ABSTRACT
Non-small cell lung cancer (NSCLC) is one of the most prevalent tumors with high incidence and mortality across the globe. Recently, increasing studies have demonstrated that circular RNAs (circRNAs) exert outstanding functions in NSCLC progression. Notwithstanding, we are still in the dark about the function and exact mechanism of circ-PITX1, a newly discovered circRNA. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) confirmed the profile of circ-PITX1 in NSCLC tissues and adjacent normal tissues. Gain- and loss- of function assay verified the impact of circ-PITX1 and miR-30e-5p on the proliferation, invasion, and migration of NSCLC cells (H1975 and A549). Bioinformatics analysis corroborated the downstream mechanisms of circ-PITX1. Dual-luciferase reporter gene assay and RNA immunoprecipitation (RIP) examined the interactions between circ-PITX1 and miR-30e-5p, miR-30e-5p and ITGA6. The protein levels of ITGA6, PI3K, AKT were determined by Western blot. circ-PITX1 was substantially up-regulated in NSCLC tissues and cells, and circ-PITX1 up-regulation was correlated with NSCLC patients’ poor survival. Functionally, circ-PITX1 overexpression or miR-30e-5p inhibition markedly facilitated proliferation, migration, invasion, epithelial-mesenchymal transition (EMT), reduced apoptosis, and enhanced ITGA6/PI3K/AKT expression in NSCLC cells, whereas circ-PITX1 knockdown or miR-30e-5p up-regulation resulted in the opposite results. Mechanistically, circ-PITX1 acted as a sponge of miR-30e-5p, which targeted the 3ʹuntranslated region (UTR) of ITGA6. Knockdown of circ-PITX1 or overexpressing miR-30e-5p reduced ITGA6/PI3K/AKT axis. circ-PITX1 modulates the miR-30e-5p/ITGA6 axis to boost NSCLC progression, hence functioning as an oncogene.
KEYWORDS: NSCLC, progression, circ- PITX1, miR-30e-5p, ITGA6
1. Introduction
Non-small cell lung cancer (NSCLC), the most prevailing lung cancer that accounts for around 80% of all relevant cases, is a leading contributor to cancer-related death across the world [1]. Death caused by NSCLC can be largely attributed to recurrent metastasis, while there is still a lack of accurate indicators to evaluate NSCLC progression or effective targeted drugs for NSCLC treatment [2,3]. Therefore, probing the mechanisms of NSCLC is critical to identifying new therapeutic targets and developing more effective therapies.
Circular RNAs (circRNAs) are a new type of non-coding RNAs that are abundant in eukaryotes [4]. Also, circRNAs are a class of trashy by-products in the course of gene transcription. Nevertheless, circRNAs can act as a decoy molecule, a scaffold molecule, or a guide molecule to modulate gene expression, thereby influencing the pathophysiological processes of diverse diseases [5–8]. Emerging circRNAs have been uncovered to present aberrant expression in tumors and affect tumor development. For instance, hsa-circRNA-103809 exhibits pronounced expression in lung cancer tissues and bolsters lung cancer cell proliferation and invasion [9]. Circ-0026344, down-regulated in colorectal cancer, can suppress colorectal cancer progression when it gets overexpressed [10]. Various circRNAs have been studied in NSCLC. For instance, circSLC8A1 and circFARSA are up-regulated in NSCLC tissues and function as prospective prognostic biomarkers and therapeutic targets of NSCLC [11,12]. Additionally, an increasing number of circRNAs have been confirmed to regulate NSCLC development [13]. circ-PITX1, situated at chr5:134363423–134369964, was first identified by Julia Salzman, et al. in A549 cells [14]. Nonetheless, the function of circ-PIXT1 in NSCLC remains poorly understood.
MicroRNAs (miRNAs) are small non-coding molecules that modulate the expression of over 60% of human genes. Abnormally expressed miRNAs regulate tumor proliferation, apoptosis, migration, and metastasis, pertaining to human cancer pathogenesis [15]. Fundamentally, miRNAs regulate gene expression by specifically combining with the 3ʹ-untranslated region (3ʹ-UTR) of target mRNAs, thus modulating tumor cells’ biological behaviors [16]. For instance, miR-224-5p targets the PIK3R3/AKT3 pathway to participate in uveal melanoma cell proliferation, invasion, and migration [17]. miR-30e-5p, a member of the miR-30 family, regulates the Sirt1/JAK/STAT3 signaling mediated by USP22, serving as an inhibitor in NSCLC [18]. These reports hint that miR-30e-5p has the potential to mediate NSCLC treatment.
Integrin subunit alpha 6 (ITGA6), an integrin receptor, contributes to cell-to-cell adhesion [19] and exerts a function in inflammation and fibrosis [20]. ITGA6 has been revealed as a modulator to influence tumor cell proliferation, migration, and invasion. For instance, Twist2, a member of the basic helix-loop-helix (bHLH) family, modulates the profiles of ITGA6 and CD44 to exacerbate kidney cancer cells’ proliferation, migration, and invasion and cramp their apoptosis [21]. What’s more, higher ITGA6 expression predicts a poor prognosis of pancreatic cancer, while ITGA6 down-regulation attenuates cancer cell invasion and metastasis [22]. Interestingly, p53-elicited miR-30e-5p targets ITGA6 and ITGB1 to repress invasion and metastasis, displaying its anti-tumor function in colorectal cancer [23]. Nonetheless, the function of miR-30e-5p-mediated ITGA6 in NSCLC has not been investigated.
Here, we examined the interaction among circ-PITX1, miR-30e-5p, and ITGA6 in NSCLC cells so as to reveal their underlying mechanisms in NSCLC development. We discovered that circ-PITX1 was up-regulated in NSCLC tissues and cell lines. Moreover, circ-PITX1 accelerated the malignant phenotypes of NSCLC cells. Mechanistically, circ-PITX1 sponged miR-30e-5p as a competitive endogenous RNA (ceRNA), which then targeted ITGA6. Overall, this work has disclosed a novel molecular mechanism of the circ-PITX1-miR-30e-5p-ITGA6 axis in NSCLC and provided new theoretical references for treating NSCLC.
2. Materials and methods
2.1. Clinical sample collection
Tumor tissues and adjacent non-tumor tissues of 40 patients with primary NSCLC who underwent surgical resections in the Tianjin First Central Hospital from January 2014 to January 2015 were collected. All patients were diagnosed with NSCLC by pathologists. None of the patients had been subjected to any chemotherapy or radiotherapy prior to the surgery. The adjacent non-tumor tissues were at least 3 cm away from the tumor margin, with no cancer cells discovered in them. All the specimens were removed and immediately stored in liquid nitrogen at -196°C for subsequent analysis. This study had received the green light from the ethics committee of Tianjin First Central Hospital, and all patients involved signed the informed consent.
2.2. Cell culture
Human NSCLC cell lines (H1975 and A549) and the normal lung epithelial cell line BEAS-2B were ordered from the Chinese Academy of Sciences (Shanghai, China). The cells were cultured with a DMEM-F12 medium (Thermo Fisher HyClone, Utah, USA) incorporating 10% FBS (Thermo Fisher Scientific, MA, USA) in an incubator with 5% CO2 at 37℃. The medium was substituted every 2 days, and the cells were passed every 2 or 3 days. The following experiments were implemented when the cells achieved about 70–90% abundance.
2.3. Cell transfection
H1975 and A549 cells in the logarithmic growth phase were inoculated onto 60-mm culture plates at 1 × 107 cells/well and incubated at 37°C with 5% CO2 for 24 hours prior to transfection. GenePharma (Shanghai, China) was responsible for the design of the siRNA specifically targeting circ-PITX1 (si-circ-PITX1), siRNA specifically targeting ITGA6 (si-ITGA6), circ-PITX1 overexpression plasmids, and circ-PITX1 negative controls (vector or si-NC). miR-30e-5p mimics, miR-30e-5p inhibitors, and their negative controls (miR-NC and NC-in) were supplied by RiboBio (Guangzhou, China). Lipofectamine 3000 (Thermo Fisher Scientific, IL, USA) was adopted to transfect the above expressing vectors into NSCLC cells in line with the supplier’s instructions. The transfection validity was examined by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) 48 hours following the transfection.
2.4. RNA extraction and qRT-PCR analysis
Total RNA was extracted out of tissues or cultured cells with the use of the TRIzol reagent (Invitrogen, Shanghai, China). The Cytoplasmic and Nuclear RNA Purification Kit (Norgen Biotek, Cat. 21000) was employed for the purification of the cytoplasmic and nuclear RNA obtained from the cultured NSCLC cells. The First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Rockford, IL, USA) was taken to reversely transcribe the total RNA into cDNA. The LightCycler FastStart DNA MasterPlus SYBR Green I kit (Roche Diagnostics, Burgess Hill, UK) was applied for qRT-PCR as instructed by the manufacturer. The 2−ΔΔCT method was utilized to calculate the relevant profiles of circ-PITX1, miR-30e-5p, and ITGA6. Primers for qRT-PCR are as follows: circ-PITX1 Forward: 5ʹ- GTGCCAGTAGCAATAACTGCC-3ʹ, Reverse: 5ʹ-CAGAAGATGGCACTGGTAGGT-3ʹ; miR-30e-5p Forward: 5ʹ-GTCTCCCAGAUAAUGCG-3ʹ, Reverse: 5ʹ-CGTTCAGGGTCC-3ʹ; ITGA6 Forward: 5ʹ-TCATGGATCTGCAAATGGAA-3ʹ, Reverse: 5ʹ-GGCGGAGGTCAATTCTGTTA-3ʹ; GAPDH Forward: 5ʹ-AGGTCGGTGTGAACGGATTTG; Reverse: 5ʹ-GGGGTCGTTGATGGCAACA-3ʹ; U6 Forward: 5ʹ-TCTTCGTCATCACATATACTAAAAT-3ʹ, Reverse:5ʹ- CTCTTCACGAATTTTCGTGTCAT -3ʹ.
2.5. Cell couting kit-8 (CCK-8) assay
H1975 and A549 cells in the logarithmic phase of each group were counted following trypsinization and centrifugation. Then the cells were inoculated into 96-well plates, with 100 μl of cell suspension (2 × 104 cells/mL) administered to each well. Twenty-four hours after incubation, the culture medium was discarded and substituted by a 90 μL complete medium in company with 10 μL of CCK-8 solution (Abcam, Shanghai, China). Subsequent to one hour’s incubation, a microplate reader was adopted to check the absorbance value of each well at 450 nm.
2.6. Transwell assay
Before the Transwell assay was conducted, the bottoms of the Transwell chambers were coated with Matrigel (Becton, Dickinson and Company) overnight. H1975 and A549 cells in the logarithmic growth phase were harvested, with the cell density adjusted to 1 × 105/mL using an FBS-free DMEM-F12 medium. The cell suspension was given to the upper Transwell compartment, with 500 μL of a complete medium supplemented with 20% FBS administered to the lower chamber of the 24-well plate, for 24 hours’ incubation at 37°C with 5% CO2. As the Transwell compartments were taken out, the cells on the bottoms of them were cleaned using cotton swabs. The cells in the plates were immobilized with 95% ethanol at room temperature for 30 minutes, dyed with crystal violet for 20 minutes, and flushed in water 3 times. An inverted microscope was introduced to observe the cells and capture their images. In the invasion experiment, the steps were the same as those in the migration assay barring the use of Matrigel for coating the bottoms of Transwell chambers. The experiment of each group was implemented in triplicate.
2.7. Western blot
The tissues and cells were lysed employing RIPA lysis buffer (Beyotime, Wuhan, China) on ice for 20 minutes and then centrifuged at 14000 rpm and 4°C for 20 minutes. Following centrifugation, the supernatant was harvested, and the total protein was quantified with the BCA protein quantitative kit (Beyotime, Wuhan, China). With the concentration adjusted, the protein samples were boiled for 5 minutes for denaturation. Subsequent to SDS-polyacrylamide gel electrophoresis (SDS-PAGE), the protein was electrically transferred onto PVDF membranes. After that, TBST solution containing 3% BSA was applied to seal the membranes at room temperature for 1 hour, which were then incubated along with the diluted primary antibodies at 4°C overnight. The next morning, the membranes were flushed with TBST 5 times (3 minutes each) and incubated along with the diluted horseradish peroxidase (HRP)-tagged anti-rabbit secondary antibody (concentration 1:300) for an hour. They were then rinsed in TBST another 3 times, 10 minutes each. Western blot reagent (Invitrogen) was exploited for color development. All the primary antibodies utilized in this study were bought from Abcam, covering the Anti-ITGA6 antibody (ab181551, 1:1500), anti-PI3K(ab32089, 1:1000), anti-PI3K (phospho Y458) (ab278545, 1:1000), anti-AKT(ab8805, 1:500), anti-AKT (phospho T308) (ab38449, 1:1000), Anti-E-cadherin antibody (ab40772, 1:1000), Anti-Bax antibody (ab32503, 1:1000), Anti-Bcl-2 antibody (ab32124, 1:1000), Anti-C-caspase-3 antibody (ab32042, 1:1000), Anti-GAPDH antibody (ab9485, 1:1000), Anti-N-cadherin antibody (ab207608, 1:1000), Anti-Vimentin antibody (ab16700, 1:1000), and anti-GAPDH (ab9485, 1:500).
2.8. Colony formation assay
Colony formation assay evaluated NSCLC cell proliferation. NSCLC cells were collected, and 500 cells were seeded onto a 6-well plate. Following ten days of incubation, the cells in the plate were flushed with PBS three times, immobilized with ethyl alcohol for 30 seconds, and dyed using crystal violet. Finally, the number of colonies was calculated with the naked eye after PBS washing.
2.9. RNA immunoprecipitation (RIP)
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) was taken for RIP. NSCLC cells H1975 and A549, transfected along with miR-30e-5p or miR-NC, were lysed with RIP Lysis buffer incorporating the protease inhibitor cocktail and RNase inhibitor. Cell lysates were extracted and incubated along with the antibodies against Ago2 (Millipore) or rabbit IgG-coated beads at 4°C overnight. After the use of proteinase K buffer for protein removal, the immunoprecipitated RNA was extracted using TRIzol. qRT-PCR examined circ-PITX1 and ITGA6 enrichment.
2.10. Dual-luciferase reporter assay
The targeting correlation between miR-30e-5p and circ-PITX1, miR-30e-5p and ITGA6 was verified by the dual-luciferase reporter assay. The sequences of wild-type circ-PITX1 (circ-PITX1-WT) and ITGA6 (ITGA6-WT) or Mutant circ-PITX1 (circ-PITX1-MT) and ITGA6 (ITGA6-MT) without the binding sites of miR-30e-5p were amplified and slotted into pmirGLO dual-luciferase vectors. Next, H1975 and A549 cells were transfected together with the above vectors and miR-30e-5p or miR-NC for 48 hours’ incubation. The Dual-Luciferase Reporter Assay System (Promega Corp.) was introduced to examine the dual-luciferase activity as instructed by the supplier. The experiment of each group was done in triplicate.
2.11. In-vivo experiments
Twenty female nude mice on a BALB/c background, 4 to 5 weeks of age, were supplied by Shanghai Jiesijie experimental animal Co., Ltd. The mice were reared under specific pathogen-free (SPF) housing. H1975 cells were transfected along with circ-PITX1 overexpression plasmids and their negative controls (Vector). Then, 1 × 107 cells suspended in 100 μL of PBS buffer were subcutaneously administered to the animals, which were further kept under SPF conditions. Within the following 5 weeks, the tumor volume in vivo was gauged every week employing a digital caliper as per the formula V = 0.5 × (W2 × L), where “V” is defined as the volume, “W” as the minor axis, and L as the major axis of the measurement. Five weeks later, the mice were sacrificed, with their tumors resected, weighed, and photographed. The expression of in the tumor tissues ITGA6 was detected by immunohistochemistry using Anti-ITGA6 antibody (ab181551, 1:100) as primary antibody [24]. This experiment had received the imprimatur from the Animal Ethics Committee of Tianjin First Central Hospital.
2.12. Statistical analysis
The statistical software SPSS17.0 (SPSS Inc., Chicago, IL, USA) was introduced for analysis, with the measurement statistics presented as mean ± standard deviation (x ± s). T test was taken for comparison between two groups, and the analysis of variance was adopted to compare multiple means. The Chi-square test was done for fourfold table data analysis. The Kaplan-Meier method and log-rank test analyzed the overall survival difference between patients with a high or low profile of circ-PITX1. P < 0.05 was regarded statistically meaningful.
3. Results
3.1. The profile and significance of circ-PITX1 in NSCLC tissues and cells
To understand the function of circ-PITX1 in NSCLC, we implemented qRT-PCR to check circ-PITX1 expression in NSCLC tissues and cell lines. It turned out that circ-PITX1 expression was dramatically elevated in NSCLC tissues as opposed to normal non-tumor tissues (Figure 1a, p < 0.001). With the increase in the TNM stage, there was also an uplift in circ-PITX1’s levels (Figure 1b). NSCLC patients with up-regulated circ-PITX1 manifested poorer overall survival (Figure 1c), higher TNM stages, and earlier local lymph node metastasis (Table 1). More interestingly, circ-PITX1’s level was substantially heightened in human NSCLC cells (H1975 and A549) as compared with normal lung epithelial cells BEAS-2B (Figure 1d). These findings revealed that circ-PITX1 functioned as an oncogene in NSCLC.
Figure 1.
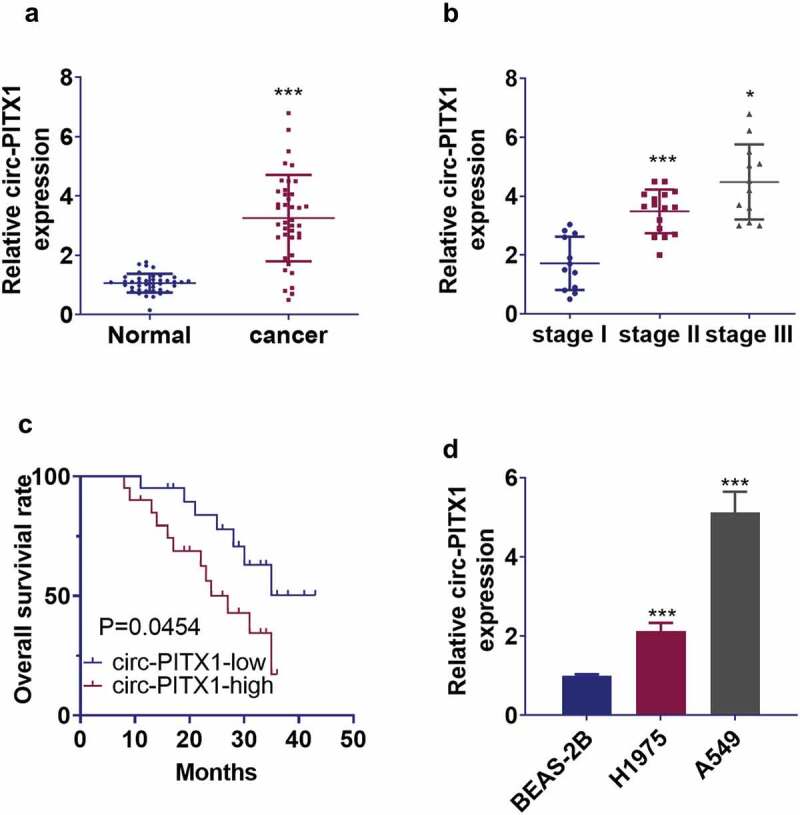
The expression and significance of circ-PITX1 in NSCLC tissues and cells. (a) qRT-PCR determined circ-PITX1 expression in 40 NSCLC tissues and in the 40 paired normal non-tumor tissues, ***P < 0.001. (b) The level of circ-PITX1 in NSCLC tissues with different TNM stages, *P < 0.05, **P < 0.01. (c) The Kaplan-Meier method and log-rank test evaluated the overall survival differences between NSCLC patients with high or low levels of circ-PITX1 expression. (d) qRT-PCR checked the profile of circ-PITX1 in human NSCLC cell lines (H1975 and A549) and normal lung epithelial cells BASE-2B, ***P < 0.001 vs. the BASE-2B group.
3.2. The impact of circ-PITX1 on the malignant phenotypes of NSCLC cells
To dig deeper into the function of circ-PITX1 in NSCLC, we performed gain- and loss- of function assays to examine circ-PITX1 expression in H1975 and A549 cell lines, respectively. CCK-8 and colony formation assays measured cell proliferation, indicating that circ-PITX1 overexpression facilitated NSCLC cell proliferation, whereas circ-PITX1 down-regulation led to proliferation inhibition (Figure 2a–c). As testified by TUNEL outcomes, circ-PITX1 overexpression restrained TUNEL-positive cell numbers, whereas down-regulation of circ-PITX1 raised the number of TUNEL-positive cells in H1975 and A549 cells (Figure 2d). WB examined the expression of apoptosis-related proteins (Bax, Bcl-2, and c-Caspase3) in H1975 and A549 cells. As a result, circ-PITX1 overexpression blocked the expression of the pro-apoptotic proteins Bax and c-Caspase3 and facilitated the expression of the apoptosis-inhibitory protein Bcl-2 in H1975 and A549 cells, while down-regulation of circ-PITX1 led to the opposite result (Figure 2e). Tranwell assay was adopted to monitor cell migration and invasion, while Western blot was utilized to confirm the protein profiles of EMT markers (E-cadherin, Vimentin, and N-cadherin). Notably, circ-PITX1 overexpression enhanced cell migration and invasion, down-regulated the epithelial marker E-cadherin, and up-regulated interstitial markers Vimentin and N-cadherin (Figure 2f–h). On the contrary, circ-PITX1 down-regulation markedly reduced NSCLC cells’ migration, invasion, and EMT. Therefore, circ-PITX1 was involved in NSCLC development via boosting cell proliferation and metastasis.
Figure 2.
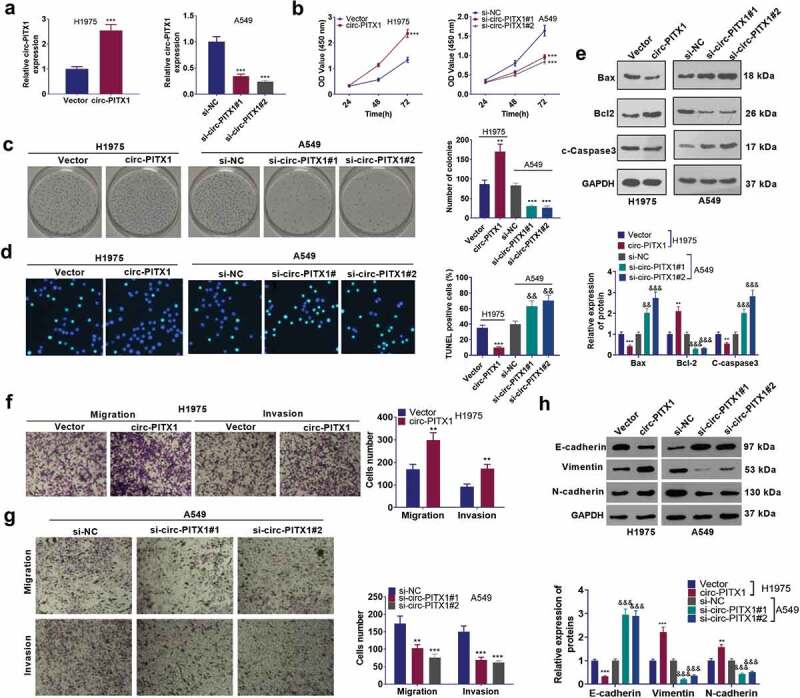
The influence of circ-PITX1 on the malignant phenotypes of NSCLC cells. (a) H1975 and A549 cell lines were taken for the construction of the circ-PITX1 overexpression and down-regulation cell models. B and C. CCK-8 (b) and colony formation assay (c) examined cell proliferation, NS P > 0.05, **P < 0.01 ***P < 0.001 vs. the vector group or the si-vector group. (d) TUNELgauged cell apoptosis. (e) Western blot tested the expression of apoptosis-related proteins (Bax, Bcl-2, and c-Caspase3). f and g. Tranwell tracked cell migration (f) and invasion (g). (h) Western blot determined the protein levels of EMT markers (E-cadherin, Vimentin, and N-cadherin). **P < 0.01, ***P < 0.001 (vs.Vector group), &&P < 0.01, &&&P < 0.001 (vs.si-NC group). n = 3.
3.3. The influence of circ-PITX1 on NSCLC cells in vivo
To grasp the function of circ-PITX1 in NSCLC progression, we resorted to in-vivo experiments. It was discovered that circ-PITX1 overexpression considerably accelerated NSCLC cell growth (Figure 3(a–c)). We also determined the profiles of EMT markers in the in-vivo tumors and uncovered that E-cadherin was down-regulated, whereas Vimentin and N-cadherin were up-regulated in the circ-PITX1 overexpression group (vs. the vector group) (Figure 3d). Additionally, WB data identified that up-regulation of circ-PITX1 curbed the expression of Bax and c-Caspase3 and heightened the Bcl-2 expression in tumor tissues versus the Vector group (Figure 3e). These discoveries supported that circ-PITX1 was an oncogene in NSCLC.
Figure 3.
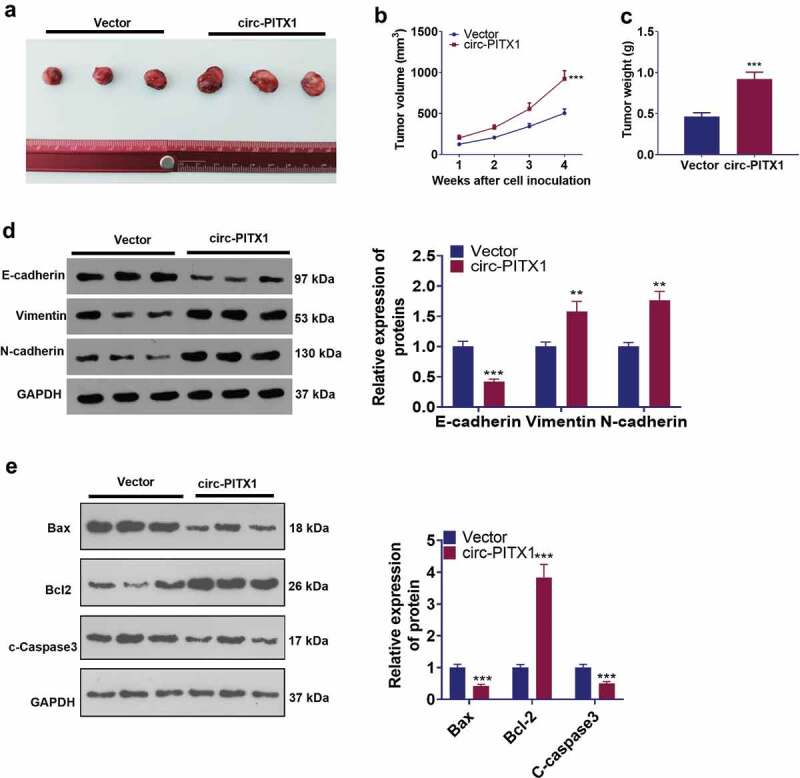
The impact of circ-PITX1 on NSCLC cells in vivo. H1975 cells were transfected along with circ-PITX1 overexpression plasmid or the vector (NC) and then subjected to in-vivo experiments. (a) The images of tumors in the two groups. b and c. The tumor volume (b) and weight (c) were calculated, **P < 0.01, ***P < 0.001 vs. the vector group. d and e. Western blot verified the protein levels of EMT markers (E-cadherin, Vimentin, and N-cadherin) and apoptosis-related proteins (Bax, Bcl-2, and c-Caspase3). **P < 0.01, ***P < 0.001 (vs.Vector group). n = 3.
3.4. Circ-PITX1 bolstered ITGA6 expression
We analyzed ITGA family members expression in LUSC (Lung squamous cell carcinoma) through the online GEPIA database (http://gepia.cancer-pku.cn/) [25]. The data showed that ITGA6 and ITGA11 was significantly upregulated in LUSC tissues compared with those in normal tissues (sup Figure 1). As indicated by the analysis result from Kaplan-Meier Plotter (http://kmplot.com/analysis/) [26], higher level of ITGA5 and ITGA6 predict poorer overall survival of lung cancer patients (sup Figure 2). The data from GEPIA and the Human Protein atlas (http://gepia.cancer-pku.cn/detail.php) unveiled that ITGA6 presented a negative expression in normal lung tissues but exhibited moderate-to-strong expression in lung tissues, mainly situated at the cytomembrane of lung cancer cells (Figure 4(a–b)). A higher level of ITGA6 was correlated with the poorer overall survival and first progression of lung cancer patients (http://kmplot.com/analysis/) (Figure 4c). Those statistics further demonstrated that ITGA6 exerted an oncogenic function in NSCLC. Next, the mRNA and protein levels of ITGA6 in NSCLC cells with up-regulated circ-PITX1 or down-regulated circ-PITX1 were determined. The outcomes denoted that circ-PITX1 boosted ITGA6 expression at both the mRNA and protein levels (Figure 4d–e). For investigating the downstream mechanism of ITGA6, we analyzed ITGA6-related genes in lung cancer through LinkedOmics (http://linkedomics.org/login.php) [27]. Using its online Gene Set Enrichment Analysis (GSEA) tool, we found that ITGA6 positively regulates PI3K/AKT pathway (sup Figure 3a–b). Moreover, PIK3CA and AKT1 have positive relationship with ITGA6 in terms of their expression in LUSC (http://gepia.cancer-pku.cn/) (sup Figure 3c). We implemented WB, which corroborated that up-regulating circ-PITX1 boosted the phosphorylation of PI3K and Akt, while down-regulating circ-PITX1 choked the phosphorylation of PI3K and Akt in NSCLC cells (Figure 4f). Finally, we employed IHC and uncovered that up-regulating circ-PITX1 elevated the ITGA6 expression in tumor tissues (compared with vector group) (Figure 4g). Hence, we substantiated that circ-PITX1 might boost NSCLC via enhancing ITGA6 expression.
Figure 4.
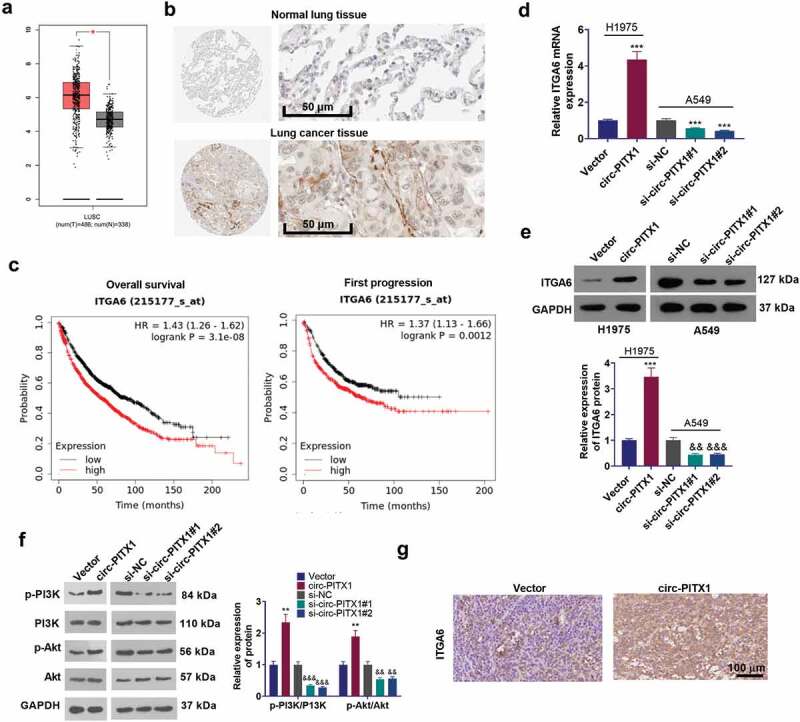
Circ-PITX1 enhanced ITGA6’s expression. (a) The GEPIA (http://gepia.cancer-pku.cn/detail.php) database determined ITGA6’s expression in lung squamous cell carcinoma (LUSC). b–c. The Human Protein atlas (http://gepia.cancer-pku.cn/detail.php) confirmed ITGA6 expression in normal lung tissues or lung cancer tissues. A higher level of ITGA6 intricately pertained to the poorer overall survival of lung cancer patients. D-F. RT-PCR and Western blot measured the mRNA and protein levels of ITGA6, p-PI3K and p-Akt in NSCLC cells up-regulating circ-PITX1 or down-regulating circ-PITX1. G: IHC testified ITGA6 expression in tumor tissues. **P < 0.01 (vs.Vector group). &&P < 0.01, &&&P < 0.001 (vs.si-NC group), n = 3.
3.5. Circ-PITX1 sponged miR-30e-5p
To probe the molecular mechanism between circ-PITX1 and ITGA6, we predicted the underlying miRNA of circ-PITX1 or ITGA6 with the help of Starbase (http://starbase.sysu.edu.cn/). Venn’s diagram displayed that eight miRNAs were discovered. miR-30e-5p has been substantiated to target ITGA6 [23]. To further examine the binding correlation between circ-PITX1 and miR-30e-5p, ITGA6 and miR-30e-5p, we implemented the dual-luciferase reporter gene assay and disclosed that miR-30e-5p mimics vigorously dampened the luciferase activity of H1975 cells transfected along with the circ-PITX1-WT or ITGA6-WT luciferase vectors (Figure 5b–c) but exerted no remarkable influence on H1975 cells transfected with the circ-PITX1-MT or ITGA6-MT luciferase vectors (Figure 5d). RIP assay confirmed that circ-PITX1 and ITGA6 were more enriched by miR-30e-5p in the anti-Ago2 group (Figure 5e–f). The affinities between miR-30e-5p’s level and the overall survival status of lung cancer patients were examined. As a result, a lower level of miR-30e-5p correlated with the worse survival of lung cancer patients (Figure 5g). In short, miR-30e-5p was a sponge of circ-PITX1 and targeted ITGA6, and miR-30e-5p might play a potential part in NSCLC development.
Figure 5.
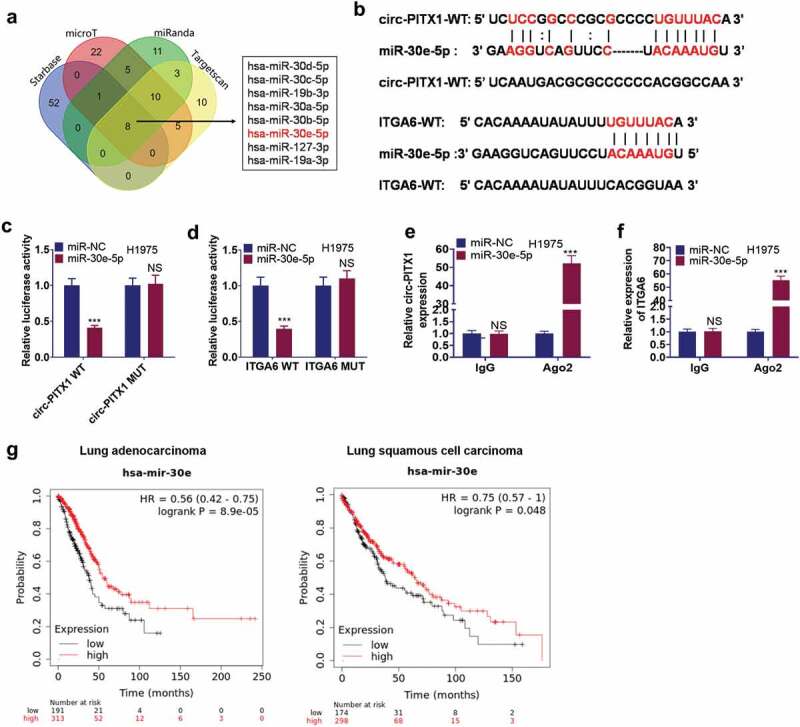
Circ-PITX1 sponged miR-30e-5p. (a) miR-30e-5p was a candidate miRNA target of circ-PITX1 and ITGA6 as predicted by Starbase (http://starbase.sysu.edu.cn/). (b) The binding sites between miR-30e-5p and circ-PITX1, miR-30e-5p and ITGA6. c–f. Dual-luciferase reporter gene assay (c–d) and RIP assay (e–f) examined the binding correlation between miR-30e-5p and circ-PITX1, miR-30e-5p and ITGA6. (g). K-M plotter (http://kmplot.com/analysis/) revealed that a lower level of miR-30e-5p was relevant to the poorer survival of lung cancer patients. NS P > 0.05, ***P < 0.001.
3.6. miR-30e-5p regulated NSCLC development
To investigate the function of miR-30e-5p in NSCLC development, we transfected H1975 cells along with miR-30e-5p mimics and transfected A549 cells with miR-30e-5p inhibitors (Figure 6a). CCK8, colony formation assay, TUNEL staining, Transwell assay, and Western blot evaluated the function of miR-30e-5p in NSCLC cells’ proliferation, apoptosis, invasion, migration, and EMT. As a result, miR-30e-5p up-regulation mitigated H1975 cells’ proliferation, intensified apoptosis, curbed cell migration, invasion, and EMT. In contrast, miR-30e-5p inhibition contributed to opposites results (Figure 6b–g). Finally, we evaluated the expression of Bax, Bcl-2, C-caspase2 and the ITGA6/PI3K/Akt pathways in the cells using WB. The outcome displayed that up-regulating miR-30e-5p enhanced the expression of Bax and c-Caspase3 and declined the expression of Bcl-2, ITGA6 and phosphorylation of PI3K and Akt in cells, while inhibition of miR-30e-5p exerted the opposite effect (Figure 6h,i). These findings signified that miR-30e-5p attenuated the profile of ITGA6 to display its tumor-suppressing function in NSCLC.
Figure 6.
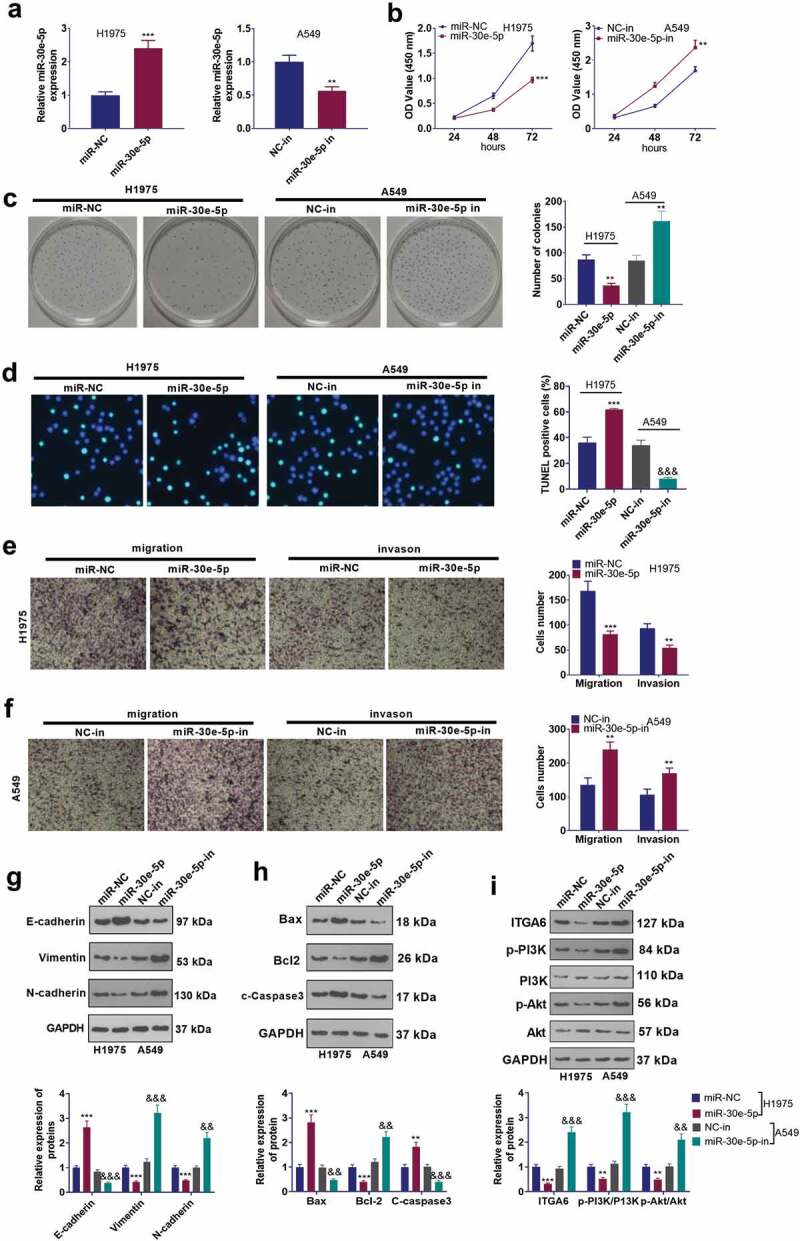
miR-30e-5p exerted an inhibitory function in NSCLC development. (a) H1975 and A549 cells were transfected along with miR-30e-5p mimics or the miR-30e-5p inhibitor, with miR-30e-5p’s level confirmed by RT-PCR. B and C. CCK-8 (b) and colony formation assay (c) examined cell proliferation, NS P > 0.05, *P < 0.05, **P < 0.01. ***P < 0.001 vs. the miR-NC group or the si-NC group. (d) TUNEL assay checked cell apoptosis. E and F. Tranwell tracked cell migration (e) and invasion (f). * P < 0.05, **P < 0.01, ***P < 0.001. g–i. Western blot measured the protein levels of EMT markers (g), apoptosis-related proteins (h), ITGA6, p-PI3K and p-Akt (i). **P < 0.01, ***P < 0.001(vs.miR-NC group). &&P < 0.01, &&&P < 0.001(vs.NC-in group). n = 3.
3.7. Circ-PITX1 modulated NSCLC cell proliferation and metastasis by sponging miR-30e-5p
Considering the correlation between miR-30e-5p and circ-PITX1 in NSCLC cells, we performed rescue experiments to better understand the function of the circ-PITX1/miR-30e-5p axis in NSCLC. By contrast to the circ-PITX1 group, miR-30e-5p mimics barely influenced circ-PITX1’s level, whereas circ-PITX1 overexpression hampered miR-30e-5p’s level (Figure 7a–b). miR-30e-5p up-regulation repressed ITGA6’s level and curbed the phosphorylation of PI3K and Akt, while the expression of these molecules was enhanced by circ-PITX1 (Figure 7c). Furthermore, NSCLC cells’ malignant behaviors were monitored. It turned out that overexpression of circ-PITX1 exacerbated NSCLC cell proliferation, curbed apoptosis, and heightened migration, invasion, and EMT. Surprisingly, miR-30e-5p impeded cells’ proliferation, amplified apoptosis, and hindered migration, invasion, and EMT in the circ-PITX1 group, which were all elicited by circ-PITX1 overexpression (Figure 7d–i). These outcomes disclosed that circ-PITX1 sponged miR-30e-5p to modulate NSCLC cell proliferation and metastasis.
Figure 7.
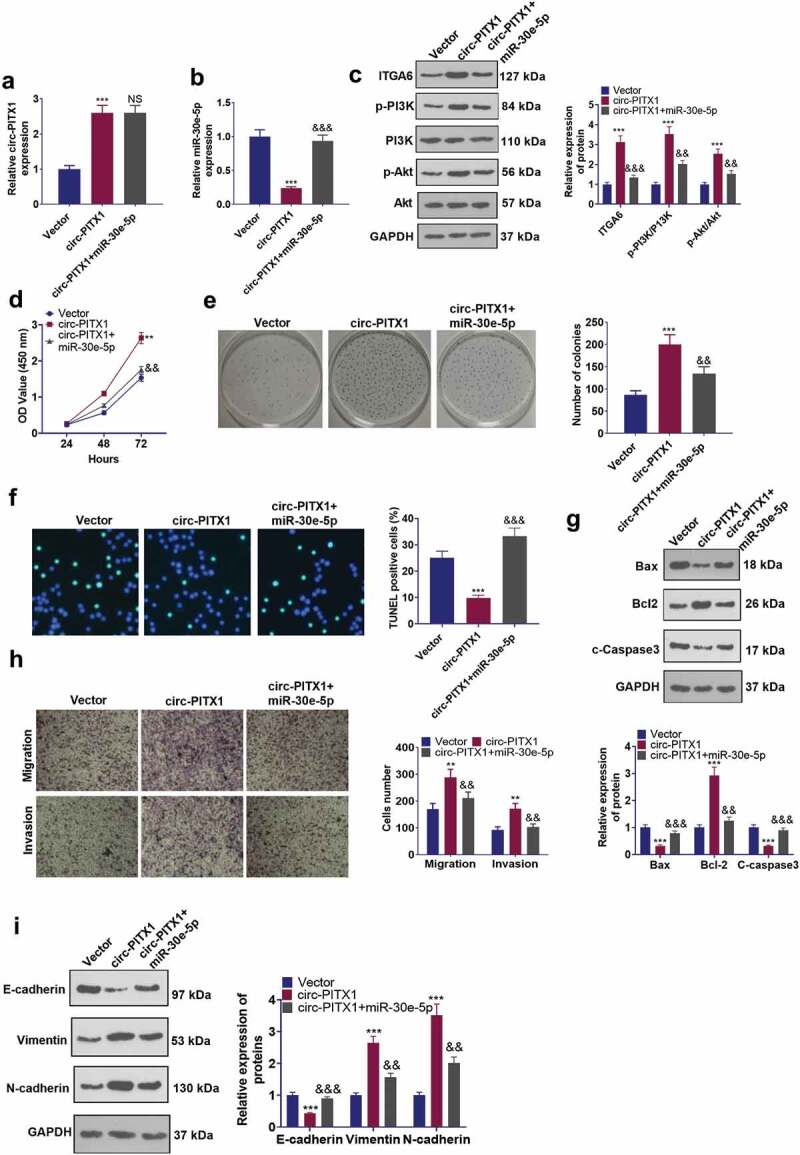
Circ-PITX1 sponged miR-30e-5p to modulate NSCLC cell proliferation and metastasis. a and b. H1975 cells were transfected together with miR-30e-5p mimics and/or circ-PITX1 overexpression plasmid. RT-PCR confirmed the levels of circ-PITX1 (a) and miR-30e-5p (b). (c). Western blot checked ITGA6, p-PI3K and p-Akt expression. d and e. CCK-8 (d) and colony formation assay (e) monitored cell proliferation. (f) TUNEL staining was adopted to examine the TUNEL-positive cell number. (g) WB determined the profiles of Bax, Bcl-2 and c-Caspase3. (h) Tranwell assay tracked cell migration and invasion. (i) Western blot determined the protein levels of EMT markers (E-cadherin, Vimentin, and N-cadherin). **P < 0.01, ***P < 0.001 (vs.Vector group), && P < 0.01, &&&P < 0.001 (vs.circ-PITX1 group), n = 3.
3.8. Circ-PITX1 exerted oncogenic effects via the ITGA6/PI3K/Akt pathway
To further assay the function of the ITGA6/PI3K/Akt pathway in NSCLC, we administered si-ITGA6 and PI3K inhibitor (Wortmannin) to H1975 cells overexpressing circ-PITX1 or knocking down miR-30e-5p, respectively, for 24 hours. The ITGA6/PI3K/Akt pathway expression in H1975 cells was tested using WB. The data displayed that inhibition of ITGA6 choked ITGA6 expression and abated phosphorylation of PI3K and Akt in H1975 cells versus the circ-PITX1 group or the miR-30e-5p-in group. In parallel, the application of PI3K inhibitors resulted in the attenuation of PI3K and Akt phosphorylation in H1975 cells versus the circ-PITX1 group or the miR-30e-5p-in group, but it had little impact on ITGA6 expression (Figure 8a,b).
Figure 8.
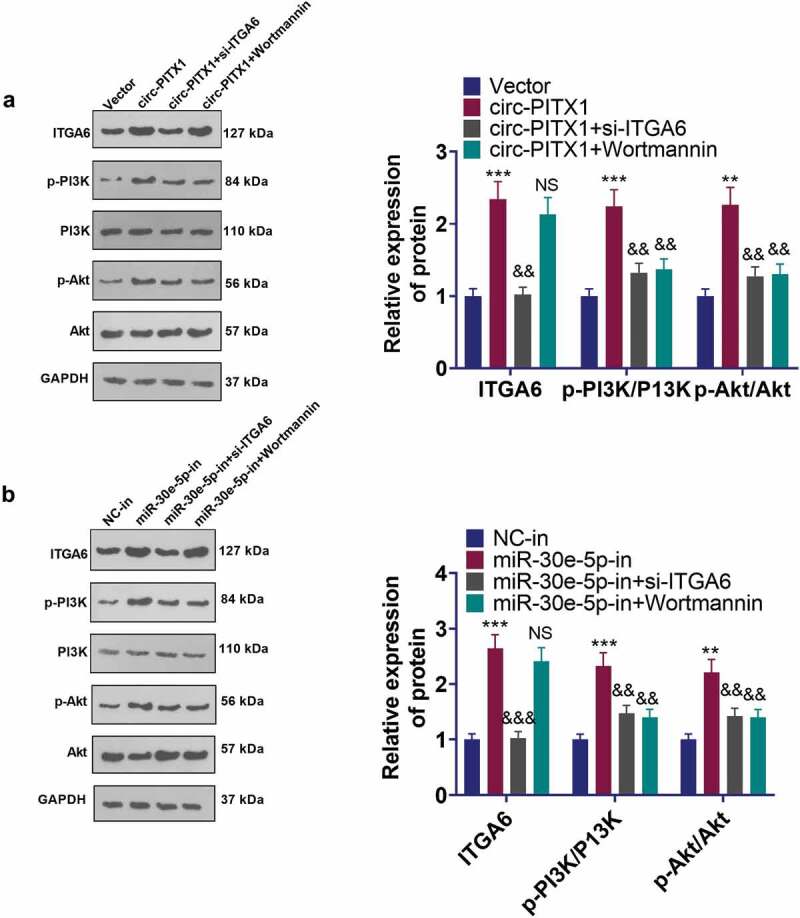
Circ-PITX1 exerted carcinogenic effects via the ITGA6/PI3K/Akt pathway. H1975 cells overexpressing circ-PITX1 or knocking down miR-30e-5p were handled with si-ITGA6 or dealt with the PI3K inhibitor (Wortmannin) for 24 hours, respectively. a and b: WB tested the expression of the ITGA6/PI3K/Akt pathway in H1975 cells. ns P > 0.05, ** P < 0.01, ***P < 0.001 (vs. Vector or NC-in group). && P < 0.01, &&&P < 0.001 (vs. circ-PITX1 or miR-30e-5p-in groups). n = 3.
4. Discussion
With continuous explorations in NSCLC growth and metastasis, many circRNAs are discovered to be associated with NSCLC development [28]. Here, we uncovered that circ-PITX1 was notably up-regulated in NSCLC tissues and cells. Further experiments confirmed the novel circ-PITX1/miR-30e-5p/ITGA6 axis in NSCLC development.
circRNAs, a type of non-coding RNAs widely consisting in human cells, and emerging studies have demonstrated that circRNAs present an aberrant expression in tumors and are entangled in tumor occurrence and development [29]. Abnormally expressed circRNAs can be used as significant indicators for the early diagnosis of tumors, including lung cancer [30,31]. Moreover, the exploration of the circRNA mechanism in tumors can offer new insights into NSCLC treatment by inhibiting tumor growth and chemoresisance. For instance, circARHGAP10 presents a higher expression and bears a relation to the underwhelming prognosis of NSCLC patients, and circARHGAP10 down-regulation abates GLUT1 to suppress glycometabolism in NSCLC cells [32]. circRNA_103762 down-regulation potentiates NSCLC cells to chemotherapeutic drugs via enhancing CHOP’s expression [33]. circ-PITX1, a novel circRNA up-regulated in glioma, facilitates glioma cells’ growth, migration, and invasion and curbs their apoptosis to exert an outstanding function in glioma development regulation [34,35]. Here, we discovered that circ-PITX1 was also up-regulated in NSCLC and markedly associated with the poorer outcomes of NSCLC patients. circ-PITX1 functioned as an oncogene via boosting NSCLC cells’ proliferation, migration, invasion, and EMT, and inhibiting cell apoptosis, indicating that circ-PITX1 could serve as an efficient diagnosis index and treatment target for NSCLC.
With the advances of deep sequencing technologies, more miRNAs have been discovered, and their important functions in tumor progression have also been corroborated [36,37]. Mechanistically, altered miRNAs in tumors affect gene expression and induce abnormal activation of tumor-associated signaling pathways by targeting specific genes at the transcription or post-transcriptional level [38,39]. Many miRNAs present an abnormal expression in NSCLC and exert a significant function [40]. miR-30a, b, c, d, e-5p, five miRNAs belonging to the miR-30 sub-family, all play a part in NSCLC by modulating cell proliferation, drug resistance, migration, and invasion [41–45]. Here, our statistics displayed that miR-30e-5p overexpression could considerably impede NSCLC cells’ proliferation and metastasis, and miR-30e-5p knockdown was associated with the poorer survival of NSCLC patients, which further verified the anti-tumor function of miR-30e-5p in NSCLC.
Prior studies have exhibited that circRNAs interact with miRNAs as ceRNAs, thus influencing tumor progression [46]. For instance, hsa-circ-001895 sponges miR-296-5p to modulate clear cell renal cell carcinoma progression [47]. circ-ARHGAP-10 sponges miR-150-5p to influence NSCLC proliferation, differentiation, and metastasis [32]. Here, we uncovered that circ-PITX1 manifested negative affinities with miR-30e-5p in NSCLC cells. Besides, circ-PITX1 could function as a miRNA sponge to hinder miR-30e-5p in NSCLC. The rescue experiments also demonstrated miR-30e-5p up-regulation partly inverted the promoting function elicited by circ-PITX1 in NSCLC. Our work signified that circ-PITX1 dampened miR-30e-5p to modulate NSCLC progression.
Integrin subunit alpha family contains a group of genes, and their encoded proteins form a cell-surface receptor for collagen and laminin, thus getting involved in cell-cell adhesion [48]. ITGA proteins is abnormally expressed in several tumors, and function an oncogenic role in tumor progression by modulating cell cycle progression and cell differentiation, influencing tumor cell proliferation, invasion, migration, and apoptosis [49–51]. ITGA6, a member of ITGAs, is a valuable prognostic factor in the context of many cancers like ovarian cancer [52]. Downregulating ITGA6 is a promising method in treating tumors [53]. For example, microRNA-3940-5p in exosomes derived from mesenchymal stem cells targets ITGA6 to cramp colorectal cancer metastasis remarkably [54]. Here, we discovered that ITGA6, distinctly up-regulated in NSCLC tissues, predicted a poorer survival rate of NSCLC patients. circ-PITX1 drove up ITGA6’s expression. Given that circ-PITX1 and miR-30e-5p were both marked mediators in NSCLC cells’ EMT, we believed that the circ-PITX1/miR-30E-5p axis could modulate cells’ metastasis and EMT via ITGA6.
PI3K/AKT pathway activation is an outstanding characteristic in lung cancer, and this pathway plays a prominent role in controlling tumor cell proliferation, metastasis, stemness maintenance, chemoresistance [55,56]. Repressing PI3K/AKT pathway is a promising method in treating lung cancer [57]. Interestingly, ITGA6 has been confirmed to induce PI3K/AKT pathway activation [58]. Targeting ITGA6 by miR-143-3p represses tumor growth and angiogenesis via the PI3K/AKT pathway in gallbladder carcinoma [59]. Here, we also observed that circ-PITX1 activates PI3K/AKT pathway activation, accompanied with ITGA6 overexpression. Downregulating ITGA6 or inhibiting PI3K/AKT pathway reversed circPITX1-mediated or miR-30e-5p inhibitor-mediated PI3K/AKT pathway activation.
To conclude, our study has, for the first time, confirmed that circ-PITX1 could enhance NSCLC cells’ proliferation, metastasis, and EMT via the miR-30E-5p/ITGA6/PI3K/AKT axis (Figure 9). Notwithstanding, the function of ITGA6 modulated by the circ-PITX1/miR-30e-5p axis in NSCLC development, particularly in regulating metastasis, requires more experiments. Collectively, these findings provide a better understanding of NSCLC and new substances for its diagnosis and treatment.
Figure 9.
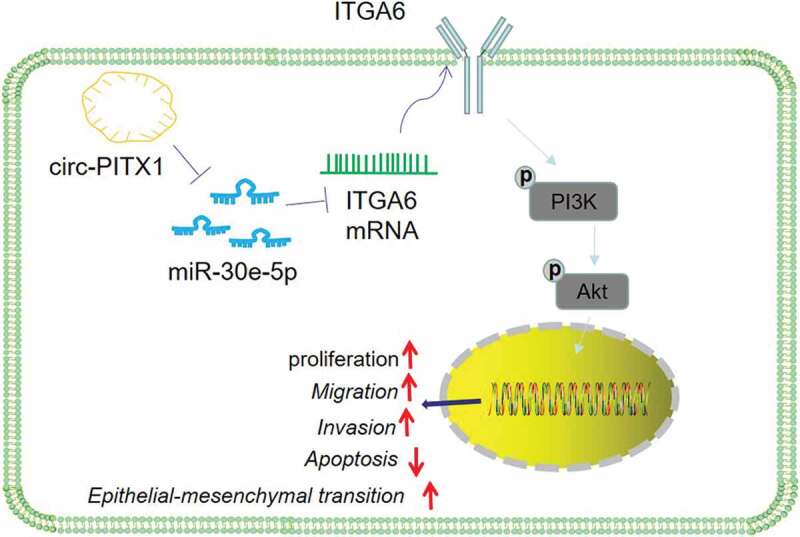
The schematic diagram. circ-PITX1 is upregulated in NSCLC, and it targets miR-30e-5p and upregulates ITGA6/PI3K/AKT pathway, thus regulating the proliferation, invasion, migration, and apoptosis of NSCLC cells.
Supplementary Material
Funding Statement
This study is supported by Tianjin Natural Science Foundation Project (No.20 JCYBJC 00600).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Ethics statement
Our study was approved by the Ethics Committee of Tianjin First Central Hospital.
Data availability statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries [published correction appears in CA. Cancer J Clin. 2020. Jul;70(4):313]. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- [2].Reck M, Popat S, Reinmuth N, et al. Metastatic non-small-cell lung cancer (NSCLC): ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii27–iii39. [DOI] [PubMed] [Google Scholar]
- [3].Guo X, Shi J, Wen Y, et al. Increased high-mobility group A2 correlates with lymph node metastasis and prognosis of non-small cell lung cancer. Cancer Biomark. 2018;21(3):547–555. [DOI] [PubMed] [Google Scholar]
- [4].Patop IL, Wüst S, Kadener S.. Past, present, and future of circ RNAs. EMBO J. 2019;38(16):e100836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang H, Zhang Q, Cui W, et al. Circ_0004018 suppresses cell proliferation and migration in hepatocellular carcinoma via miR-1197/PTEN/PI3K/AKT signaling pathway. Cell Cycle. 2021;20(20):2125–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zheng YL, Song G, Guo JB, et al. Interactions among lncRNA/circRNA, miRNA, and mRNA in musculoskeletal degenerative diseases. Front Cell Dev Biol. 2021;9:753931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yang H, Wang H, Shang H, et al. Circular RNA circ_0000950 promotes neuron apoptosis, suppresses neurite outgrowth and elevates inflammatory cytokines levels via directly sponging miR-103 in Alzheimer’s disease [published correction appears in Cell Cycle. Cell Cycle. 2019. Dec;18(24):3603. 2019;18(18):2197–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kristensen Lasse S, Andersen Maria S, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. [DOI] [PubMed] [Google Scholar]
- [9].Liu W, Ma W, Yuan Y, et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. [DOI] [PubMed] [Google Scholar]
- [10].Yuan Y, Liu W, Zhang Y, et al. CircRNA circ_0026344 as a prognostic biomarker suppresses colorectal cancer progression via microRNA-21 and microRNA-31. Biochem Biophys Res Commun. 2018;503(2):870–875. [DOI] [PubMed] [Google Scholar]
- [11].Yong W, Deng S, Tan Y, et al. Circular RNA circSLC8A1 inhibits the proliferation and invasion of non-small cell lung cancer cells through targeting the miR-106b-5p /FOXJ3 axis. Cell Cycle. 2021. Nov 1;1–10. published online ahead of print. DOI: 10.1080/15384101.2021.1995968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hang D, Zhou J, Qin N, et al. A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med. 2018;7(6):2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang Y, Li H, Lu H, et al. Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. Onco Targets Ther. 2019;12:7055–7065. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [14].Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression [published correction appears in PLoS Genet. PLoS Genet. 2013. Dec;9(12). 2013;9(9):e1003777. DOI: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Khan Abdul Q, Ahmed Eiman I, Elareer Noor R, et al. Role of miRNA-Regulated cancer stem cells in the pathogenesis of human malignancies. Cells. 2019;8:undefined. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rupaimoole R, Calin George A, Lopez-Berestein G, et al. miRNA deregulation in cancer cells and the tumor microenvironment.[J]. Cancer Discov. 2016;6(3):235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Jianchang L, Xiuming L, Li C, et al. miR-224-5p inhibits proliferation, migration, and invasion by targeting PIK3R3/AKT3 in uveal melanoma. J Cell Biochem. 2019;120(8):12412–12421. [DOI] [PubMed] [Google Scholar]
- [18].Xu G, Cai J, Wang L, et al. MicroRNA-30e-5p suppresses non-small cell lung cancer tumorigenesis by regulating USP22-mediated Sirt1/JAK/STAT3 signaling. Exp Cell Res. 2018;362(2):268–278. [DOI] [PubMed] [Google Scholar]
- [19].Rubenstein CS, Gard JMC, Wang M, et al. Gene editing of α6 integrin inhibits muscle invasive networks and increases cell-cell biophysical properties in prostate cancer. Cancer Res. 2019;79(18):4703–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yuan J, Li P, Pan H, et al. miR-542-5p attenuates fibroblast activation by targeting integrin α6 in silica-induced pulmonary fibrosis. Int J Mol Sci. 2018;19(12):3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zhang HJ, Tao J, Sheng L, et al. Twist2 promotes kidney cancer cell proliferation and invasion by regulating ITGA6 and CD44 expression in the ECM-receptor interaction pathway. Onco Targets Ther. 2016;9:1801–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wu Y, Tan X, Liu P, et al. ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Exp Cell Res. 2019;379(1):30–47. [DOI] [PubMed] [Google Scholar]
- [23].Laudato S X, Patil N, Abba ML, et al. P53-induced miR-30e-5p inhibits colorectal cancer invasion and metastasis by targeting ITGA6 and ITGB1. Int J Cancer. 2017;141(9):1879–1890. [DOI] [PubMed] [Google Scholar]
- [24].Li R, Qiu X, He M, et al. METTL3-mediated mature miR-497-5p/195-5p inhibits trophoblast migration and invasion by targeting WWP1 in preeclampsia. Cell Cycle. 2021. Sep 30;1–16. [published online ahead of print]. DOI: 10.1080/15384101.2021.1982527. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [25].Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Győrffy B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput Struct Biotechnol J. 2021;19:4101–4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wei H, Li L, Zhang H, et al. Circ-FOXM1 knockdown suppresses non-small cell lung cancer development by regulating the miR-149-5p/ATG5 axis. Cell Cycle. 2021;20(2):166–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Verduci L, Strano S, Yarden Y, et al. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. [DOI] [PubMed] [Google Scholar]
- [31].Ma D, Qin Y, Huang C, et al. Circular RNA ABCB10 promotes non-small cell lung cancer progression by increasing E2F5 expression through sponging miR-584-5p. Cell Cycle. 2020;19(13):1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jin M, Shi C, Yang C, et al. Upregulated circRNA ARHGAP10 predicts an unfavorable prognosis in NSCLC through regulation of the miR-150-5p/GLUT-1 axis. Mol Ther Nucleic Acids. 2019;18:219–231. DOI: 10.1016/j.omtn.2019.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xiao G, Huang W, Zhan Y, et al. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP). J Clin Lab Anal. 2020;34(6):e23252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lv X, Wang M, Qiang J, et al. Circular RNA circ-PITX1 promotes the progression of glioblastoma by acting as a competing endogenous RNA to regulate miR-379-5p/MAP3K2 axis. Eur J Pharmacol. 2019;863:172643. [DOI] [PubMed] [Google Scholar]
- [35].Zhan L, Mu Z, Yang M, et al. Elevation of circ-PITX1 upregulates interleukin 17 receptor D expression via sponging miR-518a-5p and facilitates cell progression in glioma. J Cell Biochem. 2019;120(10):16495–16502. [DOI] [PubMed] [Google Scholar]
- [36].Bahubeshi A, Tischkowitz M, Foulkes WD. Foulkes William D,miRNA processing and human cancer: DICER1 cuts the mustard. Sci Transl Med. 2011;3(111):111ps46. [DOI] [PubMed] [Google Scholar]
- [37].Di LG, Croce CM. Croce Carlo M,miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23(1):3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gareev I, Beylerli O, Liang Y, et al. The role of MicroRNAs in therapeutic resistance of malignant primary brain tumors. Front Cell Dev Biol. 2021;9:740303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Oh-Hohenhorst SJ, Lange T. Role of metastasis-related microRNAs in prostate cancer progression and treatment. Cancers (Basel). 2021;13(17):4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Huang Y, Feng G. MiR-423-5p aggravates lung adenocarcinoma via targeting CADM1 [published online ahead of print]. Thorac Cancer. 2020. Nov 18. DOI: 10.1111/1759-7714.13745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang F, Meng F, Wong SCC, et al. Combination therapy of gefitinib and miR-30a-5p may overcome acquired drug resistance through regulating the PI3K/AKT pathway in non-small cell lung cancer. Ther Adv Respir Dis. 2020;14:1753466620915156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cheng Y, Yang S, Shen B, et al. Molecular characterization of lung cancer: a two-miRNA prognostic signature based on cancer stem-like cells related genes. J Cell Biochem. 2020;121(4):2889–2900. [DOI] [PubMed] [Google Scholar]
- [43].Zhou Y, Shi H, Du Y, et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging (Albany NY). 2019;11(18):7386–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zeng Q, Dai Y, Duan C, et al. Long noncoding RNA POU3F3 enhances cancer cell proliferation, migration and invasion in non-small cell lung cancer (adenocarcinoma) by downregulating microRNA-30d-5p. BMC Pulm Med. 2020;20(1):185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang S, Li G, Liu C, et al. miR-30e-5p represses angiogenesis and metastasis by directly targeting AEG-1 in squamous cell carcinoma of the head and neck. Cancer Sci. 2020;111(2):356–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Song J, Lu Y, Sun W, et al. Changing expression profiles of lncRNAs, circRNAs and mRNAs in esophageal squamous carcinoma. Oncol Lett. 2019;18(5):5363–5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chen Z, Xiao K, Chen S, et al. Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 2020;111(2):713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nurzat Y, Su W, Min P, et al. Identification of therapeutic targets and prognostic biomarkers among integrin subunits in the skin cutaneous melanoma microenvironment. Front Oncol. 2021;11:751875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Liu X, Tian H, Li H, et al. Derivate Isocorydine (d-ICD) suppresses migration and invasion of hepatocellular carcinoma cell by downregulating ITGA1 expression. Int J Mol Sci. 2017;18(3):514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Li H, Wang Y, Rong SK, et al. Integrin α1 promotes tumorigenicity and progressive capacity of colorectal cancer. Int J Biol Sci. 2020;16(5):815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Li B, Jiang J, Assaraf YG, et al. Surmounting cancer drug resistance: new insights from the perspective of N6-methyladenosine RNA modification. Drug Resist Updat. 2020;53:100720. [DOI] [PubMed] [Google Scholar]
- [52].Zhu T, Chen R, Wang J, et al. The prognostic value of ITGA and ITGB superfamily members in patients with high grade serous ovarian cancer. Cancer Cell Int. 2020;20(1):257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang G, Fang J, Shi Q, et al. Screening of molecular markers of induced chemotherapy in supraglottic laryngeal squamouscell carcinoma. World J Otorhinolaryngol Head Neck Surg. 2020;6(1):34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Li T, Wan Y, Su Z, et al. Mesenchymal stem cell-derived exosomal microRNA-3940-5p inhibits colorectal cancer metastasis by targeting integrin α6. Dig Dis Sci. 2020. Jul 15. [published online ahead of print]. DOI: 10.1007/s10620-020-06458-1 [DOI] [PubMed] [Google Scholar]
- [55].Pérez-Ramírez C, Cañadas-Garre M, Molina MÁ, et al. PTEN and PI3K/AKT in non-small-cell lung cancer. Pharmacogenomics. 2015;16(16):1843–1862. [DOI] [PubMed] [Google Scholar]
- [56].Iksen PS, Pongrakhananon V, Pongrakhananon V. Targeting the PI3K/AKT/mTOR signaling pathway in lung cancer: an update regarding potential drugs and natural products. Molecules. 2021;26(13):4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tan AC. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac Cancer. 2020;11(3):511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yuan M, Xie F, Xia X, et al. UNC5C-knockdown enhances the growth and metastasis of breast cancer cells by potentiating the integrin α6/β4 signaling pathway. Int J Oncol. 2020;56(1):139–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Jin YP, Hu YP, Wu XS, et al. miR-143-3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis. 2018;9(2):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.


