to play a role during development and disease regulating gene expression and DNA metabolism [27–30]. Methylated cytosines are recognized by methyl-CpG binding domain (MBD) containing proteins, such as MBD1, 2, 4, and methyl-CpG binding protein 2 (MeCP2) [28,31–33]. MBD1 and MeCP2 were found to tether the SUV39H1-HP1 complex to methylated DNA [34–37]. MeCP2 level increases during cellular differentiation [37,38]. Accordingly, MeCP2 plays a role in the maturation of the brain and multiple mutations (including missense, nonsense, and gene duplication events) within the MeCP2 gene were found in patients with the neurological disorder Rett syndrome (RTT; OMIM: #312,750) [39–43]. In addition to the methyl-CpG binding domain (MBD), MeCP2 was shown to contain a transcriptional repression domain (TRD) responsible for the recruitment of histone deacetylase complexes to chromatin [44]. Subsequently, the TRD has been narrowed down to the NCoR/SMRT interacting domain (NID) [45]. Expression of the MBD fused to the NID was shown to be sufficient to rescue Rett syndrome-like phenotype in mouse models [46]. Albeit MeCP2 has been generally described as a transcriptional repressor, evidence suggests that it may also act as a transcriptional activator [47]. Importantly, MeCP2 deficiency results in only mild gene expression changes [48]. It is also not understood how the several MeCP2 gene deletion, mutations, and multiplication lead to the neurological Rett syndrome-like phenotype. In addition to and/or instead of a transcriptional regulation role, accumulating evidence points to a structural chromatin role of MeCP2 in Rett syndrome [32,49–56]. MeCP2 was previously shown to accumulate at heterochromatin compartments within the nucleus [57] and to promote the heterochromatin clustering (fusion) during cellular differentiation in a level-dependent manner [24,58]. These MeCP2 enriched heterochromatin aggregates were shown to be dynamic compartments with fusion events taking place when pericentromeric heterochromatin from different chromosomes came into contact in interphase cells and, after mitotic cell division, the aggregates could rebuild themselves up again [24]. These data clearly indicated the importance of MeCP2 in heterochromatin organization and dynamics. Multiple RTT-related MeCP2 mutations have been reported to compromise the heterochromatin clustering function in part due to their lower binding to the heterochromatin [49,59].
Structural analysis by circular dichroism spectroscopy has shown that MeCP2 protein is highly unstructured and mostly the MBD exhibits a stable structure [60]. Unstructured regions in proteins are amongst the common features shared by multiple proteins that mediate subcellular compartmentalization via LLPS [61]. In addition, MeCP2 has multivalent interactions including: DNA, methylated cytosine (5mC), RNA, histones, HP1s, MDB2, N-CoR/SMRT and itself (reviewed in [32]). Multivalency has also been shown to play a role in LLPS of various disordered proteins, promoting the assembly and maintenance of membraneless organelles [62,63]. In vitro, MeCP2 was shown to promote the condensation of NAs by forming highly condensed ellipsoidal particles and oligomeric suprastructures as analyzed by electron microscopy [64]. This was independent of DNA methylation and relying upon regions downstream of MBD [65–67]. These facts could in part explain how nonsense mutations of MeCP2 that lead to a truncated protein stopping after the MBD lead to severe RTT symptoms and highlight the importance of these unstructured regions of MeCP2 [32]. Recently, purified MeCP2 together with NAs was also shown to form liquid droplets in vitro that could be compromised by RTT mutations [68,69]. Yet, it is unclear which factor(s) determine the LLPS properties of MeCP2 and how the MeCP2 molecules diffusion and binding properties change when inside the droplets formed at various conditions and mimicking the physiology situation.
Here, we address these questions by establishing a minimal system for MeCP2 phase separation in vitro and compare it with cellular systems tuned to mimicking the in vivo conditions and MeCP2 levels. We define the minimal requirements for MeCP2 LLPS and evaluate the effects of DNA and DNA methylation. Furthermore, we map the regions required for LLPS in vitro and for heterochromatin compartmentalization in cells. With this, we find that MeCP2 self-interaction is essential for compartmentalization. In parallel, we measure the MeCP2 wild type and mutant mobility in the different conditions in droplets and in cells and define the relevant parameters and binding modalities. Altogether, these data allow to draw a model of MeCP2-dependent chromatin condensate formation via LLPS and the role of MeCP2 concentration, DNA as well as methylated DNA in the establishment and growing of these compartments and the consequences of each for MeCP2 mobility.
Materials and methods
Plasmids
All plasmids used in this study and the source references are listed in Table S1.
Bacterial expression plasmids
The pTYB1 vector was used for fusion protein purification, because it utilizes the inducible self-cleavage activity of intein to separate the target protein (MeCP2) from the affinity tag (chitin-binding domain, CBD).
The pTYB1-MeCP2wt (pc1294) plasmid [64] was modified to generate the GFP-tagged version pTYB1-GFP-MeCP2. The GFP-MeCP2 coding sequence was amplified by PCR from pEG-MeCP2 (pc1208) [70] using a primer pair containing NdeI (NEB, R0111S) for the forward primer and EcoRI (NEB, R0101S) for the reverse primer (Table S2). Then, the GFP-MeCP2 amplified coding sequence was digested with NdeI/EcoRI and ligated into the NdeI/EcoRI digested pTYB1-MeCP2wt vector. Using either pTYB1-MeCP2wt or pTYB1-GFP-MeCP2 as templates, the Q5 site-directed mutagenesis strategy was adopted following the standard protocol (NEB, E0554S) using the primer pairs listed in Table S2 (from IDT) to generate the truncated versions R168X or R255X without or with GFP tag respectively, followed by ligation, transformation of E. coli cells Top 10 (Table S4), and DNA sequencing.
Insect expression plasmids
The MeCP2-GFP was also expressed and purified from the SF9 insect cells. The Bac-To-Bac baculovirus expression system (Invitrogen) was used. The generation of the full-length MeCP2 tagged with GFP at the C-terminus (pFB-MeCP2G, pc1571) was described before [49,71].
Mammalian expression plasmids
The pGBP (pc2203) plasmid was generated from pGBPLacImR (pc1378) [71]. First, the GBP (GFP-binding protein) was released through BglII and EcoR restriction enzymes and then inserted into the mammalian expression vector pDNA3 (Invitrogen) digested with the compatible BamHI (NEB, R3136M) and EcoRI (NEB, R0101S) restriction enzymes. pEG-MeCP2-R168X and pEG-MeCP2-R255X truncations were engineered using the Q5 site-directed mutagenesis with the pEG-MeCP2 (pc1208) vector as the PCR template as described above. pMaSat-miRFP703 (pc3988) was generated by replacing mRFP1 from pMaSat-mRFP (pc2063) [72] with miRFP703 from pLifeAct-miRFP703 (pc3378) [73] using MfeI (NEB, R3589L) and AgeI (NEB, R3552L) restriction sites. pCAG-MeCP2-IB (pc2635) was generated by amplification of the mouse MeCP2 cDNA, inserted into pCAG-IRES blast plasmid (pc1234) [74] and confirmed by sequencing and immunostaining.
Protein purification and analysis
Protein purification from insect cells
The protein production and purification from SF9 cells (Invitrogen) was performed as described before [49,71]. In brief, Sf9 cells (Table S3) were cultivated in EX-CELL 420 Serum-Free Medium (24,420 G-10L, Sigma-Aldrich) supplemented with 10% fetal calf serum (FCS) at 27°C in a rotary shaker. For the recombinant baculovirus production, the SF9 cells were transfected using poly(ethylenimine) (PEI) (Polysciences, 23,966) following the manufacturer’s instruction. Baculovirus (P1 stock) in medium was collected by centrifugation and the supernatant containing the virus was used to infect new SF9 cells to get higher titers of baculovirus (P2). This step was repeated once to generate enough baculovirus (P3). For the recombinant protein production, the new SF9 cells were infected with the virus (P3) and incubated at 27°C for 4 days. Cell pellets containing GFP-MeCP2 were collected by centrifugation at 1200 rpm for 5 min and stored at −80°C.
Cell pellets were thawed on ice and lysed with cold PARP buffer (0.025 M Tris HCl pH 8, 1 M NaCl, 0.05 M glucose, 0.01 M EDTA, 0.2% Tween 20, 0.2% NP-40 substitutive and protease inhibitors (phenylmethylsulfonyl fluoride (PMSF) (Carl Roth, 6367.1), 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) (Sigma-Aldrich, A1421,0100), E64 (Sigma-Aldrich, E3132), pepstatin A (Sigma-Aldrich, P5318)) with further sheared by syringe treatment. The lysate was cleared by centrifugation and loaded onto GFP-binding protein coupled to beads [75]. After binding, the beads were washed with PARP buffer and PBS (137 mM NaCl, 2.7 mM KCl, 1 mM Na2HPO4 and 1 mM KH2PO4), proteins were eluted using 4 M MgCl2 and the buffer was exchanged to PBS. Protein concentration was determined by comparison to the BSA standard on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Protein purification from bacteria
(GFP tagged) human MECP2 protein and truncations carrying the C-terminal intein-CBD were expressed in BL21(DE3) E. coli cells (Table S4). Expression was induced with 0.5 mM IPTG (Sigma-Aldrich, I6758-10 G) at room temperature (RT) overnight. Subsequently, the cell lysates were prepared by pelleting and resuspending the bacteria in lysis buffer (20 mM Tris-HCl pH 8.5, 500 mM NaCl, 0.25% Triton X-100 and protease inhibitors PMSF, AEBSF, E64, and pepstatin A), followed by sonication on ice and centrifugation at 15,000 rpm for 30 min. The cleared lysates were incubated with 2 ml chitin beads (NEB, S6651S) at 4°C with rotation for 3 h to allow CBD-chitin binding. Then beads were washed and treated in benzonase buffer (20 mM Tris-HCl, pH 8.5, 2 mM MgCl2, 20 mM NaCl, 0.1 mM PMSF) with benzonase (MERCK, 70,746–3, 1:2000 dilution) at 37°C for 4 h, followed by washing and treatment in DNase buffer (20 mM Tris-HCl pH 8.5, 50 mM KCl, 2 mM MgCl2) with DNase I (4 µg/ml), RNase A (0.2 µg/µl) at 37°C for 25 min to remove DNA and RNA contaminants. Finally, proteins were eluted by cleavage at 4°C for two days in cleavage buffer (20 mM Tris-HCl pH 8.5, 500 mM NaCl) with 50 mM DTT (Sigma-Aldrich, D9779-5 G). The eluted fraction was concentrated using Amicon® Ultra Centrifugal Filters (MERCK), aliquoted, flash frozen, and stored at −80°C in storage buffer (20 mM Tris-HCl, pH 8.5, 300 mM NaCl).
Protein quantification and characterization
Protein concentrations were determined using Pierce™ 660 nm Protein Assay Kit (Thermo Fisher Scientific, 22,660) following the manufacturer’s instruction. In brief, 10 µl of BSA standard (Thermo Fisher Scientific, 23,208), proteins and storage buffer (blank) were mixed with 150 µl Protein Assay Reagent in the 96-well microtest plate (SARSTEDT, 82.1581.001) and incubated at room temperature for 5 min. Three replicates for each condition were performed. The absorbance at 660 nm was measured using a plate reader Infinite 200 (TECAN). The blank-corrected absorbance was calculated by subtracting the average absorbance of the blank. The standard curve was generated by plotting the average blank-corrected absorbance for each BSA standard versus the relative concentrations (µg/µl). The protein concentrations were calculated according to the standard curve using the blank-corrected measurements.
2 µg and 10 µg protein were loaded separately onto a SDS-PAGE gel and 15% Tris-borate EDTA polyacrylamide gel. The gels were stained with coomassie (to detect the proteins) and ethidium bromide (EtBr; to detect the potential contamination with nucleic acids) separately after electrophoresis. The SDS-PAGE gels after coomassie staining were captured by colorimetric trans-illumination imaging using the Amersham Imager 600 (Table S5) equipped with white light trans-illumination following the manufacturer’s instruction. The Tris-borate EDTA PAGE gels after EtBr staining were imaged using the VWR genosmart (Table S5) UV trans-illumination system.
DNA templates and methylation
Synthesis of short DNA templates for binding assay
Forty-two bp dsDNA was synthesized by primer extension using the large (Klenow) fragment of E. coli DNA polymerase I (NEB, M0210 L) as described before [75,76]. In brief, the longer CG-up/MG-up and shorter Fill-in-647 N (Table S2) were annealed by slowly cooling down to 37°C from 95°C in NEB buffer 2 (50 mM NaCl, 10 mM Tris–HCl, 10 mM MgCl2, 1 mM dithiothreitol; New England Biolabs). The short CG/MG-down were extended by adding 1 mM dATP, dGTP, dTTP, and 1 mM dCTP (Carl Roth) or 0.1 mM dmCTP (Jena Bioscience, NU-1125S) and Klenow fragment polymerase, followed by incubating for 1 h at 37°C to generate the double-strand oligos with or without CpG methylation.
20 bp dsDNA with or without cytosine methylation was generated by resuspending and mixing the Cy5-MG/CG-up and MG/CG-down (Table S2) to a final concentration of 10 mM in a solution containing 20 mM Tris pH 8.5 and 150 mM NaCl, followed by heating at 95°C for 2 min and gradual cooling by switching off the thermomixer.
Synthesis of long DNA templates for phase separation assay
The DNA used for the phase separation assay with different lengths and methylation levels were synthesized by PCR using Q5 polymerase (NEB, M0491S) as described before [72,76]. In brief, pUC18-MINX plasmid (Table S1) was applied as a template and different reverse (Rev) primers (Table S2) were used to amplify DNA of different lengths. The 800 bp DNA with cytosine methylation was synthesized by replacing the dCTP with dmCTP in the PCR mixture. The 800 bp DNA with CpG methylation was obtained with the CpG methyltransferase M.SssI (NEB, M0226S) after PCR and followed by DNA purification from agarose gel according to the manufacturer’s instructions. Briefly, 1 µg of purified 800 bp DNA product was mixed with 160 µM SAM (S-adenosyl-methionine; NEB, B9003S), methylated by 4 units M.SssI for 4 h at 37°C in the 1 x NEB buffer 2.
DNA methylation assay
Methylation was determined with methylation-sensitive restriction enzymes HpaII (NEB, R0171S) and its methylation insensitive isoschizomers MspI (NEB, R0106S). In brief, 150 ng DNA was incubated with MspI or HpaII (control with no enzyme) at 37°C for 2 h before being loaded to 15% Tris-borate EDTA PAGE. After electrophoresis, the gel was stained with EtBr for 4 min and washed with water. The images were captured by the Amersham Imager 600 equipped with RGB fluorescence (Cy5-20 bp DNA) and the VWR genosmart UV trans-illumination system.
Mononucleosome isolation
HEK293T cells were cultured as described below. The mononucleosomes were isolated as described before [15]. In brief, 3 × 107 HEK293T cells (Table S3) were resuspended in 1 ml of hypotonic buffer (10 mM Tris-HCl pH 8, 10 mM KCl, 1.5 mM MgCl2, 1 mM DTT and protease inhibitor 2 mM PMSF) containing 0.1% Triton-X 100 on ice for 5 min and centrifuged at 1000 × g at 4°C to obtain intact nuclei. Nuclei were washed 5x with a buffer (10 mM Tris-HCl, pH 7.4, 300 mM NaCl) to remove the chromatin-bound proteins. Nuclei were then resuspended in 800 µl of MNase digestion buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM CaCl2, 0.1% NP-40, and protease inhibitors) supplemented with 40 U/ml MNase (NEB, M0247S) and incubated at 37°C for 5 min. The digestion was inactivated by a 5x stop buffer containing 10 mM Tris-HCl, pH 7.4, 710 mM NaCl, and 7.5 mM EDTA. Mononucleosome extracts were cleared by centrifugation at 500 g for 15 min at 4°C. After nucleosome isolation, the buffer was exchanged to the conditions for assaying MeCP2 phase separation using Zeba™ spin desalting columns (Thermo Fisher Scientific). The quality and quantity were determined on an agarose gel after isolating DNA from the mononucleosome extracts and the histones were analyzed on a SDS-PAGE gel stained with coomassie blue.
In vitro phase separation and microscopy analysis
Proteins were firstly thawed on ice, centrifuged at 14,000 rpm, 4°C for 10 min to remove all aggregates. Phase separation properties were evaluated in solutions (20 mM Tris-HCl pH 8.5) with various salt, protein (only untagged MeCP2 or MeCP2 mixed with 1% GFP-MeCP2), crowding agents, and DNA concentrations after incubation for 45 min at RT.
To check the droplet morphology, phase separation samples were loaded onto chambers made of double-sided tapes and sealed with coverslips. Fluorescence and differential interference contrast (DIC) images were taken using a Nikon Eclipse TiE2 microscope equipped with a Plan Apo λ 40x objective or a Nikon Ti-E microscope equipped with a CFI Planapochromat VC 20x objective (Table S5). All images were processed and analyzed using ImageJ.
The biophysical properties of droplets were quantified using ImageJ according to the fluorescent images from the phase separation assay. Droplets were identified and segmented using a FFT/bandpass filter to decrease the background and fluorescence intensity-based thresholding which was manually adjusted. Droplets with an area >0.1 µm2 were considered. The aspect ratio of droplets was calculated by the ratio of the minimal Feret diameter to the maximal Feret diameter.
Turbidity assay
20 µl phase separation solutions with various conditions were prepared as above and transferred to the 384-well plate with an optically clear bottom (PerkinElmer, 6,007,550). The phase separation was done by incubating at RT for 45 min, followed by absorbance measurement at 340 nm at room temperature using a plate reader Infinite 200 (TECAN).
Droplet sedimentation assay
The MeCP2 droplets formed by incubation at RT for 45 min were sedimented by centrifugation at 14,000 rpm for 15 min at RT. The top half of the supernatants were applied to a 12% SDS-PAGE gel, which was stained with Coomassie for 1–2 h after electrophoresis and subsequently washed with destaining buffer (100 ml acetic acid, 100 ml ethanol, and 500 ml H2O) overnight. The image was taken using an Amersham Imager (Table S5), quantitatively analyzed by Adobe photoshop and plotted.
In-droplet and in-solution MeCP2 quantification
Standard curve
Solutions containing gradient concentrations of GFP-MeCP2 were loaded onto chambers made of double-sided tapes and sealed with coverslips. The images were taken using a Nikon Eclipse TiE2 microscope equipped with Plan Apo λ 40X objective (Table S5). The mean fluorescence intensities of free GFP-MeCP2 were measured using ImageJ and plotted versus the corresponding known GFP-MeCP2 concentrations to generate the standard curves.
In-droplet and in-solution MeCP2 quantification
Purified GFP-MeCP2 was mixed with unlabeled MeCP2 in a molar ratio of 1:99 and diluted to a final concentration of 80 µM in buffer (20 mM Tris-HCl, pH 8.5, 300 mM NaCl). In vitro phase separation was done by incubating at RT for 45 min at various conditions. To quantify the protein concentration in droplets, droplet mixtures were then moved to chambers made of double-sided tapes and sealed with coverslips. To quantify the protein concentration in solution, droplets were sedimented by centrifugation at 14,000 rpm for 15 min at RT, the upper clear supernatants were transferred to new PCR tubes and mixed by pipetting before being transferred to chambers. The images were taken by a Nikon Eclipse TiE2 microscope equipped with a Plan Apo λ 40x objective (Table S5). Mean fluorophore intensity in droplets or solutions was measured using ImageJ, the protein concentrations were calculated relative to the standard curves.
Pull-down interaction assay
The immobilization of untagged human MeCP2 and truncations with intein-CBD were generated as described above. Briefly, the human MeCP2 or truncations fused with intein-CBD were bound to the chitin beads by incubating the bacteria extracts with chitin beads. Then the beads were treated with nuclease to remove nucleic acid contaminants as described above. 25 µl clean beads (control) and beads with immobilized MeCP2 or truncations were transferred to cold 1.5 ml tubes, washed with PBS supplemented with 125 mM NaCl, 0.05% NP-40, and protease inhibitors PMSF, AEBSF, E64 and pepstatin A for twice, and incubated with 300 µl 0.1 µg/µl corresponding GFP tagged full-length MeCP2 or truncations in PBS supplemented with 125 mM NaCl, 0.05% NP-40, and protease inhibitors for 90 min with rotation at 4°C. Finally, the beads were collected by centrifugation, washed three times using the PBS supplemented with 125 mM NaCl, 0.05% NP-40, and protease inhibitors and followed by adding 50 µl 1X protein loading buffer (1% SDS, 25 mM Tris pH 6.8, 5% glycerol, 50 mM DTT and 0.005% bromophenol blue), and boiling at 95°C for 5 min.
Samples were then loaded to SDS-PAGE gel, electrophoresis was performed at 90 V for 100 min followed by semi-dry transfer at 25 V for 35 min. Then, the membranes were blocked with 3% low-fat milk in PBS for 30 min at RT, incubated with rabbit anti-GFP antibody (Table S6) at 4°C overnight on a rotary shaker, and followed by anti-rabbit IgG conjugated with Cy3 for 1 h at RT. The fluorescence signals were detected using an Amersham Imager (Table S5).
Microscale thermophoresis assay
The microscale thermophoresis (MST) assay was conducted using the Monolith NT.115 (NanoTemper) following the manufacturer’s instruction. To check if MeCP2 is capable of self-interaction, 10 µl 200 or 300 µM GFP tagged MeCP2 or truncations were mixed with equal volumes of untagged MeCP2 gradients or truncations in buffer containing 20 mM Tris-HCl and 150 mM NaCl. To check the binding affinity of MeCP2 with DNA, 10 µl 100 µM short oligos labeled with ATTO-647 N and Cy5 with or without methylation (Table S2) were mixed with equal volumes of untagged MeCP2 gradients or truncations in buffer containing 20 mM Tris-HCl and 150 mM NaCl. Then the mixtures were transferred into Monolith NTTM capillaries and the fluorescent changes to microscopic temperature gradients were measured by Monolith NT.115 using 20% (for self-interaction assay) or 40–50% (for protein–DNA interaction) excitation power. The dissociation constants (Kd) were calculated using MO. Affinity Analysis software.
Mammalian cell culture and transfection
All cell lines were free of mycoplasma contamination and listed in Table S3.
Human embryonic kidney (HEK) 293 T cells were authenticated by STR profiling. HEK293T cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 50 µg/ml gentamicin (Sigma-Aldrich Chemie GmbH, G1397).
Mouse cell lines were counterstained with 1 µg/ml 4′,6-diamidino-2-phenylindole (DAPI) and visually inspected for the presence of chromocenters.
MEF-P (P53−/−, methylation proficient) and MEF-PM (P53−/− DNMT1n/m, methylation deficient) mouse embryonic fibroblast were cultured in DMEM high glucose (Sigma-Aldrich Chemie GmbH, D6429) supplemented with 15% FCS, 1x L-glutamine (Sigma-Aldrich Chemie GmbH, G7513), and 1 µM gentamicin (Sigma-Aldrich Chemie GmbH, G1397). The MEF-PM cells were stained with anti-5mC specific antibody and showed no signal compared to the wild-type counterpart (MEF-P) (Fig. S6).
C2C12 myoblasts cells were tested for the ability to generate differentiated myotubes. C2C12 mouse myoblast cells were cultured in DMEM high glucose (Sigma-Aldrich Chemie GmbH, D6429) supplemented with 20% FCS, 1x L-glutamine (Sigma-Aldrich Chemie GmbH, G7513), and 1 µM gentamicin (Sigma–Aldrich Chemie GmbH, G1397).
Transfection of all cell lines was performed using either Neon Transfection System (Thermofisher) or AMAXA nucleofector (Lonza). Cells were harvested for Western blot/FACS sorting, used for live (single-molecule) cell microscopy or fixed for immunofluorescence staining 20 h after transfection if not stated differently.
Quantification of MeCP2 levels
Flow cytometry
C2C12 myoblasts transiently transfected with pMeCP2G (pc1121) were harvested 20 h after transfection, resuspended in PBS and separated according to their transfection level by fluorescence-activated cell sorting (FACS) on the S3 Cell Sorter (Bio-Rad Laboratories) with a 488 nm laser and a 525 ± 30 nm filter. Cells were plotted for log10 GFP sum intensity and divided into 40 bins. The first 11 bins were defined as negative cells by comparison to the untransfected C2C12 control cells. Cells in bins 13 to 21 were defined as low expressing, cells in bin 24 to 32 as high expressing (plots with the categories negative, low and high are shown in Figure 3(h)). Low and high MeCP2 expressing cells were collected as pellets for Western blot.
Figure 3.
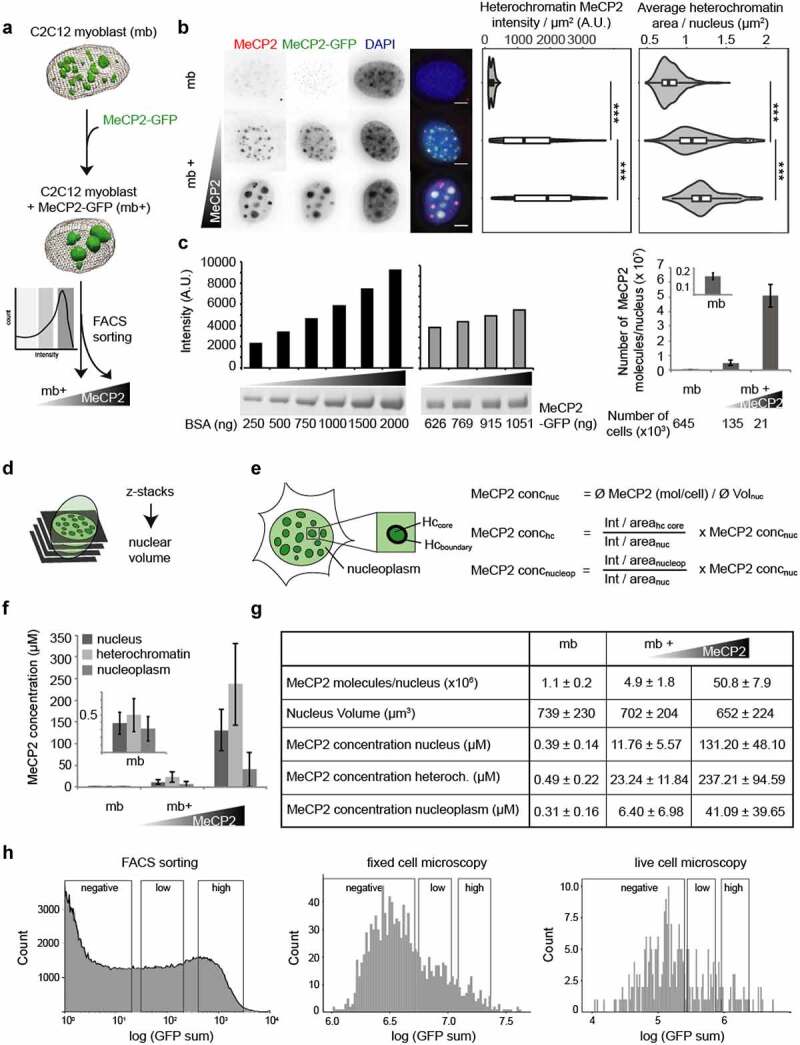
Validation and calibration of a cellular system mimicking in vivo MeCP2 physiological behavior.
(A) Scheme of the experiment: C2C12 myoblast (mb-) were transfected with a plasmid encoding for MeCP2-GFP. After 20 h, transfected cells (mb+) were sorted into two categories, low and high expressing, according to the GFP intensity using a Fluorescence-activated cell sorting (FACS).(B) Immunofluorescence staining showing MeCP2 levels in mouse myoblasts before and after transfection of MeCP2-GFP. Scale bars = 5 µm. Boxplots show the MeCP2 heterochromatin mean intensity and the mean heterochromatin cluster area of untransfected, low and high MeCP2 expressing myoblasts of three independent replicates (***p < 0.001, Wilcoxon test).(C) Quantification of total MeCP2 in mouse myoblasts. The concentration of MeCP2-GFP standard was determined by SDS-PAGE and coomassie staining in comparison to a BSA standard series. The MeCP2 standard was used to quantify the MeCP2 protein level in untransfected, low and high expressing FACS sorted mouse myoblasts by Western blot against MeCP2 for three independent replicates (average values ± standard deviation). Full gels and Blots are shown in Fig. S3.(D) Scheme of the calculation of the nuclear volume based on thresholded 0.2 µm z-stacks.(E) Scheme of the cell segmentation and the calculations of the MeCP2 concentration in the nucleus and its subcompartments heterochromatin and nucleoplasm. Nucleus concentration (concnuc) was obtained from total protein amount from Western blot divided by nucleus area, subcompartment concentration (conchc or concnucleop) were calculated from the intensity ratios between the correspondent subcompartments (for conchc, we used the core region, nucleoplasm for concnucleop) versus nucleus and nucleus concentration.(F) Average MeCP2 concentrations in the nucleus and its subcompartments heterochromatin and nucleoplasm in untransfected, low and high expressing mouse myoblasts (average ± standard deviation).(G) Table showing the results of total MeCP2 quantification (as average ± standard deviation) in untransfected, low and high transfected myoblasts. Calculated values of number of molecules, nucleus volume (µm3) and MeCP2 concentration (µM) for each condition. Low mb+ present similar number of molecules to those calculated in mouse brain cells ([83]).(H) Instrument-independent calibration of the quantification of MeCP2. To allow comparison of intensities between instruments, we classified the cells using a population curve after transfection plotting counts versus log of the GFP intensity, considering three windows: negative, defined from the measurements obtained from untransfected cells, and then dividing the positively transfected cells in 29 bins, in which 2–10 correspond to low MeCP2 levels and 13–21 to high MeCP2 levels.
Western blot
Cell pellets collected from FACS (for low and high MeCP2 expressing cells) or directly from culture (for untransfected cells) were lysed in 1 M lysis buffer (0.025 M Tris, 1 M NaCl, 0.9 g glucose, 0.01 M EDTA, 0.2% Tween 20, 0.2% NP-40 substitutive) supplemented with protease inhibitors PMSF, AEBSF, E64 and pepstatin A, mechanically disrupted and boiled at 95°C in Laemmli buffer (2% SDS, 50 mM Tris, 10% Glycerol, 0.01% bromophenol blue, 100 mM DTT). Samples were run on 8% SDS-PAGE and transferred to nitrocellulose membrane. After blocking with 5% low-fat milk in PBS the membranes were incubated with anti-MeCP2 rat monoclonal antibodies 4H7 and 4G10 cell culture supernatants from hybridoma cell lines overnight followed by anti-rat IgG Cy3 secondary antibody diluted 1:1000 for 1 h (Table S6). Fluorescent signals were detected using Amersham Imager (Table S5). The bands on the Western blots were quantified using ImageJ by selecting single lanes, plotting intensities along the lanes, selecting the peaks corresponding to the bands of interest and measuring the bands relative intensity (area under the peak).
Immunostaining
For immunostaining, cells were grown on gelatin-coated coverslips and fixed with ice-cold methanol for 6 min. After washing cells were permeabilized with 0.5% Triton, washed with PBST (0.01% Tween-20), blocked with 0.1% fish skin gelatin for 20 min and incubated with primary antibody anti-MeCP2 rabbit polyclonal (Table S6) 1:250 for 2 h. After washing with PBST (0.1% Tween-20), cells were incubated 1 h with secondary antibody anti-rabbit IgG Cy5 (Table S6) 1:400, followed by another washing step with PBST (0.1% Tween-20). Samples were counterstained with 1 µg/ml DAPI and mounted in Mowiol.
Imaging and image analysis
The immunofluorescence stainings of fixed C2C12 cells were imaged using an Axiovert 200 microscope (Table S5) with 63x Plan-Apochromat 1.4 NA oil immersion Ph3 objective. Segmentation of nuclei, heterochromatin and intensity measurements were performed in ImageJ 2.0 (https://imagej.nih.gov/ij/). Heterochromatin segmentation was based on normalization of pixel intensities in relation to a local maximum intensity. First, individual pixel intensities were calculated in squares of 30 × 30 pixels, being the maximum of these 900 pixels the local maximum. To avoid that ‘dark squares’ became thresholded, only the pixels with intensities higher than 8/42 of the cell maximum (higher local maximum on the cell) were considered for further steps. Then, thresholding was applied by giving a value 1 (white) to pixels with intensities higher than 21/42 (for heterochromatin compartments) or 37/42 (for heterochromatin core) of the local maximum intensity. These thresholded images were used to generate individual and total heterochromatin (core) ROIs, that were subsequently subtracted to generate the nucleoplasm ROI for each nuclei. The full script is available in omero (http://cc-omero.bio.tu-darmstadt.de/webclient/?show=project-352) and TUdatalib (https://doi.org/10.48328/tudatalib-672).
To categorize the cells in an equivalent manner as the cells used in Western blot, we measured the total intensity of the nucleus and applied an equivalent binning system as in the FACS sorting.
Additional imaging of the same samples was performed on a confocal microscope Leica TCS SPE-II equipped with a 63x/1.30 ACS APO Oil CS 0.17/E,0.16 objective (Table S5) as stacks with 0.2 µm z interval. For this subset of images, 3D segmentation of nuclei and heterochromatin compartments was performed using Volocity software. Briefly, nuclei segmentation was done based on the 1 µg/ml DAPI channel by finding objects, dilating three times, filling holes, eroding three times and choosing objects by size. Violin plots and Wilcoxon significance tests were done using R. Bar diagrams were prepared in Microsoft Excel.
Quantification of MeCP2 concentrations
To calculate MeCP2 nuclear concentration, we used the number of molecules obtained in Western blot and the volumes of the nuclei obtained from the confocal images. These concentration values together with the ratios of mean intensity in the compartments in comparison to the whole nucleus were used to calculate the local MeCP2 concentrations in nuclear subcompartments. Although intensity ratios obtained from compartment volumes and compartment areas were comparable, the information used here derives from the areas as the sample number was larger.
DNA methylation rescue assay
Rescue assay
MEF-PM cells were cultured as described above on gelatin-coated coverslips. To rescue the DNA methylation deficient phenotype, cells were transfected using the Neon transfection system with pGEMT1L (pc1031) and pCAG-GMT3b-IB (pc1269), together with pMaSat-GBP (pc2469, to target GFP-tagged proteins to constitutive heterochromatin) and pCAG-MeCP2-IB (pc2635). As a control, cells were exposed to Neon transfection without DNA. Cells were further cultured for 48 or 72 h prior to fixation with 3.7% formaldehyde for 20 min.
Immunostaining
For detection of 5mC levels, the immunostaining protocol described in [76] was used. In short, a permeabilization with 0.5% Triton for 20 min was performed on the fixed cells, followed by incubation with ice-cold methanol for 5 min. Cells were washed three times with PBST (0.01% Tween 20) and treated with 10 µg/mL RNase A for 30 min at 37°C. Cells were washed again three times with PBST (0.01% Tween 20) and unspecific antibody binding was blocked with 0.2% fish skin gelatine for 30 min at 37°C. The primary mouse monoclonal anti-5mC (Table S6) (1:250 in 2% BSA) antibody was incubated for 70 min at 37°C during DNase I (2000 U/mL in DNase I buffer) treatment. Additionally, rabbit polyclonal anti-MeCP2 (Table S6) (1:250) was applied. To stop the enzymatic reaction, cells were washed with PBSTE (0.01% Tween 20, 1 mM EDTA) three times for 0, 5 and 10 min, respectively. Afterward, a secondary antibodies mix (anti-rabbit IgG Cy3 1:400 and anti-mouse IgG Cy5 1:200 in 4% BSA) was applied for 45 min. Cells were washed with 0.01% PBST three times for 0, 5, and 10 min, respectively. Samples were counterstained with 1 µg/ml DAPI for 15 min, mounted with vectashield and sealed with nail polish.
Imaging and image analysis
Immunofluorescence stainings were imaged on a confocal microscope (Leica TCS SPE-II) equipped with a 63x/1.30 ACS APO oil immersion objective (Table S5).
Segmentation of nuclei, heterochromatin and intensity measurements were performed in ImageJ 2.0 using a self-made macro as described above.
Single-molecule microscopy and analysis
Time lapse image acquisition
Single-molecule movies were taken at RT in a single-molecule microscope setup previously described in [77] (Table S5). In live cell microscopy, the background was partially prevented/contrast was improved, by using highly inclined laminar optical sheet illumination (HILO) [78], while wide field mode was used for the in vitro minimal system. The laser intensity was no more than 2.5 mW/cm2 at the sample and the acquisition was not longer than 1 h per sample to avoid possible DNA damage and/or overheating of the cells. Every experiment was performed at 20 ms exposure time.
Data analysis
The analysis was guided by a self-made macro running on FIJI [79] divided into four main parts: I) processing of the movies and generating the ROIs (nucleus contour, droplet border, droplet boundary) necessary for the analysis; II) TrackMate [80] run with pre-set parameters and obtain the spot coordinates; III) Use TrackMate coordinates to calculate track parameters and location of the tracks in relation with the ROIs and sort the tracks for the step-size analysis (see below); IV) Generate the final images based on the coordinates and statistics obtained in III. The script is available in omero (http://cc-omero.bio.tu-darmstadt.de/webclient/userdata/?experimenter=-1) or TUdatalib (https://doi.org/10.48328/tudatalib-672).
In brief, input movies were categorized into ‘overview’ (short, 200–500 frames that will be then resliced to 1 frame by averaging intensity) or ‘single-molecule’ (3000–15,000 frames, from which the pre-bleached frames are removed) and processed differently. Overview pictures were either not processed, or a Gaussian blur filter (2 pixel) and background subtraction (10 pixel) were applied. Single-molecule movies were processed using Gaussian blur (1 pixel), background subtracted (5 pixel) and a manual threshold to eliminate the most obvious noise was done by modifying the brightness/contrast minimal value. This processing allowed us to run TrackMate in similar conditions for the same protein in different experiments.
Next, we sorted the tracks in four (heterochromatin core, heterochromatin boundary, nucleoplasm and crossing tracks) categories. The sorting was based on ROIs generated from the overview image corresponding to MaSat, which was thresholded to generate the heterochromatin compartments. For the heterochromatin boundary, one pixel to the inside and one pixel to the outside of the heterochromatin segmentation line was used, thus creating a two pixel-wide heterochromatin border region surrounding the core heterochromatin region. Nucleoplasm ROI was obtained as the subtraction of the corresponding heterochromatin regions from the nuclear ROI. For sorting, spot coordinates of the tracks were obtained from TrackMate and the presence of the tracks in the ROIs corresponding to either only heterochromatin, or heterochromatin core or boundary was checked. Once the tracks were sorted, spots were relocated as a movie into a pre-established mesh (contour coordinates recognized in the MATLAB-derived software) pattern generated in Oufti [81], so only one spot is appearing per frame and mesh. Tracks were again generated with TrackMate, this time exported as a track file (xml) suitable for SMTracker. The effects of relocation were tested by generating meshes for the tracks in situ and showed no differences to the relocated in SMTracker when calculating diffusion coefficient or population weight.
The track file from the second TrackMate run and the Oufti mesh pattern were then imported into SMTracker [82], where step size analysis was performed. This analysis determined the dynamic populations, including the diffusion coefficient (D) and the percentage of each population using two different methods (square displacement and Gaussian-mixture model). Square displacement model calculates the D and populations for each condition individually, including a statistical model for the best fitting model (defined as the simplest model that can explain the data obtained) and the residuals (differences between the different models and the original data). While in the Gaussian-mixture model all the steps from all conditions are used to obtain the D and then the populations are fitted individually for each condition. In this study, however, the square displacement model was used to define the number of populations, while the Gaussian-mixture model was used to both define the D in the individual and comparison conditions.
Statistics
All sample size, statistic evaluations and significance tests are provided in Tables S7-S21.
Results
MeCP2 forms condensates with characteristics of liquid-like droplets in physiological concentrations
Proteins undergoing LLPS commonly contain intrinsically disordered regions (IDRs), which form multivalent interactions based on electrostatic and/or hydrophobic interactions. Thus, we first analyzed the physicochemical properties of the MeCP2 amino acid sequence. We found that IDRs are present both upstream and downstream of the conserved structured MBD within MeCP2 as depicted in the PONDR prediction plot (Figure 1(a)). Next, we purified recombinant human MeCP2 using a bacterial expression system based on a chitin binding domain and an intein motif. In this manner, soluble MeCP2 is obtained, which after protein splicing yields untagged MeCP2 [64]. Concomitantly, GFP-tagged MeCP2 was produced and purified in a similar manner. MeCP2 contains a large number of basic amino acids, leading to an overall positive charge and high isoelectric point (pI, pH 10.56). This characteristic is also responsible for a strong non-sequence specific interaction between MeCP2 and DNA. As a result, during MeCP2 protein purification DNA in addition to methylated DNA are often co-purified. As nucleic acids have been reported to promote LLPS, DNaseI and benzonase were applied to remove the nucleic acids from MeCP2 purifications. This was important to establish the minimal conditions and the ability of MeCP2 alone to form liquid-like droplets in vitro. As shown in Figure 1(b), purified recombinant human untagged as well as GFP-tagged MeCP2 were free of detectable DNA contamination. We, then, used these preparations to analyze whether MeCP2 could by itself phase separate.
Figure 1.
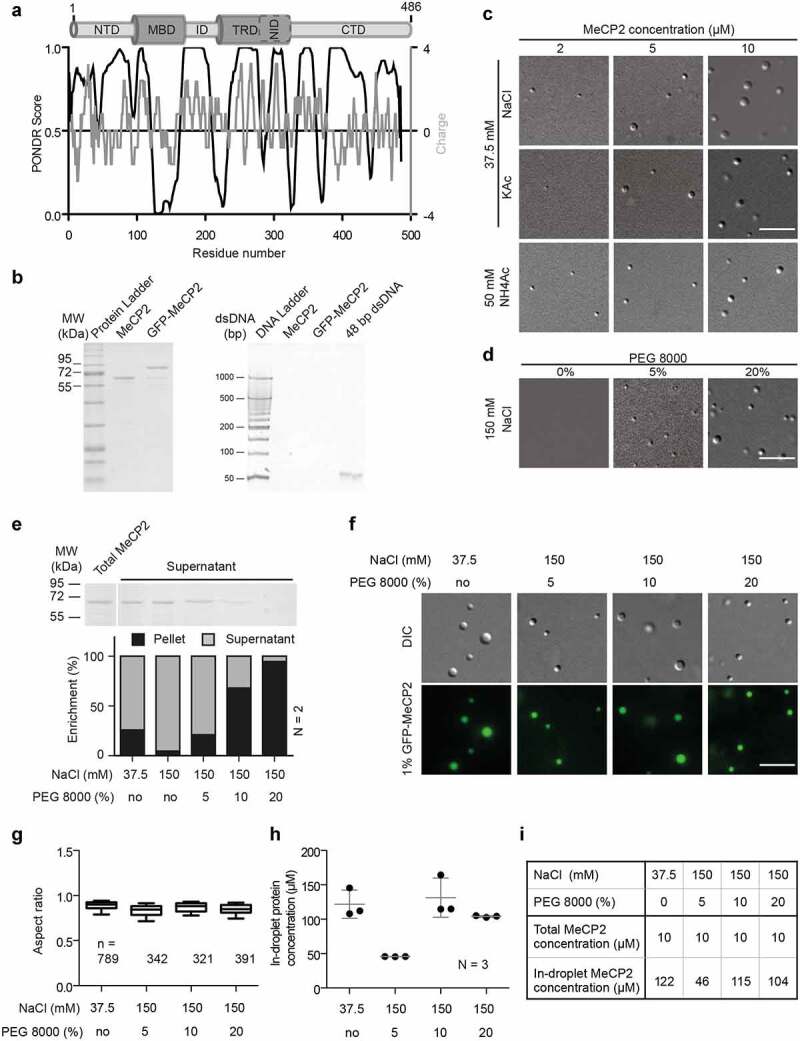
Purified MeCP2 forms liquid-like droplets in physiologically crowding environments.
(A) Analysis of human MeCP2 protein sequence. Top: Schematic overview of human MeCP2 structure. NTD: N-terminal domain; MBD: methyl binding domain; ID: intervening domain; NID: N-CoR interacting domain; CTD: C-terminal domain; TRD: transcriptional repression domain. Bottom black line: PONDR prediction (http://www.pondr.com/) of MeCP2 ordered/disordered regions, >0.5 is considered disordered. Bottom gray line: protein charge, >0 means positively charged (https://www.bioinformatics.nl/cgi-bin/emboss/charge). The Isoelectric point (PI) of MeCP2 is predicted 10.56 using INNOVAGEN (https://pepcalc.com/). Amino acid labeling is according to human MeCP2 isoform 1.(B) Validation of MeCP2 purity. The MeCP2 and GFP-MeCP2 proteins were expressed in bacteria by IPTG induction, purified using chitin beads and eluted by DTT. The final protein concentrations were measured by Pierce™ 660 nm Protein Assay Reagent. 2 µg and 10 µg purified protein were then used for SDS polyacrylamide gel electrophoresis and tris borate EDTA polyacrylamide gel electrophoresis, respectively. Left: SDS polyacrylamide gel electrophoresis of purified human MeCP2 and GFP-MeCP2 followed by Coomassie staining. 2 µg each lane. Right: tris borate EDTA polyacrylamide gel electrophoresis of purified human MeCP2 and GFP-MeCP2 followed by ethidium bromide (EtBr) staining. 10 µg each lane. ~140 ng 42 bp DNA was used as positive control.(C) DIC images of MeCP2 phase separated droplets. The in vitro phase separation assay performed at different protein concentrations in buffers containing low concentrations of monovalent cations (37.5 mM NaCl, KCl and NH4Ac). The mixtures were transferred to chambers made of double-sided tapes and sealed with coverslips 45 min after incubation at room temperature. The droplets were observed using a Nikon Eclipse TiE2 microscope equipped with differential interference contrast (DIC) microscopy. Scale bars = 10 µm.(D) DIC images of MeCP2 phase separated droplets in the presence of crowder, PEG 8000. The in vitro phase separation was done by incubating at room temperature for 45 min and the droplets were observed using the Nikon Eclipse TiE2 microscope equipped with differential interference contrast microscopy. NaCl: 150 mM, MeCP2: 10 µM. Scale bars = 10 µm.(E) Quantification of MeCP2 distribution in solution (supernatant) and in droplets (pellets). 10 µM MeCP2 was applied for the phase separation assay in different conditions. The in vitro phase separation assay was done by incubating the mixtures at room temperature for 45 min. Then droplets were pelleted by centrifugation at 12,000 rpm for 10 min at room temperature. The top half of the supernatant was transferred to new tubes for SDS polyacrylamide gel (SDS-PAGE) electrophoresis, followed by coomassie staining and subsequent quantitative analysis. Top: Coomassie staining result of the supernatants after SDS-PAGE electrophoresis. Bottom: Quantitative analysis for SDS-PAGE gel above. Replicates (N) = 2.(F) Fluorescence images of MeCP2 phase separated droplets. The 1% GFP-MeCP2 was mixed with 99% untagged MeCP2 (molar ratio) and applied for the in vitro phase separation assay in different conditions by incubating 45 min at room temperature. Both fluorescent images (GFP) and DIC images were taken under the Nikon Eclipse TiE2 microscope equipped with differential interference contrast (DIC) microscopy. Total MeCP2 concentration: 10 µM. Scale bar = 10 µm.(G-H) Droplet aspect ratio (G) and in-droplet protein concentration (H) for experiments in F.The GFP channel was applied for droplet segmentation by bandpass filter and threshold based on the mean intensity in/out droplets. Droplets with size > 0.1 µm2 were identified and droplet parameters were measured. The aspect ratio (G) was calculated as the ratio of maximal Feret diameter to minimal Feret diameter. The in-droplet MeCP2 concentration (H) was measured by the GFP intensity inside the droplets. For details, see Fig. S2. n: Number of droplets. Replicates (N) = 3. n: number of droplets.(I) Mean values of aspect ratio and in-droplet MeCP2 concentration from (G-H).
Based on the MeCP2 levels in the mouse brain (6 × 106 molecules per nucleus) [83] and the size of the nuclei (200–800 µm3, 500 µm3 was taken as average) [84], we calculated the MeCP2 concentration (~s10-50 µM in mouse brain cells). We found that MeCP2 alone in the range of physiological concentrations (~10 µM) formed distinct spherical condensates at low salt concentration independently of the cation used in the buffer (Na+, K+, NH4+). The number and size of the condensates increased with increasing protein concentration (Figure 1(c), Fig. S1A) but decreased with increasing salt concentration till no droplet formed at physiological salt conditions (150 mM salt) (Fig. S1A). The results were further validated by analyzing the turbidity of the solution as a measure of droplet formation and phase separation (Fig. S1B, Table S18), which increased at higher protein concentration and lower salt concentration. Time-lapse imaging showed that these condensates could fuse into bigger ones (Fig. S1C, Movie S1), indicating that the MeCP2 condensates are highly dynamic and liquid-like. This is reminiscent of the fusion of MeCP2 labeled heterochromatin domains in cells as we have previously shown [24]. Surprisingly, the addition of low concentrations of 1,6-hexanediol, a chemical used to disrupt hydrophobic interactions in LLPS, did not show a significant influence on droplet morphology (Fig. S1D). Only at high concentrations of 1,6-hexanediol, in addition to liquid-like spherical droplets (red ellipse), irregular aggregates (yellow ellipse) could also be observed (Fig. S1D). Thus, the MeCP2 LLPS is probably mainly driven by electrostatic interactions rather than hydrophobic interactions. Bio-macromolecules are commonly used to mimic nuclear crowding environments and were shown to promote LLPS. We found that in physiological salt conditions both PEG 8000 and dextran T150 could promote LLPS of MeCP2 in a concentration-dependent manner (Figure 1(d), Fig. S1E). The MeCP2 levels in the buffer phase (supernatant (S)) and in the droplets were quantified by droplet sedimentation assay (Fig. S2A). The result showed that more MeCP2 molecules were concentrated in the droplets with higher concentrations of PEG 8000 (Figure 1(e), Table S7), indicating a decreased MeCP2 concentration threshold for LLPS when in crowding environments. Yet, the crowders themselves were not enriched inside the protein droplets (Fig. S1F).
The droplet properties were further quantitatively analyzed. Untagged MeCP2 mixed with 1% GFP-tagged MeCP2 was applied for LLPS in the conditions described above. We found that all droplets were labeled with GFP (Figure 1(f)) and highly spherical (Figure 1(g), Table S7), further suggesting that they are liquid-like. We then quantified the MeCP2 concentration in the droplets and in solution at low salt concentration and in the absence of crowders. The MeCP2 concentration was quantified based on the GFP intensity inside the droplets and in solution interpolated from a standard curve of GFP-MeCP2 in solution (Fig. S2B-D, Table S19). Firstly, 10 µM MeCP2 was incubated with 37.5 mM NaCl in the absence of crowders and the in-droplet and in-solution concentrations were calculated. Results of the MeCP2 concentration in droplets (121.7 ± 20.5 µM) and in the solution (8.6 ± 0.5 µM) revealed a 14-fold enrichment of MeCP2 in droplets following LLPS (Figure 1(h), Fig. S2E, Table S7, Table S19). Yet, the MeCP2 concentration in solution did not decrease significantly (8.6 ± 0.3 µM compared to the total initial concentration 10 µM) probably because only a small fraction (~25%) of MeCP2 underwent LLPS in this condition (Fig. S1A-C, Figure 1(e)). Then, we analyzed the MeCP2 concentration in the droplets at physiological salt (150 mM) and protein (10 µM) conditions with various crowder concentrations. With increasing crowder concentration, the MeCP2 concentration in the droplets increased from 45.63 ± 0.11 µM at 5% PEG 8000 to 104.01 ± 1.27 µM at 20% PEG 8000 (Figure 1(h), Table S7). This means that the (crowding) environment influences the MeCP2 distribution. In vivo, MeCP2 was reported to be 9-fold enriched at heterochromatin compartments in NIH3T3 fibroblast cells and 5-fold enriched in Pmi28 myoblast cells [23,37], which is of a similar magnitude to the MeCP2 enrichment we found in the droplets upon phase separation.
In conclusion, purified MeCP2 showed the ability to form liquid-like droplets by itself in physiological protein concentrations, which we defined as the minimal MeCP2-LLPS system.
MeCP2 moves freely and homogeneously inside liquid droplets
With the minimal LLPS system established, we investigated whether and how LLPS impacts MeCP2 dynamics in addition to its local concentration. For that, we used a high-resolution single-molecule tracking approach adding a small amount of fluorescent MeCP2 (GFP-MeCP2) to the untagged MeCP2 solution (Figure 2(a)). We performed single-molecule microscopy by illuminating the settled down LLPS droplets with a laser and used the pre-bleached image to determine the LLPS contour (Figure 2(b)). For single-molecule tracking, fluorescence bleaching was stopped when about 10–20 spots per time point were detected in TrackMate. Then, the track coordinates were imported to SMTracker to determine the number of populations, diffusion coefficients, and population weights as described in methods. To make the comparison between conditions clearer, we removed the model curves for each condition in the square displacement graphs (Figure 2(d)), and left only the differences between data and model (residuals), which indicate the simplest model statistically accepted (Figure 2(c), lower graphs, Table S8). We could observe movement of MeCP2 molecules mostly inside the LLPS-droplets (Figure 2(b)), but not outside. To explain the lack of tracks outside, we calculated the theoretical diffusion coefficient (D) for GFP-MeCP2 in salt solution, using the Einstein-Stokes relationship for 3D diffusion (Equation (1)).
| (1) |
Figure 2.
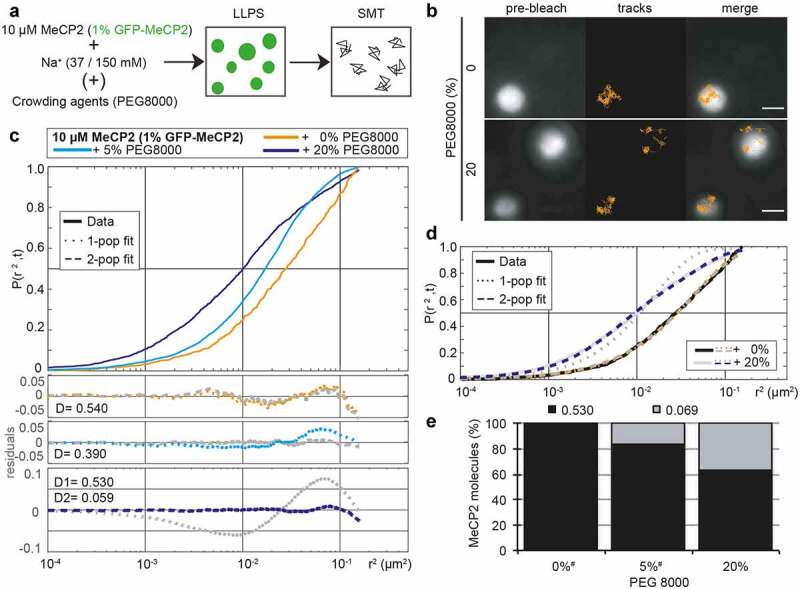
MeCP2 is confined in the droplets but diffuses freely and homogeneously inside.
(A) Scheme of the experiment: 10 µM MeCP2 containing 1% GFP-MeCP2 was incubated in presence of Na+ (37 mM when no crowders are present, 150 mM in any other case) and in absence or presence of crowding agents (PEG 8000), allowing the formation of LLPS droplets for 45 min at room temperature prior to the imaging in the single-molecule microscope to obtain MeCP2 tracks.(B) Exemplary GFP-MeCP2 single-molecule tracks in MeCP2 LLPS droplets. The pre-bleach image allows the recognition of the droplet contour. Only tracks containing at least four spots are shown and used for the analysis. Scale bars = 2 µm.(C) Square displacement cumulative distribution function for the MeCP2 tracks obtained in single-molecule tracking on MeCP2 LLPS droplets, indicating the probability of a track to be included in a concrete circle with radius r (r2). Bottom panels correspond to the residuals of the comparison of the data versus models for one or two population(s), being the colored one the statistically acceptable simplest model according to the Bayesian Information Criterion, for which the diffusion coefficients (D) are given (in µm2s−1).(D) Graphical illustration of the meaning of the residual, as square displacement cumulative function of the data of 0% and 20% PEG from C, together with the corresponding cumulative functions for the 1 (dotted line) and 2 (dashed line) for each condition. The residuals correspond to the difference between the model lines to the data.(E) Predicted population weight in a 2 population model, considering the same D for all conditions for better comparison using Gaussian-mixture model. # indicates that for these conditions, the prediction is forced as the best fitting model is 1 population.
We needed to make some assumptions, as a globular conformation for MeCP2 and a temperature (T) of 25°C. For this assumed parameters, we used calculated dynamic density (η) for water (0.89 mPa s, from IAPWS [The International Association for the Properties of Water and Steam] R12-08) and NaCl buffers of 40 mM (0.903 mPa s) or 150 mM (0.9237 mPa s) from [85], as well as an estimation of the density (ρ) of a globular protein as 1430 kg m3 from Quilling and Matters, 2000 [86], together with the known Avogadro (NA, 6.022·1023) and Boltzmann (kB, 1.381·10−23) constants and molecular weight (Mw) of GFP-MeCP2 of 80,053.30 Da. Taking all these values, the stocking radius of a MeCP2 molecule (Rs, Equation (2)) is 2.18·10−8 m.
| (2) |
Altogether, the D results in 8.73, 8.60 and 8.41 µm2 s−1 in water, 40 mM NaCl, and 150 mM, respectively. These values are far higher than the maximum D that can be calculated in our settings for tracking. The latter consider a maximum movement within a 200 nm radius every 20 ms in 4 consecutive frames, allowing a maximum D of 2.5 µm2 s−1 obtained from the slope of the regression of the MSD curve (y = 10x-0.2).
In absence of any other component, MeCP2 showed an isotropic diffusion fitted to a one-population model with a D of 0.540 µm2 s−1 (Figure 2(c), orange line, Table S8), which is far below the maximum D for our setup and approximately 6% of the theoretical diffusion of MeCP2 in the buffer used. The addition of crowders (20%, Figure 2(b), down) led to an anisotropic MeCP2 movement, as it required at least a second population to statistically explain the data (Figure 2(c), dark blue line, Table S8). From these 2populations, one diffuses in a similar fashion as in the absence of crowders (0.530 µm2 s−1), while the second population is almost 10-fold more constrained with a D of 0.059 µm2 s−1. Lower amount of crowders (5%, light blue line), showed an intermediate effect as, despite being statistically explained by a 1-population model, the residuals showed a deviation of the data. Forcing the Gaussian-mixture model analysis to define 2 populations confirmed this observation. Hence, in the absence of crowders, 99.9% of MeCP2 molecules showed a fast diffusion while 5% crowders raised the slow diffusing population to 16% and 20% crowders raised the slow population to 36% of MeCP2 molecules (Figure 2(e)).
In summary, we found that MeCP2 exhibited constrained and homogenous mobility within the phase-separated droplets and addition of crowding agents yielded a second slower mobility population.
Comparison of MeCP2 concentrations in vitro and in vivo
Abnormal MeCP2 dosage can cause neuronal dysfunction and Rett-like phenotypes, and overexpression as well as loss of MeCP2 were reported to show these phenotypes [40,87–90]. In the mouse brain, as mentioned above, the MeCP2 average cellular concentration was reported to be ~10 µM. Hence, for the phase separation assays, we used concentrations around this value to stay in the physiological range. For this reason, we wanted to use the same range throughout all experiments in vitro and in cells. Thus, we developed a procedure to calibrate the MeCP2 concentration in individual cells and subcellular compartments. Moreover, we established the conditions that allow us to apply these calculations across different measurement platforms. This ensured that the measurements and data obtained at different systems in cells and in vitro could be directly combined and mimicked the physiological conditions in vivo.
Therefore, we estimated the MeCP2 concentration in mouse myoblasts transiently transfected with MeCP2-GFP and compared the total levels to in vivo levels in the mouse brain reported previously [83]. Myoblasts virtually have low levels of MeCP2 protein, making them a perfect system to manipulate the levels of MeCP2 and measure the effects [24].
First, we transfected the cells with a plasmid coding for GFP-tagged MeCP2 and FACS sorted them into three categories according to MeCP2 levels 20 hours after transfection: negative (no transfection), low MeCP2 and high MeCP2 (Figure 3(a), Fig. S3). The sorted cells were collected for absolute protein quantification on Western blot. To start, the concentration of purified MeCP2-GFP was determined by comparison to a BSA standard row on Coomassie stained SDS-PAGE. Then, the MeCP2-GFP standard was used to determine the total number of MeCP2 molecules in the FACS sorted cells by Western blot and detection with antibodies against MeCP2 (Figure 3(c), Fig. S3, Table S10). The total number of MeCP2 molecules calculated per lane was divided by the number of cells lysed and loaded in the respective lane. The untransfected myoblasts had 1.1 ± 0.2 million MeCP2 molecules per cell, thus 4.5 times less MeCP2 as low transfected cells with 4.9 ± 1.8 million molecules per cell. The high expressing cells had 50.8 ± 7.9 million per cell, i.e., 10 times more than low expressing cells. In physiological terms, low expressing cells emulate the protein amount of 6 million molecules per cell reported for unsorted mouse brain cells including glia and neurons [83], while high expressing cells have about three times higher levels than reported for mouse brain neurons (16 million per cell [83],).
Next, we wanted to apply the FACS gates and the corresponding protein levels to microscopy images (Figure 3(h)). Therefore, we fixed MeCP2-GFP expressing myoblasts 20 hours after transfection and performed immunofluorescence staining against MeCP2 and counterstained with the DNA dye 1 µg/ml DAPI. The cells were imaged on a widefield fluorescence microscope (Figure 3(b)) and nuclei, heterochromatin fractions, heterochromatin core and edge regions were segmented based on 1 µg/ml DAPI channel intensities (see Figure 3(e)). Based on the nucleus GFP intensity, the cells were binned into untransfected, low and high expressing using the same binning system as for the FACS sorting. The MeCP2 mean intensity in the heterochromatin area reflected these classification bins and the average heterochromatin area increased significantly with higher MeCP2 levels (Figure 3(b), Table S9), which is in line with our previous studies [24].
To obtain the MeCP2 concentration needed for comparison to in vitro protein amounts, we took z-stack images of fixed cells, segmented the nuclei and calculated the nuclear volumes (Figure 3(d), Table S9). Nuclear volume of untransfected cells was 739 ± 230 µm3 and remained similar in MeCP2 transfected cells, showing just a small reduction correlating with increasing levels of MeCP2 (702 ± 204 µm3 and 652 ± 224 µm3 for low and high MeCP2 levels, respectively) (Figure 3(g), Table S10). Using the numbers of molecules per cell nucleus and the volume of the nuclei, we calculated MeCP2 concentrations of 0.39 ± 0.14 µM for untransfected, 11.76 ± 5.57 µM for low and 131.20 ± 48.10 µM for high expressing cells (Figure 3(e)-G, Table S9). Thus, the above mentioned concentration of 10 µM MeCP2 used in the in vitro phase separation experiments is within the physiological range.
As MeCP2 is enriched at the heterochromatin compartments, we were interested in the local MeCP2 concentration at heterochromatin and in the nucleoplasm outside of heterochromatin (nucleus area minus heterochromatin area). Based on the segmentation of nucleus and heterochromatin, we could calculate the concentration in these regions by multiplying the MeCP2 concentration of the whole nucleus with the ratios of MeCP2 mean intensity in heterochromatin core versus nucleus and nucleoplasm versus nucleus (Figure 3(e-g), Table S10). The numbers show that the MeCP2 accumulation is level dependent, as it increases from 1.6 fold for untransfected cells to 3.6 fold in low and 5.8 fold in high expressing cells.
Overall, with this strategy we can calculate in absolute terms the concentrations of MeCP2 across measurement platforms and with subnuclear resolution. In addition, the MeCP2 amount in low-expressing cells corresponds to physiological protein levels and can be used to mimic the in vivo situation whereas the high expressing cells are still within the range of some neuronal cell types.
Increasing MeCP2 level leads to a reduced mobility of MeCP2 molecules in live cells
We next investigated the influence of the MeCP2 concentration and compartmentalization on its dynamics in cells by single-molecule tracking using the calibration system described above to categorize the cells based on MeCP2 concentration (Figure 3(h)).
In cells with both low and high MeCP2 levels, MeCP2 movement within the whole nucleus (Fig. S4A) was heterogenous presenting three dynamic populations (Figure 4(a)). Two of the populations were related with what we identified in LLPS droplets with D being 0.520 and 0.095 µm2 s−1, respectively, and a third population arised with D of 0.023 µm2 s−1. The latter population corresponds to the summatory effects of limited precision on the localization of the individual molecules and the chromatin mobility itself, and is hereafter called static (Figure 4(b), Table S11). Similar values had been reported before for proteins that are known to bind tightly to DNA, such as histone H2A (D = 0.032 µm2 s−1 [91],) or p53 after irradiation of cells (D = 0.014 µm2 s−1 [92],). Overall, increasing MeCP2 concentration led to a general slow down of MeCP2 movement (Figure 4(a)-B, Table S11) by increasing in a similar ratio the slow (0.095 µm2 s−1) and the static population. To test whether the dynamic changes we observed in MeCP2 could be a result of a change in heterochromatin morphology and/or size, we simultaneously tracked a synthetic polidactyl zinc finger domain protein, MaSat [93] in high versus low MeCP2 expressing cells. This artificial protein recognizes major satellite DNA repeats, which are enriched in mouse heterochromatin [24] but there are no known protein interactions and, therefore, serves as a live marker for heterochromatin [72,94]. In contrast to MeCP2, MaSat mobility was unaffected by MeCP2 concentration (Figure 4(a-c), Table S11).
Figure 4.
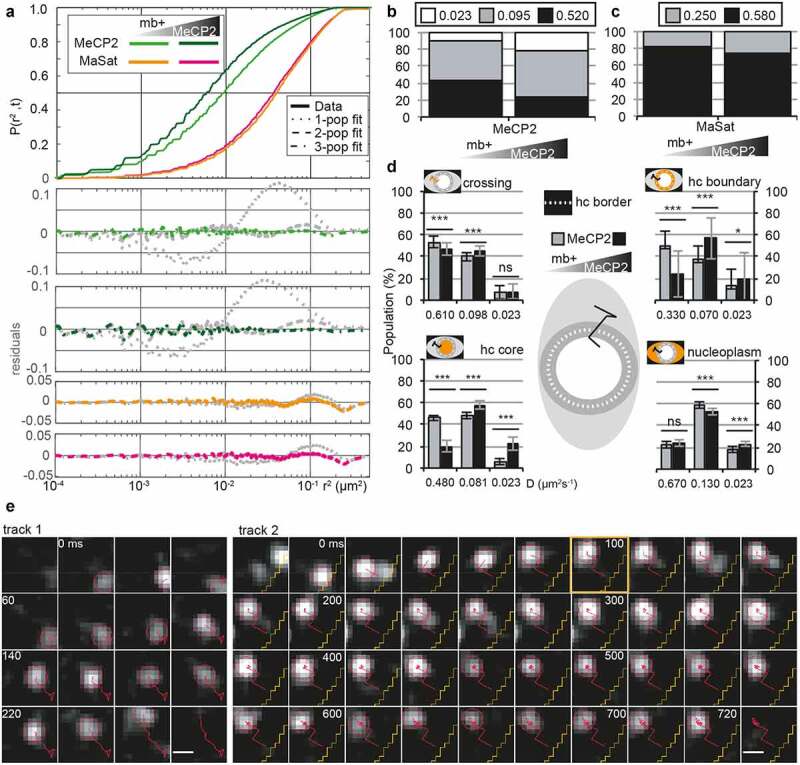
MeCP2 in heterochromatin exhibits three mobility populations depending on its location and concentration.
C2C12 myoblasts were transfected with pEG-MeCP2 and pMaSat-mRFP or pMaSat-miRFP703, seeded on glass slides and incubated 16–20 h in DMEM 20% FBS 5% CO2. During image acquisition, cells were maintained in PBS. Images were processed in ImageJ and tracked using TrackMate, sorting out the detected tracks with less than 5 spots per track.(A) Square displacement curves and best fit model for MeCP2 and MaSat tracks in transfected myoblasts with low and high levels of MeCP2 defined by the calibration system (Figure 3(h)). Upper panel shows the cumulative square displacement curve and the down panel the residues when comparing the data to the model for one (dotted line), two (dashed line) or three (line-dashed) populations as described in Figure 2.(B and C) Gaussian-mixture model comparison of MeCP2 (B) or MaSat (C) populations between low- and high-MeCP2 transfected myoblast considering all tracks in the cell and equaling the D for each population between conditions.(D) Gaussian-mixture model comparison of MeCP2 populations in tracks sorted according to its position in the cell: crossing tracks, heterochromatin boundary (hc boundary), heterochromatin core (hc core) and nucleoplasm, as stated in the middle representation, considering the levels of MeCP2 according to the calibration system (Figure 3(h)). ns, not significant; *, p < 0.05; **, p < 0.001; ***, p < 0.0001.(E) Exemplary tracks (red lines) in 1.1 × 1.1 µm sections every 20 ms. Track 1: mixed movement between short and long steps to cover a long distance of almost 1 µm in the nucleoplasm; track 2: mixed movement between short and long steps until the molecule is trapped (at 100 ms, orange square) and becomes static until it bleached within heterochromatin. Yellow lines: heterochromatin-euchromatin border. Scale bar = 500 nm.
To dissect the influence of the subnuclear compartmentalization on the dynamic populations we selected representative tracks where we could analyze the movement of a unique molecule in detail (Figure 4(e), Fig. S4B-C). Tracks that covered long distances (Figure 4(e), track 1) were found mostly in the nucleoplasm and, less frequently, in the heterochromatin core. These tracks combined long with short steps that could represent the fast and slow D, respectively. As we described for the LLPS droplets (Figure 2(c, e)), this could be an intrinsic effect of the crowdedness in the nucleus in the otherwise free diffusion of the MeCP2 molecules. In contrast, static tracks were found all around the nucleus, with special abundance in the heterochromatin, particularly in the heterochromatin boundary. Interestingly and different to the other movement behaviors, static tracks tended to stay static until photobleached (Fig. S4C, tracks 6, 7). Indeed, when a moving track became static (Figure 4(e), track 2, orange mark), it could not become mobile even at very long times (620 ms).
In essence, higher MeCP2 level led to a slowed down mobility, suggesting non-saturable binding modalities at MeCP2 physiological concentration (11 µM). Furthermore, we observed a static MeCP2 population that was not identified in the in vitro minimal system and which could reflect (methylated) DNA binding.
MeCP2 mobility suggests the presence of a border between heterochromatin and nucleoplasm
We further investigated MeCP2 mobility by comparing the effect of MeCP2 concentration in the different compartments of the cell nucleus. Based on direct observations, in order to better analyze the static MeCP2 molecules we defined a region of 200 nm around the heterochromatin border as the boundary, thus classifying the tracks into four categories: heterochromatin core, heterochromatin boundary, nucleoplasm; and crossing (Figure 4(d), Table S11). Interestingly, the increased static population occurred in the heterochromatin core, with a 2-fold increase with respect to the low MeCP2 concentration (Figure 4(d), down left panel, Table S11).
Secondly, we analyzed the diffusion coefficients. We noted a high variability in the fastest population independent of the MeCP2 level, which almost doubled in crossing (0.610 µm2 s−1) and nucleoplasm (0.670 µm2 s−1) tracks with respect to the tracks in the heterochromatin boundary (0.330 µm2 s−1) and core (0.480 µm2 s−1). A similar change in the diffusion was observed for the slow population, but only between nucleoplasm (0.130 µm2 s−1) and boundary (0.070 µm2 s−1). The static population remained constant among compartments (Figure 4(d), Table S11).
Thirdly, we analyzed individual tracks. We observed that tracks crossing the boundary between heterochromatin and nucleoplasm exhibited long steps (Fig. S4B, panels 4, 5), while tracks with shorter steps near the boundary resulted in the molecule bouncing off it (Fig. S4B, panel 8).
Altogether, both diffusion coefficient and individual tracks pointed to the existence of a barrier that affects the freely and confined diffusion of MeCP2. The finding that MeCP2 could diffuse across the heterochromatin-euchromatin border further confirmed that the heterochromatin compartment is liquid-like. Furthermore, the MeCP2 concentration mainly affects the motion within heterochromatin with more MeCP2 leading to more static MeCP2 molecules, suggesting an increase of methylated DNA binding.
DNA promotes the liquid-liquid phase separation of MeCP2
We next investigated the influence of DNA on in vitro MeCP2 phase separation properties. Firstly, template DNA was synthesized with different lengths (Fig. S5A-B) and labeled with the DNA dye DRAQ5 to detect the DNA distribution during MeCP2 LLPS. By mixing template DNA with MeCP2 in the physiological salt condition (150 mM NaCl) in which MeCP2 alone does not form droplets (Fig. S1A), we found that MeCP2 could form liquid-like droplets with longer DNA (800, 1600, or 3000 bp) but not with short (380 bp) DNA (Figure 5(a)) together with an enrichment of DNA (DRAQ5) in the droplets. This indicates a multi-valency induced phase separation property of MeCP2 with DNA. The DRAQ5 signals were further applied for droplet segmentation and quantitative analysis. Longer and higher concentrations of DNA were shown to promote the formation of more and bigger droplets (Figure 5(a)-B, Table S12). Thus, DNA promoted (switched on) de novo phase separation of MeCP2 in physiological conditions in a DNA length and concentration-dependent manner, highlighting the importance of multiple ionic interactions among MeCP2 and DNA molecules for LLPS.
Figure 5.
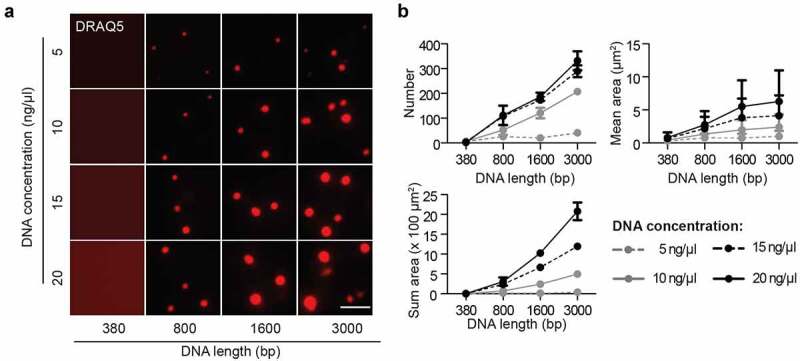
DNA promotes the liquid-liquid phase separation of MeCP2.
The synthesized DNA was labeled with DRAQ5. The in vitro phase separation assay at different conditions was done by incubation at room temperature for 45 min. Then, the mixtures were transferred to chambers made of double-sided tapes and sealed with coverslips. The fluorescent and DIC images were taken using the Nikon Eclipse TiE2 microscope. MeCP2: 3 µM, NaCl: 150 mM, no PEG.(A) Fluorescent images of MeCP2 droplets in the presence of DRAQ5 labeled DNA with different concentration and length. Scale bar = 10 µm.(B) Quantification of size, area and number of droplets from (A). The red channel was applied for droplet segmentation by bandpass filter and threshold based on the mean intensity in/out droplets. Droplets with size >0.1 µm2 were considered and droplet parameters were measured.≥3 images were taken for each condition. The droplets number and sum droplet area per image and mean droplet area were plotted with mean ± SD (standard deviation).
DNA methylation restricts the size of MeCP2 droplets
As MeCP2 was originally recognized as a methyl-cytosine binding protein [28], we then explored whether specific mCpG-MeCP2 interaction plays a role in MeCP2 LLPS. As the 800 bp template DNA could already promote the LLPS of MeCP2 under physiological conditions, we further generated 800 bp DNA templates with methylated cytosines. CpG methylated DNA (mCpG) was generated by the M.SssI methyltransferase, validated with the methylation-sensitive enzyme HpaII (Fig. S5C-D), labeled with DRAQ5, and introduced into the in vitro phase separation assay of MeCP2 (mixed with 1% GFP-MeCP2). We found that both unmethylated (CpH) and CpG methylated DNA could be incorporated into the MeCP2 droplets (Fig. S6A). The GFP signals were used for droplet segmentation. Quantification of droplet size showed that both protein and DNA promoted the LLPS of MeCP2 in a concentration-dependent manner (Figure 6(a), Fig. S6C, Table S13). Surprisingly, quantification of droplet size and numbers showed that the addition of mCpG DNA to the mixture restricted the increase in droplet size in most conditions tested (Figure 6(a)-B, Table S13) compared to the unmethylated DNA.
Figure 6.
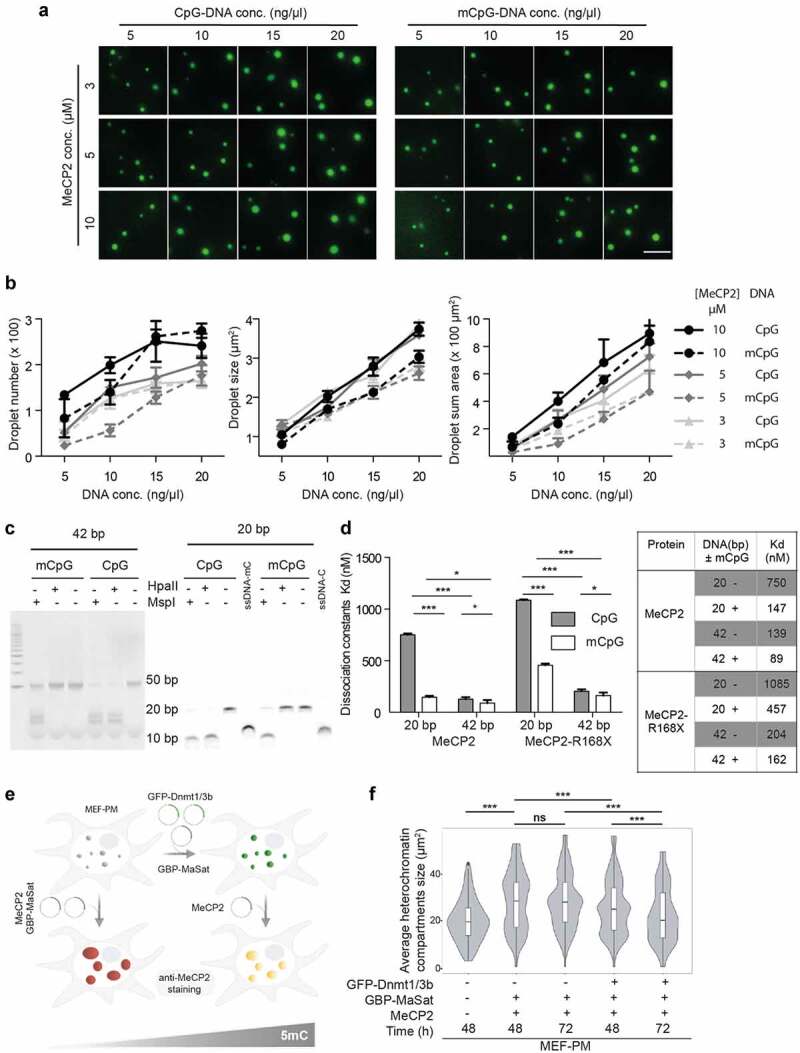
Cytosine methylation restricts droplet growth in vitro and in vivo.
(A) Fluorescent images showing the MeCP2 phase property in the presence of methylated and unmethylated DNA. The synthesized 800 bp DNA was labeled with DRAQ5. 1% GFP-MeCP2 was mixed with 99% untagged MeCP2. The in vitro phase separation assay at different conditions was done by incubation at room temperature for 45 min. Then, the mixtures were transferred to chambers made of double-sided tapes and sealed with coverslips. The fluorescent and DIC images were taken using the Nikon Eclipse TiE2 microscope. MeCP2 and DNA concentrations (conc.) are as mentioned. NaCl: 150 mM, no PEG. Representative images of the GFP channel are shown. Scale bar = 10 µm.(B) Graphs showing the influence of mCpG on the LLPS properties of MeCP2 based on droplet size, sum area and number from (C). Droplets were segmented by a bandpass filter and thresholded based on the mean GFP intensities in/out droplets. The droplet parameters were measured and plotted. conc.: concentration.(C) DNA methylation detection. 1 µl of 10 µM 42 bp and 20 bp DNA with and without CpG methylation was treated with HpaII and MspI respectively for 2 h ar 37°C and then applied to tris borate EDTA polyacrylamide gel electrophoresis. The gel was stained with ethidium bromide and imaged.(D) Left: MST analysis of the dissociation constants of MeCP2 and RTT related truncation MeCP2-R168X with methylated or unmethylated DNA synthesized in (C) (see also Fig. S6G). Student t-test was applied, ns (not significant), P > 0.05; *, p < 0.05; **, p < 0.001; ***, p < 0.0001. Right: Table showing the mean values of the dissociation constant (Kd).(E) Methylation rescue experiment in cells. MEF-PM cells (with low level of DNA methylation) were transfected with GBP-MaSat, untagged MeCP2 and/or GFP-Dnmt1/3b. The impact on heterochromatin clustering induced by methylation increase and/or MeCP2 clustering was analyzed by fixation after 48 or 72 h and confocal imaging. Mock-treated MEF-PM cultured for 48 h prior fixation served as a control.(F) Quantitative average heterochromatin compartments size comparison in MEF-PM with and without targeted heterochromatin methylation. ns, not significant; *, p < 0.05; **, p < 0.001; ***, p < 0.0001.
Recently, MeCP2 was found to bind methylated cytosine at non-CpG sites (mCpH, H = A,G,C). mCpH was shown to increase during early brain development, in parallel with MeCP2 postnatal accumulation. MeCP2 interactions with noncanonical cytosine methylation was shown to be important for normal brain function [63,95,96]. Therefore, we generated DNA with all cytosines methylated (5mC) by replacing all dCTP with dmCTP (Fig. S5C-D). Then, we tested whether including non-canonical DNA methylation influences the phase separation properties of MeCP2. Similar to the canonical CpG methylation, we found that fully cytosine methylated DNA was incorporated into the MeCP2 droplets and exhibited a concentration-dependent effect on MeCP2 phase separation (Fig. S6A-C). Moreover, MeCP2 formed smaller and fewer droplets with all cytosine methylated DNA than with unmethylated DNA (Fig. S6D), which was also the case for CpG methylated DNA (Figure 6(b), Table S13). Hence, independent of the methylation context, cytosine methylation counteracts the increase of MeCP2 droplet number and size in comparison to unmodified DNA.
As the DNA is wrapped around a core of proteins (histone octamer) in eukaryotic cell nuclei, we wondered if nucleosomal DNA still plays a role in the phase separation properties of MeCP2. Thus, we first isolated the mononucleosomes from cultured HEK293T human cells by treating the cell nuclei with MNase (Fig. S6E). Then, we checked if mononucleosomes could affect the LLPS of MeCP2 and found that the addition of mononucleosomes could also promote the formation of bigger droplets with higher MeCP2 enrichment (Fig. S6F, Table S20). The result is consistent with our previous observations found in the presence of naked DNA regardless of cytosine methylation. In conclusion, DNA enhances MeCP2 phase separation, and this is restrained by cytosine methylation.
To measure the binding affinities of MeCP2 to (methyl)-DNA, we applied the microscale thermophoresis (MST) technique. Using 20 and 42 bp template DNA with single mCpG methylation or no methylation (Figure 6(c)), we found that MeCP2 had higher binding affinities for methylated and longer DNA. This is due to, on the one hand, specific 5mC-MeCP2 interaction and, on the other hand, cumulative multivalent nonspecific DNA-MeCP2 interaction. With the longer DNA, adding methylation did not affect the binding affinity. With the shorter DNA, adding methylation increased the binding affinity (Figure 6(d), Table S14). We interpret this as, in the longer template, the multiple MeCP2-DNA interactions dominate the population KD, whereas in the shorter template with less possible MeCP2-DNA interactions, the 5mC-MeCP2 interaction becomes more prominent. Accordingly, MeCP2-R168X, an RTT-related nonsense mutation lacking all regions downstream of the MBD that are responsible for DNA interaction, showed an overall decreased binding affinity with DNA (Figure 6d), Fig. S6G, Table S14).
To check whether the surprising reduction of LLPS droplet size produced by DNA methylation occurs in vivo, we used a cell system in which the global DNA methylation is drastically reduced, i.e., mouse embryonic fibroblasts deficient in Dnmt1 function (MEF-PM, Table S3). As shown in Fig. S6H-I (Table S21), MEF-PM showed reduced 5mC compared to the MEF-P, in which the Dnmt1 function is normal. We then rescued the DNA methylation levels by targeting ectopically expressed GFP-Dnmt1 and GFP-Dnmt3b to the heterochromatin regions using MaSat-GBP as described before [59], and compared the effects of MeCP2 expression in the rescued and not rescued MEF-PM (Figure 6(e)). Consistent with the in vitro data, restoring 5mC levels reduced the total heterochromatin area of the cells in the presence of MeCP2 (Figure 6(f), Table S15)
In summary, the cumulative weak DNA-MeCP2 interaction enhances the MeCP2 LLPS, while the stronger specific 5mC-MeCP2 restricts the growth of MeCP2 droplets, both in vitro and in cells.
DNA methylation reduces MeCP2 mobility
We used the minimal system to emulate the three populations observed in cells. For this purpose, we measured the effect of DNA and methyl-DNA in the absence and presence of crowders on MeCP2 dynamics. To our surprise, MeCP2 mobility after the addition of DNA (Figure 7(a), green line) was still isotropic. However, in the presence of crowders, we measured an enhancement in the arising of the second (slow) population, as it was statistically defined with the lesser amount of PEG 8000. However, the presence of 5mC, disturbed MeCP2 isotropic mobility (Figure 7(a), light blue line). A closer look into the populations showed that, indeed, in both cases the arising of the slow population was causing the anisotropy (Figure 7(c), Table S16). Only in the presence of crowders and DNA methylation we were able to observe the static population of MeCP2 and a residual profile similar to the one obtained in the cellular system (Figure 7(a), dark blue line versus Figure 4(a) and B). The lack of a static population in the absence of crowders could be explained as a result of the overall movement of the DNA within the droplet, which would get reduced in the presence of crowders allowing the visualization of the MeCP2 bound to it. In any case, the mobility of MeCP2 was as slow as the one we measured in the heterochromatin, suggesting the influence of other factors not considered in our minimal system. Altogether, these data point to the role of 5mC binding by MeCP2 in creating specific long-lasting association with DNA (static population) but also enhancing the ability of MeCP2 to bind DNA in a non-sequence specific manner (slower population).
Figure 7.
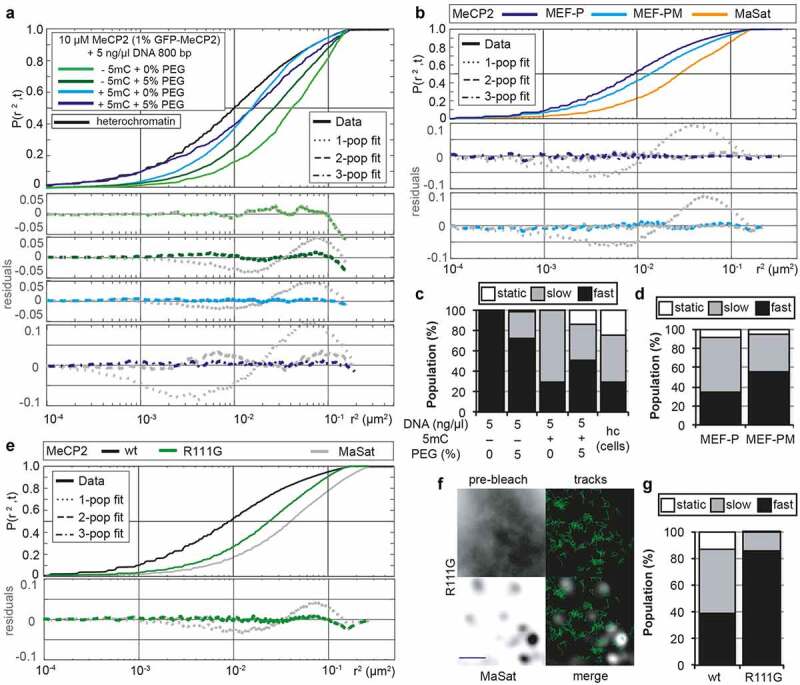
DNA and methyl-DNA addition to MeCP2 LLPS emulates the three mobility populations observed in heterochromatin in cells.
(A) 10 µM MeCP2 containing 1% GFP-MeCP2 was mixed with of 5 ng/µl 800 bp (methyl-)DNA with or without crowding agents (5% PEG 8000) in a buffer containing 150 mM NaCl and incubated 45–60 min at room temperature before imaging. GFP-MeCP2 tracks were obtained using TrackMate, sorting out tracks with less than 4 spots and analyzed in SMTracker. Square displacement cumulative distribution of the steps is shown as described in Figure 2. A curve containing the square displacement analysis of the tracks located in heterochromatin in myoblasts with low MeCP2 from Figure 4 has been added for comparison.(B) Mouse embryonic fibroblast proficient (MEF-P) or deficient (MEF-PM) in DNA methylation were transfected with pEG-MeCP2 and pMaSat-mRFP or pMaSat-miRFP703 using AMAXA transfection and seeded on glass slides in DMEM 15% FBS at 37°C, 5% CO2 for 16–20 h. During image acquisition, cells were maintained in PBS. MeCP2 tracks were obtained in TrackMate, sorting out the tracks containing less than 5 spots and analyzed using SMTracker. Square displacement curves and best fit model for MeCP2 is shown as described in Figure 2.(C) Effect of (methyl)-DNA in MeCP2 populations fixed in Gaussian-mixture model with the diffusion coefficient obtained for live cells (fast: 0.520; slow: 0.095; static: 0.023). Last column belongs to the sorted tracks in low MeCP2 myoblast for the heterochromatin compartment as shown in Figure 4.(D) Comparison of the population variation with the same diffusion coefficients forcing three population model in Gaussian-mixture model in cells deficient (MEF-PM) or proficient (MEF-P) in 5mC maintenance.(E) C2C12 myoblasts were transfected with of pEG-MeCP2R111G and pMaSat-mRFP using AMAXA transfection and seeded on glass slides in DMEM 20% FBS at 37°C, 5% CO2. During acquisition, cells were maintained in PBS. MeCP2 tracks were obtained in TrackMate, sorting out the tracks containing less than 5 spots and analyzed using SMTracker. Square displacement curves and best fit model for MeCP2 and MaSat is shown as described in Figure 2. The data from Figure 4(a) corresponding to low levels of MeCP2 has been added for reference. The residuals showed the 2-population model as the simplest fitting model according to Bayesian Information Criterion for the R111G mutant.(F) Exemplary cell for MeCP2 R111G mutant, which showed no enrichment in heterochromatin neither in the pre-bleach image nor in the track overview. Scale bar = 2 µm.(G) MeCP2 wt versus R111G comparison of the population variation with the same diffusion coefficients forcing a three population model in the Gaussian-mixture model. The third population is completely absent in the R111G mutant.5mC: DNA with cytosine methylation.
To further test this functional cross-talk in vivo, we used two different approaches in cells. Firstly, we used the previously mentioned MEF-PM cells with lower DNA methylation level and compared the mobility of MeCP2 to the mobility in the parental MEF-P cells. Visualization of the tracks indicated that neither MeCP2 nor MaSat tracks location was altered in the MEF-PM cells (Fig S7A), as both showed a preferential heterochromatin location. The overall mobility of MeCP2 was increased in MEF-PM cells (Figure 7(b), upper panel), and a closer look at the dynamic populations resulted in a statistically preferred 2-population model, in which the static population is the one missing relative to the MEF-P cells and the myoblasts (Figure 7(b) down panel, Figure 4(a)). This change was inherent to MeCP2 function and not to any changes in the physiology or morphology of the heterochromatin compartment as MaSat mobility was unaltered (Fig. S7B, S7C). Same results were found when using the MeCP2 RTT mutant R111G, a residue located in the MBD domain. This mutant MeCP2 has been extensively studied and has lost the ability to bind 5mC leading to its lack of heterochromatin enrichment in cells [49]. Ectopic targeting to heterochromatin, though, has demonstrated that this mutant is not impaired in the ability to cluster heterochromatin in cells [59]. As expected, both pre-bleached images and track distribution showed a pattern totally different from the wild-type MeCP2 and heterochromatin binding could not be discerned (Figure 7(f)). R111G mobility was much faster than the wild-type protein (Figure 7(e), upper panel) and the most prominent observation was the presence of only 2-populations as the best fit model (Figure 7(e), down panel). Moreover, we saw a drastic reduction not only in the static, but also in the slow population compared to the wild-type MeCP2 (Figure 7(g), Table S16), confirming the direct implication of the MBD binding to both mobility populations. Similar results were found in the MEF-PM though the reduction of the static population was not so drastic when a 3-population fit was enforced (Figure 7(d) versus Figure 7(g), Table S16). This can be explained by the presence of residual DNA methylation and/or the presence of other substrates for MBD binding, such as histones modifications [97]
MeCP2 liquid-like droplet formation requires self-oligomerization
RTT-related MeCP2 nonsense mutations frequently occur in the intervening domain (ID) and transcriptional repression domain (TRD). Previously, we reported that the ID-TRD is responsible for homo-dimerization of MeCP2 [71]. As LLPS can be induced by weak homo/hetero-interactions, we examined if the ID-TRD driven self-interaction is required for MeCP2 LLPS.
Firstly, as the MeCP2 RTT mutation R168X lacks the ID-TRD while the R255X retains partial ID-TRD (Figure 8(a), top), we examined whether these nonsense mutations retain the ability of self-interaction by in vitro pull-down assay and MST assay. The in vitro pull-down experiment was done by incubating the immobilized MeCP2 truncations with purified GFP tagged MeCP2 truncations, followed by Western blot analysis (Figure 8(a), bottom, 8B, Fig. S8 A-B). The GFP-MeCP2-R255X and the GFP-MeCP2 were pulled down by MeCP2-R255X and MeCP2 separately, while the GFP-MeCP2-R168X was not pulled down by MeCP2-R168X (Figure 8(b)).
Figure 8.
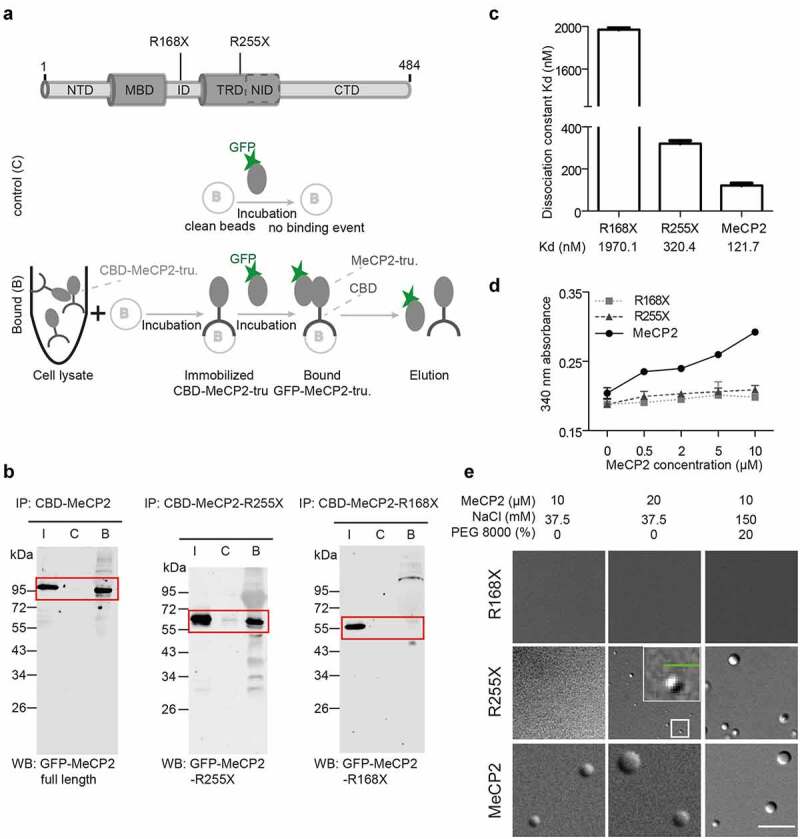
Oligomerization of MeCP2 is a prerequisite for liquid-liquid phase separation in vitro and heterochromatin clustering in vivo.
(A) Top: Schematic graph showing two representative nonsense mutations in the MeCP2 gene in patients with Rett syndrome. Bottom: Schematic graph showing the in vitro pull-down experiments. Shortly, the GFP tagged proteins were purified and stored in a buffer containing 0.5 M NaCl using chitin beads (Fig. S8A-B). The untagged MeCP2-CBD and MeCP2-truncations (CBD-MeCP2-tru.) were purified using immobilized chitin beads by incubating the bacterial lysates with the beads for 3 h at 4°C with rotation. The pull-down assay was conducted by incubating purified GFP-MeCP2 or truncations in solution with immobilized MeCP2 or truncations in PBS with 150 mM NaCl and 0.05% NP-40. The bound ‘B’ fractions were released by adding 40 µl loading buffer and boiling, followed by Western blot for further analysis. CBD: chitin-binding domain. Negative control: clean beads were incubated purified MeCP2 and truncations.(B) Western blot of in vitro pull-down experiment, corresponding to Figure 8(a). Samples from pull-down assay were detected using anti-GFP antibody. Predicted signals are highlighted by red boxes. Input ‘I’ control: purified GFP-MeCP2 full length or truncations. IP: immobilized proteins. WB: Western blot.(C) Dissociation constant (KD) values measured by microscale thermophoresis assay (MST). See Fig. S8C for details.(D) Turbidity changes of the full-length MeCP2 and RTT-related nonsense mutations with increasing protein concentration. The phase separation was done by incubating proteins of various concentrations in a buffer containing 37.5 mM NaCl at room temperature for 45 min in a 384-well plate. The absorbance was measured at λ = 340 nm at room temperature. Absorbance was plotted as mean ± SD.(E) DIC images showing the phase separation properties of MeCP2 full length and truncations at different conditions. The in vitro phase separation assay at the conditions indicated was done by incubating at room temperature for 45 min. The DIC images were taken using the Nikon Eclipse TiE2 microscope. The boxed region is shown at higher magnification. Scale bar (green) = 3 µm; Scale bar (white) = 10 µm.
We, then, quantified the self-interaction strength of full-length MeCP2 and the two truncations by MST assay. The GFP tagged MeCP2 and the two truncations were incubated with corresponding untagged proteins and the dissociation constants (Kd) were calculated (Figure 8(c), S8C, Table S17). We found that MeCP2-R255X had a higher dissociation constant (Kd) (320.36 ± 14.35 nM) than the MeCP2 (121.66 ± 11.00 nM), while the shortest truncation MeCP2-R168X showed a micromolar scale dissociation constant (1970 ± 19.91 nM). These data indicate that the shorter nonsense mutation MeCP2-R168X is not capable of self-interaction, the MeCP2-R255X is capable of self-interaction but with a lower binding affinity than the MeCP2. In summary, the amino acid sequence from position 168 to 254 is responsible and essential for MeCP2 self-interaction, which could be enhanced by the downstream carboxy-terminal region.
Secondly, we checked the phase separation properties of MeCP2-R168X and MeCP2-R255X compared to the MeCP2 using the turbidity assay and the phase separation droplet assay. We registered the absorbance changes at 340 nm of MeCP2 and truncations at low salt conditions (37.5 mM NaCl). The turbidity of MeCP2 increased with higher protein concentration as shown in Fig. S1B, while there was no turbidity change for MeCP2 R168X and R255X (Figure 8(d), Table S17). Then, we analyzed the phase separation properties of the two MeCP2 truncations and found that R168X failed to form liquid-like droplets in all tested conditions, while the MeCP2 R255X could form liquid-like droplets at higher protein concentrations or in the presence of crowding agents (Figure 8(e)).
In conclusion, self-interaction is essential for MeCP2 LLPS. Both self-interaction and LLPS are enhanced by the MeCP2 C-terminus.
Discussion
We have previously shown that MeCP2 clusters heterochromatin compartments in cells in a dose-dependent manner, and these subnuclear compartments are dynamic and can fuse over time [24]. Here, we demonstrated that purified MeCP2 alone was able to form liquid-like phase separated compartments in vitro and self-oligomerization was the minimal requirement for MeCP2 LLPS (Figure 9(a)). This is consistent with in vivo experiments, which demonstrate that the very severe RTT mutant R168X (or the MBD alone) [98,99] is less enriched in heterochromatin and is more mobile [100,101]. As increasing salt concentration negatively correlated with droplet size but 1,6-hexanediol did not influence it, we defined electrostatic interactions as the main binding force for oligomerization that drives MeCP2-mediated LLPS [102] (Figure 9(b)). Self-electrostatic interactions were also sufficient to reduce the mobility in an isotropic manner within the droplet to a diffusion coefficient of 0.5 µm2 s−1. In cells, this D corresponded to the fastest population, which is 20–40 times slower than a size-equivalent GFP-oligomer (9.1–20 µm2 s−1) [103,104] and in concordance with previous results with endogenous MeCP2 in neurons (1 µm2 s−1) [54]. As electrostatic self-interactions were the basis for MeCP2 LLPS, in physiological salt concentration purified MeCP2 was not able to form droplets. Addition of crowding agents, though, allowed MeCP2 LLPS (Figure 9(b)). In biological systems, high densities of macromolecules (protein, nucleic acids, etc.) not only perform function-related activities but also play a role via a nonspecific ‘excluded volume’ effect [105]. The macromolecular crowding could increase the local concentration of targets [106], influencing biochemical reactions (binding, enzymatic activities) [47] and restricting molecular diffusion [107]. In that regard, the eukaryotic cell is a heterogeneous system with various crowding states. Estimates of 50–400 mg/ml macromolecules in the cell yield a 5–40% crowded environment [108], with 100 mg/ml protein and 50 mg/ml chromatin in the cell nucleus resulting in around 15% crowded environment [105,109,110] and 110 mg/ml macromolecules in the nucleoplasm making the nucleoplasm around 11% crowded [111]. We have shown previously that DNA compaction within the cell nucleus, corresponding to increased molecular crowding, affected local mobility of molecules [19].
Figure 9.
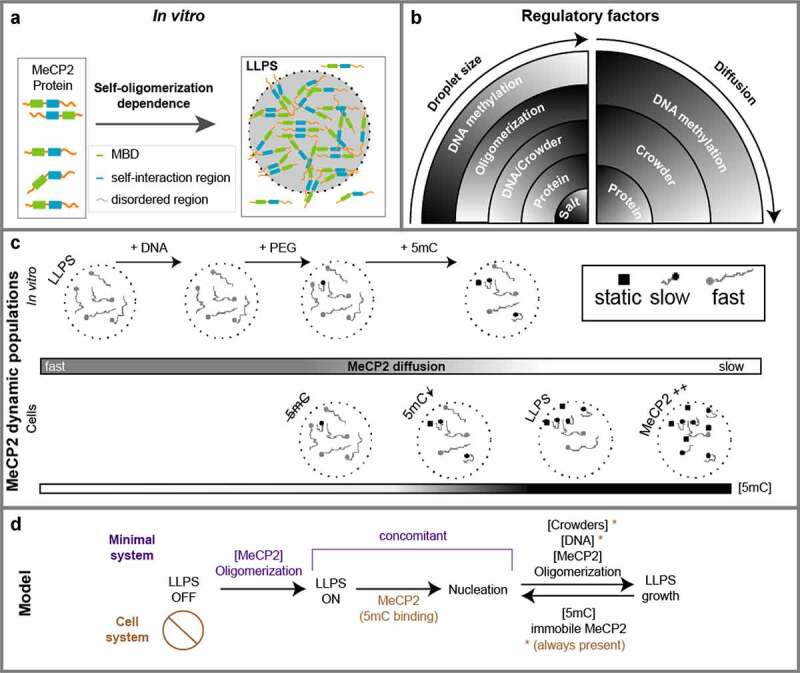
Summary of factors and conditions affecting MeCP2 LLPS and diffusion properties leading to a model comparing minimal and cell systems.
(A) Schematic chart depicting the conditions required for MeCP2 LLPS in vitro. MeCP2 is composed of the structured MBD domain, which is surrounded by the highly disordered amino acid sequences including the self-interaction domain (amino acids 168 to 254). Purified MeCP2 molecules locally condense to form a liquid-like spherical phase (LLPS). Self-interaction drives the formation of liquid-like droplets.(B) Regulatory factors for MeCP2 LLPS and dynamics. The droplet size is positively regulated by protein, crowders and unmethylated DNA concentration and negatively regulated by DNA methylation and salt concentration. MeCP2 mobility is reduced by increased concentration of protein, crowders and methylated DNA, but not affected by unmethylated DNA.(C) Representation of MeCP2 dynamic populations in in vitro and in vivo LLPS. MeCP2 alone presents an isotropic (fast) mobility in LLPS and addition of DNA does not alter it. In crowded environments (e.g., PEG), the appearance of a slow MeCP2 population in vitro reflects the situation of the MeCP2 moving in absence of 5mC binding interactions in vivo. With an increasing amount of 5mC the static MeCP2 population increment. Overall, both slow and static MeCP2 populations increase together with MeCP2 concentrations.(D) Model of MeCP2 driven LLPS. In vitro, MeCP2 in solution will not form LLPS droplets at ionic conditions preventing its self-interaction (LLPS OFF). Lowering the salt concentration allows MeCP2 oligomerization and starts the LLPS process (LLPS ON; nucleation). These MeCP2 droplets grow in size by addition of crowders or DNA and this growth is constrained by 5mC binding consistent with the appearance of a static MeCP2 population. In vivo, heterochromatin formation per se is a multifactorial redundant process within a crowded environment and, thus, the LLPS OFF step can not be captured. MeCP2, depending on its 5mC binding, becomes enriched at heterochromatin (nucleation). Similar to the minimal in vitro system, the subsequent growth of the heterochromatin compartments depends on MeCP2 oligomerization and concentration whereas 5mC limits the heterochromatin compartment size.
In addition, DNA is commonly considered an important factor for LLPS of multiple proteins involved in DNA replication, repair, and chromatin architecture [112]. Consistent with recent reports, we found that both crowding agents and DNA promoted MeCP2 LLPS in a concentration-dependent manner (Figure 9(b)) [68,113,114]. Diffusion measurements, however, unveiled different mechanisms for droplet size increase. While crowders increased MeCP2 concentration in droplets by reducing the volume available for movement, subsequently reducing its diffusion, DNA allowed more electrostatic (hetero-) interactions, which did not affect MeCP2 mobility (Figure 9(b)). Combination of DNA and crowders, though, reduced further MeCP2 mobility. As DNA is a much larger molecule than MeCP2, crowding agents directly and more pronouncedly affect its ability to move. Therefore, the MeCP2 electrostatically interacting with DNA would show slower diffusion. Alternatively, the DNA may also contribute to the crowding effect.
To be able to directly compare the different in vitro and in vivo measurements and use physiologically relevant MeCP2 concentrations [83], we carefully tuned all our assays and platforms based on a calibrated relationship between fluorescence intensity and concentration across measuring systems. To avoid technical variations of the intensity due to changes in illumination and/or detection systems, we based the intensity numbers on population distributions in which the negative MeCP2 level was obtained for each platform and MeCP2 positive cells were sorted into equivalent windows of fluorescence intensity. These corresponded to the sorting windows used to sort cells with different MeCP2 levels that were used to biochemically quantify the absolute amounts of MeCP2 molecules and concentrations per cell. As a result, a similar and physiologically related MeCP2 concentration (~10 µM) was applied for phase separation and for single-molecule tracking both in vivo and in vitro.
With these conditions, we measured the influence of DNA methylation on MeCP2 condensation properties and dynamics in vivo and in vitro. Surprisingly, DNA methylation inhibited the growth of MeCP2 condensates (droplets in vitro and heterochromatin in vivo). In vivo, using methyl cytosine deficient cells and RTT mutants, we showed that, on the one hand, DNA methylation was required for MeCP2 nucleation at heterochromatin needed for its subsequent clustering. On the other hand, DNA methylation also restricted heterochromatin compartment size (Figure 9(b)). This dual effect could be one possible explanation for the great variability of effects of gain/loss of MeCP2 reported in different neurons [50]. Other in vitro studies addressed the effects of DNA methylation on LLPS and reported a sum increase in droplets [68,113,114]. The apparent inconsistency to our results, can be explained by the fact that while we base this conclusion on measuring individual droplet compartment size to directly relate to the size of heterochromatin compartments within the cell nucleus, the other authors measured the sum of all the droplets. The latter measures whether DNA methylation promotes LLPS altogether and not whether the individual compartments can grow in size to the same extent. By comparing to the equivalent in vivo situation, e.g., using cells with lower DNA methylation and RTT mutants of MeCP2 deficient on 5mC binding, we found our measurement and effect to be a good representation of heterochromatin compartmentalization dynamics in cells.
Single-molecule tracking analysis of MeCP2 allowed us to define three different mobility populations: a fast, ‘free’ diffusion defined by a diffusion coefficient of about 0.5 µm2 s−1; a slowed, confined but mobile population dominated by a D in the range of 0.1 µm2 s−1; and a static population with a D of 0.023 µm2 s−1. The latter is reminiscent of other molecules that tightly bind DNA such as transcription factors [92] or histones [91]. Importantly, we could find the three populations both in vitro and in vivo. Quantification of MeCP2 binding affinity with itself (121.7 nM) and with DNA (138.7 nM) yielded a similar binding affinity, suggesting that they may influence mobility similarly. Thus, the MeCP2 mobility populations derived from self-interaction and from protein-DNA interaction separately are not distinguishable in our measurements and appear as one population. In addition, both in vitro and in vivo studies indicated that the static population corresponds to the tighter binding of the MBD to 5mC on DNA (binding affinity 89.3 nM) (Figure 9(c)). Finally, the intermediate slow MeCP2 population could arise by a combination of nonspecific DNA binding and reduction of the space available for diffusion by macromolecular crowding producing a confinement. Altogether, the different parameters involved in compartmentalization interplay with each other to affect mobility of MeCP2. Hence, when we abrogate MeCP2-5mC binding using the R111G mutant in cells, we affected not only the static population but also decreased the slow population (Figure 9(c)). This indicates that MeCP2-5mC binding also contributes to the low-affinity MeCP2-DNA binding interactions. The long dwell time of MeCP2 binding to 5mC likely allows: i) other MeCP2 domains to interact with DNA ii) to attract (via dimerization) other MeCP2 molecules to the DNA and iii) to attract other MeCP2 molecules stimulating oligomerization. In fact, previous studies showed that proteins can promote DNA binding of other factors by direct interaction [115] or even in the absence of interaction [116].
Based on our findings, we propose a model in which heterochromatin compartmentalization can be subdivided into four steps (Figure 9(d)). A first state (LLPS off), means that the conditions are not suitable for a liquid–liquid phase separation or compartmentalization. A second state is the formation of LLPS compartments (LLPS ON). The third state is the enrichment of MeCP2 in the compartments (nucleation). The fourth state is the growth of the LLPS compartments. The crowded cellular environment and the presence of multiple redundant factors involved in heterochromatin LLPS (e.g., HP1 [2–4,15] and histones [5]) does not permit the visualization of a condition in which these compartments are not formed (first state). In the minimal system, though, we were able to distinguish these two steps, based on the ability of MeCP2 to oligomerize and an appropriate environment to allow the electrostatic interactions needed. The second and third states happen concomitantly in the minimal system, as LLPS formation takes place only when MeCP2 is enriched in the droplets. In cells, though, MeCP2 depends on the MBD domain and 5mC binding to associate with the heterochromatin and be enriched in those compartments and, therefore, these two states are distinguishable. Albeit the three states are not distinguishable in both in vitro and in vivo, they rely on common parameters: MeCP2 concentration and oligomerization. The fourth state, compartment growth, is modulated in a multifactorial manner. Increasing MeCP2 concentration (and, therefore, its oligomerization), crowdedness and DNA positively modulate the compartment growth. In contrast, immobilization of MeCP2 by DNA methylation negatively modulates compartment growth. Importantly, mutations of MeCP2 found in Rett syndrome patients (e.g., R111G or R133C, which are 5mC binding deficient) were shown to affect binding to heterochromatin (second to third state) and concomitant clustering of heterochromatin (third to fourth state), while other mutations (e.g., P101H, 5mC binding proficient) were shown to affect only the latter [59].
In summary, we establish in vitro and in vivo the parameters determining the LLPS properties of MeCP2 and how MeCP2 diffusion and binding properties change when inside the droplets as well as in cells and how these contribute to heterochromatin compartment organization and kinetics.
Supplementary Material
Acknowledgments
We thank Alexander Rapp for support with image analysis. We are grateful to Christopher L. Woodcock and Shinichi Kudo for the kind gift of plasmids.
Funding Statement
H.Z. received a fellowship of the China Scholarship Council. This research was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) grants [CA 198/10-1 project number 326470517 and CA 198/16-1 project number 425470807 to M.C.C.; HL 721/18-1 project number 425470807 and SFB 1064 project number 213249687 to H.L].
Highlights
Purified MeCP2 forms liquid-like compartments in vitro through ionic self-interaction
Crowding and DNA promote MeCP2 phase separation and slows down its mobility
DNA methylation restricts MeCP2 droplet size and mobility in vitro and in vivo
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data and custom written scripts for single-molecule tracking, droplet, chromocenter segmentation and analysis are available from the OMERO open microscopy environment public repository (http://cc-omero.bio.tu-darmstadt.de/webclient/userdata/?experimenter=-1)). It is also available through the library of the Technical University of Darmstadt (TUdatalib) (https://doi.org/10.48328/tudatalib-672).
All biological materials will be made available upon request from the lead corresponding author M. Cristina Cardoso (E-mail: cardoso@bio.tu-darmstadt.de).
Author contributions
Conceptualization, H.Z., H.R., H.L. and M.C.C.; Methodology, H.Z., H.R., B.B., W.Q., A.L., M.M. and T.M.; Investigation, H.Z., H.R., A.S., K.G., W.Q., B.B. and M.E.; Formal Analysis, H.Z., H.R., and A.S.; Resources, H.L. and M.C.C; Funding Acquisition, H.L. and M.C.C; Visualization, H.Z., H.R., A.S. and K.G.; Writing - Original Draft, H.Z., H.R.; Writing - Review & Editing, H.Z., H.R., A.S., K.G. and M.C.C.
Supplementary material
Supplemental data for this article can be accessed here
References
- [1].Leonhardt H, Rahn H-P, Cardoso MC.. Functional links between nuclear structure, gene expression, DNA replication, and methylation. Critic Rev Eukaryotic Gene Exp. 1999;9. [DOI] [PubMed] [Google Scholar]
- [2].Strom AR, Emelyanov AV, Mir M, et al. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Larson AG, Elnatan D, Keenen MM, et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sanulli S, Trnka MJ, Dharmarajan V, et al. HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature. 2019;575:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gibson BA, Doolittle LK, Schneider MWG, et al. Organization of chromatin by intrinsic and regulated phase separation. Cell. 2019;179:470–484. e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Weber SC. Evidence for and against liquid-liquid phase separation in the Nucleus. Noncoding RNA. 2019;5:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kilic S, Felekyan S, Doroshenko O, et al. Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1α. Nat Commun. 2018;9:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bryan LC, Weilandt DR, Bachmann AL, et al. Single-molecule kinetic analysis of HP1-chromatin binding reveals a dynamic network of histone modification and DNA interactions. Nucleic Acids Res. 2017;45:10504–10517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].de Koning APJ, Gu W, Castoe TA, et al. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384–e1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Solovei I, Thanisch K, Feodorova Y. How to rule the nucleus: divide et impera. Curr Opin Cell Biol. 2016;40:47–59. [DOI] [PubMed] [Google Scholar]
- [11].Falk M, Feodorova Y, Naumova N, et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature. 2019;570:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brown KE, Guest SS, Smale ST, et al. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. [DOI] [PubMed] [Google Scholar]
- [13].Bertulat B, De Bonis ML, Della Ragione F, et al. MeCP2 dependent heterochromatin reorganization during neural differentiation of a novel Mecp2-deficient embryonic stem cell reporter line. PLoS One. 2012;7:e47848–e47848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Penagos-Puig A, Furlan-Magaril M. Heterochromatin as an important driver of genome organization. Front Cell Dev Biol. 2020;8:982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Qin W, Stengl A, Ugur E, et al. HP1β carries an acidic linker domain and requires H3K9me3 for phase separation. Nucleus. 2021;12:44–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tsouroula K, Furst A, Rogier M, et al. Temporal and spatial uncoupling of DNA double strand break repair pathways within mammalian heterochromatin. Mol Cell. 2016;63:293–305. [DOI] [PubMed] [Google Scholar]
- [17].Chiolo I, Minoda A, Colmenares SU, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Peters AH, O’Carroll D, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. [DOI] [PubMed] [Google Scholar]
- [19].Grünwald D, Martin RM, Buschmann V, et al. Probing intranuclear environments at the single-molecule level. Biophys J. 2008;94:2847–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nozaki T, Imai R, Tanbo M, et al. Dynamic organization of chromatin domains revealed by super-resolution live-cell Imaging. Mol Cell. 2017;67:282–293.e287. [DOI] [PubMed] [Google Scholar]
- [21].Becker JS, Nicetto D, Zaret KS. H3K9me3-dependent heterochromatin: barrier to cell fate changes. Trends Genet. 2016;32:29–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Montavon T, Shukeir N, Erikson G, et al. Complete loss of H3K9 methylation dissolves mouse heterochromatin organization. Nat Commun. 2021;12:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Müller‐Ott K, Erdel F, Matveeva A, et al. Specificity, propagation, and memory of pericentric heterochromatin. Mol Syst Biol. 2014;10:746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Brero A, Easwaran HP, Nowak D, et al. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J Cell Biol. 2005;169:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Erdel F, Rademacher A, Vlijm R, et al. Mouse heterochromatin adopts digital compaction states without showing hallmarks of HP1-driven liquid-liquid phase separation. Mol Cell. 2020;78:236–249. e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Narlikar GJ, Myong S, Larson D, et al. Is transcriptional regulation just going through a phase? Mol Cell. 2021;81:1579–1585. [DOI] [PubMed] [Google Scholar]
- [27].Rausch C, Zhang P, Casas-Delucchi CS, et al. Cytosine base modifications regulate DNA duplex stability and metabolism. Nucleic Acids Res. 2021;49:12870–12894 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lewis JD, Meehan RR, Henzel WJ, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. [DOI] [PubMed] [Google Scholar]
- [29].Edwards JR, Yarychkivska O, Boulard M, et al. DNA methylation and DNA methyltransferases. Epigenetics Chromatin. 2017;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Petryk N, Bultmann S, Bartke T, et al. Staying true to yourself: mechanisms of DNA methylation maintenance in mammals. Nucleic Acids Res. 2021;49:3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Ludwig AK, Zhang P, Cardoso MC. Modifiers and readers of DNA modifications and their impact on genome structure, expression, and stability in disease. Front Genet. 2016;7:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Schmidt A, Zhang H, Cardoso MC. MeCP2 and Chromatin Compartmentalization. Cells. 2020;9:878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Nan X, Tate P, Li E, et al. DNA methylation specifies chromosomal localization of MeCP2. Mol Cell Biol. 1996;16:414–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Fujita N, Watanabe S, Ichimura T, et al. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003;278:24132–24138. [DOI] [PubMed] [Google Scholar]
- [35].Fuks F, Hurd PJ, Wolf D, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. [DOI] [PubMed] [Google Scholar]
- [36].Lunyak VV, Burgess R, Prefontaine GG, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. [DOI] [PubMed] [Google Scholar]
- [37].Agarwal N, Hardt T, Brero A, et al. MeCP2 interacts with HP1 and modulates its heterochromatin association during myogenic differentiation. Nucleic Acids Res. 2007;35:5402–5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Song C, Feodorova Y, Guy J, et al. DNA methylation reader MECP2: cell type-and differentiation stage-specific protein distribution. Epigenetics Chromatin. 2014;7:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Amir RE, Van Den Veyver IB, Wan M, et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. [DOI] [PubMed] [Google Scholar]
- [40].Guy J, Hendrich B, Holmes M, et al. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27:322–326. [DOI] [PubMed] [Google Scholar]
- [41].Guy J, Gan J, Selfridge J, et al. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Krishnaraj R, Ho G, Christodoulou J. RettBASE: rett syndrome database update. Hum Mutat. 2017;38:922–931. [DOI] [PubMed] [Google Scholar]
- [43].Ehrhart F, Jacobsen A, Rigau M, et al. A catalogue of 863 Rett-syndrome-causing MECP2 mutations and lessons learned from data integration. Sci Data. 2021;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. [DOI] [PubMed] [Google Scholar]
- [45].Lyst MJ, Ekiert R, Ebert DH, et al. Rett syndrome mutations abolish the interaction of MeCP2 with the NCoR/SMRT co-repressor. Nat Neurosci. 2013;16:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tillotson R, Selfridge J, Koerner MV, et al. Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature. 2017;550:398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chahrour M, Jung SY, Shaw C, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tudor M, Akbarian S, Chen RZ, et al. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc Natl Acad Sci U S A. 2002;99:15536–15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Agarwal N, Becker A, Jost KL, et al. MeCP2 Rett mutations affect large scale chromatin organization. Hum Mol Genet. 2011;20:4187–4195. [DOI] [PubMed] [Google Scholar]
- [50].Ito-Ishida A, Baker SA, Sillitoe RV, et al. MeCP2 levels regulate the 3d structure of heterochromatic foci in mouse neurons. J Neurosci. 2020;40:8746–8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Baker SA, Chen L, Wilkins AD, et al. An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell. 2013;152:984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gulmez Karaca K, Brito DVC, Oliveira AMM,et al. MeCP2: a critical regulator of chromatin in neurodevelopment and adult brain function. Int J Mol Sci. 2019;20:4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Marano D, Fioriniello S, Fiorillo F, et al. ATRX contributes to MeCP2-mediated pericentric heterochromatin organization during neural differentiation. Int J Mol Sci. 2019;20:5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Piccolo FM, Liu Z, Dong P, et al. MeCP2 nuclear dynamics in live neurons results from low and high affinity chromatin interactions. Elife. 2019;8:e51449–e51449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Imaizumi Y, Feil R. Emerging chromatin structural roles of the methyl-CpG binding protein MeCP2. Epigenomics. 2021;13:405–409. [DOI] [PubMed] [Google Scholar]
- [56].Vincent JB, Ausió J. MeCP2: latest insights fundamentally change our understanding of its interactions with chromatin and its functional attributes. Bioessays. 2021;43:e2000281–e2000281. [DOI] [PubMed] [Google Scholar]
- [57].Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. [DOI] [PubMed] [Google Scholar]
- [58].Bertulat B, De Bonis ML, Della Ragione F, et al. MeCP2 dependent heterochromatin reorganization during neural differentiation of a novel MeCP2-deficient embryonic stem cell reporter line. PLoS ONE . 2012;7:e47848 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Casas-Delucchi CS, Becker A, Bolius JJ, et al. Targeted manipulation of heterochromatin rescues MeCP2 Rett mutants and re-establishes higher order chromatin organization. Nucleic Acids Res. 2012;40:e176–e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Adams VH, McBryant SJ, Wade PA, et al. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J Biol Chem. 2007;282:15057–15064. [DOI] [PubMed] [Google Scholar]
- [61].Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Banani SF, Lee HO, Hyman AA, et al. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol. 2017;18:285–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Jang HS, Shin WJ, Lee JE, et al. CpG and non-CpG methylation in epigenetic gene regulation and brain function. Genes (Basel). 2017;8:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Georgel PT, Horowitz-Scherer RA, Adkins N, et al. Chromatin compaction by human MeCP2: assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. [DOI] [PubMed] [Google Scholar]
- [65].Yusufzai TM, Wolffe AP. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 2000;28:4172–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Nikitina T, Ghosh RP, Horowitz-Scherer RA, et al. MeCP2-chromatin interactions include the formation of chromatosome-like structures and are altered in mutations causing Rett syndrome. J Biol Chem. 2007;282:28237–28245. [DOI] [PubMed] [Google Scholar]
- [67].Ghosh RP, Horowitz-Scherer RA, Nikitina T, et al. MeCP2 binds cooperatively to its substrate and competes with histone H1 for chromatin binding sites. Mol Cell Biol. 2010;30:4656–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Wang L, Hu M, Zuo M-Q, et al. Rett syndrome-causing mutations compromise MeCP2-mediated liquid–liquid phase separation of chromatin. Cell Res. 2020;30:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kucherenko MM, Shcherbata HR. miRNA targeting and alternative splicing in the stress response–events hosted by membrane-less compartments. J Cell Sci. 2018;131:jcs202002–jcs202002. [DOI] [PubMed] [Google Scholar]
- [70].Kudo S, Nomura Y, Segawa M, et al. Heterogeneity in residual function of MeCP2 carrying missense mutations in the methyl CpG binding domain. J Med Genet. 2003;40:487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Becker A, Allmann L, Hofstätter M, et al. Direct homo-and hetero-interactions of MeCP2 and MBD2. PLoS One. 2013;8:e53730–e53730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Herce HD, Schumacher D, Schneider AFL, et al. Cell-permeable nanobodies for targeted immunolabelling and antigen manipulation in living cells. Nat Chem. 2017;9:762–771. [DOI] [PubMed] [Google Scholar]
- [73].Shcherbakova DM, Baloban M, Emelyanov AV, et al. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun. 2016;7:12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Chen T, Ueda Y, Dodge JE, et al. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Rothbauer U, Zolghadr K, Muyldermans S, et al. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. [DOI] [PubMed] [Google Scholar]
- [76].Ludwig AK, Zhang P, Hastert FD, et al. Binding of MBD proteins to DNA blocks Tet1 function thereby modulating transcriptional noise. Nucleic Acids Res. 2017;45:2438–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Babel L, Kruse L, Bump S, et al. Lipid-rafts remain stable even after ionizing radiation induced disintegration of β1 integrin containing focal adhesions. BMC Res Notes. 2017;10:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. [DOI] [PubMed] [Google Scholar]
- [79].Schindelin J, Arganda-Carreras I, Frise E, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Tinevez J-Y, Perry N, Schindelin J, et al. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. [DOI] [PubMed] [Google Scholar]
- [81].Paintdakhi A, Parry B, Campos M, et al. Oufti: an integrated software package for high-accuracy, high-throughput quantitative microscopy analysis. Mol Microbiol. 2016;99:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Rösch TC, Oviedo-Bocanegra LM, Fritz G, et al. SMTracker: a tool for quantitative analysis, exploration and visualization of single-molecule tracking data reveals highly dynamic binding of B. subtilis global repressor AbrB throughout the genome. Sci Rep. 2018;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Skene PJ, Illingworth RS, Webb S, et al. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37:457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Wegiel J, Flory M, Kuchna I, et al. Neuronal nucleus and cytoplasm volume deficit in children with autism and volume increase in adolescents and adults. Acta Neuropathol Commun. 2015;3:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhang H-L, Han S-J. Viscosity and density of water+ sodium chloride+ potassium chloride solutions at 298.15 K. J Chem Eng Data. 1996;41:516–520. [Google Scholar]
- [86].Quillin ML, Matthews BW. Accurate calculation of the density of proteins. Acta Crystallogr D Biol Crystallogr. 2000;56:791–794. [DOI] [PubMed] [Google Scholar]
- [87].Chen RZ, Akbarian S, Tudor M, et al. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27:327–331. [DOI] [PubMed] [Google Scholar]
- [88].Collins AL, Levenson JM, Vilaythong AP, et al. Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet. 2004;13:2679–2689. [DOI] [PubMed] [Google Scholar]
- [89].Van Esch H, Bauters M, Ignatius J, et al. Duplication of the MECP2 region is a frequent cause of severe mental retardation and progressive neurological symptoms in males. Am J Hum Genet. 2005;77:442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Samaco RC, Fryer JD, Ren J, et al. A partial loss of function allele of methyl-CpG-binding protein 2 predicts a human neurodevelopmental syndrome. Hum Mol Genet. 2008;17:1718–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Zhen CY, Tatavosian R, Huynh TN, et al. Live-cell single-molecule tracking reveals co-recognition of H3K27me3 and DNA targets polycomb Cbx7-PRC1 to chromatin. eLife. 2016;5:1–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Loffreda A, Jacchetti E, Antunes S, et al. Live-cell p53 single-molecule binding is modulated by C-terminal acetylation and correlates with transcriptional activity. Nat Commun. 2017;8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lindhout BI, Fransz P, Tessadori F, et al. Live cell imaging of repetitive DNA sequences via GFP-tagged polydactyl zinc finger proteins. Nucleic Acids Res. 2007;35:e107–e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Rausch C, Weber P, Prorok P, et al. Developmental differences in genome replication program and origin activation. Nucleic Acids Res. 2020;48:12751–12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Chen L, Chen K, Lavery LA, et al. MeCP2 binds to non-CG methylated DNA as neurons mature, influencing transcription and the timing of onset for Rett syndrome. Proc Nat Acad Sci. 2015;112:5509–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Tillotson R, Cholewa-Waclaw J, Chhatbar K, et al. Neuronal non-CG methylation is an essential target for MeCP2 function. Mol Cell. 2021;81:1260–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Thambirajah AA, Ng MK, Frehlick LJ, et al. MeCP2 binds to nucleosome free (linker DNA) regions and to H3K9/H3K27 methylated nucleosomes in the brain. Nucleic Acids Res. 2012;40:2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Schanen C, Houwink EJF, Dorrani N, et al. Phenotypic manifestations of MECP2 mutations in classical and atypical Rett syndrome. Am J Med Genet Part A. 2004;126:129–140. [DOI] [PubMed] [Google Scholar]
- [99].Neul JL, Fang P, Barrish J, et al. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70:1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Schmiedeberg L, Skene P, Deaton A, et al. A temporal threshold for formaldehyde crosslinking and fixation. PLoS One. 2009;4:e4636–e4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Kumar A, Kamboj S, Malone BM, et al. Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo. J Cell Sci. 2008;121:1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Alberti S, Gladfelter A, Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Hihara S, Pack C-G, Kaizu K, et al. Local nucleosome dynamics facilitate chromatin accessibility in living mammalian cells. Cell Rep. 2012;2:1645–1656. [DOI] [PubMed] [Google Scholar]
- [104].Dross N, Spriet C, Zwerger M, et al. Mapping eGFP oligomer mobility in living cell nuclei. PLoS One. 2009;4:e5041–e5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Richter K, Nessling M, Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim Biophys Acta-Mol Cell Res. 2008;1783:2100–2107. [DOI] [PubMed] [Google Scholar]
- [106].Homouz D, Perham M, Samiotakis A, et al. Crowded, cell-like environment induces shape changes in aspherical protein. Proc Nat Acad Sci. 2008;105:11754–11759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Dix JA, Verkman AS. Crowding effects on diffusion in solutions and cells. Annu Rev Biophys. 2008;37:247–263. [DOI] [PubMed] [Google Scholar]
- [108].Biswas S, Kundu J, Mukherjee SK, et al. Mixed macromolecular crowding: a protein and solvent perspective. ACS Omega. 2018;3:4316–4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell. 2005;16:202–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Hancock R. Seminars in cell & developmental biology. Packing of the polynucleosome chain in interphase chromosomes: evidence for a contribution of crowding and entropic forces. Elsevier; 2007,18;668–675 [DOI] [PubMed] [Google Scholar]
- [111].Rippe K. Dynamic organization of the cell nucleus. Curr Opin Genet Dev. 2007;17:373–380. [DOI] [PubMed] [Google Scholar]
- [112].Laflamme G, Mekhail K. Biomolecular condensates as arbiters of biochemical reactions inside the nucleus. Commun Biol. 2020;3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Li CH, Coffey EL, Dall’Agnese A, et al. MeCP2 links heterochromatin condensates and neurodevelopmental disease. Nature. 2020;586:440–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Fan C, Zhang H, Fu L, et al. Rett mutations attenuate phase separation of MeCP2. Cell Discov. 2020;6:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Mivelaz M, Cao A-M, Kubik S, et al. Chromatin fiber invasion and nucleosome displacement by the Rap1 transcription factor. Mol Cell. 2020;77:488–500. e489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Grand RS, Burger L, Gräwe C, et al. BANP opens chromatin and activates CpG-Island-regulated genes. Nature. 2021;596:133–137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and custom written scripts for single-molecule tracking, droplet, chromocenter segmentation and analysis are available from the OMERO open microscopy environment public repository (http://cc-omero.bio.tu-darmstadt.de/webclient/userdata/?experimenter=-1)). It is also available through the library of the Technical University of Darmstadt (TUdatalib) (https://doi.org/10.48328/tudatalib-672).
All biological materials will be made available upon request from the lead corresponding author M. Cristina Cardoso (E-mail: cardoso@bio.tu-darmstadt.de).


