ABSTRACT
To investigate the function of lncRNA HOXD-AS1 in cervical squamous cell carcinoma (CESC) and the underlying mechanism. The expressions of HOXD-AS1 and FRRS1 were analyzed on the online software GEPIA based on CESC-related information in The Cancer Genome Atlas (TCGA). Cervical cancer cells (SiHa and Hela) were accordingly transfected with pCDNA3.1-HOXD-AS1, sh-HOXD-AS1, sh-FRRS1 or pCDNA3.1-ELF1. After cell transfection, CCK-8, EDU and flow cytometry were applied for measurement of cell vitality, quantity and apoptosis, respectively. The relationship between HOXD-AS1 and FRRS1 was predicted on the online software LncMap and further verified by RNA binding protein immunoprecipitation. Nude mice were injected with stabilized SiHa cells transfected with sh-HOXD-AS1 to assess the tumorigenic ability of HOXD-AS1 in vivo. Immunohistochemistry detected the expression of the proliferation marker Ki-67. The levels of HOXD-AS1, ELF1 and FRRS1 were measured in vivo and in vitro. HOXD-AS1 and FRRS1 were overexpressed in CESC. After transfection of sh-HOXD-AS1, sh-ELF1 or sh-FRRS1, the proliferation of SiHa and Hela cells was inhibited and their apoptosis was promoted; while HOXD-AS1 overexpression had opposite effects on CESC development. Co-transfection of sh-FRRS1 and pCDNA3.1-HOXD-AS1 could abolish the tumor suppressive effect of FRRS1 knockdown. HOXD-AS1 elevated the level of FRRS1 by binding ELF1. Furthermore, HOXD-AS1 contributed to the CESC growth in mouse models. HOXD-AS1 promotes CESC by up-regulating FRRS1 via ELF1.
KEYWORDS: LncRNA HOXD-AS1, ELF1, FRRS1, cervical squamous cell carcinoma, SiHA
Introduction
Cervical cancer ranks the fourth devastating gynecologic malignancy worldwide [1]. The incidence and mortality of cervical cancer are much higher in low- and middle-resource regions compared with that in developed countries [2]. Cervical squamous cell carcinoma (CESC) is the dominant type of invasive cervical carcinomas, which accounts for about three quarters of all cervix cancers [3]. Papanicolaou (Pap) test has been the standard approach for cervical cancer diagnosis over the years, but it is constrained by the test sensitivity and significant proportion of deficient samples [4]. As human papillomavirus (HPV) plays a critical role in the majority of cervical malignancies, primary HPV screening has surfaced as a promising method for preventing cervical cancer [5]. However, the HPV vaccination coverage is greatly limited due to the high expense of the current vaccines [6]. Definitive radiation therapy combined with cisplatin-based chemotherapy is currently deemed as the standard treatment for invasive cervical cancer [7]. Considering the current high incidence and mortality of cervical malignancies worldwide, there is ample room for improvements of diagnosis and treatment of this common cancer.
Long non-coding RNAs (lncRNAs) are defined as a class of no protein-coding RNA molecules which are over 200 bases in length [8]. LncRNAs function in diverse biological and pathological processes through mechanisms including chromatin reprogramming, cis regulation at enhancers, and post-transcriptional regulation of mRNA processing [9]. More and more lncRNAs have been discovered to be aberrantly expressed in cancer development and act as regulators and hallmarks of diverse cancers [10]. Collected evidence shows that lncRNAs are also involved in cervical cancer. For instance, lncRNA OIP5-AS1 contributed to metastasis of cervical cancer by up-regulating SMAD3 via targeting miR–143–3p [11]. Baicalein downregulated the expression of lncRNA BDLNR to suppress cervical cancer via the PI3K/Akt signaling pathway [12].
LncRNA HOXD-AS1 is a gene located between the HOXD1 and HOXD3 genes in the HOXD cluster and implicated in the development and progression of various cancers including cervical cancer [13]. A recent study showed that HOXD-AS1 was overexpressed in cervical cancer and promoted tumor cell migration and invasion by increasing the expression of FGF2 via miR–877–3p [14]. As lncRNAs can bind to distal regulatory elements and recruit protein complexes to modulate gene expression in different biological conditions [15], we further searched for the downstream targets of HOXD-AS1 in regulating cervical cancer. E74 like ETS transcription factor 1 (ELF1) is a transcription factor that activates genes involved in tumorigenesis and promotes the development of several cancers [16–20]. Ferric chelate reductase 1 (FRRS1) is an enzyme that reduces ferric to ferrous iron before its transport from the endosome to the cytoplasm [21]. Iron in the cytoplasmic labile iron pool is metabolically active and finally utilized in various physiological processes such as DNA synthesis and mitochondrial oxidative metabolism or stored by cytoplasmic ferritin that is highly expressed in cancers [22]. Therefore, FRRS1 is a potential tumor regulator. Through analysis of cervical cancer-related gene information in The Cancer Genome Atlas (TCGA), the online software GEPIA revealed that HOXD-AS1 and FRRS1 were overexpressed and positively related in CESC. Furthermore, the online prediction software LncMAP suggested that HOXD-AS1 might modulate the expression level of FRSS1 by targeting transcription factor ELF1. The present study is designed to validate the interaction between HOXD-AS1 and FRRS1 in CESC.
Materials and methods
Cell culture and transfection
Normal human cervical epithelial cells and cervical cancer cells (SiHa and Hela) were all procured from the American Type Culture Collection (ATCC) (Manassas, VA, USA). These cells were cultivated in RPMI-1640 medium containing 10% fetal bovine serum (FBS) and 100 μg/mL penicillin/streptomycin in a moist incubator which was maintained at 37°C with 5% CO2.
Cell transfection was performed in SiHa cells and Hela cells via Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA). All the plasmids for cell transfection were obtained from GenePharma (Shanghai, China). The cells were accordingly divided into the following groups: pCDNA3.1 group (transfected with pCDNA3.1), over-HOXD-AS1 group (transfected with pCDNA3.1-HOXD-AS1), sh-ELF1 group (transfected with sh-ELF1), sh-HOXD-AS1 (transfected with sh-HOXD-AS1), sh-FRRS1 group (transfected with sh-FRRS1), sh-FRRS1+ over-HOXD-AS1 group (transfected with pCDNA3.1-HOXD-AS1 and sh-FRRS1), and Control group. Following experiments were conducted after 24 h of transfection.
CCK-8 assay
The cells were transferred into a 96-well plate at a density of 5 × 103 cells/well and cultured for 24 h. Then the cells were added with CCK-8 reagent and incubated at 37°C for 4 h. The optical density (OD) was measured at 450 nm by a microplate reader. Each reaction was set with three dupilcates. The cell survival rate = (OD of the experimental group/OD of the control group) × 100%.
EDU staining
The cells were transferred into a 96-well plate at a density of 1 × 105 cells/well and incubated with 100 μL of 50 μM EDU (Sigma-Aldrich, Merck KGaA, Darmstadt, Germany) at 37°C for 4 h. The cells were fixed in 4% formaldehyde for 30 min before glycine neutralization. After that, the cells were incubated with 100 μL of 1 × Apollo reaction mixture (Ribbio, China) for 30 min and subjected to 0.5% TritonX-100 infiltration for 10 min. DAPI was used to stain cell nuclei. The cells were observed under a fluorescence microscope (Olympus, Tokyo, Japan).
Flow cytometry
After the single cell suspension was centrifuged at 2000 r/min, the sediments obtained were subjected to two PBS washes and resuspended in binding buffer. The cell suspension (195 μL, containing about 1 × 105 cells) was incubated with 5 μL of Annexin-V-FITC and PI in the dark for 10 min. The apoptosis rate was measured by a flow cytometer (BD Biosciences, Suzhou, China).
RNA binding protein immunoprecipitation (RIP)
The cells were lysed in RIPA buffer containing protease inhibitors. The cell lysate was centrifuged at 12,000 rpm for 15 min after which the supernatant was discarded. The protein concentration was measured using a BCA kit. The total protein extract was diluted to 2 μg/μL and incubated with ELF1 antibody (1:500, sc-133096, Santa Cruz, Dallas, USA) at 4°C overnight. The expression of ELF1 was measured by Western blotting and the expression of HOXD-AS1 was measured by PCR.
qRT-PCR
The total RNA was obtained from cells or tissues using TRIzol® reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instruction. The RNA was quantified by a spectrophotometer (Shimadzu, Kyoto, Japan) and reverse transcribed into cDNA using a cDNA synthesis kit (Toyobo, Osaka, Japan). The quantitative real-time PCR was conducted using SYBR Green PCR Kit (Toyobo, Osaka, Japan) and the gene sequences were determined by PRISM 7300 detection system (Applied Biosystems, CA). Below were the PCR parameters: 5 min of pre-denaturation at 95°C and 40 cycles of denaturation (10 s) at 95°C, annealing (10 s) at 60°C and extension (20 s) at 72°C. The 2−∆∆Ct method was applied for relative quantification of gene expression. The primer sequences are exhibited in Table 1.
Table 1.
Primer sequences
| Name of primer | Sequences |
|---|---|
| HOXD-AS1 | F: 5ʹ-CGCCCTTTCTGACCTGCTTA-3’ |
| R: 5ʹ-TGGCAGTCGTCTGGACATTC-3’ | |
| ELF1 | F: 5ʹ-GGAAACACAGCAGGTGCAAG-3’ |
| R: 5ʹ-GGAATCTGGTCGTGGTGGTT-3’ | |
| FRRS1 | F: 5ʹ-AACGTTACCTCCCGTTTCCC-3’ |
| R: 5ʹ-TTTACTGGGGCCGCTCATTT-3’ | |
| GAPDH | F: 5ʹ-TCTTGTGCAGTGCCAGCCT-3’ |
| R: 5ʹ-TGAGGTCAATGAAGGGGTCG-3’ |
F, forward; R, reverse
Western blot
Cells or tissues were immersed in RIPA buffer containing protease inhibitors and phosphatase inhibitors and then centrifuged at 4°C, 13,000 rpm for 30 min. The supernatant was collected for measurement of protein concentration by a BCA kit and the proteins were separated by SDS-PAGE. After that, the proteins were transferred onto a PVDF membrane and incubated with primary rabbit antibodies of GAPDH (1:10000, ab181602, abcam, Cambridge, UK), ELF1 (1:500, sc-133096, Santa Cruz, Dallas, USA) and FRRS1 (1:1000, orb183772, Biorbyt, Cambridge, UK) at 4°C overnight. After three PBST washes, the proteins were incubated with secondary antibody for 30 min. Finally, the membrane was washed in PBST for 4 times and added with ECL for color development. The blots were photographed in a chemiluminescence imaging system (GE Healthcare, Beijing, China).
Animal experiments
Animal experiments set in this study conformed to the guidelines published by the Animal Protection and Management Committee of Xiangya Hospital Central South University. Nude BALB/C-A mice aged four weeks were purchased from Hunan SJA Laboratory Animal Co., Ltd (Changsha, China). The mice were fed on standard food and water in a pathogen-free environment with a 12 h day and 12 h night shift over one week. The mice were subcutaneously injected with 1 × 106 stabilized SiHa cells (Hanheng, Shanghai, China) transfected with sh-HOXD-AS1 or untreated SiHa cells on the right back; each group consisted of six mice. The mice were euthanatized four weeks after injection to obtain the tumor and the tumor weight was thereafter measured.
Immunohistochemistry (IHC)
The tumor tissues were fixed in formalin and then made into 4 μm paraffin sections. The sections were deparaffinized in xylene and hydrated in gradient alcohol. Then the tissues were treated with 10 mM of citrate (pH 6.0) for antigen retrieval and 3% hydrogen peroxide for endogenous peroxidase confinement. The tissues were then incubated with Ki-67 antibody (2 µg/mL, ab15580, Abcam, Cambridge, UK) at 4°C overnight. After three PBS washes, the tissues were incubated with secondary antibody at room temperature for 1 h. After three PBS washes, the tissues were subjected to color development using DAB for 1 ~ 3 min and stained in hematoxylin for 3 min. Finally, the tissues were dehydrated, permeabilized and mounted. Ki-67+ cells were presented in yellow or brown. For each slide, five fields were randomly selected and observed under a microscope (200x). The percentage of Ki-67+ cells = the number of yellow or brown cells/the number of blue cells × 100%. The experiment was repeated three times.
Statistical analysis
Statistics were analyzed in GraphPad Prism 7.0 and presented as mean ± SD (standard deviation). T method was applied for comparison between two groups and one-way analysis of variance was for multiple group comparisons. A difference with P < 0.05 was deemed to have statistical significance.
Results
HOXD-AS1 and FRRS1 are overexpressed and positively related in CESC
Based on analysis of CESC-related genes in TCGA, high levels of HOXD-AS1 and FRRS1 were found in CESC tissues compared to adjacent tissues (Figure 1(a,b), P < 0.05). Meanwhile, the expressions of these two genes were positively related (Figure 1(c), p < 0.05). Furthermore, HOXD-AS1 and FRRS1 were discovered to be highly expressed in different cervical cancer cell lines compared to normal human cervical epithelial cells (Figure 1(d-f), P < 0.05).
Figure 1.
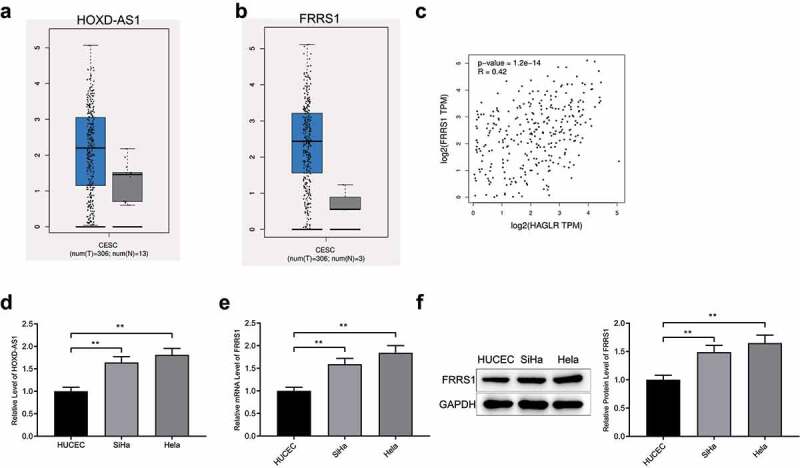
The expressions of HOXD-AS1 and FRRS1 in CESC Notes: (a-b) The expressions of HOXD-AS1 and FRRS1 in CESC detected in online software GEPIA via analysis of TCGA database; (c) the correlation analysis of the expressions of HOXD-AS1 and FRRS1 in CESC; (d) qRT-PCR detected the expression of HOXD-AS1 in different cervical cancer cell lines and normal human cervical epithelial cells; (e-f) the level of FRRS1 in normal and cancer cervical cells measured by Western blotting. Data are presented as mean ± SD (standard deviation). **P < 0.01; CESC, cervical squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
HOXD-AS1 enhances the proliferation of cervical cancer cells
After transfection of pCDNA3.1-HOXD-AS1, sh-HOXD-AS1 or the negative controls, HOXD-AS1 was successfully overexpressed or knocked down in cervical cancer cells (SiHa and Hela) (Figure 2(a), p < 0.05). FRRS1 was up-regulated in the over-HOXD-AS1 group while down-regulated in the sh-HOXD-AS1 group compared to the Control group (Figure 2(b,c), P < 0.05).
Figure 2.
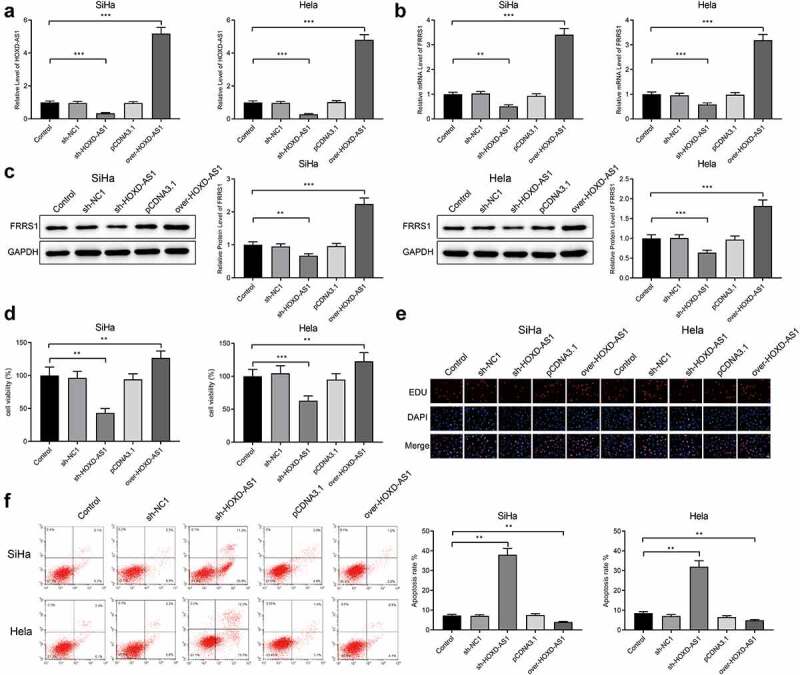
HOXD-AS1 enhances the proliferation of cervical cancer cells Notes: The cervical cancer cells (SiHa and Hela) were transfected with pCDNA3.1-HOXD-AS1, sh-HOXD-AS1 or the negative controls. (a) The expression of HOXD-AS1 assessed by qRT-PCR; (b-c) the expression of FRRS1 detected by qRT-PCR and Western blotting; (d) the cell vitality measured by CCK-8; (e) the quantity of proliferated cells showed by EDU staining; (f) apoptosis rate assessed by flow cytometry. Data are presented as mean ± SD (standard deviation). **P < 0.01, ***P < 0.001; CESC, cervical squamous cell carcinoma.
Subsequently, the proliferative ability of SiHa cells and Hela cells was assessed. CCK-8 assay detected that the cell vitality was enhanced in the over-HOXD-AS1 group while impeded in the sh-HOXD-AS1 group compared to the Control group (Figure 2(d), p < 0.05). Furthermore, EDU staining showed that the over-HOXD-AS1 group had more newborn cells compared to the Control group while the cell proliferation in the sh-HOXD-AS1 group was impaired (Figure 2(e)). Flow cytometry detected reduced apoptosis in the over-HOXD-AS1 group and increased apoptosis in the sh-HOXD-AS1 group compared to the Control group (Figure 2(f), p < 0.05). From the above, HOXD-AS1 positively regulates the expression of FRRS1. HOXD-AS1 can also promote proliferative capacity of cervical cancer cells and further suppress cell apoptosis.
FRRS1 enhances the proliferation of cervical cancer cells
The SiHa cells and Hela cells were transfected with sh-FRRS1 or co-transfected with sh-FRRS1+ pCDNA3.1-HOXD-AS1 to explore the function of FRRS1. FRRS1 was successfully inhibited after the transfection of sh-FRRS1; the expression of FRRS1 was increased in the sh-FRRS1+ over-HOXD-AS1 group compared to the sh-FRRS1 group (Figure 3(a,b), P < 0.05). CCK-8 assay detected that the cell vitality was hindered in the sh-FRRS1 group compared to the Control group while enhanced in the sh-FRRS1+ over-HOXD-AS1 group compared to the sh-FRRS1 group (Figure 3(c), p < 0.05). From the results of EDU staining, cells were decreased in the sh-FRRS1 group compared to the Control group while increased in the sh-FRRS1+ over-HOXD-AS1 group compared to the sh-FRRS1 group (Figure 3(d)). Flow cytometry showed that the sh-FRRS1 group had a higher apoptosis level than the Control group and the apoptosis level was reduced in the sh-FRRS1+ over-HOXD-AS1 group compared to the sh-FRRS1 group (Figure 3(e), p < 0.05). The above results indicate that FRRS1 knockdown inhibits the proliferative ability of cervical cancer cells and enhances the cell apoptosis. HOXD-AS1 overexpression can abolish the suppressive effect of FRRS1 knockdown on CESC.
Figure 3.
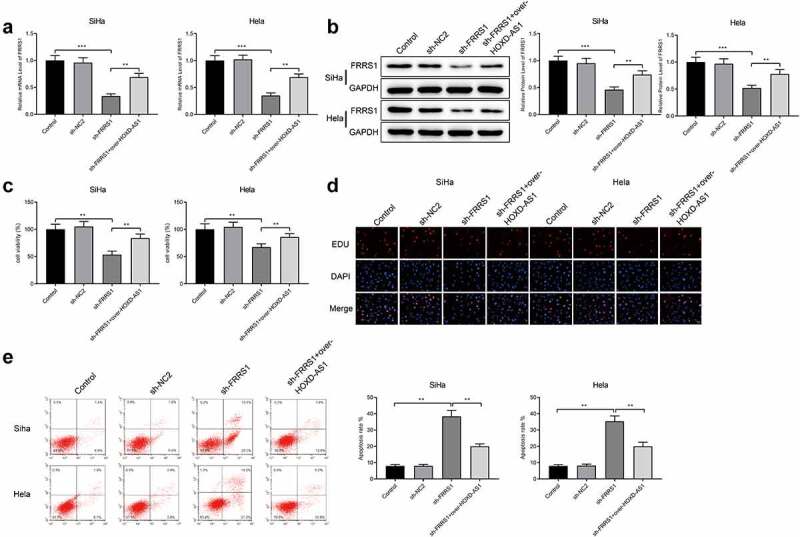
FRRS1 enhances the proliferation of cervical cancer cells Notes: After the SiHa cells and Hela cells were transfected with sh-FRRS1 or co-transfected with sh-FRRS1+ pCDNA3.1-HOXD-AS1, (a-b) qRT-PCR and Western blotting measured the expression of FRRS1; (c) CCK-8 detected the cell vitality; (d) EDU staining assessed the proliferative ability of the cervical cancer cells; (e) flow cytometry assessed the apoptosis rate. Data are presented as mean ± SD (standard deviation). **P < 0.01, ***P < 0.001; CESC, cervical squamous cell carcinoma.
HOXD-AS1 regulates the expression of FRSS1 through ELF1 in CESC
The downstream target of HOXD-AS1 was predicted on the online software LncMAP (http://bio-bigdata.hrbmu.edu.cn/LncMAP) (Figure 4(a)). LncMAP suggested that HOXD-AS1 might regulate the expression of FRSS1 through ELF1 (Figure 4(b)). In the starBase database (http://starbase.sysu.edu.cn/), HOXD-AS1 and ELF1 were weakly correlated in CESC (Figure 4(c), r = 0.104, P < 0.05). The GEPIA2 database (http://gepia2.cancer-pku.cn/) showed there was a significant positive correlation between ELF1 and FRRS1 in CESC (Figure 4(d), r = 0.51, P < 0.05). Moreover, we verified the binding between HOXD-AS1 and ELF1 through RIP assay (Figure 4(e)).
Figure 4.
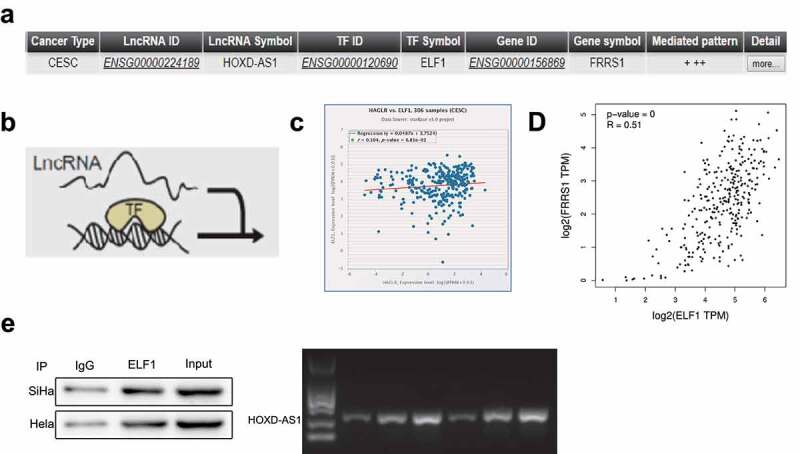
HOXD-AS1 regulates the expression of FRSS1 by binding ELF1 in CESC Notes: (a) The regulatory mechanism between HOXD-AS1 and FRRS1 predicted by the online software LncMap; (b) the schematic diagram of this regulating process. (c) The correlation analysis of HOXD-AS1 and ELF1 in CESC by the starBase database. (d) The correlation analysis of ELF1 and FRRS1 in CESC by the GEPIA2 database. (e) RIP assay was performed to verify the binding of HOXD-AS1 to ELF1. Data are presented as mean ± SD (standard deviation). CESC, cervical squamous cell carcinoma; RIP, RNA binding protein immunoprecipitation.
ELF1 acts on the proliferation and apoptosis of cervical cancer cells by up-regulating FRSS1
SiHa cells and Hela cells were transfected with sh-ELF1. ELF1 was underexpressed in the cervical cancer cells (Figure 5(a,b)), indicating successful transfection of sh-ELF1. FRSS1 was down-regulated in the sh-ELF1 group compared to the Control group (Figure 5(c,d), P < 0.05). CCK-8 suggested that the sh-ELF1 group had lower cell vitality than the Control group (Figure 5(e), p < 0.05). According to the results of EDU and flow cytometry, the cell proliferation was impeded and apoptosis rate was increased in the sh-ELF1 group compared to the Control group (Figure 5(f,g), P < 0.05). Taken together, knockdown of ELF1 suppresses FRRS1 expression to inhibit the proliferative capacity and promote the apoptosis of cervical cancer cells.
Figure 5.
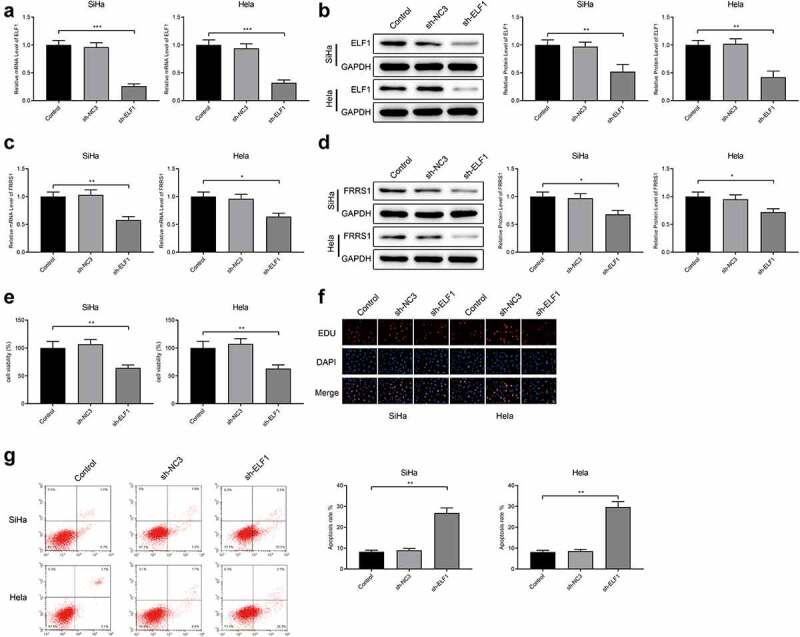
ELF1 regulates FRSS1 to promote the proliferation and apoptosis of cervical cancer cells Notes: After SiHa cells and Hela cells were transfected with sh-ELF1, (a-d) qRT-PCR and Western blotting detected the expressions of ELF1 and FRRS1; (e) CCK-8 measured the cell vitality; (f) EDU staining revealed the quantity of proliferated cells; (g) flow cytometry tested the apoptosis rate. Data are presented as mean ± SD (standard deviation). *P < 0.05, **P < 0.01, ***P < 0.001.
HOXD-AS1 accelerates the proliferation of cervical cancer cells in mouse models
The nude mice were subcutaneously injected with stabilized SiHa cells which were transfected with sh-HOXD-AS1. This group of mice was named the KD-HOXD-AS1 group. The measurement showed that HOXD-AS1 was down-regulated in the KD-HOXD-AS1 group (Figure 6(a), p < 0.05). The expression of FRRS1 was also lowered down in the KD-HOXD-AS1 group compared to the Normal group (Figure 6(b,c), P < 0.05). The tumors of the KD-HOXD-AS1 group were smaller than those of the Normal group (Figure 6(d), p < 0.05). Furthermore, the Ki-67 level in the tumors was decreased in the KD-HOXD-AS1 group compared to the Normal group (Figure 6(e), p < 0.05). Taken together, HOXD-AS1 promotes the CESC growth in vivo.
Figure 6.
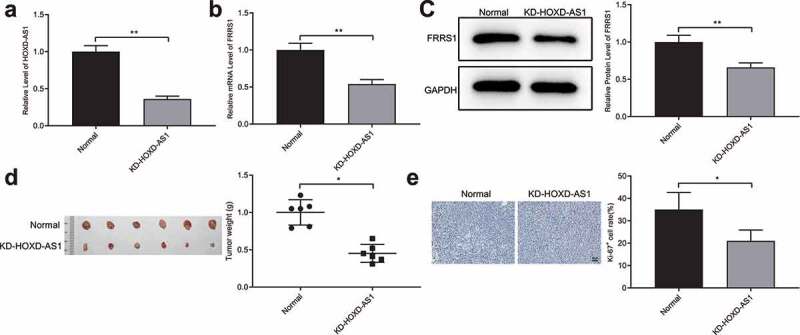
HOXD-AS1 accelerates the proliferation of cervical cancer cells in vivo Notes: After the nude mice were subcutaneously injected with stabilized SiHa cells transfected with sh-HOXD-AS1, (a) qRT-PCR detected the level of HOXD-AS1 in the tumor tissues; (b-c) qRT-PCR and Western blotting measured the level of FRRS1; (d) the weight measurement of tumors; (e) IHC assessed the level of Ki-67 in the tumors. Data are presented as mean ± SD (standard deviation). *P < 0.05, **P < 0.01; CESC, cervical squamous cell carcinoma; IHC, immunohistochemistry.
Discussion
In spite of established screening programs for detecting precancerous changes in the cervix, cervical cancer remains one of the deadliest cancers in global females [23]. The promising HPV vaccination still has a long way to go before reaching the goal of eliminating cervical cancer [24]. Patients with cervical cancer in low-source regions are usually diagnosed at the advanced stage with restricted treatment options and poor prognosis [25]. The survival has been improved over decades for patients with early-staged and locally advanced cervical cancer especially those with stage II and III tumors, but no significant survival improvements have been seen in patients with metastatic cervical carcinomas [26]. This study is aimed at searching for new therapeutic targets for cervical cancer and uncovers a novel molecular mechanism in this disease.
HOXD-AS1 and FRRS1 were found overexpressed in CESC compared with adjacent normal tissues based on analysis of TCGA data in the online software GEPIA. These two genes were positively related in CESC. In in vitro settings, HOXD-AS1 and FRRS1 were overexpressed in two different cervical cancer cell lines (SiHa and Hela) compared to HUCECs. In the following experiments, the impacts of dysregulated HOXD-AS1 and FRRS1 on CESC were investigated through cell transfection. The results indicated that overexpressed HOXD-AS1 promoted the proliferative capacity of cervical cancer cells as well as the expression level of FRRS1 while restrained the cell apoptosis. On the other hand, FRRS1 inhibition impeded the progression of CESC and overexpressed HOXD-AS1 abolished the tumor suppressive effect of FRRS1 inhibition.
HOXD-AS1 has been authenticated as a carcinogenic regulator in different cancers. For instance, HOXD-AS1 down-regulated miR-147a to promote the growth of non-small cell lung cancer [27]. HOXD-AS1 contributed to the proliferation, migration and invasion of ovarian cancer cells via the miR-608/FZD4 axis [28]. As for cervical cancer, Hu et al. claimed that HOXD-AS1 was significantly up-regulated in cancer cells and promoted tumor growth by activating the Ras/ERK signaling pathway [29]. In addition to the oncogenic effect, HOXD-AS1 could also enhance chemoresistance of cisplatin-resistant cervical cancer cells [30]. Linton et al. applied the microarray technology to examine gene expression in archival formalin-fixed paraffin-embedded tissues and FRRS1 was found to have prognostic significance for cancers [31]. A recent study demonstrated that FRRS1 was consistently overexpressed in multimorbidity for asthma, rhinitis and dermatitis [32]. To date, evidence for the specific role of FRRS1 in cervical cancer or any other pathological conditions is rare.
The online software LncMAP predicted that HOXD-AS1 might mediate the expression of FRRS1 by targeting ELF1 and the binding of HOXD-AS1 to ELF1 was verified by a RIP assay. To uncover the function of ELF1 in CESC, we inhibited the expression of ELF1 in cervical cancer cells. ELF1 inhibition down-regulated the level of FRRS1 in cervical cancer cells and inhibited the cell proliferation while promoted the apoptosis. ELF1, belonging to the Ets transcription factor family, acts as a vital regulator of the hematopoietic system [33]. Nicol et al. claimed that ELF1 was significantly increased in HPV+/HIV− women with high grade cervical intraepithelial neoplasia and tumor [34]. Moreover, ELF1 increased the expression of human Pygopus 2 that was required for the growth of transformed HPV-infected human cervical cancer cells [35]. Evidence for the expression and effect of ELF1 in cancers is still insufficient. Future research could be focused on the specific function of this tumor-related transcription factor.
We also set animal experiments to validate the role of HOXD-AS1 in vivo. FRRS1 was down-regulated post HOXD-AS1 knockdown. The tumor size in mice treated with sh-HOXD-AS1 was smaller than that of untreated mice. The proliferation-related protein Ki-67 was also decreased by HOXD-AS1 inhibition. Taken together, HOXD-AS1 can promote the CESC growth in mice.
HOXD-AS1 improves proliferative capacity of cervical cancer cells and inhibits cell apoptosis through activation of FRRS1 by targeting transcription factor ELF1. HOXD-AS1 shows great potential as a molecular indicator and therapeutic target for cervical cancer. Understanding of the mechanism underlying the effect of HOXD-AS1 may also broaden the prospects of cervical cancer therapy and therefore relieve the burden of women with this disease.
Acknowledgments
Thanks for all the contributors.
Funding Statement
Thanks for the grants from the Natural Science Foundation of Hunan Province (Grant No. 2019JJ40490) and the Clinical Research Project of Xiangya Hospital (Grant No. 2016L06).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Cohen PA, Jhingran A, Oaknin A, et al. Cervical cancer. Lancet. 2019;393(10167):169–182. [DOI] [PubMed] [Google Scholar]
- [3].Small W Jr., Bacon MA, Bajaj A, et al. Cervical cancer: a global health crisis. Cancer. 2017;123(13):2404–2412. [DOI] [PubMed] [Google Scholar]
- [4].Marth C, Landoni F, Mahner S, et al. Cervical cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28:iv72–iv83. [DOI] [PubMed] [Google Scholar]
- [5].Rizzo AE, Feldman S.. Update on primary HPV screening for cervical cancer prevention. Curr Probl Cancer. 2018;42(5):507–520. [DOI] [PubMed] [Google Scholar]
- [6].Roden RBS, Stern PL. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat Rev Cancer. 2018;18(4):240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Marchetti C, Fagotti A, Tombolini V, et al. Survival and toxicity in neoadjuvant chemotherapy plus surgery versus definitive chemoradiotherapy for cervical cancer: a systematic review and meta-analysis. Cancer Treat Rev. 2020;83:101945. [DOI] [PubMed] [Google Scholar]
- [8].Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47(3):199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839:1097–1109. [DOI] [PubMed] [Google Scholar]
- [11].Chen X, Xiong D, Yang H, et al. Long noncoding RNA OPA-interacting protein 5 antisense transcript 1 upregulated SMAD3 expression to contribute to metastasis of cervical cancer by sponging miR–143–3p. J Cell Physiol. 2019;234(4):5264–5275. [DOI] [PubMed] [Google Scholar]
- [12].Yu X, Yang Y, Li Y, et al. Baicalein inhibits cervical cancer progression via downregulating long noncoding RNA BDLNR and its downstream PI3K/Akt pathway. Int J Biochem Cell Biol. 2018;94:107–118. [DOI] [PubMed] [Google Scholar]
- [13].Li L, Wang Y, Zhang X, et al. Long non-coding RNA HOXD-AS1 in cancer. Clin Chim Acta. 2018;487:197–201. [DOI] [PubMed] [Google Scholar]
- [14].Chen S, Li K. HOXD-AS1 facilitates cell migration and invasion as an oncogenic lncRNA by competitively binding to miR–877–3p and upregulating FGF2 in human cervical cancer. BMC Cancer. 2020;20(1):924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zhang G, Lan Y, Xie A, et al. Comprehensive analysis of long noncoding RNA (lncRNA)-chromatin interactions reveals lncRNA functions dependent on binding diverse regulatory elements. J Biol Chem. 2019;294(43):15613–15622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheng M, Zeng Y, Zhang T, et al. Transcription factor ELF1 activates MEIS1 transcription and then regulates the GFI1/FBW7 axis to promote the development of glioma. Mol Ther Nucleic Acids. 2021;23:418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hu M, Li H, Xie H, et al. ELF1 transcription factor enhances the Progression of glioma via ATF5 promoter. ACS Chem Neurosci. 2021;12(7):1252–1261. [DOI] [PubMed] [Google Scholar]
- [18].Qiao C, Qiao T, Yang S, et al. SNHG17/miR-384/ELF1 axis promotes cell growth by transcriptional regulation of CTNNB1 to activate Wnt/beta-catenin pathway in oral squamous cell carcinoma. Cancer Gene Ther. 2021; DOI: 10.1038/s41417-021-00294-9. [DOI] [PubMed] [Google Scholar]
- [19].Wang L. ELF1-activated FOXD3-AS1 promotes the migration, invasion and EMT of osteosarcoma cells via sponging miR–296–5p to upregulate ZCCHC3. J Bone Oncol. 2021;26:100335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wang L, Tang D, Wu T, et al. ELF1-mediated LUCAT1 promotes choroidal melanoma by modulating RBX1 expression. Cancer Med. 2020;9(6):2160–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vargas JD, Herpers B, McKie AT, et al. Stromal cell-derived receptor 2 and cytochrome b561 are functional ferric reductases. Biochim Biophys Acta. 2003;1651(1–2):116–123. [DOI] [PubMed] [Google Scholar]
- [22].Wang Y, Yu L, Ding J, et al. Iron Metabolism in Cancer. Int J Mol Sci. 2018;20(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Campos NG, Castle PE, Wright TC Jr., et al. Cervical cancer screening in low-resource settings: a cost-effectiveness framework for valuing tradeoffs between test performance and program coverage. Int J Cancer. 2015;137(9):2208–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sundstrom K, Elfstrom KM. Advances in cervical cancer prevention: efficacy, effectiveness, elimination? PLoS Med. 2020;17(1):e1003035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ojesina AI, Lichtenstein L, Freeman SS, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506(7488):371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wright JD, Chen L, Tergas AI, et al. Population-level trends in relative survival for cervical cancer. Am J Obstet Gynecol. 2015;213(5):670 e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Wang Q, Jiang S, Song A, et al. HOXD-AS1 functions as an oncogenic ceRNA to promote NSCLC cell progression by sequestering miR-147a. Onco Targets Ther. 2017;10:4753–4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wang Y, Zhang W, Wang Y, et al. HOXD-AS1 promotes cell proliferation, migration and invasion through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res. 2018;8(1):170–182. [PMC free article] [PubMed] [Google Scholar]
- [29].Hu YC, Wang AM, Lu JK, et al. Long noncoding RNA HOXD-AS1 regulates proliferation of cervical cancer cells by activating Ras/ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(22):5049–5055. [DOI] [PubMed] [Google Scholar]
- [30].Chi C, Mao M, Shen Z, et al. HOXD-AS1 exerts oncogenic functions and promotes chemoresistance in cisplatin-resistant cervical cancer cells. Hum Gene Ther. 2018;29(12):1438–1448. [DOI] [PubMed] [Google Scholar]
- [31].Linton KM, Hey Y, Saunders E, et al. Acquisition of biologically relevant gene expression data by Affymetrix microarray analysis of archival formalin-fixed paraffin-embedded tumours. Br J Cancer. 2008;98(8):1403–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lemonnier N, Melen E, Jiang Y, et al. A novel whole blood gene expression signature for asthma, dermatitis, and rhinitis multimorbidity in children and adolescents. Allergy. 2020;75(12):3248–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Choi HJ, Geng Y, Cho H, et al. Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood. 2011;117(6):1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Nicol AF, Pires AR, de Souza SR, et al. Cell-cycle and suppressor proteins expression in uterine cervix in HIV/HPV co-infection: comparative study by tissue micro-array (TMA). BMC Cancer. 2008;8(1):289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tzenov YR, Andrews PG, Voisey K, et al. Human papilloma virus (HPV) E7-mediated attenuation of retinoblastoma (Rb) induces hPygopus2 expression via Elf-1 in cervical cancer. Mol Cancer Res. 2013;11(1):19–30. [DOI] [PubMed] [Google Scholar]


